ABSTRACT
Akkermansia muciniphila is a mucin-degrading bacterium found in the human gut and is often associated with positive human health. However, despite being detected by as early as 1 month of age, little is known about the role of Akkermansia in the infant gut. Human milk oligosaccharides (HMOs) are abundant components of human milk and are structurally similar to the oligosaccharides that comprise mucin, the preferred growth substrate of human-associated Akkermansia. A limited subset of intestinal bacteria has been shown to grow well on HMOs and mucin. We therefore examined the ability of genomically diverse strains of Akkermansia to grow on HMOs. First, we screened 85 genomes representing the four known Akkermansia phylogroups to examine their metabolic potential to degrade HMOs. Furthermore, we examined the ability of representative isolates to grow on individual HMOs in a mucin background and analyzed the resulting metabolites. All Akkermansia genomes were equipped with an array of glycoside hydrolases associated with HMO deconstruction. Representative strains were all able to grow on HMOs with various efficiencies and growth yields. Strain CSUN-19, belonging to the AmIV phylogroup, grew to the highest level in the presence of fucosylated and sialylated HMOs. This activity may be partially related to the increased copy numbers and/or the enzyme activities of the α-fucosidases, α-sialidases, and β-galactosidases. This study examines the utilization of individual purified HMOs by Akkermansia strains representing all known phylogroups. Further studies are required to examine how HMO ingestion influences gut microbial ecology in infants harboring different Akkermansia phylogroups.
IMPORTANCE Human milk oligosaccharides (HMOs) are the third most abundant component of breast milk and provide several benefits to developing infants, including the recruitment of beneficial bacteria to the human gut. Akkermansia strains are largely considered beneficial bacteria and have been detected in colostrum, breast milk, and young infants. A. muciniphila MucT, belonging to the AmI phylogroup, contributes to the HMO deconstruction capacity of the infant. Here, using phylogenomics, we examined the genomic capacities of four Akkermansia phylogroups to deconstruct HMOs. Indeed, each phylogroup contained differences in their genomic capacities to deconstruct HMOs, and representative strains of each phylogroup were able to grow using HMOs. These Akkermansia-HMO interactions potentially influence gut microbial ecology in early life, a critical time for the development of the gut microbiome and infant health.
KEYWORDS: Akkermansia muciniphila, human milk oligosaccharides, fucosylated HMO, sialylated HMO, HMO utilization, Akkermansia phylogroups, glycoside hydrolase (GH), GH29, GH95
INTRODUCTION
Akkermansia muciniphila is a mucin-degrading specialist that colonizes the mucus layer of the human gastrointestinal tract (1). Paradoxically, Akkermansia also promotes mucus production by enhancing the differentiation of gut epithelial cells, thereby influencing mucosal homeostasis (2). Numerous positive associations have been observed between this bacterial lineage and human health. In adults, a decreased abundance of Akkermansia is associated with metabolic impairments (3), ulcerative colitis (4), and inflammatory bowel disease (5). In infants, a decrease in mucosal residents such as Akkermansia is associated with a compromised immune system and the development of atopic dermatitis (6).
The mechanisms by which A. muciniphila benefits human health appear to be directly linked to its ecological niche along the human gastrointestinal tract. Specifically, A. muciniphila bacteria colonize the oxic-anoxic interface of the mucus layers adjacent to host epithelial cells where they degrade host-produced mucins (7). Mucins are the main structural components of mucus and are composed of polypeptide chains rich in serine, threonine, and proline residues that are O-linked to a variety of oligosaccharides (8). These oligosaccharide side chains are comprised of N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc), and galactose and are capped with N-acetylneuraminic acid (Neu5Ac) (sialic acid), fucose, or sulfate. Akkermansia can utilize mucins as its sole carbon and nitrogen source, generating metabolites such as acetate, succinate, and propionate in the presence of vitamin B12 (9, 10). Co-occurring members of the gut microbiome convert some of the acetate produced to butyrate (11). Together, these organic acids fuel colonocytes and act as signaling molecules helping to maintain an overall anti-inflammatory tone in the gut (12). In addition to producing anti-inflammatory metabolites, A. muciniphila produces an extracellular surface protein, encoded by Amuc_1100, that interacts directly with Toll-like receptors on host epithelial cells (13, 14). This interaction results in the production of specific anti-inflammatory cytokines, including interleukin-10 (IL-10), which leads to an improvement in overall gut barrier function (13).
Building upon previous work by Guo and colleagues (15), we recently performed a comparative genomic analysis of 75 Akkermansia genomes to define the genomic and functional landscape of this lineage. This analysis identified at least four distinct phylogroups, AmI to AmIV, with A. muciniphila MucT belonging to the AmI phylogroup. Additionally, this work showed that the Akkermansia phylogroups had differing functional potentials, including de novo biosynthesis of vitamin B12 by members of the AmII and AmIII phylogroups (10).
Continuing to explore the genomic and metabolic diversity of human-associated Akkermansia, we next wanted to determine if host-produced glycans, other than those in mucin, could support the growth of various Akkermansia phylogroups. Because of the compositional and structural similarities between the oligosaccharides found in mucin and human milk, we focused on human milk oligosaccharides (HMOs) (8, 16, 17). Human milk contains 5 to 15 g/L HMOs, of which 50 to 80% are fucosylated and 10 to 20% are sialylated (16). Although HMOs are present in milk as a pool of over 200 diverse structures, they are composed of only five monosaccharides: glucose, galactose, fucose, GlcNAc, and sialic acid (16). These oligosaccharides contain a lactose core at the reducing end that is extended with building block monosaccharides via glycosidic linkages. In human milk, fucose can be attached via α1-2, α1-3, and α1-4 linkages, and sialic acid can be attached via α2-3 and α2-6 linkages. Simple, abundant, and routinely studied HMO structures include lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), 2′-fucosyllactose (2′-FL), 3-fucosyllactose (3-FL), 6′-sialyllactose (6′-SL), and 3′-sialyllactose (3′-SL) (18).
The oligosaccharides found in human milk are not digestible by the developing infant and reach the intestine intact (19). Once there, HMOs have a variety of functions, including providing protection from pathogens, playing a role in the modulation of gut epithelial cells, and enriching for a beneficial microbiota (20–22). Several studies have screened HMO consumption by various intestinal commensals and have identified a limited group of bacteria, primarily Bifidobacterium and select Bacteroides species, with this ability (23–25). One Akkermansia strain, belonging to phylogroup AmI (i.e., A. muciniphila MucT), has recently been shown to grow on human milk and select HMOs using a repertoire of glycoside hydrolase (GH) enzymes (26). In this study, we expand our understanding of this HMO-degrading capacity of human-associated Akkermansia beyond the one phylogroup. We hypothesized that Akkermansia strains from different phylogroups will differ in their abilities to metabolize HMOs, and these differences are related to their genomic composition. To investigate the ability of Akkermansia to grow on select HMOs, we first took a comparative genomics approach focusing on the presence and abundance of genes coding for glycoside hydrolase enzymes known to be involved in HMO catabolism. We then performed comparative growth experiments and demonstrated the robust growth of one representative strain from each of the four phylogroups in a basal medium supplemented with five individual pure HMOs, in a background of mucin, thus simulating the carbon sources available in the infant gut environment. These findings expand the known metabolic capabilities of human-associated Akkermansia and point to further functional differences among the genomically distinct phylogroups.
RESULTS
Newly isolated Akkermansia strains differ in their phylogenomic characteristics.
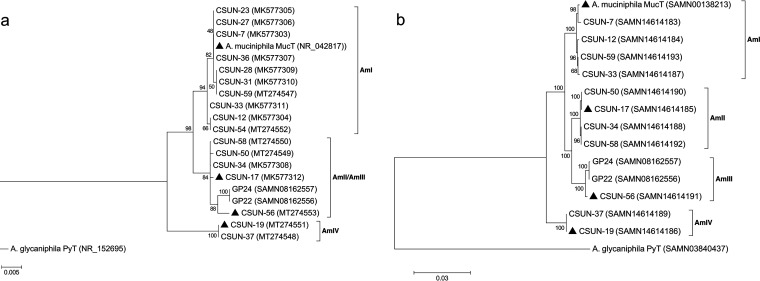
In total, 17 human-associated Akkermansia strains were isolated from healthy adults, 10 from males and 7 from females (see Table S2 in the supplemental material). Phylogenetic analyses of the nearly complete 16S rRNA gene sequences from each isolate revealed three well-supported clades, with the AmIII phylogroup nested within the AmII phylogroup (Fig. 1a). At least one isolate was obtained from the four known human-associated phylogroups (10). Ten of the 17 isolates treed within the AmI phylogroup, followed by 4 in AmII, 2 in AmIV, and 1 in AmIII.
FIG 1.
Phylogenetic relationship of Akkermansia isolates based on nearly full-length 16S rRNA gene sequences (a) and concatenation of 49 ribosomal protein-coding genes obtained from draft genomes (b). Both trees are rooted using the only other named species of the genus, Akkermansia glycaniphila PyT. Isolates with triangles were used in HMO growth experiments. GP22 and GP24 in the AmIII phylogroup are from Guo and colleagues (15) and are included because only one AmIII representative is available in our culture collection. Both trees were generated in MEGA7 (68) using the maximum likelihood method, and numbers at the nodes indicate bootstrap values for 100 replicates. The tree in panel a was generated considering only unambiguously aligned nucleotide positions (n = 1,305). For panel b, a total of 7,327 amino acid positions across 49 protein-coding genes were used. Both trees are drawn to scale, with branch lengths measured in the number of substitutions per site. GenBank accession numbers are in parentheses.
Using the 16S rRNA tree as a guide, we selected 11 of the isolates spanning each phylogroup for genomic sequencing. The characteristics of these draft genomes are presented in Table 1. Of the new isolates, draft genome sizes ranged from 2.86 Mb (CSUN-56 [AmIII]) to 3.15 Mb (CSUN-19 [AmIV]), with 2,658 to 3,111 coding sequences (CDs), respectively, compared with a 2.67-Mb genome size and 2,576 CDs in A. muciniphila MucT. Across phylogroups, approximately 52% of CDs could be assigned a function, on average. The resolution of the AmIII phylogroup was improved by phylogenomic analysis that included 49 protein-coding genes (Fig. 1b).
TABLE 1.
Genomic properties of 11 human-associated Akkermansia isolatesa
| Strain (phylogroup) | No. of contigs | GC content (%) | Genome length (bp) | No. of CDs | No. of tRNAs | No. of rRNAs | No. of hypothetical proteins | No. of proteins with assignments | No. of EC no. assignments | No. of GO assignments | No. of KEGG pathways |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. muciniphila MucT (AmI) | 1 | 55.8 | 2,664,102 | 2,576 | 53 | 3 | 1,072 | 1,504 | 620 | 529 | 459 |
| Akkermansia CSUN-7 (AmI) | 52 | 55.1 | 2,875,736 | 2,880 | 50 | 3 | 1,377 | 1,503 | 623 | 530 | 464 |
| Akkermansia CSUN-12 (AmI) | 56 | 55.3 | 2,810,203 | 2,823 | 50 | 3 | 1,307 | 1,516 | 632 | 538 | 465 |
| Akkermansia CSUN-33 (AmI) | 49 | 55.3 | 2,833,117 | 2,853 | 50 | 3 | 1,307 | 1,546 | 633 | 541 | 469 |
| Akkermansia CSUN-59 (AmI) | 65 | 55.2 | 2,942,175 | 3,010 | 50 | 3 | 1,472 | 1,538 | 628 | 535 | 466 |
| Akkermansia CSUN-17 (AmII) | 29 | 58.2 | 2,999,178 | 2,856 | 49 | 3 | 1,354 | 1,502 | 641 | 542 | 474 |
| Akkermansia CSUN-34 (AmII) | 87 | 57.8 | 3,024,116 | 2,949 | 47 | 3 | 1,451 | 1,498 | 640 | 538 | 473 |
| Akkermansia CSUN-50 (AmII) | 23 | 58.2 | 3,005,559 | 2,842 | 49 | 3 | 1,365 | 1,477 | 632 | 533 | 470 |
| Akkermansia CSUN-58 (AmII) | 71 | 57.8 | 3,087,515 | 2,988 | 49 | 3 | 1,489 | 1,499 | 637 | 535 | 472 |
| Akkermansia CSUN-56 (AmIII) | 48 | 58.5 | 2,860,685 | 2,658 | 48 | 3 | 1,246 | 1,412 | 612 | 518 | 462 |
| Akkermansia CSUN-19 (AmIV) | 89 | 56.6 | 3,149,202 | 3,111 | 49 | 3 | 1,656 | 1,455 | 628 | 535 | 472 |
| Akkermansia CSUN-37 (AmIV) | 72 | 56.7 | 3,142,630 | 3,077 | 49 | 3 | 1,631 | 1,446 | 624 | 531 | 469 |
For comparison, the fasta sequence of Akkermansia muciniphila MucT was downloaded from GenBank (accession number CP001071.1) and analyzed identically to the new isolates. CDs, coding sequences; EC, Enzyme Classification; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
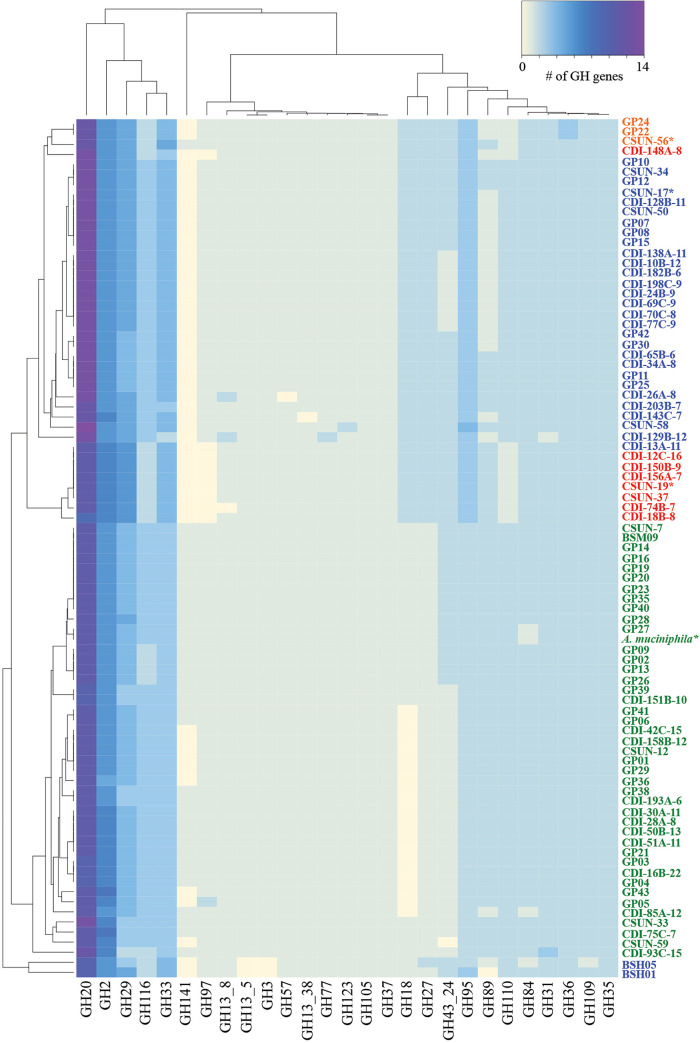
To investigate the carbohydrate-degrading potential of the Akkermansia strains, 85 genomes, including the 11 isolates from this study, were annotated against the CAZy database (27) using dbCAN (28, 29). We first took a global look at all annotated GH families and found significantly fewer GH annotations in genomes from the AmI phylogroup than in those from the other phylogroups (χ2 = 55.128; P < 0.0001 [by a Kruskal-Wallis test]) (Fig. S2). Furthermore, we identified consistent similarities and differences in the complements of GH annotations within and between each phylogroup (Fig. 2). With a few minor exceptions, these similarities and differences in GH counts resulted in the clustering of genomes into their respective phylogroups as evidenced by the dendrogram along the y axis in Fig. 2.
FIG 2.
Human-associated Akkermansia strains possess different complements of glycoside hydrolase (GH) genes potentially impacting their carbohydrate-degrading capabilities. The heat map shows the counts of different GH families present in the draft genomes of 85 total Akkermansia genomes. Genomes labeled with “CSUN” prefixes are isolates from this work, while the “CDI” genomes are from metagenome-assembled genomes (10), and the “GP” or “BSM” genomes are from isolates from Guo and colleagues (15). Each genome is colored by phylogroup affiliation. Green, AmI; blue, AmII; orange, AmIII; red, AmIV. Only three genomes (CDI-148A-8, BSH05, and BSH01) tree outside their phylogroup affiliation based on the GH content. Genomes with asterisks were used in the HMO growth experiments.
Next, since we were interested in the ability of Akkermansia to degrade HMOs, we focused on HMO-associated GH families previously identified in other organisms (26, 30–33). With this approach, we identified differences in the copy numbers of several GH families that are associated with the degradation of HMO glycans: α-fucosidases, α-sialidases, β-galactosidases, and N-acetyl β-hexosaminidases (Table 2). Most of these genes were also found to possess a signal peptide (Data Set S1), which is indicative of encoding extracellular enzymes (34, 35). Of note was the high number of GH20 genes compared with any other GH gene in all the genomes. The numbers of putative α-fucosidases (GH29, GH95, and GH141) and N-acetyl β-hexosaminidases (GH18, GH20, GH84, and GH109) also varied across phylogroups (lowest for AmI, including the strain tested here, A. muciniphila MucT). Of the four strains investigated for HMO catabolic capacity in this study, the CSUN-19 (AmIV phylogroup) and CSUN-56 (AmIII) strains showed 9 fucosidase annotations, compared with 8 for CSUN-17 (AmII) and 7 for A. muciniphila MucT (AmI) (Table 2).
TABLE 2.
Copy numbers of several human milk oligosaccharide-associated glycoside hydrolase families in representative strains from the different Akkermansia phylogroups
| Glycoside hydrolase family | Enzyme activity(ies) | Copy no. (avg copy no.) |
|||
|---|---|---|---|---|---|
| A. muciniphila MucT (AmI) | CSUN-17 (AmII) | CSUN-56 (AmIII) | CSUN-19 (AmIV) | ||
| GH2 | β-Galactosidase (or similar) | 6 (6.3) | 6 (6) | 6 (6) | 7 (6.9) |
| GH16 | β-Galactosidase (or similar) | 3 (2.9) | 3 (2.9) | 2 (2) | 2 (2) |
| GH18 | Chitinase; endo-β-N-acetylglucosaminidase (or similar) | 1 (0.5) | 2 (1.9) | 2 (2) | 2 (2) |
| GH20 | β-Hexosaminidase; lacto-N-biosidase; β-N-acetylglucosaminidase | 11 (11) | 13 (12.8) | 12 (12) | 11 (11) |
| GH29 | α-l-Fucosidase | 4 (3.8) | 5 (4.7) | 5 (5) | 6 (5.9) |
| GH33 | Sialidase (or similar) | 3 (3) | 4 (3.9) | 5 (4.3) | 4 (3.9) |
| GH35 | β-Galactosidase; exo-β-glucosaminidase | 2 (2) | 2 (2) | 2 (2) | 2 (2) |
| GH84 | N-Acetyl β-glucosaminidase; hyaluronidase | 1 (1.9) | 2 (2) | 2 (2) | 2 (2) |
| GH95 | α-l-Fucosidase; α-l-galactosidase | 2 (2) | 3 (3) | 3 (3) | 3 (3) |
| GH109 | α-N-Acetylgalactosaminidase; β-N-acetylhexosaminidase | 2 (2) | 2 (2) | 2 (2) | 2 (2) |
| GH141 | α-l-Fucosidase; xylanase | 1 (0.8) | 0 (0) | 1 (0.3) | 0 (0) |
The four Akkermansia strains show strain-dependent growth and utilization of HMOs.
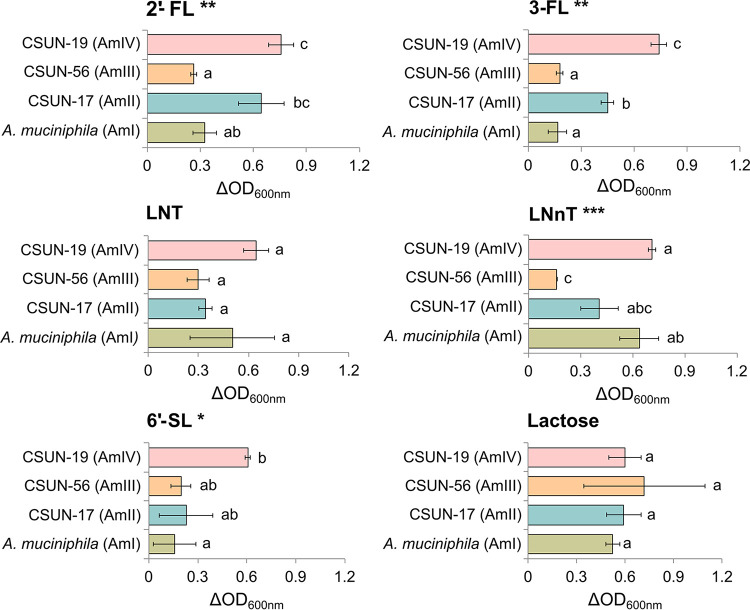
One representative of each of the four phylogroups was tested for its ability to grow on HMO in the presence of mucin. After 48 h of incubation, all strains tested grew to higher optical densities (ODs) in HMO (or lactose)-supplemented mucin medium than in medium lacking HMOs (Fig. 3). Growth yields varied across strains on media with 2′-FL, 3-FL, LNnT, and 6′-SL but not LNT or lactose (P < 0.05 by analysis of variance [ANOVA]). Post hoc comparisons revealed that strain CSUN-19, representing the AmIV phylogroup, showed the greatest growth in comparison to the other strains, with significant increases compared with A. muciniphila MucT in 2′-FL, 3-FL, and 6′-SL and with CSUN-56 in 2′-FL, 3-FL, and LNnT (Fig. 3).
FIG 3.
A representative strain from each of the four Akkermansia phylogroups was incubated in mucin-containing medium alone or supplemented with 20 mM individual human milk oligosaccharides or lactose. The experiment was conducted in triplicate and repeated at least two times. The difference in the OD600 from the growth in mucin-containing medium alone was used to plot the bacterial growth for each strain. Values are expressed as averages ± standard deviations. ANOVAs reveal significant effects (**, P < 0.01; *, P < 0.05) with the substrates 2′-fucosyllactose (2′-FL), 3-fucosyllactose (3-FL), lacto-N-neotetraose (LNnT), and 6′-sialyllactose (6’-SL) but not with lacto-N-tetraose (LNT) and lactose. Pairwise comparisons within each substrate using Tukey’s honestly significant difference test reveal significant differences between the phylogroups (P < 0.05); means showing letters in common are not significantly different.
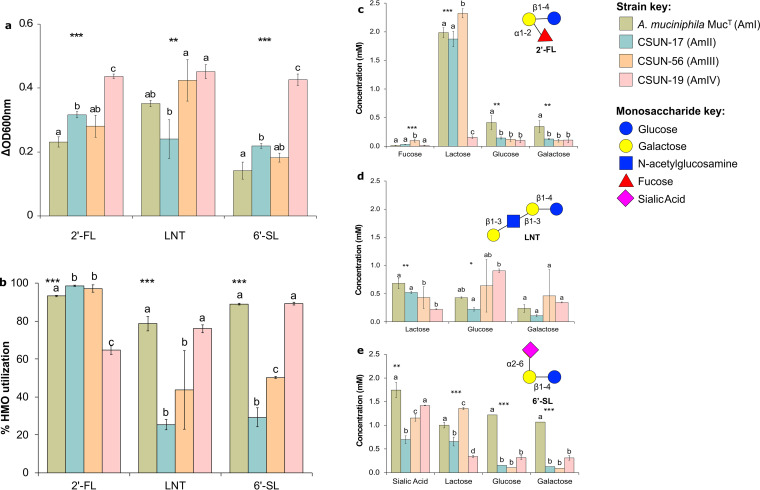
To confirm HMO utilization, we measured the concentrations of HMOs (2′-FL, LNT, and 6′-SL) and their sugar constituents (except GlcNAc for LNT) before and after 48 h of incubation (Fig. 4a and b). In addition to the differences in growth yields, the differences in the percentages of HMO utilized also varied across strains (P < 0.05). For 2′-FL, strains representing the AmI, AmII, and AmIII phylogroups utilized >93% of the available HMO, while CSUN-19 (AmIV) utilized just over 64% despite having the highest growth yield as measured by the change in the OD at 600 nm (OD600). Nearly all of the fucose liberated from 2′-FL was removed from the medium within 48 h by all the strains, while the lactose backbone accumulated in the culture medium of all strains except CSUN-19 (AmIV) (Fig. 4c). The degradation of LNT ranged from 25.4 to 78.6% across the tested strains, with CSUN-17 (AmII) utilizing the least and A. muciniphila MucT (AmI) utilizing the most. In contrast to growth on 2′-FL, most of the lactose from LNT was consumed across strains (Fig. 4d). Similar to LNT, there was a wide range of 6′-SL utilizations across strains (P < 0.001), ranging from 29.3% (CSUN-17 [AmII]) to 89.2% (CSUN-19 [AmIV]). In the case of 6′-SL, CSUN-19 showed the greatest growth, while A. muciniphila MucT showed the least growth, and yet the percentage of the substrate utilized (80%) showed no significant difference and was significantly higher than the ∼50% and ∼30% utilizations seen with CSUN-56 and CSUN-17, respectively. In all strains, sialic acid accumulated in the culture media and was not consumed when liberated from 6′-SL (Fig. 4e).
FIG 4.
A representative strain from each of the four Akkermansia phylogroups was incubated in mucin-containing medium alone or supplemented with 4 mM individual human milk oligosaccharides (HMOs) or lactose. The experiment was conducted in triplicate and repeated three times. (a) The difference in growth in HMO-supplemented medium from the growth in mucin-containing medium alone was used to plot the bacterial growth for each strain. (b) The concentrations of the original substrate analyzed were used to calculate the percentage of the HMO utilized. (c to e) Concentrations of the metabolites obtained after the deconstruction of 2′-fucosyllactose (2′-FL) (c), lacto-N-tetraose (LNT) (d), and 6′-sialyllactose (6′-SL) (e) expressed as averages ± standard deviations. Statistical analysis revealed significant effects between the substrates (a and b) and strains (b to d) (***, P < 0.001; **, P < 0.01; *, P < 0.05). Pairwise comparisons using Tukey’s honestly significant difference test were also performed, with a P value of <0.05, and means showing letters in common are not significantly different.
DISCUSSION
Akkermansia strains are largely considered beneficial members of the human gut microbiome and are currently of significant interest for their therapeutic potential (36). Until recently, however, all research involving these promising bacteria focused on a single species, A. muciniphila MucT, belonging to the AmI phylogroup. Here, we continue to build upon recent work by us and others describing the genomic and functional diversity within this lineage (10, 37). Specifically, we show that genomically diverse strains possess different complements of glycoside hydrolase (GH) genes that encode enzymes catalyzing the deconstruction of HMOs into constituent mono- and disaccharides. Furthermore, this study demonstrates that four different Akkermansia strains representing the four known phylogroups can deconstruct all the major types of HMOs, with this biological activity varying across strains. These differences in genomic and functional traits of human-associated Akkermansia strains, along with the diversity in the substrates that are presented to the gut bacteria in the form of breast milk or supplemented infant milk formula, potentially impact how and when Akkermansia strains colonize the human gastrointestinal tract. Thus, since HMO-Akkermansia interactions are strain specific, the corresponding pattern of early colonization with human-associated Akkermansia and the ensuing competitive advantage could also be strain specific in infants ingesting HMOs. Akkermansia bacteria are key contributors to the infants’ glycan-metabolizing capacity by as early as 4 months of age (33) and may therefore play a critical role in establishing a foundation of metabolic fitness in the naive microbiome. Taken together, these findings expand the known metabolic niche and interaction network of Akkermansia in the human gut early in life.
Bacterial growth studies have demonstrated that relatively few gut bacteria grow well on HMOs, the exceptions being bifidobacteria and select Bacteroides strains, both of which are dominant members of the infant gut (24, 25). Both bifidobacteria and Bacteroides employ an array of glycoside hydrolases, including fucosidases (GH29 and GH95), sialidases (GH33), galactosidases (GH2 and GH16), lacto-N-biosidases (GH20), and hexosaminidases (GH20), to deconstruct HMO linkages (38–44). Our phylogenomic characterization of the Akkermansia genomes shows that the various strains from the four Akkermansia phylogroups possess a wealth of these same gene annotations, albeit in differing abundances, that could be used for the deconstruction of either HMO or mucin. Genomes of the AmII, AmIII, and AmIV phylogroups were found to contain a higher number of genes encoding GH18, GH29, and GH95 than the genomes of the AmI phylogroup. This is consistent with findings by Becken and colleagues for AmII and AmIV (AmIII was not studied there) (37). They also found that the AmI phylogroup may be phylogenetically divided into two subclades, AmIa and AmIb, with A. muciniphila MucT belonging to subclade AmIa. Using this framework, they found differences in the complements of GH genes between AmIa and AmIb. Specifically, the AmIb subclade had few GH29-encoding genes and no GH18 genes compared to the AmIa subclade. This explains the lower average number of GH18 and GH29 genes in the AmI phylogroup that we observed and suggests that differences in carbohydrate catabolism likely exist within Akkermansia phylogroups.
Bifidobacteria employ two major strategies to hydrolyze HMOs (31, 42). Infant-associated Bifidobacterium infantis, Bifidobacterium breve, and Bifidobacterium longum primarily consume HMOs by employing intracellular glycoside hydrolases to deconstruct the HMO structures (41, 42, 45–47). Using an alternative strategy, Bifidobacterium bifidum extracellularly processes HMO via an array of membrane-associated glycoside hydrolases (31, 48). Bacteroides spp. harbor polysaccharide utilization loci (PULs) that encode a diverse array of glycosidases capable of breaking down host-produced and plant-derived polysaccharides (44, 49). Bacteroides bacteria are hypothesized to bind HMOs on the cell surface followed by the hydrolysis of the HMOs and the import of the resultant oligosaccharides for further breakdown. They co-opt their mucin utilization PULs to deconstruct and utilize HMOs with various efficiencies depending on the strain. Bacteroides fragilis is the most efficient in preferring HMOs with a high degree of polymerization and nonfucosylated HMOs over fucosylated HMOs (24) and even utilizes the sialic acid generated after the deconstruction of sialylated HMOs (44). Akkermansia does not have the typical PUL genomic organization seen in Bacteroides but it appears to harness extracellular GHs either in the periplasmic space or outside the cell altogether to cleave monosaccharides or disaccharides from mucin or HMOs (9, 26). In agreement, the majority of our GH annotations included signal peptide sequences indicative of export outside the cytoplasmic membrane. The extracellular cleavage of HMO (and mucin) results in the liberation of monosaccharides and disaccharides, which enables cross-feeding by other members of the gut microbiome (11). In the context of the infant gut, this cross-feeding could help facilitate colonization by new members of the gut community that are encountered as infants grow and consume new foods, aiding in the maturation of the gut microbiome in the early years of life (50).
In addition to cross-feeding on sugars liberated from host substrates, members of the gut microbiome feed off fermentation waste products produced by Akkermansia (26, 51, 52). In the case of fucosylated substrates such as 2′-FL, a distinct metabolite of fucose fermentation is 1,2-propanediol (26). Several bacteria, including both beneficial (Lactobacillus spp. and Eubacterium hallii) and pathogenic (Salmonella) bacteria, can grow on 1,2-propanediol in a vitamin B12-dependent manner (51, 52). Given our recent work showing that the AmII and AmIII phylogroups synthesize vitamin B12 (10), these findings indicate the possibility of Akkermansia-driven syntrophic interactions that are likely phylogroup specific. This is particularly relevant as the gut microbiome of exclusively breastfed infants has a decreased capacity for the de novo synthesis of vitamin B12 compared with formula-fed infants (53). Therefore, an understanding of which Akkermansia strain is already present in a host may influence the outcome of any microbiome or dietary intervention.
Several studies have detected Akkermansia in the stool of infants as early as 1 month after birth, in most 1-year-olds (54), and even in human colostrum and milk (55, 56), demonstrating that it colonizes the gut early in life and providing a possible route of inoculation. Two separate studies found direct associations between the abundances of Akkermansia and fucosylated HMO in human milk, suggesting that fucosylated HMO may help enrich for Akkermansia in the gut of the infant (57, 58). Here, we show that fucosylated HMOs support robust growth across all strains of Akkermansia. However, growth varied by strain, suggesting potential differences in growth and metabolic efficiencies across strains. When grown on 2′-FL, the liberated fucose was rapidly depleted from the culture medium, while the lactose component accumulated in the culture medium (except for CSUN-19), suggesting a general preference for fucose over lactose in Akkermansia. Cleavage of fucose from HMOs (and mucin) is mediated by fucosidases belonging to the GH29 and GH95 families (31, 38, 49), both of which were found in all four Akkermansia strains. GH141, a putative fucosidase or xylanase, was also observed in some of our AmI and AmIII genomes in this study. Kostopoulos et al. recently demonstrated that a GH29 gene product (encoded by Amuc_0010) in A. muciniphila MucT had relatively poor catalytic activity against 2′-FL, suggesting that 2′-FL was not the preferred substrate for this enzyme (26). It should be cautioned that A. muciniphila MucT belongs to subclade AmIa within the AmI phylogroup (37), hence the HMO utilization characteristics of this strain may not represent the capabilities of the whole AmI phylogroup. Overall, A. muciniphila MucT has four GH29 gene annotations, two of GH95, and one of GH141, and all these GH families could potentially encode enzymes that are involved in the degradation of fucosylated HMOs containing the α1-2 linkage. The numbers of these same GH families also varied across phylogroups, potentially leading to the differences in growth efficiencies that we observed. Given the prominent role of fucosylated HMOs in modulating the microbiome and enhancing health, and given that the concentrations of 2′-FL along with lacto-N-fucopentaose were highest during early lactation (59), the diversity of fucosidases available in each strain makes Akkermansia a potential candidate for further investigation in the field of infant-associated probiotics.
Sialyl oligosaccharides are associated with many benefits to neonates and infants (60, 61). For example, Charbonneau and colleagues demonstrated that the concentration of sialylated HMOs in breast milk correlated with growth in healthy Malawian infants (61). Furthermore, gnotobiotic mammals receiving fecal microbiota from infants with stunted growth and supplementation with sialylated bovine milk oligosaccharides showed improved growth (measured as weight gain and bone mass), with their gut microbiota developing metabolic fitness evidenced by an increase in genes related to energy metabolism (61). Sialic acid is an essential component of brain gangliosides and plays important roles in neuronal development, memory formation, and cognition (60). Three weeks of dietary supplementation with 3′-SL or 6′-SL administered to day-old piglets increased the ganglioside-bound sialic acid in the brains of the piglets, thus providing essential nutrients for brain growth and neurodevelopment (62). With regard to Akkermansia and sialylated HMOs, all four Akkermansia strains showed enhanced growth on 6′-SL and were able to deconstruct this sialylated oligosaccharide, but the growth yield and the percentage of the substrate degraded varied significantly across strains. These differences in yield and degradation did not align with the sialidase (GH33) gene copy number. For example, strain CSUN-56 representing AmIII has 5 sialidase annotations and exhibited relatively poor growth, with little degradation of 6′-SL. This incongruence between the bacterial gene number of a GH metabolizing a substrate and the physiological response to that substrate indicates the need to examine the transcription of the GHs and the enzyme kinetics of the associated GHs involved in the complete deconstruction of substrates and their transport into the cell. However, the accumulation of sialic acid in spent medium after growth on 6′-SL in all strains agrees with previous reports of Akkermansia lacking the nan operon for the import and consumption of sialic acid (26). The sialic acid released from the nonreducing end of the sugars enables access to the remaining oligosaccharides while also potentially encouraging the outgrowth of sialic acid-metabolizing, abundantly present commensal species such as B. fragilis, Faecalibacterium prausnitzii, Ruminococcus gnavus, and members of the Lactobacillus and Bifidobacterium genera (17, 63, 64). Several species of Enterobacteriaceae such as Escherichia coli and Salmonella enterica also thrive in a sialic acid-rich gut environment, with their fitness and virulence being directly proportional to their ability to metabolize sialic acid (63). Interestingly, though, studies in piglets demonstrated that supplementation with 6′-SL enhanced colonic bacteria such as Collinsella aerofaciens, Ruminococcus, Faecalibacterium, and Prevotella spp. while suppressing Enterobacteriaceae, Enterococcaceae, Lachnospiraceae, and Lactobacillales (62). Given the vulnerability of the infant population and the immaturity of the gut microbiome in early life, identifying the metabolic fate of sialic acid and the interaction between Akkermansia and sialic acid-metabolizing commensals and potential pathogens warrants further investigation.
Akkermansia strains are adapted to robust growth on mucin due to their habitation in the gut epithelial mucosa (65). Furthermore, HMOs, which are resistant to host digestive enzymes, are presented to the colonic microbiota in a mucin-rich background of the infant gut (66). We therefore included mucin in our HMO utilization experiments, and growth in the non-HMO mucin control served as a reference point to quantify HMO utilization. However, it is recognized that Akkermansia can grow in a mucin-deficient medium supplemented with GlcNAc, threonine, and tryptone (9). GlcNAc is a requirement for growth as Akkermansia does not express the enzyme required for the conversion of fructose-6-phosphate to glucosamine-6-phosphate, an essential component of the cell wall peptidoglycan (65). GlcNAc was thus added to the basal growth medium by Kostopoulos and colleagues while investigating HMO utilization by Akkermansia muciniphila MucT (26). We speculate that Akkermansia may grow exclusively using GlcNAc-containing HMOs such as LNT or LNnT, provided that the amino acid sources are added to the growth medium. However, since our current technique precluded analysis of GlcNAc, further growth experiments and chemical analyses are required to confirm this prediction.
In conclusion, human-associated Akkermansia strains can utilize a variety of host-derived HMOs for growth in vitro in a strain-dependent manner. This implies that the prebiotic effects of HMOs will depend on the resident strain of Akkermansia present in an individual. When grown on HMO, Akkermansia liberates sugars and produces fermentation products that can fuel other members of the gut microbiome. Together, these findings lay the foundation for future work examining the molecular mechanisms of HMO deconstruction by diverse strains of Akkermansia and how these activities influence interactions with the human host and other members of the gut microbiome in a strain-dependent manner.
MATERIALS AND METHODS
Recruitment and sampling.
Fecal samples used for Akkermansia isolations were obtained from 17 consenting healthy adults as previously described by Kirmiz et al. (10) under protocol number 1516-146, with approval from the Institutional Review Board at California State University, Northridge. Samples were refrigerated (4°C) and inoculated into culture medium (see below) within 24 h of collection.
Bacterial isolation and identification.
Akkermansia isolation and identification were conducted as previously described (10). Briefly, 5 mL of anaerobic basal mucin medium (BMM) containing 0.5% (vol/vol) mucin (see Table S1 in the supplemental material) was inoculated with fecal swabs in serum tubes, and a 10-fold serial dilution of up to 10−6 or 10−7 was performed for each sample. Cultures were incubated at 37°C for up to 5 days, and those with oval cells in pairs were further diluted in broth medium and/or transferred to BMM agar until purity could be verified microscopically using a Zeiss Axioskop instrument or as single colonies on BMM agar. For identification, genomic DNA was extracted using the DNeasy UltraClean microbial isolation kit (Qiagen Inc., MD, USA), and the nearly full-length 16S rRNA gene was amplified using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGA-3′) with the GoTaq Hot Start colorless master mix (Promega Corp., Madison, WI, USA). PCR was performed using an Eppendorf Vapo Protect Mastercycler Pro S 6325 system (Hamburg, Germany) and included an activation/denaturation step at 95°C for 3 min; 30 cycles of 95°C for 45 s, 45°C for 1 min, and 72°C for 1 min 45 s; and a final extension step at 72°C for 7 min, followed by a hold at 4°C. PCR products were purified (QIAquick PCR purification kit; Qiagen Inc.) and sequenced using either the 8F or 1492R primer on an ABI Prism 3730 DNA sequencer (Laragen Sequencing and Genotyping, Culver City, CA). If sequences were pure and positively matched to A. muciniphila in GenBank by BLAST analysis, the nearly full-length 16S rRNA gene was sequenced with additional primers (515F [GTGCCAGCMGCCGCGGTAA], 806R [GGACTACHVGGGTWTCTAAT], and 8F or 1492R). Sequences associated with each isolate were then assembled in Geneious 7.1.3 and imported into ARB (67). In ARB, sequences were manually aligned with secondary structure constraints against the 16S rRNA gene sequence of A. muciniphila MucT. To determine phylogroup affiliation based on 16S rRNA gene sequences, each isolate was added to our in-house database of Akkermansia 16S rRNA gene sequences as previously described (10, 15). Masked alignments were exported from ARB and imported into Kumar and colleagues’ (68) MEGA7, where phylogenetic reconstruction was performed using the maximum likelihood approach.
Genome sequencing, assembly, annotation, and phylogenomics.
Eleven Akkermansia isolates were selected for genome sequencing across three different sequencing efforts. DNAs from strains CSUN-7 and CSUN-12 were sequenced according to the Illumina sequencing protocol described previously by Oliver and colleagues (69). To obtain enough DNA for this sequencing protocol, four 5-mL cultures grown overnight in BMM were extracted as described above, and extracts were pooled and concentrated using ethanol precipitation with 3 M sodium acetate. Illumina sequencing libraries were then prepared as described previously by Oliver and colleagues. DNA from strains CSUN-17, CSUN-19, CSUN-33, and CSUN-34 were sequenced according to methods described previously by Parker and colleagues (70). For both this and the following sequencing efforts, enough quality genomic DNA was obtained from a single 5-mL culture of each isolate grown in BMM and extracted as described above. The DNAs from the remaining isolates (CSUN-37, CSUN-50, CSUN-56, CSUN-58, and CSUN-59) were sequenced on an Illumina NextSeq 550 platform (2 by 150 bp) by the Microbial Genome Sequencing Center (Pittsburgh, PA, USA).
For assembly and annotation, paired fastq files for each isolate were submitted to PATRIC (v 3.6.3) (71) for their comprehensive genome analysis workflow that uses Unicycler (72) to assemble genomes and RASTtk (73) for annotation. For comparison, the nucleotide sequence file for A. muciniphila MucT ATCC BAA-835 (BioSample accession number SAMN00138213) was downloaded from GenBank and annotated identically to the novel isolate genomes, also using tools in PATRIC. To investigate the carbohydrate-degrading potential of each Akkermansia phylogroup, the assembled contigs of the new isolates (n = 11) were combined with 74 publicly available Akkermansia genomes (10, 15, 74) and submitted to the dbCAN meta server for CAZyme annotation (27–29). dbCAN uses three tools, HMMER (75), DIAMOND (76), and Hotpep (77), for automated carbohydrate-active enzyme (CAZyme) annotation. Annotations were considered only if they matched with at least two of the three tools. Individual count files were tabulated and compiled using a custom python script to generate a frequency table for all genomes (n = 85). The resulting table was sorted and trimmed to include only glycoside hydrolase (GH) annotations, and a heat map was constructed in R (78) using the heatmap.2 function in the gplots library (79). Cluster dendrograms in the heat map were calculated using average linkage hierarchical clustering based on Bray-Curtis dissimilarity matrices calculated using the vegan package, also in R (80). To determine if there were differences in the numbers of GH predictions between phylogroups, a Kruskal-Wallis test (kruskal.test) followed by Dunn’s test (dunn.test, method=‘bonferroni’) were performed in R.
For phylogenomic analysis, amino acid sequences of 49 ribosomal protein-coding genes (81) were extracted and concatenated from assembled genomes using the phylogenomics workflow in anvi’o (82). The concatenated fasta file was then imported into MEGA7 (68) and aligned using MUSCLE (83), and a phylogenetic tree was made using the maximum likelihood method (84) with 100 bootstraps.
HMO growth experiments.
To determine if Akkermansia strains could grow using HMOs, we performed a series of growth experiments in a customized medium prepared by increasing the concentrations of threonine and tryptone (TT) in BMM (9). This medium, referred to here as BMM-TT (Table S1), was supplemented with individual HMOs before inoculation with the chosen Akkermansia strains. Five HMOs were tested, namely, 2′-FL, 3-FL, LNT, LNnT, and 6′-SL (Glycom, Hørsholm, Denmark). Lactose was also included in these growth experiments since it is the backbone of HMOs. Initially, one representative isolate of each phylogroup (AmI, A. muciniphila MucT; AmII, Akkermansia sp. strain CSUN-17; AmIII, Akkermansia CSUN-56; AmIV, Akkermansia CSUN-19) (Table S2) was grown overnight (18 to 24 h) in BMM at 37°C under an atmosphere of N2-CO2 (70:30, vol/vol). Cultures were then standardized to an OD600 of 0.5 in fresh BMM and used to inoculate (10%) 200 μL of BMM-TT or BMM-TT supplemented with 20 mM each HMO (or lactose) in 96-well microtiter plates (Falcon; Corning Incorporated, Corning, NY, USA) in triplicate. Wells were overlaid with 30 μL of filter-sterilized mineral oil to prevent evaporation over the 48-h incubation period. After 48 h of anaerobic (N2-CO2-H2 [80:15:5, vol/vol]) incubation at 37°C in a Bactron IV anaerobic chamber (Sheldon Manufacturing Inc., Cornelius, OR), plates were shaken for 10 s, and the OD600 was determined using a Spectramax microplate reader (Molecular Devices, San Jose, CA, USA). Growth was determined as the ΔOD600, i.e., the change in the OD600 of growth in BMM-TT supplemented with the HMOs relative to the growth in HMO-unsupplemented BMM-TT (i.e., BMM-TT + HMO OD600 − BMM-TT OD600). If the OD600 was >1.0, samples were diluted in half with fresh medium and reread. Each experiment was conducted in triplicate and repeated at least two times. To test for differences in growth across strains, we used repeated-measures analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test as appropriate. Uninoculated controls were included in each experiment and remained negative for growth.
To verify the degradation of three HMOs (2′-FL, LNT, and 6′-SL), the above-described experiments were repeated in 1.5 mL of BMM-TT supplemented with 4 mM HMO. These experiments were conducted in 24-well microtiter plates (Costar; Corning Incorporated, Corning, NY, USA) sealed with Microseal A film (Bio-Rad Laboratories Inc., Hercules, CA, USA) instead of mineral oil. Plates were incubated, and the OD600 and ΔOD600 were measured after 48 h as described above. For glycoanalytics, 0.5-mL aliquots were taken at time zero and 48 h after incubation, transferred to Eppendorf tubes, and centrifuged at 10,000 × g for 3 min at 4°C. The cell-free supernatants were stored at −20°C for glycoanalytics as described below. To compare growth, statistical analysis was conducted as described above.
HMO quantification.
Culture supernatants were collected at time zero and after 48 h of incubation to measure the degradation of 2′-FL, LNT, and 6′-SL. In addition to each parent HMO, individual sugars (with the exception of GlcNAc from LNT) of the three HMOs were also quantitatively measured using high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (85, 86). Frozen, cell-free spent culture media were thawed in a water bath, vortexed thoroughly to make a uniform mixture, and centrifuged at 7,000 × g for 5 min at 10°C, and 1 μl of the spent culture medium was injected into the HPAEC-PAD instrument for the detection of the above-mentioned sugars.
Carbohydrate analysis was done on a Dionex-ICS3000 system (Thermo Scientific, Sunnyvale, CA, USA) using a CarboPac PA-1 column (4 mm by 250 mm) attached to a Carbo PA1-guard column (4 mm by 50 mm). The detection of monosaccharides and oligosaccharides was done using standard Quad potential for carbohydrate analysis as supplied by the manufacturer. A gradient mixture of two solvents along with high-performance liquid chromatography (HPLC)-grade water was used for the optimum separation of monosaccharides and oligosaccharides present in the sample. Solvent A (water), solvent B (100 mM NaOH plus 7 mM sodium acetate [NaOAc]), and solvent C (100 mM NaOH plus 250 mM NaOAc) were used as elution solvents at a flow rate of 1.0 mL/min. Gradient mixture details are given in Table S4 in the supplemental material. Sugars were quantified by comparison with the area under the peaks from a standard mixture of fucose, galactose, glucose, 3-FL, lactose, 2′-FL, LNnT, LNT, sialic acid (Neu5Ac), 6′-SL, and 3-SL. Representative chromatograms are presented in Fig. S1. To determine the percentage of HMOs utilized, the amount remaining after 48 h of incubation was divided by the amount at time zero and multiplied by 100 [(HMO 48 h/HMO 0 h) × 100].
Data availability.
The data that support the findings of this study are openly available in the NCBI BioProject database under accession number PRJNA609771.
ACKNOWLEDGMENTS
We thank Louise Vigsnaes and Glycom A/S, Hørsholm, Denmark, for their generous donation of HMOs and their continued support of our work.
We declare no conflicts of interest.
E.L. helped conceive the project, conducted all HMO growth experiments, aided in data interpretation, and helped write the manuscript. S.G.P. performed statistical analysis, aided in data interpretation, and helped write the manuscript. N.K. helped conceive the project and write the manuscript. S.H., E.H., M.H., A.K., C.M., K.O., L.P., K.R., and P.S. helped collect samples and isolate Akkermansia strains. E.S. helped with the genomic analysis. B.C. and M.P. performed the glycoanalytics and helped with data interpretation. C.T.P. and K.C. performed genome sequencing and helped with genomic analysis. G.E.F. conceived of the project, aided in data interpretation, performed genomic and statistical analysis, and helped write the paper.
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award numbers SC2GM122620 and SC1GM136546 (awarded to G.E.F.). S.H., N.K., C.M., K.O., K.R., and P.S. were supported under grants TL4GM118977, RL5GM118975, and UL1GM118976, also from the NIGMS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
Contributor Information
Gilberto E. Flores, Email: gilberto.flores@csun.edu.
Andrew J. McBain, University of Manchester
REFERENCES
- 1.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Shin Y-C, Kim T-Y, Kim Y, Lee Y-S, Lee S-H, Kim M-N, O E, Kim KS, Kweon M-N. 2021. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut Microbes 13:1892441. 10.1080/19490976.2021.1892441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L, MICRO-Obes Consortium, Dumas M-E, Rizkalla SW, Doré J, Cani PD, Clément K. 2016. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65:426–436. 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 4.Rajilic-Stojanovic M, Shanahan F, Guarner F, de Vos WM. 2013. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 19:481–488. 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 5.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 6.Lee M-J, Kang M-J, Lee S-Y, Lee E, Kim K, Won S, Suh DI, Kim KW, Sheen YH, Ahn K, Kim B-S, Hong S-J. 2018. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol 141:1310–1319. 10.1016/j.jaci.2017.11.045. [DOI] [PubMed] [Google Scholar]
- 7.Ouwerkerk JP, van der Ark KCH, Davids M, Claassens NJ, Finestra TR, de Vos WM, Belzer C. 2016. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol 82:6983–6993. 10.1128/AEM.01641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbe C, Capon C, Coddeville B, Michalski JC. 2004. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J 384:307–316. 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP, Smidt H, Belzer C, de Vos WM. 2017. Genome-scale model and omics analysis of metabolic capacities of Akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Appl Environ Microbiol 83:e01014-17. 10.1128/AEM.01014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirmiz N, Galindo K, Cross KL, Luna E, Rhoades N, Podar M, Flores GE. 2020. Comparative genomics guides elucidation of vitamin B12 biosynthesis in novel human-associated Akkermansia strains. Appl Environ Microbiol 86:e02117-19. 10.1128/AEM.02117-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chia LW, Hornung BVH, Aalvink S, Schaap PJ, de Vos WM, Knol J, Belzer C. 2018. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van Leeuwenhoek 111:859–873. 10.1007/s10482-018-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A. 2019. Microbial regulation of organismal energy homeostasis. Nat Metab 1:34–46. 10.1038/s42255-018-0017-4. [DOI] [PubMed] [Google Scholar]
- 13.Ottman N, Reunanen J, Meijerink M, Pietila TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, Satokari R, Mercenier A, Palva A, Smidt H, de Vos WM, Belzer C. 2017. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One 12:e0173004. 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KC, Aalvink S, Martinez LO, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 15.Guo X, Li S, Zhang J, Wu F, Li X, Wu D, Zhang M, Ou Z, Jie Z, Yan Q, Li P, Yi J, Peng Y. 2017. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity [sic], and global distribution in mammalian gut microbiotas. BMC Genomics 18:800. 10.1186/s12864-017-4195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coker JK, Moyne O, Rodionov DA, Zengler K. 2021. Carbohydrates great and small, from dietary fiber to sialic acids: how glycans influence the gut microbiome and affect human health. Gut Microbes 13:1869502. 10.1080/19490976.2020.1869502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bode L, Jantscher-Krenn E. 2012. Structure-function relationships of human milk oligosaccharides. Adv Nutr 3:383S–391S. 10.3945/an.111.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 20.Garrido D, Barile D, Mills DA. 2012. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr 3:415S–421S. 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuntz S, Kunz C, Rudloff S. 2009. Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br J Nutr 101:1306–1315. 10.1017/S0007114508079622. [DOI] [PubMed] [Google Scholar]
- 22.Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. 2005. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr 135:1304–1307. 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- 23.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol 72:4497–4499. 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. 2010. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem 58:5334–5340. 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu ZT, Chen C, Newburg DS. 2013. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23:1281–1292. 10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, Boeren S, Mank M, Knol J, de Vos WM, Belzer C. 2020. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep 10:14330. 10.1038/s41598-020-71113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a Web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh T, Ojima MN, Sakanaka M, Ashida H, Gotoh A, Katayama T. 2020. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms 8:481. 10.3390/microorganisms8040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low KE, Smith SP, Abbott DW, Boraston AB. 2021. The glycoconjugate-degrading enzymes of Clostridium perfringens: tailored catalysts for breaching the intestinal mucus barrier. Glycobiology 31:681–690. 10.1093/glycob/cwaa050. [DOI] [PubMed] [Google Scholar]
- 33.Ioannou A, Knol J, Belzer C. 2021. Microbial glycoside hydrolases in the first year of life: an analysis review on their presence and importance in infant gut. Front Microbiol 12:631282. 10.3389/fmicb.2021.631282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Kim J, Kendall DA. 1996. Competition between functional signal peptides demonstrates variation in affinity for the secretion pathway. J Bacteriol 178:6658–6664. 10.1128/jb.178.23.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. 2018. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol 97:422–441. 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhai Q, Feng S, Arjan N, Chen W. 2019. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr 59:3227–3236. 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 37.Becken B, Davey L, Middleton DR, Mueller KD, Sharma A, Holmes ZC, Dallow E, Remick B, Barton GM, David LA, McCann JR, Armstrong SC, Malkus P, Valdivia RH. 2021. Genotypic and phenotypic diversity among human isolates of Akkermansia muciniphila. mBio 12:e00478-21. 10.1128/mBio.00478-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom H-J, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. 2011. An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21:437–447. 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 40.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286:11909–11918. 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S, Kurokawa K. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7:11939. 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James K, Motherway MO, Bottacini F, van Sinderen D. 2016. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep 6:38560. 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. 2016. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep 6:35045. 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrido D, Kim JH, German JB, Raybould HE, Mills D. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Moyano S, Totten SM, Garrido D, Smilowitz JT, German JB, Lebrilla CB, Mills DA. 2013. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol 79:6040–6049. 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA 107:19514–19519. 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcobal A, Sonnenburg JL. 2012. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect 18:12–15. 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laursen MF, Bahl MI, Licht TR. 2021. Settlers of our inner surface—factors shaping the gut microbiota from birth to toddlerhood. FEMS Microbiol Rev 45:fuab001. 10.1093/femsre/fuab001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971. 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engels C, Ruscheweyh H-J, Beerenwinkel N, Lacroix C, Schwab C. 2016. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol 7:713. 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collado MC, Laitinen K, Salminen S, Isolauri E. 2012. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res 72:77–85. 10.1038/pr.2012.42. [DOI] [PubMed] [Google Scholar]
- 56.Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, K DG, Kvist LJ, Otoo GE, Garcia-Carral C, Jimenez E, Ruiz L, Rodriguez JM, Pareja RG, Bode L, McGuire MA, McGuire MK. 2019. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr 6:45. 10.3389/fnut.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korpela K, Salonen A, Hickman B, Kunz C, Sprenger N, Kukkonen K, Savilahti E, Kuitunen M, de Vos WM. 2018. Fucosylated oligosaccharides in mother’s milk alleviate the effects of caesarean birth on infant gut microbiota. Sci Rep 8:13757. 10.1038/s41598-018-32037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aakko J, Kumar H, Rautava S, Wise A, Autran C, Bode L, Isolauri E, Salminen S. 2017. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef Microbes 8:563–567. 10.3920/BM2016.0185. [DOI] [PubMed] [Google Scholar]
- 59.Thurl S, Munzert M, Henker J, Boehm G, Müller-Werner B, Jelinek J, Stahl B. 2010. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br J Nutr 104:1261–1271. 10.1017/S0007114510002072. [DOI] [PubMed] [Google Scholar]
- 60.Wang B, Brand-Miller J. 2003. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr 57:1351–1369. 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 61.Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, Muehlbauer MJ, Ilkayeva O, Wu C, Struckmeyer T, Barile D, Mangani C, Jorgensen J, Fan Y, Maleta K, Dewey KG, Ashorn P, Newgard CB, Lebrilla C, Mills DA, Gordon JI. 2016. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164:859–871. 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobi SK, Yatsunenko T, Li D, Dasgupta S, Yu RK, Berg BM, Chichlowski M, Odle J. 2016. Dietary isomers of sialyllactose increase ganglioside sialic acid concentrations in the corpus callosum and cerebellum and modulate the colonic microbiota of formula-fed piglets. J Nutr 146:200–208. 10.3945/jn.115.220152. [DOI] [PubMed] [Google Scholar]
- 63.Almagro-Moreno S, Boyd EF. 2010. Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes 1:45–50. 10.4161/gmic.1.1.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Ark KCH, Aalvink S, Suarez-Diez M, Schaap PJ, de Vos WM, Belzer C. 2018. Model-driven design of a minimal medium for Akkermansia muciniphila confirms mucus adaptation. Microb Biotechnol 11:476–485. 10.1111/1751-7915.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rokhsefat S, Lin A, Comelli EM. 2016. Mucin-microbiota interaction during postnatal maturation of the intestinal ecosystem: clinical implications. Dig Dis Sci 61:1473–1486. 10.1007/s10620-016-4032-6. [DOI] [PubMed] [Google Scholar]
- 67.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliver A, Kay M, Cooper KK. 2018. Comparative genomics of cocci-shaped Sporosarcina strains with diverse spatial isolation. BMC Genomics 19:310. 10.1186/s12864-018-4635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parker CT, Cooper KK, Huynh S, Smith TP, Bono JL, Cooley M. 2018. Genome sequences of eight Shiga toxin-producing Escherichia coli strains isolated from a produce-growing region in California. Microbiol Resour Announc 7:e00807-18. 10.1128/MRA.00807-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 77.Busk PK, Pilgaard B, Lezyk MJ, Meyer AS, Lange L. 2017. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics 18:214. 10.1186/s12859-017-1625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 79.Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2009. gplots: various R programming tools for plotting data, v3.1.0. https://cran.r-project.org/web/packages/gplots/index.html.
- 80.Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O’Hara R, Simpson G, Solymos P, Stevens M, Szoecs E, Wagner H. 2017. vegan: community ecology package. https://CRAN.R-project.org/package=vegan.
- 81.Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Soll D, Podar M. 2013. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA 110:5540–5545. 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 85.Hardy MR, Townsend RR, Lee YC. 1988. Monosaccharide analysis of glycoconjugates by anion exchange chromatography with pulsed amperometric detection. Anal Biochem 170:54–62. 10.1016/0003-2697(88)90089-9. [DOI] [PubMed] [Google Scholar]
- 86.Townsend RR, Hardy MR, Cumming DA, Carver JP, Bendiak B. 1989. Separation of branched sialylated oligosaccharides using high-pH anion-exchange chromatography with pulsed amperometric detection. Anal Biochem 182:1–8. 10.1016/0003-2697(89)90708-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download AEM.01487-21-s0001.xlsx, XLSX file, 0.5 MB (557.2KB, xlsx)
Tables S1 to S4, Fig. S1 and S2. Download AEM.01487-21-s0002.pdf, PDF file, 0.6 MB (630.5KB, pdf)
Data Availability Statement
The data that support the findings of this study are openly available in the NCBI BioProject database under accession number PRJNA609771.