Abstract
Drug development for specific antiviral agents against coronavirus disease 2019 (COVID-19) is still an unmet medical need as the pandemic continues to spread globally. Although huge efforts for drug repurposing and compound screens have been put forth, only a few compounds are in late-stage clinical trials. New approaches and assays are needed to accelerate COVID-19 drug discovery and development. Here, we report a time-resolved fluorescence resonance energy transfer-based assay that detects the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid protein (NP) produced in infected cells. It uses two specific anti-NP monoclonal antibodies conjugated to donor and acceptor fluorophores that produce a robust ratiometric signal for high throughput screening of large compound collections. Using this assay, we measured a half maximal inhibitory concentration (IC50) for remdesivir of 9.3 μM against infection with SARS-CoV-2 USA/WA1/2020 (WA-1). The assay also detected SARS-CoV-2 South African (Beta, β), Brazilian/Japanese P.1 (Gamma, γ), and Californian (Epsilon, ε) variants of concern (VoC). Therefore, this homogeneous SARS-CoV-2 NP detection assay can be used for accelerating lead compound discovery for drug development and for evaluating drug efficacy against emerging SARS-CoV-2 VoC.
Keywords: SARS-CoV-2, variants of concern, TR-FRET, nucleocapsid phosphoprotein, assay development
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and has been responsible for the deadly global pandemic of coronavirus disease 2019 (COVID-19), rivaled only by the “Spanish Flu” pandemic of 1918–1919 (40–50 million deaths) and the ongoing HIV/AIDS pandemic from 1981–present (25–30 million deaths) (Visualizing the History of Pandemics, https://www.visualcapitalist.com/history-of-pandemics-deadliest/). As of June 2021, there have been over 168 million infection cases and 3.5 million deaths due to COVID-19 and these tolls are increasing daily.1 Thought to have been transmitted from animals to humans, the exact circumstances of the first infection have not been determined, but evolutionary and comparative analysis suggests a likely transmission from bats to pangolins to humans.2,3 Although the rate of mutation in the viral genome is believed to be lower than other viral pathogens, several variants of concern (VoC) have become dominant strains in different locations around the world such as the U.K. variant B.1.1.7 (Alpha, α), the South African variant B.1.135 (Beta, β), the Brazilian/Japanese variant P.1 (Gamma, γ), the Indian variant B.1.617.2 (Delta, δ), and the Californian variant B.1.427/B.1.429 (Epsilon, ε)4 (WHO EPI-WIN, Update 60 https://www.who.int/teams/risk-communication/epi-win-updates). VoC are problematic because they contain mutations in the viral spike (S) glycoprotein that enhance affinity to the host viral receptor human angiotensin converting enzyme 2 (hACE2).5,6 In addition, neutralizing antibodies (NAbs) produced by the immune response after natural infection or through vaccination have been shown to be less effective against VoC containing mutations in key regions of the S targeted by the NAbs.7,8 Hence, there is a need for rapid and accurate screen procedures to identify existing, or novel, drugs against newly emerging SARS-CoV-2 VoC.
Drug development for specific antiviral agents that combat COVID-19 is still an unmet urgent task. Several safe and effective vaccines have been approved and large-scale vaccination campaigns have already taken place since early 2021.9 However, there will still be a need for a multipronged approach to manage COVID-19 cases as highlighted by the situation with influenza virus that is responsible for a significant number of yearly infections associated with significant morbidity and mortality despite safe and effective vaccines being on place. Moreover, as long as there are large numbers of infections anywhere in the world, the risk for another resurgence of cases and emergence of virulent SARS-CoV-2 VoC remains high.
Remdesivir, an RNA-dependent RNA polymerase (RdRP) inhibitor, is currently the only drug approved for the treatment of hospitalized COVID-19 patients.10 One challenge for remdesivir is that it needs to be administered intravenously.11 In addition, the therapeutic efficacy of remdesivir is also in question.12−14 Therefore, effective antiviral agents are urgently needed for treatment of COVID-19 patients.
Optimized compound screening assays are critically important for early drug discovery and development. Currently, two main types of assays are being used for compound screens in the COVID-19 drug development area of research. One is a mechanistic approach directly targeting SARS-CoV-2 proteins including 3CLpro,15 PLpro,16 RdRP,17 and S-hACE2 binding.18 Another approach is utilizing phenotypic screens using cell-based functional assays such as viral entry,19,20 viral replication (replicon),21 and live virus infection. The live SARS-CoV-2 assay is an important tool for compound screening and evaluation of efficacy before advancing forward to in vivo model efficacy studies. To date, assays including SARS-CoV-2 cytopathic effect (CPE),22 RT-PCR, and antigen immunostaining, among others, have been used in compound screens and evaluation of drug efficacy.23 While the RT-PCR assay does not have enough screening throughput, the SARS-CoV-2 CPE assay may not identify all of the active compounds that inhibit SARS-CoV-2 replication in the screen because CPE is cell type-dependent and is an indirect readout of viral infection.
SARS-CoV-2 nucleocapsid protein (NP) is a structural protein that binds the positive strand viral genome within the nucleocapsid core.24,25 Previously, we developed an AlphaLISA assay for the measurement of SARS-CoV-2 NP as an indicator of virus replication in infected cells.26 We used the NP as a target viral protein to develop a high throughput screening (HTS) assay that could be used in a biosafety level 3 containment facility to identify promising antiviral drugs targeting SARS-CoV-2 multiplication.27 Targeting the more conserved and highly expressed NP instead of the S would prevent detection failures due to newly emerging SARS-CoV-2 VoC with S mutations and increase sensitivity, respectively.
In this work, we have developed a homogeneous time-resolved fluorescence (HTRF) assay that relies on the time-resolved fluorescence resonance energy transfer (TR-FRET) between a donor and acceptor fluorophore when they are in close proximity and proper orientation relative to each other.28,29 To specifically detect the SARS-CoV-2 NP, we conjugated two NP-specific monoclonal antibodies (MAbs) to either a donor or acceptor fluorophore that are brought into proximity when NP is present. We developed a microplate assay using recombinant NP added to cells, NP-transfected cells, and both tissue culture supernatants (TCS) and cell lysates from infected cells. The homogeneous format means that the whole well fluorescence emission ratio between the acceptor and donor fluorophores is read without washing using a plate reader with a TR-FRET module.
The assay we have developed involves a single step; it is easy to use and scalable to HTS to identify SARS-CoV-2 antivirals. The assay can identify drugs targeting any of the steps of SARS-CoV-2 life cycle, hence enabling screening efforts to identify drugs with activity against emerging VoC.
Methods
Reagents
Vero E6 cells [CRL-1586, Resource Research Identifier (RRID): CVCL0574] were purchased from American Tissue Type Collection. The following items were purchased from Corning TM: EMEM (10-009-CV), HI fetal bovine serum (FBS) (35-016-VC, and 0.25% Trypsin (25053CI). Pen/Strep (15140-122) was purchased from Gibco. Phosphate-buffered saline (PBS) (SH30256FS) was purchased from HyClone. The following items were purchased from Greiner Bio-One: white 384 well plate (781073), white half area 96-well plate (675083), and white 96-well plate (655083). The custom labeling of MAbs was performed by Columbia Biosciences. Rabbit anti-NP SARS-CoV-2 antibodies R001 (40143-R001, RRID number: AB_2827974), R004 (40143-R004, RRID number: AB_2827975), R019 (40143-R019, RRID number: AB_2827973), and R040 (40143-R040, RRID number: AB_2827976) were purchased from Sino Biological, and mouse anti-NP MAb 1C7C7 was provided by Dr. Tomas Moran and purchased from Leinco (LT7000). The following items were purchased from ThermoFisher: Goat anti-mouse HRP (A16072), goat-anti rabbit HRP (A16104), West Femto substrate (34094), Lipofectamine 2000 (11668019), and Optimem I Reduced Serum Media (31985070). Triton X-100 (100×) was purchased from Sigma Aldrich. The cOmplete ULTRA protease inhibitor (PI) (05892791001) was purchased from Roche.
Antibody Matrixing
Four rabbit MAbs and one mouse Mab specific for SARS-CoV-2 NP were labeled with europium (donor Ab) or DyLight650 (acceptor Ab). IgG antibodies R001, R004, R019, and R040 were raised against recombinant SARS-CoV nucleocapsid phosphoprotein [NP (Sino Biological, 0143-V08B)] and expressed in HEK293 cells. Mouse MAb 1C7 (1C7C7, IgG2a) was expressed in HEK293 using recombinant NP as the immunogen. The labeled MAbs were then tested in HTRF assay, in triplicate wells, using a cross-matrix assay format. The data are presented as a median TR-FRET ratio (acceptor fluorescence/donor fluorescence × 10,000).
Western Blot
After sodium dodecyl sulphate–polyacrylamide gel electrophoresis and Western blotting, the membranes were probed with anti-NP MAbs R001 and 1C7 at 1:1000 diluted in Superblock Buffer (ThermoFisher). Then antigen–antibody complexes were detected using appropriate anti-species HRP conjugates and West Femto substrate.
Vero E6 Cell Culture
African green monkey kidney epithelial cells (Vero E6; CRL-1586) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1% PSG (100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine) at 37 °C with 5% CO2.
Assay Condition Optimization Using Recombinant NP
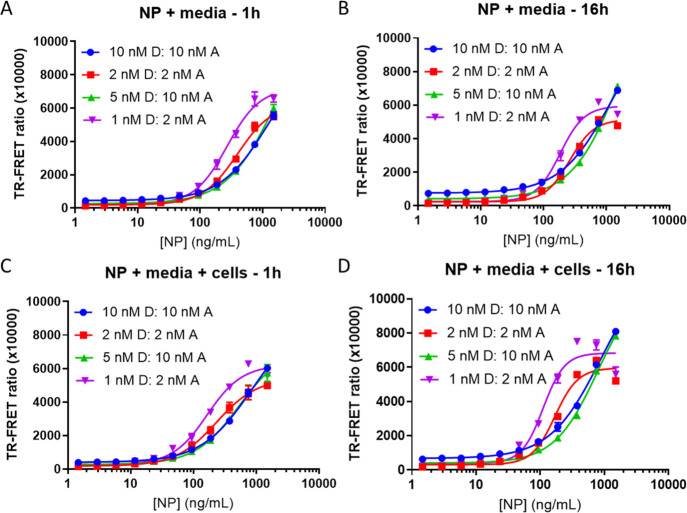
The best pair of donor and acceptor, R001-Eu (donor) and 1C7-DL650 (acceptor), was used to evaluate assay performance in cell culture media with and without cells. Recombinant untagged NP starting at 1500 ng/mL was serially diluted twofold 11 times to produce the standard curve. NP was diluted in media, and 20 μL was added to the wells followed by 10 μL of the 3× HTRF donor and acceptor diluted in 1.5% Triton X-100 and PI in the ratios indicated in Figure 2 for a total assay volume of 30 μL. In another 384-well plate, 15 μL of Vero E6 cells was plated and incubated overnight (O/N) prior to addition of 5 μL of 4× NP in media and 10 μL of 3× HTRF reagents in Triton X-100 with PI for a total assay volume of 30 μL. Plates were incubated with reagents for either 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time. Briefly, the 337 nm/620 nm excitation/emission and 337 nm/665 nm excitation/emission were measured using the dual emission HTRF optical module on the Pherastar (BMG Labtech). The ratio of emissions (665 nm/620 nm) was calculated and multiplied by 10,000.
Figure 2.
Optimization of the donor to acceptor ratio, concentration, and incubation time using media or Vero E6 cells. TR-FRET ratio signal (acceptor/donor) detection using R001-Eu and 1C7-DL650 as the donor (D) and acceptor (A) pair, respectively. SARS-CoV-2 NP was used to generate the 11-point standard curve starting from 1500 ng/mL serially diluted 1:2. HTRF reagents were incubated with NP in cell culture media only (A,B) or in media with 5000 Vero E6 cells (C,D) for either 1 h at RT or O/N at 4 °C. N = 3 wells per condition in a 384-well plate. Error bars indicate S.D.
Preparation of Viral Lysates and TCS Production
Vero-E6 cells were plated in 12-well plates at 450,000 cells/well in 1.25 mL of growth media. Cells were incubated for 24 h at 37 °C. A 250 μL aliquot of SARS-CoV-2 [USA-WA1/2020 strain (Gen Bank: MN985325.1)] was used at a multiplicity of infection (MOI) of 0.05. Cells were inoculated for 45 min at 37 °C. The supernatant was collected and pooled for 0 h TCS (Mock). A final concentration of 0.5% Triton-X 100 and 1× PI was used for all samples. TCS samples were stored at −20 °C until needed. Next, 1.5 mL of fresh media was added to each well, and cells were incubated for 24and 48 h at 37 °C. Six wells were harvested at each time point. The supernatant was collected from 6 wells each at 24 and 48 h. Samples were pooled for Mock, 24 or 48 h TCS. Samples were stored at −20 °C until needed. Lysate was collected from 6 wells each at 24 and 48 h. Cells were rinsed once with ice-cold PBS, and 250 μL/well of cell lysis buffer + PI was added to lyse cells. PI was added to the lysis buffer before lysing cells. Cells were scraped to the bottom of the well, and pooled lysates were collected into 1.5 mL tubes on ice. Tubes were vortexed for 10 s and placed on ice for 10 min. This was repeated three times. Lysates were spun down for 30 min at 13200 rpm at 4 °C and stored at −20 °C until further use.
Estimates of viral NP concentrations in infected TCS and cell lysate were conducted using standard curve interpolation within the linear range.
Testing TCS and Cell Lysates from Infected Vero E6 Cells
This TR-FRET NP assay was evaluated for performance using Vero E6 TCS and lysate in 384 well plates. Lysates and TCS were diluted in PBS + Triton-X 100 + PIs 1:15 (1×) followed by threefold dilutions. Plates were incubated for either 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time.
Evaluating 384-Well Plate Assay Robustness
Whole-plate statistics were calculated using recombinant NP at a concentration of 500 ng/mL in Vero E6 cells. 15 μL of 5000 Vero E6 cells was seeded into 384-well plates and incubated O/N. Reagents were added using a fully automated Thermo Fisher Combi Multidrop liquid dispenser. 5 μL of cell culture media with or without NP was added to columns 3–24 and columns 1–2, respectively. Next, 10 μL of 3× HTRF reagents was diluted in Triton X-100 with PI for a total assay volume of 30 μL. The TR-FRET ratio was measured at the end of each incubation time. Z-factor, coefficients of variation (CV), and signal to background (S/B) were calculated for the entire plate.
Transfection of NP into Vero E6 Cells
DNA coding sequences corresponding to SARS-CoV-2 NP were amplified from genomic RNA from SARS-CoV-2, isolate USA-WA1/2020 (BEI Resources, catalog no. NR-52285) using primer sets 5SacINP (5′- CACTGAGCTCATGTTTGTTTTTCTTGTTTTATTG-3′) and 3SmaINP (3′- CACTCCCGGGTGTGTAATGTAATTTGACTCCTTT-5′). PCR fragments were digested with SacI and SmaI and cloned into SacI- and SmaI-digested pCAGGS plasmid containing a C-terminal HA epitope tag (pCAGGS NP-HA).
20 μL of 15,000 Vero E6 cells per well was seeded into half-area 96-well plates and incubated O/N at 37 °C, 5% CO2. A plasmid encoding for SARS-CoV-2 NP (pCAGGS NP-HA) was transfected into the cells using Lipofectamine 2000 according to manufacturer’s recommendations (starting concentration 100 ng of plasmid and twofold serial dilutions). Cells were incubated with plasmid and Lipofectamine 2000 for 4-6h before replacing media with DMEM containing 2% FBS. Cells were incubated O/N at 37 °C, 5% CO2 to express NP in a volume of 20 μL. After 24 h, 10 μL of 3× HTRF reagents diluted in 1.5% Triton X-100 with PI were added to cells and incubated for 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time.
Live Virus Testing in Vero E6 Cells
A reverse time course was used to infect cells. Briefly, 20 μL of Vero E6 cells was seeded into half-area 96-well plates at 8500, 6000, and 4500 cells/well for the 48, 24, and 12 h infection durations, respectively, and incubated O/N at 37 °C, 5% CO2. 4500 cells/well were also used for the 0 h infection duration and the standard curve. Media was removed the next day, and 20 μL/well of threefold serial dilutions (starting MOI 0.25) of SARS-CoV-2 USA-WA1/2020 (BEI Resources, NR-52281) was used. Upon completion of the reverse time course, 10 μL of 3× HTRF reagents diluted in 1.5% Triton X-100 with PI was added to cells and incubated for 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time.
Demonstrating Antiviral Efficacy Using Remdesivir
20 μL of Vero E6 cells was seeded into half-area 96-well plates at 8500 cells/well and incubated O/N at 37 °C, 5% CO2. Cells were infected with 10 μL of SARS-CoV-2 USA-WA1/2020 in infection medium (DMEM with 2% FBS) at an MOI of 0.009, and cells were simultaneously treated with 10 μL of threefold serial dilutions (starting concentration of 100 μM) of remdesivir or 0.1% dimethyl sulfoxide as a vehicle control. After 24 h, 10 μL of 3× HTRF reagents diluted in 1.5% Triton X-100 with PI was added to cells and incubated for 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time.
Detecting VoC
A reverse time course was used to infect cells. 20 μL of Vero E6 cells was seeded into half-area 96-well plates at 8500 cells/well for 24 h and 6000 cells/well for 12, 8, 4, and 0 h infection durations as well as the standard curve and incubated O/N at 37 °C, 5% CO2. Media was removed the next day, and cells were infected with 20 μL/well of threefold serial dilutions (starting MOI 0.25) of SARS-CoV-2 isolates USA-WA1/2020, SARS-CoV-2/human/USA/CA-UCSF-0001C/2020 (UCSF, a gift from Dr. Charles Chiu, hCoV-19/Japan/TY7-501/2021 (BEI Resources, NR-54981), or hCoV-19/South Africa/KRISP-EC-K005321/2020 (BEI Resources, NR-54008) in infection medium (DMEM with 2% FBS) for each indicated time point. 10 μL of 3× HTRF reagents diluted in 1.5% Triton X-100 with PI was added to cells and incubated for 1 h at RT or O/N at 4 °C. The TR-FRET ratio was measured at the end of each incubation time.
NP Amino Acid Sequence Alignment
Nucleotide sequences of viral strains, SARS-CoV-2 isolate USA-WA1/2020 (accession number EPI_ISL_404895), Beta (EPI_ISL_678625), Gamma (EPI_ISL_833366), and Epsilon (EPI_ISL_2712562) were downloaded from GISAID.30 EPI_ISL_2712562 only used for alignment contains the same mutations in NP as SARS-CoV-2/human/USA/CA-UCSF-0001C/2020 used in this study. Nucleotide sequences were translated to amino acid sequences using ExPASY online software,31 and the codon frame that contained the full NP was copied to ClustalO (1.2.4)32 for multiple sequence alignment. Portions of the sequences that contained mutations were extracted and compared.
Statistical Analysis
Nonlinear regression was used to generate the curve fits. S/B calculations were made using the ratio of the TR-FRET signal at time X and the signal at time 0. For TCS and lysate NP concentration interpolation, an asymmetric sigmoidal 5PL model was used. Graphpad Prism V9.1.0 was used to generate all graphs. BioRender was used to generate the illustration in Figure 1A.
Figure 1.
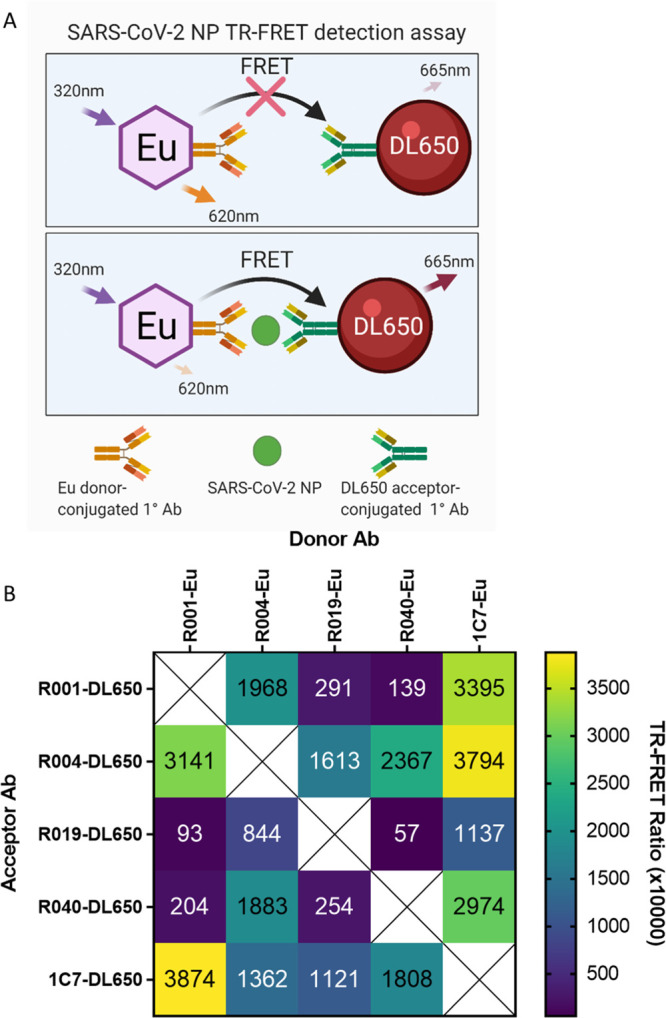
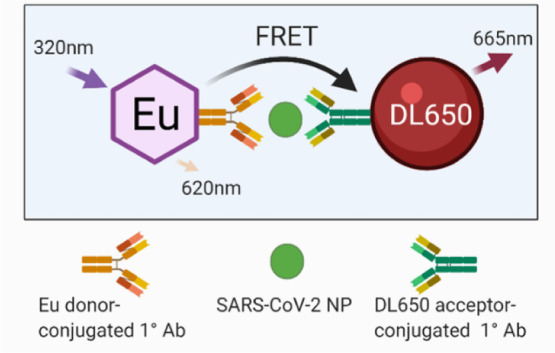
Selection of the optimal donor/acceptor antibody pair. (A) Illustration of the HTRF assay for SARS-CoV-2 NP showing the Eu donor-conjugated primary MAb and DL650 acceptor-conjugated primary MAb detecting SARS-CoV-2 and enabling FRET. (B) Five MAbs specific for SARS-CoV-2 NP labeled with europium (donor Ab) or DyLight650 (acceptor Ab). The labeled MAbs were then tested in HTRF assay, in triplicate wells using a cross-matrix assay format. The presented data are median TR-FRET counts (acceptor fluorescence/donor fluorescence X 10,000). The MAb pair (R001-Eu/1C7-DyLight650) showed the highest specific signal and was selected for further assay development.
Results
Selection of Donor and Acceptor MAb Pairs for the TR-FRET NP Assay
We sought to develop a precise, reproducible, scalable, and easy-to-use assay to detect infection and replication of SARS-CoV-2 under conditions compatible with HTS. We selected SARS-CoV-2 NP as the target viral protein to be a direct readout of viral infection and replication. We chose a FRET-based strategy using the popular HTRF system consisting of a donor and acceptor fluorophore conjugated to two SARS-CoV-2 NP MAbs, as these detection reagents offer high sensitivity and dynamic range to make HTS possible. The detection system consisted of the lanthanide fluorophore europium cryptate (Eu donor) conjugated to one NP MAb and the acceptor fluorophore DyLight 650 (DL650 acceptor) conjugated to a second NP MAb (Figure 1A). In the absence of SARS-CoV-2 NP, the distance and orientation of the donor and acceptor are incompatible with energy transfer, whereas in the presence of NP, the proximity of the donor and acceptor is close enough to allow FRET to occur. Laser excitation of Eu at 320 nm induces an emission at 620 nm. With energy transfer from the Eu donor, the DL650 acceptor produces an emission at 665 nm. The ratio of acceptor to donor emission intensity (665 nm/620 nm) upon excitation at 320 nm is calculated as the TR-FRET signal that correlates positively with the amount of NP present. In the absence of NP, the ratio is smaller and any signal present is considered background signal produced by random interactions between the donor and acceptor.
We tested several combinations of primary SARS-CoV-2 NP MAbs, and the greatest TR-FRET signal was produced by R001 coupled to the Eu donor and 1C7 coupled to the DL650 acceptor (Figure 1B). This combination was selected for further development. Both MAbs detected SARS-CoV-2 NP, as shown by Western Blot (Supporting Information Figure S1).
Evaluation of Assay Performance for Detection of Recombinant SARS-CoV-2 NP
We evaluated cell culture media, the presence of cells, reagent incubation time, the ratio of donor to acceptor, and the concentration of reagents on the TR-FRET ratio and S/B ratio in the presence of recombinant NP. In media alone with a 1 h incubation of reagents, a concentration of 1 nM donor and 2 nM acceptor produced the greatest TR-FRET ratio, was more sensitive at lower concentrations of NP, produced the largest S/B, but started to plateau at the highest concentrations of NP. The maximum ratio achieved at the highest concentration of NP (1500 ng/mL) was comparable between all reagent combinations (Figure 2A).
After O/N incubation, a hook effect appears with the lower concentration of reagents at the highest concentration of NP, suggesting an upper limit of detection between 750 and 1500 ng/mL of NP. The hook effect is defined as a signal decrease due to saturating antigen concentrations that prevent binding of both donor- and acceptor-linked antibodies to the same antigen molecule. 1 nM donor and 2 nM acceptor was saturated at 1500 ng/mL of NP, but exhibited high S/B (Figure 2B and Supporting Information Figure S2). In contrast with the low concentration combinations, the high concentrations of reagents did not saturate, suggesting a higher upper limit of detection than 1500 ng/mL of NP.
In the presence of cells, S/B was elevated (Figure 2C,D and Supporting Information Figure S2) compared to media alone. The overall greatest S/B was achieved with 1 nM donor and 2 nM acceptor (Supporting Information Figure S2), although the hook effect was more apparent for the low concentrations of reagent. The balance of sensitivity (lower limit) and dynamic range (upper limit) with S/B is an important consideration when choosing the conditions that are optimal for viral detection. The combination of 1 nM donor and 2 nM acceptor produces acceptable ratios with good sensitivity, while reducing reagent consumption, but has a reduced upper limit of detection.
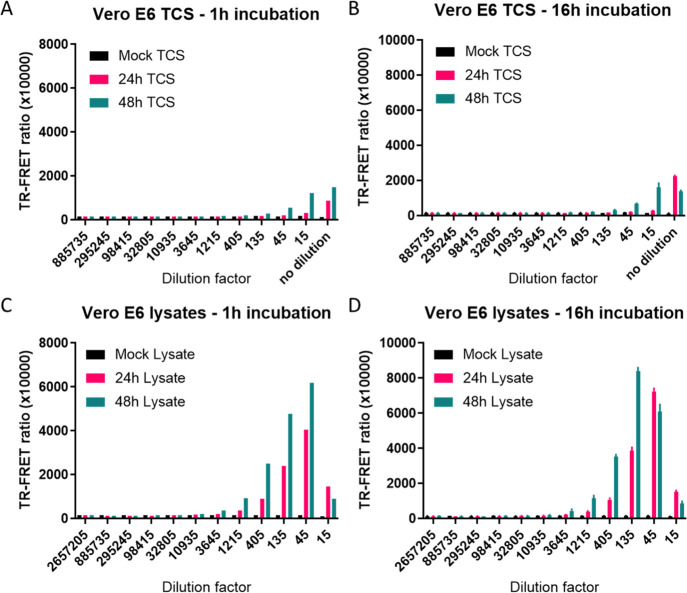
Evaluation of NP Detection in SARS-CoV-2-Infected TCS and Cell Lysates
We next evaluated the assay performance in TCS and whole cell lysates of Vero E6 cells infected with SARS-CoV-2 USA-WA1/2020 strain for 24 or 48 h using mock-infected cells as negative control. Using 1 nM donor and 2 nM acceptor, the TR-FRET ratio was greatest with cell lysate, but a low level signal was detected in TCS as well (Figure 3A–D and Supporting Information Figure S3). Interestingly, the sensitivity of NP detection in these samples increased with O/N incubation as evidenced by the increased TR-FRET ratios and hook effect at lower dilutions of lysate (Figure 3C,D and Supporting Information Figure S3). Using standard curve interpolation of the O/N incubation TR-FRET values, NP concentrations of 70.9 ± 12.3 and 1050 ± 12.3 ng/mL in TCS after 24 and 48 h were observed, respectively. The lysates exhibited larger NP concentrations of 16,380 ± 1290 and 59,520 ± 957 ng/mL after 24 and 48 h infection, respectively (Figure 3D). The data indicated that both recombinant and nascent NP produced in cells after viral infection could be detected by our TR-FRET assay.
Figure 3.
Detecting SARS-CoV-2 NP from Vero E6 TCS and cell lysate. TCS and cell lysate were collected from Vero E6 cells after infection with SARS-CoV-2 USA-WA1/2020 strain for 24 or 48 h. The TR-FRET ratio from TCS (A,C) and cell lysates (B,D) after incubating with reagents for 1 h at RT or O/N at 4 °C. TCS was diluted 1:3, and cell lysate was first diluted 15-fold followed by 1:3 dilutions. N = 3 wells in a 384-well plate. Error bars indicate S.D.
Assessing HTS Readiness of the Assay
To evaluate the suitability of the assay for HTS, we seeded Vero E6 cells into 384-well plates using an automated liquid handler (ThermoFisher Multidrop Combi) and dispensed 500 ng/mL of recombinant NP into columns 2–24 (Figure 4). Columns 1–2 were used as a negative control. After addition of 1 nM donor and 2 nM acceptor, the plate was read at 1 h and after O/N incubation at 4 °C. The assay performance was excellent with CV below 5%, an S/B above 36, and a Z-factor at or above 0.90. O/N incubation increased S/B to 50.7, compared to 36.6 after 1 h incubation, and also decreased the CV and improved the Z-factor.
Figure 4.
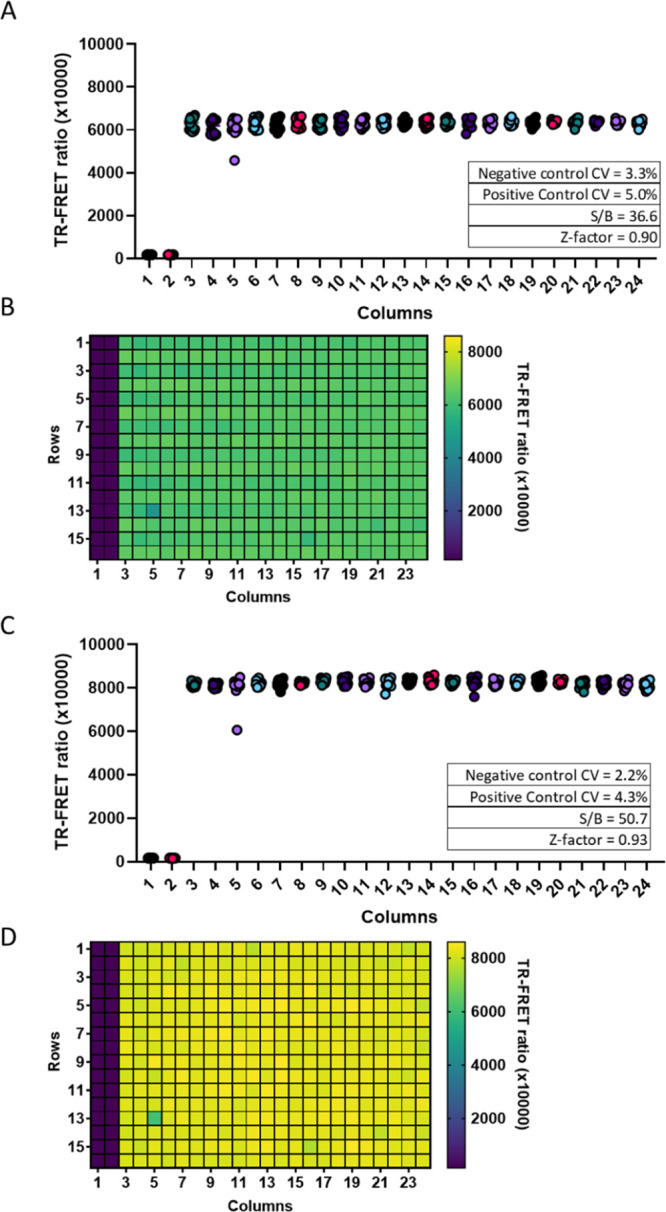
384-well plate statistics using spiked NP in Vero E6 cells. Vero E6 cells grown in a 384-well plate at 5000 cells per well were spiked with 0 or 500 ng/mL of SARS-CoV-2 NP and incubated with HTRF reagents for 1 h at RT (A,B) or O/N at 4 °C (C,D). Columns 1 and 2 contained no NP and acted as a simulated positive control for uninfected cells. Columns 3 to 24 contained 500 ng/mL of SARS-CoV-2 NP. Each column contains 16 wells. Each point represents one well. The inset shows the calculated plate statistics.
SARS-CoV-2 NP TR-FRET Assay Detects Live Virus Infection in Vero E6 Cells
To assess whether our assay reagents were able to access viral NP produced in SARS-CoV-2-infected live cells, we first transfected Vero E6 cells with different amounts of an NP expression plasmid for 24 h (Supporting Information Figure S4), followed by addition of HTRF reagents at high and low concentrations and incubation for either 1 h or O/N at 4 °C. After 1 h, the low concentration was more sensitive, and the high concentration produced larger TR-FRET ratio values with higher amounts of plasmid. O/N incubation shifted the curves leftward, increasing assay sensitivity and S/B.
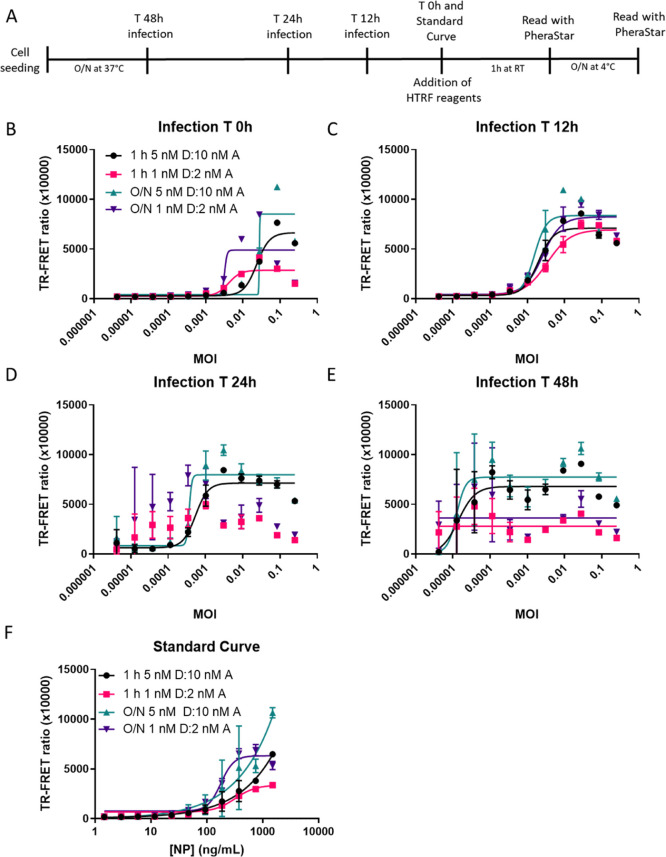
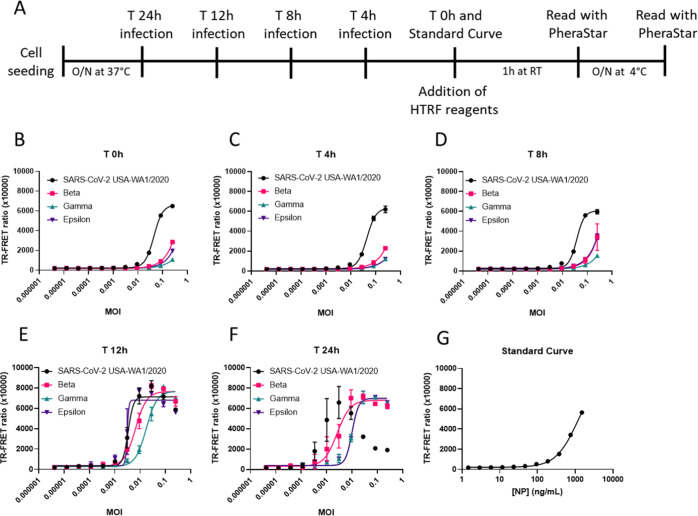
We then tested this NP TR-FRET assay using SARS-CoV-2-infected Vero-6 cells (Figure 5A). Production of SARS-CoV-2 NP in infected cells can be experimentally modulated by adjusting the MOI and infection time. To determine optimal assay conditions for the detection of SARS-CoV-2 NP in live cells, we seeded Vero E6 cells in 96-well plates and infected them with the SARS-CoV-2 USA-WA1/2020 for 48, 24, 12, and 0 h (Figure 5A). We used 1:2 ratios of the donor and acceptor and tested high (5 nM D:10 nM A) and low concentrations of reagents (1 nM D:2 nM A) with 1 h and O/N incubation. At 0 h, an MOI between 0.0003 and 0.0001 started producing a detectable signal above the background that was caused by the NP present in the virus inoculum (Figure 5B). The low concentrations of reagents were more sensitive at lower MOIs, but the high concentrations produced the larger TR-FRET signal at a higher MOI. The high concentrations of reagents exhibited a hook effect at a higher MOI than the low concentrations of reagents. At 12 h, the MOI–response curves shifted leftward toward lower MOI, suggesting that the virus was replicating and producing more NP (Figure 5C). At 24 h, the low concentrations of reagents could no longer accommodate the high levels of NP produced at high MOI and low MOI had greater variability (Figure 5D). The high concentration of reagents more accurately detected the NP produced in viral infected Vero E6 cells, and the hook effect was observed at MOI greater than 0.003. At 48 h, the viral NP concentration was saturated at all MOIs and could not be accurately detected (Figure 5E). The standard curve was added for reference (Figure 5F). Thus, an MOI of 0.009 for SARS-CoV-2 infection followed by a 24 h incubation and 5 nM donor with 10 nM acceptor was chosen as an optimized assay condition for the following experiments.
Figure 5.
HTRF assay detects NP produced by live virus infection of Vero E6. The TR-FRET ratio for SARS-CoV-2 NP detected in Vero E6 infected with the SARS-CoV-2 USA-WA1/2020 strain at different MOIs starting at 0.25 and diluted 1:3 based on a reverse time course protocol for (A) 0 h (B), 12 h (C), 24 h (D), or 48 h (E). The standard curve (F) with assay conditions equal to the 48h time point. Plates were incubated with NP HTRF reagents for 1 h at RT or O/N at 4 °C. N = 3 wells in a half-area 96-well plate. Error bars indicate S.D.
SARS-CoV-2 NP TR-FRET assay was used to assess the effect of remdesivir on NP levels in SARS-CoV-2-infected Vero E6 cells.
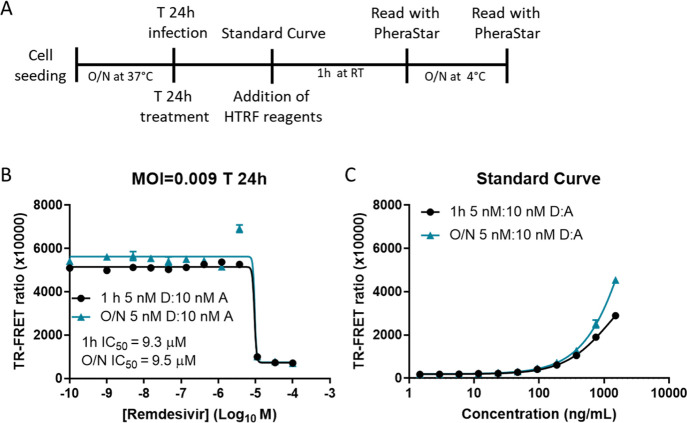
To assess the utility of our assay to determine the potencies of antiviral drugs, we examined the effect of remdesivir on NP levels in SARS-CoV-2-infected Vero E6 cells. For this, we treated SARS-CoV-2 USA-WA1/2020-infected (MOI 0.009) Vero E6 cells with remdesivir at the time of infection (Figure 6A). After 24 h, 5 nM donor and 10 nM acceptor were added to the cells and the TR-FRET ratio was measured after 1 h and O/N incubation. We observed a half maximal inhibitory concentration (IC50) of 9.3 and 9.5 μM with 1 h and O/N incubation, respectively (Figure 6B). The standard curve was added for reference (Figure 6C). Remdesivir completely inhibited SARS-CoV-2 replication as determined by the very low TR-FRET ratio at the highest concentration of drug. Residual NP from the initial inoculation likely accounted for a low amount of signal seen at the highest concentration of drug.
Figure 6.
NP HTRF assay confirmation of remdesivir inhibition of SARS-CoV-2 replication. The TR-FRET ratio for SARS-CoV-2 NP in Vero E6 infected with the SARS-CoV-2 USA-WA1/2020 strain at an MOI of 0.009 based on protocol (A) for 24 h (B). Remdesivir was added at a starting concentration of 100 μM and serially diluted 1:3. The standard curve (C) using recombinant SARS-CoV-2 NP. Plates were incubated with NP HTRF reagents for 1 h at RT or O/N at 4 °C. N = 3 wells in a half-area 96-well plate. Error bars indicate S.D.
SARS-CoV-2 NP HTRF Assay Detects VoC
Mutations in response to selective pressures driving SARS-CoV-2 evolution are heavily weighted toward the viral S protein because of its importance in cell entry and evasion of immune responses. Given that our assay depended on the detection of NP, highly mutated NPs may prevent MAb binding and would jeopardize the use of the assay. To examine whether this represented a problem for the detection of SARS-CoV-2 VoC, we infected Vero E6 cells with the SARS-CoV-2 USA-WA1/2020, Beta, Gamma, and Epsilon variants, which contained mutations in the S glycoprotein and NP, for 24, 12, 8, 4, and 0 h (Figure 7A, Supporting Information Figure S5). We aligned the NP amino acid sequences (GISAID database) from the viruses used in this study and found several mutations including P80R (Gamma), R203K (Gamma, Epislon), G204R (Gamma), T205I (Beta, Gamma), and M234I (Epsilon) (Supporting Information Figure S5). At the end of the reverse time course, 5 nM donor and 10 nM acceptor were added to the wells and incubated O/N at 4 °C. The TR-FRET readings suggested that the assay could detect the VoC with the S glycoprotein mutations and could accommodate differences in the NP amino acid sequence as well. The increase in the TR-FRET ratio was first observed at 8 h, and the curve was further shifted leftward at 12 and 24 h, indicating increasing amounts of NP production with a lower MOI (Figure 7B–F). The standard curve was added for reference (Figure 7G). Interestingly, SARS-CoV-2 USA-WA1/2020 had the largest TR-FRET ratio at early timepoints and at later timepoints exhibited the hook effect with high MOIs. The VoC exhibited low, but detectable TR-FRET ratios at early timepoints, and the hook effect was less apparent with the three VoC at high MOIs. Altogether, the results suggested that the assay could be used to detect SARS-CoV-2 VoC and has the potential to be used in HTS to identify potent antiviral compounds and biologics against these and potentially future SARS-CoV-2 VoC.
Figure 7.
NP HTRF assay detects Beta, Gamma, and Epsilon VoC. The TR-FRET ratio for SARS-CoV-2 NP in Vero E6 infected with the USA-WA1/2020 Beta, Gamma, and Epsilon strains using protocol (A) at MOIs starting at 0.25 and diluted 1:3 for 0 h (B), 4 h (C), 8 h (D), 12 h (E), and 24 h (F). The standard curve (G) with assay conditions equal to the 24 h time point. Assay completed with a reverse time course protocol; O/N incubation at 4 °C with HTRF reagents (5 nM D/10 nM A). N = 3 wells in a half-area 96-well plate. Error bars indicate S.D.
Discussion
For early drug discovery and development, compound screening assays such as the one described herein are critical for lead identification and evaluation of molecules for further development. In this work, we demonstrated the robust and reliable detection of SARS-CoV-2 NP using the TR-FRET assay format in live virus-infected cells. This was further validated by the confirmation of efficacy of the well-characterized reference compound remdesivir. We also demonstrated that this assay can also be used to measure virus replication and could be used in the future to evaluate activities of antiviral agents against emergent SARS-CoV-2 VoC.
This TR-FRET assay employs two specific MAbs to detect SARS-CoV-2 NP. Because the TR-FRET signals are produced when both MAbs bind to the same NP, the assay specificity is usually high. The use of time-resolved fluorescence instead of flash fluorescence significantly increases assay sensitivity and reduces false positives in HTS due to compound autofluorescence. The ratiometric readout reduces well-to-well variations in assay plates that may be caused by different cell densities or reagent dispensing errors.
This assay development work reveals several key insights into the appropriate detection of SARS-CoV-2 NP produced during viral replication in host cells. One important aspect is the balance between sensitivity at the lower limit of detection, the dynamic range at the upper limit of detection, and the S/B that vary depending on the cell number, MOI, and incubation time after the virus infection. At very low MOI, the number of viral particles added to each well can be more variable than at high MOI, where more virus particles per unit volume are evenly distributed to host cells. In this TR-FRET assay, the signal positively correlates with MOI or incubation time after virus inoculation because of the expression of SARS-CoV-2 NP. At high MOI, the hook effect may become prominent because NP is expressed at higher levels. Because the concentrations of two labeled MAbs are fixed, the TR-FRET signal reaches a maximal value and then decreases with the further increases of MOI or incubation time due to high antigen (NP) concentrations that prevent binding of both the donor and acceptor to the same antigen molecule. This unusual effect of reduced TR-FRET signals at a higher MOI and longer incubation times (more viral replication cycles occur) is caused by the hook effect, but can be avoided with assay optimization that identifies an optimized assay condition. In this assay, an MOI of 0.009 and 24 h infection time produced a clean MOI–FRET ratio curve when using the high 1:2 concentration of donor to acceptor. However, other concentrations and ratios may work better in different contexts such as different types of cells and different SARS-CoV-2 strains. For HTS, reagent conservation is key to keeping assay costs down and increasing the number of compounds to be screened. We demonstrated that relatively low concentrations of reagents produced a robust assay signal for an HTS campaign.
In comparison with our previously developed AlphaLISA assay, the AlphaLISA assay is more sensitive and can detect NP at single digit ng/mL with the conditions that we tested. This is advantageous when the concentration of NP is within this range such as early timepoints during infection or when using cell lines that may be less permissive to viral infection and replication. The HTRF assay performance loses sensitivity at concentrations lower than 100 ng/mL. However, saturation of the signal and the hook effect occur at higher concentrations of NP for HTRF than for AlphaLISA. This is advantageous when high amounts of NP need to be detected. Because assay requirements and conditions may vary, AlphaLISA or HTRF may be more suitable in certain conditions. Thus, the assay format should be carefully considered.
We wanted to enable the detection of SARS-CoV-2 in cell culture for HTS in BSL-3 facilities and developed complementary technologies for two main reasons. First, not all research groups will have HTRF or AlphaLISA detection technologies. Producing both assays broadens the relevance of our work to a larger audience. Second, the assay technologies can be used as confirmation screens for one another if one is used as a primary screen. Two assays using different readouts reporting the same result provides a substantial level of confidence that the hit is not an artifact. Comparing the assay formats, AlphaLISA depends on a singlet oxygen to be produced and some compounds may block the AlphaLISA reaction from taking place. On the other hand, the HTRF reagent on the other hand may suffer compound properties that could interfere with the FRET between the donor and acceptor fluorophores. The HTRF assay can be used as a simultaneous add and read assay, while AlphaLISA depends on a sequential addition of the assay components. The drug discovery toolkit benefits researchers when it is versatile and can be applied to different situations.
The differences we observed in the detection of NP from the different VoC are interesting and could be due to several factors. First, the tropism of a virus for a particular cell line can be enhanced by adapting the virus to the cell line.33 Differences in the number of passages of different virus strains may cause the variations compared to the SARS-CoV-2 USA-WA1/2020 strain that we used in this study. Another possibility is that the NP produced by different VoC may have mutations that lead to different binding affinities of labeled MAbs used in this assay, causing changes in the FRET between the donor and acceptor.34−36 An analysis of the mutations suggests minimal changes to the NP for these mutants with one P80R mutation in the nucleotide binding region in the N-terminal domain of Gamma. The other mutations are all in disordered regions of the middle of the protein. Nonetheless, each SARS-CoV-2 VoC was detected and exhibited both a time-dependent and MOI-dependent increase in the TR-FRET ratio, enabling researchers to utilize this assay for the evaluation of antiviral agent efficacies against VoC, as well as HTS campaigns to identify new antiviral compounds. Compounds that are pan-active against SARS-CoV-2 VoC would be the most attractive candidates for further development.
In contrast to previously described CPE assays for SARS-CoV-2-infected cells, which depend on a cell-killing effect, the TR-FRET assay we described in the present paper measures NP produced in cells with active SARS-CoV-2 replication, without being dependent on cell death. Thus, this assay may be more biologically relevant because not all cell types are killed by SARS-CoV-2.22,37 The CPE may not be seen in all human cells in vivo. It should be noted that only a small portion of the hits found in an HTS using the TR-FRET-based Zika virus nonstructural protein 1 assay was confirmed by CPE assay (unpublished data). We believe that this TR-FRET SARS-CoV-2 NP assay will have a broad application in BSL-3 settings for lead compound identification in HTS and evaluations of antiviral therapeutics in emerging VoC.
Acknowledgments
We thank Dr. Charles Chiu, M.D./Ph.D., Director, UCSF-Abbott Viral Diagnostics and Discovery Center at UCSF School of Medicine for the isolate SARS-CoV-2/human/USA/CA-UCSF-0001C/2020. This research was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH. We thank the development group at Columbia Biosciences for their assistance in reagent development.
Glossary
Abbreviations
- TR-FRET
time resolved fluorescence resonance energy transfer
- NP
nucleocapsid phosphoprotein
- S
spike protein
- VoC
variant of concern
- NAbs
neutralizing antibodies
- MAbs
monoclonal antibodies
- MOI
multiplicity of infection
- CPE
cytopathic effect
- HTS
high throughput screening
- TCS
tissue culture supernatant
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.1c00182.
Determination of NP concentration in viral samples; NP HTRF assay to detect recombinant SARS-CoV-2 NP; NP HTRF assay to detect SARS-CoV-2 NP in TCS and cell lysates; NP HTRF assay to detect transiently transfected SARS-CoV-2 NP; and sequence alignment of VoC Beta, Gamma, and Epsilon compared with SARS-CoV-2 USA-WA1/2020 (PDF)
Author Contributions
Initial concept—K.G., C.Z.C., L.M.S., and W.Z.; experiments—K.G., D.M.V., B.N.T., and J.C.T.; preparation of reagents—T.M., K.C., and C.Y.; figure preparation—K.G. and D.M.V.; manuscript writing—K.G., D.M.V., and W.Z.; manuscript editing—K.G., D.M.V., L.M.S., and W.Z.
The authors declare no competing financial interest.
Supplementary Material
References
- Dong E.; Du H.; Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.; Wu Q.; Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2. 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.; Chen W.; Chen J.-P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A. S.; Hodcroft E. B. Genetic Variants of SARS-CoV-2-What Do They Mean?. JAMA, J. Am. Med. Assoc. 2021, 325, 529–531. 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Plante J. A.; Liu Y.; Liu J.; Xia H.; Johnson B. A.; Lokugamage K. G.; Zhang X.; Muruato A. E.; Zou J.; Fontes-Garfias C. R.; Mirchandani D.; Scharton D.; Bilello J. P.; Ku Z.; An Z.; Kalveram B.; Freiberg A. N.; Menachery V. D.; Xie X.; Plante K. S.; Weaver S. C.; Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S.; Ye C.; Rathnasinghe R.; Stadlbauer D.; Krammer F.; Simon V.; Martinez-Sobrido L.; García-Sastre A.; Schotsaert M.; SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021, 2, e283–e284. 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Liu Y.; Liu J.; Zhang X.; Zou J.; Fontes-Garfias C. R.; Xia H.; Swanson K. A.; Cutler M.; Cooper D.; Menachery V. D.; Weaver S. C.; Dormitzer P. R.; Shi P.-Y. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621. 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Creech C. B.; Walker S. C.; Samuels R. J. SARS-CoV-2 Vaccines. JAMA, J. Am. Med. Assoc. 2021, 325, 1318–1320. 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- Young C. A. Remdesivir gains FDA approval as first treatment for COVID-19. Pharm. Today 2021, 27, 16–17. 10.1016/j.ptdy.2020.12.007. [DOI] [Google Scholar]
- Wang Y.; Zhou F.; Zhang D.; Zhao J.; Du R.; Hu Y.; Cheng Z.; Gao L.; Jin Y.; Luo G.; Fu S.; Lu Q.; Du G.; Wang K.; Lu Y.; Fan G.; Zhang Y.; Liu Y.; Ruan S.; Liu W.; Jaki T.; Hayden F. G.; Horby P. W.; Cao B.; Wang C. Evaluation of the efficacy and safety of intravenous remdesivir in adult patients with severe COVID-19: study protocol for a phase 3 randomized, double-blind, placebo-controlled, multicentre trial. Trials 2020, 21, 422. 10.1186/s13063-020-04352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang D.; Du G.; Du R.; Zhao J.; Jin Y.; Fu S.; Gao L.; Cheng Z.; Lu Q.; Hu Y.; Luo G.; Wang K.; Lu Y.; Li H.; Wang S.; Ruan S.; Yang C.; Mei C.; Wang Y.; Ding D.; Wu F.; Tang X.; Ye X.; Ye Y.; Liu B.; Yang J.; Yin W.; Wang A.; Fan G.; Zhou F.; Liu Z.; Gu X.; Xu J.; Shang L.; Zhang Y.; Cao L.; Guo T.; Wan Y.; Qin H.; Jiang Y.; Jaki T.; Hayden F. G.; Horby P. W.; Cao B.; Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H.; Tomashek K. M.; Dodd L. E.; Mehta A. K.; Zingman B. S.; Kalil A. C.; Hohmann E.; Chu H. Y.; Luetkemeyer A.; Kline S.; Lopez de Castilla D.; Finberg R. W.; Dierberg K.; Tapson V.; Hsieh L.; Patterson T. F.; Paredes R.; Sweeney D. A.; Short W. R.; Touloumi G.; Lye D. C.; Ohmagari N.; Oh M.-d.; Ruiz-Palacios G. M.; Benfield T.; Fätkenheuer G.; Kortepeter M. G.; Atmar R. L.; Creech C. B.; Lundgren J.; Babiker A. G.; Pett S.; Neaton J. D.; Burgess T. H.; Bonnett T.; Green M.; Makowski M.; Osinusi A.; Nayak S.; Lane H. C.; Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H.; Peto R.; et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Xu M.; Chen C. Z.; Guo H.; Shen M.; Hu X.; Shinn P.; Klumpp-Thomas C.; Michael S. G.; Zheng W. Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening. ACS Pharmacol. Transl. Sci. 2020, 3, 1008–1016. 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Du X.; Duan Y.; Pan X.; Sun Y.; You T.; Han L.; Jin Z.; Shang W.; Yu J.; Guo H.; Liu Q.; Wu Y.; Peng C.; Wang J.; Zhu C.; Yang X.; Yang K.; Lei Y.; Guddat L. W.; Xu W.; Xiao G.; Sun L.; Zhang L.; Rao Z.; Yang H. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell 2021, 12, 877–888. 10.1007/s13238-021-00836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Guo S.; Yi D.; Li Q.; Ma L.; Zhang Y.; Wang J.; Li X.; Guo F.; Lin R.; Liang C.; Liu Z.; Cen S. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res. 2021, 190, 105078. 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson Q. M.; Wilson K. M.; Shen M.; Itkin Z.; Eastman R. T.; Shinn P.; Hall M. D. Targeting ACE2-RBD Interaction as a Platform for COVID-19 Therapeutics: Development and Drug-Repurposing Screen of an AlphaLISA Proximity Assay. ACS Pharmacol. Transl. Sci. 2020, 3, 1352–1360. 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Z.; Xu M.; Pradhan M.; Gorshkov K.; Petersen J. D.; Straus M. R.; Zhu W.; Shinn P.; Guo H.; Shen M.; Klumpp-Thomas C.; Michael S. G.; Zimmerberg J.; Zheng W.; Whittaker G. R. Identifying SARS-CoV-2 Entry Inhibitors through Drug Repurposing Screens of SARS-S and MERS-S Pseudotyped Particles. ACS Pharmacol. Transl. Sci. 2020, 3, 1165–1175. 10.1021/acsptsci.0c00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov K.; Susumu K.; Chen J.; Xu M.; Pradhan M.; Zhu W.; Hu X.; Breger J. C.; Wolak M.; Oh E. Quantum Dot-Conjugated SARS-CoV-2 Spike Pseudo-Virions Enable Tracking of Angiotensin Converting Enzyme 2 Binding and Endocytosis. ACS Nano 2020, 14, 12234–12247. 10.1021/acsnano.0c05975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Quan S.; Xu M.; et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2025866118 10.1073/pnas.2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Z.; Shinn P.; Itkin Z.; Eastman R. T.; Bostwick R.; Rasmussen L.; Huang R.; Shen M.; Hu X.; Wilson K. M.; Brooks B. M.; Guo H.; Zhao T.; Klump-Thomas C.; Simeonov A.; Michael S. G.; Lo D. C.; Hall M. D.; Zheng W. Drug Repurposing Screen for Compounds Inhibiting the Cytopathic Effect of SARS-CoV-2. Front. Pharmacol. 2021, 11, 592737. 10.3389/fphar.2020.592737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr Z. A.; Gorshkov K.; Chen C. Z.; Zheng W. Drug Discovery Strategies for SARS-CoV-2. J. Pharmacol. Exp. Ther. 2020, 375, 127–138. 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W.; Liu G.; Ma H.; Zhao D.; Yang Y.; Liu M.; Mohammed A.; Zhao C.; Yang Y.; Xie J.; Ding C.; Ma X.; Weng J.; Gao Y.; He H.; Jin T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q.; West A. M. V.; Silletti S.; Corbett K. D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901. 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov K.; Chen C. Z.; Xu M.; Carlos de la Torre J.; Martinez-Sobrido L.; Moran T.; Zheng W. Development of a High-Throughput Homogeneous AlphaLISA Drug Screening Assay for the Detection of SARS-CoV-2 Nucleocapsid. ACS Pharmacol. Transl. Sci. 2020, 3, 1233–1241. 10.1021/acsptsci.0c00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L.; Tigabu B.; White E. L.; Bostwick R.; Tower N.; Bukreyev A.; Rockx B.; LeDuc J. W.; Noah J. W. Adapting high-throughput screening methods and assays for biocontainment laboratories. Assay Drug Dev. Technol. 2015, 13, 44–54. 10.1089/adt.2014.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergin E.; Dogan A.; Parmaksiz M.; Elçin A.; Elçin Y. Time-Resolved Fluorescence Resonance Energy Transfer [TR-FRET] Assays for Biochemical Processes. Curr. Pharm. Biotechnol. 2016, 17, 1222–1230. 10.2174/1389201017666160809164527. [DOI] [PubMed] [Google Scholar]
- Degorce F.; Card A.; Soh S.; Trinquet E.; Knapik G. P.; Xie B. HTRF: A technology tailored for drug discovery - a review of theoretical aspects and recent applications. Curr. Chem. Genomics 2009, 3, 22–32. 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S.; Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E.; Gattiker A.; Hoogland C.; Ivanyi I.; Appel R. D.; Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolskiy A. A.; Grishchenko I. V.; Yudkin D. V. Cell Cultures for Virology: Usability, Advantages, and Prospects. Int. J. Mol. Sci. 2020, 21, 7978. 10.3390/ijms21217978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A.; Sarkar M. C.; Raheja H.; Biswas N. K.; Chakraborti S.; Singh A. K.; Ghosh S.; Sarkar S.; Patra S.; Mondal R. K.; Ghosh T.; Chatterjee A.; Banu H.; Majumdar A.; Chinnaswamy S.; Srinivasan N.; Dutta S.; DAS S. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: Possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J. Biosci. 2020, 45, 76. 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K.; Steininger P.; Ziegler R.; Steinmann J.; Korn K.; Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Eurosurveillance 2020, 25, 2001650. 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.; Tian C.; Liu P.; Guo D.; Zheng W.; Huang X.; Zhang Y.; Liu L. Effects of SARS-CoV-2 mutations on protein structures and intraviral protein-protein interactions. J. Med. Virol. 2021, 93, 2132–2140. 10.1002/jmv.26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira H. G. The cytopathic effect of animal viruses. Adv. Virus Res. 1961, 8, 245–285. 10.1016/s0065-3527(08)60687-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.