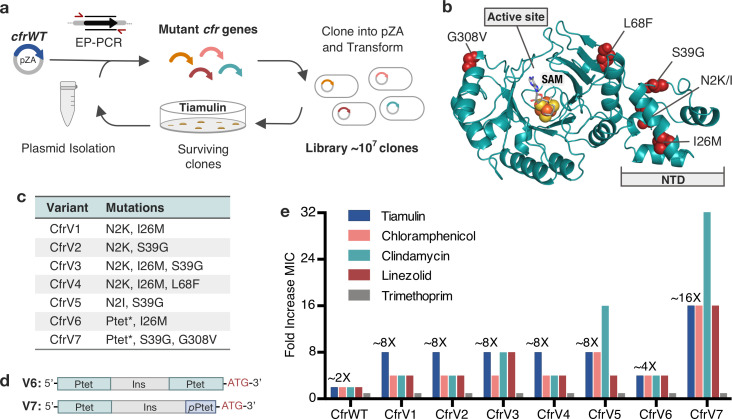
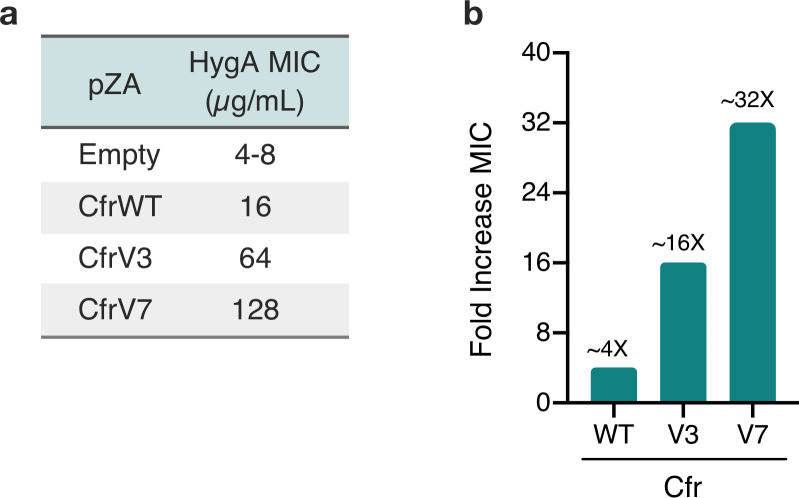
Figure 1. Evolved variants of Cfr exhibit improved resistance to PhLOPSA ribosome antibiotics.
(a) Evolution of Cfr under selection by the PTC-targeting antibiotic tiamulin. (b) Cfr homology model based on RlmN generated by I-TASSER server (Yang and Zhang, 2015) with mutagenic hotspots in red. N-terminal domain (NTD) is labeled. Active site denoted by S-adenosylmethionine (SAM, gray) and [4Fe-4S] cluster (orange). (c) Evolved variants containing Cfr mutations were selected for further study. Ptet* indicates alterations to promoter sequence. (d) Promoter architecture of CfrV6 and CfrV7 where pPtet designates a partial Ptet promoter sequence and Ins designates a variable insertion sequence. (e) Fold improvement in MIC resistance value for PhLOPSA antibiotics and trimethoprim compared to empty pZA vector control determined from three biological replicates by microbroth dilution method. Trimethoprim is a negative control antibiotic that does not target the ribosome. LZD testing was performed against Escherichia coli BW25113 lacking efflux pump, acrB. Numerical MIC values are displayed in Figure 1—source data 1.