Abstract
The latest severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant Omicron (B.1.1.529) has ushered panic responses around the world due to its contagious and vaccine escape mutations. The essential infectivity and antibody resistance of the SARS-CoV-2 variant are determined by its mutations on the spike (S) protein receptor-binding domain (RBD). However, a complete experimental evaluation of Omicron might take weeks or even months. Here, we present a comprehensive quantitative analysis of Omicron’s infectivity, vaccine-breakthrough, and antibody resistance. An artificial intelligence (AI) model, which has been trained with tens of thousands of experimental data points and extensively validated by experimental results on SARS-CoV-2, reveals that Omicron may be over ten times more contagious than the original virus or about 2.8 times as infectious as the Delta variant. Based on 185 three-dimensional (3D) structures of antibody-RBD complexes, we unveil that Omicron may have an 88% likelihood to escape current vaccines. The Food and Drug Administration (FDA)-approved monoclonal antibodies (mAbs) from Eli Lilly may be seriously compromised. Omicron may also diminish the efficacy of mAbs from AstraZeneca, Regeneron mAb cocktail, Celltrion, and Rockefeller University. However, its impacts on GlaxoSmithKline’s sotrovimab appear to be mild. Our work calls for new strategies to develop the next generation mutation-proof SARS-CoV-2 vaccines and antibodies.
Graphical Abstract

1. Introduction
On November 26, 2021, the World Health Organization (WHO) announced a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant Omicron (B.1.1.529), as a variant of concern (VOC). This variant carries an unusually high number of mutations, 32, on the spike (S) protein, the main antigenic target of antibodies generated by either infections or vaccination. In comparison, the devastating Delta variant has only 5 S protein mutations, which posed a high potential global risk and has spread internationally. Therefore, the “panic button” has been pushed in several cases worldwide, and many countries have enacted travel restrictions to prevent the rapid spread of the Omicron variant.
The mutations on the Omicron variant are widely distributed on multiple proteins of SARS-CoV-2 such as NSP3, NSP4, NSP5, NSP6, NSP12, NSP14, S protein, envelope protein, membrane protein, and nucleocapsid protein. The focus is the mutations on the S protein receptor-binding domain (RBD) for the potential impact on infectivity and antibody resistance caused by this new variant. This is due to the fact that the RBD located on the S protein facilitates the binding between the S protein and the host angiotensin-converting enzyme 2 (ACE2). Such S-ACE2 binding helps the SARS-CoV-2 enter the host cell and initiates the viral infection process. Several studies have shown that the binding free energy (BFE) between the S RBD and the ACE2 is proportional to the viral infectivity [1-5]. As such, an antibody that binds strongly to the RBD would directly neutralize the virus [6-8]. Indeed, many RBD binding antibodies are generated by the human immune response to infection or vaccination. Monoclonal antibodies (mABs) targeting the S protein, particularly the RBD, are designed to treat viral infection. As a result, any mutation on the S protein RBD would cause immediate concerns about the efficacy of existing vaccines, mAbs and the potential of reinfection.
The global panic brought by the emergence of the Omicron variant drives the scientific community to immediately investigate how much this new variant could undermine the existing vaccines and mAbs. However, relatively reliable experimental results from experimental labs will take a few weeks to come out. Therefore, an efficient and reliable in-silico analysis is imperative and valuable for such an urgent situation. Here, we present a comprehensive topology-based artificial intelligence (AI) model called TopNetmAb [9, 10] to predict the BFE changes of S and ACE2/antibody complexes induced by mutations on the S RBD of the Omicron variant. The positive BFE change induced by a specific RBD mutation indicates its potential ability to strengthen the binding of an S protein-ACE2/antibody complex, while a negative BFE change suggests a likely capacity to reduce the binding strength of an S protein-ACE2/antibody complex.
The TopNetmAb model that we proposed has been extensively validated over the past 1.9 years [10, 11]. Initially, in early 2020, we applied our TopNetmAb model to successfully predict that residues 452 and 501 ”have high chances to mutate into significantly more infectious COVID-19 strains” [9]. Such findings have been confirmed due to the emergency of multiple variants such as Alpha, Beta, Gamma, Delta, Theta, Lambda, Mu, and Omicron that carry L452R/Q and N501Y mutations. In April 2021, we provided a list of 31 RBD mutations that may weaken most of the binding to antibodies, such as W353R, I401N, Y449D, Y449S, P491R, P491L, Q493P [10]. Notably, experimental results have also shown that mutations at residues Y449, E484, Q493, S494, Y505 might enable the virus to escape antibodies [12]. Meanwhile, in the same work, we also revealed that variants found in the United Kingdom and South Africa in late 2020 may strengthen the binding of the RBD-ACE2 complex, which is consistent with the experimental results [13]. Later on, we provided a list of most likely vaccine escape RBD mutations predicted by TopNetmAb, such as S494P, Q493L, K417N, F490S, F486L, R403K, E484K, L452R, K417T, F490L, E484Q, and A475S [14], and mutations such as S494P, K417N, E484K/Q, and L452R are all detected in the variants of concern and variants of interest denounced by WHO. Last but not least, the correlation between the experimental deep mutational data [15] and our AI-predicted RBD-mutation-induced BFE changes for all possible 3686 RBD mutations on the RBD-ACE2 complex is 0.7, which indicates the reliability of our TopNetmAb model predictions [11]. As a baseline, one may keep in mind that experimental deep mutational results for SARS-COV-2 PPIs from 2 different labs only have a correlation of 0.67 [15, 16].
This work aims to analyze how the RBD mutations on the Omicron variant will affect the viral infectivity and efficacy of existing vaccines and antibody drugs. Fifteen Omicron RBD mutations, including S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, N501Y, and Y505H, are studied in this work. Additionally, three-dimensional (3D) structures of RBD-ACE2 complex and 185 antibody-RBD complexes, including many mAbs, are examined to understand the impacts of Omicron RBD mutations. We reveal that Omicron may be over ten times more contagious than the original SARS-CoV-2, more infectious than any other named variants, and over twice as infectious as the Delta variant, mainly due to its RBD mutations N440K, T478K, and N501Y. Additionally, Omicron has a high potential to disrupt the binding of most 185 antibodies with the S protein, mainly due to its RBD mutations K417N, E484A, and Y505H, indicating its stronger vaccine-breakthrough capability than the Delta or any other named variants. We have also unveiled that Omicron may seriously reduce the efficacy of the Eli Lilly mAb cocktail because of Omicron RBD mutations K417N, E484A, and Q493R. The Regeneron mAb cocktail may be impaired by Omicron RBD mutations K417N and E484A, and G446S. The efficacy of AstraZeneca mAb cocktail tixagevimab and cilgavimab may be moderately reduced by Omicron RBD mutation Q498R. Celltrion antibody Regdanvimab may be disrupted by Omicron RBD mutations E484A, Q493R, and Q498R. Omicron RBD mutation E484A may also disrupt Rockefeller University mAbs. However, Omicron’s impacts on GlaxoSmithKline’s mAb are predicted to be mild.
We stated in an earlier work that “we anticipate that as a complementary transmission pathway, vaccine-breakthrough or antibody-resistant mutations, like those in Omicron, will become a dominating mechanism of SARS-CoV-2 evolution when most of the world’s population is either vaccinated or infected” [17]. Our present finding shows it is high time to develop a new generation of vaccines and mAbs that will not be prone to viral mutations.
2. Results
2.1. Infectivity
The Infectivity of SARS-CoV-2 is mainly determined by the binding affinity of the ACE2 and RBD complex, although the furin cleavage site plays a crucial role as well [18]. Omicron has three mutations at the furin cleavage site and 15 mutations on the RBD, suggesting a significant change in its infectivity. Due to natural selection, the virus enhances its evolutionary advantages at the RBD either by mutations to strengthen the ACE2-RBD binding affinity or by mutations to escape antibody protection [19]. Since the virus has optimized its Infectivity in human cells, one should not expect a dramatic increase in the viral infectivity by any single mutation. An effective infection pathway is for the virus to have multiple RBD mutations to accumulatively enhance its infectivity, which appears to be the case for Omicron.
This work analyzes the infectivity of Omicron by examining the BFE changes of the ACE2 and S protein complex induced by 15 Omicron RBD mutations. Figure 1a illustrates the binding complex of ACE2 and S protein RBD. Most of the RBD mutations are located near the binding interface of ACE2 and RBD, except for mutations G339D, S371L, S373P, and S375F. Omicron-induced BFE changes are depicted in Figure 1b. Overall, mutations significantly increase the BFE changes, which strengthens the binding affinity of the ACE2-RBD complex and makes the variant more infectious. This result indicates that Omicron appears to have followed the infectivity-strengthening pathway of natural selection [21].
Figure 1:
Illustration of the Omicron RBD and ACE2 interaction, RBD mutation-induced BFE changes. a. The 3D structure of the ACE2 and RBD complex (PDB: 6M0J[20]). Omicron mutation sites are labeled. b. Omicron mutation-induced BFE changes. Positive changes strengthen the binding between ACE2 and S protein, while negative changes weaken the binding. c. A comparison of predicted mutation-induced BFE changes for few variants.
The infectivity-strengthening mutations N440K, T478K, and N501Y enhance the BFEs by 0.62, 1.00, and 0.55 kcal/mol, respectively. Among them, T478K is one of two RBD mutations in the Delta variant, while N501Y is presented on many prevailing variants, including Alpha, Beta, Gamma, Theta, and Mu. Notably, mutation Y505H induces a small negative BFE change of −0.20kcal/mol. All other mutations, particular those four mutations that are far away from the ACE2 and RBD binding interface, cause little or no BFE changes. Figure 1c gives a comparison of Omicron with a few other named variants, i.e., Alpha, Beta, Gamma, Delta, Theta, Kappa, and Mu. The BFE changes indicate that Omicron is more infectious than other named variants. Specifically, the accumulated BFE change is 2.60kcal/mol, suggesting a 13-fold increase in the viral Infectivity. In comparison, Omicron is about 2.8 times as infectious as the Delta (i.e., BFE change: 1.57kcal/mol for Delta).
2.2. Vaccine breakthrough
Vaccination has been proven to be the most effective means for COVID-19 prevention and control. There are four types of vaccines, i.e., virus vaccines, viral-vector vaccines, DNA/RNA vaccines, and protein-based vaccines [22]. Essentially, the current COVID-19 vaccines in use mainly target to the S protein [23]. The 32 amino acid changes, including three small deletions and one small insertion in the spike protein, suggest that Omicron may be induced by antibody resistance [17]. As a result, these mutations may dramatically enhance the variant’s ability to evade current vaccines.
In general, it is essentially impossible to accurately characterize the full impact of Omicron’s S protein mutations on the current vaccines in the world’s populations. First, different types of vaccines may lead to different immune responses from the same individual. Additionally, different individuals characterized by race, gender, age, and underlying medical conditions may produce different sets of antibodies from the same vaccine. Moreover, the reliability of statistical analysis over populations may be limited because of the inability to fully control various experimental conditions.
This work offers a molecule-based data-driven analysis of Omicron’s impact on vaccines through a library of 185 known antibody and S protein complexes. We evaluate the binding free energy changes induced by 15 RBD mutations on these complexes to understand the potential impact of Omicron’s RBD mutations to vaccines. To ensure reliability, our study does not include a few known antibody-S protein complexes that are far away from the RBD, such as those in the N-terminal domain (NTD), due to limited experimental data in our antibody library [10, 11].
Figures 2a, b1, and b2 depict the Omicron RBD mutation-induced BFE changes of 185 known antibody and RBD complexes. Overall, Omicron RBD mutations can significantly change the binding pattern of known antibodies. Positive changes strengthen the binding between antibody and RBD complexes, while negative changes weaken the binding. In the color bar, the largest negative change is more significant than the largest positive change, indicating more severe disruptive impacts. In general, there are more negative BFE changes than positive ones, as shown in Figure 2, indicating the Omicron mutations favor the escape of current vaccines.
Figure 2:
Illustration of Omicron mutation-induced BFE changes of 185 available antibody and RBD complexes and an ACE2-RBD complex. Positive changes strengthen the binding, while negative changes weaken the binding. a Heat map for 12 antibody and RBD complexes in various stages of drug development. Gray color stands for no predictions due to incomplete structures. b1 Heat map for ACE2/antibody and RBD complexes. b2 and b3 Heat map for antibody and RBD complexes.
Among 15 RBD mutations, K417N, also part of the Beta variant that originated in South Africa, causes the most significant disruption of known antibodies. Notably, E484A is another mutation that leads to overwhelmingly disruptive effects to many known antibodies. It is worthy of mentioning that most of E484A’s disruptive effects are complementary to those of K417N, which makes Omicron more effective in vaccine breakthroughs. The third disruptive mutation is Y505H. It is also able to weaken many known antibody and RBD complexes.
Mutation G339D creates a mild impact on various antibody-RBD complexes. One of the reasons is that it locates pretty far away from the binding interfaces of most known antibodies. Its change from a non-charged amino acid to a negatively charged amino acid induces mostly favorable bindings among many antibody-RBD complexes. S371L, S373P, and S375F are other mutations that have mild impacts due to their locations.
For a comparison, ACE2 is also included in Figure 2b1. The impact of Omicron on ACE2 is significantly weak, indicating the SARS-CoV-2 has already optimized its binding with ACE2, and there is a relatively limited potential for the virus to improve its infectivity. However, due to the increase in the vaccination rate, variants can become more destructive to vaccines in years to come [17].
Figure 2a gives a separated plot of the impacts of Omicron on a few mAbs. Similarly, there are dramatic reductions in their efficacy. A more specific discussion is given in the next section.
Figure 3 provides the analysis of variant mutation-induced BFE changes of 185 antibody-RBD complexes induced by Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu mutations. From Figure 3a1, it is clear that most complexes have negative accumulated BFE changes, indicating Omicron may disrupt most antibody-RBD binding complexes. In contrast, Delta’s distribution focuses on a smaller domain as shown in Figure 3e1. The BEF changes are essentially distributed around zero, suggesting Delta RBD mutations may not disrupt most known antibody-RBD binding complexes. The distributions of Beta and Gamma respectively in Figures 3c1 and d1 also indicate potential antibody-RBD binding complex disruption.
Figure 3:
Analysis of variant mutation-induced BFE changes of 185 antibody and RBD complexes. a1, b1, c1, d1, e1, f1, and g1 The distributions (counts) of accumulated BFE changes induced by Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu mutations respectively for 185 antibody and RBD complexes. Overall, there are more complexes that are weakened upon RBD mutations that complexes that are strengthened. a2, b2, c2, d2, e2, f2, and g2 The numbers (counts) of antibody-RBD complexes regarded as disrupted by Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu mutations respectively under different thresholds ranging from 0 kcal/mol, −0.3 kcal/mol, to <−3 kcal/mol.
It becomes very subtle to judge whether a mutation would disrupt an antibody and RBD complex as Omicron involves multiple vaccine-escape RBD mutations, which may generate multiple cancellations for each antibody-RBD complex over different mutations. It is useful to focus on disruptive mutations, i.e, mutations leading to negative BFE changes. Therefore, we previously have used −0.3 kcal/mol as a threshold to judge whether a mutation disrupts an antibody-RBD complex, which would give us a total of 163 disrupted antibody-RBD complexes as shown in Figure 3a2, suggesting a rate of 0.88 (i.e., 163/185) for potential vaccine breakthrough. As a comparison, Delta has 70 counts and a rate of 0.37 (70/185) in a similar estimation as shown in Figure 3e2. One would have 143 and 48 disrupted antibody and RBD complexes respectively for Omicron and Delta if the threshold is increased to −0.6kcal/mol. In both cases, Omicron is over twice more likely to disrupt antibody-RBD complexes. Note that Beta and Gamma in Figures 3c2 and d2 show a similar pattern.
2.3. Antibody resistance
The assessment of Omicron’s mutational threats to FDA-approved mAbs and a few other mAbs in clinical development is of crucial importance. Our AI-based predictions of similar threats from other variants, namely Alpha, Beta, Gamma, Delta, Epsilon, and Kappa, have shown an excellent agreement with experimental data [11]. In this section, we select on a few mAbs, specifically, mAbs from Eli Lilly (LY-CoV016 and LY-CoV555), Regeneron (REGN10933, REGN10987, and REGN10933/10987), AstraZeneca (AZD1061 and AZD8895), GlaxoSmithKline (S309), Celltrion (CT-P59), the Rockefeller University (C135 and C144). Among them, mAbs from Eli Lilly, Regeneron, AstraZeneca, and GlaxoSmithKline have had FDA approval. In addition, Celltrion’s COVID-19 antibody treatment had the EU drug agency’s recommendation in November 2021. Rockefeller University’s mAbs are still in clinical trials. Our analysis focuses on disruptive RBD mutations.
2.3.0.1. Eli Lilly mAbs
Eli Lilly mAb LY-CoV555 (PDB ID: 7KMG[24]) is also known as Bamlanivimab and is used in combination with LY-CoV016 (aka Etesevimab, PDB ID: 7C01[25]). Antibody LY-CoV016 is isolated from patient peripheral blood mononuclear cells convalescing from COVID-19. It was optimized based on the SARS-CoV-2 virus. The interaction of Eli Lilly mAbs with the S protein RBD is depicted in Figure 4a. ACE2 is included as a reference, indicating both LY-CoV016 and LY-CoV555 can directly neutralize the virus. Clearly, LY-CoV555 has a competing relationship with LY-CoV016, which might complicate our predictions slightly. In this work, we carry out the analysis of Eli Lilly mAbs separately.
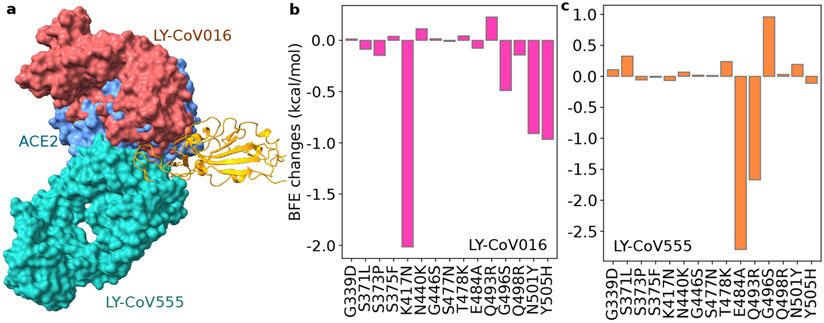
Figure 4:
Illustration of the Omicron RBD and Eli Lilly antibody interaction and RBD mutation-induced BFE changes. a The 3D structure of the ACE2 and Eli Lilly antibody complex. LY-CoV555 (PDB ID: 7KMG[24]) and LY-CoV016 (PDB ID: 7C01[25]) overlap on the S protein RBD. ACE2 is included as a reference. b Omicron mutation-induced BFE changes for the complex of RBD and LY-CoV016. c Omicron mutation-induced BFE changes for the complex of RBD and LY-CoV555.
Omicron mutation-induced BFE changes for antibody LY-CoV016 and RBD complex is given in Figure 4b. It appears that LY-CoV555 was optimized with respect to the original S protein but is sensitive to mutations. This complex may be weakened by K417N and N501Y as predicted in our earlier work [11]. New mutation Y505H may also reduce LY-CoV016’s efficacy. Overall, the complex may be significantly weakened by Omicron, leading to the efficacy reduction of Etesevimab.
The predicted BFE changes of LY-CoV555 are shown in Figure 4c. Mutation E484A induces a negative BFE change of −2.79 kcal/mol for the LY-CoV555 and RBD complex. The BFE change may translate into a dramatic efficacy reduction of 16 times for LY-CoV555, making it less competitive with ACE2 as most Omicron mutations strengthen the S protein and ACE2 binding. Similarly, Q493R may also reduce the efficacy by about 5 times. However, G496S may enhance the binding of the complex. The impacts of other mutations are mild. Therefore, Omicron is expected to reduce LY-CoV555 efficacy significantly. A previous study indicated that LY-CoV555 is prone to the E484K mutation presented in Beta and Gamma variants, for which the Eli Lilly mAb cocktail was taken off the market for many months in 2021.
Although LY-CoV555 and LY-CoV016 might slightly complement, they are both prone to Omicron mutation-induced efficacy reduction. We predict that the Eli Lilly mAb cocktail would be retaken off the market had Omicron become a prevailing variant in the world.
2.3.0.2. Regeneron mAbs
Regeneron mAbs REGN10933 and REGN10987 (aka Casirivimab and Imdevimab, respectively) are an FDA-approved antibody cocktail (PDB ID: 6XDG[26]) against COVID-19. Their 3D structure in complex with the S protein RBD is depicted in Figure 5a. ACE2 is inclused as a reference. Unlike the Eli Lilly mAb cocktail, the Regeneron mAbs do not overlap each other and bind to different parts of the RBD. Our 3D alignment shows that the antibody REGN10987 does not directly compete with ACE2 on their binding interfaces with the RBD, but still spatially conflict with ACE2. As a result, REGN10987 can directly neutralize the virus but is less sensitive to infectivity-induced RBD mutations. In contrast, REGN10933 overlaps with ECE2 both spatially and on the RBD binding interface. Consequently, REGN10933 is prone to infectivity-induced RBD mutations.
Figure 5:
Illustration of the Omicron RBD and Regeneron antibody interaction and RBD mutation-induced BFE changes. a The 3D structure of the ACE2 and Regeneron antibody complex. REGN10987 and REGN10933 do not overlap on the S protein RBD (PDB ID: 6XDG[26]). ACE2 is included as a reference. b Omicron mutation-induced BFE changes for the complex of RBD and REGN10933. c Omicron mutation-induced BFE changes for the complex of RBD and REGN10987. d Omicron mutation-induced BFE changes for the complex of RBD, REGN10933, and REGN10987.
Figure 5b plots our AI predicted BFE changes of the REGN10987-RBD complex. There are mixed responses to various Omicron mutations. Although G446K and K417N induce a negative BFE change, many other mutations may enhance the binding of the complex.
Omicron-induced BFE changes of the REGN10933-RBD complex is given in Figure 5c. Apparently, K417N and E484A induce BFE changes of −1.08 and −0.86 kcal/mol, respectively. However, most other Omicron mutations may strengthen the binding of the complex.
It is interesting to study how the two Regeneron mAbs are affected by Omicron when they are combined. Figure 5d shows the BFE changes of the complex induced by various Omicron mutations. We note that amplitudes of both positive and negative BEF changes have significantly reduced. However, Omicron RBD mutations K417N, G446S, and E484A may still weaken the cocktail binding to the RBD. We predict that Omicron will have a negative impact on the Regeneron cocktail efficacy.
2.3.0.3. AstraZeneca mAbs
AstraZeneca mAbs are designed as a cocktail of tixagevimab (AZD8895, PDB ID: 7L7D) and cilgavimab (AZD1061 PDB ID: 7L7E) as in Figure 6a. AZD8895 compete with ACE2 for the same binding interface and thus, is able to directly neutralize the virus. However, it is also prone to the infection-induced RBD mutations. Figure 6b AZD8895 can be slightly weakened by Q493R and K417N. In contrast, AZD1061 can be significantly disrupted by G477N as shown in 6c. Omicron mutation Q493R can also lead to the binding affinty reduction of the RBD and AZD1061 complex. Omicron impacts on AstraZeneca cocktail are slightly alleviated as shown in 6d. Since mutation Q493R affects both AZD8895 and AZD1061, it may give rise to a significant BFE reduction and disrupt the efficacy of the cocktail.
Figure 6:
Illustration of the Omicron RBD and AstraZeneca antibody interaction and RBD mutation-induced BFE changes. a The 3D structure of the ACE2 and AstraZeneca antibody complex. AZD1061 and AZD8895 do not overlap on the S protein RBD (PDB ID: 7L7E[27]). ACE2 is included as a reference. b Omicron mutation-induced BFE changes for the complex of RBD and AZD8895. c Omicron mutation-induced BFE changes for the complex of RBD and AZD1061. d Omicron mutation-induced BFE changes for the complex of RBD, AZD8895, and AZD1061.
2.3.0.4. Other mAbs
Celltrion’s antibody CT-P59 (aka Regdanvimab, PDB ID: 7CM4) is used to be a cocktail with CT-P63, for which we do not have its 3D structure. Figure 7a shows that antibody CT-P59 binds the RBD in a completing region with ACE2 and thus, might play a more important role than CT-P63 in combating the virus. Figure 7b shows that mutations E484A, Q493R, and Q498R respectively lead to BFE changes of −1.49, −2.82, and −1.0 kcal/mol for the CT-P59-RBD complex. These disruptive effects may be slightly offset by a positive BFE change of 1.71 kcal/mol due to mutation N501Y, which was reported in our earlier work [11]. The impacts of other mutations are relatively mild. Overall, CT-P59 may still be impaired by Omicron. Previously, we have shown that CT-P59 is prone to L452R in Delta and Q439R and S494P [11]. Due to the lack of the CT-P63 structure, we cannot provide an inclusive estimation for Celltrion’s cocktail but would recommend caution toward the use of Celltrion’s Regdanvimab in the wake of Omicron infections.
Figure 7:
Illustration of the Omicron RBD and other antibodies and RBD mutation-induced BFE changes. a Antibody CT-P59 in reference with ACE2. b BFE changes of Omicron mutation-induced on the binding of CT-P59 and RBD. c Antibody C135 in reference with ACE2. d BFE changes of Omicron mutation-induced on the binding of C135 and RBD. *: no results due to incomplete structure of C135. e Antibody C144 in reference with ACE2. f BFE changes of Omicron mutation-induced on the binding of C144 and RBD. g Antibody S309 in reference with ACE2. h BFE changes of Omicron mutation-induced on the binding of S309 and RBD.
We also analyze Rockefeller University antibodies C135 (PDB ID: 7K8Z) and C144 (PDB ID: 7K90), whose binding complexes with the RBD are given in Figures 7c and e, respectively. Antibody C135 has a relatively small region of interface with RBD and does not overlap with ACE2. Our earlier study indicates that C135 is prone to R346K and R346S mutations [11]. Mutation S317L induces a BFE change of −0.63 kcal/mol, indicating a relatively weak negative impact on C135’s efficacy. In contrast, antibody C144 share part of its binding domain with ACE2 and has more dramatic responses to Omicron mutations (see Figure 7f). Our earlier study indicates that the efficacy of C144 can be significantly reduced by E484K in the Delta variant [11]. Mutation E484A may cause a BFE change of −1.27kcal/mol. Therefore, we predict that the efficacy of C144 may be also undermined by Omicron RBD mutations.
Finally, we study antibody S309 (PDB ID: 6WPS) which is the parent antibody for Sotrovimab developed by GlaxoSmithKline and Vir Biotechnology, Inc. The alignment of S309 with ACE2 was given an earlier study [28] is also presented in Figure 7g. Since S309 does not overlap with ACE2 both spatially and on the RBD binding interface, infectivity-induced mutations will not affect S309 very much. Figure 7h shows that Omicron-induced BFE changes are from −0.47kcal/mol to 0.39kcal/mol. Therefore, Omicron may have minor impacts on S309.
3. Data, methods, and validity
Data.
To deliver an accurate and reliable machine learning model, dataset collection is of paramount importance among other steps. Both the BFE changes and next-generation sequencing enrichment ratios indicate the mutation-induced effects on protein-protein interactions (PPIs) binding affinities. Our methods integrate these two types of datasets to improve the prediction accuracy [10, 11]. Considering the urgency of COVID-19, the scattered SARS-CoV-2 data concerning BFE changes are reported inconsistently, while the sequencing enrichment ratios data is relatively easy to obtain but consistently has particular protein-protein interaction problems. The method is set up based on the BFE change dataset, SKEMPI 2.0 [29], together with SARS-CoV-2 related datasets. These datasets are obtained from the mutational scanning on ACE2 binding to the S protein RBD [30], the mutational scanning on RBD binding to ACE2 [15, 16], and the mutational scanning on RBD binding to CTC-445.2 and on CTC-445.2 binding to RBD[15]. We have also collected a library of 185 3D structures of antibody-RBD complexes [11].
Methods.
Our deep learning model for predicting BFE changes induced by mutations is constructed in two main steps. Firstly, once 3D structures of PPI complexes are obtained, mathematical features and biochemical/biophysical features are extracted. Biochemical/biophysical features provide the chemical and physical information, such as surface areas, partial charges, Coulomb interactions, van der Waals interaction, electrostatics, etc. Mathematical features, including the element-specific and site-specific persistent homology (algebraic topology), are implemented to simplify the structural complexity of PPI complexes [9, 31]. Second, a deep learning algorithm, artificial neural networks (ANNs), is constructed to tackle the massive features and mutational scanning data for predictions [11], which is available at TopNetmAb. Notice that our early model was constructed by integrating convolutional neural networks (CNNs) with gradient boosting trees (GBTs) and was trained with a large dataset of 8,338 PPI entries from the SKEMPI 2.0 dataset [29], which had already achieved a high accuracy [31]. In the following, the idea of persistent homology will be introduced briefly, which plays the key role of the feature processing. Moreover, Supporting Information S3 and S4 present more detailed description of data pre-processing, feature generation, and machine learning methods.
Recent years have seen a booming development of topological data analysis in a wide variety of scientific and engineering problems, whose main workhorse is persistent homology[32, 33]. By using persistent homology, molecular atoms can be modeled as a set of point cloud. Vertices, edges, faces, etc. can be treated as simplices with their collections to be simplicial complexes . For a simplicial complex , a chain is a finite sum of simplices as with coefficients , and the set of all chains is an group , where , 1, 2, 3. Thus, the boundary operator maps defined as , where and is a -simplex excluding . This is followed by an important property of boundary operators which is that and
| (1) |
and the -th homology group is defined by where and . The Betti numbers are defined by the ranks of -th homology group . This, in practice, is counting holes in -dimension, such as reflects the number of connected components, gives the number of loops, and is the number of cavities. Then, persistent homology can be devised to track Betti numbers along a filtration in order to describe the topological, spatial, and geometry information, which generates features for machine learning.
Validity.
In more recent work [10, 11, 19], with the help of the aforementioned deep mutational datasets associated with SARS-CoV-2, our predictions are highly consistent with experimental data. The predictions for the binding of CTC-445.2 and S protein RBD were compared with experimental data with a Pearson correlation of 0.7 [11, 15]. In the same work [11], the predictions of emerging mutations on clinical trial antibodies had a Pearson correlation of 0.8 with the natural log of experimental escape fractions [34]. In addition, the predicted mutation-induced BFE changes on L452R and N501Y for the ACE2-RBD complex have a near perfect correlation with experimental luciferase data [11, 35].
Our TopNetmAb model assumes that the RBD mutations are independent, which is very reasonable for Delta and other variants as they involve only one, two, or three isolated RBD mutations. As shown in Figure 1a, adjacent Omicron mutations S477N and T478K are dependent on each other. Similarly, S373P is just one residue away from S371L and S375F and is deemed to be depending on its neighbors. However, these three mutations are pretty far away from the ACE2 binding interface and play a less important role in our predictions. Mutation G496S and Q498R are also one amino acid apart, albeit their predicted BFE change amplitudes are very small. Overall, we expect a larger error in our prediction of Omicron infectivity compared to our earlier successful predictions [10, 11, 14]. However, we are still confident that the predicted trend of the Omicron infectivity change is correct. Figure 3 shows that the most severe antibody disruptions are not obtained from the inter-dependent mutations (i.e., S371L, S373P, S375F, S477N, T478K, G496S, and Q498R), suggesting the predicted trend of antibody disruptions is still valid. The reliability and accuracy of our assumption for Omicron are to be validated by experimental data, which may become available in a few weeks.
4. Note added in proof
After the publication of our manuscript in ArXiv on December 1 2021 [36], two sets of experimental results have been released recently [37, 38]. The first set of experimental results are the sensitivity of serum samples from COVID-19 convalescent patients [37], and the second set of experimental results are about antibody evasion impacted by the Omicron variant[38].
Figure 8a provides a comparison of accumulated BFE changes for variants Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu. For each antibody-RBD complex, the accumulated negative BFE change is obtained by the summation over RBD mutations (e.g., 15 mutations for Omicron and 2 for Delta) with positive BFE changes being set to zero, so that only disruptive effects are compared. Therefore, there are 185 accumulated BFE changes for each variant. The mean value of these 185 values is used to computed the fold of affinity reduction, which can be compared for different variants against the original variant (BFEchangeaverage = 0). It appears that Omicron is near 14 folds as capable as Delta and near 5 folds as capable as Gamma to escape vaccines.
Figure 8:
Comparison of predicted variant vaccine breakthrough potential with experimental data. a Accumulated negative BFE changes induced by Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu mutations respectively for 185 antibody-RBD complexes. For each variant, the number on the top is the fold of affinity reduction computed by e−BFEchangeaverage, where BFEchangeaverage, denoted by a circle, is the mean value of 185 antibody-RBD negative BFE changes. b The comparison of neutralization activity against Omicron, Alpha, Beta, Delta, Gamma, Lambda, and Mu variants based on 28 convalescence sera [37]. For each variant, the number on the top is the ratio of neutralization ED50 compared to the reference strain D614G.
In Figure 8b, the sensitivity of 28 serum samples from COVID-19 convalescent patients infected with SARS-CoV-2 original strain was tested against pseudotyped Omicron, Alpha, Beta, Gamma, Delta, Lambda, and Mu [37]. The mean neutralization ED50 of these sera against Omicron decreased about 8.4 folds compared to the D614G reference strain. In contrast, Delta neutralization capability decreased 1.6 fold. Both our prediction and serum experiment indicate that Omicron has the highest capability to evade vaccines. The overall correlation between our prediction and experiment is 0.9.
Figure 9 illustrates the comparison of BFE changes prediction and the experimental data of fold change in IC50 compared with wild type [38]. The BFE changes for each antibody are calculated by the peak negative BFE changes. From the figure, it is shown that only predictions for antibodies from Regeneron do not highly match to the experimental data but the rest of predictions are perfectly consistent with the experimental results.
Figure 9:
Comparison of predicted antibody breakthrough potential with experimental data [38]. Colors indicate three different range. For BFE changes, dark red: BFE changes ≤ −2kcal/mol, median red: −2kcal/mol < BFE changes ≤ −1kcal/mol, and light red: BFE changes > − 1kcal/mol. For fold changes, dark red: fold changes < −1000, median red: −1000 ≤ fold changes < −100, and light red: fold changes ≥ −100.
5. Conclusion
The identification of Omicron as a variant of concern (VOC) by the World Health Organization (WHO) has triggered countries around the world to put in place of travel restrictions and precautionary measures. At this moment, the scientific community knows little about Omicron’s infectivity, vaccine breakthrough, and antibody resistance. Since the spike (S) protein, particularly, its receptor-binding domain (RBD), plays a vital role in viral infection, it has been a key target of vaccines and antibody drugs. Therefore, the study of Omicron’s 15 RBD mutations can lead to valuable understanding of Omicron’s infectivity, vaccine breakthrough, and antibody resistance.
Based on a well-tested and experimentally confirmed deep learning model trained with tens of thousands of experimental data, we investigate the impacts of Omicron’s RBD mutations to its infectivity. We show that Omicron is about ten times more infectious than the original virus or about 2.8 times as infectious as the Delta variant. Using the structures of 185 known antibody-RBD complexes, we reveal that Omicron’s vaccine-escape capability is near 14 times as high as that of the Delta variant. We unveil that Omicron may completely abolish Eli Lilly antibody cocktail. Omicron RBD mutations may also compromise monoclonal antibodies (mAbs) from Regeneron, AstraZeneca, Celltrion, and Rockefeller University. However, mAbs from GlaxoSmithKline might not be affected much. Our results call for the development of a new generation of vaccines and mAbs that will not be easily affected by viral mutations.
Supplementary Material
Acknowledgment
This work was supported in part by NIH grant GM126189, NSF grants DMS-2052983, DMS-1761320, and IIS-1900473, NASA grant 80NSSC21M0023, Michigan Economic Development Corporation, MSU Foundation, Bristol-Myers Squibb 65109, and Pfizer.
Footnotes
Data and model availability
The structural information of 185 antibody-RBD complexes with their corresponding PDB IDs and the results of BFE changes of PPI complexes induced by Omicron mutations can be found in Section S2 of the Supporting Information. The analysis of observed SARS-CoV-2 RBD mutations is available at Mutaton Analyzer. The TopNetTree model is available at TopNetmAb. The detailed methods can be found in the Supporting Information S3 and S4. The validation of our predictions with experimental data can be located in Supporting Information S5.
Supporting information
The supporting information is available for
S1 Supplementary figures: BFE changes of 185 antibodies induced by mutations R346K, K417T, L452R/Q, E484K/Q, F490S occurred in prevailing variants
S2 Supplementary data: The Supplementary_Data.zip contains two files: the BFE changes of antibodies disrupted by Omicron mutations and the list of antibodies with corresponding PDB IDs
S3 Supplementary data pre-processing and feature generation methods
S4 Supplementary machine learning methods
S5 Supplementary validation: validations of our machine learning predictions with experimental data
References
- (1).Li W; Shi Z; Yu M; Ren W; Smith C; Epstein JH; Wang H; Crameri G; Hu Z; Zhang H, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [DOI] [PubMed] [Google Scholar]
- (2).Qu X-X; Hao P; Song X-J; Jiang S-M; Liu Y-X; Wang P-G; Rao X; Song H-D; Wang S-Y; Zuo Y, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem 2005, 280, 29588–29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Song H-D; Tu C-C; Zhang G-W; Wang S-Y; Zheng K; Lei L-C; Chen Q-X; Gao Y-W; Zhou H-Q; Xiang H, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A 2005, 102, 2430–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hoffmann M; Kleine-Weber H; Schroeder S; Krüger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu N-H; Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Walls AC; Park Y-J; Tortorici MA; Wall A; McGuire AT; Veesler D Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang C; Li W; Drabek D; Okba NM; van Haperen R; Osterhaus AD; van Kuppeveld FJ; Haagmans BL; Grosveld F; Bosch B-J A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun 2020, 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yu F; Xiang R; Deng X; Wang L; Yu Z; Tian S; Liang R; Li Y; Ying T; Jiang S Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther 2020, 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Li C; Tian X; Jia X; Wan J; Lu L; Jiang S; Lan F; Lu Y; Wu Y; Ying T The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Transduct. Target. Ther 2021, 6, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen J; Wang R; Wang M; Wei G-W Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol 2020, 432, 5212–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen J; Gao K; Wang R; Wei G-W Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem. Sci 2021, 12, 6929–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chen J; Gao K; Wang R; Wei G-W Revealing the threat of emerging SARS-CoV-2 mutations to antibody therapies. J. Mol. Biol 2021, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Alenquer M; Ferreira F; Lousa D; Valério M; Medina-Lopes M; Bergman M-L; Gonçalves J; Demengeot J; Leite RB; Lilue J, et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS pathogens 2021, 17, e1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dupont L; Snell LB; Graham C; Seow J; Merrick B; Lechmere T; Maguire TJ; Hallett SR; Pickering S; Charalampous T, et al. Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern. Nat. Microbiol 2021, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang R; Chen J; Gao K; Wei G-W Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics 2021, 113, 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Linsky TW; Vergara R; Codina N; Nelson JW; Walker MJ; Su W; Barnes CO; Hsiang T-Y; Esser-Nobis K; Yu K, et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 2020, 370, 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Starr TN; Greaney AJ; Hilton SK; Ellis D; Crawford KH; Dingens AS; Navarro MJ; Bowen JE; Tortorici MA; Walls AC, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang R; Chen J; Wei G-W Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J. Phys. Chem. Lett 2021, 12, 11850–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhang L; Mann M; Syed Z; Reynolds HM; Tian E; Samara NL; Zeldin DC; Tabak LA; Ten Hagen KG Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. U.S.A 2021, 118, e2109905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang R; Chen J; Hozumi Y; Yin C; Wei G-W Emerging vaccine-breakthrough SARS-CoV-2 variants. arXiv preprint arXiv:2103.08023 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lan J; Ge J; Yu J; Shan S; Zhou H; Fan S; Zhang Q; Shi X; Wang Q; Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [DOI] [PubMed] [Google Scholar]
- (21).Chen J; Wang R; Wei G-W Review of the mechanisms of SARS-CoV-2 evolution and transmission. ArXiv 2021. [Google Scholar]
- (22).Callaway E. The race for coronavirus vaccines: a graphical guide. Nature 2020, 580, 576. [DOI] [PubMed] [Google Scholar]
- (23).Dai L; Gao GF Viral targets for vaccines against COVID-19. Nat. Rev. Immunol 2021, 21, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jones BE; Brown-Augsburger PL; Corbett KS; Westendorf K; Davies J; Cujec TP; Wiethoff CM; Blackbourne JL; Heinz BA; Foster D, et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med 2021, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shi R; Shan C; Duan X; Chen Z; Liu P; Song J; Song T; Bi X; Han C; Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [DOI] [PubMed] [Google Scholar]
- (26).Hansen J; Baum A; Pascal KE; Russo V; Giordano S; Wloga E; Fulton BO; Yan Y; Koon K; Patel K, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dong J; Zost SJ; Greaney AJ; Starr TN; Dingens AS; Chen EC; Chen RE; Case JB; Sutton RE; Gilchuk P, et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol 2021, 6, 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chen J; Wang R; Wei G-W Review of the mechanisms of SARS-CoV-2 evolution and transmission. arXiv preprint arXiv:2109.08148 2021. [Google Scholar]
- (29).Jankauskaitė J; Jiménez-Garcıa B; Dapkūnas J; Fernández-Recio J; Moal IH SKEMPI 2.0: an updated benchmark of changes in protein–protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics 2019, 35, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chan KK; Dorosky D; Sharma P; Abbasi SA; Dye JM; Kranz DM; Herbert AS; Procko E Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang M; Cang Z; Wei G-W A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat. Mach. Intell 2020, 2, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zomorodian A; Carlsson G Computing persistent homology. Discrete Comput Geom 2005, 33, 249–274. [Google Scholar]
- (33).Edelsbrunner H; Harer J, et al. Persistent homology-a survey. Contemp. Math 2008, 453, 257–282. [Google Scholar]
- (34).Starr TN; Greaney AJ; Addetia A; Hannon WW; Choudhary MC; Dingens AS; Li JZ; Bloom JD Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021, 371, 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Deng X; Garcia-Knight MA; Khalid MM; Servellita V; Wang C; Morris MK; Sotomayor-González A; Glasner DR; Reyes KR; Gliwa AS, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. MedRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chen J; Wang R; Gilby NB; Wei G-W Omicron (B. 1.1. 529): Infectivity, vaccine breakthrough, and antibody resistance. arXiv preprint arXiv:2112.01318 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhang L; Li Q; Liang Z; Li T; Liu S; Cui Q; Nie J; Wu Q; Qu X; Huang W, et al. The significant immune escape of pseudotyped SARS-CoV-2 Variant Omicron. Emerging Microbes & Infections 2021, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liu L. et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. bioRxiv 2021, DOI: 10.1101/2021.12.14.472719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.