Abstract
Salmonella Typhimurium is widely distributed in food. It can colonise the gastrointestinal tract after ingestion, causing lamina propria edema, inflammatory cell infiltration, and mucosal epithelial decomposition. A high-fat diet (HFD) can induce an inflammatory response, but whether HFD can increase the infection level of S. Typhimurium is unknown. We established a model of Salmonella enterica subsp. enterica serovar Typhimurium strain ATCC 13311 ATCC 13311 infection in healthy adult mice with a maintenance diet (MD) or HFD to explore the effect of Lactiplantibacillus plantarum 1201 intervention on S. Typhimurium ATCC 13311 colonization and its protective effects on mice. HFD exacerbated the infection of S. Typhimurium ATCC 13311, while the intervention of L. plantarum 1201 effectively mitigated this process. L. plantarum 1201 can reduce the colonies of S. ATCC 13311 in the intestines and tissues; and reduce intestinal inflammation by down-regulating the level of TLR4/NF-κB pathway related proteins in serum and the expression of related inflammatory factors in the colon and jejunum. Since L. plantarum 1201 can inhibit the colonization of S. Typhimurium ATCC 13311 and relieve inflammation in HFD, current research may support the use of L. plantarum 1201 to prevent S. Typhimurium infection.
Keywords: Lactiplantibacillus plantarum 1201, Salmonella Typhimurium ATCC 13311, high-fat diet, intestinal infection, colonization, inflammation
1. Introduction
With the continuous development of society, we are consuming ever more high-fat foods, especially those of animal origin, including beef [1], pork [2], poultry [3] and turkey [4]. Salmonella Typhimurium has been found on these foods, which can cause disease. According to statistics, more than 90% of bacterial-related food poisoning cases worldwide are caused by Salmonella and Campylobacter [5]. However, non-animal products, such as fresh vegetables and fruits, fruit juices and spices, are also associated with infection [6,7,8]. Because of this, is there any relationship between high-fat diet (HFD) and S. Typhimurium infection? S. Typhimurium can evade the host’s natural immune system, replicate in the host and cause disease [9]. Humans infected with S. Typhimurium show loss of appetite, vomiting and diarrhea, and may even develop sepsis [10]. At present, there is no way to control S. Typhimurium without side effects. Probiotic bacteria are reported to prevent the adherence, establishment and invasion of specific enteropathogenesis [11]. Several mechanisms have been proposed: contribution to mucosal barrier function, competitive exclusion, modulation of the immune response, coaggregation to pathogens, decreasing luminal pH via the production of lactic acid and secretion of specific compounds such as bacteriocins [12,13,14]. Therefore, we may start research from the direction of intestinal flora and colonization.
Studies have found that probiotics can colonise the gastrointestinal tract, supplement beneficial flora, maintain the balance of intestinal bacteria, and competitively inhibit the colonization of pathogenic bacteria. In addition, probiotics have an antagonistic effect on pathogenic bacteria in the intestinal tract and in food [15]. Some probiotics can produce beneficial metabolites, such as organic acids or hydrogen peroxide and natural antibiotics, forming an environment that inhibits or kills harmful bacteria. These acids can lower the pH value of the animal’s intestinal tract, effectively inhibit the growth of pathogenic bacteria, and create conditions for the growth of lactic acid bacteria. Lactobacillus, one of the most widely used probiotics, can form a biological barrier by regularly colonizing on mucous membranes, skin and other surfaces or between cells, thereby preventing the colonization of pathogenic microorganisms and inhibiting the survival of pathogenic bacteria. Limosilactobacillus reuteri can maintain the balance of pathogens and probiotics by neutralizing Porphyromonas gingivalis, thereby inhibiting inflammation and promoting wound healing [16]. Lacticaseibacillus rhamnosus can inhibit the colonization of Vibrio parahaemolyticus in the intestine [17]. Lactobacillus gasseri can inhibit the colonization of Campylobacter jejuni in the gastrointestinal tract [18]. After attaching to the cell surface, Lactobacillus can effectively control the reattachment of Salmonella bacteria to cells [19]. L. plantarum 1201 was isolated from Fuzhou preserved pickles. The antimicrobial activities experiment proved that L. plantarum 1201 has a better inhibitory ability against Salmonella Typhimurium which meant L. plantarum 1201 might inhibit the S. Typhimurium ATCC 13311 infections. Therefore, in this study, a mouse model of S. Typhimurium ATCC 13311 infection was constructed to investigate whether L. plantarum 1201 could inhibit S. Typhimurium ATCC 13311 infections, aiming to provide a new perspective for the treatment of S. Typhimurium infection.
2. Materials and Methods
2.1. Lactiplantibacillus Strains and Culture Conditions
L. plantarum 1201 was culture under anaerobic conditions at 37 °C in sterile deMan, Rogosa, Sharpe broth (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China). Subsequently, cells were harvested, centrifuged at 5000× g for 5 min at 25 °C, and washed in sterile phosphate buffer saline (PBS). L. plantarum 1201 isolated from fermented acid beans is deposited in the China Center for Type Culture Collection with the collection number CCTCC M 2021050.
2.2. Animals and Experimental Design
Eight-week-old specific pathogen free (SPF) female KM mice (Hunan Slack Jingda Experimental Animal Co., Ltd.) were assigned to cages, unloaded from the car in random order, and then randomly assigned into six groups. They were raised in the animal facility of Nanchang University, Jiangxi Province, under standard conditions with a light/dark cycle of 12 h. All mice were provided with ad libitum access to food and water. All experimental procedures are in accordance with the guidelines of the National Institutes of Health and were approved by the local animal health and use committee of Nanchang University. All pure feeds of the control group (MD group) and the experimental group were purchased from the China Nutrition Animal Feed High-Tech Co. Ltd. After 1 week of acclimatization in the sterile mouse room, the mice in the MD group, MD2 group and MD3 group (n = 8) were given a normal diet and normal drinking water, while the mice in the HFD group, HFD2 group and HFD3 group (n = 8) were maintained on an HFD (60% calories from fat) and normal drinking water. Then, the MD and HFD groups (n = 8) were intra-gastrically administered with 200 μL of 1 × PBS, and the MD2 and HFD2 groups (n = 8) were intra-gastrically administered with 100 μL of 1 × PBS and 100 μL of 1 × 108 CFU/mL S. Typhimurium ATCC 13311, MD3 and HFD3 groups (n = 8) were intra-gastrically administered with 100 µL 1 × 108 CFU/mL L. plantarum 1201 and 100 µL 1 × 108 CFU/mL S. Typhimurium ATCC 13311. Mouse feces were collected at 3 h, 6 h, 24 h, 48 h, 72 h, and 96 h after gavage with strain. After 4 days of intra-gastric administration, the mice were euthanized with ether. The mouse serum, spleen, mesenteric lymph nodes (mLN), colon, cecum, jejunum and colon contents were collected in a sterile centrifuge tube, respectively, and stored at −80 °C for subsequent experiments.
2.3. Determination of Infection in Mice
S. Typhimurium ATCC 13311 was incubated with 20 μg/mL Hoechst 33342 for 15 min [20]. All the samples including fecal, spleen, mLN, jejunum and colon contents were collected and weighed, then they were homogenized in PBS with grinding beads in a grinder. The homogenate was diluted gradually and spread on Salmonella Shigella (SS) agar selection medium [21]. After incubating for 24 h at 37 °C under aerobic conditions, we counted the number of colonies. The number of S. Typhimurium in the sample was calculated, and then the bacterial load of S. Typhimurium per gram of sample was calculated. The fluorescence intensity of the mouse fecal homogenate 6 h after gavage was observed under an inverted microscope.
2.4. Hematoxylin-Eosin Staining and Histopathological Damage Scores
Fresh colon tissues were collected and soaked in 10% formalin tissue fixative solution. After the alcohol was dehydrated, the tissue blocks were sequentially transparent, waxed, encased, sliced, and baked in turn. The conventional dewaxing process is to sequentially put paraffin sections into xylene I (10 min)—xylene II (10 min)—ethanol I (5 min)—ethanol II (5 min)—95% alcohol (3 min)—90% alcohol (3 min)—80% alcohol (2 min)—70% alcohol (2 min). Then, the slices were soaked in distilled water and washed for 2 min, and stained with hematoxylin and eosin (HE), anhydrous ethanol I 5 min, anhydrous ethanol II 5 min, xylene I in 5 min, and xylene II 5 min, then dried to a neutral gum sealing piece, and finally observed microscopy. Pathological score was determined by analysing four markers of inflammation: (1) submucosal oedema, (2) polymorphonuclear granulocyte infiltration into the lamina propria, (3) number of goblet cells harbouring mucus-filled vacuoles and (4) epithelial integrity. Pathological scores ranged from 0 to 13 arbitrary units (0–3, no or minimal inflammation; 4–6, slight inflammation; 7–9, moderate inflammation; 10–13, profound inflammation) [22,23].
2.5. Analysis of Inflammatory Levels in Intestine of the Mice
High-quality RNA was isolated from frozen colon and jejunum tissue using the TaKaRa RNA extraction kit (Takara, Otsu, Japan), and then used for cDNA synthesis with a transcriptor cDNA kit (Takara, Otsu, Japan) according to the manufacturer’s protocol. The mRNA levels of the pro-inflammatory factors (IFN-γ, IL-1β, TNF-α, IL-6, IL-17A), anti-inflammatory factors (IL-10, IL-22, TGF-β) and NF-κB inflammation pathway-related genes (NF-κB, IκB-α) in the colon and jejunum tissue were measured using real-time PCR. Three-step PCR reaction procedure: 5 µL SYBR Green, 0.8 µL primer (10 μM), 1 µL cDNA (1000 ng/μL) plus ddH2O supplement 10 µL system. Reaction conditions were: preheat at 95 °C for 30 s, cycling stage: denaturation at 95 °C for 5 s; 59 °C annealing for 1 min; 72 °C extension for 30 s, 40 cycles, and then cooling to 65 °C for 5 s. The primer sequence information is shown in the Supplementary Materials (online supplementary Table S1).
2.6. Enzyme-Linked Immunosorbent Assay (ELISA) Detection of TLR4/NF-κB Inflammation Pathway in Serum of the Mice
The protein levels of TLR4, MyD88, IKKβ, IκB-α and NF-κB in mouse serum were determined by the double antibody sandwich method according to the steps of the ELISA kit (MEIMIAN, Jiangsu, China). After being left overnight at 4 °C, the freshly collected plasma was centrifuged at 3000 r/min for 10 min, and the serum was collected for testing.
2.7. S. Typhimurium Inhibition Assay
The agar diffusion assay was used to detect the inhibitory effect of L. plantarum 1201 on S. Typhimurium ATCC 13311. In short, 200 μL aliquot of S. Typhimurium ATCC 13311 (adjusted to 108 CFU/mL, 107 CFU/mL, 106 CFU/mL) as an indicator microorganism was spread on LB agar. After the bacterial solution was dry, we put the Oxford cup on the LB plate. Then, 200 μL of the supernatant of L. plantarum 1201 was cultured overnight, and MRS broth with pH = 3.5 and fresh MRS broth with pH = 7 were added to Oxford cups (with an outer diameter of 6.8 ± 0.1 mm, an inner diameter of 6.0 ± 0.1 mm, and a height of 10.0 ± 0.1 mm). The plate was incubated at 37 °C for 36 h under aerobic conditions, and the diameter of the inhibition zone was measured.
2.8. Statistical Analysis
Data analysis was carried out using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). One-way ANOVA was used for multiple comparisons, and all results are expressed as an average of S.E.M. The double-tailed unpaired Student t test was used for statistical evaluation. A p value of 0.05 was considered statistically significant.
3. Results
3.1. High-Fat Diet (HFD) Promotes S. Typhimurium ATCC 13311 Colonization while L. plantarum 1201 Inhibits It
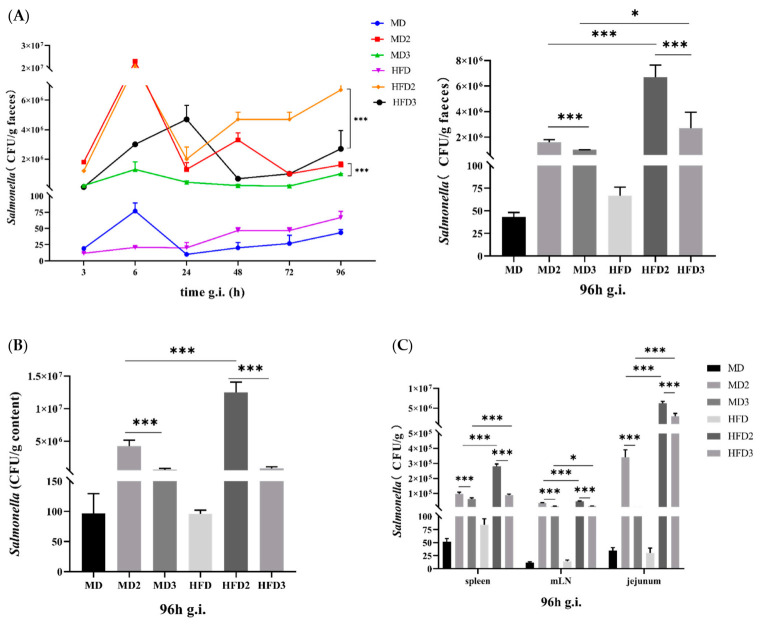
We used SS selective agar medium to detect the colonization of S. Typhimurium. The levels of S. Typhimurium in the feces of the mice in the MD2, MD3, HFD2 and HFD3 groups that received S. Typhimurium were higher than those in the MD and HFD groups. The HFD2 group had the highest S. Typhimurium content. Interestingly, the relative colony number of the continuous high-fat diet group was higher than that of the normal diet group (Figure 1A–B). This is in agreement with a previous discovery [24]. Similarly, the relative colony number of S. Typhimurium in the organs of mice 96 h after intra-gastric administration also showed the same situation (Figure 1C). It is speculated that HFD can promote the colonization of S. Typhimurium, and L. plantarum 1201 has an inhibitory effect on the colonization of S. Typhimurium ATCC 13311.
Figure 1.
High-fat diet (HFD) promotes S. Typhimurium ATCC 13311 colonization while L. plantarum 1201 inhibits it. (A) statistical graph of S. Typhimurium content in mouse feces after gavage and the content of S. Typhimurium in mouse feces after intra-gastric administration for 96 h; (B) statistical graph of the content of S. Typhimurium in the colon content of mice after intra-gastric administration for 96 h; (C) statistical graph of the content of S. Typhimurium in mouse organs after intra-gastric administration for 96 h. * p < 0.05; *** p < 0.001; paired two-tailed t-test. MD: normal diet; MD2: normal diet and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; MD3: normal diet, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; HFD: high-fat diet (HFD); HFD2: HFD and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; HFD3: HFD, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; mLN, mesenteric lymph node; g.i., gavage infection.
3.2. L. plantarum 1201 Alleviates the Intestinal Injury Induced by S. Typhimurium ATCC 13311
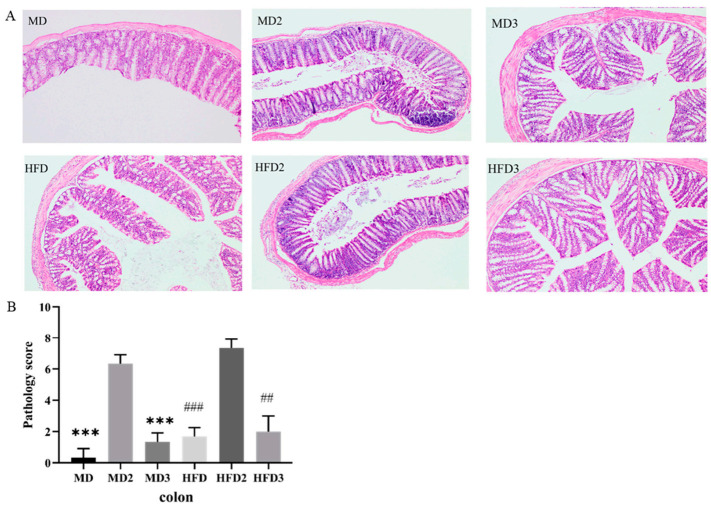
Colonization of S. Typhimurium ATCC 13311 usually leads to intestinal pathophysiology. By comparing the sections of the colon of mice with hematoxylin-eosin staining and pathological scores, we observed that S. Typhimurium ATCC 13311 caused inflammation in the colon, and a high-fat diet could exacerbate this inflammatory response (Figure 2A,B). Compared with the MD group, the MD2 group mice that received a normal diet and received the S. Typhimurium ATCC 13311 gavage showed significant intestinal inflammation, while the HFD2 group that received a high-fat diet and gavaged S. Typhimurium ATCC 13311 showed more serious intestinal inflammation, with close arrangement and abnormal morphology of the intestinal villi and the number of abnormal goblet cells. However, in the MD3 and HFD3 groups treated with L. plantarum 1201, the number of abnormal goblet cells in mice decreased and the intestinal margin became clear, the intestinal villi were closely arranged, and the inflammation was relieved (Figure 2A,B). Therefore, it is speculated that HFD promoted the damage of S. Typhimurium ATCC 13311 to the intestinal tract, while L. plantarum 1201 reduced the intestinal inflammation induced by S. Typhimurium ATCC 13311.
Figure 2.
L. plantarum 1201 alleviates the intestinal injury induced by S. Typhimurium ATCC 13311. (A) hematoxylin-eosin staining (HE) pathological section of mouse colon tissue; (B) Colon histopathological score chart. *** p < 0.001; ## p < 0.01; ### p < 0.001; * is compared with MD2, # is compared with HFD2; paired two-tailed t-test. MD: normal diet; MD2: normal diet and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; MD3: normal diet, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; HFD: HFD; HFD2: HFD and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; HFD3: HFD, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201.
3.3. L. plantarum 1201 Relieves Intestinal Inflammation through the TLR4/NF-κB Inflammatory Pathway
The levels of Toll-like receptor TLR4, myeloid differentiation factor MyD88, I-κB kinase IKK-β, nuclear transcription factor inhibitor IκB-α, and nuclear transcription factor NF-κB in serum were detected with an ELISA kit. At the same time, the mRNA expression levels of NF-κB, IκB-α in jejunum and cecum were detected. Compared with the MD group, the TLR4, MyD88, IKK-β, and NF-κB protein levels of the serum of the MD2 group and the HFD2 group were increased, and the HFD2 group showed a more significant increase in protein expression. However, in the MD3 and HFD3 groups that were given L. plantarum 1201, the protein expression level of TLR4, MyD88, IKK-β, NF-κB was down-regulated. In contrast, the protein expression level of IκB-α was down-regulated in the MD2 and HFD2 groups, but recovered in the MD3 and HFD3 groups (Figure 3B). The expression levels of NF-κB and IκB-α mRNA detected by qPCR also showed similar changes to those of protein levels (Figure 3A). It can be speculated that the colonization of S. Typhimurium ATCC 13311 caused inflammation by affecting the expression of related proteins in the TLR4/NF-kB inflammation pathway. HFD aggravated this change, and L. plantarum restored the inflammation pathway to inhibit inflammation.
Figure 3.
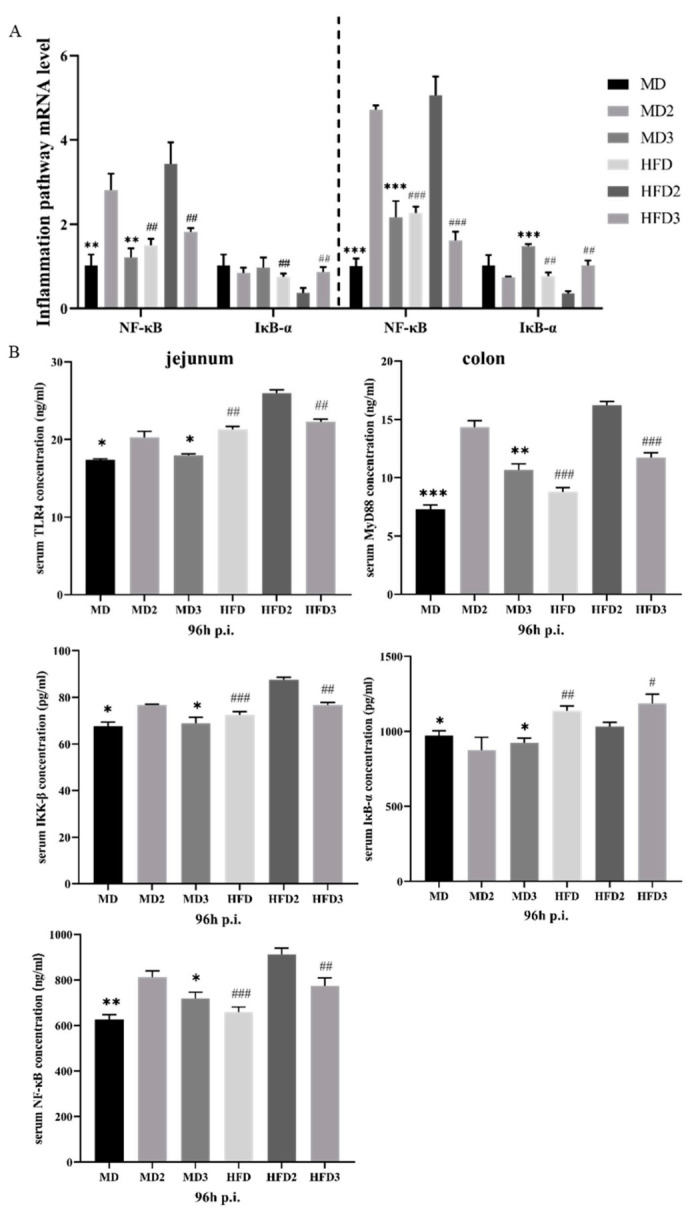
L. plantarum 1201 relieves intestinal inflammation through the TLR4/NF-κB inflammatory pathway. (A) mRNA expression of NF-kB inflammation pathway in colon and jejunum of mice in different groups; (B) concentration of TLR4/NF-κB pathway protein in mouse serum. * p < 0.05; ** p < 0.01; *** p < 0.001; # p < 0.05; ## p < 0.01; ### p < 0.001; * is compared with MD2, # is compared with HFD2; paired two-tailed t-test. MD: normal diet; MD2: normal diet and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; MD3: normal diet, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; HFD: HFD; HFD2: HFD and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; HFD3: HFD, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; p.i., post infection.
3.4. L. plantarum 1201 Restores the Secretion of Intestinal Inflammatory Factors Caused by S. Typhimurium ATCC 13311 Colonization
We detected the mRNA expression levels of interferon-γ, transforming growth factor TGF-β, tumor necrosis factor TNF-α, and interleukin IL-1β, IL-6, IL-10, IL-22, and IL-17A in jejunum and colon tissues. The results showed that the colonization of S. Typhimurium ATCC 13311 resulted in an increase in the mRNA expression levels of IL-1β, IL-6, IL-10, IL-22, IL-17A, TGF-β, TNF-α (Figure 4A,B). Therefore, we hypothesized that the colonization of S. Typhimurium ATCC 13311 would lead inflammation of the jejunum and colon. HFD aggravated this change and L. plantarum 1201 had different degrees of relief effects on it. Compared with the increased mRNA levels of inflammatory factors in the MD2 and HFD2 groups after ingesting S. Typhimurium ATCC 13311, under the intervention of L. plantarum 1201, the inflammatory mRNA expression levels in the MD3 and HFD3 groups were consistent with those in the MD and HFD groups, indicating that L. plantarum 1201 can inhibit intestinal inflammation caused by S. Typhimurium.
Figure 4.
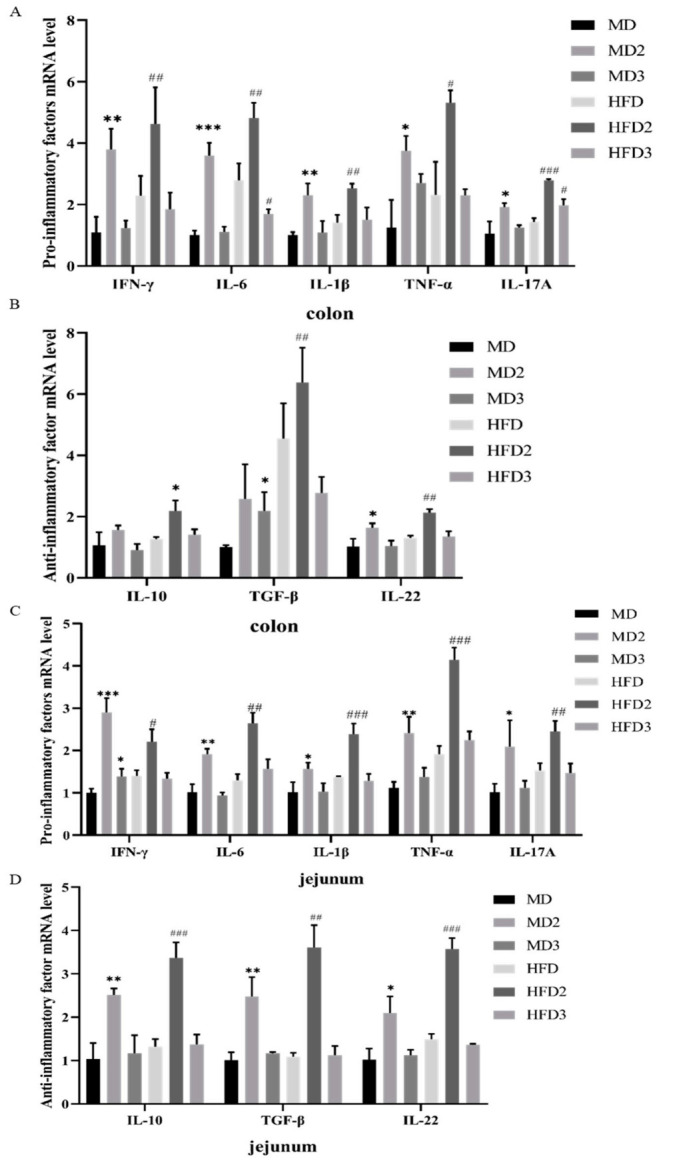
L. plantarum 1201 recovers the expression disorder of inflammatory factors caused by S. Typhimurium ATCC 13311. (A) mRNA expression levels of pro-inflammatory factors in mouse colon; (B) mRNA expression levels of anti-inflammatory factor in mouse colon; (C) mRNA expression levels of pro-inflammatory factors in mouse jejunum; (D) mRNA expression levels of anti-inflammatory factors in mouse jejunum. * p < 0.05; ** p < 0.01; *** p < 0.001; # p < 0.05; ## p < 0.01; ### p < 0.001; * is compared with MD, # is compared with HFD; paired two-tailed t-test. MD: normal diet; MD2: normal diet and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; MD3: normal diet, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201; HFD: HFD; HFD2: HFD and 1 × 108 CFU/mL S. Typhimurium ATCC 13311; HFD3: HFD, 1 × 108 CFU/mL S. Typhimurium ATCC 13311 and 1 × 108 CFU/mL L. plantarum 1201.
3.5. L. plantarum 1201 Inhibits S. Typhimurium ATCC 13311 Growth Activity
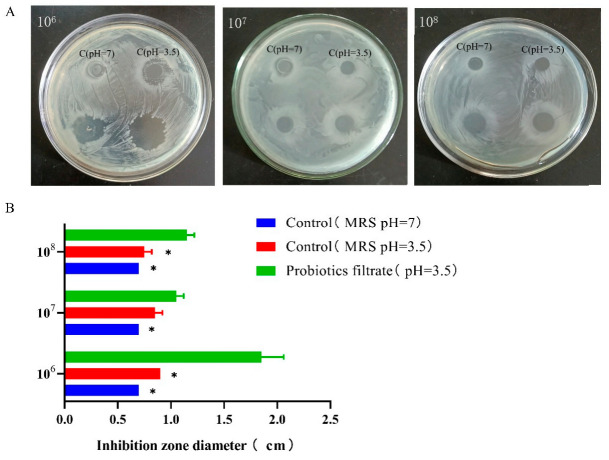
Compared with the MRS medium control with pH = 7, the supernatant of L. plantarum 1201 showed significant anti-S. Typhimurium ATCC 13311 activity, and had the best antibacterial effect on S. Typhimurium ATCC 13311 at 106 CFU/mL (Figure 5). In order to verify whether L. plantarum inhibits the growth of S. Typhimurium ATCC 13311 by producing an acidic environment, a set of MRS medium controls with pH = 3 was added. Compared with the MRS medium with pH = 7, the antibacterial effect of the MRS medium with pH = 3 was increased, but it was significantly lower than the fermentation supernatant of L. plantarum 1201 (Figure 5B). These results indicate that L. plantarum 1201 can significantly inhibit the growth activity of S. Typhimurium ATCC 13311, and this inhibition is not only performed by acid production.
Figure 5.
L. plantarum 1201 inhibits the growth of S. Typhimurium ATCC 13311. (A) Intuitive diagram of inhibition zone; (B) Bar graph of inhibition zone. * p < 0.05; paired two-tailed t-test.
4. Discussion
At present, HFD has been proven to promote inflammation [25,26]. With HFD, the incidence of intestinal inflammatory diseases has gradually increased. Obese mice showed a special immune response during oral infection with Salmonella [27] and HFD can exacerbate inflammatory bowel disease [28]. The intake of high-fat meat protein diets resulted in the impairment of the colon barrier through mucus suppression, downregulation of tight junctions, and gut inflammation in mice [29]. In this study, HFD mice showed higher colonization of S. Typhimurium, more serious intestinal inflammation and more secretion of pro-inflammatory factors. These demonstrated that HFD can promote S. Typhimurium ATCC 13311 infection and cause more severe inflammation.
The colonization of S. Typhimurium ATCC 13311 affects the balance of human intestinal flora and induces inflammation. How to inhibit S. Typhimurium infection has become a key research direction of social concern. In previous studies, probiotics have been found to have an inhibitory effect on S. Typhimurium. Bacillus probiotics can inhibit the activity of S. Typhimurium [30]. Bifidobacterium can down-regulate the gene expression of S. Typhimurium pathogenicity islands 1 and 2 [31]. High-yielding L. casei with linoleic acid can limit the growth and activity of S. Typhimurium [32]. L. plantarum can reduce the levels of S. Typhimurium in the livers and spleens of mice [33]. However, the mechanism by which L. plantarum inhibits S. Typhimurium infection is still unclear. In this study, we discussed the inhibition of S. Typhimurium ATCC 13311 infections by L. plantarum 1201.
In vivo, the first direct contact of Salmonella with a host is the adhesion to the surface of epithelial cells, and then it colonises the gastrointestinal tract [34]. This event is a prerequisite for the subsequent steps in pathogenesis that lead to mucosal infection, systemic spread and disease. Inhibition of the invasion of Salmonella into epithelial cells is the first step in disease prevention [35]. Our animal experiment results show that L. plantarum 1201 can reduce the colonization of S. Typhimurium ATCC 13311 in mice. This protective effect is similar to previous research. Mixed probiotics can inhibit the colonization of C. jejuni in the intestine [36]. L. casei LC2W can inhibit the colonization of E. coli O157:H7 in vivo through an in vivo imaging system [37]. B. clausii and L. reuteri can inhibit the colonization of C. difficile in the human intestine [38]. In summary, our research found that HFD can promote the colonization of S. Typhimurium ATCC 13311 in the host, while the protective effect of L. plantarum 1201 is achieved by reducing it.
The TLR4/NF-κB pathway is a common inflammation pathway induced by Gram-negative bacteria. Toll-like receptors are the main receptors for the innate immune system to recognize pathogenic microorganisms and play an important role in the innate immune response [39]. TLR4 is an important receptor for the transmembrane signal transduction of pathogenic microorganisms. It is closely related to the body’s innate immunity against infection and plays an important role in the early recognition of invading pathogenic microorganisms by the immune system [40]. In the inflammatory response, NF-κB is the most critical transcriptional regulatory factor. It plays an important role in normal cell metabolism and immune response, and participates in the MyD88-dependent signaling pathway in the TLR4 signaling pathway [41]. Many studies have shown that S. Typhimurium can activate NF-κB, thereby causing inflammation [42,43,44]. It has also been found that probiotics can inhibit the NF-κB pathway activated by S. Typhimurium. L. plantarum JSA22 can inhibit the activation of NF-κB induced by S. Typhimurium and reduce the secretion of IL-8 [45]. Our conclusions are consistent with the literature [46]. In our study, when mice ingested a large amount of S. Typhimurium ATCC 13311, TLR4 was activated and bound with MyD88, which, in turn causes the phosphorylation of serine residues at specific sites of IκB-α. Then, IκB-α is ubiquitinated and degraded, while NF-κB is released and transferred to the nucleus, inducing the production of inflammatory factors and promoting intestinal inflammation. HFD exacerbated this inflammatory response. However, the intervention of L. plantarum 1201 inhibited the colonization of S. Typhimurium, thereby inhibiting the activation of this inflammatory pathway.
Previous studies have shown that after TLR4 activates the TLR4/NF-κB signaling pathway, it will cause an immune response against microbial infections and release various inflammatory factors, such as TNF-α [47]. Inflammatory factors play an important role in the infection process of S. Typhimurium. In the case of inflammation, necrosis, or immune cells stimulated by tumor cell antigens, the secretion of IL-6 increases, which reflects an increase in bacterial colonization in tissues [48,49]. In contrast, IFN-γ and IL-17A have been shown to help the host resist S. Typhimurium. The increased secretion of these two cytokines reflects the enhanced protective immune response regulated by the microflora in mice [50,51]. CD4+ T cells can generate the transforming growth factor TGF-β, and TGF-β can induce the differentiation of Th17 cells or Treg cells to produce the inflammatory factor IL-17. Therefore, TGF- β has a role in inflammatory response and immune regulation [52]. TNF-α is a pro-inflammatory cytokine produced by specific cells under inflammatory stimulation and has a variety of biological activities [53]. It can expose the nuclear localization sequence of NF-κB by mediating the phosphorylation and ubiquitination of IκB. NF-κB then translocates into the nucleus and binds to the NF-κB site in the nucleus to initiate gene transcription and induce the massive release of cytokines including TNF-α. The released cytokines can further activate NF-κB, so that the positive feedback makes the inflammatory response progressively amplified [54]. IL-10 is another key cytokine for S. Typhimurium infection. The increase in IL-10 will improve the body’s immune tolerance, prevent S. Typhimurium from being eliminated, and trigger the spread of the system [55]. Interleukin-1β is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis [56]. IL-22 can effectively regulate inflammation, especially in inflammation caused by pathogenic bacteria and prevent the colonization and spread of S. Typhimurium in the intestine by maintaining the integrity of the epithelial barrier, thereby limiting bacterial growth and reducing inflammation [57,58].
From the qPCR results, we can see that S. Typhimurium colonization can promote the mRNA expression of inflammation-related cytokines, enhance the tissue’s inflammatory response and damage the body, which can promote the occurrence of intestinal inflammation. The situation in combination with an HFD is more pronounced. However, L. plantarum 1201 has significant alleviating effects on the inflammation and body damage caused by S. Typhimurium infection.
5. Conclusions
S. Typhimurium ATCC 13311 can activate the TLR4/NF-κB inflammatory pathway after colonization, promote the abnormal secretion of inflammatory factors, and cause intestinal inflammation. HFD can facilitate this process. L. plantarum 1201 can inhibit S. Typhimurium ATCC 13311 infections by inhibiting its colonization and inflammation pathways (Figure 6). It is expected that the L. plantarum 1201 can be used as a new target to protect human health from infection of S. Typhimurium through diet.
Figure 6.
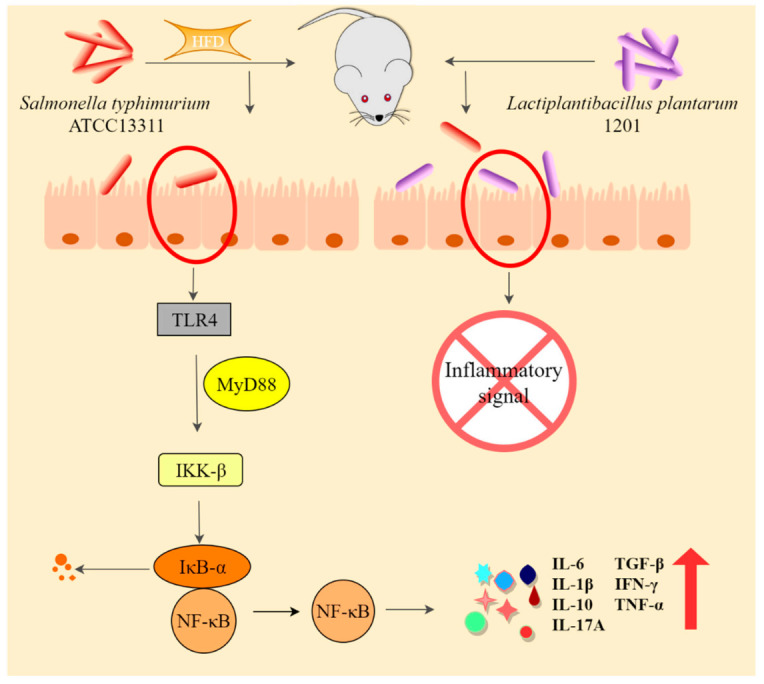
HFD exacerbates the intestinal infection of S. Typhimurium ATCC 13311, while L. plantarum 1201 inhibits it.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods11010085/s1, Table S1: Primer sequences for quantitative Real-Time polymerase chain reaction.
Author Contributions
Z.R. Methodology, Investigation, Data curation, Writing—original draft. L.P. Methodology, Data curation, Formal analysis. S.C. Software, Validation. Y.P. Software, Validation. H.L. Software, Data curation. H.W. Software, Data curation. C.W. Methodology, Data curation, Conceptualization, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science foundation of China (81760102 and 31770133). We sincerely thank the Editor and reviewers for their contributions and suggestions.
Institutional Review Board Statement
The study was approved by the Ethics Committee of the Institutional Animal Care and Use Committee of Nanchang Royo Biotech Co. Ltd. (RYE2020061101) and conducted according to the Guide for the Care and Use of Laboratory Animals (China).
Data Availability Statement
The data presented in this study are available in the article and its supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wells S., Fedorka-Cray P., Dargatz D., Ferris K., Green A. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Prot. 2001;64:3–11. doi: 10.4315/0362-028X-64.1.3. [DOI] [PubMed] [Google Scholar]
- 2.Malorny B., Hoorfar J. Toward standardization of diagnostic PCR testing of fecal samples: Lessons from the detection of Salmonellae in pigs. J. Clin. Microbiol. 2005;43:3033–3037. doi: 10.1128/JCM.43.7.3033-3037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carli K., Unal C., Caner V., Eyigor A. Detection of salmonellae in chicken feces by a combination of tetrathionate broth enrichment, capillary PCR, and capillary gel electrophoresis. J. Clin. Microbiol. 2001;39:1871–1876. doi: 10.1128/JCM.39.5.1871-1876.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayak R., Kenney P., Keswani J., Ritz C. Isolation and characterisation of Salmonella in a turkey production facility. Br. Poult. Sci. 2003;44:192–202. doi: 10.1080/0007166031000088370. [DOI] [PubMed] [Google Scholar]
- 5.Thorns C. Bacterial food-borne zoonoses. Rev. Sci. Tech. 2000;19:226–239. doi: 10.20506/rst.19.1.1219. [DOI] [PubMed] [Google Scholar]
- 6.Jain S., Bidol S., Austin J., Berl E., Elson F., Williams M., Deasy M., Moll M., Rea V., Vojdani J., et al. Multistate outbreak of Salmonella Typhimurium and saintpaul infections associated with unpasteurized orange Juice—United States, 2005. Clin. Infect. Dis. 2009;48:1065–1071. doi: 10.1086/597397. [DOI] [PubMed] [Google Scholar]
- 7.Sivapalasingam S., Friedman C., Cohen L., Tauxe R. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 2004;67:2342–2353. doi: 10.4315/0362-028X-67.10.2342. [DOI] [PubMed] [Google Scholar]
- 8.Vojdani J., Beuchat L., Tauxe R. Juice-associated outbreaks of human illness in the United States, 1995 through 2005. J. Food Prot. 2008;71:356. doi: 10.4315/0362-028X-71.2.356. [DOI] [PubMed] [Google Scholar]
- 9.Matamouros S., Miller S.S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochim. Biophys. Acta-Biomembr. 2015;1848:3021–3025. doi: 10.1016/j.bbamem.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poppe C., Smart N., Khakhria R., Johnson W., Spika J., Prescott J. Salmonella typhimurium DT104: A virulent and drug-resistant pathogen. Can. Vet. J. La Rev. Vet. Can. 1998;39:559–565. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Zou Y., Wang J., Ma H., Wang S. The protective effects of 2′-Fucosyllactose against E. Coli O157 infection are mediated by the regulation of gut microbiota and the inhibition of pathogen adhesion. Nutrients. 2020;12:1284. doi: 10.3390/nu12051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernet-Camard M., Liévin V., Brassart D., Neeser J., Servin A., Hudault S. The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl. Environ. Microbiol. 1997;63:2747–2753. doi: 10.1128/aem.63.7.2747-2753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soledad B., Juan E., Fernando V., Covadonga B. Adherence of human vaginal Lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 1998;66:1985–1989. doi: 10.1128/iai.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coconnier M.-H., Liévin V., Lorrot M., Servin A. Antagonistic activity of Lactobacillus acidophilus LB against intracellular Salmonella enterica serovar Typhimurium infecting human enterocyte-like Caco-2/TC-7 cells. Appl. Environ. Microbiol. 2000;66:1152–1157. doi: 10.1128/AEM.66.3.1152-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Lai F., Chen Z. Research progress on the mechanism of action and antibacterial properties of probiotics. Agric. Prod. Process. 2011;9:65–68. [Google Scholar]
- 16.Han N., Jia L., Guo L., Su Y., Luo Z., Du J., Mei S., Liu Y. Balanced oral pathogenic bacteria and probiotics promoted wound healing via maintaining mesenchymal stem cell homeostasis. Stem Cell Res. Ther. 2020;11:61. doi: 10.1186/s13287-020-1569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran S., Munoz C., Elola L., Quintana J., Segovia C., Trombert A. The expression of heterologous MAM-7 in Lactobacillus rhamnosus reduces its intrinsic capacity to inhibit colonization of pathogen Vibrio parahaemolyticus in vitro. Biol. Res. 2016;49:2. doi: 10.1186/s40659-015-0064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama K., Nakazato A., Ueno S., Seto Y., Kakuda T., Takai S., Yamamoto Y., Mukai T. Cell surface-associated aggregation-promoting factor from Lactobacillus gasseri SBT2055 facilitates host colonization and competitive exclusion of Campylobacter jejuni. Mol. Microbiol. 2015;98:712–726. doi: 10.1111/mmi.13153. [DOI] [PubMed] [Google Scholar]
- 19.Hai D., Lu Z., Huang X., Lv F., Bie X. In vitro screening of chicken-derived Lactobacillus strains that effectively inhibit Salmonella colonization and adhesion. Foods. 2021;10:569. doi: 10.3390/foods10030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Han Y., Yang R., Zhao X. Condition optimization and application of FM4-64 and Hoechst dyes in live bacterial membranes and nucleoid marker positioning. Acta Microbiol. Sin. 2015;55:1068–1073. [PubMed] [Google Scholar]
- 21.Ruiz J., Núñez M., Lorente I., Pérez J., Simarro E., Gómez J. Performance of six culture media for isolation of Salmonella species from stool samples. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 1996;15:922–926. doi: 10.1007/BF01690509. [DOI] [PubMed] [Google Scholar]
- 22.Zappa M., Doblas S., Cazals-Hatem D., Milliat F., Lavigne J., Daniel F., Jallane A., Garteiser P., Vilgrain V., Ogier-Denis E., et al. Quantitative MRI in murine radiation-induced rectocolitis: Comparison with histopathological inflammation score. NMR Biomed. 2018;31:e3897. doi: 10.1002/nbm.3897. [DOI] [PubMed] [Google Scholar]
- 23.Sahami S., Wildenberg M., Koens L., Doherty G., Martin S., D’Haens G., Cullen G., Bemelman W., Winter D., Buskens C. Appendectomy for therapy-refractory ulcerative colitis results in pathological improvement of colonic inflammation: Short-term results of the PASSION study. J. Crohns Colitis. 2019;13:165–171. doi: 10.1093/ecco-jcc/jjy127. [DOI] [PubMed] [Google Scholar]
- 24.Wotzka S., Kreuzer M., Maier L., Arnoldini M., Nguyen B., Brachmann A., Berthold D., Zund M., Hausmann A., Bakkeren E., et al. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat. Microbiol. 2019;4:2164–2174. doi: 10.1038/s41564-019-0568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryyti R., Hämäläinen M., Peltola R., Moilanen E. Beneficial effects of lingonberry (Vaccinium vitis-idaea L.) supplementation on metabolic and inflammatory adverse effects induced by high-fat diet in a mouse model of obesity. PLoS ONE. 2020;15:e0232605. doi: 10.1371/journal.pone.0232605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q., Liu X., Zhang Y., Song Y., Ma X., Shi Y., Li X.L. plantarum, L. fermentum, and B. breve beads modified the intestinal microbiota and alleviated the inflammatory response in high-fat diet-fed mice. Probiotics Antimicrob. Proteins. 2020;12:535–544. doi: 10.1007/s12602-019-09564-3. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez-Orozco R., Franco R., Pérez V., Ramírez E., Hernández L., López B. Diet-induced obese mice exhibit altered immune responses to early Salmonella Typhimurium oral infection. J. Microbiol. Biotechnol. 2018;56:673–682. doi: 10.1007/s12275-018-8083-6. [DOI] [PubMed] [Google Scholar]
- 28.Lee J., Cevallos S., Byndloss M., Tiffany C., Olsan E., Butler B., Young B., Rogers A., Nguyen H., Kim K., et al. High-fat diet and antibiotics cooperatively impair mitochondrial bioenergetics to trigger dysbiosis that exacerbates pre-inflammatory bowel disease. Cell Host Microbe. 2020;28:273–284.e6. doi: 10.1016/j.chom.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain M., Umair I., Ahmad M., Khan I., Brohi S., Shah A., Shinwari K., Zhao D., Xu X., Zhou G., et al. Meat proteins in a high-fat diet have a substantial impact on intestinal barriers through mucus layer and tight junction protein suppression in C57BL/6J mice. Food Funct. 2019;10:6903–6914. doi: 10.1039/C9FO01760G. [DOI] [PubMed] [Google Scholar]
- 30.Kawarizadeh A., Pourmontaseri M., Farzaneh M., Hosseinzadeh S., Ghaemi M., Tabatabaei M., Pourmontaseri Z., Pirnia M. Interleukin-8 gene expression and apoptosis induced by Salmonella Typhimurium in the presence of Bacillus probiotics in the epithelial cell. J. Appl. Microbiol. 2020;131:449–459. doi: 10.1111/jam.14898. [DOI] [PubMed] [Google Scholar]
- 31.Bayoumi M., Griffiths M. Probiotics down-regulate genes in Salmonella enterica serovar typhimurium pathogenicity islands 1 and 2. J. Food Prot. 2010;73:452–460. doi: 10.4315/0362-028X-73.3.452. [DOI] [PubMed] [Google Scholar]
- 32.Tabashsum Z., Peng M., Bernhardt C., Patel P., Carrion M., Rahaman S., Biswas D. Limiting the pathogenesis of Salmonella Typhimurium with berry phenolic extracts and linoleic acid overproducing Lactobacillus casei. J. Microbiol. Biotechnol. 2020;58:489–498. doi: 10.1007/s12275-020-9545-1. [DOI] [PubMed] [Google Scholar]
- 33.Tsai C., Liang H., Yu B., Hsieh C., Hwang C., Chen M., Tsen H. The relative efficacy of different strain combinations of lactic acid bacteria in the reduction of populations of Salmonella enterica Typhimurium in the livers and spleens of mice. FEMS Immunol. Med. Microbiol. 2011;63:44–53. doi: 10.1111/j.1574-695X.2011.00826.x. [DOI] [PubMed] [Google Scholar]
- 34.Ofek I., Sharon N. Adhesins as lectins: Specificity and role in infection. Curr. Top. Microbiol. Immunol. 1990;151:91–113. doi: 10.1007/978-3-642-74703-8_5. [DOI] [PubMed] [Google Scholar]
- 35.Darwin K., Miller V. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 1999;12:405–428. doi: 10.1128/CMR.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan V. The role of probiotics in the inhibition of Campylobacter jejuni colonization and virulence attenuation. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:1503–1513. doi: 10.1007/s10096-015-2392-z. [DOI] [PubMed] [Google Scholar]
- 37.Wang G., Zhang Y., Song X., Xia Y., Lai P., Ai L. Lactobacillus casei LC2W can inhibit the colonization of Escherichia coli O157:H7 in vivo and reduce the severity of colitis. Food Funct. 2019;10:5843–5852. doi: 10.1039/C9FO01390C. [DOI] [PubMed] [Google Scholar]
- 38.Mills J., Rao K., Young V. Probiotics for prevention of Clostridium difficile infection. Curr. Opin. Gastroenterol. 2018;34:3–10. doi: 10.1097/MOG.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim K., Staudt L. Toll-Like receptor signaling. Cold Spring Harb. Perspect. Biol. 2013;5:a011247. doi: 10.1101/cshperspect.a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W., Liu M., Cao J., Yang P., Zheng X., Dong B., Lu J. Role of TLR4 in process of colonic inflammation recovery induced by LPS. Chin. J. Pathophysiol. 2017;33:336–343. [Google Scholar]
- 41.Zhu H., Bian C., Yuan J., Chu W., Xiang X., Chen F., Wang C., Feng H., Lin J. Curcumin attenuates acute inflammatory injury by inhibiting the TLR4/MyD88/NF-κB signaling pathway in experimental traumatic brain injury. J. Neuroinflamm. 2014;11:59. doi: 10.1186/1742-2094-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos M., Zambrano S., Tiérrez A., Bianchi M., Agresti A., García D. Single-cell analyses reveal an attenuated NF-κB response in the Salmonella-infected fibroblast. Virulence. 2017;8:719–740. doi: 10.1080/21505594.2016.1229727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong N., Xue C., Zhang L., Zhang T., Wang C., Bi C., Shan A. Oleanolic acid enhances tight junctions and ameliorates inflammation in Salmonella typhimurium-induced diarrhea in mice via the TLR4/NF-κB and MAPK pathway. Food Funct. 2020;11:1122–1132. doi: 10.1039/C9FO01718F. [DOI] [PubMed] [Google Scholar]
- 44.Van W., Fricke F., Herhaus L., Gupta J., Hötte K., Pampaloni F., Grumati P., Kaulich M., Sou Y., Komatsu M., et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2017;2:17066. doi: 10.1038/nmicrobiol.2017.66. [DOI] [PubMed] [Google Scholar]
- 45.Eom J., Song J., Choi H. Protective Effects of a Novel Probiotic Strain of Lactobacillus plantarum JSA22 from Traditional Fermented Soybean Food Against Infection by Salmonella enterica Serovar Typhimurium. J. Microbiol. Biotechnol. 2015;25:479–491. doi: 10.4014/jmb.1501.01006. [DOI] [PubMed] [Google Scholar]
- 46.Wei H., Yin L., Feng S., Wang X., Yang K., Zhang A., Zhou H. Dual-parallel inhibition of IL-10 and TGF-β1 controls LPS-induced inflammatory response via NF-κB signaling in grass carp monocytes/macrophages. Fish Shellfish Immunol. 2015;44:445–452. doi: 10.1016/j.fsi.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Guo Y., Chen S., Li Y., Fan M., Sun Y., Li W., Shi Q. TLR4 activation with LPS inhibits BMP9-induced osteogenic differentiation of immortalized mouse embryonic fibroblasts. Basic Clin. Med. 2017;37:25–31. [Google Scholar]
- 48.Hong D., Angelo L., Kurzrock R. Interleukin-6 and its receptor in cancer: Implications for translational therapeutics. Cancer. 2010;110:1911–1928. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 49.Thiemann S., Smit N., Roy U., Lesker T., Galvez E., Helmecke J., Basic M., Bleich A., Goodman A., Kalinke U., et al. Enhancement of IFN-γ production by distinct commensals ameliorates Salmonella-Induced Disease. Cell Host Microbe. 2017;21:682–694. doi: 10.1016/j.chom.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Sharipov F., Kalempa D. Developmentally regulated intestinal expression of IFN-γ and Its target genes and the age-specific response to Enteric Salmonella infection. J. Immunol. 2005;175:1127–1136. doi: 10.4049/jimmunol.175.2.1127. [DOI] [PubMed] [Google Scholar]
- 51.Raffatellu M., Santos R., Verhoeven D., George M., Wilson R., Winter S., Godinez I., Sankaran S., Paixao T., Gordon M., et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yun S., Kim S., Kim E. The molecular mechanism of transforming growth factor-β signaling for intestinal fibrosis: A mini-review. Front. Pharmacol. 2019;10:162. doi: 10.3389/fphar.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenstiel P., Sina C., Franke A., Schreiber S. Towards a molecular risk map-recent advances on the etiology of inflammatory bowel disease. Semin. Immunol. 2009;21:334–345. doi: 10.1016/j.smim.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Gunther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M., Hedrick S., Tenzer S., Neurath M. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiennimitr P., Winter S., Winter M., Xavier M., Tolstikov V., Huseby D., Sterzenbach T., Tsolis R., Roth J., Baumler A. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H., Li Y., Zuo Z., Han Q.J., Zhang J., Zhang C. Mutual regulation between intestinal macrophages and CD4+ T cells enhances resistance to Salmonella typhimurium infection; Proceedings of the 9th National Immunology Academic Conference of the Chinese Society of Immunology Assembly; Jinan, China. 18–21 October 2014. [Google Scholar]
- 57.Raffatellu M., George M., Akiyama Y., Hornsby M., Nuccio S., Paixao T., Butler B., Chu H., Santos R., Berger T., et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keir M., Yi Y., Lu T., Ghilardi N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 2020;217:e20192195. doi: 10.1084/jem.20192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article and its supplementary materials.