ABSTRACT
The improvement of genome sequencing techniques has brought to light the biosynthetic potential of actinomycetes due to the large number of gene clusters they present compared to the number of known compounds. Genome mining is a recent strategy in the search for novel bioactive compounds, which involves the analysis of sequenced genomes to identify uncharacterized natural product biosynthetic gene clusters, many of which are cryptic or silent under laboratory conditions, and to develop experimental approaches to identify their products. Owing to the importance of halogenation in terms of structural diversity, bioavailability, and bioactivity, searching for new halogenated bioactive compounds has become an interesting issue in the field of natural product discovery. Following this purpose, a screening for halogenase coding genes was performed on 12 Streptomyces strains isolated from fungus-growing ants of the Attini tribe. Using the bioinformatics tools antiSMASH and BLAST, six halogenase coding genes were identified. Some of these genes were located within biosynthetic gene clusters (BGCs), which were studied by construction of several mutants for the identification of the putative halogenated compounds produced. The comparison of the metabolite production profile of wild-type strains and their corresponding mutants by ultrahigh-performance liquid chromatography-UV and high-performance liquid chromatography-mass spectrometry allowed us the identification of a novel family of halogenated compounds in Streptomyces sp. strain CS147, designated colibrimycins.
IMPORTANCE Genome mining has proven its usefulness in the search for novel bioactive compounds produced by microorganisms, and halogenases comprise an interesting starting point. In this work, we have identified a new halogenase coding gene that led to the discovery of novel lipopetide nonribosomal peptide synthetase/polyketide synthase (NRPS/PKS)-derived natural products, the colibrimycins, produced by Streptomyces sp. strain CS147, isolated from the Attini ant niche. Some colibrimycins display an unusual α-ketoamide moiety in the peptide structure. Although its biosynthetic origin remains unknown, its presence might be related to a hypothetical inhibition of virus proteases, and, together with the presence of the halogenase, it represents a feature to be incorporated in the arsenal of structural modifications available for combinatorial biosynthesis.
KEYWORDS: halogenases, lipopeptides, α-ketoamide, biosynthetic gene clusters, polyketides, nonribosomal peptides
INTRODUCTION
In the 1950s, decades following the discovery of penicillin in 1928, the number of antibiotics discovered increased exponentially, and many infectious diseases almost disappeared (1). However, after this golden age, microbial resistance to antibiotics increased exponentially, while classical screening methods became inefficient and the rate of discovery of novel compounds and new drugs introduced into the market decreased, increasing the costs of the studies and reducing the funding in this field (1). Furthermore, old pathogens are reappearing and new pathogens are emerging, and this, together with the increasing incidence of cancer and the urgent need of more effective and specific antitumor drugs, has intensified the search for novel bioactive compounds.
Natural products (NPs) and their derivatives make a total of 65% of new drugs approved by the U.S. FDA (Food and Drug Administration) since 1981 to 2014 (2) due to their chemical diversity, biochemical specificity, binding efficiency, and high affinity in the interaction with biological systems (3). Among them, most of the bioactive microbial NPs are produced by Actinobacteria (1), a broad and heterogeneous group of filamentous and aerobic Gram-positive bacteria with high G+C content (4, 5). Within this group, the genus Streptomyces provides 80% of the already known actinomycete secondary metabolites (6).
New strategies in the search for novel NPs are required due to the dramatic decrease in the number of bioactive compounds discovered. Genome sequencing has revealed that in the genus Streptomyces, 5 to 10% of the genome is dedicated to secondary metabolism (7). The production of secondary metabolites is determined by biosynthetic gene clusters (BGCs), many of which are silent under standard laboratory conditions, and their products remain undiscovered, representing a great potential for the search of novel bioactive NPs (8). Genome mining involves the identification of uncharacterized BGCs within the genomes of sequenced organisms, sequence analysis of the enzymes encoded by them, and the experimental identification of their products using different molecular biology techniques (9).
Among others, one approach to study the presence of putatively novel BGCs is to determine the presence of specific structural genes as indicators (e.g., halogenases). Many bioactive NPs are halogenated compounds, for instance, the antibiotics vancomycin and chloramphenicol, the antitumor rebeccamycin, or the antifungal pyrrolnitrin (10). The halogen moiety is important for their specificity, effectivity, and stability, increasing their bioavailability and bioactivity (11). In many cases, such as vancomycin, the absence of this moiety causes a great reduction of activity (12).
Halogenases introduce halogen atoms into a range of compounds, such as indoles, phenols, terpenes, peptides, or polyketides (10). Brominated metabolites predominate in the marine environment due to the higher bromide ion content, while chlorinated metabolites predominate in the terrestrial environment (13). Some halogenases, as is the case of some FADH2-dependent halogenases, can accept both chloride or bromide (14), while others perform bromination over chlorination, although few halogenases accept only bromide ions (15). There are several types of halogenases classified according to different criteria, such as their enzymatic mechanism, cofactors, or preferred substrates. According to the cofactor they use, halogenases commonly involved in the halogenation of NPs in microorganisms can be divided into three types, SAM-dependent, non-heme/Fe(II)-oxoglutarate-dependent, and FAD-dependent (FDHs), that commonly halogenate tryptophan (Trp) but also a range of substrates, such as pyrroles and aliphatic activated compounds (10–12, 14, 16–18). Trp halogenases are also classified as Trp 5-, 6-, and 7-halogenases, which have key conserved residues that determine the difference in regioselectivity (19). The different types of halogenases have characteristic conserved motifs useful in bioinformatic analysis for the detection of halogenase coding genes, such as the N-terminal GXGXXG and the C-terminal GWXWXIP of FDHs (10); FPRYHIGES, GVGEATIP, DLFIDCSGFR, and FVEPLSSG specific for Trp halogenases (20–22); or NYDRHLDXXXL, PKYPGDEGTDWHQ, and GFFGYDY in non-heme/Fe(II)-oxoglutarate-dependent halogenases (23).
Symbiotic relationships between actinomycetes and insects constitute an environment in which the search for new NPs can be carried out (5, 24, 25). In the relationship established between Attini tribe leaf-cutting fungus-growing ants and Pseudonocardia, bacteria produce antifungal compounds inhibiting Escovopsis sp. fungi, which infects the Basidiomycota fungi that the ants cultivate as a food source (25–27). There are also other bacterial species associated with the ants in the same ecological niche. In this work, we searched for novel halogenated secondary metabolites in a collection of 12 Streptomyces strains isolated from the surface of Attini ants (Acromyrmex octospinosus) in Lambayeque (Perú), using bioinformatics tools to identify halogenases within new BGCs. Analysis by ultrahigh-performance liquid chromatography-UV (UPLC-UV), high-performance liquid chromatography (HPLC)-high-resolution mass spectrometry (HRMS), nuclear magnetic resonance (NMR), and dereplication of metabolic profiles from several constructed mutants led to the discovery of novel halogenated NRPS-PKS products, which we have designated colibrimycins.
RESULTS
Bioinformatics mining for halogenase genes.
We ran BLAST (Basic Local Alignment Search Tool) analysis against the genome sequences of 12 Streptomyces sp. strains, CS014, CS057, CS065a, CS081a, CS090a, CS113, CS131, CS147, CS149, CS159, CS207, and CS227, using as a query several well-characterized halogenases (belonging to different types), leading to the identification of six putative halogenase coding genes (Table 1).
TABLE 1.
Halogenases detected by bioinformatic analysis in Streptomyces sp. strainsa
| Strain | Gene | Annotation | Halogenase model/% similarity | Conserved motif | Size (no. of amino acids) | Cluster no./type |
|---|---|---|---|---|---|---|
| CS081a | DBP18-RS02065 | Chlorinating enzyme | SyrB2: 55 | NYDRHLDXXXL, PKYPGDEGTDWHQ, GFFGYDY | 323 | 4: NRPS Butyrolactone |
| CS113 | B9W62- 33020 | Halogenase (FAD-dep) | PrnC: 41 | GGGIGG | 585 | |
| ClmS: 22 | WFWVIP | |||||
| CS147 | DBP21-RS21585 | Tryptophan halogenase | RebH: 38 | GWIWTIP, GGGTAGWM | 547 | 15: NRPS-ladderane |
| PyrH: 55 | DLFIDCTGFR | |||||
| PrnA: 39 | GVGEATF, FVEPLESTG | |||||
| CS159 | B9W64-RS22640 | Hypothetical protein | PrnC: 43 | GSGIAG | 562 | 17: PKS II |
| ClmS: 25 | GWLWIIP | |||||
| CS159 | B9W64-31600 | Halogenase (FAD-dep) | PrnC: 43 | GGGIGG | 585 | |
| ClmS: 23 | WLWVIP | |||||
| CS207 | DBP22-13900 | Halogenase (FAD-dep) | PrnC: 42 | GGGIGG | 585 | |
| ClmS: 24 | WFWVIP |
Accession numbers (UniProtKB): Q8KHZ8 for RebH, A4D0H5 for PyrH, P95480 for PrnA, P95482 for PrnC, and A0A6H0CYP4-9ACTN for CmlS.
SalI, which participates in salinosporamide A biosynthesis in Salinispora tropica (28), was used as a model of SAM-dependent halogenases; SyrB2, involved in syringomycin biosynthesis by Pseudomonas syringae (29), was used as a model of non-heme/Fe(II)-oxoglutarate-dependent halogenases; PrnC and CmlS, involved in pyrrolnitrin (30) and chloramphenicol biosynthesis (31), respectively, were used as representatives of enzymes halogenating pyrroles and aliphatic activated compounds. RebH, PyrH, and PrnA, from rebeccamycin (32), pyrroindomycin (22), and pyrrolnitrin biosynthetic pathways (33), were used as representatives of Trp halogenases (Table 1).
In the analysis of the 12 genomes screened, no significant homology was found using SalL SAM-dependent halogenase, and only one similar to SyrB2 non-heme/Fe(II)-oxoglutarate-dependent halogenase was detected in strain CS081a. The five remaining genes identified corresponded to FAD-dependent halogenases, four of which exhibited similarity with FAD-dependent CmlS and PrnC: two in CS159, one in CS113, and one in CS207. The remaining FAD-dependent halogenase, detected in CS147, showed high similarity to Trp halogenases RebH, PyrH, and PrnA.
From a bioinformatic perspective, in addition to the percentages of similarity with other halogenases, the presence of conserved motifs typical from FAD-dependent, Trp, or non-heme/Fe(II)-oxoglutarate-dependent halogenases seemed to confirm that the genes identified code for the expected enzymes (Table 1). Furthermore, similar to other halogenase-containing clusters, FAD-oxidase-like genes were detected close to those encoding the putative halogenase (18), and the deduced protein sizes were similar to that of other halogenases, being approximately 500 amino acids in the case of FAD-dependent halogenases (18) and 300 in the case of non-heme/Fe(II)-oxoglutarate-dependent halogenases (23).
Bioinformatic analysis of BGCs containing halogenases.
The three FAD-dependent halogenase genes identified by bioinformatic analysis in strains CS113, CS159, and CS207 were not located within any BGC identified by anti-SMASH under commonly established parameters. However, in the case of CS113, analysis of their upstream and downstream regions to identify their surrounding genes revealed the presence of some regulatory genes and biosynthetic structural features characteristic of secondary metabolism BGCs. In the case of halogenases identified in strains CS159 and CS207, their surrounding genes are more likely involved in primary metabolism. According to anti-SMASH analysis, the remaining halogenases from CS081a, CS159, and CS147 strains were located in BGCs.
Cluster 4 from Streptomyces sp. strain CS081a is an 80-kb NRPS-butyrolactone BGC in which a non-heme Fe(II)-α-oxoglutarate-dependent type halogenase was located (Table 1). Based on its genetic organization (see Fig. S1A and Table S1 at https://figshare.com/s/526a9777225b6750159c) and their homologous BGCs in other actinomycete strains (see Fig. S1B at https://figshare.com/s/526a9777225b6750159c), the cluster might extend from orf19 to orf35, and it does not show similarities to any already-characterized BGC. Thus, this BGC could determine the production of a putative halogenated nonribosomal peptide not characterized in this work.
Cluster 17 from Streptomyces sp. strain CS159 is a 40-kb type II PKS BGC that probably codes for the biosynthetic route of a type II halogenated and glycosylated polyketide based on the presence of the FAD-dependent halogenase coding gene, one gene encoding a glycosyltransferase, and the beta-ketoacyl-synthase, reductase, cyclase, and dehydratase domains (see Fig. S2A, Table S2 at https://figshare.com/s/526a9777225b6750159c). According to the homologies predicted by antiSMASH, with a 22% of similarity to the arixanthomycin BGC (34), the product determined by this BGC might be related to pentangular polyphenols, a group of cyclized polyketide synthesized by type II PKS BGC with a central conserved region that include a cyclase and two type II PKS genes, which is also present in cluster 17 from CS159 (see Fig. S2B at https://figshare.com/s/526a9777225b6750159c). There are examples of chlorinated pentangular polyphenols, such as xantholipin B or allocyclinones. Their corresponding BGCs (35, 36) contain halogenase genes homologous to the FAD-dependent halogenase located at cluster 17 in CS159.
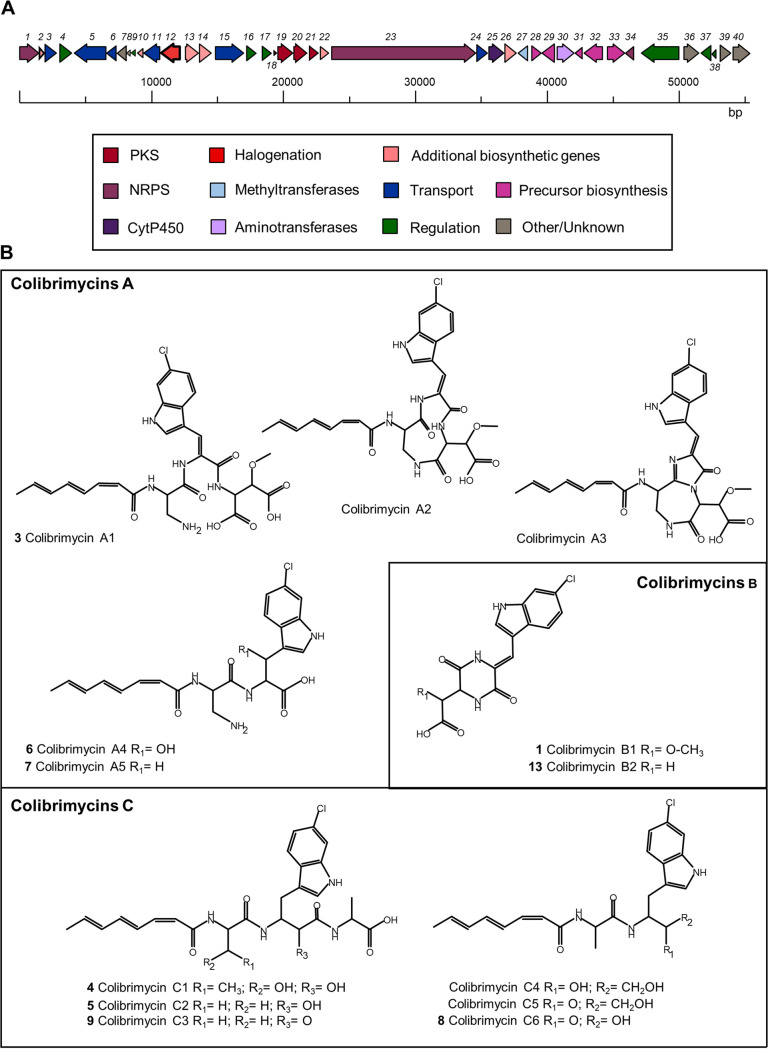
Cluster 15 from Streptomyces sp. strain CS147 is a 40-kb BGC composed of 40 open reading frames (ORFs). Although annotation provided by antiSMASH indicates that this cluster is an NRPS-ladderane BGC, its genetic composition indicates it is an NRPS-PKS cluster (Fig. 1A, Table S3 at https://figshare.com/s/526a9777225b6750159c). It comprises one NRPS consisting of three modules, of which the first condensation domain is a C-starter type domain that might condense the polyketide chain synthesized by a type II PKS adjacent system, as previously described in several lipopeptide antibiotic BGCs (37). It also contains some additional biosynthetic activities, such as cytochrome P450, methyltransferase, aminotransferase, additional AMP-binding and thioesterase standing-alone modules, the Trp halogenase gene (Table 1), and several genes encoding enzymes presumably involved in tryptophan metabolism. Although it is a smaller BGC, cluster 15 from CS147 has certain similarities with the BGCs of some cyclodepsipeptide type compounds, such as skyllamycin or athratumycin, in particular with respect to the NRPS (cbm23), type II PKS (cbm19-22), and cytochrome P450 (cbm25). Furthermore, cbm28 to cbm33, coding for enzymes related to tryptophan metabolism, have homologous genes in the ulleungmycin BGC (see Fig. S3 at https://figshare.com/s/526a9777225b6750159c).
FIG 1.
(A) Genetic organization of the BGC 15 from Streptomyces sp. strain CS147 (cbm1-40). The halogenase coding gene (cbm12, in red) identified in this study is highlighted. (B) Structures of different colibrimycins synthesized by Streptomyces sp. strain CS147. Compounds are numbered according to Fig. 2.
Search for halogenated compounds: identification of colibrimycins, novel halogenated compounds produced by Streptomyces sp. strain CS147.
After several attempts to identify the products synthesized by the clusters containing the detected halogenases, several mutants were obtained in the genes encoding halogenases, structural and/or regulatory genes, but NPs determined by cluster 17 in Streptomyces sp. strain CS159 and cluster 4 in Streptomyces sp CS081a were not detected under our standard cultivation conditions.
However, disruption of the NRPS gene (cbm23) and the overexpression of the gene encoding the SARP regulator (cbm16) in cluster 15 from Streptomyces sp. strain CS147 led to the identification of novel chlorinated compounds.
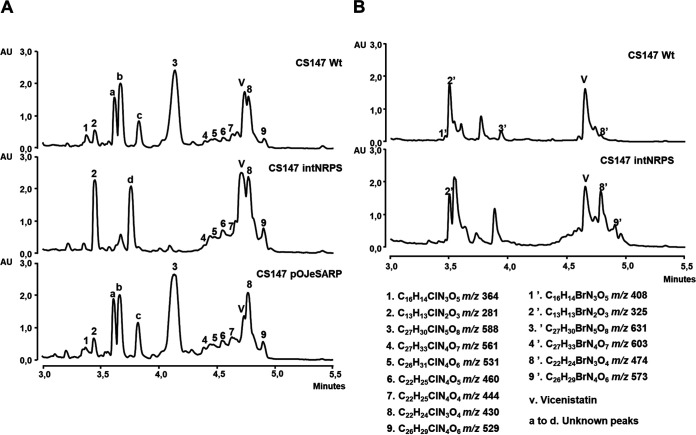
Analysis by UPLC-UV (MaxPlot) and HPLC-MS of secondary metabolites produced by these strains compared to the wild-type Streptomyces sp. strain CS147 grown in R5A-Cl medium and extracted with butanol (Fig. 2A) showed some differential peaks, with masses ranging from m/z 364 to 588 and the same UV spectra (with maxima of around 230 and 290 nm) observed in the wild-type strain but not in the mutant CS147intNRPS (peaks 1 and 3), and some peaks existed in both strains but were significantly increased in the mutant (peaks 2, 4, 5, 6, 7, 8, and 9). A pattern corresponding to a slight increase in the production of peaks 3 to 9 among several replicates was observed in the CS147pOJeSARP strain, indicating the putative role of SARP as a positive regulator. Dereplication analysis of samples from the wild-type and the NRPS mutant strain showed that the products that correspond to these peaks were chlorinated compounds not present in our in-house dereplication libraries (38) or in the Dictionary of Natural Products (39), with the exception of peak 2, corresponding to N-acetyl-6-chlorotryptophan.
FIG 2.
(A) UPLC analysis and MaxPlot (ranging from 210 to 410 nm) of butanol extracts of the wild-type strain (Wt) compared to the cbm23 mutant (CS147intNRPS) and the overexpression of the SARP regulator (cbm16), CS147pOJeSARP. Extracts were obtained from 6-day cultures in R5A-Cl. (B) Brominated analogues of colibrimycins analyzed by UPLC (MaxPlot) in butanol extracts from the wild-type strain (Wt) and the mutant altered in cbm23 (CS147intNRPS) after 6 days of cultivation in R5A-Br medium.
Addition of extra NaCl to R5A medium was carried out to test a putative increase in the production of those compounds, because Cl is limiting in R5A. Analysis by UPLC of butanol extracts at 6 days of R5A and R5A-Cl production of wild-type strain revealed that production was higher when 1.5% NaCl was added (see Fig. S4 at https://figshare.com/s/526a9777225b6750159c).
There was a peak next to peak 8 (labeled v) corresponding to vicenistatin, a glycosylated compound produced in strain CS147 (40). The BGC responsible for the biosynthesis of vicenistatin corresponds to cluster 14, which is located immediately upstream of cluster 15, responsible for the biosynthesis of colibrimycins. As vicenistatin is produced under the same conditions as colibrimycin and it coelutes with peak 8, mutations leading to variations in colibrimycin production also led to variations in the observation of vicenistatin levels. There were certain unidentified peaks (a to d) (Fig. 2A) between retention times 3.5 and 4 min. The suggested molecular formula for peak a is C15H22O7 without coincidences in the DPN; peak b corresponds to an unknown peak UV without positive electrospray ionization (ESI+); peak c is related to the adduct M-H2O+H+ of the component C27H30ClN5O8 (M+H+ = 588.1856; M+NH4+ = 605.2121), corresponding to colibrimycin A1 (peak 3); and peak d, with suggested molecular formula C12H13NO, has coincidences in the DPN with streptazone E and 3-(2-methylpropylidene)-2-indolinone (E and Z). These preliminary UPLC, HRMS, and dereplication analyses did not indicate a clear relationship with the colibrimycin pathway, and they were not further studied.
Compounds from peaks 1 and 3 to 9 were purified and their structures were elucidated by NMR, HRMS, and tandem MS (MS/MS) analyses (see data at https://figshare.com/s/526a9777225b6750159c), determining that they correspond to a family of related chlorinated compounds, designated colibrimycins. They share a chemical pattern consisting of a two-/three-amino-acid peptide chain, including a chlorinated tryptophan residue, bound to an eight-carbon polyunsaturated polyketide chain (except peak 1, consisting of a diketopiperazine-type structure) (Fig. 1B).
Three types of compounds were observed depending on their amino acid composition. Colibrimycin A compounds contain diamino propanoic acid as the first amino acid and aspartic acid as the third amino acid (in case it exists, since intermediates containing only two amino acids were observed). Colibrimycin C compounds contain alanine as the first and third amino acids (in case it is present, since intermediates containing only two amino acids were also observed in this case). The chlorinated tryptophan residue in this type of colibrimycin has turned into a β-amino acid with an extra carbonile group of ketone nature between the carboxylic acid group of the second amino acid and the amino group of the third amino acid. A third type of diketopiperazine (DKP) compound formed by the cyclization of the dipeptide 6-Cl-tryptophan-aspartic acid was observed and designated colibrimycin B.
Colibrimycin A1 (compound 3) was the compound with the highest nominal mass, m/z 588 [M+H]+, detected in the wild-type strain, indicating it is the final compound of the biosynthetic pathway. This compound showed a high instability under the acidic conditions necessary for its purification, experiencing sequential cyclizations with concomitant dehydrations (likely acid catalyzed), rendering colibrimycins A2, m/z 570 [M+H]+, and A3, m/z 552 [M+H]+.
Colibrimycins C4 and C5 were purified from larger-scale cultures as they coelute with compounds 4 to 7 and, therefore, are not represented in the chromatograms.
A detailed description of the structural elucidation of the different colibrimycins identified in this work can be found in the supplemental material at https://figshare.com/s/526a9777225b6750159c.
Brominated analogues and halogenase regioselectivity.
The ability of the halogenase Cbm12 to incorporate bromide instead of chloride during colibrimycin biosynthesis was tested by substituting the chlorinated components present in the R5A culture medium for brominated components (R5A-Br). The generation of brominated analogues (Fig. 2B, peaks 1′ to 4′, 8′, and 9′) of chlorinated compounds 1 to 4, 8, and 9 was verified by the analysis of UV and mass spectra of these compounds (see Fig. S5 at https://figshare.com/s/526a9777225b6750159c). LC-HRMS analysis ratified these results. These results indicate that the Trp halogenase Cbm12 can accept bromide if chloride is not available in the culture medium.
Cmb12 has been annotated in antiSMASH analysis as a Trp 7-halogenase, and it showed similarities to other Trp 7-halogenases according to BLAST. However, in all halogenated colibrimycins identified, the tryptophan moiety is halogenated in position 6. More detailed studies of conserved amino acid residues compared with those of other characterized Trp halogenases with different regioselectivities indicate that Cbm12 is, in fact, a Trp 6-halogenase. The AV and LPP conserved residues, identified in other studies (19) as determinant for regioselectivity in Trp 6-halogenases, are observed instead of the FET residues from Trp 5-halogenases, and neither is present in Trp 7-halogenases (see Fig. S6 at https://figshare.com/s/526a9777225b6750159c).
Role of different Streptomyces sp. strain CS147 BGC 15 genes in colibrimycin biosynthesis.
To verify the role of a halogenase gene (cbm12) as well as the participation of several enzymes coded by genes present in cluster 15 in the biosynthesis of the corresponding halogenated compounds, regulation process, or precursor supply, the colibrimycin production of several mutants compared to the wild-type strain was analyzed by UPLC.
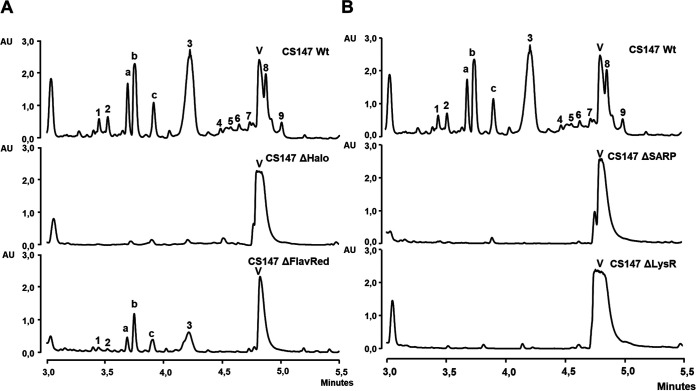
Colibrimycins were not detected in R5A-Cl medium butanol extracts of CS147ΔHalo, and their production was significantly reduced in the mutant of flavin reductase gene cbm10 (CS147ΔFlavRed). In the last case, only peaks 1 to 3 were detected but not minor peaks 4 to 9 (Fig. 3A).
FIG 3.
(A) UPLC analysis (MaxPlot) of butanol extracts of 6 days of R5A-Cl production of wild-type strain (Wt) compared to the mutants altered in cbm12 (CS147ΔHalo) and cbm10 (CS147ΔFlavRed). (B) UPLC analysis (MaxPlot) of butanol extracts of 6 days of R5A-Cl production of the wild-type strain (Wt) compared to the mutants altered in cbm16 (CS147ΔSARP) and cbm17 (CS147ΔLysR).
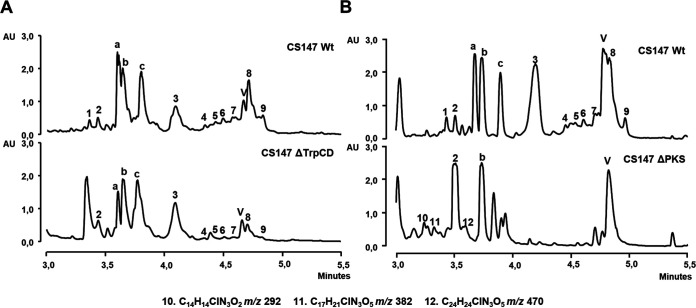
With respect to regulation, inactivation of regulator SARP and LysR coding genes (cbm16 and cbm17, respectively) completely abrogated colibrimycin production (Fig. 3B). However, inactivation of genes related to tryptophan metabolism, trpCD (cbm28-29), only led to a small reduction in their production (Fig. 4A).
FIG 4.
(A) UPLC analysis (MaxPlot) of butanol extracts of 6 days of R5A-Cl production of wild-type strain (Wt) compared to the mutants altered in cbm28 and cbm29 (CS147ΔTrpCD). (B) UPLC analysis (MaxPlot) of butanol extracts of 6 days of R5A-Cl production of wild-type strain (Wt) compared to the mutant altered in cbm18-21 (CS147ΔPKS). Differential peaks detected and their molecular formulas, obtained by dereplication analysis, are indicated.
With respect to the lipoic moiety, genes encoding PKS (cbm18-20) were inactivated. The mutant lacking the PKS coding genes (CS147ΔPKS) showed a totally different chromatographic profile (Fig. 4B). Some differential peaks detected in CS147ΔPKS were determined by mass spectrometry and dereplication analysis, showing them to be chlorinated compounds (see Fig. S7 at https://figshare.com/s/526a9777225b6750159c). However, they did not correspond to any colibrimycin identified in the wild-type strain, with peak 2, N-acetyl 6-chlorotryptophan, being the only one detected. Those compounds had smaller masses than most of the colibrimycins previously identified, and their deduced molecular formulas did not correlate with the peptide part alone lacking the characteristic lipidic chain.
In addition, genes encoding aminotransferase (cbm30), methyltransferase (cbm27), and cytochrome P450 (cbm25) were inactivated. Mutant CS147ΔAmino only showed a reduction in colibrimycin production (Fig. 5).
FIG 5.
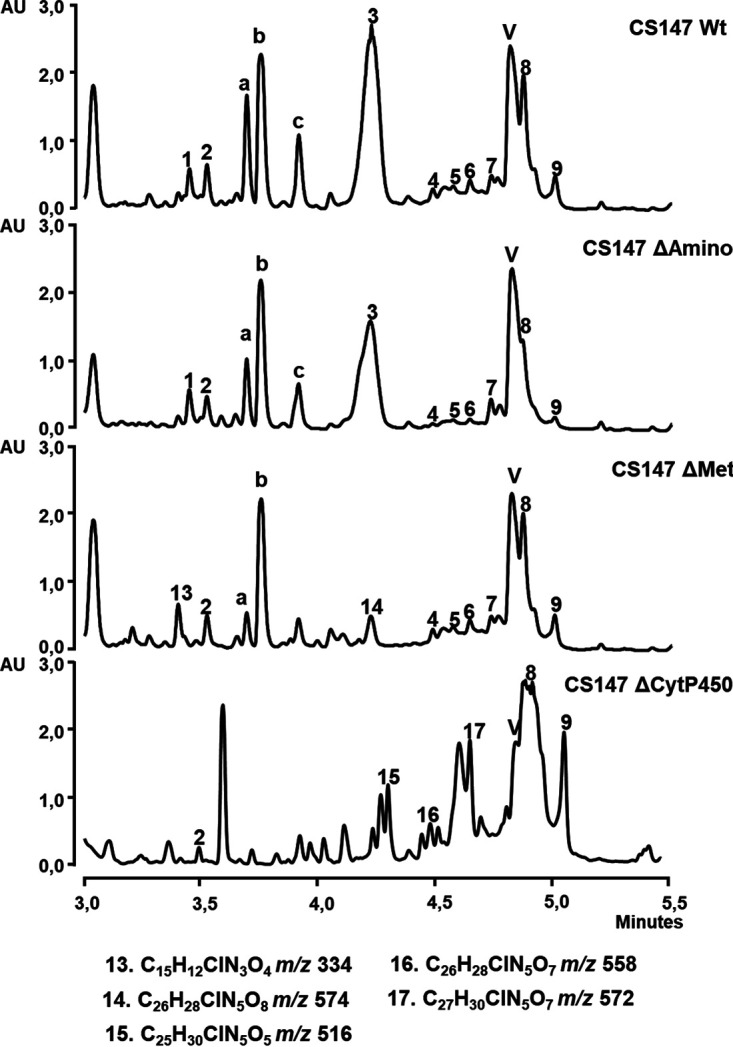
UPLC analysis (MaxPlot) of butanol extracts of 6 days of R5A-Cl production of wild-type strain (Wt) compared to the mutants altered in cbm30 (CS147ΔAmino), cbm27 (CS147ΔMet), and cbm25 (CS147ΔCytP450). Differential peaks detected and their molecular formula obtained by dereplication analysis are indicated.
In the case of the mutant lacking methyltransferase Cbm27, CS147ΔMet, peaks 1 and 3 were not produced, while peaks 13 (molecular formula C15H12ClN3O4, m/z 334) and 14 (molecular formula C26H28ClN5O8, m/z 574) were detected (see Fig. S8 at https://figshare.com/s/526a9777225b6750159c). Those peaks had masses consistent with the lack of a methyl group with respect to peaks 1 (colibrimycin B1, molecular formula C16H14ClN3O5, m/z 364) and 3 (colibrimycin A1, molecular formula C27H30ClN5O8, m/z 588), respectively. Larger-scale production experiments showed that they were already produced by the wild-type CS147 but in a lower quantity than that by CS147ΔMet. Colibrimycin B2 (peak 13) was purified, and its structural elucidation revealed the lack of the methoxyl moiety (-OCH3) (see data at https://figshare.com/s/526a9777225b6750159c) (Fig. 1B).
Inactivation of the cytochrome P450 coding gene cbm25 led to the overaccumulation of many peaks corresponding to colibrimycin-related compounds at retention times between 4 and 5 min. Some of those produced in greater amounts are peaks 15 (C25H30ClN5O5, m/z 516), 16 (C26H28ClN5O7, m/z 558), and 17 (C27H30ClN5O7, m/z 572) (see Fig. S9 at https://figshare.com/s/526a9777225b6750159c). Their coelution and reactivity hindered their purification, but at least as far as compound 16 is concerned, according to high-resolution mass spectrometry results and the determined molecular formula, it would fit with a putative compound 3 (colibrimycin A1) without the methoxyl (-OCH3) group.
Restoration of the wild-type phenotype in each mutant when complementing the mutation with the respective ORF was verified by UPLC (see Fig. S10 at https://figshare.com/s/526a9777225b6750159c).
In vitro bioactivity assays.
Under our test conditions, the colibrimycin family of compounds showed neither antifungal activity against Candida albicans nor antibacterial activity against Gram-positive bacteria Staphylococcus aureus and Micrococcus luteus or Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa. In addition, colibrimycins did not show cytotoxicity against different human tumor cell lines tested: colon adenocarcinoma (HT29), non-small-cell lung cancer (A549), breast adenocarcinoma (MDA-MB-231), promyelocytic leukemia (HL-60), and pancreatic cancer (CAPAN-1) (see Table S4 at https://figshare.com/s/526a9777225b6750159c).
DISCUSSION
Six halogenase coding genes were identified by bioinformatic analyses of 12 sequenced genomes of Streptomyces species isolated from the ecological niche of Attini tribe leaf-cutting ants. The study of the Trp-halogenase located in cluster 15 of strain CS147 allowed the identification of colibrimycins, a novel family of halogenated compounds.
Colibrimycins have an eight-carbon polyketide chain with three double bonds that remains invariable in all characterized compounds. Its biosynthesis might be initiated by 3-oxoacyl-ACP-synthase α (cbm19) and subsequently elongated and reduced by the 3-oxoacyl-ACP-synthase β (cbm20) and 3-oxoacyl-ACP-reductase (cbm21), respectively. It then would be condensed to the first amino acid of the nascent peptide chain by the C-starter type domain located in the first NRPS module, as occurs during the biosynthesis of other lipopeptides (37). This biosynthetic hypothesis would be supported by the absence of colibrimycins or products comprising only the peptide moiety in the mutant CS147ΔPKS lacking the PKS coding genes cbm18-21, which instead produces some chlorinated products with smaller masses than colibrimycins identified so far. Those compounds could correspond to truncated versions of the tripeptide in which other smaller fatty acids from the primary metabolism are incorporated.
Regarding the peptide chain, colibrimycin NRPS is constituted by three modules. The mutant in the NRPS coding gene (cbm23), CS147intNRPS, was generated by disruption resulting in the inactivation of the third module (see Fig. S11 at https://figshare.com/s/526a9777225b6750159c). Colibrimycins A1 to A3 and B, containing aspartic acid in their structures, are not produced by this mutant, while colibrimycin C (with alanine as the third amino acid) and intermediate colibrimycins A and C (with only two amino acids) are produced. This indicates that the third module is responsible for the incorporation of aspartic acid. The second module, the only one for which there was a clear prediction regarding the incorporation of aromatic amino acids (according to antiSMASH data), would incorporate 6-chlorotryptophan. The first module would incorporate alanine (colibrimycin C) or 2,3-diaminopropionic acid (colibrimycin A). Cbm13 and Cbm14 have high similarity to 2,3-diaminopropionate biosynthesis proteins SbnB and SbnA (41), which could be responsible for the biosynthesis of that precursor. On the other hand, the presence of threonine as the first amino in minor compound colibrimycin C1 indicates the promiscuity of the first module to incorporate several types of precursors. Additionally, the ability of CS147intNRPS mutant to biosynthesize colibrimycins despite the inactivation of the third module (including the thioesterase domain) might be due to the involvement of an additional thioesterase encoded by another gene present in the BGC (cbm34).
As mentioned before, colibrimycin C contains an alanine as a third amino acid instead of aspartic acid. The possible participation of the additional adenylation domain encoded by cbm1, the role of which has not yet been defined, was evaluated as a candidate for adding the third alanine. However, preliminary data indicated that its inactivation does not seem to have an effect in this regard (data not shown). Otherwise, an iterative behavior of the NRPS might explain the incorporation of this additional alanine, the incorporation of aspartic acid being the result of the canonical work of the NRPS, and, hypothetically, the incorporation of alanine might be the result of a new operation of the first module. Additionally, the presence of domain TIGR01720, of unknown function, between the second and third NRPS modules led us to consider the possibility that it is involved somehow in this iterative operation of the NRPS. This domain appears in several NRPS systems involved in the biosynthesis of compounds, such as skyllamycin or glycopeptides (see Fig. S12 at https://figshare.com/s/526a9777225b6750159c), and it is located between the epimerization domain of one module and the condensation domain of the next, as is the case in the colibrimycin NRPS. However, currently it has only been suggested that it carries out some postcondensation modifications.
In relation to the above-described comments, attention was drawn to the presence of an α-ketoamide moiety in colibrimycin C, located between tryptophan and the second alanine. This type of unusual amino acid with an α-ketone group has been observed in several compounds showing inhibition of virus proteases. In those compounds, the α-ketoamide moiety seems to have a critical role for this type of bioactivity (42–44). Some biosynthetic mechanisms have been proposed for the formation of α-keto-β-amino acids. For instance, the formation of α-ketoamide has been attributed to an intermediate α-ketoacid in the case of complestatin (45), to auxiliary oxygenases in the case of the polyketide rapamycin (46), or to hybrid PKS/NRPS systems plus additional oxidase domains in the case of nonribosomal peptides such as myxoprincomide (47) and jahnellamides (48). Therefore, the presence of this type of enzyme was evaluated in BGC 15 of strain CS147 and in the whole genome using BLAST homology analysis, not finding any PKS/NRPS systems or other genes that could encode auxiliary oxygenase or oxidase type enzymes to which the biosynthesis of the α-ketoamide could be attributed. Furthermore, the possibility that some of the genes present in the cluster related to tryptophan metabolism could lead to α-keto-β-amino acid tryptophan derivatives was also evaluated due to the structures observed in colibrimycin C4 and C5, not finding among them genes that might encode the enzymes with the expected functionality.
Furthermore, the possible role of cytochrome P450 together with the methyltransferase (cbm25 and cbm27, respectively) in such modifications of tryptophan was assessed. However, there is no precedent, as far as we know, regarding this type of enzymatic activity associated with cytochrome P450, and the analysis of the production of the mutants in these genes does not indicate its role in the generation of this moiety.
A second possibility for the formation of the α-ketone group involves the previously mentioned iterative activity of colibrimycin NRPS, behavior that would be responsible for the incorporation of an additional amino acid. This extra amino acid might then be removed, resulting in the ketone group. Examples of such enzymatic activity responsible for unusual ketone groups have been described previously in the biosynthesis of some ribosomal peptides (RiPPs), such as the enzyme system PlpXY, which posttranslationally excises a previously incorporated amino acid generating an α-keto-β-amino amide moiety (49) (see Fig. S13 at https://figshare.com/s/526a9777225b6750159c).
Colibrimycin B has a different diketopiperazine (DKP)- or cyclodipeptide (CDP)-type structure. Diketopiperazines can be the result of the action of cyclodipeptide synthetases (CDPSs), small enzymes that use aminoacyl-tRNAs as substrates to catalyze the formation of peptide bonds between two amino acids. However, both CDPSs and NRPSs are responsible for the biosynthesis of cyclodipeptides in nature, and the pathways mediated by NRPSs are predominant in fungi and bacteria and may even be the result of spontaneous nonenzymatic cyclization (50). In the case of cyclodipeptides synthesized by NRPSs, they can be biosynthesized by specialized NRPSs such as brevianamide F (51), but they can also be generated as a result of premature release of nonribosomal peptides, such as cyclomarazines (52). The latter are generated during the synthesis of the cyclic heptapeptide cyclomarin A, presumably due to the activity of a type II thioesterase leading to the premature release of the dipeptide N-1,1-dimethylallyl-tryptophan-α-hydroxyleucine. Enzymatic activities such as methyltransferases, or cytochrome P450, participate in the additional modifications of those peptides.
Therefore, according to these data, the association of colibrimycin B with an NRPS-like pathway such as the colibrimycin cluster is plausible, given that these compounds are likely synthesized secondarily in the colibrimycin biosynthetic pathway. This would be supported not only by the structure of this compound itself, formed by 6-Cl-tryptophan and O-methylated aspartic acid (amino acids that are part of the structure of colibrimycins A1 to A3), but also due to the absence of diketopiperazine production in the mutants CS147intNRPS, CS147ΔSARP, CS147ΔLysR, and CS147ΔHalo, which would confirm the relationship of this compound with the colibrimycin biosynthesis pathway.
With respect to the halogenation process, compounds related to colibrimycins identified in this work are tryptophan chlorinated. Mutant CS147ΔHalo lacking the halogenase coding cbm12 was unable to biosynthesize any of the halogenated colibrimycins or N-acetyl-6-chlorotryptophan, which indicates that 6-chlorotryptophan is the substrate for the NRPS second module. In addition, it indicates that the halogenation of the tryptophan residue is strictly required for the biosynthesis of colibrimycins or somehow influences its efficiency when that step does not occur, since nonhalogenated versions have been observed. Genetic complementation of the halogenase gene led to the recovery of the producing phenotype. Additionally, since flavin reductase genes are usually located in close vicinity to halogenase genes due to their activity in reducing the FAD required for the halogenation mechanism, inactivation of flavin reductase-coding cbm10 (mutant strain CS147ΔFlavRed) was carried out to evaluate its role. Behavior of this mutant was as expected, similar to that of the mutant CS147ΔHalo: a decrease in the biosynthesis of halogenated compounds was seen, although in this case a certain level of production of colibrimycins B1 (compound 1) and A1 (compound 3) and N-acetyl-6-chlorotryptophan (compound 2) was observed. Complementation of the mutant with the intact flavin reductase gene resulted in the expected production increase of these compounds. According to these results, flavin reductase might be necessary for the halogenation step and, consequently, for the biosynthesis pathway, although other flavin reductases could be replacing its function, since there remains a low level of production of colibrimycins in the mutant.
Even though databases identify the halogenase from CS147 as a Trp 7-halogenase, conserved residues typical from Trp 6-halogenases, detected in more exhaustive analyses, are in concordance with the chlorination at position 6 observed in all colibrimycins. Also related to halogenation, the observation of brominated analogues produced in a bromine broth strongly point to the putative involvement of this halogenase in the incorporation of bromide instead of chloride when the latter is not available in the medium. Although protein purification and in vitro experiments should be carried out to demonstrate this hypothesis, the apparent absence of other possible genes encoding halogenases in the Streptomyces sp. strain CS147 genome that could intervene in this halogenation, or the lack of other halogenated compounds in the strain under the same conditions, makes quite probable the implication of halogenase Cbm12 in the bromination process.
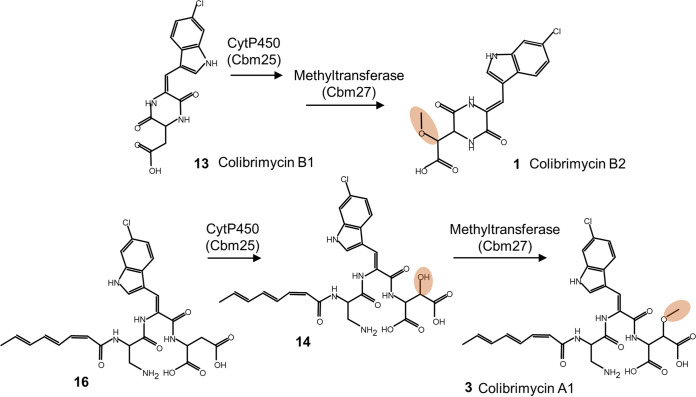
Regarding the role of methyltransferase and cytochrome P450, the presence of colibrimycins lacking the methoxyl (O-CH3) or methyl (-CH3) groups when inactivating their corresponding ORFs (cbm27 and cbm25, respectively) indicate their involvement in the hydroxylation of the aspartic acid in colibrimycins A and B by the cytochrome and subsequent O-methylation by the methyltransferase (Fig. 6). On the other hand, the double α-β bond in tryptophan could be the result of the reduction of the -OH group observed in colibrimycin A4, but we have no data to explain this hydroxylation for the moment. The small effect of the inactivation of the aminotransferase coding gene cbm30 and genes cbm28 and cbm29, involved in tryptophan metabolism, indicate they do not have an indispensable role in colibrimycin biosynthesis as long as they could be replaced by other genes from primary metabolism.
FIG 6.
Proposed participation of cytochrome P450 (Cbm25) and methyltransferase (Cbm27) during the biosynthesis of colibrimycins.
With respect to the regulation of BGC 15, the total absence of colibrimycin peaks 1 to 9 in extracts from the CS147ΔSARP mutant strain would confirm that the removed gene encodes a pathway-specific activator. This result is in consonance with the slight increase observed in the production of colibrimycins in the strain overexpressing SARP-coding cbm16, CS147pOJeSARP. Genetic complementation of the mutant strain with the intact cbm16 under the control of the ermE* promoter using pSETEcH resulted in the recovery of the wild-type level of colibrimycin production. The inability to complement the mutant of the regulator LysR (the phenotype of which was also a noncolibrimycin producer), except when the two regulators, SARP and LysR, were expressed together also supported the important role of the SARP protein.
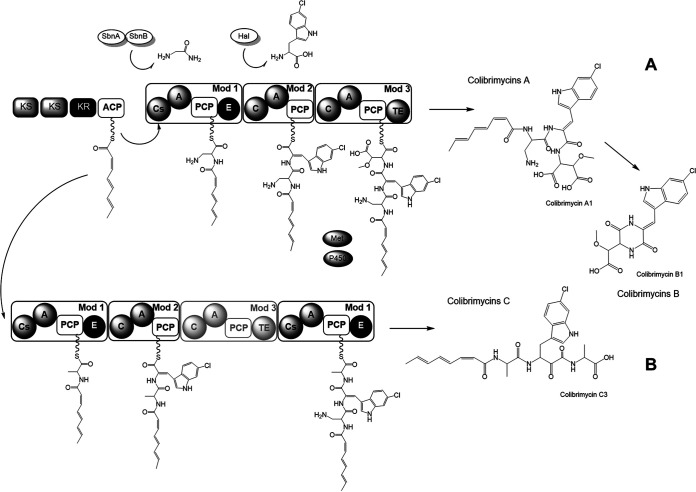
Therefore, the chemical structure of characterized colibrimycins and production analyses of mutant strains indicate that there are two alternative pathways during colibrimycin biosynthesis (Fig. 7). Both would begin with the biosynthesis of the fatty acid moiety by the PKS and its subsequent incorporation into the nascent peptide chain thanks to the C-starter-type condensation domain present in the first module of the NRPS. Subsequently, in the first alternative (Fig. 7A), the first module of the NRPS would incorporate 2,3-diaminopropionic acid, which would be synthesized by the SbnAB activities encoded by cbm13-14. Next, the second module would incorporate tryptophan chlorinated by the action of the Cbm12 halogenase. Finally, the third module would incorporate aspartic acid, which might be hydroxylated and O-methylated thanks to the successive action of cytochrome P450 (Cbm25) and methyltransferase (Cbm27). As a result of this pathway, colibrimycin A would be synthesized and, from their degradation and subsequent cyclization, then be transformed into colibrimycin B. This route would be the main one in the wild-type strain. Colibrimycins A4 and A5, with only two amino acids, would be the intermediates of this route.
FIG 7.
Proposed alternative pathways in the biosynthesis of colibrimycins. (A) Biosynthesis of colibrimycins A and B as the main outcome in the wild-type strain. (B) Biosynthesis of colibrimycin C as a second alternative in CS147 Wt and the only pathway in CS147intNRPS.
The second alternative (Fig. 7B), minoritarian in the wild-type strain, is the only alternative existing in the CS147intNRPS strain due to the inactivation of the third module, and it should be responsible for the biosynthesis of colibrimycin C. It would involve the incorporation of alanine by the first NRPS module followed by the condensation of Cl-tryptophan by the second module. Finally, a hypothetical iterative operation of the NRPS would take place so that the first module would work, again incorporating a second alanine as the third amino acid. Colibrimycins C4, C5, and C6, with only two amino acids, would be intermediates in this route.
In summary, the search for halogenase-coding genes using genome sequence analysis led to the identification of several halogenases in a collection of Streptomyces species isolated from the same ecological niche of Attini ants. The study of one of these halogenases in Streptomyces sp. strain CS147 allowed the identification of colibrimycins, novel hybrid PKS-NRPS tryptophan-halogenated compounds whose biosynthetic pathway might be activated by a cluster-specific SARP regulator. The role of these compounds remains unknown, as they did not exhibit any antibiotic or cytotoxic activity, although the presence of an α-ketoamide group in some of them might be related to hypothetical activity as inhibitors of virus proteases.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A collection of 12 Streptomyces species strains (CS collection) (40) was used in this work. Briefly, 60 Atinni ants (Acromyrmex octospinosus) were collected from the region of Lambayeque in Perú by Carlos Sialer (40). The present work using the CS collection has been done with explicit permission from the collector. The ants were heated in a water bath at 45°C for 30 min and then incubated in nutrient agar plates at 25 to 28°C for 6 to 8 days for the isolation and identification of several actinomycetes strains, using macroscopic and microscopic characteristics of the colonies.
Medium A (MA) plates (53) were used for sporulation during 7 days at 30°C. Intergeneric conjugation was carried out according to reference 54 using mannitol soy medium (MS) (55). Escherichia coli strains were grown at 37°C in 2× tryptone-yeast medium supplemented with the appropriate antibiotic.
For metabolite production, spores were grown for 1 day at 30°C and 250 rpm in 250-ml flasks containing 50 ml of tryptic soy broth (TSB; Oxoid) and then inoculated into 250-ml flasks containing 50 ml of R5A medium (56) supplemented with NaCl 1.5% (R5A-Cl medium) to a final optical density at 600 nm (OD600) of 0.2, so cell growth should be equivalent between cultures in order to compare them. A variation of R5A medium (R5A-Br) in which MgCl2 · 6H2O was replaced by MgBr2 · 6H2O and supplemented with an extra 1.5% NaBr was used for the production of brominated compounds.
Antibiotics were used to supplement the culture medium when necessary: apramycin (100 μg/ml for E. coli, 50 μg/ml for Streptomyces), kanamycin (25 μg/ml for E. coli, 200 μg/ml for Streptomyces), chloramphenicol (25 μg/ml), ampicillin (100 μg/ml), hygromycin (200 μg/ml), thiostreptone (for Streptomyces, 25 μg/ml in solid cultures and 5 μg/ml in liquid cultures), and nalidixic acid (50 μg/ml).
All bacterial strains used in this work are listed in Table 2. All plasmid constructs, sequences, and microbial strains reported in this research are available and in compliance with current laws.
TABLE 2.
Strains used in this work
| Microorganism | Genotype | Use | Reference or source |
|---|---|---|---|
| Escherichia coli | |||
| DH5α | F− φ80lacZΔM15Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Cloning | ThermoFisher Scientific |
| ET12567/pUZ8002 | dam13::Tn9 dcm6 hsdM hsdR recF143 zjj201::Tn10 galK2 galT22 ara14 lacY1 xyl5 leuB6 thi1 tonA31 rpsL136 hisG4 tsx78 mtli glnV44 F− | Intergeneric conjugation | 54 |
| Streptomyces sp. | |||
| CS003, CS014, CS030, CS044a, CS052a, CS052b, CS057, CS065a, CS074a, CS081a, CS084, CS088c, CS090a, CS092, CS109, CS113, CS115, CS119, CS123, CS131, CS149, CS170, CS174, CS159, CS207, CS215, CS217, CS227, CS147 | Wild-type strains | 40 | |
| CS147intNRPS | C15 cbm23::pOJ260 | This work | |
| CS147pOJeSARP | C15::pOJ260P ermEp-cbm16 | ||
| CS147ΔHalo | C15 Δcbm12::aac(3)IV | ||
| CS147ΔHalo pSETEcH Halo | C15 Δcbm12::aac(3)IV ermEp-cbm12 | ||
| CS147ΔFlavRed | C15 Δcbm10::aac(3)IV | ||
| CS147ΔFlavRed pSETEcH FlavRed | C15 Δcbm10::aac(3)IV ermEp-cbm12 | ||
| CS147ΔSARP | C15 Δcbm16::aac(3)IV | ||
| CS147ΔSARPeSARP | C15 Δcbm16::aac(3)IV ermEp-cbm16 | ||
| CS147ΔLysR | C15 Δcbm17::aac(3)IV | ||
| CS147ΔLysRpSETEcHeSARP+LysR | C15 Δcbm17::aac(3)IV ermEp-cbm17 | ||
| CS147ΔTrpCD | C15 Δcbm28,29::aac(3)IV | ||
| CS147ΔPKS | C15 Δcbm18-21::hygR | ||
| CS147 ΔPKS pSETETcPKS | C15 Δcbm18-21::hygR ermEp-cbm18-21 | ||
| CS147ΔAmino | C15 Δcbm30::aac(3)IV | ||
| CS147ΔMet | C15 Δcbm27::aac(3)IV | ||
| CS147ΔMet pSETETcMet | C15 Δcbm27::aac(3)IV ermEp-cbm27 | ||
| CS147ΔCytP450 | C15 Δcbm25::aac(3)IV | ||
| CS147ΔCytP450pSETEcHCytP450 | C15 Δcbm25::aac(3)IV ermEp-cbm25 |
DNA manipulation and vectors.
DNA manipulations were performed according to standard procedures for E. coli (57) and Streptomyces (58).
For DNA amplification to perform plasmid constructions, Herculase II Fusion high-fidelity polymerase (Agilent Technologies) was used according to the manufacturer’s indications with the following PCR conditions: initial denaturation at 98°C for 5 min, 10 touchdown cycles including denaturation at 98°C for 20 s, annealing at melting point (Tm) for 20 s, and elongation at 72°C for 30 s/1 kb; 25 cycles of denaturation 98°C for 20 s, annealing Tm for 20 s, and elongation at 72°C for 30 s/1 kb; and a final elongation at 72°C for 3 min.
DNA amplification to verify plasmid constructions and mutants were carried out using DreamTaq polymerase (ThermoFisher Scientific) according to the manufacturer’s procedures, and PCR conditions were initial denaturation at 99°C for 2 min; 10 touchdown cycles comprised of 10 cycles of denaturation at 95°C for 30 s, 10 annealing cycles for 30 s, equally varying from the highest oligonucleotide Tm to smallest oligonucleotide Tm, 10 cycles of elongation at 72°C for 60 s/1 kb; 25 cycles of denaturation at 95°C for 30 s, annealing Tm for 30 s, and elongation at 72°C for 60 s/1 kb; and a final elongation at 72°C for 5 min.
All plasmids used in this work are listed in Table 3. pSETEcH was generated by cloning a PstI-EcoRV fragment containing a hygromycin cassette from pLHyg (59) into pSETEc (60) digested with SacI to remove an apramycin cassette (cohesive ends were removed by Pfx polymerase).
TABLE 3.
Plasmids used in this work
| Plasmid | Characteristic(s) | Use | Reference or source |
|---|---|---|---|
| pSETEcH | Integrating vector hyg, oriTRK2, intΦC31, attPΦC31, ermEp | Gene overexpression | This work |
| pSETEc | Integrating vector aac(3)IV, oriTRK2, intΦC31, attPΦC31, ermEp | Gene overexpression | 60 |
| pSETETc | Integrating vector aac(3)IV, tsr, oriTRK2, intΦC31, attPΦC31, ermEp | Gene overexpression | 60 |
| pOJ260P | Suicide vector lacZ, Rep pUC, aac(3)IV, oriTRK2, ermEp | Gene overexpression | 59 |
| pOJ260 | Suicide vector lacZ, Rep pUC, aac(3)IV, oriTRK2 | Gene disruption | 65 |
| pUO9090 | aac(3)IV km | Replacement cassette construction | 66 |
| pUK21Hyg | hygR | Replacement cassette construction | 40 |
| pHZ1358 | bla, tsr, oriTRK2 | Gene replacement | 67 |
| pOJ260PeSARP | Carries homology region for cbm16 C15 CS147 overexpression under control of ermEp, lacZ, Rep pUC, aac(3)IV, oriTRK2 | cbm16 C15 CS147 overexpression | This work |
| pOJ260NRPS | Carries homology region for cbm23 C15 CS147 disruption, lacZ, rep pUC, aac(3)IV, oriTRK2 | cbm23 C15 CS147 disruption | This work |
| pUO9090ΔHalo | Carries upstream and downstream flanking regions of cbm12 C15 CS147 at both sides of aac(3)IV, km | Provides cbm12 C15 CS147 replacement cassette | This work |
| pHZ1358ΔHalo | Carries cbm12 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm12 C15 CS147 replacement with aac(3)IV | This work |
| pSETEcHeHalo | Carries cbm12 C15 CS147 under control of ermEp, hyg, oriTRK2, intΦC31, attPΦC31 | cbm12 C15 CS147 overexpression and complementation | This work |
| pUO9090ΔFlavRed | Carries upstream and downstream flanking regions of cbm10 C15 CS147 at both sides of aac(3)IV, km | Provides cbm10 C15 CS147 replacement cassette | This work |
| pHZ1358ΔFlavRed | Carries cbm10 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm10 C15 CS147 replacement with aac(3)IV | This work |
| pSETETceFlavRed | Carries cbm10 C15 CS147 under control of ermEp, aac(3)IV, tsr, oriTRK2, intΦC31, attPΦC31 | cbm10 C15 CS147 complementation | This work |
| pUO9090ΔSARP | Carries upstream and downstream flanking regions of cbm16 C15 CS147 at both sides of aac(3)IV, km | Provides cbm16 C15 CS147 replacement cassette | This work |
| pHZ1358ΔSARP | Carries cbm16 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm16 C15 CS147 replacement with aac(3)IV | This work |
| pSETEcHeSARP | Carries cbm16 C15 CS147 under control of ermEp, hyg, oriTRK2, intΦC31, attPΦC31 | cbm16 C15 CS147 complementation | This work |
| pUO9090ΔLysR | Carries upstream and downstream flanking regions of cbm17 C15 CS147 at both sides of aac(3)IV, km | Provides cbm17 C15 CS147 replacement cassette | This work |
| pHZ1358ΔLysR | Carries cbm17 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm17 C15 CS147 replacement with aac(3)IV | This work |
| pSETEcHeLysR+SARP | Carries cbm16-17 C15 CS147 under control of ermEp, hyg, oriTRK2, intΦC31, attPΦC31 | cbm17 C15 CS147 complementation | This work |
| pUO9090ΔTrpCD | Carries upstream and downstream flanking regions of cbm28-29 C15 CS147 at both sides of aac(3)IV, km | Provides cbm28,29 C15 CS147 replacement cassette | This work |
| pHZ1358ΔTrpCD | Carries cbm28-29 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm28,29 C15 CS147 replacement with aac(3)IV | This work |
| pUK21HygΔPKS | Carries upstream and downstream flanking regions of cbm18-21 C15 CS147 at both sides of aac(3)IV, km | Provides cbm18-21 C15 CS147 replacement cassette | This work |
| pHZ1358ΔPKS | Carries cbm18-21 C15 CS147 replacement cassette, hygR, bla, tsr, oriTRK2 | cbm18-21 C15 CS147 replacement with hygR | This work |
| pSETETcePKS | Carries cbm18-21 C15 CS147 under control of ermEp, aac(3)IV, tsr, oriTRK2, intΦC31, attPΦC31 | cbm18-21 C15 CS147 complementation | This work |
| pUO9090ΔAmino | Carries upstream and downstream flanking regions of cbm30 C15 CS147 at both sides of aac(3)IV, km | Provides cbm30 C15 CS147 replacement cassette | This work |
| pHZ1358ΔAmino | Carries cbm30 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm30 C15 CS147 replacement with aac(3)IV | This work |
| pUO9090ΔMet | Carries upstream and downstream flanking regions of cbm27 C15 CS147 at both sides of aac(3)IV, km | Provides cbm27 C15 CS147 replacement cassette | This work |
| pHZ1358ΔMet | Carries cbm27 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm27 C15 CS147 replacement with aac(3)IV | This work |
| pSETETceMet | Carries cbm27 C15 CS147 under control of ermEp, aac(3)IV, tsr, oriTRK2, intΦC31, attPΦC31 | cbm27 C15 CS147 complementation | This work |
| pUO9090ΔCytP450 | Carries upstream and downstream flanking regions of cbm25 C15 CS147 at both sides of aac(3)IV, km | Provides cbm25 C15 CS147 replacement cassette | This work |
| pHZ1358ΔCytP450 | Carries cbm25 C15 CS147 replacement cassette, aac(3)IV, bla, tsr, oriTRK2 | cbm25 C15 CS147 replacement with aac(3)IV | This work |
| pSETEcHeCytP450 | Carries cbm25 C15 CS147 under control of ermEp, hyg, oriTRK2, intΦC31, attPΦC31 | cbm25 C15 CS147 complementation | This work |
Genome sequencing.
Genomic DNAs were extracted using the standard phenol-chloroform-isoamyl alcohol method described in reference 54.
The CS014, CS065a, CS081a, CS090a, CS131, CS147, and CS207 chromosomes were sequenced using Illumina MiSeq sequencing technology from a 2 × 300 bp insert TruSeq PCR-free library (paired-end reads) by the Department of Biochemistry at the University of Cambridge (United Kingdom). The sequencing reads were processed and assembled using default parameters in Newbler assembler software, version 2.9. Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (https://submit.ncbi.nlm.nih.gov/subs/genome/) (61). Strains CS057, CS113, CS149, CS159, and CS227 were previously sequenced (40) using the same technology stated above.
BLAST analysis of halogenases was carried out using the 12 sequenced Streptomyces sp. CS strains: CS014 (NZ_QBHV00000000.1), CS057 (NZ_NEVF00000000.1), CS065a (NZ_QBHW00000000.1), CS081a (NZ_QBHX00000000.1), CS090a (NZ_QBHY00000000.1), CS113 (NZ_NEVC00000000.1), CS131 (NZ_QBHZ00000000.1), CS147 (NZ_QBIA00000000.1), CS149 (NZ_PVZY00000000.1), CS159 (NZ_NEVD00000000.1), CS207 (NZ_QBIB00000000.1), and CS227 (NZ_NEVE00000000.1).
Bioinformatic analysis of halogenases.
Database searching and sequence analysis of the genomes in previously sequenced strains was performed using the bioinformatics software antiSMASH 6.0 (62).
Several proteins representing different types of halogenases were used as a query in the search for halogenases in the genome of the latter strains: SalL SAM-dependent halogenase, SyrB2 nonheme/Fe(II)-oxoglutarate-dependent halogenase, RebH, PyrH, and PrnA tryptophan FAD-dependent halogenases, PrnC pyrrol FAD-dependent halogenase, and CmlS aliphatic activated compound FAD-dependent halogenase.
Analysis of tryptophan halogenase regioselectivity was carried out through the study of conserved amino acid residues by multiple-sequence alignment using ClustalW2. The halogenase from cluster 15 of Streptomyces sp. strain CS147 was compared with several characterized Trp 6-halogenases (Th-Hal and SttH), Trp 5-halogenase (PyrH), and Trp7-halogenase (RebH).
Generation of mutant strains by gene disruption.
For CS147intNRPS strain construction by homologous recombination, an internal region of the NRPS gene was amplified by PCR using genomic DNA of the CS147 wild-type strain as the template, Herculase II Fusion high-fidelity DNA polymerase, and oligonucleotides listed in Table 4 according to the scheme represented in Fig. S14 at https://figshare.com/s/526a9777225b6750159c. PCR amplification products were gel purified (GeneJet gel extraction kit from Thermo Scientific) and digested with the correspondent restriction enzymes (appropriate restriction enzyme sites were added to the oligonucleotide sequences) to be introduced in pOJ260 vector (Table 4). The constructed plasmids were introduced in Streptomyces by intergeneric conjugation with E. coli ET12567/pUZ8002. Apramycin-resistant mutants were confirmed by PCR (see Fig. S15A at https://figshare.com/s/526a9777225b6750159c) using oligonucleotides listed in Table 5 and by sequencing of the amplification products.
TABLE 4.
Oligonucleotides used for gene disruption
| Oligonucleotide | Sequence, 5′–3′ | Restriction site | Use |
|---|---|---|---|
| intNRPS147FW | GCTCTAGAGTCTATGTGCTCGACCGTCA | XbaI | cbm23 C15 CS147 disruption |
| intNRPS147RV | CGGAATTCTCGGTGCTGAAGTTGAACAG | EcoRI |
TABLE 5.
Oligonucleotides used for mutant verification
| Oligonucleotide | Sequence 5′–3′ | Size (bp) of mutant | Mutant | |
|---|---|---|---|---|
| NRPS147IFW | CCCTCGTCGACCTGATCAAC | 2,339 | CS147intNRPS | |
| M13RV | CAGGAAACAGCTATGAC | |||
| NRPS147DRV | TCCAGTTCGGCGTACGTCAG | 2,347 | ||
| M13FW | GTAAAACGACGGCCAG | |||
| ermEFW | GTAAAACGACGGCCAG | 2,202 | CS147pOJeSARP | |
| eLysR147DRV | TGGATCCGACCGGCAGATAG | |||
| ΔHalo147IFW | AGTGCTTCCTGTGTCATC | 1,275 | CS147ΔHalo | |
| ApraRV | CGGATGCAGGAAGATCAAC | |||
| ΔHalo147DRV | CTCGATCACTCCGAAGAC | 1,554 | ||
| AprFW | TCAGCTTCTCAACCTTGG | |||
| ΔFlavRed147DRV | CGATGGCCGTGTGGG | 1,371 | CS147ΔFlavRed | |
| AprFW | TCAGCTTCTCAACCTTGG | |||
| ΔFlavRed147IFW | GTCATCGAACGCTGCT | 944 | ||
| AprRV | CGGATGCAGGAAGATCAAC | |||
| ΔSARP147IFW | CTGGTTCTGGGCAGAATTTG | 984 | CS147ΔSARP | |
| ApraRV | CGGATGCAGGAAGATCAAC | |||
| ΔSARP147DRV | GGACGGCAATGCCAATTC | 1,308 | ||
| ApraFW | TCAGCTTCTCAACCTTGG | |||
| ΔLysR147IFW | CGCGTCCTGAACAAG | 2,433 | CS147ΔLysR | |
| Apra Seq I | GAAGCTGACCGATGAG | |||
| ΔLysR147DRV | TACGGCACGGTCTAC | 2,748 | ||
| Apra Seq D | ACATTGTGGCGACAGC | |||
| TrpCDDRV | CTGCCGCCGTTCAAG | 2,773 | CS147ΔTrpCD | |
| Apr Seq I | GAAGCTGACCGATGAG | |||
| TrpCDIFW | GCGGCCGATCATGAC | 2,587 | ||
| Apr Seq D | ACATTGTGGCGACAGC | |||
| PKS147DRV | CGAGGTAGTCGGTGAAG | 3,403 | CS147ΔPKS | |
| HygFW | ACCGGCCGTGCGGAATTAAG | |||
| PKS147IFW | TCTCGCTGACGTGTG | 3,611 | ||
| HygRV | AGTTCCTCCGGATCGGTGAAG | |||
| AminoDRV | ACTTCCTCGCGATGG | 1,479 | CS147ΔAmino | |
| AprFW | TCAGCTTCTCAACCTTGG | |||
| AminoIFW | ACGACCGGATGAACC | 981 | ||
| AprRV | CGGATGCAGGAAGATCAAC | |||
| MetDRV | GTCCGGTGTGGTCTAC | 1,879 | CS147ΔMet | |
| AprFW | TCAGCTTCTCAACCTTGG | |||
| MetIFW | AAACTCGCAGGTGATCG | 979 | ||
| AprRV | CGGATGCAGGAAGATCAAC | |||
| CytP450IFW | CGTGCGTGTGAAGTG | 2,407 | CS147ΔCytP450 | |
| Apr Seq I | GAAGCTGACCGATGAG | |||
| CytP450DRV | TCGGCCGCATTCATC | 2,624 | ||
| Apr Seq D | ACATTGTGGCGACAGC | |||
| M13 FW | GTAAAACGACGGCCAG | Verification of regions AB, CD in pUO9090 | ||
| M13 RV | CAGGAAACAGCTATGAC | |||
| Apra Seq I | GAAGCTGACCGATGAG | |||
| Apra Seq D | ACATTGTGGCGACAGC | |||
| Apra Seq I | GAAGCTGACCGATGAG | Verification of regions AB, CD in pHZ1358 | ||
| Apra Seq D | ACATTGTGGCGACAGC | |||
| pHZ1358 I FW | GTGCGCGTGATTGC | |||
| pHZ1358 D RV | CGCCGGACAACGTG |
Generation of mutant strains by gene replacement.
CS147ΔHalo, CS147ΔSARP, CS147ΔFlavRed, and CS147ΔLysR mutants were constructed by gene replacement cloning 2-kb 5′ and 3′ flanking regions of the gene of interest at both sides of the apramycin gene using pUO9090 and pHZ1358 plasmids according to reference 63. Briefly, 5′ and 3′ flanking regions of the gene of interest were amplified by PCR using genomic DNA of CS147 wild-type strain as the template, Herculase II Fusion high-fidelity DNA polymerase, and oligonucleotides with the appropriate enzyme restriction sites added in their sequences listed in Table 6 and according to the scheme presented in Fig. S16 at https://figshare.com/s/526a9777225b6750159c. These regions were amplified by PCR, digested with the correspondent enzymes, and introduced in the pUO9090 vector at both sides of the aac(3)IV resistance gene. The cassette containing 5′ and 3′ flanking regions of the gene of interest and the antibiotic resistance gene was extracted by SpeI digestion and introduced in pHZ1358 digested with XbaI. Apramycin-resistant mutants were verified by PCR (see Fig. S15C to K at https://figshare.com/s/526a9777225b6750159c) with oligonucleotides listed in Table 5 and by sequencing of the PCR amplification products.
TABLE 6.
Oligonucleotides used for gene replacement
| Oligonucleotide | Sequence, 5′–3′ | Restriction site | Use |
|---|---|---|---|
| ABdHalo147FW | CGGAATTCATCTCCGGCACGTGG | EcoRI | cbm12 C15 CS147 replacement (upstream region) |
| ABdHalo147RV | CCCAAGCTTCGCCCACGATAACGAC | HindIII | |
| CDdHalo147FW | GATATCTACTTCTCCCGGATGC | EcoRV | cbm12 C15 CS147 replacement (downstream region) |
| CDdHalo147RV | GCGGCCGCACGGTCAGCAGTTCC | NotI | |
| ABdFlavRed147FW | GAAGATCTGACACGCTGCCCAAC | BglII | cbm10 C15 CS147 replacement (upstream region) |
| ABdFlavRed147RV | CGGAAGCTTGGTAGCCGCCAGTGC | HindIII | |
| CDdFlavRed147FW | TTCAGATATCCCGCCGCTCCTCTAC | EcoRV | cbm10 C15 CS147 replacement (downstream region) |
| CDdFlavRed147RV | GCTCTAGATGATGGCGGAGGGTG | XbaI | |
| ABdSARP147FW | TACGAATTCTGCACCCAGGCAAGG | EcoRI | cbm16 C15 CS147 replacement (upstream region) |
| ABdSARP147RV | CCCAAGCTTCGAGAGCGCACCCAG | HindIII | |
| CDdSARP147FW | TATAGATATCGCACCGGCTTCCTGAC | EcoRV | cbm16 C15 CS147 replacement (downstream region) |
| CDdSARP147RV | GCTCTAGAACCGGTGAGGATTCCC | XbaI | |
| ABdLysR147FW | TACGAATTCCGATCCCGGAGATGC | EcoRI | cbm17 C15 CS147 replacement (upstream region) |
| ABdLysR147RV | CCCAAGCTTTCGGGCATGTCGTAG | HindIII | |
| CDdLysR147FW | GCCGGATATCTTCGCCGAGTTGAGC | EcoRV | cbm17 C15 CS147 replacement (downstream region) |
| CDdLysR147RV | GCTCTAGAACGGTGTTCGGGAAG | XbaI | |
| ABdTrpCD147FW | CCCAAGCTTCCGAGCGCATCCAT | BglII | cbm28,29 C15 CS147 replacement (upstream region) |
| ABdTrpCD147RV | GAAGATCTCCGCAACGTCTCTACTC | HindIII | |
| CDdTrpCD147FW | CTGGATCCTTCGGGCCAGTTGCG | BamHI | cbm28,29 C15 CS147 replacement (downstream region) |
| CDdTrpCD147RV | GCTCTAGAACTCCGGCCATGTCG | XbaI | |
| ABdPKS147FW | TATCTAGAACGCTTCTCGCACTG | XbaI | cbm18-21 C15 CS147 replacement (upstream region) |
| ABdPKS147RV | GCGGCCGCGGCCGCGTCCACCTG | NotI | |
| CDdPKS147FW | AAGCTTACCGGCCAGGTCTTC | HindIII | cbm18-21 C15 CS147 replacement (downstream region) |
| CDdPKS147RV | GGTACCCTGCCTCGCCCTTGTC | KpnI | |
| ABdAmino147FW | CGGGTACCACGGAAGGCTGACATC | KpnI | cbm30 C15 CS147 replacement (upstream region) |
| ABdAmino147RV | CCCAAGCTTTGCAGGGCCATCGG | HindIII | |
| CDdAmino147FW | TTCAGATATCGCGGCGGGATTCCAC | EcoRV | cbm30 C15 CS147 replacement (downstream region) |
| CDdAmino147RV | TATCTAGACGGCCTGGCACATGG | XbaI | |
| ABdMet147FW | TAGGTACCCTGCCGCCGTTCAAG | KpnI | cbm27 C15 CS147 replacement (upstream region) |
| ABdMet147RV | CCCAAGCTTATCCGCTGACGGAAGG | HindIII | |
| CDdMet147FW | TTCCTTACATATGCGCCCGGGAACCG | NdeI | cbm27 C15 CS147 replacement (downstream region) |
| CDdMet147RV | TAGGATCCACCGGCGGCAGCTTT | BamHI | |
| ABdCytP450147FW | GAATTCTTGCGGCTGGTGAGTC | EcoRI | cbm25 C15 CS147 replacement (upstream region) |
| ABdCytP450147RV | AAGCTTCGGGAGCAACGGTCT | HindIII | |
| CDdCytP450147FW | GATATCCCGCAACGTCTCTACTCC | EcoRV | cbm25 C15 CS147 replacement (downstream region) |
| CDdCytP450147RV | TCTAGAGTGACGCCCTATGCC | XbaI |
Plasmid construction for gene overexpression and complementation.
To overexpress the main biosynthetic operon of structural and regulatory genes (including SARP and LysR transcriptional regulator, PKS, and NRPS genes), the CS147pOJeSARP mutant was generated by cloning an approximately 2-kb fragment sense ATG codon of cbm16 (SARP) regulator under ermE*p control into pOJ260p. PCR was carried out using genomic DNA of the CS147 wild-type strain as the template, Herculase II Fusion high-fidelity DNA polymerase, and oligonucleotides with appropriate enzyme restriction sites added in their sequences listed in Table 7 according to the scheme represented in Fig. S17 at https://figshare.com/s/526a9777225b6750159c. Apramycin-resistant mutants were verified by PCR (see Fig. S15B at https://figshare.com/s/526a9777225b6750159c) using oligonucleotides listed in Table 5 and by sequencing of the PCR amplification products.
TABLE 7.
Oligonucleotides used for gene overexpression
| Oligonucleotide | Sequence 5′–3′ | Restriction site | Use |
|---|---|---|---|
| eSARP/LysR147FW | AACTGCAGCGGCGACCGATTACGGG | PstI | cbm16-17 C15 CS147 overexpression |
| eSARP/LysR147RV | GCTCTAGACCCCAGTTGGGAACCGTCGA | XbaI |
For genetic complementation of mutants in halogenase, flavin reductase, SARP, and LysR regulator genes (CS147ΔHalo, CS147ΔOxid, CS147ΔSARP, and CS147ΔLysR, respectively), the complete ORF of each gene (and both ORFs coding LysR and SARP regulators in the case of CS147ΔLysR mutant) were amplified by PCR and cloned under ermE*p control into pSETEcH (halogenase, SARP, and LysR regulators) or pSETETc (flavin reductase gene) using oligonucleotides and restriction enzyme sites added, with their sequences listed in Table 8 according to the scheme represented in Fig. S18 at https://figshare.com/s/526a9777225b6750159c.
TABLE 8.
Oligonucleotides used for gene complementation
| Oligonucleotide | Sequence, 5′–3′ | Restriction site | Use |
|---|---|---|---|
| eHalo147FW | TCTAGAACCGGAATCTGTGGAG | XbaI | cbm12 C15 CS147 complementation |
| eHalo147RV | GATATCCCGCATGTCCTTTCG | EcoRV | |
| eFlavRed147FW | GCTCTAGAAGCAGCGTTCGATGAC | XbaI | cbm10 C15 CS147 complementation |
| eFlavRed147RV | TATGAATTCGATGGCCGTGTGGG | EcoRI | |
| eSARP147FW | CGCTCTAGAACGGCGACCGATTACG | XbaI | cbm16 C15 CS147 complementation |
| eSARP147RV | TGCAGATATCGGCGGCGTCAGGAAG | EcoRV | |
| eSARP147FW | GCTCTAGACGGCGACCGATTACG | XbaI | cbm17 C15 CS147 complementation |
| eLysR147RV | TATGATATCTCAGGGCCTGGATCCG | EcoRV | |
| ePKSFW | GCTCTAGATCTGCCGCATGACTAC | XbaI | cbm18-21 C15 CS147 complementation |
| ePKSRV | GCCGAATTCCGCTTCCCGGATTCAG | EcoRI | |
| eMet147FW | GCTCTAGATCGGCCGCATTCATCC | XbaI | cbm27 C15 CS147 complementation |
| eMet147RV | TATGAATTCTCAGGCGTCGGTTCC | EcoRI | |
| eCytP450FW | TATCTAGAATGACCACGCAGCAGAC | XbaI | cbm25 C15 CS147 complementation |
| eCytP450RV | TACGAATTCCGCCCGCCATGTCAG | EcoRI |
Thiostreptone- or hygromycin-complemented Streptomyces strains were verified by PCR with oligonucleotides listed in Table 5.
Analysis of metabolites by UPLC and HPLC-MS.
Secondary metabolites produced in R5A-Cl medium at 3, 6, and 9 days were extracted from 1-ml whole cultures for 2 h with one equal volume of three different solvents: ethyl acetate, butanol, and ethyl acetate with 1% formic acid, centrifuged at 16,400 relative centrifugal force to take the upper organic phase, dried in vacuum in a SpeedVac, and resuspended in 60 μl of methanol.
Samples were then analyzed by reversed-phase chromatography in Acquity UPLC I-class equipment (Waters) using a BEH C18 column (1.7 μm, 2.1 by 100 mm; Waters). The injected volume was 10 μl, and the mobile phase was composed of solvent A, 100% acetonitrile, and B, milliQ water with 0.1% trifluoroacetic acid (TFA); a 0.5 ml/min flow rate and 35°C column temperature were set. The gradient used for samples elution was 100% to 10% of acetonitrile in 1 min, 10% to 100% acetonitrile in 7 min, and 2 min in 100% acetonitrile.
For HPLC-MS spectrometry, samples were extracted and prepared as described above and then analyzed by an Alliance HPLC chromatographic system coupled to a ZQ4000 mass spectrometer (Waters) with a Sunfire C18 column (3.5 μm, 2.1 by 150 mm; Waters). The mobile phase was the same as that described above, the flow rate was 0.25 ml/min, and the gradient used for sample elution was 4 min at 10% A and 90% B, 10% to 88% A in 26 min, and 10 min at 100% A. The mass analysis was carried out by positive ESI with 3-kV capillary voltage and 20-V cone voltage.
Mass values from HPLC-MS were correlated with the peaks detected by UPLC-UV analysis by comparison between UV spectra and retention time values. Reanalysis of the purified compounds in both the UPLC and HPLC systems confirmed the match between the peaks observed with each chromatographic technique.
HRMS and dereplication analysis.
Cultures (2 ml) extracted as described above were resuspended in 100 μl methanol and then analyzed with a Zorbax SB-C8 column (3.5 μm, 2.1 by 30 mm) in an Agilent 1200 rapid-resolution HPLC coupled with a maXis from a Bruker mass spectrometer. The sample volume injected was 2 μl, and the mobile phase was composed of solvent A, 90:10 milliQ water-acetonitrile, and solvent B, milliQ 90:10 milliQ water-acetonitrile, both with 13 mM ammonium formate and 0.01 trifluoroacetic acid (TFA). Samples were eluted with a 0.3-ml/min flow rate, and the gradient used was 90% to 0% of solvent A/10% to 100% solvent B in 6 min, 0% solvent A/100% solvent B in 2 min, 0% to 90% solvent A/100% to 100% solvent B in 0.1 min, and 90% solvent A/10% solvent B for 9.1 min. The mass spectrometer was set in ESI mode with 4-kV capillary voltage, 11 L/min, 200°C, and 2.8 × 105 Pa. The retention time together with the exact mass (and the derived molecular formula) were used as criteria to search the in-house database from Fundación MEDINA (38) and the Dictionary of Natural Products (39).
Purification of compounds.
For isolation of compounds 1 to 9, supernatant from 2-L R5A-Cl 6-day cultures was filtrated and then retained by 10-g Sep-Pac Vac C18 cartridge (Waters) conditioned with methanol and milliQ water. The sample held in the cartridge was then fractionated using a collector by elution with an Alliance HPLC preparative chromatographic system (Waters) using as mobile phase solvents methanol and milliQ water, a flow rate of 5 ml/min, and a gradient of 0% to 100% methanol in 55 min. Fractions containing compounds of interest were dried in vacuum in a Rotavapor and resuspended in 2 ml of methanol.
Purification of compounds was carried out by reversed-phase chromatography with the same Alliance HPLC equipment as described above, using a SunFire C18 column (10 μm, 10 by 150 mm; Waters), and isocratic conditions with a flow rate of 5 ml/min and a column temperature of 35°C, using different ratio of solvents A, acetonitrile, and B, milliQ water plus TFA, depending on the compound (acetonitrile 25% for colibrimycin B, 35% for colibrimycins A1 to A3, 40% for colibrimycins C, A4, and A5). TFA was used, ranging between 0.05 and 0.1% depending on the compound (0.05% for colibrimycins A1 to A3 and B, 0.1% for colibrimycins C, A4, and A5).
Peaks containing the desired compounds were collected by a collector setup with the appropriate retention time and lyophilized.
Structural elucidation of each compound was carried out by ESI-time of flight high-resolution mass spectrometry, MS/MS, and NMR spectroscopy. HRMS was employed to determine the molecular formulas based on the experimental accurate masses and the corresponding isotopic distribution. Connectivity of the compounds was established based on one-dimensional (1D) and 2D NMR analyses alongside the fragmentation pattern observed in the MS/MS experiments. HRMS spectra were collected as previously indicated in “HRMS and dereplication analysis,” above. MS/MS of each compound was obtained in the same Bruker maXis high-resolution mass spectrometer by selecting the molecular ion [M+H]+ for fragmentation, using N2 as collision-induced dissociation (CID) gas and the automatically interpolated collision energy provided by the instrument for each parent ion. NMR spectra were recorded in dimethyl sulfoxide (DMSO)-d6 at 24°C on a Bruker AVANCE III-500 (500 and 125 MHz for 1H and 13C NMR, respectively) equipped with a 1.7-mm TCI MicroCryoProbe, using the residual solvent signal as the internal reference (1H 2.50 and 13C 39.5 for DMSO-d6). A description of the structural elucidation of the new compounds alongside their HRMS, MS/MS, and NMR spectra is included in Fig. S19 to S100 and Tables S5 to S16 at https://figshare.com/s/526a9777225b6750159c.
In vitro bioactivity assays.
Solid medium bioassays were performed for the analysis of antimicrobial activity of the purified compounds against Gram-negative E. coli and Pseudomonas aeruginosa, Gram-positive Staphylococcus aureus and Micrococcus luteus, and the fungus Candida albicans.
Bacterial strains were grown in 250-ml flasks containing 50 ml of TSB medium at 37°C and 250 rpm to a final OD600 of 0.4 to 0.6. YPD medium (10 g/L yeast extract, 10 g/L peptone, 20 g/L dextrose) and 30°C were used for Candida albicans. Once this optical density was reached, 150 μl of microorganism (300 μl in the case of M. luteus) was added to 15 ml of medium (TSA or YPD for bacterial and yeast strains, respectively) with 1.5% agar tempered at 50°C and plated. Once solidified, the discs previously impregnated with 20 μl of the compound at different concentrations (10, 25, 50, 75, and 100 μg/ml) were placed, and plates were incubated for 1 day.
The cytotoxicity of compounds was tested against the following human tumor cell lines: colon adenocarcinoma (HT29), non-small-cell lung cancer (A549), breast adenocarcinoma (MDA-MB-231), promyelocytic leukemia (HL-60), and pancreatic cancer (CAPAN-1). The mouse embryonic fibroblast cell line NIH/3T3 was used as a control to evaluate cytotoxicity against nonmalignant cells. These analyses were carried out as described previously (63).
Data availability.
The colibrimycin biosynthetic gene cluster has been deposited at the Minimum Information about a Biosynthetic Gene Cluster (MIBiG) repository (64) under the accession number BGC0002100.
ACKNOWLEDGMENTS
This research was supported by grants of the Spanish Ministry of Economy and Competitiveness (MINECO) (BIO2015-64161-R to J.A.S.) and MCIU/AEI/FEDER, UE (RTI2018-093562-B-I00 to J.A.S and C.O). The NMR spectrometer used in this work was purchased via a grant for scientific and technological infrastructures from the Ministerio de Ciencia e Innovación (PCT-010000-2010-4).
We acknowledge the technical support provided by Servicios Científico-Técnicos de la Universidad de Oviedo.
Contributor Information
Carlos Olano, Email: olanocarlos@uniovi.es.
Isaac Cann, University of Illinois at Urbana-Champaign.
REFERENCES
- 1.Bérdy J. 2012. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot 65:385–395. 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi Y, Nakashima T. 2018. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics 7:45. 10.3390/antibiotics7020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves C, Silva J, Pinteus S, Gaspar H, Alpoim MC, Botana LM, Pedrosa R. 2018. From marine origin to therapeutics: the antitumor potential of marine algae-derived compounds. Front Pharmacol 9:777. 10.3389/fphar.2018.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wink J, Mohammadipanah F, Hamedi J. 2017. Biology and bio/technology of Actinobacteria. Springer International Publishing, Cham, Switzerland. 10.1007/978-3-319-60339-1. [DOI] [Google Scholar]
- 5.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clément C, Ouhdouch Y, van Wezel GP. 2016. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80:1–43. 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demain AL, Sanchez S. 2009. Microbial drug discovery: 80 years of progress. J Antibiot 62:5–16. 10.1038/ja.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olano C, Méndez C, Salas JA. 2011. Molecular insights on the biosynthesis of antitumour compounds by actinomycetes. Microb Biotechnol 4:144–164. 10.1111/j.1751-7915.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N, Hwang S, Lee Y, Cho S, Palsson B, Cho BK. 2019. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J Microbiol Biotechnol 29:667–686. 10.4014/jmb.1904.04015. [DOI] [PubMed] [Google Scholar]
- 9.Olano C, García I, González A, Rodríguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA. 2014. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner C, El Omari M, König GM. 2009. Biohalogenation: nature’s way to synthesize halogenated metabolites. J Nat Prod 72:540–553. 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- 11.Smith DR, Grüschow S, Goss RJ. 2013. Scope and potential of halogenases in biosynthetic applications. Curr Opin Chem Biol 17:276–283. 10.1016/j.cbpa.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Karabencheva-Christova TG, Torras J, Mulholland AJ, Lodola A, Christov CZ. 2017. Mechanistic insights into the reaction of chlorination of tryptophan catalyzed by tryptophan 7-halogenase. Sci Rep 7:17395. 10.1038/s41598-017-17789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Pée KH. 2001. Microbial biosynthesis of halometabolites. Arch Microbiol 175:250–258. 10.1007/s002030100263. [DOI] [PubMed] [Google Scholar]
- 14.Brown S, O’Connor SE. 2015. Halogenase engineering for the generation of new natural product analogues. ChemBioChem 16:2129–2135. 10.1002/cbic.201500338. [DOI] [PubMed] [Google Scholar]
- 15.Neubauer PR, Widmann C, Wibberg D, Schröder L, Frese M, Kottke T, Kalinowski J, Niemann HH, Sewald N. 2018. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS One 13:e0196797. 10.1371/journal.pone.0196797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann CS, Fujimori DG, Walsh CT. 2008. Halogenation strategies in natural product biosynthesis. Chem Biol 15:99–109. 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Senn HM. 2014. Insights into enzymatic halogenation from computational studies. Front Chem 2:98. [PMC] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Pée KH, Patallo EP. 2006. Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl Microbiol Biotechnol 70:631–641. 10.1007/s00253-005-0232-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Kim J, Kim H, Kim E, Jeong H, Choi K, Kim B. 2020. Characterization of a tryptophan 6‐halogenase from Streptomyces albus and its regioselectivity determinants. ChemBioChem 21:1446–1452. 10.1002/cbic.201900723. [DOI] [PubMed] [Google Scholar]
- 20.Hornung A, Bertazzo M, Dziarnowski A, Schneider K, Welzel K, Wohlert SE, Holzenkämpfer M, Nicholson GJ, Bechthold A, Süssmuth RD, Vente A, Pelzer S. 2007. A genomic screening approach to the structure-guided identification of drug candidates from natural sources. ChemBioChem 8:757–766. 10.1002/cbic.200600375. [DOI] [PubMed] [Google Scholar]
- 21.Liao L, Chen R, Jiang M, Tian X, Liu H, Yu Y, Fan C, Chen B. 2016. Bioprospecting potential of halogenases from Arctic marine actinomycetes. BMC Microbiol 16:34. 10.1186/s12866-016-0662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehner S, Kotzsch A, Bister B, Süssmuth RD, Méndez C, Salas JA, van Pée KH. 2005. A regioselective tryptophan 5-halogenase is involved in pyrroindomycin biosynthesis in Streptomyces rugosporus LL-42D005. Chem Biol 12:445–452. 10.1016/j.chembiol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Fullone MR, Paiardini A, Miele R, Marsango S, Gross DC, Omura S, Ros-Herrera E, Bonaccorsi di Patti MC, Laganà A, Pascarella S, Grgurina I. 2012. Insight into the structure-function relationship of the nonheme iron halogenases involved in the biosynthesis of 4-chlorothreonine–Thr3 from Streptomyces sp. OH-5093 and SyrB2 from Pseudomonas syringae pv. syringae B301DR. FEBS J 279:4269–4282. 10.1111/febs.12017. [DOI] [PubMed] [Google Scholar]
- 24.Blodgett JAV, Oh DC, Cao S, Currie CR, Kolter R, Clardy J. 2010. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc Natl Acad Sci USA 107:11692–11697. 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Currie CR. 2001. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol 55:357–380. 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 26.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704. 10.1038/19519. [DOI] [Google Scholar]
- 27.Currie CR, Stuart AE. 2001. Weeding and grooming of pathogens in agriculture by ants. Proc Biol Sci 268:1033–1039. 10.1098/rspb.2001.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. 2009. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci USA 106:12295–12300. 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rugg G, Senn HM. 2017. Formation and structure of the ferryl [Fe=O] intermediate in the non-haem iron halogenase SyrB2: classical and QM/MM modelling agree. Phys Chem Chem Phys 19:30107–30119. 10.1039/c7cp05937j. [DOI] [PubMed] [Google Scholar]
- 30.Hohaus K, Altmann A, Burd W, Fischer I, Hammer PE, Hill DS, Ligon JM, van Pée KH. 1997. NADH-dependent halogenases are more likely to be involved in halometabolite biosynthesis than haloperoxidases. Angew Chem Int Ed Engl 36:2012–2013. 10.1002/anie.199720121. [DOI] [Google Scholar]
- 31.Piraee M, White RL, Vining LC. 2004. Biosynthesis of the dichloroacetyl component of chloramphenicol in Streptomyces venezuelae ISP5230: genes required for halogenation. Microbiology (Reading) 150:85–94. 10.1099/mic.0.26319-0. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. 2002. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem Biol 9:519–531. 10.1016/S1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 33.Hammer PE, Hill DS, Lam ST, Van Pée KH, Ligon JM. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl Environ Microbiol 63:2147–2154. 10.1128/aem.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HS, Brady SF. 2014. Arixanthomycins A–C: phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem Biol 9:1267–1272. 10.1021/cb500141b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz JCS, Maffioli SI, Bernasconi A, Brunati C, Gaspari E, Sosio M, Wellington E, Donadio S. 2017. Allocyclinones, hyperchlorinated angucyclinones from Actinoallomurus. J Antibiot (Tokyo) 70:73–78. 10.1038/ja.2016.62. [DOI] [PubMed] [Google Scholar]
- 36.Wu S, Huang T, Xie D, Wo J, Wang X, Deng Z, Lin S. 2017. Xantholipin B produced by the stnR inactivation mutant Streptomyces flocculus CGMCC 4.1223 WJN 1. J Antibiot (Tokyo) 70:90–95. 10.1038/ja.2016.60. [DOI] [PubMed] [Google Scholar]
- 37.Chooi YH, Tang Y. 2010. Adding the lipo to lipopeptides: do more with less. Chem Biol 17:791–793. 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Victoria I, Martín J, Reyes F. 2016. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med 82:857–871. 10.1055/s-0042-101763. [DOI] [PubMed] [Google Scholar]
- 39.Buckingham J. 2017. Dictionary of natural products on USB. Version 29.2. CRC Press, Boca Raton, FL. https://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml. [Google Scholar]
- 40.Malmierca MG, González-Montes L, Pérez-Victoria I, Sialer C, Braña AF, Salcedo R, Martín J, Reyes F, Méndez C, Olano C, Salas JA. 2018. Searching for glycosylated natural products in actinomycetes and identification of novel macrolactams and angucyclines. Front Microbiol 9:39. 10.3389/fmicb.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobylarz MJ, Jason CG, Shin-Ichi JT, Dushyant KR, David EH, Michael EPM. 2014. Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor. Chem Biol 21:379–388. 10.1016/j.chembiol.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Asano T, Matsuoka K, Hida T, Kobayashi M, Kitamura Y, Hayakawa T, Iinuma S, Kakinuma A, Kato K. 1994. Novel retrovirus protease inhibitors, RPI-856 A, B, C and D, produced by Streptomyces sp. AL-322. J Antibiot (Tokyo) 47:557–565. 10.7164/antibiotics.47.557. [DOI] [PubMed] [Google Scholar]
- 43.Kaysser L. 2019. Built to bind: biosynthetic strategies for the formation of small-molecule protease inhibitors. Nat Prod Rep 36:1654–1686. 10.1039/c8np00095f. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. 2020. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368:409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu HT, Hubbard BK, Shah AN, Eide J, Fredenburg RA, Walsh CT, Khosla C. 2001. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc Natl Acad Sci USA 98:8548–8553. 10.1073/pnas.151246498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory MA, Hong H, Lill RE, Gaisser S, Petkovic H, Low L, Sheehan LS, Carletti I, Ready SJ, Ward MJ, Kaja AL, Weston AJ, Challis IR, Leadlay PF, Martin CJ, Wilkinson B, Sheridan RM. 2006. Rapamycin biosynthesis: elucidation of gene product function. Org Biomol Chem 4:3565. 10.1039/b608813a. [DOI] [PubMed] [Google Scholar]
- 47.Cortina NS, Krug D, Plaza A, Revermann O, Müller R. 2012. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl 51:811–816. 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 48.Plaza A, Viehrig K, Garcia R. 2013. Jahnellamides, r-keto-β-methionine-containing peptides from the terrestrial Myxobacterium Jahnella sp.: structure and biosynthesis. Org Lett 15:5882–5885. 10.1021/ol402967y. [DOI] [PubMed] [Google Scholar]
- 49.Morinaka BI, Lakis E, Verest M, Helf MJ, Scalvenzi T, Vagstad AL, Sims J, Sunagawa S, Gugger M, Piel J. 2018. Natural noncanonical protein splicing yields products with diverse β-amino acid residues. Science 359:779–782. 10.1126/science.aao0157. [DOI] [PubMed] [Google Scholar]
- 50.Mishra A, Choi J, Choi SJ, Baek KH. 2017. Cyclodipeptides: an overview of their biosynthesis and biological activity. Molecules 22:1796. 10.3390/molecules22101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maiya S, Grundmann A, Li S, Turner G. 2006. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem 7:1062–1069. 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 52.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. 2008. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc 130:4507–4516. 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 53.Sánchez L, Braña AF. 1996. Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology 142:1209–1220. 10.1099/13500872-142-5-1209. [DOI] [PubMed] [Google Scholar]
- 54.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics, 2nd ed. John Innes Centre, Norwich, England. [Google Scholar]
- 55.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. 1989. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol 31:272–277. 10.1007/BF00258408. [DOI] [Google Scholar]
- 56.Fernández E, Weißbach U, Reillo CS, Braña AF, Méndez C, Rohr J, Salas JA. 1998. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol 180:4929–4937. 10.1128/JB.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. University of Texas Southwestern Medical Center, Dallas, TX. [Google Scholar]
- 58.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 59.Olano C, Wilkinson B, Sanchez C, Moss S, Sheridan R, Math V, Weston A, Brana A, Martin C, Oliynyk M. 2004. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem Biol 11:87–97. 10.1016/S1074-5521(03)00297-7. [DOI] [PubMed] [Google Scholar]
- 60.Cano-Prieto C, García-Salcedo R, Sánchez-Hidalgo M, Braña AF, Fiedler HP, Méndez C, Salas JA, Olano C. 2015. Genome mining of Streptomyces sp. Tü 6176: characterization of the nataxazole biosynthesis pathway. ChemBioChem 16:1461–1473. 10.1002/cbic.201500153. [DOI] [PubMed] [Google Scholar]
- 61.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malmierca MG, Pérez-Victoria I, Martín J, Reyes F, Méndez C, Olano C, Salas JA. 2018. Cooperative involvement of glycosyltransferases in the transfer of amino sugars during the biosynthesis of the macrolactam sipanmycin by Streptomyces sp. strain CS149. Appl Environ Microbiol 84:e01462-18. 10.1128/AEM.01462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, Selem-Mojica N, Alanjary M, Robinson SL, Lund G, Epstein SC, Sisto AC, Charkoudian LK, Collemare J, Linington RG, Weber T, Medema MH. 2019. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res 48:D454–D458. 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 66.Peláez AI, Ribas-Aparicio RM, Gomez A, Rodicio MR. 2001. Structural and functional characterization of the recR gene of Streptomyces. Mol Genet Genomics 265:663–672. 10.1007/s004380100460. [DOI] [PubMed] [Google Scholar]
- 67.Sun Y, He X, Liang J, Zhou X, Deng Z. 2009. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl Microbiol Biotechnol 82:303–310. 10.1007/s00253-008-1793-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The colibrimycin biosynthetic gene cluster has been deposited at the Minimum Information about a Biosynthetic Gene Cluster (MIBiG) repository (64) under the accession number BGC0002100.