Abstract
DNA sequences of high guanine (G) content have the potential to form G quadruplex (G4) structures. A more complete understanding about the biological functions of G4 DNA requires the investigation about how these structures are recognized by proteins. Here, we conducted exhaustive quantitative proteomic experiments to profile the interaction proteomes of G4 structures by employing different sequences of G4 DNA derived from the human telomere and the promoters of c-MYC and c-KIT genes. Our results led to the identification of a number of candidate G4-interacting proteins, many of which were discovered here for the first time. These included three proteins that can bind to all three DNA G4 structures and 78 other proteins that can bind selectively to one or two of the three DNA G4 structure(s). We also validated that GRSF1 can bind directly and selectively toward G4 structure derived from the c-MYC promoter. Our quantitative proteomic screening also led to the identification of a number of candidate “antireader” proteins of G4 DNA. Together, we uncovered a number of cellular proteins that exhibit general and selective recognitions of G4 folding patterns, which underscore the complexity of G4 DNA in biology and the importance of understanding fully the G4-interaction proteome.
Keywords: G quadruplex, quantitative proteomics, SILAC, nucleic acid-binding protein, gene regulation
Graphical Abstract
INTRODUCTION
Regions of genomic DNA with contiguous runs of guanines exhibit the ability to fold into non-B-form secondary structures known as guanine quadruplexes (G4).1 The G4 structures are assembled from multiple G-tetrads stacked upon one another, where a monovalent cation, primarily K+ or Na+, further stabilizes the G tetrad structure.2
Bioinformatic and experimental studies have revealed the widespread occurrence of G4 structures in the human genome. In this vein, computational analyses uncovered more than 300,000 putative G4-forming motifs in the human genome.3–5 With the use of a G4 structure-specific antibody (BG4) and fluorescence microscopy analysis, Biffi et al.6 revealed the presence of G4 structures in chromosomal DNA of human cells. Moreover, chromatin immunoprecipitation using BG4 followed by next-generation sequencing (ChIP-Seq) analyses led to the discovery of approximately 10,000 G4 structure sites in chromatin of cultured human cells.6–8 These G4 structure sites are enriched at loci of important biological relevance and regulatory functions, including more than 2000 gene promoters and telomeric regions.6,7
DNA G4 structures have been shown to assume important roles in many biological processes, including DNA replication, transcription, alternative polyadenylation, and maintenance of genomic stability.9–14 In this vein, promoter sequences with the ability to fold into G4 structures are of particular importance owing to the potential roles of these G4s in gene regulation. For instance, the nuclease hypersensitivity element III1, which is found within the promoter of c-MYC oncogene and regulates 85–90% of its transcriptional activity, harbors a G4 motif.15 Likewise, the c-KIT proto-oncogene contains two different G4 sequence motifs upstream to its core promoter, and these G4 structures are involved in regulating the expression of the c-KIT gene.16,17 Moreover, a recent study showed that G4 structure can remotely regulate gene expression by enabling DNA looping.13 Apart from gene promoters, the human telomere is known to fold readily into G4 structure,18–20 which modulates telomere integrity.21
Many proteins, including nucleolin, Pif1, PARP1, SLIRP, SUB1, Rif1, VEZF1, WRN, and YY1, were found to interact with G4 structures.13,14,22–30 We reason that a better understanding about how DNA G4 structures function in gene regulation and human diseases entails a systematic investigation about how these structures are recognized by cellular proteins. Additionally, since the loop sizes and primary DNA sequence for each G4 are unique, we reason that cells may also be equipped with proteins that interact selectively with only certain G4-folding pattern(s).
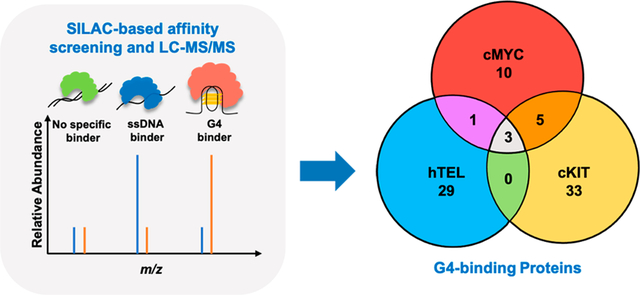
In this study, we conducted an exhaustive quantitative proteomics-based interaction screening using three pairs of DNA probes that are capable or incapable of folding into G4 structures (Figure 1 and Table S1). Through these experiments, we identified more than 80 candidate G4-binding proteins (Table S2 and Figure 2). Interestingly, some of these proteins display preferential binding to all three G4 structures than their corresponding mutated sequences, whereas others interact uniquely with certain G4 structures.
Figure 1.
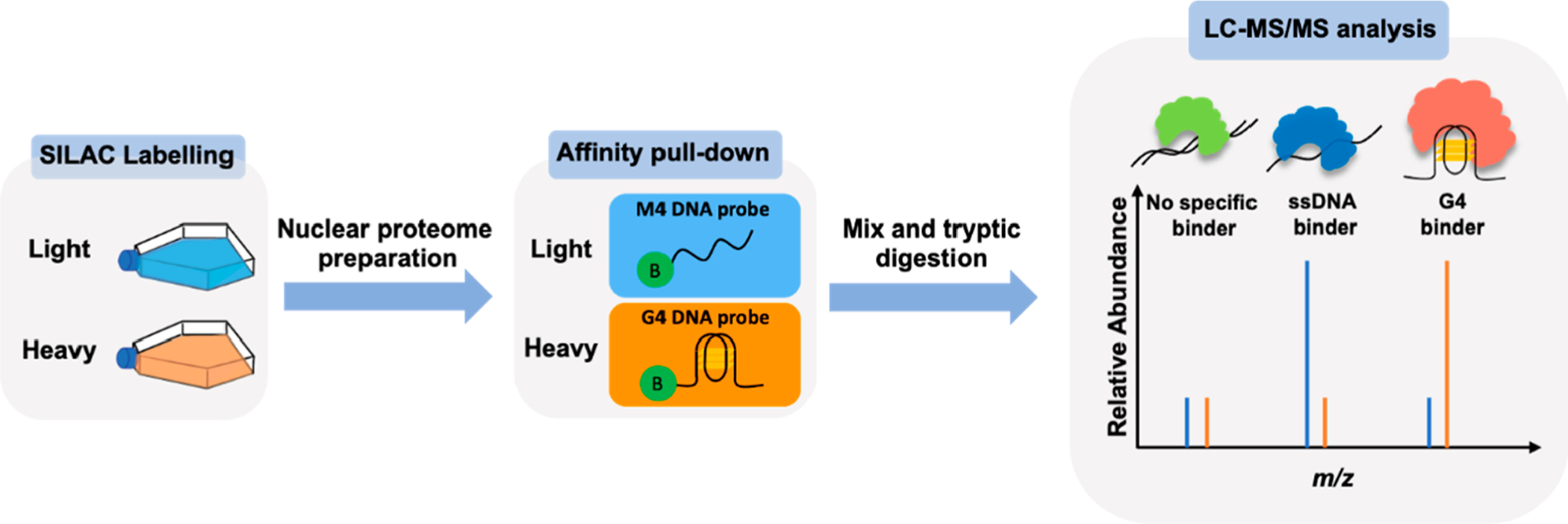
Experimental workflow for the identification of novel DNA G4-binding proteins. Shown in the scheme is a reverse SILAC-labeling experiment, where the heavy- and light-labeled nuclear protein lysates are incubated with 5′-biotinylated G4 DNA probe and the corresponding single-stranded DNA probe (M4), respectively. The “B” in the green circle denotes 5′-biotin labeling.
Figure 2.
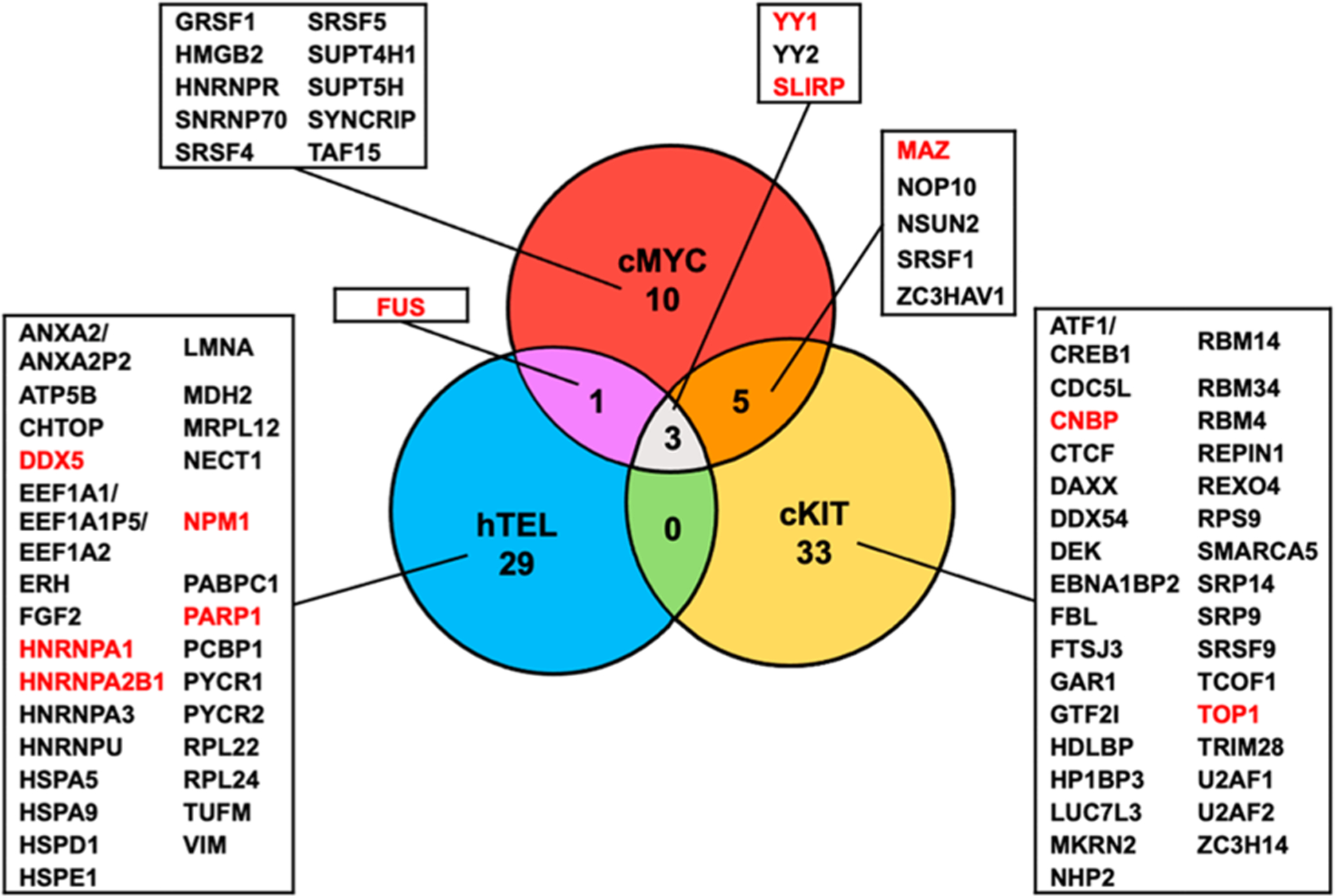
Venn diagram displaying the overlap in interacting proteins among the three G4 folding patterns studied. Candidate G4-binding proteins identified from SILAC-based affinity screening are listed. Among the identified candidate G4-binding proteins, unique peptides were detected for YY1; the peptides detected for YY2 are shared with YY1. Common peptides were detected for ATF1 and CREB1; for ANXA2 and ANXA2P2; and for EEF1A1, EEF1A1P5, and EEF1A2. Proteins highlighted in red are known to bind to DNA G4 structures.
RESULTS
To discover systematically novel G4-interacting proteins and to assess their binding specificities, we employed three G4 DNA probes derived from the G-rich sequences of the human telomere and the promoters of c-KIT and c-MYC genes, and these sequences were previously characterized by solution-phase NMR studies to adopt well-defined G4 foldings in vitro.31–33 We also obtained the corresponding mutated probes incompetent in G4 folding.30 The proper folding of the G4-containing probes and the inabilities of the mutant probes in G4 folding were confirmed by circular dichroism (CD) measurements, as described elsewhere.30 In this vein, the sequences derived from the c-KIT and c-MYC promoters form parallel G4 folding topology, whereas that from the human telomere exhibits a G4 folding pattern with both parallel and antiparallel strands.30
Equal amounts of the heavy- and light-labeled nuclear proteomes, which were obtained from stable isotope labeling by amino acids in cell culture (SILAC),34 were passed through streptavidin columns immobilized with biotin-conjugated G4 DNA and the corresponding mutated sequence (M4), respectively, which we designate as the reverse experiment (Figure 1). In this vein, by culturing cells in a medium in which arginine and lysine are replaced with their stable isotope-labeled counterparts, i.e., [13C6]-l-arginine and [13C6,15N2]-l-lysine, SILAC facilitates the labeling of the proteome with these heavy-labeled amino acids during protein synthesis. To remove potential experimental bias arising from incomplete SILAC labeling, we also conducted the forward experiment, where the light- and heavy-labeled nuclear proteomes were passed through streptavidin columns immobilized with biotin-conjugated G4 and M4 DNA probes, respectively.35 In total, we conducted a minimum of 4 independent (2 forward and 2 reverse) SILAC-based interaction screening experiments for each pair of G4/M4 probes (Table S1).
After incubation with the nuclear protein lysate, the DNA-bound avidin beads were washed, and the proteins captured on the beads were eluted, combined, trypsin-digested, and subjected to LC-MS/MS analysis, as detailed in the Supporting Information. By performing this experiment on multiple G4 folding patterns, we could achieve a quantitative comparison about the binding selectivities of candidate proteins toward the three G4 structures.
We were able to identify many proteins exhibiting preferential binding toward G4 probes over the mutated single-stranded DNA probes (Figure 2, Tables S2–S3). We employed a stringent criterion for considering a protein to be a G4-binding protein, where the protein needs to be enriched on the G4 over the corresponding M4 probes in both forward and reverse SILAC experiments with an average G4/M4 ratio being greater than 1.5. With this criterion, we identified 41, 19, and 33 proteins that can bind preferentially to the G4 sequences derived from the promoters of c-KIT and c-MYC genes and the human telomere, respectively, over their mutant counterparts (Figure 2 and Tables S2–S3). In this context, it is worth noting that the SILAC-based proteomic approach provides a quantitative measure about relative, but not absolute, binding affinities of proteins toward G4 over the corresponding M4 probes. The method, therefore, does not offer insights into the relative binding affinities of different G4-binding proteins toward any specific G4 probe employed in this study.
Among these proteins, 11 were previously described to interact directly with G4 DNA structures, including DDX5, MAZ, NPM1, etc., where known DNA G4-binding proteins are highlighted in red in Figure 2.36–46 Interestingly, we also observed several proteins (e.g., FUS and SRSF1) that were previously characterized as RNA G4-binding proteins.47,48 Aside from proteins that bind specifically to all three G4 structures, i.e., SLIRP, YY1, and YY2 (Figure 2 and Table S2), we identified a number of proteins that bind exclusively to one or two of the G4 structures, e.g., NSUN2 (to c-MYC and c-KIT G4 structures) and GRSF1 (to c-MYC G4 structure) (Figures 2–3 and Table S2). In this vein, it is worth noting that more comprehensive proteomic data sets are employed in the present study; hence, the SILAC ratios for YY1 and SLIRP proteins differ slightly from our previously published results.13,30
Figure 3.
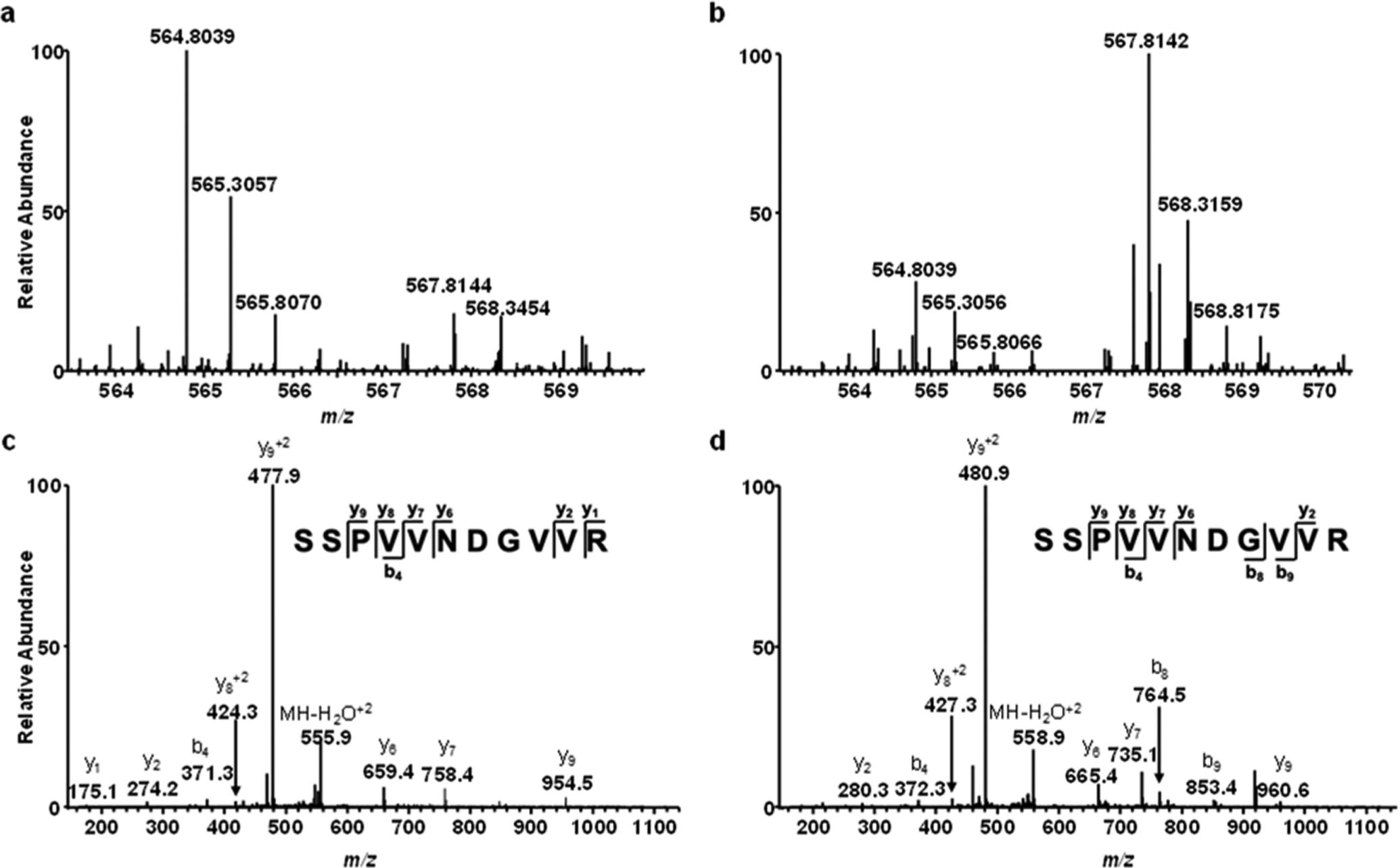
GRSF1 binds preferentially to G4 structures derived from the promoter of the c-MYC gene. (a,b) ESI-MS showing the [M + 2H]2+ ions of light and heavy arginine-containing peptide SSPVVNDGVVR with monoisotopic m/z values of ~564.8 and 567.8, respectively, obtained from forward (a) and reverse (b) SILAC-based interaction screening experiments. (c,d) MS/MS for the [M+2H]2+ ions of the light (c) and heavy (d) arginine-containing peptide, SSPVVNDGVVR, derived from GRSF1.
The results from our quantitative proteomics-based interaction screening showed that GRSF1 binds selectively to the G4 structure derived from the c-MYC promoter, but not that from the human telomere or c-KIT promoter (Figures 2–3 and Table S2). Thus, we next asked whether this protein can bind directly and selectively to the G4 structure derived from the promoter of the c-MYC gene by using fluorescence anisotropy measurements. It turned out that GRSF1 indeed displays strong and selective binding toward the c-MYC G4 probe over the corresponding mutated probe, as manifested by the Kd values of 59 nM and 1.28 μM for the G4 and M4 probes, respectively (Figure 4). On the other hand, we did not detect obvious binding of the protein toward G4 structure derived from the human telomere or c-KIT promoter (Data not shown).
Figure 4.
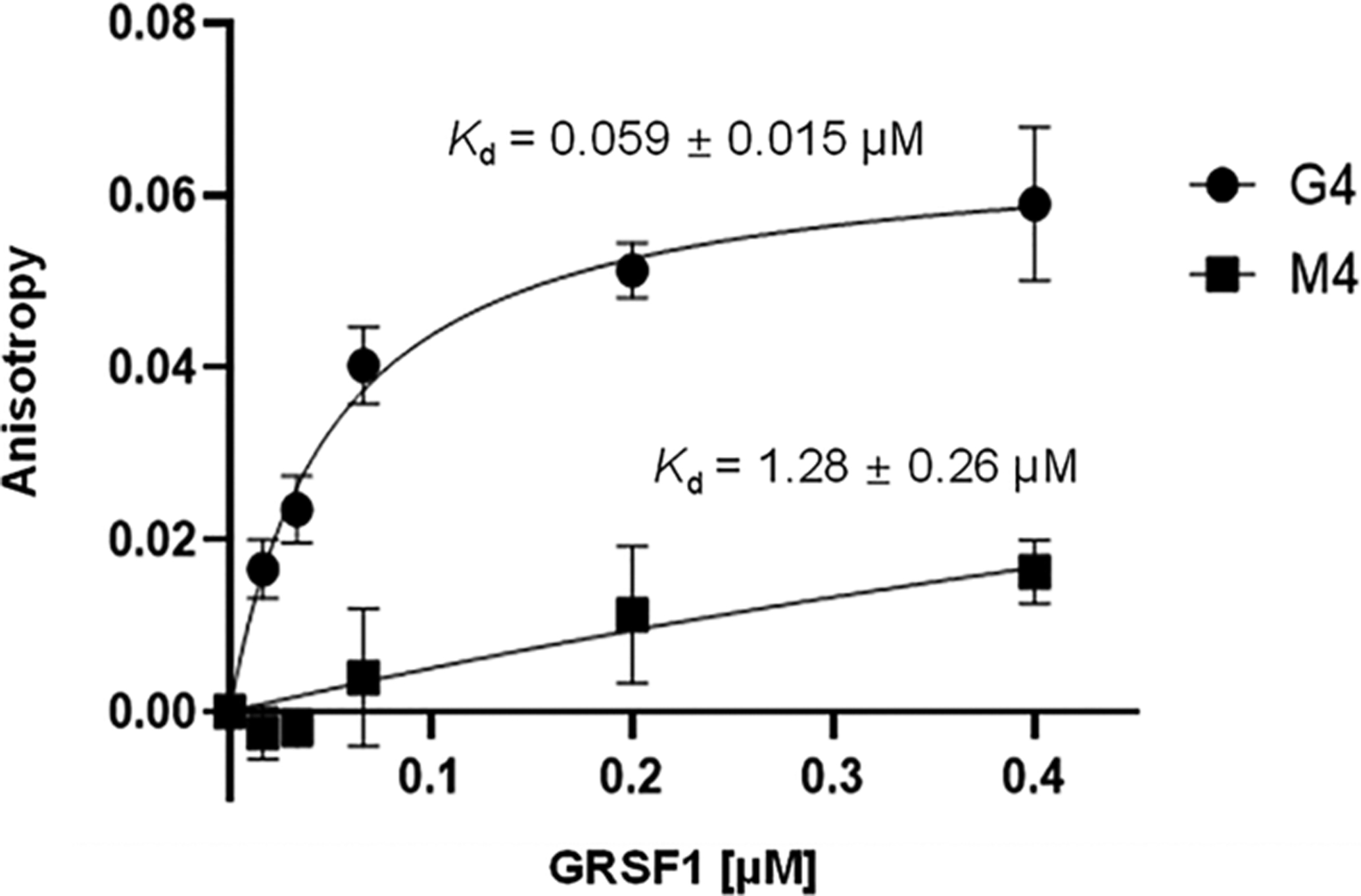
Fluorescence anisotropy for measuring the Kd values for the binding of the GRSF1 protein toward G4 structures derived from the promoter of c-MYC gene.
Our proteomic data also led to the discovery of a number of proteins that bind more strongly to M4 over the corresponding G4 DNA probes (Figure 5 and Table S3). Interestingly, several of these proteins (BLM, nucleolin, HNRNPA2, and SUB1) were previously characterized as DNA G4-binding proteins, and TARDBP was characterized as a RNA G4-binding protein (Figure 5).23,24,49–52 Along this line, BLM was shown to unfold G4 DNA, which may explain its preferential binding toward M4 over G4 DNA probes.50 While these proteins were shown to bind to G4 DNA, it will be important to examine their relative affinities in binding to G4 DNA vs mutated single-stranded DNA that cannot fold into G4 structure. It will also be important to assess the functions of these proteins in modulating the biology of G4 DNA.
Figure 5.
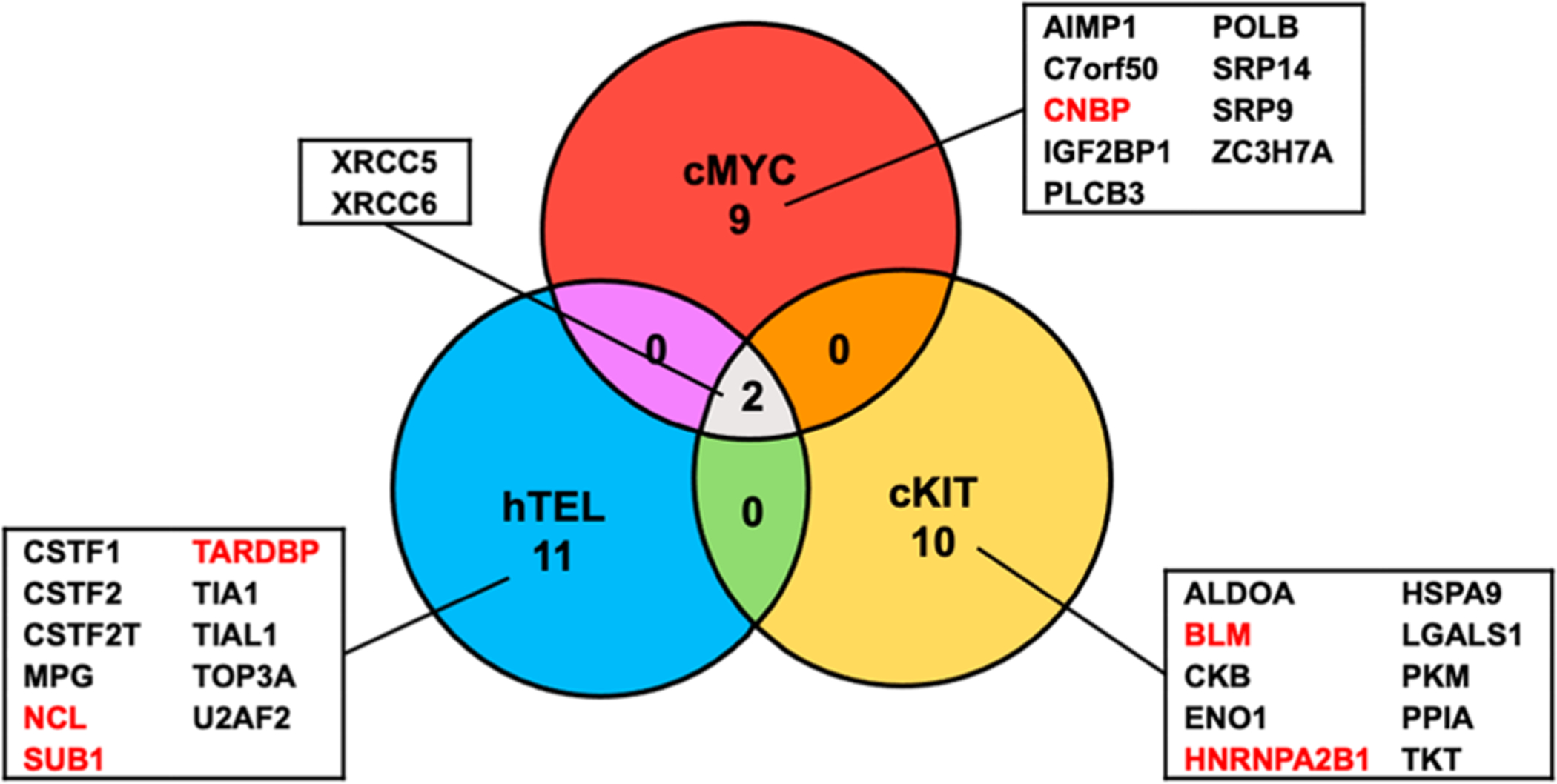
Venn diagram displaying the overlap in proteins that bind more strongly to mutated single-stranded DNA probes (M4) over the corresponding G4 DNA probes derived from human telomere (hTEL) and the promoters of c-MYC and c-KIT genes. Putative antireader proteins for G4 DNA are listed. Proteins highlighted in red are known to bind to DNA G4 structures.
Our proteomic data revealed that CNBP binds preferentially with M4 over G4 probe derived from the c-MYC promoter, whereas it binds more strongly to G4 over M4 probe derived from the c-KIT promoter. The exact reason for the different selectivities of this protein in binding with G4 vs M4 probes derived from these two promoter sequences is not clear, though we reason that the nucleobases not involved in G tetrad formation and G4 folding (i.e., those residing in the loop regions of the G4 structure) may also modulate the differential interactions between CNBP and G4/M4 probes. In this respect, it is worth noting that purified CNBP protein was found to be capable of unfolding G4 structures, where the protein binds more strongly to unfolded than folded G4 DNA derived from the c-MYC promoter, but exhibits similar affinities to unfolded and folded G4 DNA derived from the c-KIT promoter.45
Our proteomic data showed that XRCC5 and XRCC6, a.k.a. Ku86 and Ku70, respectively, display stronger binding to M4 over G4 probes derived from all three sequences. These two proteins form a heterodimer and function in the repair of DNA double-strand breaks through the nonhomologous end-joining pathway.53 At first glance, this seems to be incongruent with the heterodimer’s ability in binding with and sliding along broken ends of DNA at the DSB sites.53 However, Yuan et al.54 showed that the Ku complex can also bind directly with single-stranded DNA, albeit at a lower affinity than that with the corresponding double-stranded DNA. In addition, Shao et al.55 revealed that the Ku heterodimer can bind to RNA and assume an important function in 18S rRNA processing. Thus, the Ku complex’s ability in binding with single-stranded nucleic acids may contribute to its preferential interaction with single-stranded M4 DNA over the corresponding folded G4 DNA.
DISCUSSION
Although in vitro formation of G4 structure has been known for decades, only recently has the widespread presence of these DNA structures in human cells come to light. 6,7 Previous studies also suggested the functions of G4 structures in many different biological processes. In this vein, G4 sequence motifs are highly enriched in genomic regions of biological importance, e.g., promoters of genes.7 Hence, it is important to have a more complete understanding about how G4s are sensed by cellular proteins and how these proteins modulate the biological functions of G4 DNA. Many studies have attempted to address this question using diverse approaches and have led to the discovery of a number of proteins that bind directly and strongly with G4 DNA. High-resolution mass spectrometry-based techniques are particularly well suited to explore the interaction proteomes of G4 DNAs, and they were previously employed for the identification of a diverse set of G4 DNA-binding proteins.23,51 Different folding patterns have been described for G4 DNA; nevertheless, many of these prior interactome studies only employed one G4 folding pattern. Given the high structural diversity of G4 folding patterns, we aimed to expand this knowledge by comparing directly the interaction proteomes of three unique G4 folding patterns. Interestingly, we discovered three proteins that can bind specifically and recognize G4 structures derived from all three G-rich sequences (Figure 2).
Among the identified G4-binding proteins, we recently validated that SLIRP and YY1 can bind directly to all three G4 structures with low-nanomolar binding affinity in vitro, and ChIP-Seq results showed that SLIRP and YY1 can also bind with G4 structures in chromatin.13,30 It will be important to assess whether YY2, a closely related protein of YY1, can bind directly with G4 DNA structures and to explore the biological functions of such interactions.
Aside from the generic G4-binding proteins, our method allowed for the discovery of proteins that specifically recognize selected G4 structure(s). We also revealed that purified GRSF1 protein can bind strongly and selectively to G4 structures derived from the G4 sequence contained in the c-MYC promoter, but not to that derived from the human telomere or c-KIT promoter (Figure 4).
In summary, we identified, from exhaustive SILAC-based quantitative proteomic experiments, many novel putative G4-binding proteins that recognize all G4 folding patterns and/or selected G4 folding patterns. We further demonstrated that GRSF1 can interact directly and selectively with G4 DNA derived from c-MYC promoter region with high affinity in vitro. Hence, our study revealed that G4-binding proteins hold the ability to differentiate and selectively bind only certain G4-folding patterns, which may bear a significant impact on understanding the biological functions of G4 DNA. Moreover, the large number (~80) of candidate G4 DNA-binding proteins identified in the present study constitute an important resource for the research community to assess the functions of these proteins in the biology of G4 DNA. Our work also revealed a number of putative “antireader” proteins for G4 DNA, which calls for the assessment of the functions of these proteins in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R35 ES031707).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.1c00603
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00603.
Methods. Table S1: The DNA sequences employed for the affinity pull-down of cellular proteins that can bind to G4 DNA. Table S2: A complete list of G4-binding proteins identified from the SILAC-based interaction screening and their G4/M4 ratios. Table S3: A list of peptide ratios of c-KIT, c-MYC, and hTEL G4-binding proteins. Table S4: A complete list of G4-antireader proteins identified from the SILAC-based interaction screening and their G4/M4 ratios. Table S5: A list of peptide ratios of c-KIT, c-MYC and hTEL G4-antireader proteins. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD026794 (PDF)
The authors declare no competing financial interest.
Contributor Information
Zi Gao, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Preston Williams, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Lin Li, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Yinsheng Wang, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States;.
REFERENCES
- (1).Gellert M; Lipsett MN; Davies DR Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A 1962, 48, 2013–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Burge S; Parkinson GN; Hazel P; Todd AK; Neidle S Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 2006, 34, 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Huppert JL; Balasubramanian S Prevalence of quadruplexes in the human genome. Nucleic Acids Res 2005, 33, 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Todd AK; Johnston M; Neidle S Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res 2005, 33, 2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bedrat A; Lacroix L; Mergny JL Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res 2016, 44, 1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Biffi G; Tannahill D; McCafferty J; Balasubramanian S Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem 2013, 5, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hänsel-Hertsch R; Beraldi D; Lensing SV; Marsico G; Zyner K; Parry A; Di Antonio M; Pike J; Kimura H; Narita M; Tannahill D; Balasubramanian S G-quadruplex structures mark human regulatory chromatin. Nat. Genet 2016, 48, 1267–1272. [DOI] [PubMed] [Google Scholar]
- (8).Spiegel J; Cuesta SM; Adhikari S; Hansel-Hertsch R; Tannahill D; Balasubramanian S G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol 2021, 22, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gray LT; Vallur AC; Eddy J; Maizels N G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol 2014, 10, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bochman ML; Paeschke K; Zakian VA DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet 2012, 13, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ribeyre C; Lopes J; Boulé JB; Piazza A; Guédin A; Zakian VA; Mergny JL; Nicolas A The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 2009, 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cogoi S; Xodo LE G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res 2006, 34, 2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Li L; Williams P; Ren W; Wang MY; Gao Z; Miao W; Huang M; Song J; Wang Y YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol 2021, 17, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Li L; Williams P; Gao Z; Wang Y VEZF1-guanine quadruplex DNA interaction regulates alternative polyadenylation and detyrosinase activity of VASH1. Nucleic Acids Res 2020, 48, 11994–12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Siddiqui-Jain A; Grand CL; Bearss DJ; Hurley LH Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bejugam M; Sewitz S; Shirude PS; Rodriguez R; Shahid R; Balasubramanian S Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc 2007, 129, 12926–12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bejugam M; Gunaratnam M; Müller S; Sanders DA; Sewitz S; Fletcher JA; Neidle S; Balasubramanian S Targeting the c-Kit Promoter G-quadruplexes with 6-Substituted Indenoisoquinolines. ACS Med. Chem. Lett 2010, 1, 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sundquist WI; Klug A Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- (19).Dai J; Punchihewa C; Ambrus A; Chen D; Jones RA; Yang D Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res 2007, 35, 2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schaffitzel C; Berger I; Postberg J; Hanes J; Lipps HJ; Plückthun A In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Paeschke K; Simonsson T; Postberg J; Rhodes D; Lipps HJ Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol 2005, 12, 847–854. [DOI] [PubMed] [Google Scholar]
- (22).Mendoza O; Bourdoncle A; Boulé JB; Brosh RM; Mergny JL G-quadruplexes and helicases. Nucleic Acids Res 2016, 44, 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gonzalez V; Guo K; Hurley L; Sun D Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem 2009, 284, 23622–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gao J; Zybailov BL; Byrd AK; Griffin WC; Chib S; Mackintosh SG; Tackett AJ; Raney KD Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem. Commun 2015, 51, 7242–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kanoh Y; Matsumoto S; Fukatsu R; Kakusho N; Kono N; Renard-Guillet C; Masuda K; Iida K; Nagasawa K; Shirahige K; Masai H Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol 2015, 22, 889–897. [DOI] [PubMed] [Google Scholar]
- (26).Soldatenkov VA; Vetcher AA; Duka T; Ladame S First evidence of a functional interaction between DNA quadruplexes and poly(ADP-ribose) polymerase-1. ACS Chem. Biol 2008, 3, 214–219. [DOI] [PubMed] [Google Scholar]
- (27).Pagano B; Margarucci L; Zizza P; Amato J; Iaccarino N; Cassiano C; Salvati E; Novellino E; Biroccio A; Casapullo A; Randazzo A Identification of novel interactors of human telomeric G-quadruplex DNA. Chem. Commun. (Cambridge, U. K.) 2015, 51, 2964–2967. [DOI] [PubMed] [Google Scholar]
- (28).Zhang T; Zhang H; Wang Y; McGown LB Capture and identification of proteins that bind to a GGA-rich sequence from the ERBB2 gene promoter region. Anal. Bioanal. Chem 2012, 404, 1867–1876. [DOI] [PubMed] [Google Scholar]
- (29).Sanders CM Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J 2010, 430, 119–128. [DOI] [PubMed] [Google Scholar]
- (30).Williams P; Li L; Dong X; Wang Y Identification of SLIRP as a G quadruplex-binding protein. J. Am. Chem. Soc 2017, 139, 12426–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang Y; Patel DJ Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- (32).Ambrus A; Chen D; Dai J; Bialis T; Jones RA; Yang D Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res 2006, 34, 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Phan AT; Kuryavyi V; Burge S; Neidle S; Patel DJ Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc 2007, 129, 4386–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (35).Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (36).Arimondo PB; Riou JF; Mergny JL; Tazi J; Sun JS; Garestier T; Hélène C Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res 2000, 28, 4832–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Marchand C; Pourquier P; Laco GS; Jing N; Pommier Y Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J. Biol. Chem 2002, 277, 8906–8911. [DOI] [PubMed] [Google Scholar]
- (38).Takahama K; Takada A; Tada S; Shimizu M; Sayama K; Kurokawa R; Oyoshi T Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol 2013, 20, 341–350. [DOI] [PubMed] [Google Scholar]
- (39).Wu G; Xing Z; Tran EJ; Yang D DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 20453–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Xodo L; Paramasivam M; Membrino A; Cogoi S Protein hnRNPA1 binds to a critical G-rich element of KRAS and unwinds G-quadruplex structures: implications in transcription. Nucleic Acids Symp. Ser. (Oxf) 2008, 52, 159–160. [DOI] [PubMed] [Google Scholar]
- (41).Wang F; Tang ML; Zeng ZX; Wu RY; Xue Y; Hao YH; Pang DW; Zhao Y; Tan Z Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 20413–20418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Federici L; Arcovito A; Scaglione GL; Scaloni F; Lo Sterzo C; Di Matteo A; Falini B; Giardina B; Brunori M Nucleophosmin C-terminal leukemia-associated domain interacts with G-rich quadruplex forming DNA. J. Biol. Chem 2010, 285, 37138–37149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Edwards AD; Marecki JC; Byrd AK; Gao J; Raney KD G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res 2021, 49, 416–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cogoi S; Shchekotikhin AE; Xodo LE HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res 2014, 42, 8379–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).David AP; Pipier A; Pascutti F; Binolfi A; Weiner AMJ; Challier E; Heckel S; Calsou P; Gomez D; Calcaterra NB; Armas P CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res 2019, 47, 7901–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Fan JH; Bochkareva E; Bochkarev A; Gray DM Circular dichroism spectra and electrophoretic mobility shift assays show that human replication protein A binds and melts intramolecular G-quadruplex structures. Biochemistry 2009, 48, 1099–1111. [DOI] [PubMed] [Google Scholar]
- (47).Yagi R; Miyazaki T; Oyoshi T G-quadruplex binding ability of TLS/FUS depends on the beta-spiral structure of the RGG domain. Nucleic Acids Res 2018, 46, 5894–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).von Hacht A; Seifert O; Menger M; Schutze T; Arora A; Konthur Z; Neubauer P; Wagner A; Weise C; Kurreck J Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res 2014, 42, 6630–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lopez CR; Singh S; Hambarde S; Griffin WC; Gao J; Chib S; Yu Y; Ira G; Raney KD; Kim N Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res 2017, 45, 5850–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Mohaghegh P; Karow JK; Brosh RM Jr.; Bohr VA; Hickson ID The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res 2001, 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wang F; Tang ML; Zeng ZX; Wu RY; Xue Y; Hao YH; Pang DW; Zhao Y; Tan Z Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 20413–20418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ishiguro A; Kimura N; Watanabe Y; Watanabe S; Ishihama A TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells 2016, 21, 466–481. [DOI] [PubMed] [Google Scholar]
- (53).Grundy GJ; Moulding HA; Caldecott KW; Rulten SL One ring to bring them all–the role of Ku in mammalian nonhomologous end joining. DNA Repair 2014, 17, 30–38. [DOI] [PubMed] [Google Scholar]
- (54).Yuan Y; Britton S; Delteil C; Coates J; Jackson SP; Barboule N; Frit P; Calsou P Single-stranded DNA oligomers stimulate error-prone alternative repair of DNA double-strand breaks through hijacking Ku protein. Nucleic Acids Res 2015, 43, 10264–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Shao Z; Flynn RA; Crowe JL; Zhu Y; Liang J; Jiang W; Aryan F; Aoude P; Bertozzi CR; Estes VM; Lee BJ; Bhagat G; Zha S; Calo E DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis. Nature 2020, 579, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


