Abstract
Background
Blood‐based DNA methylation patterns are linked to types of diseases. FKBP prolyl isomerase 5 (FKBP5), a protein cochaperone, is known to be associated with the inflammatory response, but the regulatory mechanisms by leukocyte FKBP5 DNA methylation in patients with dilated cardiomyopathy (DCM) remain unclear.
Methods and Results
The present study enrolled patients with DCM (n=31) and age‐matched and sex‐matched control participants (n=43). We assessed FKBP5 CpG (cytosine‐phosphate‐guanine) methylation of CpG islands at the 5′ side as well as putative promoter regions by methylation‐specific quantitative polymerase chain reaction using leukocyte DNA isolated from the peripheral blood. FKBP5 CpG methylation levels at the CpG island of the gene body and the promoter regions were significantly decreased in patients with DCM. Leukocyte FKBP5 and IL‐1β (interleukin 1β) mRNA expression levels were significantly higher in patients with DCM than in controls. The protein expressions of DNMT1 (DNA methyltransferase 1) and DNMT3A (DNA methyltransferase 3A) in leukocytes were significantly reduced in patients with DCM. In vitro methylation assay revealed that FKBP5 promoter activity was inhibited at the methylated conditions in response to immune stimulation, suggesting that the decreased FKBP5 CpG methylation was functionally associated with elevation of FKBP5 mRNA expressions. Histological analysis using a mouse model with pressure overload showed that FKBP5‐expressing cells were substantially infiltrated in the myocardial interstitium in the failing hearts, indicating a possible role of FKBP5 expressions of immune cells in the cardiac remodeling.
Conclusions
Our findings demonstrate a link between specific CpG hypomethylation of leukocyte FKBP5 and DCM. Blood‐based epigenetic modification in FKBP5 may be a novel molecular mechanism that contributes to the pathogenesis of DCM.
Keywords: biomarker, dilated cardiomyopathy, DNA methylation, epigenetics, FKBP5
Subject Categories: Biomarkers, Cardiomyopathy, Epigenetics
DNA methylation is an epigenetic mechanism involving the covalent transfer of a methyl group to cytosine within the context of the cytosine‐phosphate‐guanine (CpG), thereby modifying the function and expressions of the genes. Blood‐based DNA methylation patterns are linked to types of diseases. Recently, it has been reported that epigenetic upregulation of FKBP prolyl isomerase 5 (FKBP5), a protein cochaperone that is acutely induced by stress, is associated with nuclear factor‐κB (NF‐κB)–driven inflammation. 1 Idiopathic dilated cardiomyopathy (DCM) is a myocardial disease characterized by progressive contractile dysfunction and ventricular dilatation, and persistent myocardial inflammation contributes to ventricular remodeling. 2 Peripheral leukocytes are recruited into the myocardial tissue and play important roles in the inflammatory response, but the regulatory mechanisms by DNA methylation in patients with DCM remain unclear.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
In the present study, we prospectively enrolled the patients with DCM (n=31) with symptomatic stage C/D heart failure and age‐matched and sex‐matched control participants (n=43) who had no history of any cardiovascular diseases at Fukushima Medical University Hospital between March 2018 and November 2019. DCM was diagnosed according to American College of Cardiology/American Heart Association guidelines based on echocardiography, coronary angiography, and endomyocardial biopsies by excluding other types of cardiomyopathies. Clinical data including laboratory tests and echocardiography were obtained with our standard methods. 2 The protocols were approved by the institutional ethical committee of Fukushima Medical University (approval number 29348). All participants provided written informed consent.
We isolated DNA from the leukocytes in the peripheral blood using a QuickGene DNA whole blood kit (Kurabo). A total of 200 ng of DNA was subjected to bisulfite conversion by using an EZ DNA Methylation‐Lightning Kit (Zymo Research) to convert unmethylated cytosines into uracils while methylated cytosines remain unchanged. Methylation‐specific quantitative polymerase chain reaction (PCR) was performed using the bisulfite‐treated DNA by the THUNDERBIRD SYBR qPCR Mix (Toyobo) in the CFX Connect Real‐Time PCR System (Bio‐Rad). We evaluated the methylation levels of 3 CpG sites of FKBP5 at the 5′ side using methylated and unmethylated specific primers (Figure [A]) 3 as follows: CpG‐1, methylation‐specific forward 5′‐TTTTAGTTTTTCGAGTAGTTGGGAC‐3′ and reverse 5′‐ATTCGACCAAATAAAATAACTCACG‐3′, unmethylation‐specific forward 5′‐TTTAGTTTTTTGAGTAGTTGGGATG‐3′ and reverse 5′‐TCAACCAAATAAAATAACTCACACC‐3′; CpG‐2, methylation‐specific forward 5′‐AAATTGGTTTTCGAGGTGGGATTTTTTTAGTTTCGAT‐3′ and reverse 5′‐TAAGTTAGTTGATTTTTTATTTGTATCGTTTTTTTGAA‐3′, unmethylation‐specific forward 5′‐AAATTGGTTTTTGAGGTGGGATTTTTTTAGTTTTGAT‐3′ and reverse 5′‐TAAGTTAGTTGATTTTTTATTTGTATTGTTTTTTTGAA‐3′; CpG‐3, 5′‐ATGATAGGTTTCGTTTCGTTTTC‐3′ and reverse 5′‐CTAAAATCCGATTATCCCGAC‐3′, unmethylation‐specific forward 5′‐GGATGATAGGTTTTGTTTTGTTTTT‐3′ and reverse 5′‐CCCTAAAATCCAATTATCCCAAC‐3′. The percentages of methylation levels were calculated by 100/[1+2ΔCt (methylation−unmethylation)]. 4 The primer specificities were validated using universally methylated DNAs (EpiScope Methylated HCT116 gDNA, Takara Bio) and unmethylated DNAs (EpiScope Unmethylated HCT116 DKO gDNA; Takara Bio). RNA isolation and reverse transcription–quantitative PCR was performed as previously described 5 by using the following primers: FKBP5 (human), forward 5′‐AGAGCTTCGAAAAGGCCAAAG‐3′ and reverse 5′‐CGCCTGCATGTATTTGCCTC‐3′; FKBP5 (mouse), forward 5′‐GCAACGGTAAAAGTCCACCTG‐3′ and reverse 5′‐TCCCAATCGGAATGTCGTGG‐3′; IL‐1β (interleukin 1β; human), forward 5′‐TTCGAGGCACAAGGCACAA‐3′ and reverse 5′‐TGGCTGCTTCAGACACTTGAG‐3′; GAPDH (human), forward 5′‐CCATGTTGCAACCGGGAAG‐3′ and reverse 5′‐GCCCAATACGACCAAATCAGAG‐3′; GAPDH (mouse), 5′‐ATGACAACTTTGTCAAGCTCATTT‐3′ and reverse 5′‐GGTCCACCACCCTGTTGCT‐3′. Samples were run in duplicates. For in vitro methylation assays, fragments of FKBP5 CpG sites containing CpG‐2 and CpG‐3 (GeneBank: NC_000006.12, position 35688298 to 35689546) were subcloned into luciferase reporter pGL3‐Basic (Promega) at the BglII and HindIII restriction sites. 1 , 6 The FKBP5‐luciferase reporter constructs were treated with CpG methyltransferase M. SssI (New England Biolabs) or buffer alone. RAW264.7 cells (American Type Culture Collection) were transfected with either unmethylated or methylated FKBP5‐luciferase plasmids together with pNL1.1.TK[Nluc/TK] as a control vector using Lipofectamine 3000 (Thermo Fisher Scientific). The cells were incubated with lipopolysaccharide (Wako), and the luciferase reporter activity was determined by Nano‐Glo Dual Luciferase Reporter Assay System (Promega). Western blot analysis was performed as described 5 using the following antibodies: DNMT1 (DNA methyltransferase 1; Cell Signaling Technology), DNMT3A (DNA methyltransferase 3A; Cell Signaling Technology), and GAPDH (Proteintech). For the animal study, mice aged 8 to 10 weeks were subjected to transverse aortic constriction or sham surgeries as previously described. 7 Optimal cutting temperature–embedded left ventricular samples were fixed by 4% paraformaldehyde and then stained with an anti‐FKBP5 antibody (Proteintech) and 4´,6‐diamidino‐2‐phenylindole. All animal studies were approved by Fukushima Medical University Animal Research Committee (approval number 2019026).
Figure 1. Decreases in leukocyte FKBP5 DNA methylation in patients with dilated cardiomyopathy.
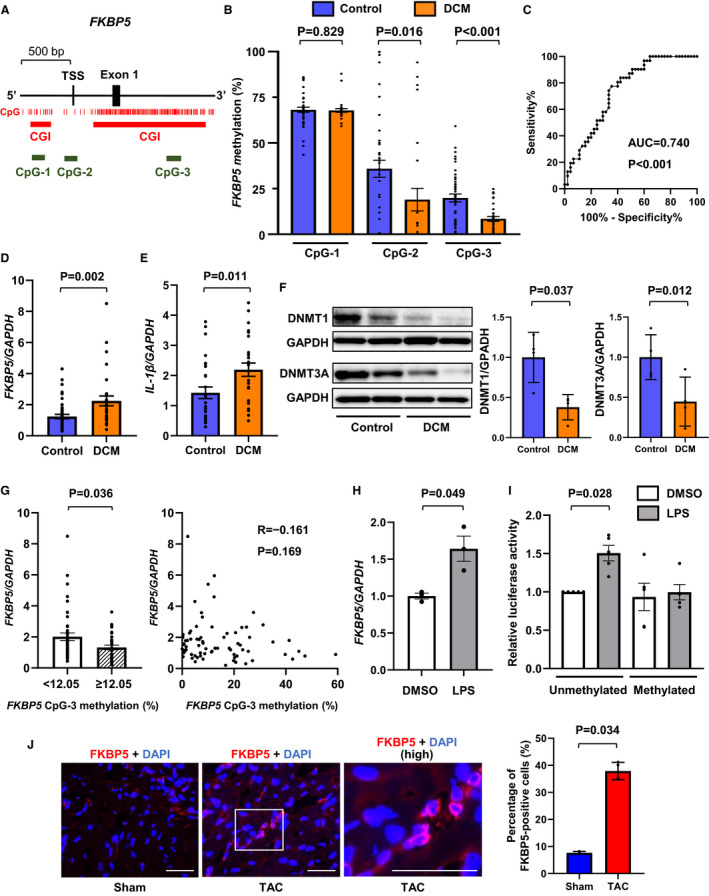
A, Schematic illustration of CpGs of FKBP5 at the 5′ side including TSS and 5′ untranslated region (Exon 1). Individual CpG, CpG island, and the positions for CpG methylation analyzed in the present study are indicated. B,FKBP5 methylation levels of each CpG in peripheral leukocytes in patients with DCM (n=31) in comparison with age‐matched and sex‐matched controls (n=43). C, Receiver operating characteristic curve to determine patients with DCM from controls by FKBP5 CpG‐3 methylation levels. D and E,FKBP5 (D) and IL‐1β mRNA (E) expression levels in leukocytes. The data were normalized by GAPDH. F, DNMT1 and DNMT3A expressions in leukocytes by Western blotting (n=4 in each). The data were normalized by GAPDH and expressed over the controls. G (left), FKBP5 mRNA levels in patients with <12.05% FKBP5 CpG‐3 methylation were significantly higher than in those with ≥12.05% using the cut‐off value from C. G (right), Scatterplot of FKBP5 mRNA and FKBP5 CpG‐3 methylation levels. H, mRNA expressions of FKBP5 were increased in response to immune stimulation. RAW264.7 cells were treated with 100 nmol/L LPS for 6 hours. The data were normalized by GAPDH. I, Effect of methylation on FKBP5 gene promoter by a dual luciferase reporter assay. The pGL3‐basic vector containing the sequences of FKBP5 CpG‐2 and CpG‐3 (TSS‐214/1052) was methylated by the CpG methyltransferase. The methylated or unmethylated reporter constructs were transfected with pNL1.1.TK[Nluc/TK] as a control vector in RAW264.7 cells. At 24 hours after transfection, the cells were incubated with 100 nmol/L LPS for 24 hours, and the relative luciferase activity was determined by the ratio of firefly luciferase to Nano luciferase activity (n=5). J, The percentage of FKBP5‐expressing cell infiltration was increased in the pressure overload‐induced failing hearts in mice. Male C57BL/6 mice aged 8 to 10 weeks were subjected to sham and TAC for 4 weeks. The heart sections immunostained with an anti‐FKBP5 antibody (red) and DAPI (blue) are shown. Images in boxed areas at higher magnification are shown on the right (high). Scale bars=50 μm. The graph shows quantitative analysis of the percentage of FKBP5‐positive cells (n=3–4). The statistical comparison was performed by Mann–Whitney U test or 1‐way analysis of variance followed by multiple comparisons with the Tukey test. All data are presented as mean±SEM. AUC indicates area under the curve; bp, base pairs; CGI, CpG island; CpG, cytosine‐phosphate‐guanine; DAPI, 4´,6‐diamidino‐2‐phenylindole; DCM, dilated cardiomyopathy; DMSO, dimethyl sulfoxide; DNMT1, DNA methyltransferase 1; DNMT3A, DNA methyltransferase 3A; FKBP5, FKBP prolyl isomerase 5; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; IL‐1β, interleukin 1β; LPS, lipopolysaccharide; TAC, transverse aortic constriction; and TSS, transcription start site.
The data are expressed as mean±SD, mean±SE, median (interquartile range), or number (percentage). Comparisons of the values were performed using unpaired Student t tests, Mann–Whitney U tests, or 1‐way analysis of variance followed by multiple comparisons with the Tukey test. Categorical variables were compared using the chi‐square test with the Yates correction or the Fisher exact test. Correlations were analyzed by the Spearman correlation test. The receiver operating characteristic curve was constructed to determine patients with DCM from controls. A multivariable linear regression analysis was used to determine the factors related to the FKBP5 methylation level. A value of P<0.05 was considered statistically significant. All data were analyzed using Statistical Package for Social Sciences version 26 software (SPSS Inc).
Results
Comparisons of clinical characteristics between patients with DCM and controls are shown in Table 1. Among patients with DCM, the age was 57.2±16.0 years, 80.6% were men, the left ventricular ejection fraction was 32.3±16.0%, and BNP (B‐type natriuretic peptide) was 172.0 pg/mL (median [25%, 75% quantiles: 69.9, 265.3]). There was no significant difference in age or sex between patients with DCM and control participants, although the prevalence of diabetes, dyslipidemia, and atrial fibrillation was higher in patients with DCM than in controls. As CpG island methylation plays an important role in epigenetic gene control such as transcriptional regulation, differential promoter usage, transcription elongation, and alternative splicing, 8 we assessed FKBP5 CpG methylation levels of CpG islands at the 5′ side, which is located at the upstream of the transcriptional start site (CpG‐1), and the gene body region, which is linked to the 5′ untranslated region (CpG‐3), as well as putative promoter regions 1 (CpG‐2) using leukocyte DNA isolated from the peripheral blood (Figure [A]). Interestingly, FKBP5 methylation levels at CpG‐2 and CpG‐3 were significantly lower in patients with DCM than in control participants, whereas those at CpG‐1 were not different between the 2 groups (Figure [B]). Although each CpG methylation alternation may have differential functional consequences, it is likely that the CpG‐3 was the most altered CpG of FKBP5 in patients with DCM among the investigated CpGs. Then, we performed receiver operating characteristic analysis demonstrating that FKBP5 CpG‐3 methylation significantly discriminated patients with DCM from control participants with the area under the curve of 0.740 (95% CI, 0.631–0.849; P<0.001; Figure [C]). A multivariable linear regression analysis adjusted for confounders including age, sex, body mass index, white blood cell count, hemoglobin, and C‐reactive protein demonstrated that the FKBP5 CpG‐3 methylation level was independently associated with the presence of DCM (β=–0.537; P=0.003). Correlation analyses in patients with DCM showed that FKBP5 CpG‐3 methylation levels were not associated with clinical characteristics such as age, sex, smoking, echocardiographic parameters, or BNP, suggesting that FKBP5 hypomethylation was associated with the presence of DCM independently of patient background or the severity of the disease (Table 2). FKBP5 CpG‐3 methylation level in patients with DCM was 7.300±1.299% in men and 9.670±1.742% in women, and there was no difference between them (P=0.261). Next, we evaluated mRNA levels in the peripheral leukocytes, showing that FKBP5 mRNA expression levels were significantly higher in patients with DCM than in controls (Figure [D]). In addition, IL‐1β mRNA levels were significantly increased in patients with DCM compared with controls (Figure [E]). DNMTs are involved in maintaining DNA methylation and de novo methylation. The leukocyte protein expression levels of both DNMT1 and DNMT3A were significantly decreased in patients with DCM (Figure [F]). By categorizing the patients with the cut‐off value from receiver operating characteristic analysis, FKBP5 mRNA levels in patients with <12.05% FKBP5 CpG‐3 methylation were significantly higher than in those with ≥12.05% (Figure [G]), indicating that the decreased FKBP5 methylation levels were associated with the elevated FKBP5 mRNA expressions. A negative correlation between FKBP5 CpG‐3 methylation and mRNA expression was found, but no statistical significance was reached (R=–0.161, P=0.169; Figure [G]). To assess the direct effect of FKBP5 CpG methylation on gene expression, we performed in vitro methylation assay in a RAW264.7 monocyte/macrophage cell line. FKBP5 mRNA was upregulated in response to immune stimulation by lipopolysaccharide (Figure [H]). The cells transfected with the unmethylated FKBP5‐luciferase reporter containing the sequences of FKBP5 CpG‐2 and CpG‐3 showed significant increases in promoter activity by lipopolysaccharide stimulation, whereas the methylated FKBP5‐luciferase constructs abolished the induction of the activity (Figure [I]). To know the possible role of FKBP5 of immune cells in the cardiac remodeling, histological analysis using a mouse model with pressure overload‐induced heart failure by transverse aortic constriction showed that FKBP5‐expressing cells were more substantially infiltrated among the myocardial interstitium in the failing mouse hearts than in the nonfailing hearts with a sham operation (Figure [J]).
Table 1.
Baseline Clinical Characteristics of the 2 Groups
| Controls (n=43) | DCM (n=31) | P value | |
|---|---|---|---|
| Age, y | 62.9±13.6 | 57.2±16.0 | 0.198 |
| Male participants | 32 (74.4) | 25 (80.6) | 0.728 |
| Body mass index, kg/m2 | 25.5±4.0 | 23.6±4.7 | 0.110 |
| New York Heart Association functional class, Ⅰ/Ⅱ/Ⅲ/Ⅳ | … | 0 (0.0)/5 (16.1)/21 (67.7)/5 (16.1) | … |
| Comorbidities | |||
| Hypertension | 23 (53.4) | 18 (58.0) | 0.878 |
| Diabetes | 2 (4.6) | 7 (22.5) | 0.030 |
| Dyslipidemia | 7 (16.2) | 14 (45.1) | 0.014 |
| Atrial fibrillation | 0 (0.0) | 11 (35.4) | <0.001 |
| Laboratory data | |||
| White blood cell, ×103/μL | 5.8±1.3 | 6.8±2.1 | <0.001 |
| Hemoglobin, g/dL | 14.4±1.6 | 14.3±1.5 | 0.671 |
| Platelet, ×103/μL | 218.1±45.4 | 235.4±83.3 | 0.325 |
| Aspartate aminotransferase, U/L | 25.7±12.3 | 28.4±15.4 | 0.400 |
| Alanine transaminase, U/L | 25.6±19.4 | 27.7±23.6 | 0.661 |
| Albumin, g/dL | 4.3±0.3 | 3.8±0.5 | 0.001 |
| Creatinine, mg/dL | 0.8±0.1 | 1.0±0.2 | 0.001 |
| Hemoglobin A1c, % | 5.9±0.4 | 5.7±0.6 | 0.198 |
| Uric acid, mg/dL | 5.3±1.0 | 6.8±2.0 | 0.001 |
| Low‐density lipoprotein cholesterol, mg/dL | 105.3±22.4 | 106.2±22.5 | 0.939 |
| C‐reactive protein, mg/dL | 0.10 (0.06–0.15) | 0.32 (0.08–1.09) | 0.036 |
| B‐type natriuretic peptide, pg/mL | 15.4 (10.8–50.8) | 172.5 (72.0–310.4) | <0.001 |
| Troponin I, ng/mL | … | 0.024 (0.017–0.106) | … |
| Echocardiography | |||
| Left ventricular end‐diastolic dimension, mm | 44.8±5.4 | 61.2±9.5 | <0.001 |
| Left ventricular ejection fraction, % | 65.2±3.9 | 32.3±16.2 | <0.001 |
| Medication | |||
| Renin‐angiotensin system inhibitors | 0 | 27 (87.1) | <0.001 |
| β‐blockers | 0 | 26 (83.9) | <0.001 |
| Diuretics | 0 | 21 (67.7) | <0.001 |
Data are presented as mean±SD, median (interquartile range), or number (percentage). DCM indicates patients with dilated cardiomyopathy.
Table 2.
Correlation Analyses of FKBP5 CpG‐3 Methylation and Other Parameters in Patients With Dilated Cardiomyopathy
| Factors | FKBP5 CpG‐3 methylation | |
|---|---|---|
| r | P value | |
| Age, y | 0.180 | 0.333 |
| Male participants (0=no, 1=yes) | 0.117 | 0.359 |
| Body mass index, kg/m2 | −0.255 | 0.174 |
| Laboratory data | ||
| White blood cell, ×103/μL | −0.229 | 0.223 |
| Hemoglobin, g/dL | −0.321 | 0.110 |
| Platelet, ×103/μL | −0.160 | 0.417 |
| Aspartate aminotransferase, U/L | −0.018 | 0.923 |
| Alanine transaminase, U/L | −0.284 | 0.122 |
| Albumin, g/dL | −0.177 | 0.430 |
| Creatinine, mg/dL | 0.153 | 0.410 |
| Hemoglobin A1c, % | 0.074 | 0.802 |
| Uric acid, mg/dL | −0.203 | 0.390 |
| Low‐density lipoprotein cholesterol, mg/dL | −0.299 | 0.299 |
| C‐reactive protein, mg/dL | 0.208 | 0.289 |
| B‐type natriuretic peptide, pg/mL | −0.029 | 0.875 |
| Troponin I, ng/mL | 0.185 | 0.337 |
| Echocardiography | ||
| Left ventricular end‐diastolic dimension, mm | 0.018 | 0.930 |
| Left ventricular ejection fraction, % | −0.020 | 0.913 |
CpG indicates cytosine‐phosphate‐guaninea and FKBP5, FKBP prolyl isomerase 5.
Discussion
The present study was the first to demonstrate a link between CpG hypomethylation of the leukocyte FKBP5 and DCM. Emerging evidence implicates that blood‐based biomarkers of epigenetic DNA methylation are associated with specific diseases including diabetes. Age‐related changes in DNA methylation of pancreatic islets were similarly seen in that of blood. 9 A single methylated CpG directly affects transcription factor binding and cellular regulation. 10 Likewise, we suggested here that the downregulation of specific methylated CpGs in FKBP5 might be associated with epigenetic upregulation of FKBP5 and related to the inflammatory response in the myocardium as sterile chronic inflammation is important for the pathogenesis of DCM and heart failure.
CpG methylation in promoter regions is profoundly associated with transcriptional repression, whereas the function of gene body methylation remains unknown. 8 We showed that CpG islands in FKBP5 gene are located at the gene body of the 5′ untranslated region, in addition to the upstream of the transcription start site. Although differential CpG methylation may have a different impact on transcriptional regulations, our data revealed that FKBP5 methylations at promoter regions and the gene body CpG island were similarly downregulated in patients with DCM, which functionally enhanced FKBP5 expression associated with promoter regulation. FKBP5 acts as a cochaperone that modulates glucocorticoid receptor activity in response to stressors and influences the types of cellular biological processes. 1 Epigenetic upregulation of FKBP5 promotes inflammation by activating the immune regulator NF‐κB. 1 It is known that IL‐1β, promoted through activation of NF‐κB, was increased in heart failure, and IL‐1β had a negative inotropic effect on cardiomyocytes by downregulating Ca2+ re‐uptake. 11
We found a lack of significant correlations of FKBP5 methylation levels with age or other characteristics in patients with DCM. In turn, investigation of FKBP5 methylation may be an independent biomarker for the presence of DCM. The associations of general effects of heart failure with FKBP5 CpG methylation need to be clarified. Given that FKBP5 positive‐cells were markedly observed in the murine failing hearts, FKBP5 expressions in the infiltrated cells might be involved in myocardial damage attributed to chronic inflammatory responses. Although numbers and patterns of CpGs and CpG islands are different between humans and mice, 12 differential methylation of FKBP5 orthologues in the infiltrated FKBP5‐expressing cells in the myocardium remains to be determined. Further studies are needed to validate our findings in independent larger cohorts, and the precise mechanisms and causal relationships between FKBP5 hypomethylation and DCM pathogenesis remain to be elucidated in the future.
Sources of Funding
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grant 20K17088 to Dr Wada.
Disclosures
Dr Kaneshiro belongs to a department sponsored by Biotronik Japan, Abbott Japan, and Nihon Kohden Kogyo. These companies are not associated with the contents of this study. The remaining authors have no disclosures to report.
Acknowledgments
We thank Chisato Kubo at the Office for Gender Equality Support, Fukushima Medical University, and Tomiko Miura and Shoko Sato at Department of Cardiovascular Medicine, Fukushima Medical University, for their excellent technical assistance.
For Sources of Funding and Disclosures, see page 6.
References
- 1. Zannas AS, Jia M, Hafner K, Baumert J, Wiechmann T, Pape JC, Arloth J, Ködel M, Martinelli S, Roitman M, et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF‐κB‐driven inflammation and cardiovascular risk. Proc Natl Acad Sci USA. 2019;116:11370–11379. doi: 10.1073/pnas.1816847116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Misaka T, Yoshihisa A, Yokokawa T, Sato T, Oikawa M, Kobayashi A, Yamaki T, Sugimoto K, Kunii H, Nakazato K, et al. Plasma levels of melatonin in dilated cardiomyopathy. J Pineal Res. 2019;66:e12564. doi: 10.1111/jpi.12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- 4. Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high‐throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yokokawa T, Misaka T, Kimishima Y, Wada K, Minakawa K, Sugimoto K, Ishida T, Morishita S, Komatsu N, Ikeda K, et al. Crucial role of hematopoietic JAK2V617F in the development of aortic aneurysms. Haematologica. 2021;106:1910–1922. doi: 10.3324/haematol.2020.264085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li D, Da L, Tang H, Li T, Zhao M. CpG methylation plays a vital role in determining tissue‐ and cell‐specific expression of the human cell‐death‐inducing DFF45‐like effector a gene through the regulation of Sp1/Sp3 binding. Nucleic Acids Res. 2008;36:330–341. doi: 10.1093/nar/gkm1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Misaka T, Murakawa T, Nishida K, Omori Y, Taneike M, Omiya S, Molenaar C, Uno Y, Yamaguchi O, Takeda J, et al. FKBP8 protects the heart from hemodynamic stress by preventing the accumulation of misfolded proteins and endoplasmic reticulum‐associated apoptosis in mice. J Mol Cell Cardiol. 2018;114:93–104. doi: 10.1016/j.yjmcc.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 9. Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O, Gjesing AP, Eiberg H, Tuomi T, Almgren P, et al. Blood‐based biomarkers of age‐associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. doi: 10.1038/ncomms11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang L, Chen Z, Stout ES, Delerue F, Ittner LM, Wilkins MR, Quinlan KGR, Crossley M. Methylation of a cgata element inhibits binding and regulation by GATA‐1. Nat Commun. 2020;11:2560. doi: 10.1038/s41467-020-16388-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Tassell BW, Toldo S, Mezzaroma E, Abbate A. Targeting interleukin‐1 in heart disease. Circulation. 2013;128:1910–1923. doi: 10.1161/CIRCULATIONAHA.113.003199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995 [DOI] [PMC free article] [PubMed] [Google Scholar]


