Abstract
Brain metastasis is a serious consequence of breast cancer for women as these tumors are difficult to treat and are associated with poor clinical outcomes. Preclinical mouse models of breast cancer brain metastatic (BCBM) growth are useful but are expensive, and it is difficult to track live cells and tumor cell invasion within the brain parenchyma. Presented here is a protocol for ex vivo brain slice cultures from xenografted mice containing intracranially injected breast cancer brain-seeking clonal sublines. MDA-MB-231BR luciferase tagged cells were injected intracranially into the brains of Nu/Nu female mice, and following tumor formation, the brains were isolated, sliced, and cultured ex vivo. The tumor slices were imaged to identify tumor cells expressing luciferase and monitor their proliferation and invasion in the brain parenchyma for up to 10 days. Further, the protocol describes the use of time-lapse microscopy to image the growth and invasive behavior of the tumor cells following treatment with ionizing radiation or chemotherapy. The response of tumor cells to treatments can be visualized by live-imaging microscopy, measuring bioluminescence intensity, and performing histology on the brain slice containing BCBM cells. Thus, this ex vivo slice model may be a useful platform for rapid testing of novel therapeutic agents alone or in combination with radiation to identify drugs personalized to target an individual patient’s breast cancer brain metastatic growth within the brain microenvironment.
Introduction
Breast cancer brain metastases (BCBM) develop when cells spread from the primary breast tumor to the brain. Breast cancer is the second most frequent cause of brain metastasis after lung cancer, with metastases occurring in 10–16% of patients1. Unfortunately, brain metastases remain incurable as >80% of patients die within a year after their brain metastasis diagnosis, and their quality of life is impaired due to neurological dysfunctions2. There is an urgent need to identify more effective treatment options. Monolayer two-dimensional or three-dimensional culture models are the most commonly used methods in testing therapeutic agents in the laboratory. However, they do not mimic the complex BCBM microenvironment, a major driver of tumor phenotype and growth. Although these models are useful, they do not capture the complex tumor-stromal interactions, the unique metabolic requirements, and the heterogeneity of the tumors3. To more faithfully recapitulate tumor-stromal interactions and microenvironment heterogeneity, our group and others have begun to generate organotypic brain metastasis “slice” cultures with patient-derived tumor cells (primary or metastatic) or cancer cell lines4,5,6. Compared to classical in vitro systems, this short-term ex vivo model may provide more relevant conditions for screening new therapeutics prior to preclinical assessment in large animal cohorts.
Ex vivo models have been constructed and successfully used primarily for the identification of successful treatments of various cancers. They require few days of assessment and additionally can be tailored to patient-specific drug screening. For example, human bladder and prostate cancer ex vivo tissues have shown a dose-dependent antitumor response of docetaxel and gemcitabine7. Similar colorectal carcinoma ex vivo tissues were developed to screen chemotherapeutic drugs Oxaliplatin, Cetuximab, and Pembrolizumab8. This application has been widely used in pancreatic cancer, considering the essential interaction between the stromal environment and the genotypic and phenotypic characteristics of pancreatic ductal adenocarcinoma9,10. Furthermore, such organotypic models have been developed for similar screenings in head, neck, gastric, and breast tumors11,12.
Here, an ex vivo brain slice model of xenografted breast cancer brain metastatic tumor cells in their microenvironment is being generated. Mice were intracranially injected with breast cancer brain metastatic brain trophic MDA-MB-231BR cells13 in the cerebral cortex parietal lobe- a common site of TNBC metastasis14,15 and allowed to develop tumors. Brain slices were generated from these xenografted animals and maintained ex vivo as organotypic cultures as described16,17. This novel ex vivo model allows for the analysis of BCBM cell’s growth within the brain parenchyma and can be used to test therapeutic agents and radiation effects on tumor cells within the brain microenvironment.
Protocol
This protocol was approved and follows the animal care guidelines by the Drexel University College of Medicine Institutional Animal Care and Use Committee (IACUC). Nu/Nu athymic female mice (6–8 weeks old) were used in this study.
1. Intracranial injection of tumor cells
Sterilize all equipment (tweezers, scissors, suturing scissors, hand drill) under a dry cycle of an autoclave for up to 45 min in sterilization pouches, including a sterilization indicator. If conducting surgeries on multiple animals, sterilize tools between animals using a heated bead sterilizer (up to 3x).
Anesthetize the mice according to the users’ animal care protocol (e.g., isoflurane (4% for induction, 1–2% maintenance) and apply analgesia (buprenorphine s.r.) prior to the start of surgery. Ensure optimal anesthesia depth by monitoring the heartbeat of mice until it reaches 0.5–1 beats/s.
Place the mice into a stereotaxic frame and immobilize the head using standard ear bars (Figure 1A).
Sterilize the surface of the head by repeated, alternating application (3x) of iodine swabs and 70% ethanol prior to incision.
Use a surgical scalpel to make a 1 cm midline incision through the skin at a slightly diagonal angle to the imaginary line that divides the mice’s brain into two symmetrical halves to expose the skull. Wipe any blood with a cotton swab.
Deflect the skin laterally to expose the injection site: 2 mm on the right, 1 mm forward with reference to the bregma (+2 mediolateral (+2ML), +1 rostral/caudal (+1RC).
Use a bur bit (0.73 mm) to penetrate the skull +2ML, +1RC to the bregma and drill a hole using slight pressure and twisting motion.
Use a brain injection syringe to inject 5 μL of a 100,000 MDA-MB-231BR (stably expressing GFP-Luciferase) cells/μL solution in sterile 1x PBS by initially inserting the syringe 3.5 mm in depth to the brain.
Allow the syringe to settle for 2 min. Pull it up about 1 mm, and after 2 min, slowly inject the first half of the volume.
-
Wait for another 2 min before injecting the rest, and then slowly pull the syringe 3 min after the final injection.
NOTE: Slow injection is empirical in allowing the volume to be well absorbed by the brain tissue and not create leakage after pulling the syringe, which may lead to cancer cells growing outside of the intended site.
Apply bone wax to the injection site on the skull and then use polypropylene sutures to suture the tissue. Mice will recover within 5 min after removal of anesthesia.
Monitor their behavior right after surgery for about 10 min, 3 h later and the following day to determine if special treatment, including saline injection, soft food, etc., are needed to help quick recovery.
- Monitor the tumor growth via bioluminescence imaging on the imaging system.
- For this, first, turn on the oxygen and isoflurane on the imaging system. Allow both to be distributed to the separate chamber outside of the imaging box.
- Then, turn on the software, initialize the instrument and choose the appropriate option for visualizing and capturing the whole mouse body.
Place the mice in the separate chamber with isoflurane and allow the heartbeat to reach 0.5–1 beat/s. Inject the mice intraperitoneally with 200 μL of 30 mg/mL luciferin.
Transfer the mice stomach down to the imaging chamber’s noselet supplied with oxygen and isoflurane. Lock the chamber, and choose 2 min time exposure to start imaging.
Use the ROI (region of interest) tool of the software to quantify the bioluminescent signal of the tumor.
To analyze the tumor size, choose the circle shape tool, fit it around the tumor shape and size. Measure the ROI and report the total readings.
-
Allow solid tumor growth for 12–14 days (or until luciferase reaches about 7.0 × 106 units) (Figure 1B) before generating brain slices.
NOTE: Increasing the number of cells to be injected will lead to faster tumor growth; therefore, the generation of brain slices can occur earlier than 12 days. On the other hand, a bigger tumor will lead to more GFP+ slices if allowed to grow for 14 days. After 14 days, mice’s health begins to deteriorate rapidly; hence animal health monitoring should be increased to once daily after the 10-day time point, and any mice displaying signs of pain and/or discomfort should be used for generating the slices.
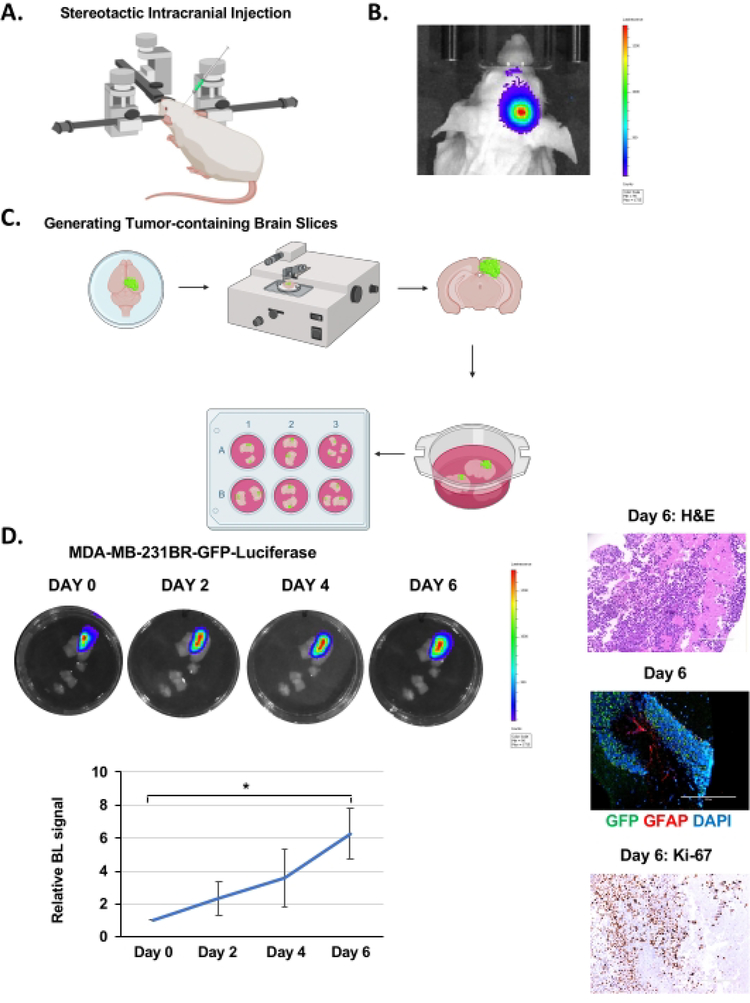
Figure 1. Intracranial injection, brain slice generation, and BCBM growth ex vivo.
(A) Schematic representation of mouse during intracranial injection under a stereotactic instrument. (B) Representative images of bioluminescent detection of tumors from 4–6 weeks old Nu/Nu mice injected with 5 × 105 MDA-MB-231BR-GFP-Luciferase cells 12 days post-injection. (C) Schematic representation of the generation of ex vivo mouse brain slices from (B). (D) Representative images depicting tumor growth in organotypic brain slices derived from mice intracranially injected with MDA-MB-231BR-GFP-Luciferase cells detected via bioluminescence over 6 days. H&E & Ki-67, GFP (tumor cells) GFAP (reactive astrocytes) DAPI (nucleus) staining of a representative brain slice with tumor (image magnification 20x, scale bar: 200 μm). Quantification of tumor growth represents bioluminescence signal relative to day 0 (n = 3). Student’s t-test reported as mean ± SEM, * p < 0.05. Please click here to view a larger version of this figure.
2. Generate brain slices
NOTE: Perform the steps after brain isolation in a sterile laminar flow hood. It is typically possible to generate 35–40 individual slices containing tumor cells (luciferase signal) from one mouse injected with MDA-MB-231BR cells (Figure 1C).
Sterilize all the surgical tools in an autoclave. Place tissue slicer in the sterile laminar flow hood and clean all tools and instruments with 70% ethanol.
Following 12–14 days after intracranial MDA-MB-231BR cell injection, anesthetize the mice by placing a paper towel wetted with isofluorane in the cage. Wait until the mouse’s heartbeat lowers to 0.5–1 beat/s and decapitate the mice.
Remove and place their brains rapidly (within 45 s) into ice-cold (4 °C) sucrose-ACSF composed of the following: 280 mM sucrose, 5 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 20 mM glucose, 10 mM HEPES, 5 mM Na+-ascorbate, 3 mM thiourea, 2 mM Na+, 29 mM pyruvate; pH=7.3.
Using a sharp, sterile scalpel or razor blade, cut away excess parts of the brain that do not contain any tumor from all sides, including the bottom part of the brain.
-
Bring the shape of the brain to a non-perfect cube to sit flat on the ACSF wetted filter paper on a 60 mm dish lid to facilitate slicing.
NOTE: Extra tissue of the brain where the tumor is unlikely to have grown is removed prior to slicing to allow generation of mostly GFP+ slices and create a shape of the brain that is stable on the surface of the instrument and will not be perturbed by the vibrations caused by the slicing.
Place several sheets of pre-wet (with ACSF) filter paper circles onto the cutting platform and place the blocked tissue atop this.
-
Set the cut size on the instrument to 2 or 2.5 units on the provided ruler to slice the tissue into 200–250 μm sections.
NOTE: Slices up to 350 μm or as thin as 100 μm are viable. For slices <200 μm, use a vibratome or compresstome.
Use a paintbrush (sable) to transfer brain slices into a dish containing sucrose ACSF and separate the slices under a light microscope using 27 G needles.
-
Identify GFP positive slices under the fluorescent microscope and transfer them to a new 60 mm dish containing 2–3 mL of adult slice media (Neurobasal medium A, 2% B12 supplement, 1% N2 supplement, 1% glutamine, 0.5% glucose, 10 U/mL penicillin, and 100 μg/mL streptomycin) using a sterile 1 mL broken off pipette (wide opening).
NOTE: The brain slices that contain tumor cells growing over the brain surface (maybe due to injection cell leakage, etc.) are excluded.
Transfer 3–5 slices onto each 30 mm, 0.4 μm pore size tissue culture insert in 6-well plates at a distance that does not allow growing into each other.
Remove the excess medium from the surface of the insert using a P1000 pipette, add 1 mL of adult mouse slice medium underneath each insert and equilibrate the media in the incubator prior to slicing.
Incubate the tissues at 37 °C, 5% CO2, and 95% humidity for 24 h before imaging.
3. IVIS imaging of slices
NOTE: Perform luciferase and inhibitor addition steps in a sterile laminar flow hood.
Pipette 5 μL of 30 mg/mL luciferin solution into the medium underneath the inserts by lifting the insert with forceps and placing them back into the well after addition of luciferin to the media in the well.
Place the 6-well plate with the lid inside the imaging chamber of the instrument below the stationary camera and lock the chamber door.
Open the software and initialize the instrument.
Choose camera settings that allow visualization and capturing of only one well per image. Move the stage up(FOV of 5 cm) and place the well that must be imaged directly under the camera.
Set the imaging time to most appropriate depending on the intensity of the luciferase that the tumor will emit, which can vary from 10 s to 5 min. Start with setting it to 10 s and then increase if the signal is low but keep it consistent for all time points and conditions until the end of the experiment (Figure 1D).
Use the ROI tool (circle shape tool), fit it around the tumor shape and size, measure the ROI, and report the total readings to quantify the bioluminescent signal of each tumor on the slice.
Bring the plate back to the sterile laminar flow hood, use forceps to slightly lift the insert from the well.
Aspirate the medium, replace it with 1 mL of fresh medium and place the insert back in the well.
For experiments where the slices are being treated with various compounds such as inhibitors, metabolites, etc., prepare the appropriate concentration of the reagent in 1.2 mL of brain slice media in a 1.5 mL tube and then transfer 1 mL to the well containing the slice.
Repeat this process every 48 h for the duration of the study.
4. Live imaging of tumor growth in ex vivo brain slices
NOTE: Supply inserts with enough medium (1.5 mL) to last the length of the experiment as it is not possible to add additional medium during the live imaging.
Place the 6-well plate on the automated microscope plate holder inside the incubator (37 °C, 5% CO2, and 95% humidity).
Open the software, turn on brightfield on the first quadrant to adjust the exposure needed to view the slices. To find the position (coordinates “xy”) of the slice, navigate with “xy” on the second quadrant to control the coordinates “xyz” of the microscope.
Select 6-well plate as the labware used for this experiment. Select the ROI map editor and register that position by selecting a new ROI, add and save that ROI.
Repeat the same steps for all the other regions of interest in other wells. After having added all the ROIs, save all the ROIs.
Under Protocol, select Clear Existing and select Time-Lapse under Acquisition, followed by selecting the channels of interest (in this case GFP).
Turn off Autofocus in the Focus Setup and manually adjust to the appropriate focus for all the slices. Finally, adjust the Brightfield Intensity and Fluorescent Channel Excitation and Exposure Time to the appropriate level.
Set the protocol to acquire images every 15 min for 48 h.
Save and run the protocol. At the finish of the experiment, the saved file will contain both the brightfield and GFP images captured for every ROI.
To combine the images in a video, rename all images of interest with the same name followed by a binary number to organize them in the order they were captured (e.g., ROI1 (1), RO1 (2) ...)
Open ImageJ and import the images by clicking on File > Import > Image Sequence. Select the file names that appear in a list, and write the content of the files’ name in the File Name Contains box that will appear to identify those files selected for the video.
Save the combined images under File > Save As > AVI.
5. MTS assay and immunohistochemistry of ex vivo brain tissue
1. MTS ASSAY
Excise the slices by cutting the tissue culture membrane underneath each slice along the edges of the tissue and transfer the tissue, still attached to the membrane, into a 96-well plate.
Add the MTS reagent mixed in a 1:5 ratio with culture media and 0.01% triton (1x) to allow penetration of the solution in the tissue, in a total volume of 120 μL in each well.
Use 4% Paraformaldehyde (PFA) as an agent to render brain slices inviable, and thus a positive control for this experiment. Immerse the brain slice of interest in 4% PFA for 1 h at RT (room temperature) or overnight at 4 °C before proceeding with the MTS assay.
Allow the MTS reagent to react on the slices for 4 h, remove the slices from the wells and measure the absorbance at 490 nm for all conditions, including an empty well with no slice as a reference.
Report the readings as a function of tissue slice area, measured with a digital caliper ruler.
2. Immunohistochemistry preparation
Immerse the brain slices attached to the membrane insert surface in 10% formalin overnight at 4 °C.
Perform embedding, processing, blocking and generate unstained slides of the tissue. Use standard immunohistochemistry staining protocol to stain for H&E, Ki-67, gamma-H2AX, CC3, GFAP, GFP, DAPI
6. Irradiation of tumor in ex vivo brain slices
-
Irradiate the tumor with 6 Gy (310 kVp x-rays).
NOTE: The units entered to get the total dose of 6 Gy are 3522 units after considering R units (measured using Victoreen electrometer) and corrections for the thimble, air density, and cGy/R as previously described18. The exposure time is 130.9 s.
Use 0.25 mm Cu +1 mm Al added filtration, 50 cm SSD, 125 cGy per min.
Perform dosimetry by an in-the-beam ionization chamber calibrated against a primary standard.
Make corrections daily for humidity, temperature, and barometric pressure.
Representative Results
MDA-MB-231BR-GFP-Luciferase cells were intracranially injected into the right hemisphere of 4–6 week old Nu/Nu mice as explained above (Figure 1A) and were allowed to grow for 12–14 days, during which time tumor growth was monitored by bioluminescence imaging (Figure 1B). We injected 100,000 cancer cells intracranially as reported by other groups19, but it’s possible to inject as low as 20,000 cell20. Following brain slice generation (Figure 1C), as described above, slices were imaged on the IVIS to determine tumor presence via bioluminescent imaging after 24 h incubation. This is considered as day 0. Tumor growth was monitored by quantifying bioluminescent signaling every 48 h, which indicated progressive growth over 6 days (Figure 1D). The H&E staining and GFP-positive fluorescence confirm the presence of a tumor in the brain slice (Figure 1D). We were also able to detect staining of reactive astrocytes, visualized by staining for Glial Fibrillary Acidic Protein (GFAP)-positive cells, visible between the cluster of cancer cells (GFP-positive) (Figure 1D) similar to what is seen in vivo21. Unlike models that inoculate cancer cells ex vivo into a mouse brain slice22, this model contains reactive astrocytes which is an important aspect of the BCBM tumor microenvironment. We also detect increased Ki-67 staining confirming cancer cells are highly proliferative (Figure 1D).Thus, tumor cells within the brain parenchyma can survive and proliferate within a brain slice ex vivo for a number of days and also contain tumor-associated stroma.
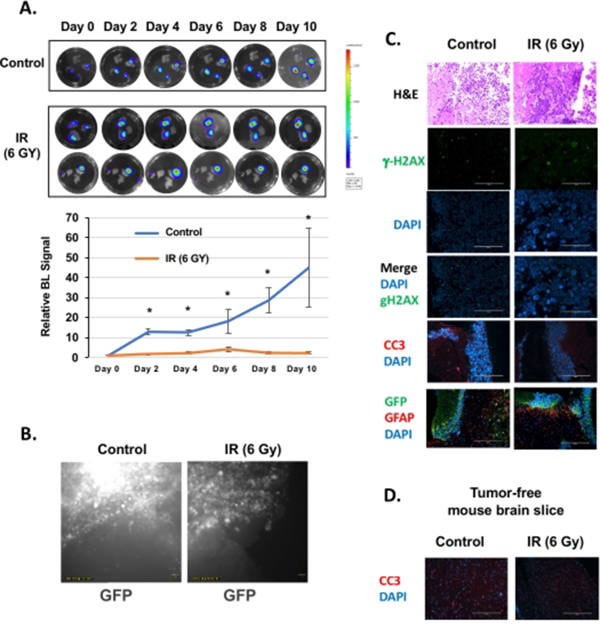
Irradiation (IR) is one of the first lines of treatment for patients that present with brain metastases in the clinic. To understand how the ex vivo slice model would respond to such treatment, brain slices containing MDA-MB-231BR-GFP Luciferase tumors were exposed to 6Gy irradiation. Tumors in brain slices that were not exposed to irradiation continued growing ex vivo, while tumors exposed to irradiation showed a stalled growth (Figure 2A). Brain slices containing MDA-MB-231BR-GFP-Luciferase tumors were also monitored via live imaging for 48 h after day 4 to visualize tumor growth in the brain microenvironment following IR treatment (Figure 2B). Consistent with bioluminescent imaging, control cancer cells are seen rapidly growing as GFP intensity increases and a number of cells are seen invading the brain parenchyma (Video 1). In contrast, cancer cells in brain slices treated with 6Gy IR are cytostatic, and GFP intensity is maintained (Video 2). H&E staining confirms smaller tumors in IR-treated cells (Figure 2C). We also detected large multinucleated cancer cells and an increase in staining of DNA damage marker g-H2AX in IR treated cancer cells (Figure 2C). No increase in apoptosis in IR treated slices was detected, suggesting that IR may reduce proliferation (Figure 2C). Nevertheless, an increase in staining in reactive astrocytes was detected, visualized by staining for GFAP-positive cells, which are visible around the cluster of cancer cells (GFP-positive) treated by IR (Figure 2C). This suggests that this ex vivo model may also be useful in understanding how some tumor-stroma components respond to treatments. Additionally, we did not detect any apoptosis caused by IR treatment of tumor-free brain mouse slices (Figure 2D).
Figure 2. Treatment of BCBM organotypic brain slices with radiation.
(A) Representative images depicting tumor growth in organotypic brain slices derived from mice intracranially injected with MDA-MB-231BR-GFP-Luciferase cells detected via bioluminescence. Tumors are exposed to no irradiation or 6 Gy irradiation (one dose) and allowed to grow for the indicated days. Quantification of tumor growth at indicated day (control. n = 3; 6Gy, n=3). Student’s t-test reported as mean ± SEM. * p < 0.05.(B) Brain slices were imaged for 48 h every 15 min. Representative images of videos of non-irradiated and irradiated samples indicate tumor growth. (C) Slices, as treated in (A), were fixed and assayed for H&E, g-H2AX, DAPI, cleaved-Caspase-3 (CC3), and GFP staining (image magnification 20x, scale bar: 200 μm). (D) Tumor-free mouse brain slices were treated as in (A) and were fixed and assayed for CC3 and DAPI (image magnification 20x, scale bar: 200 μm). Please click here to view a larger version of this figure.
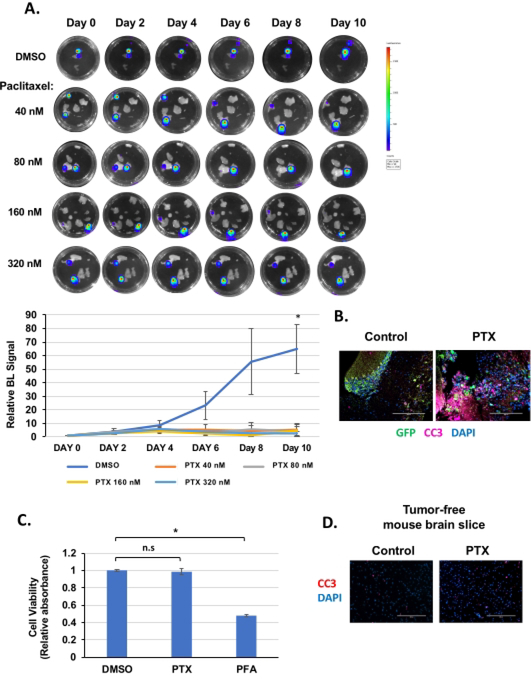
We also tested whether MDA-MB-231BR preformed tumors in brain slices would respond to chemotherapy. The brain slices containing MDA-MB-231BR tumors were treated to different concentrations of paclitaxel (Figure 3A), a chemotherapeutic drug used in breast cancer patients23. Treatment decreased the tumor’s size as quantified by bioluminescent signaling, but this inhibition was only significant following day 10 of treatment (Figure 3A). This may be due to a heterogeneous response to paclitaxel ex vivo. For example, in the 40 nM treated well, one slice containing a tumor (top) is reduced while the tumor (bottom) in the same well seems to be growing. Also, some of the smaller tumors seemed to be more sensitive to paclitaxel and exhibited reduced bioluminescence, while the larger tumors seemed refractory (Figure 3A, 80 nM treatment). Consistent with reduced tumor size, an increase in apoptosis was detected in paclitaxel-treated tumor cells compared to controls (Figure 3B). Importantly, using a viability MTS assay as adapted from Mews et al. to incorporate the use of triton24, we tested that inhibitor treatment did not alter brain tissue viability (in the absence of tumor) (Figure 3C). Consistent with this result, we did not detect and increase apoptosis as measured by cleaved caspase 3 staining in paclitaxel-treated tumor-free mouse brain slice (Figure 3D). These data suggest that this model may be helpful in understanding the chemotherapeutic response and resistance of BCBM within a brain microenvironment. Due to the nature of the model, an ex vivo setting that conserves the brain-tumor properties and interaction, its essentiality stands in providing primary evidence of the response of BCBM to chemotherapeutic reagents and thus eliminate drugs that do not shrink the tumor in this model from future in vivo trials, which are still essential in validating the effect of drugs that provide positive results in this model, the systemic response in the organism, and the ability to cross the blood-brain barrier.
Figure 3. Treatment of BCBM organotypic brain slices with paclitaxel.
(A) Brain slices derived from mice intracranially injected with MDA-MB-231BR-GFP-Luciferase cells treated with DMSO or following treatment of paclitaxel at 40 nM, 80 nM, 160 nM and 320 nM for 10 days, replenishing the medium every 48 h and allowed to grow for the indicated days (image magnification 20x, scale bar: 200 μm). Quantification of tumor growth at indicated day (control n = 6, Paclitaxel n = 3) (bottom). Student’s t-test reported as mean ± SEM. * p < 0.05. (B) Ex vivo brain slice as treated in (A) with either DMSO or 320 nM paclitaxel were collected, fixed, and assayed for GFP, CC3, and DAPI (image magnification 20x, scale bar: 200 μm). (C) Ex vivo brain slice as treated in (A) (without tumor) with either DMSO or 320 nM paclitaxel were collected at day 10 and analyzed for cell viability (MTS assay). As a positive control, slices were treated with paraformaldehyde (PFA) that renders the brain slices inviable (n = 3) Student’s t-test reported as mean ± SEM. * p < 0.05. (D) Ex vivo brain slice as treated in (A) (without tumor) with either DMSO or 320 nM paclitaxel were collected, fixed, and assayed for CC3 and DAPI (image magnification 20x, scale bar: 200 μm). Please click here to view a larger version of this figure.
Video 1. Live imaging of BCBM organotypic brain slices. Brain slices derived from mice intracranially injected with MDA-MB-231BR-GFP-Luciferase cells were allowed to grow for 4 days and then placed under a live image microscope for 48 h where images were taken every 15 min and combined to create a video using ImageJ. Please click here to download this Video.
Video 2. Live imaging of irradiated BCBM organotypic brain slices. Brain slices derived from mice intracranially injected with MDA-MB-231BR-GFP-Luciferase cells were irradiated the day after slices were generated and 4 days later were imaged every 15 min for 48 h. Please click here to download this Video.
Discussion
This study establishes a novel ex vivo brain culture method for explanted xenograft brain tumors. We show that BCBM cells MDA-MB-231BR cells intracranially injected in the brain of mice can survive and grow in ex vivo brain slices. The study also tested intracranially injected U87MG glioblastoma (GBM) cells and also found that these cancer cells survive and grow in brain slices (data not shown). We believe this model can be expanded beyond BCBM and GBM to other cancers that readily metastasize to the brain, including lung cancer and melanoma. Intracranial injections of breast cancer brain trophic cells were chosen since it is a simple and fast method to generate brain tumors and is regarded as a suitable model for brain macrometastases25. However, similar tumor slice models can be developed from either intracardiac inoculation or orthotopic injections into primary fat pads. Optimization of the culture conditions by growing adult brain slices in serum-free medium and supplying fresh medium every 48 h provided better preservation of the tissue viability and survival and growth of tumors. The 0.4 μm inserts utilized under hyperoxic conditions of the incubator serve as structural support for the ex vivo brain slices and also provide a connection to the artificial supply of nutrients and oxygen to the tissue and tumor cells in the absence of blood supply which seems to be adequate for maintaining tissue viability for at least ten days in culture. The live imaging represents tumor growth as a function of GFP signal increase. However, the software has a threshold of maximal GFP luminosity that saturates the signal and does not allow for proper identification of single-cell motility in the brain microenvironment. Future modification of the microscopic-software signal detection will ameliorate the ability to accurately report the temporal increase in GFP signal as a function of tumor growth.
This model also allows for the monitoring of tumor growth and viability through immunohistochemical staining. Importantly, this model will allow for rapid testing of the efficacy of radiation sensitivity with or without small molecule inhibitors of tumors grown in the brain microenvironment. We show that this model is amenable to both treatments with radiation and chemotherapeutics. In addition, this system may be utilized for omics studies such as proteomics, metabolomics, and genomics to further understand cancer growth and response to treatments within the brain microenvironment. Furthermore, testing the potential relapse of tumors upon removing inhibitors after a shorter exposure would allow further understanding of chemoresistant pathways unique to cancer cells within the brain microenvironment.
In summary, we have developed a novel ex vivo brain slice of xenograft tumor tissue culturing that will monitor tumor growth and interaction with the brain tumor microenvironment and for rapid screening of therapeutic agents to block tumor growth in the brain.
Acknowledgments
We want to thank Julia Farnan, Kayla Green, and Tiziana DeAngelis for their technical assistance. This work was supported in part by the Pennsylvania Commonwealth Universal Research Enhancement Grant Program (MJR, JGJ), UO1CA244303 (MJR), R01CA227479 (NLS), R00CA207855 (EJH), and W.W. Smith Charitable Trusts (EjH).
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/62617.
Disclosures
Authors have no financial conflicts to declare.
References
- 1.Watase C et al. Breast cancer brain metastasis-overview of disease state, treatment options and future perspectives.Cancers (Basel). 13 (5), (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niikura N et al. Treatment outcomes and prognostic factors for patients with brain metastases from breast cancer of each subtype: a multicenter retrospective analysis.Breast Cancer Research and Treatment. 147 (1), 103–12 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Fong EL et al. Heralding a new paradigm in 3D tumor modeling.Biomaterials. 108, 197–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker JJ et al. A human glioblastoma organotypic slice culture model for study of tumor cell migration and patient-specific effects of anti-invasive drugs.Journal of Visualized Experiments: JoVE. 125, 53557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang HN et al. Coculture system with an organotypic brain slice and 3D spheroid of carcinoma cells.Journal of Visualized Experiments: JoVE. 80, 50881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohensee I et al. PTEN mediates the cross talk between breast and glial cells in brain metastases leading to rapid disease progression.Oncotarget. 8 (4), 6155–6168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Merbel AF et al. An ex vivo Tissue culture model for the assessment of individualized drug responses in prostate and bladder cancer.Frontiers in Oncology. 8, 400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin SZ et al. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer.BMC Cancer. 19 (1), 1030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orimo A, Weinberg RA Stromal fibroblasts in cancer: a novel tumor-promoting cell type.Cell Cycle. 5 (15), 1597–601 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Lim CY et al. Organotypic slice cultures of pancreatic ductal adenocarcinoma preserve the tumor microenvironment and provide a platform for drug response.Pancreatology. 18 (8), 913–927 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Gerlach MM et al. Slice cultures from head and neck squamous cell carcinoma: a novel test system for drug susceptibility and mechanisms of resistance.British Journal of Cancer. 110 (2), 479–488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koerfer J et al. Organotypic slice cultures of human gastric and esophagogastric junction cancer.Cancer Medicine. 5 (7), 1444–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmieri D et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain.Cancer Research. 67 (9), 4190–4198 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Kyeong S et al. Subtypes of breast cancer show different spatial distributions of brain metastases.PLoS One. 12 (11), e0188542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengel K et al. Attributes of brain metastases from breast and lung cancer.International Journal of Clinical Oncology. 18 (3), 396–401 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Jackson JG et al. Neuronal activity and glutamate uptake decrease mitochondrial mobility in astrocytes and position mitochondria near glutamate transporters.Journal of Neuroscience. 34 (5), 1613–1624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnan JK, Green KK, Jackson JG Ex vivo imaging of mitochondrial dynamics and trafficking in astrocytes.Current Protocols in Neuroscience. 92 (1), e94 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Simone NL et al. Ionizing radiation-induced oxidative stress alters miRNA expression.PLoS One. 4 (7), e6377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couturier CP et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy.Nature Communications. 11 (1), 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candolfi M et al. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression.Journal of Neuro-Oncology. 85 (2), 133–48 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald DP et al. Reactive glia are recruited by highly proliferative brain metastases of breast cancer and promote tumor cell colonization.Clinical & Experimental Metastasis. 25 (7), 799–810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondru N et al. An Ex Vivo Brain Slice Culture Model of Chronic Wasting Disease: Implications for Disease Pathogenesis and Therapeutic Development. Scientific Reports. 10 (1), 7640 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abu Samaan TM et al. Paclitaxel’s mechanistic and clinical effects on breast cancer.Biomolecules. 9 (12) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mewes A, Franke H, H., Singer D Organotypic brain slice cultures of adult transgenic P301S mice--a model for tauopathy studies.PLoS One. 7 (9), e45017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valiente M et al. Brain metastasis cell lines panel: A public resource of organotropic cell lines.Cancer Research. 80 (20), 4314–4323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]