Hirsch et al. found that genetic disruption of central spindle microtubule assembly in C. elegans embryos does not block cytokinesis or midbody assembly. These central spindle–independent midbodies appeared to form from astral microtubules, bundled by contractile ring constriction.
Abstract
Contractile ring constriction during cytokinesis is thought to compact central spindle microtubules to form the midbody, an antiparallel microtubule bundle at the intercellular bridge. In Caenorhabditis elegans, central spindle microtubule assembly requires targeting of the CLASP family protein CLS-2 to the kinetochores in metaphase and spindle midzone in anaphase. CLS-2 targeting is mediated by the CENP-F–like HCP-1/2, but their roles in cytokinesis and midbody assembly are not known. We found that although HCP-1 and HCP-2 mostly function cooperatively, HCP-1 plays a more primary role in promoting CLS-2–dependent central spindle microtubule assembly. HCP-1/2 codisrupted embryos did not form central spindles but completed cytokinesis and formed functional midbodies capable of supporting abscission. These central spindle–independent midbodies appeared to form via contractile ring constriction–driven bundling of astral microtubules at the furrow tip. This work suggests that, in the absence of a central spindle, astral microtubules can support midbody assembly and that midbody assembly is more predictive of successful cytokinesis than central spindle assembly.
Introduction
Cytokinesis in animal cells is driven by constriction of an actomyosin contractile ring that is positioned between the separating chromosomes in anaphase by signals from spindle microtubules (MTs; D’Avino et al., 2015; Green et al., 2012). There are two major spindle MT subtypes implicated in cytokinesis signaling: the central spindle and astral MTs (Mishima, 2016). The central spindle is an overlapping array of stable antiparallel MTs that forms at the spindle midzone in anaphase (Euteneuer and McIntosh, 1980; Mastronarde et al., 1993; Uehara and Goshima, 2010). The central spindle accumulates many essential signaling molecules required for cytokinesis and is thought to be a primary driver of cytokinesis in animal cells (Alsop and Zhang, 2003, 2004; Cao and Wang, 1996; Glotzer, 2009; Kawamura, 1977; Lee et al., 2012; Wheatley and Wang, 1996). Astral MTs are dynamic, grow circumferentially from the centrosomes toward the cell cortex, and can position the contractile ring in the absence of a central spindle (Bringmann and Hyman, 2005; Lewellyn et al., 2010; Motegi et al., 2006; Rappaport, 1961; Su et al., 2014; von Dassow et al., 2009; Werner et al., 2007). Thus, both central spindle and astral MTs promote cytokinesis (Cao and Wang, 1996; Chapa-y-Lazo et al., 2020; Harris and Gewalt, 1989; Rappaport, 1971, 1996; von Dassow et al., 2009), and evidence suggests these MT populations may function cooperatively (Baruni et al., 2008; Bringmann and Hyman, 2005; Lewellyn et al., 2010; Motegi et al., 2006; von Dassow et al., 2009).
Central spindle MTs are thought to become compacted at the spindle midzone during cytokinesis to form the midbody at the intercellular bridge (e.g., Hu et al., 2012; for review, see D’Avino and Capalbo, 2016; Glotzer, 2009). The midbody is a highly stable antiparallel MT structure, resistant to pharmacological MT disassembly and >750 atmospheric pressure (Gorbsky et al., 1990; Kreis, 1987; Mullins and McIntosh, 1982; Salmon et al., 1976; Shelden and Wadsworth, 1990). The midbody promotes abscission at the end of cytokinesis (Steigemann and Gerlich, 2009) and has been proposed to play a role in promoting cell fate and stemness (Dionne et al., 2015; Ettinger et al., 2011; Kuo et al., 2011; Peterman et al., 2019; Peterman and Prekeris, 2019; Salzmann et al., 2014). Many central spindle proteins localize to the midbody (Hirose et al., 2001; Hu et al., 2012; Mollinari et al., 2002; Zhu et al., 2005), and contractile ring constriction is required for midbody assembly and MT stabilization (Hu et al., 2012; Landino and Ohi, 2016; Straight et al., 2003). While cytokinesis can complete in the absence of a bipolar central spindle, as observed in Caenorhabditis elegans spindle defective 1 (SPD-1) disrupted embryos (Verbrugghe and White, 2004) and in cells with drug-induced monopolar spindles (Canman et al., 2003; Hu et al., 2008; Shrestha et al., 2012), it is unclear if a midbody forms in these cells. While some studies have found that central spindle proteins can localize to astral MTs at the division plane and may contribute to midbody assembly in the absence of a central spindle (Savoian et al., 1999; Su et al., 2014), others have found that astral MTs alone cannot support midbody assembly (Green et al., 2013; Uehara et al., 2016).
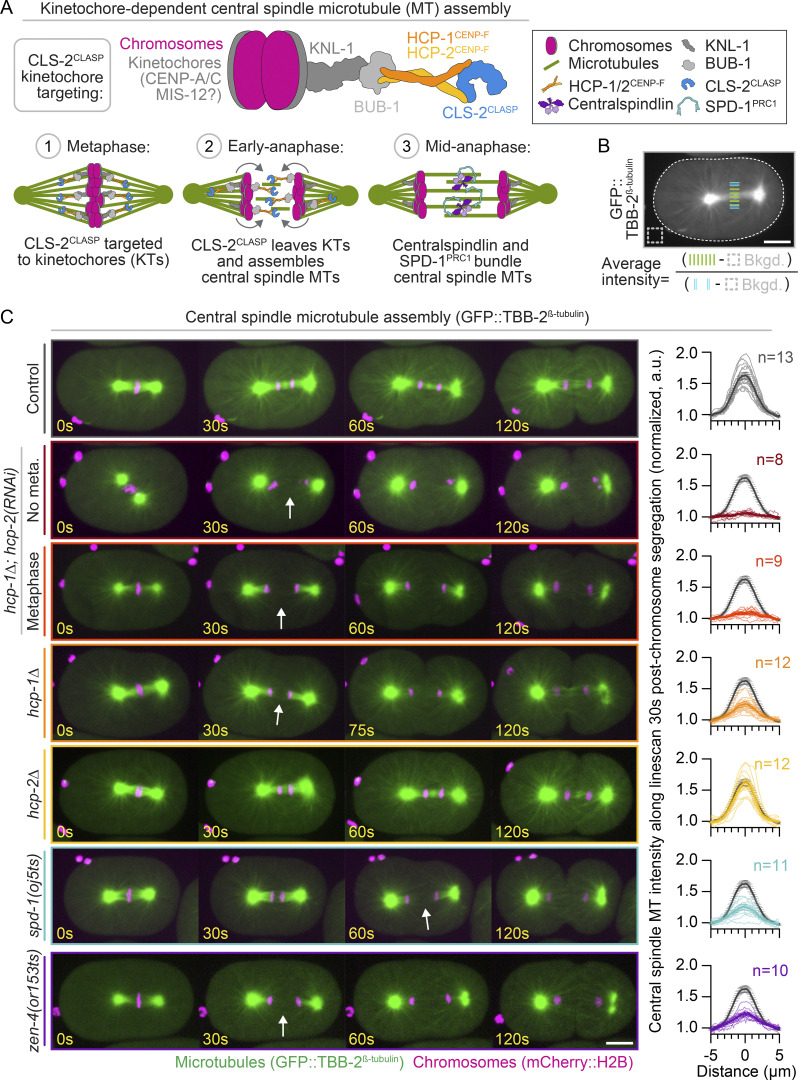
In C. elegans, central spindle MT assembly requires targeting the TOGL domain-containing protein CLS-2, a member of the cytoplasmic linker-associated protein (CLASP) family of MT-binding proteins, to the kinetochores in metaphase (Maton et al., 2015). CLS-2 kinetochore targeting requires the outer kinetochore proteins KNL-1, BUB-1, and holocentric chromosome-binding proteins 1/2 (HCP-1/2; Fig. 1 A; Maton et al., 2015). The centromere protein F (CENP-F)–like proteins HCP-1/2 bind and target CLS-2 to the kinetochores early in mitosis for accurate chromosome segregation (Cheeseman et al., 2005; Edwards et al., 2018), then to the spindle midzone after anaphase onset, where CLS-2 promotes the assembly and stabilization of central spindle MTs (Fig. 1 A; Maton et al., 2015). CLASP proteins in other systems, including fission yeast, Drosophila, and mammalian cultured cells, also stabilize central spindle MTs and contribute to cytokinesis (Bratman and Chang, 2007; Inoue et al., 2004; Liu et al., 2009; Maiato et al., 2003). Following CLS-2–mediated stabilization, central spindle MTs are bundled at the spindle midzone by the conserved MT cross-linking protein SPD-1 (protein regulating cytokinesis 1 [PRC1] in mammals) and the conserved MT bundling complex centralspindlin, composed of the GTPase-activating protein cytokinesis defective 4 (CYK-4; MgcRacGAP or Cyk4 in mammals) and the kinesin-6 zygotic epidermal enclosure defective 4 (ZEN-4; mitotic kinesin-like protein 1 [MKLP1] in mammals; Davies et al., 2015; Jantsch-Plunger et al., 2000; Mishima et al., 2002; Mollinari et al., 2002; Verbrugghe and White, 2004). Importantly, centralspindlin and other signaling molecules that localize to central spindle MTs early in anaphase (e.g., the chromosomal passenger complex) are also required for cytokinesis Canman et al., 2008; Hirose et al., 2001; Jantsch-Plunger et al., 2000; Kaitna et al., 2000; Mackay et al., 1998; Mishima et al., 2002; Mollinari et al., 2002; Romano et al., 2003; Schumacher et al., 1998; Severson et al., 2000; Speliotes et al., 2000). Thus, it has been assumed that HCP-1/2–mediated kinetochore targeting of CLS-2, which is essential for central spindle MT assembly (Maton et al., 2015), is also required for cytokinesis.
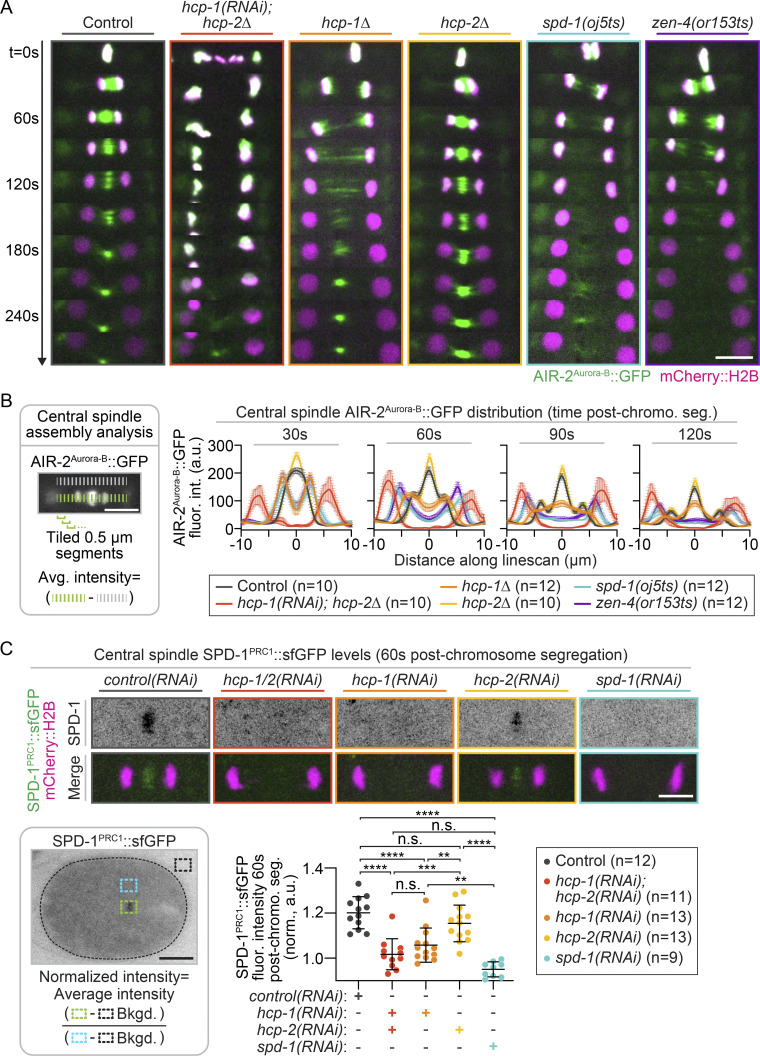
Figure 1.
HCP-1 plays a primary role in promoting central spindle MT assembly. (A) Schematic of kinetochore (KT)-dependent central spindle MT assembly in the one-cell C. elegans embryo. Top: CLS-2 recruitment to the KTs in metaphase. Bottom: Localization to the spindle midzone in anaphase, where it promotes central spindle MT assembly. (B) Schematic of line scan analysis of central spindle MT fluorescence intensity; white dashed line outlines the embryo. (C) Left: Representative time-lapse images of GFP::TBB-2β-tubulin-expressing (green) and mCherry::H2B-expressing (magenta) embryos undergoing the first mitotic cell division; see Video 4. hcp-1Δ; hcp-2(RNAi) embryos with (red) and without (maroon) chromosome bi-orientation and metaphase plate formation. White arrows, no central spindle MTs. Right: Quantification of central spindle MT intensity 30 s post-chromosome segregation on sum projections of all Z-planes. Individual (thin lines) and average (bold lines) line scans are shown for each genotype. n is listed to the right of each graph [n = 13 control(RNAi), n = 8 hcp-1Δ; hcp-2(RNAi) no metaphase plate, n = 9 hcp-1Δ; hcp-2(RNAi) with metaphase plate, n = 12 hcp-1Δ, n = 12 hcp-2Δ, n = 11 spd-1(oj5ts), and n = 10 zen-4(or153ts)]. Average levels in controls are shown on each graph for reference (gray). Error bars represent the SEM; scale bars, 10 µm.
In this study, we investigated the function of HCP-1/2–mediated CLS-2 localization in central spindle and midbody MT assembly in the one-cell C. elegans embryo. Through a combination of genetic approaches and live-cell imaging, we found that while HCP-1 and HCP-2 cooperate in CLS-2 localization, HCP-1 plays a primary role in CLS-2–mediated central spindle MT assembly. Surprisingly, we found that HCP-1/2 codisrupted embryos complete cytokinesis and form robust midbodies despite never forming a central spindle. This work suggests that astral MTs can support midbody assembly and challenges the assumption that midbody assembly is always dependent on central spindle assembly.
Results and discussion
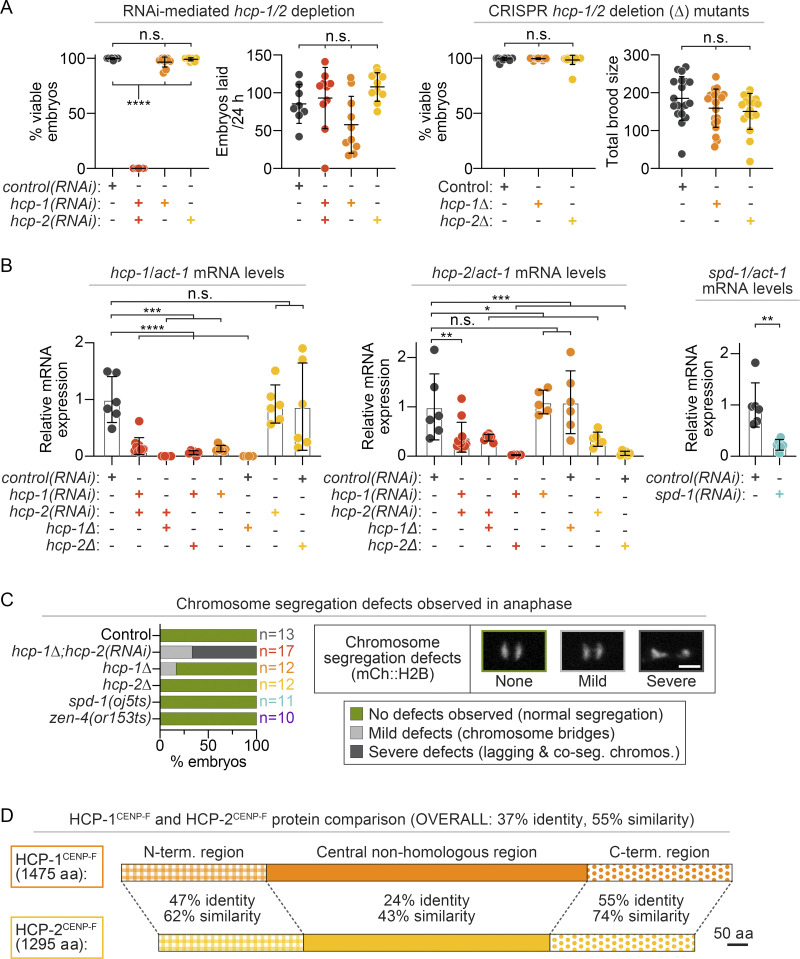
Because targeting of the CLASP family protein CLS-2 to the kinetochores and spindle midzone is required for both chromosome segregation and central spindle assembly (Cheeseman et al., 2005; Maton et al., 2015), we first probed individual roles for the CLS-2–targeting CENP-F–like proteins HCP-1/2 in dividing C. elegans one-cell embryos. While most animal genomes encode one CENP-F protein, C. elegans has two putative CENP-F–like paralogs: HCP-1 and HCP-2 (hereafter HCP-1 and HCP-2 singly or HCP-1/2 together). Single hcp-1 or hcp-2 gene disruptions by either CRISPR knockout or RNAi-mediated knock-down do not lead to significant chromosome segregation defects, embryonic lethality, or lower fecundity (Fig. S1, A–C; see also Materials and methods and Cheeseman et al., 2005; Edwards et al., 2018; Moore et al., 1999). Thus, most studies have focused on codisruption of hcp-1/2, which leads to sister chromosome cosegregation defects (Cheeseman et al., 2005; Edwards et al., 2018). Even a partial hcp-1/2 codisruption prevents central spindle MT assembly due to a failure in CLS-2 recruitment to the kinetochores and spindle midzone (Maton et al., 2015). Surprisingly, given this apparent redundancy, hcp-1 and hcp-2 share only 37% sequence identity and 55% similarity (Fig. S1 D; Moore et al., 1999), and genetic evidence suggests that they may not always function together (Hajeri et al., 2008; Tarailo et al., 2007).
Figure S1.
HCP-1 and HCP-2 single- and double-disruption effects on embryonic lethality and qRT-PCR validation. (A) Left: Quantification of embryonic viability (left) and number of embryos laid in 24 h (right) after hcp-1 and hcp-2 RNAi-mediated depletion individually and together [n = 8 control(RNAi), n = 10 hcp-1/2(RNAi), n = 10 hcp-1(RNAi), n = 10 hcp-2(RNAi)]. Right: Quantification of embryonic viability (left) and total brood size (right) for hcp-1Δ and hcp-2Δ CRISPR deletion mutants [n = 19 control, n = 20 hcp-1Δ, and n = 18 hcp-2Δ]. (B) Quantification of hcp-1 (left), hcp-2 (middle), and spd-1 (right) mRNA levels normalized to act-1 mRNA by qRT-PCR in embryos with and without RNAi-mediated knock-down. Statistical significance (A and B) was determined by one-way ANOVA with Tukey’s (A) or Dunnett’s (B) multiple comparison test; error bars represent the SD (A and B). (C) Characterization of chromosome segregation defects in embryos imaged expressing GFP::TBB-2β-tubulin and mCherry::H2B. n is listed to the right side of graphs in the indicated color for each genotype [n = 13 control, n = 17 hcp-1Δ; hcp-2(RNAi), n = 12 hcp-1Δ, n = 12 hcp-2Δ, n = 11 spd-1(oj5ts), and n = 10 zen-4(or153ts)]. Segregation defects were categorized as severe (dark gray, lagging chromosomes and chromosome cosegregation), mild (light gray, chromosome bridges), and no defects (green, normal segregation); scale bar, 10 µm. (D) Schematic depicting amino acid sequence homology between HCP-1 (top) and HCP-2 (bottom) proteins, with alignment of N- and C-termini (drawn to scale). n.s., P ≥ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
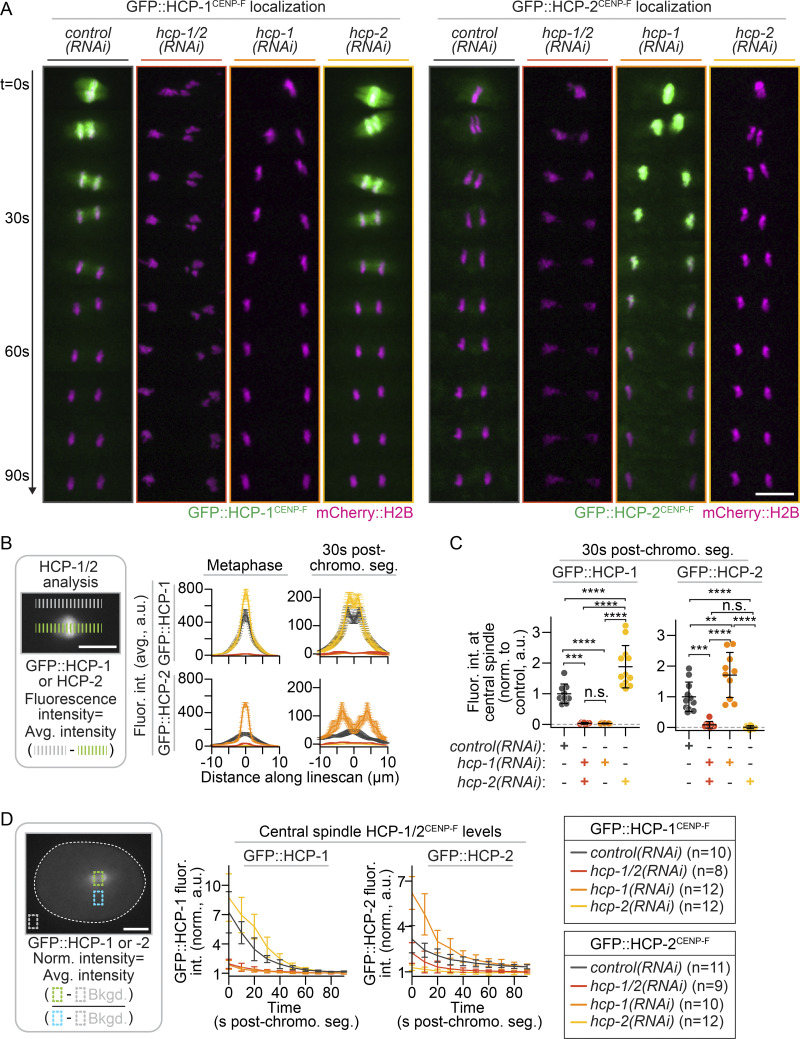
To test independent roles for each CENP-F paralog in cell division, we compared the dynamics of GFP-tagged HCP-1, HCP-2, and CLS-2 with and without RNAi-mediated depletion of hcp-1 and hcp-2 individually and together. Consistent with previous work (Cheeseman et al., 2005; Edwards et al., 2018), in control embryos, HCP-1, CLS-2, and, to a lesser extent, HCP-2 localized to kinetochores and spindle MTs in metaphase and then transiently to the spindle midzone between the dividing chromosomes upon anaphase onset (Figs. S2 and S3; and Videos 1, 2, and 3). In hcp-2(RNAi) embryos, CLS-2 levels did not differ from controls, and HCP-1 levels were dramatically increased at the kinetochores and spindle in metaphase and at the spindle midzone in anaphase (Figs. S2 and S3; and Videos 1 and 2). In hcp-1(RNAi) embryos, CLS-2 levels decreased and HCP-2 levels increased at kinetochores, but neither protein relocalized to the spindle midzone in anaphase (Figs. S2 and S3; and Videos 1, 2, and 3). In hcp-1/2(RNAi) codepleted embryos, no CLS-2, HCP-1, or HCP-2 localization was observed (Figs. S2 and S3; and Videos 1, 2, and 3). These data suggest that while HCP-1 and HCP-2 likely compensate for each other at kinetochores during chromosome attachment in metaphase, HCP-1 plays a more primary role in localizing CLS-2 to the spindle midzone for central spindle MT assembly in anaphase.
Figure S2.
CRISPR-tagged endogenous GFP::HCP-1 and GFP::HCP-2 localization. (A) Representative pseudokymographs showing midzone localization of mCherry::H2B (magenta) and endogenously tagged GFP::HCP-1 (green, left) or GFP::HCP-2 (green, right) during early central spindle assembly; see also Videos 1 and 2. Time is indicated on the left (imaging every 10 s), where t = 0 s is the metaphase just before anaphase onset. (B) Schematic of the method used for line scan analysis (left) and quantification (right) of GFP::HCP-1 (top) and GFP::HCP-2 (bottom) levels at the kinetochore (metaphase, left) and at the spindle midzone (30 s after chromosome segregation, right). Error bars represent the SEM. (C) Total levels of GFP::HCP-1 and GFP::HCP-2 at the spindle midzone (between chromosomes) 30 s after chromosome segregation in anaphase, normalized to levels in control(RNAi) embryos. Significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. (D) Schematic of the method used for analysis (left) and quantification (right) of central spindle levels of GFP::HCP-1 and GFP::HCP-2, where t = 0 s represents kinetochore levels at metaphase, and all other time points represent protein levels between chromosomes at the spindle midzone; white dashed line outlines the embryo; error bars represent the SD. Key indicates n for all data shown [GFP::HCP-1: n = 10 control(RNAi), n = 12 hcp-1(RNAi); n = 12 hcp-2(RNAi); and n = 8 hcp-1/2(RNAi); GFP::HCP-2: n = 11 control(RNAi), n = 10 hcp-1(RNAi); n = 12 hcp-2(RNAi); and n = 9 hcp-1/2(RNAi)]. Scale bars, 10 µm. n.s., P ≥ 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Figure S3.
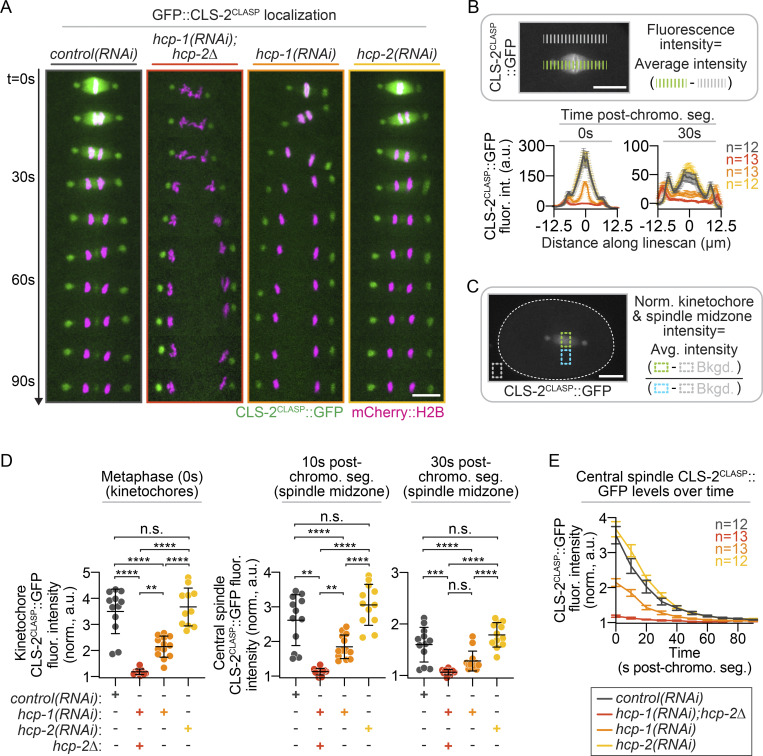
CLS-2CLASP::GFP localization at the spindle midzone with and without hcp-1/2 disruption. (A) Representative pseudokymographs of CLS-2CLASP::GFP (green) and mCherry::H2B (magenta) localization during central spindle assembly; see also Video 3. t = 0 s (imaging every 10 s) represents the time point just before chromosome segregation in anaphase [or sooner in the absence of metaphase plate assembly in hcp1/2(RNAi); see Edwards et al. (2018)]. Time (in s) after chromosome segregation is indicated on the left; scale bar, 10 µm. (B) Schematic of method used for CLS-2CLASP::GFP analysis (top), and quantification (bottom) of CLS-2CLASP::GFP fluorescence intensity along a line scan at the kinetochores (0 s, left) and spindle midzone (30 s, right) on sum projections of all Z-planes. (C) Schematic depicting the method used for CLS-2CLASP::GFP intensity quantification at the kinetochores and central spindle on sum projections of all Z-planes; white dashed line outlines the embryo. (D) Quantification of CLS-2CLASP::GFP fluorescence intensity at the kinetochores (left) and at the central spindle 10 s and 20 s after chromosome segregation (right). Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. (E) CLS-2CLASP::GFP levels over time at the kinetochores (t = 0 s) and central spindle (t = 10–90 s). n is listed to the right of the graph in the indicated color for each genotype [n = 12 control(RNAi), n = 13 hcp-1(RNAi); hcp-2Δ, n = 13 hcp-1(RNAi); and n = 12 hcp-2(RNAi)]; error bars represent the SEM; scale bars, 10 µm. n.s., P ≥ 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Video 1.
GFP::HCP-1CENP-F in C. elegans embryos. Representative control(RNAi) (left), hcp-1(RNAi) (center), and hcp-2(RNAi) (right) C. elegans one-cell embryos expressing GFP::HCP-1 (green) and mCherry::H2B (magenta). Time is relative to chromosome separation at 26°C; genotype labels are located at the top of each individual video clip. All time-lapse images are maximum projections of 11 × 1–µm Z-sections collected every 10 s. Playback rate is 5 frames per second. Scale bar (top right), 10 µm. This video corresponds to images shown in Fig. S2 A (left).
Video 2.
GFP::HCP-2CENP-F in C. elegans embryos. Representative time-lapse images of control(RNAi) (left), hcp-1(RNAi) (center), and hcp-2(RNAi) (right) C. elegans one-cell embryos expressing GFP::HCP-2 (green) and mCherry::H2B (magenta). Time is relative to chromosome separation at 26°C; genotype labels are located at the top of each individual video clip. All time-lapse images are maximum projections of 11 × 1–µm Z-sections collected every 10 s. Playback rate is 5 frames per second. Scale bar (top right), 10 µm. This video corresponds to images shown in Fig. S2 A (right).
Video 3.
CLS-2CLASP::GFP in C. elegans embryos. Representative time-lapse images of control(RNAi) (top, left), hcp-1(RNAi); hcp-2Δ (top, right), hcp-1(RNAi) (bottom, left), and hcp-2(RNAi) (bottom, right) C. elegans one-cell embryos expressing CLS-2CLASP::GFP (green) and mCherry::H2B (magenta). Time is relative to chromosome separation at 26°C; genotype labels are located at the top of each embryo in the video clip. All time-lapse images are maximum projections of 11 × 1–µm Z-sections collected every 10 s. Playback rate is 5 frames per second. Scale bar (top left), 10 µm. This video corresponds to images shown in Fig. S3 A.
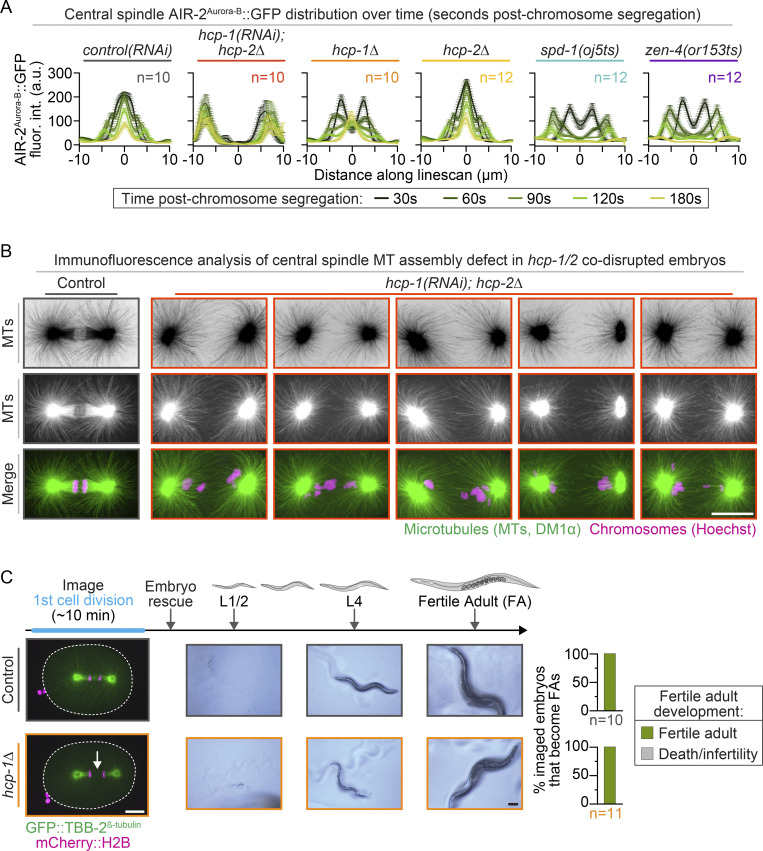
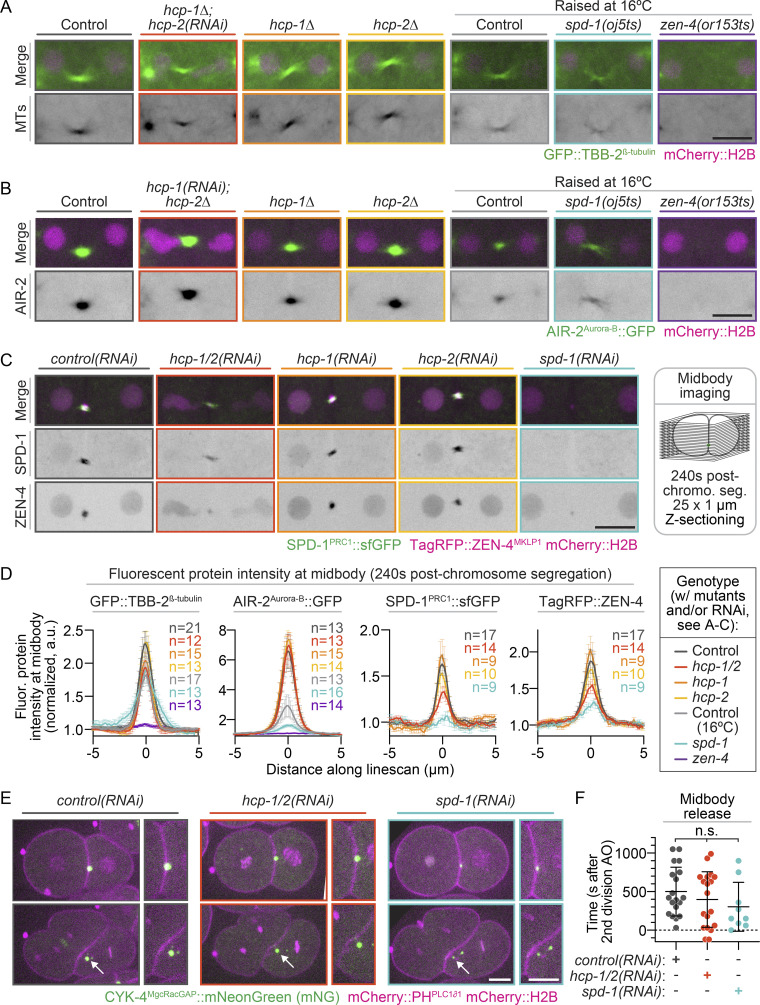
To directly test the individual roles of HCP-1/2 in central spindle assembly, we measured the spindle midzone levels of fluorescently tagged MTs (tubulin β-2 chain [TBB-2]; Lacroix et al., 2018) and two central spindle-associated proteins, Aurora/Ipl1-related kinase 2 (AIR-2; Aurora-B in mammals; Cheerambathur et al., 2019) and SPD-1 (PRC1 in mammals; Gigant et al., 2017), between the separating chromosomes 30 s (or 60 s for SPD-1::superfolder GFP [sfGFP]) after chromosome segregation. In control and hcp-2–disrupted embryos (deletion mutant or RNAi), visible central spindle assembly occurred within 30 s (or 60 s for SPD-1::sfGFP) after chromosome segregation (Figs. 1 C, 2, and S4 A; and Videos 4 and 5). As expected, no central spindle assembly occurred after disruption of the central spindle MT bundling proteins SPD-1 and ZEN-4 using temperature-sensitive (ts) mutants or in hcp-1/2 codisrupted embryos (Figs. 1 C, 2, and S4 A; and Videos 4 and 5; Maton et al., 2015; Severson et al., 2000; Verbrugghe and White, 2004). In hcp-1/2 codisrupted embryos, central spindle MTs were also not observed by immunofluorescence after fixation (Fig. S4 B; see also Maton et al., 2015). In hcp-1 disrupted embryos (deletion mutant or RNAi), central spindle assembly was delayed but eventually occurred (Figs. 1 C, 2 A, and S4 A; and Videos 4 and 5), and the embryos were viable (Fig. S4 C). Thus, HCP-1/2 function cooperatively to promote CLS-2–mediated central spindle MT assembly at the spindle midzone.
Figure 2.
HCP-1 cooperates with HCP-2 to promote robust central spindle assembly. (A) Representative pseudokymographs of AIR-2::GFP (green) and mCherry::H2B (magenta) localization throughout cell division; see Video 5. Time on the left; t = 0 s is the metaphase before anaphase onset [or chromosome segregation in hcp-1(RNAi); hcp-2Δ embryos that did not form a metaphase plate]. Images were acquired every 15 s and are shown every 30 s; scale bar, 10 µm. (B) Method used for line scan analysis of AIR-2::GFP distribution (left) and quantification (right) of average fluorescence intensity at the central spindle 30 s, 60 s, 90 s, and 120 s after chromosome segregation on sum projections of all Z-planes. Error bars represent the SEM; n is listed in the key [n = 10 control(RNAi), n = 10 hcp-1(RNAi); hcp-2Δ, n = 12 hcp-1Δ, n = 10 hcp-2Δ, n = 12 spd-1(oj5ts), and n = 12 zen-4(or153ts)]. (C) Top: Representative images of SPD-1::sfGFP (inverted contrast, top; green, bottom) and mCherry::H2B (magenta, bottom) at the central spindle 60 s after anaphase onset [or chromosome segregation in hcp-1/2(RNAi) embryos that did not form a metaphase plate]; scale bar, 5 µm. Bottom: Method used for SPD-1::sfGFP midzone analysis (left; black dashed line outlines embryo; scale bar, 10 µm) and quantification (right) at 60 s after chromosome segregation. Genotype is indicated below graphs; n is listed in the key [right; n = 12 control(RNAi), n = 11 hcp-1/2(RNAi), n = 13 hcp-1(RNAi), n = 13 hcp-2(RNAi), and n = 9 spd-1(RNAi)]. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. n.s., P ≥ 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Figure S4.
HCP-1/2 function redundantly for central spindle assembly, but HCP-1 plays a more prominent role. (A) Quantification of AIR-2Aurora-B::GFP distribution over time (30–180 s after chromosome segregation) separated by genotype; data are repeated from Fig. 2, A and B; see also Video 5. n is listed to the right side of each graph in the indicated color for each genotype [n = 10 control(RNAi), n = 10 hcp-1(RNAi); hcp-2Δ, n = 10 hcp-1Δ, n = 12 hcp-2Δ, n = 12 spd-1(oj5ts), and n = 12 zen-4(or153ts)]; error bars represent the SEM. Key indicates colors used to show time differences (bottom). (B) Representative fixed immunofluorescence samples showing MTs (inverted gray [top], gray [middle], green [bottom]) and chromosomes (magenta, bottom) in one control embryo (left) and five hcp-1(RNAi); hcp-2Δ embryos. (C) Top: Experimental timeline for postimaging viability assay. Arrows indicate timing of embryo rescue and viability/fertility assessment. Bottom: Representative images of the central spindle (left; white dashed line outlines the embryo), worm development (center), and quantification (right) of the number of fertile adult (FA; green) or dead/infertile (gray) worms. White arrows indicate the absence of timely central spindle MT assembly. n is listed below each graph in the indicated color for each genotype [n = 10 control and n = 11 hcp-1Δ]. All white scale bars, 10 µm; black scale bar in C, 100 µm.
Video 4.
GFP::TBB-2β-tubulin in C. elegans embryos. Representative time-lapse images of control (top, left), hcp-1Δ; hcp-2(RNAi) no metaphase plate (top, center), hcp-1Δ; hcp-2(RNAi) with a metaphase plate (top, right), hcp-1Δ (middle, left), hcp-2Δ (middle, center), spd-1(oj5ts) (middle, right), and zen-4(or153ts) (bottom, left) C. elegans one-cell embryos expressing GFP::TBB-2β-tubulin (green) and mCherry::H2B (magenta). Time is relative to chromosome separation at 26°C; genotype labels are located at the top of each embryo in the video clip. Two representative embryos are shown for hcp-1Δ; hcp-2(RNAi), one with and one without a metaphase plate. All time-lapse images are maximum projections of 13 × 1–µm Z-sections collected every 15 s [30 s for hcp-1Δ; hcp-2(RNAi), no metaphase]. Playback rate is 7 frames per second. Scale bar (lower left), 10 µm. This video corresponds to images shown in Fig. 1 C.
Video 5.
AIR-2Aurora-B::GFP in C. elegans embryos. Representative time-lapse images of control (top, left), hcp-1(RNAi); hcp-2Δ (top, center), hcp-1Δ (top, right), hcp-2Δ (bottom, left), spd-1(oj5ts) (bottom, center), and zen-4(or153ts) (bottom, right) C. elegans one-cell embryos expressing AIR-2Aurora-B::GFP (green) and mCherry::H2B (magenta). Time is indicated relative to chromosome separation at 26°C; genotype labels are located at the top of each embryo in the video clip. All time-lapse images are maximum projections of 12 × 1–µm Z-sections collected every 15 s. Playback rate is 7 frames per second. Scale bar (lower left), 10 µm. This video corresponds to images shown in Fig. 2 A.
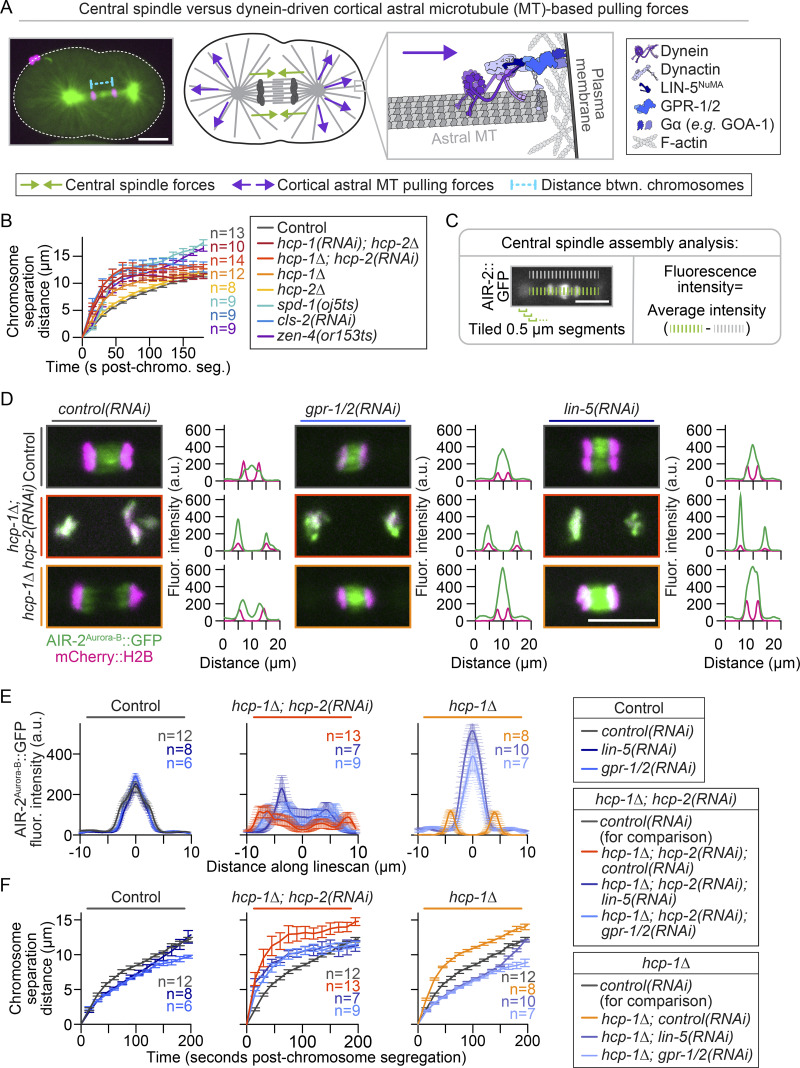
We next tested the roles of HCP-1 and HCP-2 in central spindle mechanical integrity. In C. elegans embryos, the rate of chromosome segregation is controlled by a balance of forces between central spindle MTs and dynein-driven cortical astral MT-based pulling forces (Fig. S5 A; Grill et al., 2001; Lee et al., 2015; Maton et al., 2015; Nahaboo et al., 2015; Yu et al., 2019). In the absence of a central spindle, chromosomes segregate at a faster rate due to the predominance of astral MT-based cortical pulling forces (Grill et al., 2001; Lee et al., 2015; Maton et al., 2015; Yu et al., 2019). Thus, as a readout for central spindle mechanical integrity, we measured the distance between chromosomes during chromosome segregation. As was previously reported, we found that the rate of chromosome segregation in cls-2(RNAi) and hcp-1/2 disrupted embryos was faster than that in control and hcp-2Δ embryos (Cheeseman et al., 2005; Maton et al., 2015) and similar to the segregation rates in spd-1(ts) and zen-4(ts) mutant embryos, which also lack central spindles (Fig. S5 B; Severson et al., 2000; Verbrugghe and White, 2004). Chromosome segregation in hcp-1Δ embryos was faster than in controls and hcp-2Δ embryos but slower than in hcp-1/2 codisrupted embryos (Fig. S5 B). Thus, HCP-1/2 function cooperatively to promote central spindle mechanical integrity.
Figure S5.
Reducing dynein-driven cortical astral MT-based pulling forces can rescue central spindle assembly in hcp-1Δ but not hcp-1/2 codisrupted embryos. (A) Representative image (left; white dashed line outlines the embryo) showing method used to measure chromosome segregation rates in B and schematic (center) showing the balance between central spindle versus dynein-driven cortical astral MT-based pulling forces. Inset (right) depicts the molecular players in dynein-driven cortical astral MT-based pulling forces. (B) Measurement of chromosome separation as a proxy for the mechanical integrity of the central spindle in embryos expressing mCherry::H2B and GFP::PHPLC1δ1 (see Fig. 3 A). Time is relative to chromosome segregation, and n is listed to the right of the graph in the indicated color for each genotype [n = 13 control, n = 10 hcp-1(RNAi); hcp-2Δ, n = 14 hcp-1Δ; hcp-2(RNAi), n = 12 hcp-1Δ, n = 8 hcp-2Δ, n = 9 spd-1(oj5ts), n = 9 cls-2(RNAi), and n = 9 zen-4(or153ts)]; error bars represent the SEM. (C) Schematic of method used for quantification of AIR-2Aurora-B::GFP fluorescence intensity at the central spindle in D and E. (D) Representative images at 45 s after chromosome segregation of control (top; gray outline), hcp-1Δ; hcp-2(RNAi) (middle; red outline), and hcp-1Δ (bottom; orange outline) embryos expressing AIR-2Aurora-B::GFP (green) and mCherry::H2B (magenta) with and without disruption of cortical pulling forces via RNAi-mediated depletion of LIN-5NuMA or GPR-1/2. Note that central spindle assembly is rescued upon depletion of LIN-5NuMA or GPR-1/2 in hcp-1Δ but not in hcp-1Δ; hcp-2(RNAi) codisrupted embryos, which do not assemble central spindle MTs (Maton et al., 2015). Representative individual line scans of AIR-2Aurora-B::GFP (green) and mCherry::H2B (magenta) are shown to the right of each image. (E) Quantification of AIR-2Aurora-B::GFP levels at the spindle midzone 45 s after chromosome segregation in control (left), hcp-1Δ; hcp-2(RNAi) (middle), and hcp-1Δ (right) embryos with and without RNAi-mediated depletion of LIN-5NuMA or GPR-1/2. (F) Measurement of chromosome separation with and without cortical astral MT-pulling forces (see A). Time is relative to chromosome segregation, and control(RNAi) analysis is included on all graphs for comparison. Keys (right) indicate genotype; n is listed to the right of graphs in the indicated color for each genotype [n = 12 control(RNAi), n = 8 lin-5(RNAi), n = 6 gpr-1/2(RNAi), n = 13 hcp-1Δ; hcp-2(RNAi); control(RNAi), n = 7 hcp-1Δ; hcp-2(RNAi); lin-5(RNAi), n = 9 hcp-1Δ; hcp-2(RNAi); gpr-1/2(RNAi), n = 8 hcp-1Δ; control(RNAi), n = 10 hcp-1Δ; lin-5(RNAi), n = 7 hcp-1Δ; gpr-1/2(RNAi)]; error bars represent the SEM (E and F). All fluorescence intensity analysis was done on sum projections of all Z-planes. All scale bars, 10 µm.
We also tested whether eliminating dynein-driven cortical astral MT-based pulling forces can rescue central spindle assembly after disruption of hcp-1 or hcp-1/2. In the absence of central spindle MT bundling factors such as SPD-1 or ZEN-4, central spindle assembly can be restored by disruption of cortical astral MT-based pulling forces (Lee et al., 2015; Maton et al., 2015; Nahaboo et al., 2015). In contrast, because CLS-2 is essential for central spindle MT assembly, disruption of cortical astral MT-based pulling forces cannot restore central spindle assembly in the absence of CLS-2 activity (Maton et al., 2015). G protein regulator 1/2 (GPR-1/2; LGN in mammals) and abnormal cell lineage 5 (LIN-5; nuclear mitotic apparatus protein [NuMA] in mammals) are both required for cortical dynein localization and astral MT-based pulling forces (Fig. S5 A; Lorson et al., 2000; Nguyen-Ngoc et al., 2007; Park and Rose, 2008; Srinivasan et al., 2003). We found that disruption of astral MT-based pulling forces by RNAi-mediated depletion of gpr-1/2 or lin-5 restored central spindle formation and mechanical integrity in hcp-1Δ but not in hcp-1/2 codisrupted embryos (Fig. S5, C–F). Thus, like CLS-2 (Maton et al., 2015), both HCP-1/2 are required for central spindle assembly in the absence of dynein-driven cortical astral MT-based pulling forces.
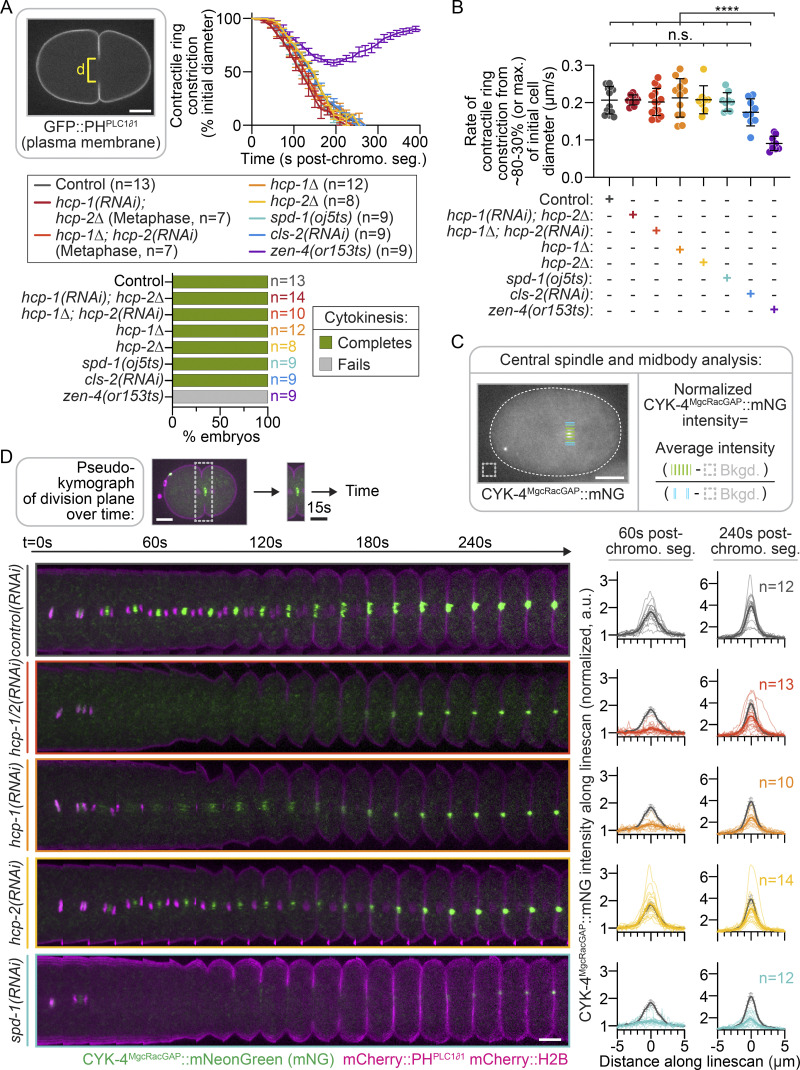
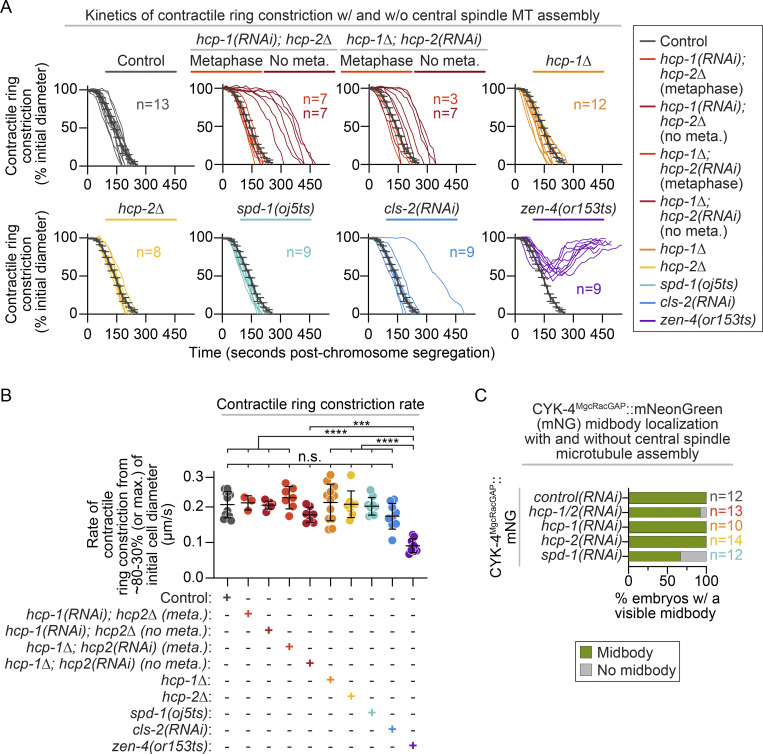
The central spindle serves as an important signaling hub that promotes contractile ring constriction during cytokinesis (Glotzer, 2009); thus, we next tested the role of CLS-2 and HCP-1/2 in the kinetics of cell division. We imaged strains expressing a plasma membrane marker (GFP::PHPLC1δ1; Audhya et al., 2005) and measured cell diameter at the division plane throughout cytokinesis (Fig. 3 A, left). As expected, zen-4(ts) embryos all initiated contractile ring constriction at a slower rate than in controls to ∼50% of cell diameter before furrow regression and cytokinesis failure (Fig. 3, A and B; and Fig. S6, A and B; Canman et al., 2008; Severson et al., 2000). In contrast, all control, hcp-1, hcp-2, spd-1, cls-2, and hcp-1/2 disrupted embryos completed cytokinesis (Fig. 3, A and B; Fig. S6, A and B; and Video 6). Although some cls-2 and hcp-1/2 disrupted embryos that did not form a metaphase plate were delayed in initiating cytokinesis after chromosome segregation, there was no significant difference in their rate of contractile ring constriction relative to control embryos (Fig. 3, A and B; Fig. S6, A and B; and Video 6). While it was previously known that SPD-1 disrupted embryos complete cytokinesis without central spindle assembly (Verbrugghe and White, 2004), this was not known for disruption of CLS-2 or codisruption of HCP-1/2. Thus, at least in the one-cell C. elegans embryo, central spindle assembly per se is not required for successful cell division.
Figure 3.
Central spindle assembly is not required for successful cytokinesis. (A) Method used for analysis (left; d, cell diameter) and quantification (right) of contractile ring constriction during cytokinesis in embryos expressing mCherry::H2B and GFP::PHPLC1δ1 (top). Time relative to anaphase onset; only hcp-1/2 disrupted embryos that formed a metaphase plate were analyzed (see Materials and methods and Fig. S3 A); error bars represent the SEM; n is indicated in the key [bottom; n = 13 control(RNAi), n = 7 hcp-1(RNAi); hcp-2Δ, n = 7 hcp-1Δ; hcp-2(RNAi), n = 12 hcp-1Δ, n = 8 hcp-2Δ, n = 9 spd-1(oj5ts), n = 9 cls-2(RNAi), and n = 9 zen-4(or153ts)]; see Video 6. Quantification of cytokinesis success (bottom). (B) Quantification of the average peak rate of contractile ring constriction between 80% and 30% of initial embryo diameter for all dividing embryos in A; genotypes indicated below graph. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. (C) Schematic of line scan analysis of CYK-4::mNG fluorescence intensity at the central spindle and midbody on sum projections of all Z-planes; white dashed line outlines the embryo. (D) Schematic of selected region shown (top) and representative time-lapse pseudokymographs of the division plane in embryos expressing CYK-4::mNG (green), mCherry::H2B, and mCherry::PHPLC1δ1 (magenta; bottom, left); time at top. t = 0 s is metaphase before chromosome segregation; maximum projection is shown; see Video 7. Right: Quantification of CYK-4::mNG fluorescence intensity along a central line scan at 60 s and 240 s after chromosome segregation to measure central spindle and midbody assembly, respectively. Individual (thin lines) and average (bold lines) line scans are shown for each genotype. n is listed to right side of the 240 s graphs [n = 12 control(RNAi), n = 13 hcp-1/2(RNAi), n = 10 hcp-1(RNAi), n = 14 hcp-2(RNAi), and n = 12 spd-1(RNAi)]. Average levels in control(RNAi) embryos are shown on each graph for reference (gray). Error bars represent the SEM; white scale bars, 10 µm; black scale bar in D, 15 s. n.s., P ≥ 0.05; ****, P < 0.0001.
Figure S6.
Cytokinesis and midbody assembly can occur in the absence of a central spindle. (A) Quantification of cytokinesis kinetics in embryos expressing mCherry::H2B and GFP::PHPLC1δ1 showing individual replicates for data shown in Fig. 3 A; see Fig. 3 A for schematic of quantification. Time is relative to chromosome segregation, and hcp-1/2 disrupted embryos that did or did not form a metaphase plate are indicated. Average cytokinesis kinetics of controls are shown on each graph as a reference (dark gray); error bars represent the SEM; see also Video 6. (B) Quantification of the average peak rate of contractile ring constriction for all dividing embryos in A between 80% and 30% of initial embryo diameter. Genotypes are indicated below graph. Data are repeated from Fig. 3 B with hcp-1/2 disrupted embryos that did or did not form a metaphase plate indicated. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. (A and B) n is indicated in key [n = 13 control, n = 7 hcp-1(RNAi); hcp-2Δ with metaphase plate, n = 7 hcp-1(RNAi); hcp-2Δ no metaphase plate, n = 3 hcp-1Δ; hcp-2(RNAi) with metaphase plate, n = 7 hcp-1Δ; hcp-2(RNAi) no metaphase plate, n = 12 hcp-1Δ, n = 8 hcp-2Δ, n = 9 spd-1(oj5ts), n = 9 cls-2(RNAi), and n = 9 zen-4(or153ts)]. (C) Bar graphs showing the percentage of embryos with a visible midbody (as assessed qualitatively) and a measurable peak in fluorescence intensity at the division site at 240 s after chromosome segregation in CYK-4MgcRacGAP::mNG embryos. Data correspond to embryos from Fig. 3 D. n is listed to the right side of bars in the indicated color for each genotype [n = 12 control(RNAi), n = 13 hcp-1/2(RNAi), n = 10 hcp-1(RNAi), n = 14 hcp-2(RNAi), and n = 12 spd-1(RNAi)]. n.s., P ≥ 0.05; ***, P < 0.001; and ****, P < 0.0001.
Video 6.
GFP::PHPLC1δ1 in C. elegans embryos. Representative time-lapse images of control (top, far left), hcp-1Δ; hcp-2(RNAi) (no metaphase plate; top, middle left), hcp-1(RNAi); hcp-2Δ (with a metaphase plate; top, middle right), hcp-1Δ (top, far right), hcp-2Δ (bottom, far left), spd-1(oj5ts) (bottom, middle left), cls-2(RNAi) (bottom, middle right), and zen-4(or153ts) (bottom, far right) C. elegans one-cell embryos expressing GFP::PHPLC1δ1 (green) and mCherry::H2B (magenta). Time is indicated relative to chromosome separation at 26°C; genotype labels are located at the top of each embryo in the video clip. All time-lapse images are a single central Z-plane (green) and a maximum projection of all Z-planes (magenta) from 15 × 2–µm Z-sections collected every 15 s. Playback rate is 7 frames per second. Scale bar (top right), 10 µm. This video corresponds to data shown in Figs. 3 A and S6 A.
Central spindle MTs are thought to become compacted during cytokinesis to form the midbody (for review, see Glotzer, 2009); yet, we often observed midbody assembly in the absence of central spindle assembly in HCP-1/2 and SPD-1 disrupted embryos (e.g., Fig. 2 A; and Videos 4 and 5). To probe this further, we imaged dividing embryos coexpressing a plasma membrane marker (mCherry::PHPLC1δ1) and CYK-4::mNeonGreen (mNG; Lee et al., 2018) and measured CYK-4 levels at the central spindle and midbody throughout cytokinesis with and without RNAi-mediated disruption of proteins essential for central spindle assembly (Fig. 3, C and D). Again, all control, hcp-1, hcp-2, spd-1, and hcp-1/2 disrupted embryos completed cytokinesis (Fig. 3 D and Video 7). Upon anaphase onset, CYK-4 localized to the central spindle in control and hcp-2 disrupted embryos (Fig. 3 D and Video 7). The CYK-4–labeled central spindle MTs then became further organized into individual MT bundles (or stem bodies; e.g., see Salmon et al., 1976) that expanded toward the cortex and appeared to become compacted into a midbody late in cytokinesis along with astral MTs near the equatorial cell cortex (Fig. 3 D and Video 7). This central spindle CYK-4 localization was delayed in hcp-1(RNAi) embryos and absent in hcp-1/2 and spd-1 disrupted embryos (Fig. 3 D and Video 7). Similar to what we observed with labeled tubulin and AIR-2 (Fig. 2 A; and Videos 4 and 5), CYK-4 often accumulated at the spindle midzone in a midbody-like structure by the end of contractile ring constriction in hcp-1/2 and spd-1 disrupted embryos despite never forming a central spindle (Figs. 3 D and S6 C; and Video 7). This suggests that central spindle assembly is not required for midbody assembly.
Video 7.
CYK-4MgcRacGAP::mNG in C. elegans embryos. Representative time-lapse images of control(RNAi) (top, left), hcp-1/2(RNAi) (top, middle), hcp-1(RNAi) (top, right), hcp-2(RNAi) (bottom, left), and spd-1(RNAi) (bottom, center) C. elegans one-cell embryos expressing CYK-4MgcRacGAP::mNG (green), mCherry::H2B (magenta), and mCherry::PHPLC1δ1 (magenta). Time is relative to chromosome separation at 26°C; genotype labels are located at the top of each embryo in the video clip. Time-lapse images are maximum projections of all Z-planes (green) or four central Z-planes (magenta) from 15 × 2–µm Z-sections collected every 15 s. Playback rate is 7 frames per second. Scale bar (lower right), 10 µm. This video corresponds to images shown in Fig. 3 D.
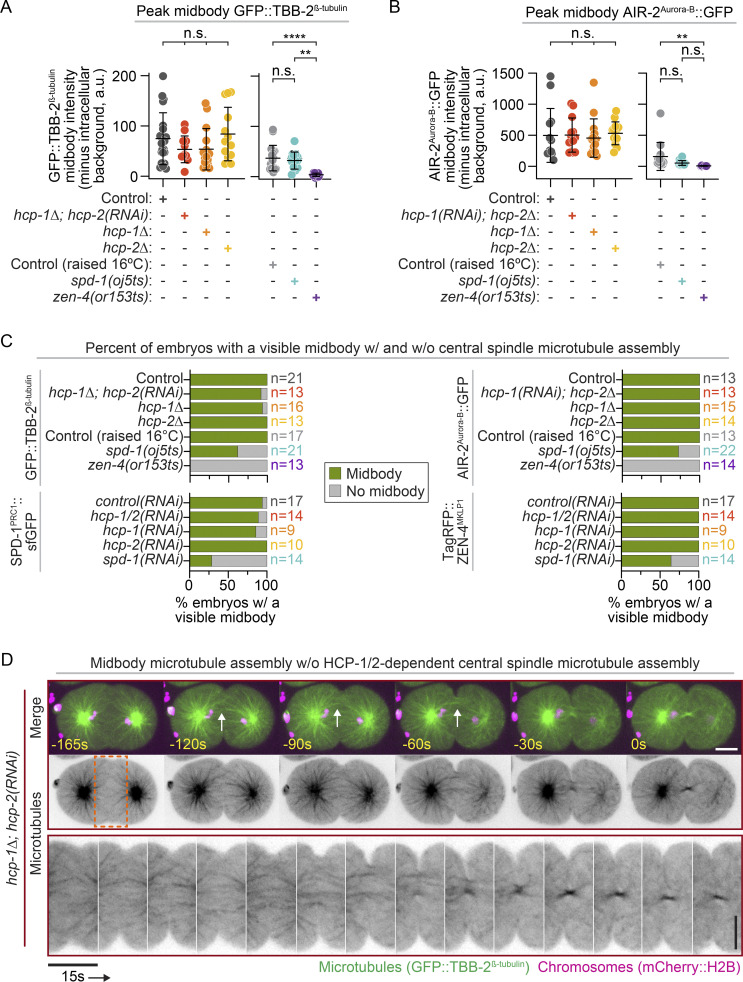
To further test the requirement of central spindle assembly for midbody assembly and to quantify the rate of midbody formation, we measured midbody levels of four additional fluorescently tagged midbody-associated proteins (TBB-2, AIR-2, SPD-1, and ZEN-4) with and without genetic disruption of central spindle assembly (Fig. 4, A–D). For mutant strains raised at 16°C (spd-1 and zen-4 ts mutants), control strains were also raised at 16°C. We imaged embryos at 240 s (or 210 s) after chromosome segregation at 26°C, when a midbody is visible in most controls. All midbody markers robustly localized to the midbody in nearly all control, hcp-1, and hcp-2 disrupted embryos, and in most hcp-1/2 disrupted embryos (Fig. 4, A–D; and Fig. S7, A–C). All fluorescently tagged midbody-associated proteins except for SPD-1 also localized to a midbody-like structure in the majority of spd-1 disrupted embryos (Fig. 4, A–D; and Fig. S7 C), though, like CYK-4 (Fig. 3 D), midbody fluorescence levels of AIR-2 and ZEN-4 were reduced (Figs. 4 D and S7 B), and MT localization occurred slightly earlier (210 s versus 240 s after chromosome separation) and was more transient than in other genotypes (Videos 4 and 5). No midbody localization was observed for any fluorescently tagged midbody-associated protein in zen-4 disrupted embryos (Fig. 4, A, B, and D; and Fig. S7 C), which did not complete cytokinesis (Fig. 3 A; Raich et al., 1998; Severson et al., 2000). Importantly, neither spd-1 nor hcp-1/2 disrupted embryos formed a central spindle, but they did form midbodies (Videos 4, 5, 7, and 8). These results are inconsistent with a model in which midbody assembly occurs solely via central spindle compaction.
Figure 4.
Functional midbody assembly is independent of central spindle assembly. (A) Representative images of GFP::TBB-2β-tubulin (green, top; inverted contrast, bottom) and mCherry:H2B (magenta, top) expressing embryos at 240 s after chromosome segregation [210 s in spd-1(oj5ts) embryos]. (B) Representative images of AIR-2::GFP-expressing (green, top; inverted contrast, bottom) and mCherry::H2B-expressing (magenta, top) embryos at 240 s after chromosome segregation [210 s in spd-1(oj5ts) embryos]. (C) Representative images of SPD-1::sfGFP-expressing (green, top; inverted contrast, bottom), TagRFP::ZEN-4–expressing, and mCherry::H2B-expressing (magenta, top; inverted contrast, bottom) embryos at 240 s after chromosome segregation (all genotypes). Schematic depicting Z-sectioning used for midbody imaging in A–C (right). (D) Line scan analysis of fluorescent protein–tagged midbody protein accumulation for cells with a visible midbody [except for zen-4(or153ts), for which all embryos were included; see Fig. S7 C] was done as for CYK-4::mNG (240 s; Fig. 3 D). n is listed on right side of graphs in the indicated color for each genotype [GFP::TBB-2β-tubulin: n = 21 control, n = 12 hcp-1Δ; hcp-2(RNAi), n = 15 hcp-1Δ, n = 13 hcp-2Δ, n = 17 control raised at 16°C, n = 13 spd-1(oj5ts), and n = 13 zen-4(or153ts); AIR-2::GFP: n = 13 control, n = 13 hcp-1(RNAi); hcp-2Δ, n = 15 hcp-1Δ, n = 14 hcp-2Δ, n = 13 control raised at 16°C, n = 16 spd-1(oj5ts), and n = 14 zen-4(or153ts); SPD-1::sfGFP and TagRFP::ZEN-4: n = 17 control(RNAi), n = 14 hcp-1/2(RNAi), n = 9 hcp-1(RNAi), n = 10 hcp-2(RNAi), and n = 9 spd-1(RNAi)]. SPD-1::sfGFP quantification is shown for all embryos with visible midbodies by TagRFP::ZEN-4; error bars represent the SEM. (E) Representative central plane images of CYK-4::mNG-expressing (green), mCherry::H2B-expressing (magenta), and mCherry::PHPLC1δ1-expressing (magenta) embryos before (top) and after (bottom) midbody release from the plasma membrane (white arrows) after abscission. Black box behind spd-1(RNAi) images is for presentation purposes. (F) Quantification of abscission timing, determined by release of the CYK-4::mNG-labeled midbody from the plasma membrane [n = 19 control(RNAi), n = 19 hcp-1/2(RNAi), and n = 9 spd-1(RNAi)]. Time relative to anaphase onset (AO) of the second (AB) cell division. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD; scale bars, 10 µm. n.s., P ≥ 0.05.
Figure S7.
Midbody assembly can occur in the absence of a central spindle via astral MT bundling. (A) Quantification of peak levels of GFP::TBB-2β-tubulin at the midbody above intracellular background; related to Fig. 4 A and the normalized line scans in Fig. 4 D. (B) Quantification of peak levels of AIR-2Aurora-B::GFP at the midbody above intracellular background; related to Fig. 4 B and the normalized line scans in Fig. 4 D. (A and B) Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test; error bars represent the SD. (C) Bar graphs showing the percentage of embryos with a visible midbody (as assessed qualitatively) and a measurable peak in fluorescence intensity at the division site. Data correspond to embryos from Fig. 4, A–D. n is listed to the right side of bars in the indicated color for each genotype [GFP::TBB-2β-tubulin: n = 21 control, n = 13 hcp-1Δ; hcp-2(RNAi), n = 16 hcp-1Δ, n = 13 hcp-2Δ, n = 17 control raised at 16°C, n = 21 spd-1(oj5ts), and n = 13 zen-4(or153ts); AIR-2Aurora-B::GFP: n = 13 control, n = 13 hcp-1(RNAi); hcp-2Δ, n = 15 hcp-1Δ, n = 14 hcp-2Δ, n = 13 control raised at 16°C, n = 22 spd-1(oj5ts), and n = 14 zen-4(or153ts); SPD-1PRC1::sfGFP and TagRFP::ZEN-4MKLP1: n = 17 control(RNAi), n = 14 hcp-1/2(RNAi), n = 9 hcp-1(RNAi), n = 10 hcp-2(RNAi), and n = 14 spd-1(RNAi)]. (D) Representative time-lapse images of GFP::TBB-2β-tubulin-expressing (green, top; inverted grayscale, middle) and mCherry::H2B-expressing (magenta) embryos undergoing midbody assembly; see also Video 8. White arrows indicate bundled astral MTs at the furrow. Time is relative to midbody assembly. Bottom: Time-lapse pseudokymographs showing GFP::TBB-2β-tubulin during midbody assembly. Image series begins 150 s before midbody assembly. A maximum projection of three Z-sections is shown. Dashed orange box (middle) indicates area selected for pseudokymograph. White scale bar and vertical black scale bar, 10 µm. Horizontal black scale, 15 s. n.s., P ≥ 0.05; **, P < 0.01; and ****, P < 0.0001.
Video 8.
Astral MTs contribute to midbody assembly in the absence of a central spindle. Representative time-lapse images of control (left) and hcp-1Δ; hcp-2(RNAi) (two examples; middle and right) C. elegans one-cell embryos expressing GFP::TBB-2β-tubulin (green, top; grayscale, middle; inverted grayscale, bottom) and mCherry::H2B (magenta, top). Time is relative to midbody assembly at 26°C; genotype labels are located at the top of the video clip. All time-lapse images are maximum projections of three Z-planes from 12 × 1–µm Z-sections collected every 15 s. Playback rate is 6 frames per second. Scale bar (top right), 10 µm. This video corresponds to images shown in Figs. 5 A and S7 D.
Previous studies in C. elegans reported that SPD-1 disrupted embryos formed midbody rings enriched in ZEN-4 but not MTs or AIR-2 (Green et al., 2013; Verbrugghe and White, 2004). Here, after spd-1 disruption, we frequently found midbodies with ZEN-4, MTs, CYK-4, and, to a lesser extent, AIR-2 (e.g., Figs. 3 D, 4, and 5; and Videos 4, 5, 7, and 8). One major difference in these studies is our use in some backgrounds of a ts mutant versus RNAi to disrupt SPD-1 function, which could have differential effects. Another difference is the AIR-2 reporter used. Green et al. used a transgenic GFP::AIR-2 strain prone to germline silencing, whereas we used an endogenously tagged CRISPR-generated AIR-2::GFP (Cheerambathur et al., 2019), which is less susceptible to germline silencing. The most significant difference is the extent of Z-sectioning during imaging. While previous studies limited Z-sectioning to the center of the ∼25–30-µm diameter one-cell worm embryo, it has been shown that contractile ring closure is nonconcentric (Maddox et al., 2007); thus, we collected 25 × 1–µm Z-sections to ensure capture of the midbody. With this analysis, we found that midbody MTs in spd-1 disrupted embryos were more transient (e.g., Figs. 1 A and 2 A; and Videos 4 and 5) and levels of fluorescently tagged ZEN-4, CYK-4, and AIR-2 at the midbody were lower and more difficult to detect than in control embryos (e.g., Figs. 3 and 4). Our results suggest a possible role for SPD-1 in midbody formation and/or stability, but because central spindle proteins still localized to the nonbundled hemispindle MTs at the spindle midzone in spd-1 but not hcp-1/2 disrupted embryos (Figs. 1 C and 2; and Videos 4 and 5; see also Lewellyn et al., 2011), we cannot rule out that hemispindle MT binding precludes midbody MT binding. Nevertheless, the presence of MTs and AIR-2 at the midbody in the spd-1 disrupted embryos suggests formation of at least a transient midbody with midbody MTs rather than just a midbody ring.
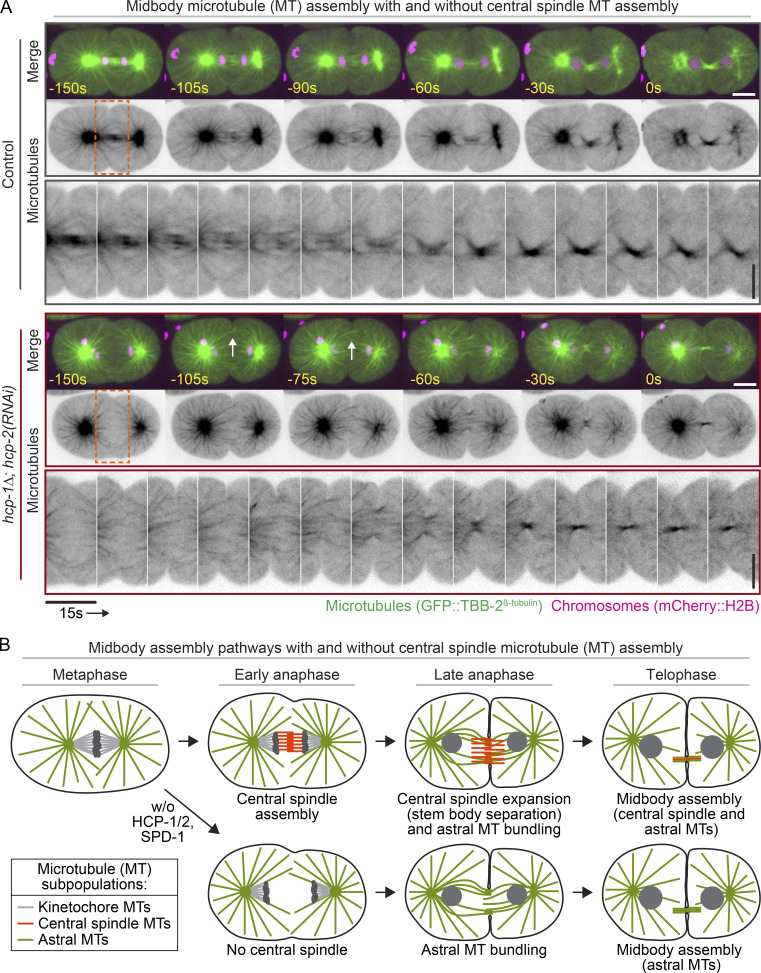
Figure 5.
Astral MTs contribute to midbody assembly in the absence of a central spindle. (A) Representative time-lapse images of GFP::TBB-2β-tubulin-expressing (green, top; inverted grayscale, middle) and mCherry::H2B-expressing (magenta) embryos undergoing midbody assembly. White arrows indicate bundled astral MTs at the furrow. Time relative to midbody assembly. Bottom: Time-lapse pseudokymographs of MTs during midbody assembly. Maximum projection of three Z-sections is shown. See Video 8. Dashed orange box indicates area selected for pseudokymograph; white and vertical black scale bars, 10 µm; horizontal black scale bar, 15 s. (B) Schematic depicting model for midbody assembly in the presence (top) or absence (bottom) of HCP-1/2– or SPD-1–dependent central spindle assembly.
We next assessed whether central spindle–independent midbodies can functionally support abscission. In C. elegans embryos, the late stage of abscission in which the midbody is released occurs after anaphase of the subsequent cell division (Green et al., 2013), so embryos coexpressing mCherry::PHPLC1δ1 and the midbody marker CYK-4::mNG were imaged through the four-cell stage. In control, hcp-1/2, and spd-1 disrupted two-cell embryos, CYK-4 was concentrated in a tight focus in the center of the cell–cell boundary, then released into the cytoplasm during abscission (Fig. 4 E). hcp-1/2 depleted embryos released midbodies from the cell–cell boundary with timing similar to that in controls and spd-1 depleted embryos (Fig. 4 F; see also Green et al., 2013). Thus, midbodies that form in the absence of a central spindle can support abscission.
To determine how these functional midbodies assemble in the absence of a central spindle, we imaged GFP::TBB-2β-tubulin-expressing embryos after furrow initiation to avoid photobleaching effects. In control embryos, both central spindle and astral MTs appeared at the division plane and became bundled near the end of cytokinesis to form a midbody (Fig. 5 A). In hcp-1/2 disrupted embryos, no central spindle formed (Fig. 1 C; Fig. 2; Fig. 3 D; Fig. 5 A; Fig. S4, A and B; Fig. S5, B–F; and Fig. S7 D; and Videos 1, 2, 3, 4, 5, 6, 7, and 8). Instead, astral MTs near the cell cortex in the division plane appeared to bundle and were compacted by the leading edge of the ingressing furrow to form the midbody (Figs. 5 A and S7 D; and Video 8). This suggests that at least in the early C. elegans embryo, both central spindle and astral MTs likely contribute to midbody assembly. This also suggests that in the absence of a central spindle, astral MTs alone can support functional midbody assembly (Fig. 5 B).
We consistently observed completion of cytokinesis and highly robust midbody assembly in HCP-1/2 codisrupted embryos (Figs. 3, 4, 5 A, S6 A, and S7 D; and Videos 4, 5, and 8), despite their never forming CLS-2–dependent central spindle MTs (Figs. 1, 2, and 3). Others have shown that asters were sufficient for furrow induction (Rappaport, 1961, 1973; Savoian et al., 1999), even in enucleated cells (Baruni et al., 2008; von Dassow et al., 2009), and that central spindle– and midbody-associated signaling proteins, such as Cyk4 and the Rho-family GTP exchange factor Ect2, localized to astral MTs and moved on asters to reach the cell cortex (Su et al., 2014). Su and colleagues suggested that in the absence of signals from the central spindle, this astral enrichment serves as an “astral simulacrum” that supports cytokinesis (Su et al., 2014). Though unlikely, it is possible that a remnant of a central spindle formed after HCP-1/2 codisruption that is not detectable by live fluorescence imaging (Figs. 1, 2, 3, 5, and S7 D), by immunofluorescence of fixed samples (Fig. S4 B), or after a reduction in cortical astral MT-based pulling forces (Fig. S5, D–F). Still, our genetic results support the simulacrum model, wherein astral MTs promote both cortical relaxation at the cell poles (e.g., see Chapa-y-Lazo et al., 2020; Harris and Gewalt, 1989; Mangal et al., 2018; Mishima, 2016; von Dassow et al., 2009) and midbody assembly, driving successful cytokinesis.
Is midbody formation a cause or consequence of successful cytokinesis? Here we find that midbody formation is closely correlated with the final stages of contractile ring constriction. While in most animal cells the midbody is required for abscission at the end of the cell cycle (Steigemann and Gerlich, 2009), the role of the midbody in promoting successful contractile ring constriction during cytokinesis is less clear. Contractile ring constriction is required for midbody assembly (Hu et al., 2012; Landino and Ohi, 2016; Straight et al., 2003), suggesting that midbody assembly may be a consequence of cytokinesis rather than a causative mediator. However, when the midbody is uncoupled from the contractile ring by expressing a Cyk4 mutant lacking the membrane-binding C1 domain, cytokinesis fails, and the midbody remains intact in the cytoplasm (Lekomtsev et al., 2012). Thus, it remains unclear if midbody formation is just a consequence of central spindle and/or astral MT compaction during contractile ring constriction. We found that both HCP-1/2 and SPD-1 disrupted embryos formed midbodies, though midbodies in SPD-1 disrupted embryos were more transient and contained a lower level of some midbody proteins. HCP-1/2 and SPD-1 disrupted embryos both completed cytokinesis with kinetics similar to that of control embryos (Fig. 3, A and B; Fig. S6, A and B; and Video 6). This suggests that the robustness of midbody assembly does not grossly influence contractile ring constriction but does not rule out a role for the midbody in promoting successful contractile ring constriction directly. Unambiguously testing whether this robust and beautiful MT structure is a cause or a consequence of successful contractile ring constriction and cytokinesis will require the development of new technologies for acute midbody assembly and/or disassembly in vivo.
Materials and methods
C. elegans strain maintenance
C. elegans were maintained on standard nematode growth medium (NGM) plates seeded with OP50 E. coli bacteria, as described previously (Brenner, 1974). All strain names and genotypes used in this study are listed in Table S1. Strains were maintained in an incubator (Binder) kept at either 16 ± 0.5°C (ts mutants and their controls) or 20 ± 0.5°C (all other strains) before imaging.
RNA-mediated interference
Exonic sequences from the desired gene were cloned in to the multiple cloning site of the L4440 vector using standard cloning techniques and then transformed into HT115 E. coli (Timmons et al., 2001). RNAi primers and template DNA for each gene are listed in Table S1. RNAi feeding bacteria were grown in Luria broth with ampicillin (100 µg/ml) for 8–16 h at 37°C. 300 µl of this culture was plated on NGM agar plates (Brenner, 1974) supplemented with 50 µg/ml ampicillin and 1 mM IPTG. These plates were grown at 37°C for 48 h. L1–L2 stage larval worms were plated on RNAi plates and incubated at 20°C for 48–72 h before dissection of adult hermaphrodites to obtain embryos.
For each relevant experiment, RNAi-mediated knock-down was confirmed by phenotypic analysis [e.g., spd-1(RNAi) always blocked central spindle assembly and led to increased chromosome segregation] and by assessing embryonic lethality in parallel: 100% embryonic lethality for hcp-1/2(RNAi), 77–100% for spd-1(RNAi), 100% for lin-5(RNAi), and 100% for gpr-1/2(RNAi). For genes that do not cause embryonic lethality (e.g., hcp-1, hcp-2 individually), synthetic embryonic lethality was assessed in parallel using the reciprocal CRISPR deletion strain (e.g., hcp-1Δ worms treated with hcp-2-targeting RNAi and vice versa [100% embryonic lethality for hcp-1Δ; hcp-2(RNAi), 100% for hcp-1(RNAi); hcp-2Δ]). For cls-2(RNAi) (Fig. 3, A and B; and Fig. S6, A and B), RNAi-mediated knock-down was confirmed by assessing embryonic lethality of individual young adult hermaphrodites, which were only dissected to obtain embryos for imaging after individual embryonic lethality was confirmed [100% embryonic lethality for cls-2(RNAi)].
When possible, we also performed quantitative analysis of the loss of fluorescently tagged protein signal (e.g., Figs. 2 C and S2) and whole-worm quantitative real-time PCR (qRT-PCR; see qRT-PCR section below). Quantitative analysis of endogenous (CRISPR-generated) fluorescently tagged HCP-1 and HCP-2 (Edwards et al., 2018) revealed endogenous GFP::HCP-1 levels at the spindle midzone in anaphase were reduced by 96% in hcp-1(RNAi) embryos and by 97% in hcp-1/2(RNAi) codepleted embryos (Fig. S2). Endogenous GFP::HCP-2 levels at the spindle midzone were reduced by 99% in hcp-2(RNAi) embryos and by 91% in hcp-1/2(RNAi) codepleted embryos (Fig. S2). Transgenic SPD-1::sfGFP levels at the spindle midzone were reduced by 109% in spd-1(RNAi) embryos [Fig. 2 C; note that spindle midzone fluorescence intensity was often lower than intracellular background fluorescence intensity in spd-1(RNAi) embryos, resulting in >100% reduction after background subtraction]. Relative fluorescently tagged protein depletion analysis was performed by measuring the integrated density within the spindle midzone region above intracellular background levels of a region of the same size and normalizing to average levels in control embryos.
qRT-PCR
To quantify knock-down efficiency of hcp-1, hcp-2, hcp-1/2, and spd-1 (Fig. S1 B) by RNAi-mediated interference at the whole-worm level in parallel to the embryonic lethality test (see above), 10 young adult worms were used per biological replicate for RNA extraction as described previously (Ly et al., 2015) using 10 µl of lysis buffer (5 mM Tris, pH 8.0 [MilliporeSigma], 0.5% Triton X-100 [MilliporeSigma], 0.5% Tween 20 [Bio-Rad Laboratories], 0.25 mM EDTA [MilliporeSigma], and 1 mg/ml proteinase K [New England Biolabs]). Samples were treated with DNase (Invitrogen), and cDNA was synthesized by the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Primers used for each gene are listed in Table S1. For quantification, standard curves were generated for each primer set, act-1 was used as a reference gene, and levels for each gene were normalized to control(RNAi) levels (empty vector). This qRT-PCR analysis (Fig. S1 B) revealed that relative to control(RNAi) worms, whole-worm hcp-1 levels were depleted by 86.4% in hcp-1(RNAi), 82% in hcp-1/2(RNAi), 99.8% in hcp-1Δ; control(RNAi), 99.99% in hcp-1Δ; hcp-2(RNAi), and 93.7% in hcp-1(RNAi); hcp-2Δ worms. Relative to control(RNAi), whole-worm hcp-2 levels were reduced by 65.6% in hcp-2(RNAi), 61.5% in hcp-1/2(RNAi), 94.1% in hcp-2Δ; control(RNAi), 97.6% in hcp-1(RNAi); hcp-2Δ, and 62.4% in hcp-1Δ; hcp-2(RNAi) worms. Whole-worm spd-1 levels were reduced by 77.6% in spd-1(RNAi) worms. We note that whole-worm qRT-PCR likely underrepresents the extent of gene knock-down in the early embryo, as some somatic worm tissues are resistant to RNAi-mediated gene knock-down (e.g., see Simmer et al., 2002).
Live-cell imaging
Live-cell imaging was performed in a room with homeostatic temperature control set to 26 ± 0.5°C for all experiments. Room temperature was monitored with two thermometers attached directly to the objective and a humidity and temperature smart sensor (SensorPush) on the microscope stage. Young gravid hermaphrodites were dissected in cooled (∼16°C) M9 buffer (Brenner, 1974), and embryos were mounted on a 2% agar pad as described previously (Davies et al., 2017; Gönczy et al., 1999; Jordan et al., 2016).
For live-cell imaging experiments, embryos (except in Fig. S4 C) were imaged on an inverted microscope (Ti; Nikon) using a spinning disk confocal unit (CSU-10; Yokogawa Electric Corporation) with Borealis (Spectral Applied Research) and a charge-coupled device camera (Orca-R2; Hamamatsu Photonics). Z-sectioning was done with a piezo-driven motorized stage (Applied Scientific Instrumentation), and focus was maintained using Perfect Focus (Nikon) before each Z-series acquisition. An acousto-optic tunable filter was used to select the excitation light of two 100-mW lasers for excitation at 491 and 561 nm for GFP and mCherry, respectively (Spectral Applied Research), and a filter wheel was used with 525/50-nm and 620/60-nm (Chroma) bandpass filters for emission wavelength (fluorescence) or analyzer (differential interference contrast [DIC]) selection (Sutter Instruments). The system was controlled by MetaMorph software (Molecular Devices). Embryos in Fig. S4 C were imaged on an inverted microscope (Ti; Nikon) using a spinning-disk confocal unit (CSU-10; Yokogawa) with Borealis (Spectral Applied Research), two 150-mW excitation lasers at 491 and 561 nm (Cairn), and an emission filter wheel (Sutter Instruments) with bandpass filters corresponding to 525/50 nm and 620/50 nm (Chroma) as described previously (Hirsch et al., 2018; Sundaramoorthy et al., 2017).
GFP::HCP-1, GFP::HCP-2 (Fig. S2 A), CLS-2CLASP::GFP (Fig. S3 A; Edwards et al., 2018), and mCherry::H2B were imaged with a 60×/1.4 NA Plan Apochromat (Plan Apo) oil immersion objective with 2 × 2 binning, and 11 × 1–µm Z-sections were collected every 10 s. For tubulin imaging (Figs. 1 C, 5 A, and S7 D), embryos coexpressing GFP::TBB-2β-tubulin and mCherry::H2B (Lacroix et al., 2018) were imaged using a 40×/1.25 NA CFI Apo Lambda S water immersion objective with no binning, and 13 × 1–µm (Fig. 1 C) or 12 × 1–µm (Figs. 5 A and S7 D) Z-sections were collected every 15 s. GFP::TBB-2β-tubulin imaging in Fig. S4 C was done with a 60×/1.4 NA Plan Apo oil immersion objective with 2 × 2 binning, and 10 × 2–µm Z-sections were collected every 15 s. In Figs. 2 A and S5 D, AIR-2Aurora-B::GFP-expressing (Cheerambathur et al., 2019) and mCherry::H2B-expressing embryos were imaged using a 40×/1.25 NA CFI Apo Lambda S water immersion objective with no binning, and 12 × 1–µm Z-sections were collected every 15 s. SPD-1PRC1::sfGFP-expressing embryos (Fig. 3 C) were imaged every 30 s with 2 × 2 binning, and 15 × 1–µm Z-sections were obtained using a 60×/1.4 NA Plan Apo oil immersion objective with 2 × 2 binning.
For imaging of cytokinetic progression (Figs. 3 A and S6 A), embryos expressing GFP::PHPLC1δ1 and mCherry::H2B (Audhya et al., 2005) were imaged with a 40×/1.3 NA CFI Plan Fluor oil immersion objective with 2 × 2 binning, and 15 × 2–µm Z-sections and a central DIC image were collected every 15 s. Embryos expressing CYK-4MgcRacGAP::mNG (Lee et al., 2018), mCherry::PHPLC1δ1 (Lee et al., 2018), and mCherry::H2B were imaged with a 60×/1.4 NA Plan Apo oil immersion objective with 2 × 2 binning, and 15 × 2–µm Z-sections were collected every 15 s (Fig. 3 D) or every 30 s for imaging midbody release (Fig. 4 E). For midbody imaging (Fig. 4, A–C), embryos were observed by DIC until anaphase onset, at which point 25 × 1–µm Z-sections were acquired every 210 s [spd-1(oj5ts)] or 120 s (all other genotypes). Embryos expressing GFP::TBB-2β-tubulin or AIR-2Aurora-B::GFP (Fig. 4, A and B) were imaged with a 40×/1.25 NA CFI Apo Lambda S water immersion objective with no binning, and strains expressing SPD-1PRC1::sfGFP and TagRFP::ZEN-4MKLP1 (Fig. 4 C) were imaged with a 60×/1.4 NA Plan Apo oil immersion objective with 2 × 2 binning.
Image analysis
All image analysis, including fluorescence intensity measurements and line scan analysis, was performed using Fiji software (National Institutes of Health; Schindelin et al., 2012) on raw images. MetaMorph software (Molecular Devices) was used for analysis presented in Fig. 3, A and B; and Fig. S6, A and B. For display purposes, a maximum projection of all Z-sections is shown unless noted otherwise. In Fig. 3 D, a maximum projection of four central Z-sections is shown; in Figs. 5 A and S7 D, a maximum projection of three Z-sections is shown for GFP::TBB-2β-tubulin. Pseudokymographs (Figs. 2 A, 3 D, 5 A, S1 E, 5 A, S2 A, S3 A, and S7 D) were generated for display from bleach-corrected images using the simple ratio method (Fiji). Images were pseudo-colored and contrast adjusted for display using Adobe Photoshop CC. Figures were prepared using Adobe Illustrator CC.
Quantification of central spindle and midbody assembly
To quantify protein distribution at the central spindle (GFP::TBB-2β-tubulin, Fig. 1 C; CYK-4MgcRacGAP::mNG, Fig. 3 D) and midbody (CYK-4MgcRacGAP::mNG, Fig. 3 D; GFP::TBB-2β-tubulin, AIR-2Aurora-B::GFP, SPD-1PRC1::sfGFP, and TagRFP::ZEN-4MKLP1, Fig. 4 D), a sum projection of all Z-sections was made. To avoid quantifying background tubulin levels, for the GFP::TBB-2β-tubulin midbody quantification only (Fig. 4 A), a sum projection of the 3 Z-sections in which the midbody was most visible (or three central slices if no midbody was visible) was used. For midbody analysis of CYK-4MgcRacGAP::mNG in Fig. 3 D, line scan analysis was done on all embryos. For midbody analysis in Fig. 4 D (GFP::TBB-2β-tubulin, AIR-2Aurora-B::GFP, and TagRFP::ZEN-4MKLP1), line scan analysis was done only on embryos that formed a visible midbody. For midbody analysis of SPD-1PRC1::sfGFP levels, line scan analysis was done in embryos where a midbody was observed with TagRFP::ZEN-4MKLP1 (Fig. 4 D). The percentage of embryos with a visible midbody (Figs. S6 C and S7 C) was scored based on a peak of protein accumulation 240 s after anaphase onset [210 s for spd-1(oj5ts)] visible to the eye at the division site of the cell and confirmed by a quantifiable peak in the measured line scan. On the frame at the indicated time point, a 10-µm line scan was drawn perpendicular to the axis of division to measure average signal in the 491-nm channel (561 nm for TagRFP::ZEN-4MKLP1), centered between the dividing chromosomes as indicated by the mCherry::H2B signal. A 5-µm × 5-µm extracellular region was measured, and the mean grayscale value of the extracellular region was subtracted from all average line scan values. Intracellular background was determined by averaging the values from pixels in the first and last 0.5 µm of the line scan. Line scans were normalized by dividing all background-subtracted values by the intracellular background value. For peak midbody levels of GFP::TBB-2β-tubulin and AIR-2Aurora-B::GFP (Fig. S7, A and B), the intracellular background value was subtracted from the raw peak value at the middle of the 10-µm line scan and individually plotted.
Quantification of spindle midzone protein distribution
To quantify distribution of proteins at the spindle midzone (AIR-2Aurora-B::GFP, Fig. 2 B; Fig. S4 A; and Fig. S5, D and E; GFP::HCP-1, GFP::HCP-2, Fig. S2 B; CLS-2CLASP::GFP, Fig. S3 B), a sum projection was made of all Z-sections, and average grayscale values of a line scan 20 × 2.54 µm (Fig. 2 B; Fig. S2 B; Fig. S4 A; Fig. S5, D and E) or 25 × 2.54 µm (Fig. S3 B) were measured on the 491-nm channel, centered on the dividing chromosomes on frames corresponding to the indicated time points (after chromosome segregation) as assessed by mCherry::H2B imaged every 15 s (Figs. 2 B and S4 A) or every 45 s (Fig. S5, D and E). mCherry::H2B distribution (Fig. S5 D) was measured in the same way in the 561-nm channel. Intracellular background was measured by generating a line scan of the same dimensions in a cytoplasmic region not containing chromosomes. Average intracellular background was subtracted from each pixel value along the line scan, and, for AIR-2Aurora-B::GFP only (Fig. 2 B; Fig. S4 A; Fig. S5, D and E), a rolling average of 0.5 µm was generated to smooth out uneven distribution within a given embryo.
Quantification of midzone protein levels
Protein levels at the spindle midzone (GFP::HCP-1 and GFP::HCP-2, Fig. S2, C and D; CLS-2CLASP::GFP, Fig. S3, D and E; SPD-1PRC1::sfGFP, Fig. 2 C) were quantified on a sum projection of all Z-sections. A box was drawn at the midzone on (metaphase) or between (anaphase) the separating chromosomes with a height of the chromosomes in a given frame and width of the span between chromosomes. In cases in which there were severe chromosome segregation defects, the box was drawn between the main chromosomal masses. The mean pixel value within the box was measured in the 491-nm channel. Areas of the same dimension were used to measure mean extracellular and intracellular background levels by measuring a region outside of the cell and within the cell but outside of the midzone. Extracellular background levels were subtracted, and midzone protein levels were normalized to intracellular background levels. For analysis over time, analysis at 0 s corresponds to the metaphase before anaphase onset, and kinetochore levels were measured rather than the midzone levels. For Fig. S2 C, normalized values were renormalized relative to the appropriate control.
Analysis of cytokinesis
Cytokinetic phenotypes were scored in embryos labeled with a plasma membrane marker (GFP::PHPLC1δ1) and a histone marker (mCherry::H2B) in which imaging began before anaphase onset and continued through anaphase onset of the next cell division or, in the case of cytokinesis failure, until complete contractile ring regression. Contractile ring diameter was measured from the time of metaphase (time point immediately before anaphase onset) until the end of contractile ring constriction (or full regression) in the Z-section in which the ring was most open and displayed as a percentage of the initial diameter over time (Fig. 3 A). For hcp-1(RNAi); hcp-2Δ and hcp-1Δ; hcp-2(RNAi) embryos, although all embryos filmed through anaphase of the following division completed cytokinesis (14 of 14 and 10 of 10 embryos), there was a large variation in timing between those that formed a metaphase plate relative to those that did not, likely due to the timing difference between chromosome separation and anaphase onset in this genotype (Edwards et al., 2018). We therefore limited our analysis of contractile ring constriction kinetics in Fig. 3 A to hcp-1/2 disrupted embryos that formed a metaphase plate and exhibited normal chromosome bi-orientation. Single traces showing individual replicates for all embryos (that did and did not form a metaphase plate) are shown in Fig. S6 A. The percentage of embryos that completed cytokinesis was scored on a maximum Z-projection for all embryos (Fig. 3 A). The average peak constriction rates (Figs. 3 B and S6 B) were plotted as µm/s when the rate of ingression peaks (between ∼80% and ∼30% of the initial cell diameter).
Analysis of chromosome segregation
Chromosome segregation was analyzed using a histone mCherry::H2B marker in embryos coexpressing GFP::PHPLC1δ1 (Fig. S5 B) or AIR-2Aurora-B::GFP (Fig. S5 F) and imaged every 15 s. The distance between chromosomes was measured for each time point acquired following metaphase. For embryos without a distinct metaphase plate, 0 s was determined as the time point preceding noticeable segregation of two chromosomal masses. Chromosome segregation data in Fig. S2 E correspond to embryos measured for cytokinetic progression (Figs. 3 A and S6 A).
Analysis of midbody release after abscission
Abscission was analyzed in embryos coexpressing CYK-4MgcRacGAP::mNG, mCherry::PHPLC1δ1 (Lee et al., 2018), and mCherry::H2B, similar to as described previously (Green et al., 2013). Control, hcp-1/2(RNAi) and spd-1(RNAi) embryos were imaged every 30 s throughout the volume of the embryo (15 × 2–µm Z-sections) before anaphase onset of the second division in the anterior (AB) cell into the four-cell embryonic stage (∼1,200 s after anaphase onset in the AB cell). Individual planes containing the midbody (CYK-4MgcRacGAP::mNG) were identified, and midbody release was assessed as the time point at which the midbody from the first cell division fully separated from the plasma membrane (mCherry::PHPLC1δ1) and was engulfed by one of the daughter cells (Fig. 4, E and F). Timing of midbody release was scored relative to anaphase onset of the AB cell using the histone marker (mCherry::H2B).
Brood size and viability analysis
For hcp-1 and hcp-2Δ CRISPR deletion mutants (Fig. S1 A, right), hcp-1Δ, hcp-2Δ, and N2 control hermaphrodites were singled as L4s onto individual mating plates (35-mm NGM agar plates seeded with 10 µl of OP50; Brenner, 1974). The individual adult hermaphrodites were transferred to a new plate every ∼24 h until they no longer produced fertilized embryos. The total number of viable larval progeny and dead embryos per hermaphrodite over the entire reproductive lifespan were quantified as total brood size. The percentage of viable larval progeny and dead embryos was quantified on the first plate 24 h after transferring off the adult worm (to allow all viable embryos to hatch) using a digital counting pen (Fisher Scientific) on a dissecting microscope (Olympus SZX16).
For hcp1/2 RNAi-mediated depletion experiments (Fig. S1 A, left), L1 stage larval worms were plated on RNAi plates and allowed to develop for ∼48 h at 20°C. L4 stage hermaphrodites were then singled onto individual mating plates, and brood size and embryonic viability over the first 24 h were quantified as described above for the CRISPR deletion mutants.
HCP-1 and HCP-2 protein sequence alignment
Pairwise sequence alignments of full-length protein, N-terminal domain, C-terminal domain, and middle of HCP-1 and HCP-2 protein sequences (Fig. S1 D) were done using EMBOSS Needle (Madeira et al., 2019; https://www.ebi.ac.uk/Tools/psa/emboss_needle). For comparison, the N-terminal domain was designated as the first 350 aa (aa 1–350 for both HCP1 and HCP-2), and the C-terminal domain was designated as the last 350 aa of each protein (aa 1126–1475 for HCP-1; aa 946–1295 for HCP-2).
Postimaging viability analysis
Following embryo dissection and imaging of the first cell division in embryos coexpressing GFP::TBB-2β-tubulin and mCherry::H2B (Lacroix et al., 2018), the coverslip was removed from the slide, and the 2% agarose pad containing the embryo was lifted from its surface using a scalpel. The agar pad was then placed embryo side down onto an NGM worm plate containing OP50 bacteria and maintained at 20°C to allow embryonic development and hatching. Images of the isolated embryos were acquired every 16–24 h from hatching and into adulthood using a MicroPublisher 3.3 real-time viewing camera (Q-Imaging) mounted on the C-mount of a dissecting microscope (Olympus SZX16) with a 120-ms exposure time using QCapture Suite Plus software (Q-Imaging) to ensure development was timely and adult reproduction was not grossly affected (Fig. S4 C). Viability was measured as the percentage of embryos that hatched and developed into fertile adult worms (Fig. S4 C).
Immunofluorescence
Worms were dissected on poly-L-lysine–coated slides, and embryos were fixed by freeze cracking and plunging into −20°C methanol for 20 min as described previously (Gönczy et al., 1999). Embryos were rehydrated in PBS, blocked in AbDil (PBS plus 2% BSA, 0.1% Triton X-100), and incubated overnight at 4°C with an FITC-labeled α-tubulin antibody diluted 1:100 (DM1α; Abcam), washed with PBST (PBS plus 0.1% Triton X-100), washed with PBST plus 2 µg/ml Hoechst, and mounted in 0.5% p-phenylenediamine, 20 mM Tris-Cl, pH 8.8, 90% glycerol. Images were acquired on a Nikon TiE microscope using a 100×/1.45 NA CFI Plan Apo Lambda oil immersion objective (Nikon), 405- and 488-nm lasers, and a CoolSNAP HQ2 charge-coupled device camera (Photometrics Scientific) with 0.2-µm Z-sectioning using a piezo-driven motorized stage (Applied Scientific Instrumentation). Maximum projections of all Z-sections containing chromosomes were generated for presentation (Fig. S4 B).
Statistical analysis
Graphs were made and statistical significance was calculated using GraphPad Prism 8. Data distribution was assumed to be normal, but this was not formally tested. For analysis of significance among datasets (except for that in Fig. S1 B), one-way ANOVA was used with Tukey’s multiple comparison test comparing all column means. For Fig. S1 B, one-way ANOVA was used with Dunnett’s multiple comparison test, comparing each column mean with control(RNAi). Error bars for line scan traces of central spindle or midbody protein distribution and cell diameter over time during cytokinesis represent the SEM. Error bars for all other analysis (brood size, viability, midzone protein accumulation, etc.) represent the SD. See Table S2 for ANOVA tables and multiple comparison tests. For P values and adjusted P values (see Table S2): n.s., P ≥ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the validation of HCP-1 and HCP-2 RNAi and deletion mutants. Fig. S2 shows the results of analysis of HCP-1 and HCP-2 function individually and together. Fig. S3 shows the results of analysis of CLS-2 localization with and without hcp-1/2 disruption. Fig. S4 shows the results of analysis of central spindle assembly with and without hcp-1 and hcp-2 disruption. Fig. S5 shows how the reduction in dynein-driven cortical astral MT-based pulling forces can rescue central spindle assembly in hcp-1Δ but not hcp-1/2 codisrupted embryos. Fig. S6 shows the results of quantitative analysis of cytokinesis and midbody assembly. Fig. S7 shows the results of quantitative analysis of midbody assembly in the absence of a central spindle. Video 1 shows GFP::HCP-1CENP-F with and without hcp-1 and hcp-2 disruption. Video 2 shows GFP::HCP-2CENP-F with and without hcp-1 and hcp-2 disruption. Video 3 shows CLS-2CLASP::GFP with and without hcp-1 and hcp-2 disruption. Video 4 shows GFP::TBB-2β-tubulin with and without central spindle assembly. Video 5 shows AIR-2Aurora-B::GFP with and without central spindle assembly. Video 6 shows GFP::PHPLC1δ1 with and without central spindle assembly. Video 7 shows CYK-4MgcRacGAP::mNG with and without central spindle assembly. Video 8 shows midbody assembly in the presence and absence of a central spindle. Table S1 lists worm strains, plasmids, and primers. Table S2 shows the results of statistical analysis.
Supplementary Material
lists worm strains, plasmids, and primers.
shows the results of statistical analysis.
Acknowledgments
We thank Tim Davies, Julia Siewert, and all members of the Canman, Dumont, and Shirasu-Hiza laboratories for their feedback, support, and advice on this work. We thank Elena Lucchetta and Caroline Connors for help with qRT-PCR setup and midbody imaging, respectively. We thank Matthew Marzo for assistance with the dynein/dynactin schematic. We thank Eva Sophia Blake and Sophia Tony-Egbuniwe for laboratory assistance. We are grateful to Karen Oegema and the Caenorhabditis Genetics Center (National Institutes of Health grant P40OD010440) for providing worm strains.
This work was funded by National Institutes of Health grant T32GM007088 (S. Hirsch), National Institutes of Health grant R01AG045842 (M. Shirasu-Hiza), National Institutes of Health grant R35GM127049 (M. Shirasu-Hiza), European Research Council consolidator grant ChromoSOMe grant 819179 (J. Dumont), National Institutes of Health grant R01GM117407 (J.C. Canman), and National Institutes of Health grant R01GM130764 (J.C. Canman).
The authors declare no competing financial interests.
Author contributions: S.M. Hirsch, F. Edwards, J. Dumont, and J.C. Canman conceived of the project. F. Edwards conducted pilot experiments and analysis on HCP-1 and HCP-2 independent function; J. Dumont performed the immunofluorescence analysis. S.M. Hirsch conducted all other experiments and data analysis. S.M. Hirsch and J.C. Canman designed all experiments. S.M. Hirsch, M. Shirasu-Hiza, J. Dumont, and J.C. Canman made intellectual contributions and wrote the manuscript. S.M. Hirsch and J.C. Canman made the figures.
References
- Alsop, G.B., and Zhang D.. 2003. Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J. Cell Biol. 162:383–390. 10.1083/jcb.200301073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop, G.B., and Zhang D.. 2004. Microtubules continuously dictate distribution of actin filaments and positioning of cell cleavage in grasshopper spermatocytes. J. Cell Sci. 117:1591–1602. 10.1242/jcs.01007 [DOI] [PubMed] [Google Scholar]
- Audhya, A., Hyndman F., McLeod I.X., Maddox A.S., Yates J.R. III, Desai A., and Oegema K.. 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171:267–279. 10.1083/jcb.200506124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruni, J.K., Munro E.M., and von Dassow G.. 2008. Cytokinetic furrowing in toroidal, binucleate and anucleate cells in C. elegans embryos. J. Cell Sci. 121:306–316. 10.1242/jcs.022897 [DOI] [PubMed] [Google Scholar]
- Bratman, S.V., and Chang F.. 2007. Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell. 13:812–827. 10.1016/j.devcel.2007.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann, H., and Hyman A.A.. 2005. A cytokinesis furrow is positioned by two consecutive signals. Nature. 436:731–734. 10.1038/nature03823 [DOI] [PubMed] [Google Scholar]
- Canman, J.C., Cameron L.A., Maddox P.S., Straight A., Tirnauer J.S., Mitchison T.J., Fang G., Kapoor T.M., and Salmon E.D.. 2003. Determining the position of the cell division plane. Nature. 424:1074–1078. 10.1038/nature01860 [DOI] [PubMed] [Google Scholar]
- Canman, J.C., Lewellyn L., Laband K., Smerdon S.J., Desai A., Bowerman B., and Oegema K.. 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 322:1543–1546. 10.1126/science.1163086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L.G., and Wang Y.L.. 1996. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell. 7:225–232. 10.1091/mbc.7.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapa-y-Lazo, B., Hamanaka M., Wray A., Balasubramanian M.K., and Mishima M.. 2020. Polar relaxation by dynein-mediated removal of cortical myosin II. J. Cell Biol. 219:e201903080. 10.1083/jcb.201903080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur, D.K., Prevo B., Chow T.L., Hattersley N., Wang S., Zhao Z., Kim T., Gerson-Gurwitz A., Oegema K., Green R., et al. 2019. The kinetochore-microtubule coupling machinery is repurposed in sensory nervous system morphogenesis. Dev. Cell. 48:864–872.e7. 10.1016/j.devcel.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., MacLeod I., Yates J.R. III, Oegema K., and Desai A.. 2005. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr. Biol. 15:771–777. 10.1016/j.cub.2005.03.018 [DOI] [PubMed] [Google Scholar]
- D’Avino, P.P., and Capalbo L.. 2016. Regulation of midbody formation and function by mitotic kinases. Semin. Cell Dev. Biol. 53:57–63. 10.1016/j.semcdb.2016.01.018 [DOI] [PubMed] [Google Scholar]
- D’Avino, P.P., Giansanti M.G., and Petronczki M.. 2015. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 7:a015834. 10.1101/cshperspect.a015834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, T., Kodera N., Kaminski Schierle G.S., Rees E., Erdelyi M., Kaminski C.F., Ando T., and Mishima M.. 2015. CYK4 promotes antiparallel microtubule bundling by optimizing MKLP1 neck conformation. PLoS Biol. 13:e1002121. 10.1371/journal.pbio.1002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, T., Sundaramoorthy S., Jordan S.N., Shirasu-Hiza M., Dumont J., and Canman J.C.. 2017. Using fast-acting temperature-sensitive mutants to study cell division in Caenorhabditis elegans. Methods Cell Biol. 137:283–306. 10.1016/bs.mcb.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Dionne, L.K., Wang X.J., and Prekeris R.. 2015. Midbody: from cellular junk to regulator of cell polarity and cell fate. Curr. Opin. Cell Biol. 35:51–58. 10.1016/j.ceb.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, F., Maton G., Gareil N., Canman J.C., and Dumont J.. 2018. BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore. eLife. 7:e40690. 10.7554/eLife.40690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger, A.W., Wilsch-Bräuninger M., Marzesco A.M., Bickle M., Lohmann A., Maliga Z., Karbanová J., Corbeil D., Hyman A.A., and Huttner W.B.. 2011. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat. Commun. 2:503. 10.1038/ncomms1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer, U., and McIntosh J.R.. 1980. Polarity of midbody and phragmoplast microtubules. J. Cell Biol. 87:509–515. 10.1083/jcb.87.2.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant, E., Stefanutti M., Laband K., Gluszek-Kustusz A., Edwards F., Lacroix B., Maton G., Canman J.C., Welburn J.P.I., and Dumont J.. 2017. Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7 prevents chromosome segregation and cytokinesis defects in oocytes. Development. 144:1674–1686.http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=000000&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20. 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P., Schnabel H., Kaletta T., Amores A.D., Hyman T., and Schnabel R.. 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144:927–946. 10.1083/jcb.144.5.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G.J., Simerly C., Schatten G., and Borisy G.G.. 1990. Microtubules in the metaphase-arrested mouse oocyte turn over rapidly. Proc. Natl. Acad. Sci. USA. 87:6049–6053. 10.1073/pnas.87.16.6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Green, R.A., Mayers J.R., Wang S., Lewellyn L., Desai A., Audhya A., and Oegema K.. 2013. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J. Cell Biol. 203:505–520. 10.1083/jcb.201306036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, S.W., Gönczy P., Stelzer E.H., and Hyman A.A.. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633. 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Hajeri, V.A., Stewart A.M., Moore L.L., and Padilla P.A.. 2008. Genetic analysis of the spindle checkpoint genes san-1, mdf-2, bub-3 and the CENP-F homologues hcp-1 and hcp-2 in Caenorhabditis elegans. Cell Div. 3:6. 10.1186/1747-1028-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, A.K., and Gewalt S.L.. 1989. Simulation testing of mechanisms for inducing the formation of the contractile ring in cytokinesis. J. Cell Biol. 109:2215–2223. 10.1083/jcb.109.5.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, K., Kawashima T., Iwamoto I., Nosaka T., and Kitamura T.. 2001. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276:5821–5828. 10.1074/jbc.M007252200 [DOI] [PubMed] [Google Scholar]
- Hirsch, S.M., Sundaramoorthy S., Davies T., Zhuravlev Y., Waters J.C., Shirasu-Hiza M., Dumont J., and Canman J.C.. 2018. FLIRT: fast local infrared thermogenetics for subcellular control of protein function. Nat. Methods. 15:921–923. 10.1038/s41592-018-0168-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.K., Coughlin M., Field C.M., and Mitchison T.J.. 2008. Cell polarization during monopolar cytokinesis. J. Cell Biol. 181:195–202. 10.1083/jcb.200711105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.K., Coughlin M., and Mitchison T.J.. 2012. Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell. 23:1024–1034. 10.1091/mbc.e11-08-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y.H., Savoian M.S., Suzuki T., Máthé E., Yamamoto M.T., and Glover D.M.. 2004. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166:49–60. 10.1083/jcb.200402052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger, V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A.A., and Glotzer M.. 2000. CYK-4: a Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404. 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, S.N., Davies T., Zhuravlev Y., Dumont J., Shirasu-Hiza M., and Canman J.C.. 2016. Cortical PAR polarity proteins promote robust cytokinesis during asymmetric cell division. J. Cell Biol. 212:39–49. 10.1083/jcb.201510063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna, S., Mendoza M., Jantsch-Plunger V., and Glotzer M.. 2000. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181. 10.1016/S0960-9822(00)00721-1 [DOI] [PubMed] [Google Scholar]
- Kawamura, K. 1977. Microdissection studies on the dividing neuroblast of the grasshopper, with special reference to the mechanism of unequal cytokinesis. Exp. Cell Res. 106:127–137. 10.1016/0014-4827(77)90249-X [DOI] [PubMed] [Google Scholar]
- Kreis, T.E. 1987. Microtubules containing detyrosinated tubulin are less dynamic. EMBO J. 6:2597–2606. 10.1002/j.1460-2075.1987.tb02550.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, T.C., Chen C.T., Baron D., Onder T.T., Loewer S., Almeida S., Weismann C.M., Xu P., Houghton J.M., Gao F.B., et al. 2011. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13:1214–1223. 10.1038/ncb2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix, B., Letort G., Pitayu L., Sallé J., Stefanutti M., Maton G., Ladouceur A.M., Canman J.C., Maddox P.S., Maddox A.S., et al. 2018. Microtubule dynamics scale with cell size to set spindle length and assembly timing. Dev. Cell. 45:496–511.e6. 10.1016/j.devcel.2018.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino, J., and Ohi R.. 2016. The timing of midzone stabilization during cytokinesis depends on myosin II activity and an interaction between INCENP and actin. Curr. Biol. 26:698–706. 10.1016/j.cub.2016.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.Y., Davies T., and Mishima M.. 2012. Cytokinesis microtubule organisers at a glance. J. Cell Sci. 125:3495–3500. 10.1242/jcs.094672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.Y., Esmaeili B., Zealley B., and Mishima M.. 2015. Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat. Commun. 6:7290. 10.1038/ncomms8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.Y., Green R.A., Gutierrez E., Gomez-Cavazos J.S., Kolotuev I., Wang S., Desai A., Groisman A., and Oegema K.. 2018. CYK-4 functions independently of its centralspindlin partner ZEN-4 to cellularize oocytes in germline syncytia. eLife. 7:e36919. 10.7554/eLife.36919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev, S., Su K.C., Pye V.E., Blight K., Sundaramoorthy S., Takaki T., Collinson L.M., Cherepanov P., Divecha N., and Petronczki M.. 2012. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 492:276–279. 10.1038/nature11773 [DOI] [PubMed] [Google Scholar]
- Lewellyn, L., Dumont J., Desai A., and Oegema K.. 2010. Analyzing the effects of delaying aster separation on furrow formation during cytokinesis in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 21:50–62. 10.1091/mbc.e09-01-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn, L., Carvalho A., Desai A., Maddox A.S., and Oegema K.. 2011. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J. Cell Biol. 193:155–169. 10.1083/jcb.201008138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Wang Z., Jiang K., Zhang L., Zhao L., Hua S., Yan F., Yang Y., Wang D., Fu C., et al. 2009. PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 284:23059–23071. 10.1074/jbc.M109.009670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson, M.A., Horvitz H.R., and van den Heuvel S.. 2000. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J. Cell Biol. 148:73–86. 10.1083/jcb.148.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly, K., Reid S.J., and Snell R.G.. 2015. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX. 2:59–63. 10.1016/j.mex.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, A.M., Ainsztein A.M., Eckley D.M., and Earnshaw W.C.. 1998. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140:991–1002. 10.1083/jcb.140.5.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, A.S., Lewellyn L., Desai A., and Oegema K.. 2007. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev. Cell. 12:827–835. 10.1016/j.devcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Madeira, F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., and Earnshaw W.C.. 2003. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 113:891–904. 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- Mangal, S., Sacher J., Kim T., Osório D.S., Motegi F., Carvalho A.X., Oegema K., and Zanin E.. 2018. TPXL-1 activates Aurora A to clear contractile ring components from the polar cortex during cytokinesis. J. Cell Biol. 217:837–848. 10.1083/jcb.201706021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N., McDonald K.L., Ding R., and McIntosh J.R.. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489. 10.1083/jcb.123.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maton, G., Edwards F., Lacroix B., Stefanutti M., Laband K., Lieury T., Kim T., Espeut J., Canman J.C., and Dumont J.. 2015. Kinetochore components are required for central spindle assembly. Nat. Cell Biol. 17:697–705. 10.1038/ncb3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, M. 2016. Centralspindlin in Rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 53:45–56. 10.1016/j.semcdb.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Mishima, M., Kaitna S., and Glotzer M.. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54. 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman J.P., Jiang W., Schoehn G., Hunter T., and Margolis R.L.. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186. 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.L., Morrison M., and Roth M.B.. 1999. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 147:471–480. 10.1083/jcb.147.3.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, F., Velarde N.V., Piano F., and Sugimoto A.. 2006. Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev. Cell. 10:509–520. 10.1016/j.devcel.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Mullins, J.M., and McIntosh J.R.. 1982. Isolation and initial characterization of the mammalian midbody. J. Cell Biol. 94:654–661. 10.1083/jcb.94.3.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahaboo, W., Zouak M., Askjaer P., and Delattre M.. 2015. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner. Mol. Biol. Cell. 26:2020–2029. 10.1091/mbc.E14-12-1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc, T., Afshar K., and Gönczy P.. 2007. Coupling of cortical dynein and Gα proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9:1294–1302. 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- Park, D.H., and Rose L.S.. 2008. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev. Biol. 315:42–54. 10.1016/j.ydbio.2007.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, E., and Prekeris R.. 2019. The postmitotic midbody: regulating polarity, stemness, and proliferation. J. Cell Biol. 218:3903–3911. 10.1083/jcb.201906148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, E., Gibieža P., Schafer J., Skeberdis V.A., Kaupinis A., Valius M., Heiligenstein X., Hurbain I., Raposo G., and Prekeris R.. 2019. The post-abscission midbody is an intracellular signaling organelle that regulates cell proliferation. Nat. Commun. 10:3181. 10.1038/s41467-019-10871-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich, W.B., Moran A.N., Rothman J.H., and Hardin J.. 1998. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell. 9:2037–2049. 10.1091/mbc.9.8.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport, R. 1961. Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 148:81–89. 10.1002/jez.1401480107 [DOI] [PubMed] [Google Scholar]
- Rappaport, R. 1971. Cytokinesis in animal cells. Int. Rev. Cytol. 31:169–214. 10.1016/S0074-7696(08)60059-5 [DOI] [PubMed] [Google Scholar]
- Rappaport, R. 1973. Cleavage furrow establishment: a preliminary to cylindrical shape change. Am. Zool. 13:941–948. 10.1093/icb/13.4.941 [DOI] [Google Scholar]
- Rappaport, R. 1996. Cytokinesis in Animal Cells. Cambridge University Press, New York, 386 pp. 10.1017/CBO9780511529764 [DOI] [Google Scholar]
- Romano, A., Guse A., Krascenicova I., Schnabel H., Schnabel R., and Glotzer M.. 2003. CSC-1: a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J. Cell Biol. 161:229–236. 10.1083/jcb.200207117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, E.D., Goode D., Maugel T.K., and Bonar D.B.. 1976. Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J. Cell Biol. 69:443–454. 10.1083/jcb.69.2.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann, V., Chen C., Chiang C.Y., Tiyaboonchai A., Mayer M., and Yamashita Y.M.. 2014. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell. 25:267–275. 10.1091/mbc.e13-09-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian, M.S., Earnshaw W.C., Khodjakov A., and Rieder C.L.. 1999. Cleavage furrows formed between centrosomes lacking an intervening spindle and chromosomes contain microtubule bundles, INCENP, and CHO1 but not CENP-E. Mol. Biol. Cell. 10:297–311. 10.1091/mbc.10.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J.M., Golden A., and Donovan P.J.. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. 10.1083/jcb.143.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., Hamill D.R., Carter J.C., Schumacher J., and Bowerman B.. 2000. The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171. 10.1016/S0960-9822(00)00715-6 [DOI] [PubMed] [Google Scholar]
- Shelden, E., and Wadsworth P.. 1990. Interzonal microtubules are dynamic during spindle elongation. J. Cell Sci. 97:273–281. 10.1242/jcs.97.2.273 [DOI] [PubMed] [Google Scholar]
- Shrestha, S., Wilmeth L.J., Eyer J., and Shuster C.B.. 2012. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol. Biol. Cell. 23:1196–1207. 10.1091/mbc.e11-12-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., Tijsterman M., Parrish S., Koushika S.P., Nonet M.L., Fire A., Ahringer J., and Plasterk R.H.. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12:1317–1319. 10.1016/S0960-9822(02)01041-2 [DOI] [PubMed] [Google Scholar]
- Speliotes, E.K., Uren A., Vaux D., and Horvitz H.R.. 2000. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 6:211–223. 10.1016/S1097-2765(00)00023-X [DOI] [PubMed] [Google Scholar]
- Srinivasan, D.G., Fisk R.M., Xu H., and van den Heuvel S.. 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 17:1225–1239. 10.1101/gad.1081203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann, P., and Gerlich D.W.. 2009. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 19:606–616. 10.1016/j.tcb.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., and Mitchison T.J.. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 299:1743–1747. 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Su, K.C., Bement W.M., Petronczki M., and von Dassow G.. 2014. An astral simulacrum of the central spindle accounts for normal, spindle-less, and anucleate cytokinesis in echinoderm embryos. Mol. Biol. Cell. 25:4049–4062. 10.1091/mbc.e14-04-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy, S., Garcia Badaracco A., Hirsch S.M., Park J.H., Davies T., Dumont J., Shirasu-Hiza M., Kummel A.C., and Canman J.C.. 2017. Low efficiency upconversion nanoparticles for high-resolution coalignment of near-infrared and visible light paths on a light microscope. ACS Appl. Mater. Interfaces. 9:7929–7940. 10.1021/acsami.6b15322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo, M., Tarailo S., and Rose A.M.. 2007. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics. 177:2525–2530. 10.1534/genetics.107.080408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., Court D.L., and Fire A.. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Uehara, R., and Goshima G.. 2010. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 191:259–267. 10.1083/jcb.201004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, R., Kamasaki T., Hiruma S., Poser I., Yoda K., Yajima J., Gerlich D.W., and Goshima G.. 2016. Augmin shapes the anaphase spindle for efficient cytokinetic furrow ingression and abscission. Mol. Biol. Cell. 27:812–827. 10.1091/mbc.E15-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe, K.J., and White J.G.. 2004. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 14:1755–1760. 10.1016/j.cub.2004.09.055 [DOI] [PubMed] [Google Scholar]
- von Dassow, G., Verbrugghe K.J., Miller A.L., Sider J.R., and Bement W.M.. 2009. Action at a distance during cytokinesis. J. Cell Biol. 187:831–845. 10.1083/jcb.200907090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, M., Munro E., and Glotzer M.. 2007. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 17:1286–1297. 10.1016/j.cub.2007.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, S.P., and Wang Y.. 1996. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135:981–989. 10.1083/jcb.135.4.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.H., Redemann S., Wu H.Y., Kiewisz R., Yoo T.Y., Conway W., Farhadifar R., Müller-Reichert T., and Needleman D.. 2019. Central-spindle microtubules are strongly coupled to chromosomes during both anaphase A and anaphase B. Mol. Biol. Cell. 30:2503–2514. 10.1091/mbc.E19-01-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Bossy-Wetzel E., and Jiang W.. 2005. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem. J. 389:373–381. 10.1042/BJ20050097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists worm strains, plasmids, and primers.
shows the results of statistical analysis.