Summary
CRISPR-Cas are adaptive immune systems that protect their hosts against viruses and other parasitic mobile genetic elements.1 Although widely distributed among prokaryotic taxa, CRISPR-Cas systems are not ubiquitous.2, 3, 4 Like most defense-system genes, CRISPR-Cas are frequently lost and gained, suggesting advantages are specific to particular environmental conditions.5 Selection from viruses is assumed to drive the acquisition and maintenance of these immune systems in nature, and both theory6, 7, 8 and experiments have identified phage density and diversity as key fitness determinants.9,10 However, these approaches lack the biological complexity inherent in nature. Here, we exploit metagenomic data from 324 samples across diverse ecosystems to analyze CRISPR abundance in natural environments. For each metagenome, we quantified viral abundance and diversity to test whether these contribute to CRISPR-Cas abundance across ecosystems. We find a strong positive association between CRISPR-Cas abundance and viral abundance. In addition, when controlling for differences in viral abundance, CRISPR-Cas systems are more abundant when viral diversity is low, suggesting that such adaptive immune systems may offer limited protection when required to target a diverse viral community. CRISPR-Cas abundance also differed among environments, with environmental classification explaining roughly a quarter of the variation in CRISPR-Cas relative abundance. The relationships between CRISPR-Cas abundance, viral abundance, and viral diversity are broadly consistent across environments, providing robust evidence from natural ecosystems that supports predictions of when CRISPR is beneficial. These results indicate that viral abundance and diversity are major ecological factors that drive the selection and maintenance of CRISPR-Cas in microbial ecosystems.
Keywords: CRISPR-Cas, bacteriophages, metagenomics, microbial ecology
Highlights
-
•
Metagenomic data from diverse ecosystems are used to analyze CRISPR prevalence
-
•
Environment type explains ∼a quarter of the variation in CRISPR-Cas abundance
-
•
There is a positive association between CRISPR-Cas abundance and viral abundance
-
•
CRISPR-Cas is more abundant when viral diversity is comparatively lower
Meaden and Biswas et al. use metagenomic data to test associations between CRISPR-Cas abundance and viral abundance and diversity across environments. There is a positive association between CRISPR-Cas abundance and viral abundance. Viral diversity is negatively correlated with CRISPR-Cas abundance when controlling for viral abundance.
Results and discussion
Variation in CRISPR-Cas abundance is partially explained by viral abundance
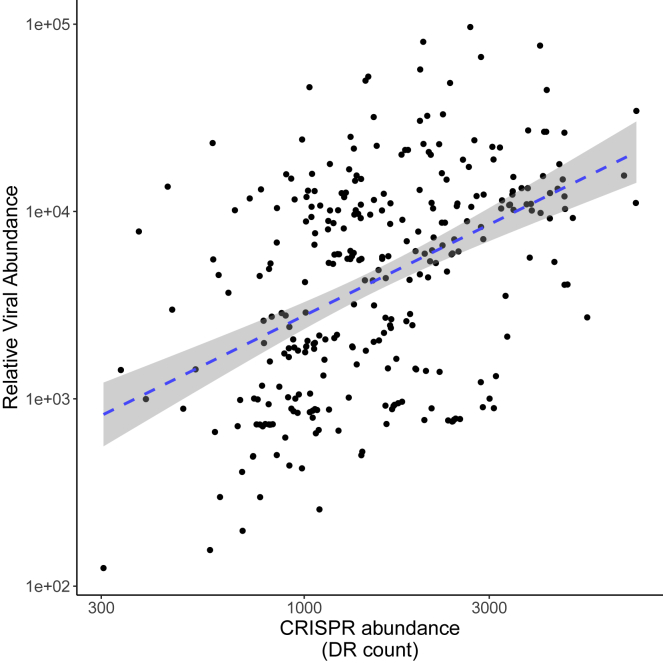
While it is well established that CRISPR-Cas immune systems can protect bacteria and archaea against viral infections under in vitro laboratory conditions, it remains unclear how important viruses are as a selective force for the maintenance of CRISPR-Cas systems in nature, as CRISPR-Cas also targets other genetic parasites, such as plasmids.11 To assess the role of viruses as a selective force for CRISPR-Cas, we first compiled a dataset of 324 metagenomes and quantified the abundance of CRISPR-Cas systems and viruses in each sample. Our analyses use all contigs classified as viral, and while the vast majority of these are of bacteriophage and prophage origin, we refer to these simply as viral for consistency (benchmarking analyses of our classifier versus existing tools are available in the supplemental information). These metagenomes vary in both CRISPR-Cas and viral abundance and therefore provided a suitable dataset to test the hypothesis that viral abundance drives selection for CRISPR-Cas (Figure 1). We found a positive correlation between viral abundance and the abundance of CRISPR-Cas systems (general linear model [GLM]; F1,313 = 81.32; p < 0.0001; Figure 1), with viral abundance explaining around 20% of the observed variation in CRISPR-Cas abundance (R2 = 0.209). We obtained qualitatively the same result when we included archaeal abundance in our model, which typically carries more CRISPR-Cas immune systems than bacteria.1 We also compared our measure of viral abundance, which is based on coverage, to measuring the total number of reads that map to viral contigs and found a near-perfect positive correlation (Pearson correlation = 0.94; Figure S1). These results strongly suggest that viruses are a fundamental selective force for the maintenance of CRISPR-Cas across diverse environmental conditions.
Figure 1.
CRISPR abundance positively correlates with viral abundance
Correlation between relative viral abundance and the read count (per million) of metagenomic reads that mapped to CRISPR array repeats across all samples. The dashed line represents the linear model fit, and shaded area represents 95% confidence interval (p < 0.0001 and R2 = 0.21).
Environmental conditions influence CRISPR-Cas abundance
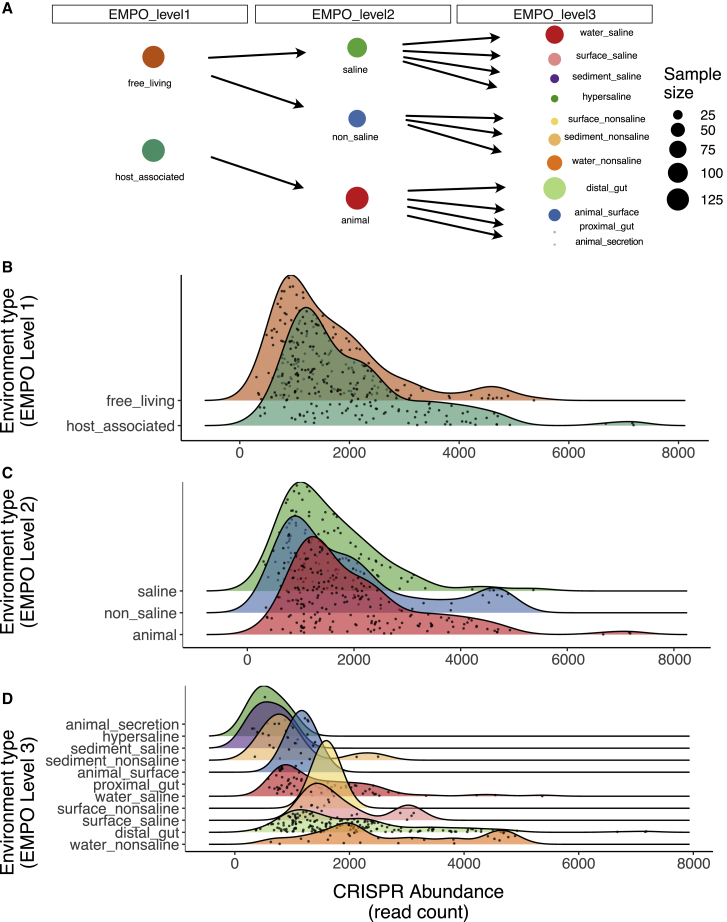
In addition to viruses being a selective force for CRISPR-Cas, ecological factors may determine when CRISPR-Cas is beneficial, and therefore, CRISPR-Cas abundance may vary across different natural environments.12 We therefore grouped samples into ecologically meaningful categories, using the Earth Microbiome Project’s sample ontology (EMPO). This framework is structured to capture two major environmental axes on which bacterial community composition orient, namely host association and salinity.13 Level 1 of the ontology classifies samples as host associated or free living; level 2 classifies samples as saline or non-saline, or animal or plant-associated; and level 3 describes microbial environments that can be grouped into levels 1 and 2 hierarchically (Figure 2A).13 These EMPO classifications highlighted the varied CRISPR-Cas and viral abundance in these different environments (Figure 2). Using this EMPO classification, we observed substantial variation in CRISPR-Cas abundance, both within and between environment types (Figure 2). For example, host-associated communities had a greater prevalence of CRISPR-Cas than free-living environments (GLM; F1,322 = 12.22; p < 0.001), although this classification only explained around 4% of the variation in CRISPR-Cas abundance (Figure 2B). In contrast, more fine-scaled classification of environments (such as gut, saline sediment, etc. as per EMPO level 3 classification; Figure 2D) explained 22% of the variation in CRISPR-Cas abundance. These results were also qualitatively the same when viral abundance was controlled for in the model (Figure S2). Taken together, these results suggest that, in addition to viruses as a key selective force for the maintenance of CRISPR-Cas, there are substantial differences in CRISPR-Cas abundance among natural environments.
Figure 2.
CRISPR abundance varies by environment
Distributions of metagenomic read counts that mapped to CRISPR arrays (read count per million that mapped to a CRISPR array predicted by CRISPRDetect v.3 from assembled contigs) grouped by environmental classification.
(A) Sample sizes and ontology are shown.
(B–D) Samples are grouped using the Earth Microbiome Project ontology (EMPO) at level 1 (B), 2 (C), or 3 (D).
Microbial community composition explains some variation in CRISPR-Cas abundance
Although we found effects of viral abundance and environmental classification on CRISPR-Cas abundance, it is plausible that these effects may be driven by differences in the microbial community composition as CRISPR-Cas prevalence can differ among taxa.1,2 We examined whether the variation in CRISPR-Cas abundance across these metagenomes might be affected by microbial community composition. We found a weak but significant relationship between CRISPR-Cas abundance and class-level community composition (PERMANOVA; F1, 313 = 12.4; p < 0.001; R2 = 0.04; permutations = 9,999; distance metric = Bray-Curtis). We next used clustering analyses to assess how well the EMPO framework levels grouped our samples based on community composition and extracted the taxa that best described differences among samples (Figure S3). In addition, we fitted CRISPR-Cas abundance to this ordination to identify “hotspots” of samples enriched with CRISPR-Cas (Figure S3). With this approach, we identified multiple groups of samples with high CRISPR-Cas abundance, supporting the notion that microbial phylogeny alone only explains a limited amount of variation in CRISPR-Cas abundance. Additional factors may contribute to the differences in CRISPR-Cas abundance across environment types. Furthermore, high rates of horizontal gene transfer (HGT) across taxa, coupled with frequent gain or loss of CRISPR-Cas, will likely reduce phylogenetic signal.
As an additional test of the influence of phylogenetic effects on our results, we assessed the impact of including archaeal abundance in our analyses. Archaea have previously been shown to be enriched for CRISPR-Cas systems;1 therefore, inclusion of archaeal abundance in our model should control for this effect. When we repeated our analysis of the correlation between CRISPR-Cas abundance and viral abundance, this time including the abundance of archaea in each sample as a covariate, we found no qualitative difference in result. Taken with the community composition analysis, these results suggests that the influence of phylogeny on our results is relatively small compared to the effect of viral abundance on the prevalence of CRISPR-Cas immune systems within a microbiome.
CRISPR-Cas and viral abundance correlate across diverse environments
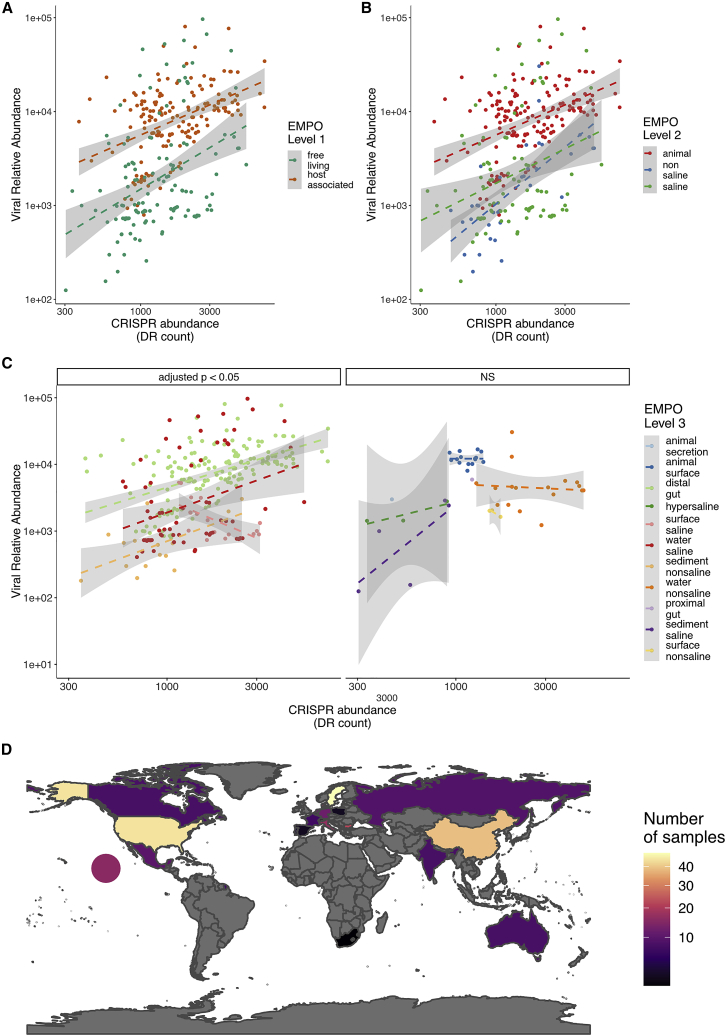
To explore to what extent the observed variation in CRISPR-Cas abundance within and between environment types is driven by variation in viral abundance, we grouped the samples by EMPO classification and quantified viral abundance for each environment type. Similar to the distributions of CRISPR-Cas abundance, we found substantial variation in viral abundance across environment types (Figure S1). The higher level classifications, EMPO levels 1 and 2, explained 30% and 31% of the variation observed, respectively, and the finer scale EMPO level 3 explained 34% of this variation in viral abundance (GLM; F1,314 = 15.78; p < 0.0001; R2 = 0.34). The relatively minor difference between classification levels suggests that the primary predictive power comes from EMPO level 1, with host-associated samples having a greater density of viruses than free-living samples. Overall, we found that the type of environment significantly predicts the abundance of viruses present, but that fine-scale classification adds little predictive power relative to high-level classification.
Although we found a significant correlation between viral abundance and CRISPR-Cas abundance, it remained unclear whether this relationship was consistent across environments. We therefore assessed whether the strength of this relationship was constant among each of our EMPO classification levels, which we modeled as an interaction in a multiple regression analysis. Strikingly, we found significant interaction effects at all EMPO levels, suggesting that the nature of the relationship between viral abundance and CRISPR-Cas abundance is, at least partly, dependent on additional environmental conditions (Data S1A; Figure 3). This was further validated by post hoc testing of the correlation between CRISPR-Cas abundance and viral abundance at each individual environment type (Data S1B). In this case, we found a consistent positive relationship at EMPO levels 1 and 2 but more varied results at level 3 (Figure 3), suggesting additional ecological factors may be playing a role in some environments. For example, when taking all EMPO level 3 classifications with more than 10 observations per group, non-saline sediments show a strong positive correlation between CRISPR-Cas abundance and viral abundance (adjusted p < 0.001; Pearson correlation = 0.71; n = 15). In contrast, non-saline water environmental samples show no significant correlation between CRISPR-Cas abundance and viral abundance (adjusted p = 1; Pearson correlation = −0.18; n = 34; all correlations can be found in Data S1C). Taken together, these results indicate that viral abundance typically correlates positively with CRISPR-Cas abundance but that the strength of this relationship is dependent on the particular environment.
Figure 3.
CRISPR abundance positively correlates with viral abundance across environments
Correlations between relative viral abundance and the read count (per million) of metagenomic reads that mapped to CRISPR array repeats per environment type.
(A and B) Environments are categorized according to the EMPO at level 1 (A) or 2 (B).
(C) Samples grouped at EMPO level 3 are divided into significant correlations or non-significant correlations (NS). Dashed lines represent linear model fits, and shaded areas represent 95% confidence intervals.
(D) The number of samples collected in each country with the circle representing samples collected in the Pacific Ocean.
See also Figures S1 and S2 and Data S1.
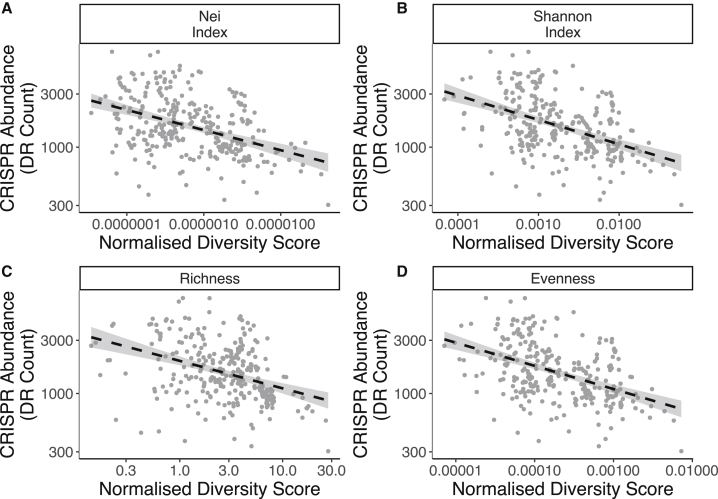
Virus diversity negatively correlates with CRISPR-Cas abundance
Both theory and in vitro experiments predict that low viral genetic diversity is also an important determinant of the benefits of CRISPR-Cas immunity.7, 8, 9, 10 This theory suggests that excess viral sequence diversity prevents the acquisition of sufficient spacer diversity to protect against the many different viruses. To test this prediction, we quantified viral diversity for each environment type and examined whether this correlated with CRISPR-Cas abundance. However, this analysis may be confounded by correlations between viral diversity and viral abundance. Indeed, in our dataset, viral diversity was strongly correlated with viral abundance (GLM; F1,295 = 208.6; p < 0.0001; R2 = 0.41). We therefore normalized the viral diversity scores by viral abundance for each sample. We then tested the correlation between CRISPR-Cas abundance and normalized viral diversity. In agreement with theory, we found a negative correlation between CRISPR-Cas abundance and normalized viral diversity for all viral diversity metrics used (Figure 4; Data S1D). The metrics used spanned multiple levels, with richness, evenness, and Shannon’s index describing inter-population diversity. By contrast, Nei’s diversity metric describes the intra-population genetic variation. Together, these results suggest that CRISPR-Cas is most effective when viral diversity is low, which supports that CRISPR-Cas immunity relies on sequence identity between spacer sequences and the viral protospacer sequence and array sizes are finite.
Figure 4.
CRISPR abundance negatively correlates with normalized viral diversity metrics
Correlations between viral diversity (normalized by viral load per sample) and CRISPR abundance (reads per million that map to CRISPR arrays). Panels represent Nei’s diversity index (A), Shannon’s index (B), contig richness (C), or contig evenness (D). (A) represents intra-contig viral diversity while (B)–(D) represent inter-contig viral diversity. Dashed lines represent linear model fits, and shaded areas represent 95% confidence intervals. See also Data S1.
Broader implications
Despite recent studies suggesting that CRISPR-Cas abundance varies across natural environments, such as soil14 and the human microbiome,15 the ecological factors that drive variation in CRISPR-Cas prevalence across natural microbial communities remained unclear.11 Furthermore, the extent of this variation across a much wider range of environments remained unexplored. We addressed this gap by using metagenomic data to quantify CRISPR array abundance within each metagenome and linked these data to the associated viral community present. We identified two key factors that predict CRISPR-Cas abundance: viral abundance and viral diversity. These factors are likely of primary importance, as the observed correlations are consistent across diverse environments. Most metagenomic studies are likely to miss a large fraction of the viral community due to biases in purification, DNA extraction, and sequencing technology;16 however, the samples in this study all fulfilled a standardized selection criteria (STAR Methods). These data suggest that relative abundance and diversity are key predictors of CRISPR-Cas prevalence. Taken together, our results show that high viral abundance and low diversity are major drivers of the selection and maintenance of CRISPR-Cas systems in nature.
There are likely factors in addition to viral abundance and diversity that contribute to CRISPR-Cas abundance in the environment because the correlations had relatively low R2 values (20% for abundance and 22% for normalized diversity). For example, many alternative phage defense systems have been described and, much like CRISPR-Cas, show scattered distributions even in closely related bacterial strains.17 The interplay and redundancy between different phage defense systems is poorly characterized and may also contribute to CRISPR-Cas distributions in different environments. Understanding the environmental parameters that select for different defenses will be crucial future research. For example, we see greater CRISPR-Cas abundance in host-associated samples over free-living samples, but it is unknown whether alternative defense mechanisms are favored in these free-living samples or whether there are fewer defenses overall.
Regarding CRISPR-Cas, multiple environmental parameters have been predicted to interact negatively with these systems. For example, aerobicity showed a negative association with CRISPR-Cas prevalence during a modeling analysis of bacterial traits,12 possibly due to an incompatibility between the requirement for non-homologous end join repair (NHEJ) in aerobic respiration and type II CRISPR-Cas sytems.18 More generally, intracellular defenses may be favored over surface receptor modifications under certain environmental conditions, as these have been shown to be subject to trade-offs with both biotic and abiotic factors.19, 20, 21 In addition, recent work has demonstrated that regulation of phage defenses, including CRISPR-Cas, is mediated by environmental conditions.22 There are also likely additional roles of plasmids that we have not examined in our analysis, as a recent longitudinal study found plasmids were targeted by CRISPR-Cas systems at 5 times the rate of phages.23 Overall, our results demonstrate that phage-mediated selection is a major driver of CRISPR-Cas prevalence, but additional biotic and abiotic complexity likely shapes the strength of this relationship.
Previous theoretical models predicted that CRISPR-Cas will be less favorable in dense and diverse viral communities.7,8 Above a threshold of phage genetic diversity, CRISPR-Cas becomes ineffective and is lost due to an associated fitness cost, and this threshold is reached more often in large viral populations.7 While these predictions seem intuitive, our results suggest that, although low viral diversity does indeed favor CRISPR-Cas, low viral density does not. It is possible that viral abundance rarely reaches levels in nature that are sufficient to preclude an effective CRISPR-Cas response, even if such densities are readily achievable in laboratory experiments.9,10,24 Future work may reveal the ecological differences between CRISPR-Cas types, as different types are likely to coevolve with viruses in fundamentally different ways.25
Genomic evidence demonstrates that CRISPR-Cas systems are frequently acquired and lost,26,27 and empirical studies show that they can be mobilized through HGT.28,29 Notably, in a previous longitudinal study, CRISPR-Cas prevalence increased through time, even in phyla that decreased in abundance,14 again suggesting high mobility and positive selection for CRISPR-Cas immunity. Consistent with high rates of HGT of CRISPR-Cas, our results demonstrate that, while phylogeny can influence the CRISPR-Cas repertoire, it is not the primary driver of selection in nature.
In summary, by quantifying the role of the viral community in shaping CRISPR-Cas abundance in complex, diverse natural communities, we found that high viral abundance but low diversity drives the selection and maintenance of CRISPR-Cas across a range of environments. Future work that embraces both the abiotic and biotic complexity of natural systems is required to further understand the prevalence of CRISPR-Cas.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Triticum aestivum genome sequence | Publicly available from GenBank. | GenBank: GCA_900519105.1 |
| Drosophila melanogaster genome sequence | Publicly available from GenBank. | GenBank: GCA_000001215.4 |
| Arabidopsis thaliana genome sequence | Publicly available from GenBank. | GenBank: GCA_000001735.1 |
| Physcomitrella patens genome sequence | Publicly available from GenBank. | GenBank: GCA_000002425.2 |
| Caenorhabdtitis elegans genome sequence | Publicly available from GenBank. | GenBank: GCA_000002985.3 |
| Heliconius melpomene | Publicly available from GenBank. | GenBank: GCA_000313835.1 |
| Metagenomic data used in this study | Publicly available from NCBI SRA database. | Accession list in Data S1F |
| Software and algorithms | ||
| BBMap | Bushnell et al.30 | https://jgi.doe.gov/data-and-tools/bbtools/ |
| MEGAHIT version 1.1.3 | Li et al.31 | https://github.com/voutcn/megahit |
| MIUMS | This study | https://github.com/ambarishbiswas/miums_v1.0 |
| Metaxa2 | Bengtsson-Palme et al.32 | https://microbiology.se/software/metaxa2/ |
| Magic-BLAST | Boratyn et al.33 | https://ncbi.github.io/magicblast/ |
| CRISPRDetect | Biswas et al.34 | https://github.com/ambarishbiswas/CRISPRDetect_3.0 |
| metaCRISPRDetect | Biswas et al.34 | https://github.com/ambarishbiswas/metaCRISPRDetect_1.0 |
| blastn | Chen et al.35 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| WGSIM | N/A | https://github.com/lh3/wgsim |
| VarScan | Koboldt et al.36 | http://dkoboldt.github.io/varscan/ |
| Metabat | Kang et al.37 | https://bitbucket.org/berkeleylab/metabat/src/master/ |
| samtools | Danecek et al.38 | http://www.htslib.org/ |
| vegan | Oksanen et al.39 | https://cran.r-project.org/web/packages/vegan/index.html |
| HMMER Version 3.2.1 | Eddy40 | http://hmmer.org/ |
| Diamond version v0.8.38.100 | Buchfink et al.41 | https://github.com/bbuchfink/diamond |
| metaGeneMark | Zhu et al.42 | https://github.com/aghozlane/spasm/tree/master/MetaGeneMark |
| VirSorter2 version 2.2.2 | Guo et al.43 | https://github.com/jiarong/VirSorter2 |
| DeepVirFinder version 1.0 | Ren et al.44 | https://github.com/jessieren/DeepVirFinder |
| cd-hit | Li and Godzik45 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| Bwa-mem | Li and Durbin46 | https://github.com/lh3/bwa |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact (S.Meaden@exeter.ac.uk).
Materials availability
This study did not generate unique reagents.
Experimental model and subject details
In October 2018, we used the SRA Advanced Search Builder and downloaded the metadata of all metagenomic samples with the properties “Platform: Illumina, Source: DNA, Access: Public, Organism: metagenome.” We then filtered out RNASeq, amplicon and treatment-specific samples which resulted in 6367 samples. However, 3243 of the samples did not have any usable “source environment” information and out of the remaining samples 1886 samples were “gut” samples collected from humans and other animals. To mitigate biases arising from skewed sample numbers from specific source environments we decided to randomly select up to 10 metagenomes per source environment (e.g., boreal lake sediment, human gut, marine sediment etc) with each metagenome containing at least 1 million reads with a minimum length of 100 nucleotides (nt) and insert size > = 150. After taxonomic profiling, samples with > 10% eukaryotic contamination were also removed, as well as samples that were no longer publicly available on 30/08/2021, resulting in a total of 324 metagenomic libraries. Paired-end metagenomic libraries (sequenced with Illumina platform) were then downloaded from the NCBI SRA database (Data S1F).
Method details
Computational pipeline
An overview of the computational pipeline is provided in Figure S4.
Metagenomic sample assembly
Libraries were processed with BBMap suite of tools for error correction30 and for each metagenomic library a representative FASTA file was created by combining both the merged and unmerged reads. A total of 324 libraries (each library containing at least 1 million reads with a minimum length of 100 nt and insert size > = 150) were selected to represent a wide range of biome diversity (Data S1F). Libraries were assembled using MegaHit (version 1.1.3)31 with default parameters and contigs with minimum length 200nt were retained. Contigs were classified as archaea, bacteria or virus using a purpose-built classification tool: MIUMS (Microbial Identification Using Marker Sequence, https://github.com/ambarishbiswas/miums_v1.0). MIUMS is designed to classify contigs based on a reference database containing protein sequence fragments highly specific to bacteria, archaea, viruses. Full details of MIUMS reference database construction and prediction process are provided in the MIUMS tool development section of the STAR Methods. Each metagenome was then subsampled to 1 million randomly selected reads with a minimum length of 100 nt.
Generation of archaeal and bacterial abundance tables
Subsampled reads were screened using metaxa232 (GSU parameters: -g ssu -f f, LSU parameters: -g lsu -f f) to generate a table of bacterial and archaeal abundances. Both the LSU and SSU based methods were used. A reference sequence database was made from contigs classified as viral by MIUMS. Subsampled reads were then mapped to the assembled contigs using Magic-BLAST33 (parameters: -no_unaligned -no_query_id_trim -perc_identity 95 -outfmt tabular). Reads with a minimum of 95% sequence identity and coverage were used.
CRISPR array identification
Accurately identifying CRISPR arrays in metagenomic data is challenging for a number of reasons. First, a large proportion of the direct repeats (DRs) identified from metagenomic contigs often show little sequence similarity to the CRISPR repeats found in published genomes and lack an isolated representative.2 Second, CRISPR arrays found in metagenomic reads are generally short (i.e., < 3 DRs) and often missing one or both flanking regions. To overcome these issues we combined information on existing, published genomes and their CRISPR arrays along with de novo extraction of putative CRISPR arrays from our assembled contigs (Figure S4). A database of metagenomic CRISPR arrays was first constructed by processing all assembled contigs with CRISPRDetect version 3 (CRISPRDetect3, https://github.com/ambarishbiswas/CRISPRDetect_3.0). CRISPRDetect version 3 was also modified to allow prediction of shorter CRISPRs (e.g., partial/broken CRISPRs with as little as 1.5 repeats). A higher CRISPR likelihood score cut-off of 4.5 was used instead of the default score cut-off of 3 to reduce potential non-CRISPR arrays. CRISPRDetect34 uses several CRISPR elements (e.g., repeats, spacers, cas genes, AT composition of flanking regions etc.) from published genomes to identify and separate true CRISPRs from other genomic repeats. In this study, a modified version of the CRISPRDetect tool was used, which uses a reference repeat database created using the cluster representative DRs from the metagenomic contigs as well as DRs found in published genomes. Predicted CRISPR arrays were checked to ensure that the total array degeneracy (i.e., number of insertion, deletion, mutation or presence of Ns in the array) was less than the total number of DRs in the array, which resulted in 51395 CRISPR arrays. Direct repeat sequences (23 to 60 nt) were extracted and clustered with cd-hit-est (parameters: -n 3 -c 0.90 -aL 0.90 -aS 0.90).45 30370 clusters were derived from 33745 unique DR sequences. Of these clusters, 22808 contained a single DR sequence while 7,562 contained multiple DR sequences. Only the multi-DR sequence containing arrays were taken forward for quantification as these represent sequences with a higher probability of containing true CRISPR arrays.
Subsampled reads were then screened against this database using metaCRISPRDetect (https://github.com/ambarishbiswas/metaCRISPRDetect_1.0), which supports rapid identification of CRISPR arrays in short reads using user-provided reference repeat database as an extension of CRISPRDetect.34 Arrays with a likelihood score > 3 were added to the existing CRISPR reference database. Subsampled reads were then mapped to the reference database using blastn35 [parameters: -task blastn-short with default parameters].
Identifying potential false positive CRISPRs
CRISPRs predicted from metagenomes are generally short, often incomplete and missing flanking regions which makes it hard to distinguish true CRISPRs from other genomic repeats. To measure how many of our identified arrays occurred in known prokaryotic genomes we compared the metagenomic CRISPR repeats against CRISPRs found in RefSeq and GenBank prokaryotic sequences (sequences published before September 9, 2019) using NCBI blast (parameters: -task blastn -word_size 11 -dust no -culling_limit 1 -num_alignments 1).47 Metagenomic DRs with > = 90% identity and > = 90% sequence coverage against RefSeq or GenBank DRs were considered as a positive match. Out of the 7562 repeat clusters 1649 were found in CRISPRs predicted from RefSeq or GenBank prokaryotic sequences. Similarly, the metagenomic repeats were screened against an in silico generated set of eukaryotic reads from 6 eukaryotic reference genomes (see Key resources table). Ten thousand 250bp paired reads were generated from each reference genome using WGSIM (https://github.com/lh3/wgsim). Eukaryotic reads were subsampled to an equal depth to remove differences due to genome size. Out of the 7562 DR clusters a total of 168 repeat clusters were found in the eukaryotic reads suggesting a high level of eukaryotic sequence contamination may increase the false positive rate of our analysis. We therefore removed samples where > 10% of reads were classified as of eukaryotic origin.
Microbial Identification Using Marker Sequence (MIUMS) tool development
MIUMS (version 1.0) utilizes a reference database of protein sequence fragments that are highly specific to their source organism. The process of constructing database is described below.
Selection of reference sequences
For the construction of a reference database of marker amino acid sequences, all amino acid sequences from 224 archaeal, 2810 bacterial and 3958 viral (4185) species (published before 1st of March 2017; Refseq release version 79; minimum sequence length of 5000 nt) were selected as the source of the protein sequences for the marker sequence database. These resulted in 155585 archaeal, 2219071 bacterial and 229026 viral amino acid sequences. In addition, a eukaryotic protein sequence database was constructed from 16179736 eukaryotic proteins. The taxonomic information of these protein sequences were collected from NCBI taxonomy files (https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/). MIUMS relies on a database of marker protein fragments constructed from RefSeq proteins, where each protein fragment contains two levels of classifications; primary (i.e., archaea, bacteria, viruses) and secondary (e.g., chromosome, plasmid, prophage and phage). During the construction of the marker protein fragment database, we used NCBI Protein search builder (https://www.ncbi.nlm.nih.gov/protein) to download the RefSeq protein accessions of all plasmid, phage and prophages (both archaeal and bacterial) using search keywords ‘plasmid’, ‘phage’ and ‘prophage’. The taxonomic details of the source protein sequences are obtained using the taxon_id associated to the protein accession. The secondary details are obtained from the protein definition and annotation using the default setting: ‘-report_dual_predictions’. Metagenomic sequences that matches marker protein fragments to one or more of these proteins (i.e., with secondary classification of plasmid, phage and prophage) are classified as such. Contigs that had a primary or secondary classification as prophage or provirus were included in our viral metrics in this study.
Construction of short protein sequence fragments
The construction of a database of marker protein sequence fragments is a multi-step process, which includes i). removal of sequence domains and ii). removal of all potential inter (super)kingdom homologous sequence regions from the target protein sequences.
The selected protein sequences from archaea, bacteria and viruses were screened with Pfam-A HMM profiles (version 30.0)48 and hmmsearch (HMMER version 3.2.1 with default parameters and–domtblout).40 Using the reported domain regions in the sequences, the protein sequences were split into multiple sequence fragments excluding the domain regions.
The sequence fragments were then concatenated into a single protein sequence database and used in an all-versus-all sequence similarity search using diamond (version v0.8.38.100; parameters:–evalue 0.001–sensitive–no-self-hits).41 By analyzing the diamond output file, longer sequence fragments which contain shorter sequence fragments were identified and further split into multiple sequence fragments followed by construction of a new sequence database. This process of identification of shorter sequence fragments continued in a cyclic manner till there were no new fragments identified.
Removal of inter superkingdom specific protein fragments
The sequence fragments were then separated into their associated superkingdom and screened against the other superkingdom specific primary protein sequences (including eukaryotic protein sequences) using diamond (parameters:–evalue 0.001–sensitive). Sequence fragments that were found to have inter (super)kingdom matches were identified and removed. This resulted in 610460 archaeal, 8312644 bacterial and 677951 viral marker protein fragments.
Assigning taxa specificity to the protein fragments
The archaeal, bacterial, and viral protein fragments were then screened against their own superkingdom specific primary source proteins using diamond (parameters:–evalue 0.001–sensitive) and the taxa specificity of each protein fragment to each of the higher taxonomic levels (i.e., phylum, class, order, family, genus and species) was determined using lowest common ancestor (LCA) algorithm from all reported diamond matches.
Taxonomic classification of metagenomic sequences
The default MIUMS runs involve three steps; i) assembly of metagenomic reads, ii) prediction of protein sequence in the assembled contigs using metaGeneMark,42 iii) screening the protein sequences against the marker protein fragment database using diamond (parameters:–evalue 0.001–sensitive). Contigs with metaGeneMark predicted proteins that contains multiple matches to the protein fragments above stringency cutoffs (e.g., overlapping length > = 21aa and sequence identity > = 40%) are assigned taxonomy based on the matched protein fragment’s taxa specificity and LCA algorithm. An output table is generated, which shows a list of classified contigs with associated taxonomy.
The raw reads (unless a subsampling of the reads were done) were mapped to the entire assembled contigs using magicblast.33 The magicblast output file is analyzed to identify all reads mapping to the classified contigs with minimum 99% identity and 99% sequence coverage. An output table is generated that shows each of those reads and their associated taxonomy.
Benchmarking of viral classification tools
In order to assess the accuracy of MIUMS for extracting viral contigs, we compared against existing tools using a test dataset of known sequences. The reference database of marker protein fragments that MIUMS V1.0 uses was constructed from sequences published before 1st of March 2017. Since then the number of newly released viral sequences in the NCBI RefSeq database has nearly doubled (5343 genomic DNA/RNA sequences published between 01-Mar-2017 and 01-June-2021; with minimum length 500 nt). To assess the performance of MIUMS against these newly published sequences; we randomly added 5000 of these viral sequences in a test dataset, comprising closed, whole genomes and contig-level assemblies. The test dataset was also supplemented with 15000 archaea, 15000 bacteria and 15000 eukaryotic sequences published during the same time period mentioned above (with minimum length cut-off of 500 nt and maximum length of 25000 nt). We also included eukaryotic sequences (randomly selected from animal, plant, fungi and protists sequences). This test dataset was analyzed with MIUMS V1.0, VirSorter2 (version 2.2.2)43 and DeepVirFinder (version 1.0)44 with default parameters. The summary outputs from the 3 tools used and their respective precision and recall scores are available in Data S1E.
DeepVirFinder reports a score between 0 to 1 for every input sequence, where a higher score (i.e., close to 1) is a strong indicator of a sequence being viral. Against a minimum score cutoff of 0.95, DeepVirFinder correctly predicted 2092 viruses and falsely predicted 4463 non-viral sequences (Archaea: 1500, Bacteria: 1058, Eukaryotes: 1905) as viruses. While reducing the minimum score-cutoff to 0.75 increases the total number of correctly predicted viral sequences to 3167, it also drastically increases the amount of false predictions to 9944 (Archaea: 3572, Bacteria: 2364, Eukaryotes: 4008). This trend continues with lower minimum score-cutoff to 0.50 (Viruses: 3988, Archaea: 6626, Bacteria: 4417, Eukaryotes: 6678) and 0.25 (Viruses: 4566, Archaea: 9765, Bacteria: 7054, Eukaryotes: 9594).
VirSorter2 generates scores between 0 to 1 (computed on both single and double stranded DNA) and reports potential viral sequences where the maximum of the two scores are > = 0.50. With a minimum score cutoff of 0.95, VirSorter2 correctly predicted 2367 viral sequences with 1317 non-viral sequences (Archaea: 97, Bacteria: 1191, Eukaryotes: 29) falsely predicted as viruses. Reducing the minimum score cutoff to 0.75 increases correctly the predicted viruses to 2798 but increases the falsely predicted non-viral sequences to 2018 (Archaea: 248, Bacteria: 1692, Eukaryotes: 78). At the default score cutoff of 0.50, VirSorter2 correctly predicted 3103 viral sequences with 2427 non-viral sequences (Archaea: 382, Bacteria: 1905, Eukaryotes: 140) falsely predicted to be viral.
In comparison, based on the main taxonomy output table, MIUMS accurately predicted 2557 viruses with a total of 32 non-viral sequences (Archaea: 0, Bacteria: 32, Eukaryotes: 0) falsely predicted to be viruses. The secondary taxonomy output table shows another 540 viral sequences correctly being predicted as potential viruses with 124 non-viral sequences (Archaea: 3, Bacteria: 122, Eukaryote: 0) falsely reported as potential viral sequences. A total of only 13 eukaryotic sequences falsely predicted as archaea, bacteria or viruses (Archaea: 0, bacteria: 12, Viruses: 1). We note that each of these tools handle prophages differently, which can affect precision and recall values, and software should be chosen based on the desired outcome i.e., inclusive or exclusive or prohages.
Quantification and statistical analysis
CRISPR array quantification
Abundance tables were generated for all spacers and direct repeat sequences by tallying the number of reads that mapped to a given DR with 100% sequence identity and coverage.
Virome diversity and abundance analysis
To assess the diversity of viruses in the different environments, we analyzed all MIUMS classified contigs from the 324 metagenomes. Across all datasets, we identified 2583 archaeal proviruses, 375179 bacterial prophages, 1218279 bacteriophages, and 174792 archaeal viruses. Per metagenome, the sub-sampled reads used for the CRISPR quantification analysis were mapped to the total set of all viral contigs with bwa mem.46 To quantify the genetic diversity of the viral community, we calculated the richness as the total number of detected viral contigs, plus the evenness and Shannon’s diversity index based on relative abundances calculated from the mean depth of coverage of all detected viral contigs. The intra-population diversity (micro-diversity) was calculated as the mean heterozygosity of viruses in the community by averaging Nei’s per-nucleotide diversity index across all detected contigs.49
where pj is the frequency of allele j at the position i of a contig of the length L, N is the number of viruses in a dataset. Single nucleotide variability was assessed with VarScan software.36
Viral abundance was calculated by first collecting the average depth values for all viral contigs in each sample using the ‘jgi_summarize_bam_contig_depths’ script from the Metabat package.37 These depth scores were then summed per sample to give an approximation of overall viral abundance relative to bacterial abundance. An additional abundance estimate of the fraction of reads mapping to viral contigs was generated by calculating the number of unmapped reads as reported by samtools idxtstats38 (See Figures S1D and S1E).
Statistical analysis
For each sample, the number of reads that mapped to a direct repeat were counted to give a measure of CRISPR array abundance per sample. Earth Microbiome Project Ontology levels were assigned using the framework previously described13 (Figure 1D) based on the associated metadata from the NCBI short read archive (SRA). In the case of bioreactor samples, a literature search was conducted to identify the original material described in the associated study (see Data S1F).
General linear models (GLM) were constructed for each of the reported correlations. In each case log10 transformations were applied to conform to model assumptions. Checks of model residuals were performed to assess model fit. Significance was determined using F-tests between null models and those containing the variable or interaction of interest.
Microbial community assessment
CRISPR is more common in archaea. Therefore, in order to minimize any phylogenetic effects deriving from high archaeal abundances in samples we estimated the number of archaeal reads in the subsampled reads using metaxa232 and converted this to relative abundance per sample. We then included this value as a fixed effect in additional GLMs to check the influence of archaeal abundance on our results. For additional phylogenetic assessments, reads classified as non-prokaryotic were removed and relative abundances were generated using the output from metaxa2 in order to assess effects of community composition. Permutational ANOVAs were performed on species abundance matrices using Bray-Curtis dissimilarity and CRISPR abundance as an explanatory variable with 9999 permutations, using the function ‘adonis’ from ‘vegan’ package.39 Visualization of clustering analyses, at the class level, was performed using non-metric multidimensional scaling (NMDS) with Bray-Curtis dissimilarity through the ‘metaMDS’ function in ‘vegan’39. Smooth surfaces were fit to these points via a generalized additive model (GAM), using either CRISPR abundance as the explanatory term, with the ‘ordisurf’ function in ‘vegan’44 (See Figure S3).
Acknowledgments
This work was funded by a grant from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (ERC-STG-2016-714478 to E.R.W.). E.R.W. was further supported by a NERC Independent Research Fellowship (NE/M018350/1). S.M. received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 842656. B.E.D. was supported by the Netherlands Organization for Scientific Research (NWO) Vidi grant 864.14.004 and European Research Council (ERC) Consolidator grant 865694: DiversiPHI. P.C.F. was supported by the former Bio-Protection Research Centre and Bioprotection Aotearoa (Tertiary Education Commission [TEC]), New Zealand.
Author contributions
Conceptualization, B.E.D., E.R.W., P.C.F., and S.E.M.; methodology, A.B., B.E.D., E.R.W., K.A., P.C.F., S.E.M., and S.M.; software, A.B., B.E.D., and K.A.; formal analysis, A.B., B.E.D., K.A., and S.M.; investigation, A.B., B.E.D., K.A., and S.M.; data curation, A.B., B.E.D., K.A., and S.M.; writing – original draft, E.R.W., P.C.F., and S.M.; writing – review & editing, A.B., B.E.D., E.R.W., K.A., P.C.F., S.E.M., and S.M.; visualization, S.M.; supervision, B.E.D., E.R.W., P.C.F., and S.E.M.; project administration, B.E.D., E.R.W., S.E.M., and P.C.F.; funding acquisition, B.E.D., E.R.W., S.E.M., S.M., and P.C.F.
Declaration of interests
The authors declare no competing interests.
Published: November 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.10.038.
Supplemental information
Interaction effects between virus abundance and CRISPR abundance across environments (A). Post-hoc testing of correlations between CRISPR abundance and viral abundance at each environmental classification (B). Pearson correlations of CRISPR abundance and viral abundance at each environmental classification (C). Associations between normalized viral diversity metrics and CRISPR abundance (D). Precision and recall analysis of viral classification tools (E). List of metagenomic samples used in this study. NCBI short read archive (SRA) accession number, metadata derived from the SRA database (sample_type, environment_biome, environment_feature, environment_material) and our classification into the Earth Microbiome Project Ontology (EMPO levels 1-3) (F).
Data and code availability
DNA sequence data are publicly available from the SRA database. Accession numbers are listed in Data S1F. No new sequence data was generated for this study. Original code is deposited in the github repositories listed in the Key resources table and statistical analysis scripts are available at https://github.com/s-meaden/Meaden_CB_2021. Code is publicly available at the time of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burstein D., Sun C.L., Brown C.T., Sharon I., Anantharaman K., Probst A.J., Thomas B.C., Banfield J.F. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 2016;7:10613. doi: 10.1038/ncomms10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pourcel C., Touchon M., Villeriot N., Vernadet J.P., Couvin D., Toffano-Nioche C., Vergnaud G. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020;48(D1):D535–D544. doi: 10.1093/nar/gkz915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mojica F.J.M., Garrett R.A. In: CRISPR-Cas Systems. Barrangou R., van der Oost J., editors. Springer; 2013. Discovery and seminal developments in the CRISPR field; pp. 1–31. [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Koonin E.V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurney J., Pleška M., Levin B.R. Why put up with immunity when there is resistance: an excursion into the population and evolutionary dynamics of restriction–modification and CRISPR-Cas. Philos. Trans. R. Soc. London B Biol. Sci. 2019;374:20180096. doi: 10.1098/rstb.2018.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iranzo J., Lobkovsky A.E., Wolf Y.I., Koonin E.V. Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR-Cas in an explicit ecological context. J. Bacteriol. 2013;195:3834–3844. doi: 10.1128/JB.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberger A.D., Sun C.L., Pluciński M.M., Denef V.J., Thomas B.C., Horvath P., Barrangou R., Gilmore M.S., Getz W.M., Banfield J.F. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput. Biol. 2012;8:e1002475. doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra E.R., van Houte S., Oyesiku-Blakemore S., Makin B., Broniewski J.M., Best A., Bondy-Denomy J., Davidson A., Boots M., Buckling A. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 2015;25:1043–1049. doi: 10.1016/j.cub.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 10.Broniewski J.M., Meaden S., Paterson S., Buckling A., Westra E.R. The effect of phage genetic diversity on bacterial resistance evolution. ISME J. 2020;14:828–836. doi: 10.1038/s41396-019-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westra E.R., Levin B.R. It is unclear how important CRISPR-Cas systems are for protecting natural populations of bacteria against infections by mobile genetic elements. Proc. Natl. Acad. Sci. USA. 2020;117:27777–27785. doi: 10.1073/pnas.1915966117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman J.L., Laljani R.M.R., Fagan W.F., Johnson P.L.F. Visualization and prediction of CRISPR incidence in microbial trait-space to identify drivers of antiviral immune strategy. ISME J. 2019;13:2589–2602. doi: 10.1038/s41396-019-0411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson L.R., Sanders J.G., McDonald D., Amir A., Ladau J., Locey K.J., Prill R.J., Tripathi A., Gibbons S.M., Ackermann G., et al. Earth Microbiome Project Consortium A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu R., Chai B., Cole J.R., Gunturu S.K., Guo X., Tian R., Gu J.D., Zhou J., Tiedje J.M. Targeted assemblies of cas1 suggest CRISPR-Cas’s response to soil warming. ISME J. 2020;14:1651–1662. doi: 10.1038/s41396-020-0635-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Münch P.C., Franzosa E.A., Stecher B., McHardy A.C., Huttenhower C. Identification of natural CRISPR systems and targets in the human microbiome. Cell Host Microbe. 2021;29:94–106.e4. doi: 10.1016/j.chom.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trubl G., Hyman P., Roux S., Abedon S.T. Coming-of-age characterization of soil viruses: a user’s guide to virus isolation, detection within metagenomes, and viromics. Soil Syst. 2020;4:23. [Google Scholar]
- 17.Bernheim A., Sorek R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 2020;18:113–119. doi: 10.1038/s41579-019-0278-2. [DOI] [PubMed] [Google Scholar]
- 18.Bernheim A., Calvo-Villamañán A., Basier C., Cui L., Rocha E.P.C., Touchon M., Bikard D. Inhibition of NHEJ repair by type II-A CRISPR-Cas systems in bacteria. Nat. Commun. 2017;8:2094. doi: 10.1038/s41467-017-02350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez C.A., Koskella B. Phage resistance evolution in vitro is not reflective of in vivo outcome in a plant-bacteria-phage system. Evolution. 2019;73:2461–2475. doi: 10.1111/evo.13833. [DOI] [PubMed] [Google Scholar]
- 20.Laanto E., Bamford J.K., Laakso J., Sundberg L.R. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS ONE. 2012;7:e53157. doi: 10.1371/journal.pone.0053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alseth E.O., Pursey E., Luján A.M., McLeod I., Rollie C., Westra E.R. Bacterial biodiversity drives the evolution of CRISPR-based phage resistance. Nature. 2019;574:549–552. doi: 10.1038/s41586-019-1662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith L.M., Jackson S.A., Malone L.M., Ussher J.E., Gardner P.P., Fineran P.C. The Rcs stress response inversely controls surface and CRISPR-Cas adaptive immunity to discriminate plasmids and phages. Nat. Microbiol. 2021;6:162–172. doi: 10.1038/s41564-020-00822-7. [DOI] [PubMed] [Google Scholar]
- 23.Martínez Arbas S., Narayanasamy S., Herold M., Lebrun L.A., Hoopmann M.R., Li S., Lam T.J., Kunath B.J., Hicks N.D., Liu C.M., et al. Roles of bacteriophages, plasmids and CRISPR immunity in microbial community dynamics revealed using time-series integrated meta-omics. Nat. Microbiol. 2021;6:123–135. doi: 10.1038/s41564-020-00794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissman J.L., Alseth E.O., Meaden S., Westra E.R., Fuhrman J.A. Immune lag is a major cost of prokaryotic adaptive immunity during viral outbreaks. Proc. Biol. Sci. 2021;288:20211555. doi: 10.1098/rspb.2021.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson B.N.J., Steens J.A., Staals R.H.J., Westra E.R., van Houte S. Coevolution between bacterial CRISPR-Cas systems and their bacteriophages. Cell Host Microbe. 2021;29:715–725. doi: 10.1016/j.chom.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Puigbò P., Makarova K.S., Kristensen D.M., Wolf Y.I., Koonin E.V. Reconstruction of the evolution of microbial defense systems. BMC Evol. Biol. 2017;17:94. doi: 10.1186/s12862-017-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Belkum A., Soriaga L.B., LaFave M.C., Akella S., Veyrieras J.B., Barbu E.M., Shortridge D., Blanc B., Hannum G., Zambardi G., et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio. 2015;6 doi: 10.1128/mBio.01796-15. e01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson B.N.J., Staals R.H.J., Fineran P.C. CRISPR-Cas-mediated phage resistance enhances horizontal gene transfer by transduction. MBio. 2018;9:e02406–e02417. doi: 10.1128/mBio.02406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varble A., Meaden S., Barrangou R., Westra E.R., Marraffini L.A. Recombination between phages and CRISPR-cas loci facilitates horizontal gene transfer in staphylococci. Nat. Microbiol. 2019;4:956–963. doi: 10.1038/s41564-019-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushnell B., Rood J., Singer E. BBMerge - accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 32.Bengtsson-Palme J., Hartmann M., Eriksson K.M., Pal C., Thorell K., Larsson D.G.J., Nilsson R.H. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 2015;15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 33.Boratyn G.M., Thierry-Mieg J., Thierry-Mieg D., Busby B., Madden T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics. 2019;20:405. doi: 10.1186/s12859-019-2996-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biswas A., Staals R.H.J., Morales S.E., Fineran P.C., Brown C.M. CRISPRDetect: a flexible algorithm to define CRISPR arrays. BMC Genom. 2016;17:356. doi: 10.1186/s12864-016-2627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Ye W., Zhang Y., Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., Weinstock G.M., Wilson R.K., Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O’hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H. 2013. Community ecology package. R package version 2(0) [Google Scholar]
- 40.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 41.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 42.Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J., Bolduc B., Zayed A.A., Varsani A., Dominguez-Huerta G., Delmont T.O., Pratama A.A., Gazitúa M.C., Vik D., Sullivan M.B., Roux S. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J., Song K., Deng C., Ahlgren N.A., Fuhrman J.A., Li Y., Xie X., Poplin R., Sun F. Identifying viruses from metagenomic data using deep learning. Quant. Biol. 2020;8:64–77. doi: 10.1007/s40484-019-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 46.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interaction effects between virus abundance and CRISPR abundance across environments (A). Post-hoc testing of correlations between CRISPR abundance and viral abundance at each environmental classification (B). Pearson correlations of CRISPR abundance and viral abundance at each environmental classification (C). Associations between normalized viral diversity metrics and CRISPR abundance (D). Precision and recall analysis of viral classification tools (E). List of metagenomic samples used in this study. NCBI short read archive (SRA) accession number, metadata derived from the SRA database (sample_type, environment_biome, environment_feature, environment_material) and our classification into the Earth Microbiome Project Ontology (EMPO levels 1-3) (F).
Data Availability Statement
DNA sequence data are publicly available from the SRA database. Accession numbers are listed in Data S1F. No new sequence data was generated for this study. Original code is deposited in the github repositories listed in the Key resources table and statistical analysis scripts are available at https://github.com/s-meaden/Meaden_CB_2021. Code is publicly available at the time of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.