ABSTRACT
Frequently, laccases are triggered during fungal cocultivation for overexpression. The function of these activated laccases during coculture has not been clarified. Previously, we reported that Gongronella sp. w5 (w5) (Mucoromycota, Mucoromycetes) specifically triggered the laccase Lcc9 overexpression in Coprinopsis cinerea (Basidiomycota, Agaricomycetes). To systematically analyze the function of the overexpressed laccase during fungal interaction, C. cinerea mycelia before and after the initial Lcc9 overexpression were chosen for transcriptome analysis. Results showed that accompanied by specific utilization of fructose as carbohydrate substrate, oxidative stress derived from antagonistic compounds secreted by w5 appears to be a signal critical for laccase production in C. cinerea. A decrease in reactive oxygen species (ROS) in the C. cinerea wild-type strain followed the increase in laccase production, and then lcc9 transcription and laccase activity stopped. By comparison, increased H2O2 content and mycelial ROS levels were observed during the entire cocultivation in lcc9 silenced C. cinerea strains. Moreover, lcc9 silencing slowed down the C. cinerea mycelial growth, affected hyphal morphology, and decreased the asexual sporulation in coculture. Our results showed that intracellular ROS acted as signal molecules to stimulate defense responses by C. cinerea with the expression of oxidative stress response regulator Skn7 and various detoxification proteins. Lcc9 takes part in a defense strategy to eliminate oxidative stress during the interspecific interaction with w5.
IMPORTANCE The overproduction of laccase during interspecific fungal interactions is well known. However, the exact role of the upregulated laccases remains underexplored. Based on comparative transcriptomic analysis of C. cinerea and gene silencing of laccase Lcc9, here we show that oxidative stress derived from antagonistic compounds secreted by Gongronella sp. w5 was a signal critical for laccase Lcc9 production in Coprinopsis cinerea. Intracellular ROS acted as signal molecules to stimulate defense responses by C. cinerea with the expression of oxidative stress response regulator Skn7 and various detoxification proteins. Ultimately, Lcc9 takes part in a defense strategy to eliminate oxidative stress and help cell growth and development during the interspecific interaction with Gongronella sp. w5. These findings deepened our understanding of fungal interactions in their natural population and communities.
KEYWORDS: Coprinopsis cinerea, interspecific interaction, laccase, oxidative stress responses, reactive oxygen species
INTRODUCTION
Interspecific interactions are critical factors involved in the ecology of fungi, as these live in complex organismal communities and often have to compete against each other for territory and to occupy the nutrient substrates [1, 2]. Fungal competition can be divided into primary resource capture for obtaining unoccupied resources, and secondary resource capture for struggling for resources already colonized by other fungi [3]. When antagonistic relations occur, fungi usually employ several strategies, including developing attack or defense mechanisms and improvements in nutrient uptake, to gain an advantage over the competitor [3–5]. Aggressors successfully occupy the primary resources by various solutions, including good dispersal, rapid spore germination, rapid mycelial extension, and the ability to utilize organic compounds available in previously unoccupied resources [2, 3]. Also, they can increase the secretion of secondary metabolites or harming enzymes, or cause peroxide accumulation, and all this may be deleterious for the cellular integrity of a competitor [3, 6]. In contrast, a defender of resources may produce extracellular and intracellular enzymes that help detoxify of such noxious compounds [2]. For fungi harboring laccase genes, their laccase activities are often found enhanced after exposure to other fungi [7–11]. For example, the basidiomycetes Pleurotus ostreatus and Agaricus bisporus increase laccase production when cocultured with Trichoderma sp., indicating that laccases play roles during fungal confrontations [8].
Laccases (p-diphenol:oxygen oxidoreductases, EC1.10.3.2) are blue multicopper-containing phenol oxidases with a broad substrate specificity range that includes diverse aromatic compounds [12]. Fungal laccases have great potentials for biotechnological applications, but the biological functions of fungal laccase have not been fully confirmed, even though thousands of papers regarding laccase have been published [13]. Fungal laccases are suggested to be synergetic with other ligninolytic enzymes and involved in the biodegradation of natural polymers since many filamentous basidiomycetes laccases can cleave bonds in non-phenolic or phenolic subunits of lignin with or without mediators (reviewed in references 14 and 15). Because heterologous expression of a fungal laccase gene in Pichia pastoris can enhance the resistance of yeast to H2O2-mediated oxidative stress [16], and deletion of most isozyme genes of laccases results in a greater sensitivity to H2O2 than in the wild-type of Podospora anserina [17], fungal laccases are also supposed to react in response to oxidative stress in the environment [11]. Other studies suggest that laccases may play an essential role in several developmental and physiological functions such as sporulation, mycelial morphology, pigment production, fruiting body formation, and virulence in plant pathogenesis. For instance, the abr2 gene product in Aspergillus fumigatus is involved in conidial pigmentation [18]. Silencing of gene lcc1 of Lentinula edodes indicates involvement in mycelial morphology [19, 20]. Upon homologous overexpression of lcc1, Hypsizygus marmoreus displays faster mycelial growth and fruiting body initiation [21]. However, the role of the strongly induced laccases during fungal interspecific interactions is still unclear.
The litter-degrading ink-cap mushroom Coprinopsis cinerea has in total 17 non-allelic laccase genes, which present one of the largest groups of laccase genes ever described for a fungus [22]. Under different culture conditions, there was a differential expression of laccase isozymes in C. cinerea. Among them, Lcc1 and Lcc5 are the main laccases secreted in different culture media [23]. Previously, we developed a fungal coculture system between the basidiomycete C. cinerea and the mucoromycete Gongronella sp. w5. The latter triggered the transcription of the almost silent laccase gene lcc9 from C. cinerea and the secretion of Lcc9 into the coculture broth, accompanied by a low expression of Lcc1 and Lcc5 [24, 25]. Based on the transcriptome analysis of C. cinerea and gene silencing analysis of lcc9, we demonstrate in this study that Lcc9 was used by C. cinerea as a defense strategy to eliminate the oxidative stress induced by Gongronella sp. w5, resulting in better growth of the C. cinerea mycelia and promoted generation of mitotic spores (oidia) during the interspecific interactions. Transcriptome analysis combined with qRT-PCR results also revealed that proteins involved in ROS elimination and the oxidative stress response regulator Skn7 were upregulated as further interspecies stress responses.
RESULTS
Overview of transcriptomes of C. cinerea during interspecific interaction with Gongronella sp. w5.
To reveal the function of the specifically activated Lcc9 [24, 25], C. cinerea mycelia, separately cocultured with Gongronella sp. w5 for 18 h and 28 h, were chosen for transcriptome analysis to get clues from the global gene-expression-pattern alterations before and after the initial lcc9 overexpression. A total of 287 million raw sequencing reads were obtained from the transcriptomics analysis of in total 6 samples (3 per time point). After removal of the low-quality reads, a total of 268 million clean reads were obtained, with average reads of 44.7 million and average data of 6,716 million reads per sample. The clean reads were mapped to the genome of C. cinerea Okayama 7. The mapping rates of each sample were more than 93%. The sequence reads matched to 11,896 coding genes in the C. cinerea Okayama 7 genome, and 1,179 DEGs were calculated to be differentially expressed when comparing the transcriptome of 28 h to that of 18 h, with 695 genes upregulated and 484 genes downregulated.
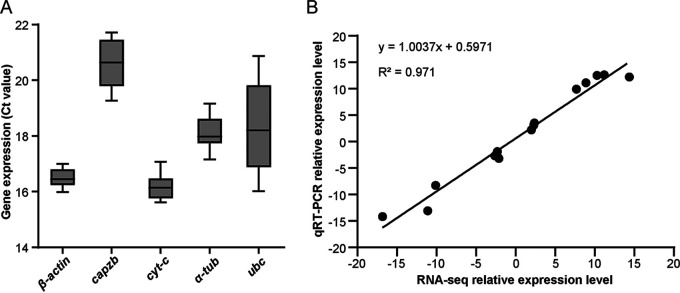
Fifteen genes, including up- and downregulated genes, were randomly chosen and tested for expression profiling using qRT-PCR to validate gene expression data obtained through RNA-seq. As a prerequisite for getting solid results, the transcriptional levels of five candidate reference genes were detected at different time points of separated cocultures and single-species cultures of C. cinerea to find a stable reference gene. The mean cycle threshold crossing-point (CT) values of the five genes ranged from 15 to 22 cycles among samples (Fig. 1A). Both the NormFinder algorithm and the BestKeeper algorithm identified β-actin (CC1G_13048, primers targeting 120 to 225 nt) as the most stable reference gene based on the NormFinder Stability value (0.037) and a higher correlation coefficient (standard deviation = 0.27, coefficient of variance = 1.66), respectively. Thus, the β-actin gene was chosen as the reference gene throughout the study. The expression values of the 15 randomly selected genes in C. cinerea mycelia in separated coculture with Gongronella sp. w5 for 28 h were then compared with 18 h. The results were consistent with the RNA-seq data (Fig. 1B), indicating the reliability and reproducibility of the sequencing results.
FIG 1.
qRT-PCR of five candidate reference genes (A) and 15 randomly chosen genes varied in RNA-seq (B). (A) The mean cycle threshold crossing-point (CT) values of β-actin, capzb (F-actin capping protein beta subunit), α-tub (tubulin), cyt-c (cytochrome c), and ubc (ubiquitin) in C. cinerea across different time points of separated cocultures and single-species cultures. (B) qRT-PCR analysis to validate the RNA-seq data. The x axis that represents the RNA-seq data was calculated by the FPKM method. The y axis indicates the qRT-PCR results by using the 2-ΔΔCt method. All experiments were performed independently in triplicate. The relative expression levels from RNA-seq and qRT-PCR obeyed a linear correlation (y = 1.0037x + 0.5971, R2 = 0.971).
KEGG pathways were used for the functional classification of DEGs to investigate whether gene pathways in C. cinerea were changed during the cocultivation. DEGs were mapped to 70 KEGG pathways (Table S1-1). The 20 significantly most enriched pathways were interpreted to be involved in the following processes: amino acid metabolism, signal sensing and transduction, carbon metabolism, energy metabolism, lipid metabolism, and fungal secondary metabolites degradation (Table S1-1).
Primary metabolic changes.
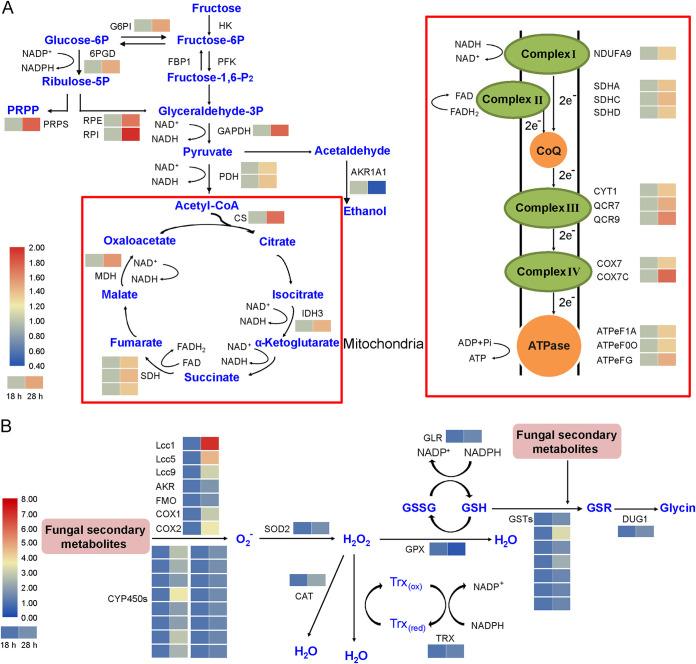
DEGs that participate in glycolysis/gluconeogenesis, citric acid cycle, and pentose phosphate metabolism pathways, including gapdh (glyceraldehyde 3-phosphate dehydrogenase, CC1G_09117), cs (citrate synthase, CC1G_12964), mdh (malate dehydrogenase, CC1G_07719), rpe (ribulose-phosphate 3-epimerase, CC1G_10281), and rpi (ribose-5-phosphate isomerase, CC1G_09808), were nearly all upregulated at 28 h compared to 18 h in separated cocultivation. Simultaneously, genes that are involved in the oxidative phosphorylation pathway were also upregulated, especially qcr9 (ubiquinol-cytochrome c reductase, CC1G_00077) in complex III and cox7 (cytochrome c oxidase subunit 7, CC1G_02587) in complex IV (Fig. 2A, Table S1-1 and S1-2).
FIG 2.
Regulation of central carbon metabolism (A), oxidative phosphorylation (A), and oxidative stress responses (B) in C. cinerea at transcriptome level under interspecific interaction with Gongronella sp. w5 in separated coculture in SAHX medium with sucrose. Expression of genes at 28 h compared to 18 h coculture was analyzed. The heatmaps located beside each function indicate the logarithm to base 10 of fold changes in transcription. Abbreviations: P, phosphate; GSH, reduced form of glutathione; GSSG, disulfide form of glutathione; Trx (dx), thioredoxin; PRPP, 5-phosphoribosyl 1-pyrophosphate.
Besides, key enzymes involved in β-oxidation breakdown of fatty acids and some amino acids into metabolites entering the citric acid cycle, such as ACAT (acetyl-CoA acetyltransferase, CC1G_14616), ECHS1 (enoyl-CoA hydratase, CC1G_02051), l-serine/threonine ammonia-lyase (CC1G_03788), and aspartate kinase (CC1G_11606), were upregulated (Table S1-1). Particularly, genes involved in tryptophan metabolism were mostly enriched in DEGs and increased in expression here. For example, aadc (aromatic-l-amino-acid decarboxylase, CC1G_02020), tdo (tryptophan 2,3-dioxygenase, CC1G_02499), echs1, acat, and cat (catalase, CC1G_09926) were all increased (Table S1-2).
Degradation of fungal secondary metabolites.
Most of the significantly upregulated KEGG pathways at 28 h of separated cocultivation were related to degradation and metabolism of toxic metabolites, cell detoxification, and signaling and stress responses (Table S1-3), indicating that C. cinerea was exposed to various secondary metabolic compounds when confronted with Gongronella sp. w5. Our previous finding by NMR of HBA (p-hydroxybenzoic acid) obtained from Gongronella sp. w5 in culture broth [25] and the further GC-MS identification of a large number and types of benzene and naphthalene compounds among other metabolites from Gongronella sp. w5 culture broth further reinforced this hypothesis (Table S2).
The degradation processes of fungal secondary metabolites in fungi are largely unknown but most likely include a series of oxidation, reduction, and acylation reactions in extracellular and intracellular environments [26–29]. Among the DEGs, 11 CYP450 (cytochrome P450) genes were upregulated during the coculture process (Fig. 2B, Table S1-3). In fact, cyp78A3 (CC1G_09431) and two cyp98A3 (cox1 and cox2, CC1G_03562 and CC1G_03564 [30]) encoding CYP subfamilies with oxidoreductases that catalyze hydroxylations of the phenolic ring of monolignols to participate in the degradation of phenolic compounds [31, 32] were 5.6, 6.7, and 11.2 times upregulated, respectively (Table S1-3). In addition, another monooxygenase of the UbiH hydroxylase family (CC1G_05962) that could modify a single position of the aromatic ring and is involved in hydroxylation of phenolic compounds [33] was 2.1 times upregulated.
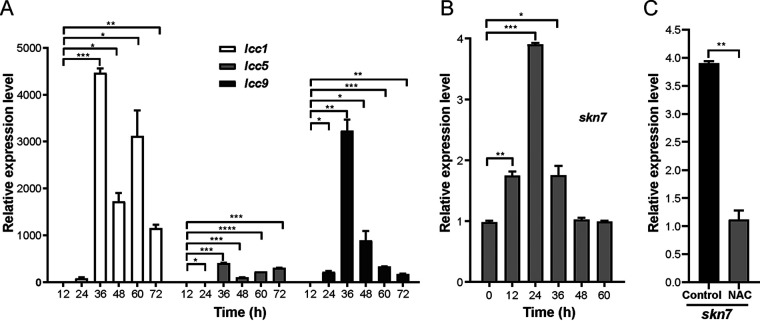
Lignin-modifying enzymes are suggested to be also involved in the degradation of fungal secondary metabolites [26, 34, 35]. According to the transcriptomic data, laccases lcc1 (CC1G_09614), lcc5 (CC1G_00690), and lcc9 (CC1G_05998) were 127.4, 1.5, and 7.7 times increased at the transcriptional level after 28 h compared to 18 h of separated cocultivation. Furthermore, as shown by qRT- PCR analysis in Fig. 3A, compared to the single-species cultures of C. cinerea, genes encoding for the three laccase isozymes Lcc1, Lcc5, and Lcc9 detected in separated coculture of C. cinerea and Gongronella sp. w5 [24, 25] were transcribed at 24 h and peaked at 36 h with hundreds to thousands fold increase. Among which, lcc1 and lcc9 were strongly induced and responded quickly. However, in contrast to lcc1 and lcc5, which were expressed up to 72 h, the expression of lcc9 decreased significantly after 60–72 h.
FIG 3.
Three laccases (A) and transcription factor Skn7 (B) were upregulated in expression in separated coculture of C. cinerea and Gongronella sp. w5 but ROS scavenger N-acetyl-l-cysteine (NAC) inhibited increase of skn7 transcription (C). C. cinerea mycelia were collected every 12 h during separated coculturing with Gongronella sp. w5. Then they were ground, extracted for RNA, and analyzed for transcriptional levels by qRT-PCR. The transcription levels of genes of monocultures without NAC (A and B) or at 0 h of separated coculture in SAHX with sucrose (C) were set as baselines. The data in panel C were detected at 24 h of separated coculture and analyzed using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Data in panels A to C show mean ± SD, n = 3.
Antibiotic molecule and siderophore production.
Fungi defend their ecological niche mainly by producing antibiotic molecules, and the sesquiterpene lagopodin was reported to be an antibacterial compound of C. cinerea [36]. The CYP450 genes cox1 and cox2, together with sesquiterpene synthase gene cop6 (CC1G_03563), form a gene cluster for lagopodin biosynthesis and oxidative modifications [30, 36–38]. In our transcriptome data, the expression of the three genes of the cluster was in parallel upregulated, between about 6 to 12 times during 18 to 28 h of coculture. Moreover, most of the genes in terpenoid backbone biosynthesis, which could positively affect lagopodin production [38], were increased in expression (Table S1-4).
Gene cpf2 (CC1G_04211) in the coprinoferrin siderophore gene cluster of C. cinerea for a putative l-ornithine-N5-monooxygenase [39] for production of N5-hydroxy-l-ornithine from ornithine as precursor of hydroxamate siderophores and competitive antimicrobial iron scavengers [40, 41], was 2.1 times upregulated. In contrast, the transcription in the cluster of genes for a non-ribosomal peptide synthetase (Cpf1, CC1G_04210) producing the coprinoferrin and for an uncharacterized cytidine deaminase-like protein (CC1G_04209 [39]) was not changed (Table S1-4).
Oxidative stress response.
Extracellular fungal bioactive compounds cause oxidative stresses in cells. Following this fact, genes encoding for oxidative stress-related proteins, including aldo/keto reductases (AKRs), superoxide dismutases (SODs), glutathione S-transferases (GSTs), and catalases (CATs), were significantly upregulated in C. cinerea during the confrontation with Gongronella sp. w5 (Fig. 2B and Table S1-3). These proteins are suggested to play roles in oxidative stress signal transduction [11, 42–44], elimination of stress, and cellular redox balance maintenance [27, 45, 46]. The proteins participate in transforming the superoxide anion radical to H2O2 (SOD2, CC1G_03559, gene localizes together with the lagopodin biosynthesis cluster), reducing H2O2 to H2O to remove oxidative stress (CAT, CC1G_09926), and catalyzing 2’-deoxyribonucleoside diphosphate and thioredoxin disulfide to form ribonucleoside diphosphate and thioredoxin (Trx, CC1G_08103; Table S1-3). Although gpx (glutathione peroxidase, CC1G_07055) was downregulated, glr (glutathione reductase, CC1G_03850) and many gsts (glutathione transferases, CC1G_06890, CC1G_10591, and CC1G_14829), which are involved in catalyzing conjugation reactions of glutathione with toxic metabolites at its sulfhydryl site, were upregulated (Table S1-3). Furthermore, the dug1 (Cys-Gly metallodipeptidase, CC1G_06968) gene, which plays a crucial role in degrading conjugated GSH [47], also increased in transcription (Fig. 2B and Table S1-3). These results indicated an increased glutathione flux in redox reactions and secondary metabolites in C. cinerea upon the confrontation with Gongronella sp. w5.
Transcription factors.
Many potential transcription factors (TFs) were among the DEGs in our transcriptome analysis, but only three were significantly upregulated, and one was significantly downregulated in expression during the separated cocultivation of C. cinerea and Gongronella sp. w5 (Table S1-5). Of these upregulated TFs, the general transcription factor tfIID (CC1G_11509) was 2.2 times more transcribed. A candidate for GATA-type transcription factor gene sreA (CC1G_14734), which is involved in metabolisms of amino acids and especially tryptophan, and in iron homeostasis and sterol metabolism [48], was 2.1 times increased in expression. A candidate of samB (CC1G_08324), encoding for an MYND-type Zinc finger protein crucial for morphogenesis [49], was also significantly upregulated. Interestingly, transcription of cbf11 (CC1G_13688) and cbf12 (CC1G_03194) for two antagonistic CSL (CBF1/Su(H)/Lag-1) paralogs was increased and decreased about 2.2 times, respectively. They were reported to positively and negatively regulate development, cell wall synthesis, as well as stress response [50].
Of the |log2FoldChange| > 0.75 TFs, one was creb1 (cAMP response element-binding protein 1, CC1G_09350), which could enforce the GSH recovering capacity by directly upregulating genes for GSH synthesis and redox cycling [51]. For |log2FoldChange| < −0.75 DEGs, there were candidates for znf1 (a C2H2 zinc-finger transcription factor, CC1G_00774) for activation of core genes in glycolysis, pyruvate metabolism, and stress responses (1.8 times downregulated) [52], for GATA-type transcription factor gene gat1 (CC1G_05900) mediating nitrogen metabolite-repression (1.7 times downregulated) [53, 54], and for hap1 (CC1G_08004) involved in ergosterol biosynthesis and efficient utilization of oxygen (1.8 times downregulated) [55] (Table S1-5).
Two TF genes were impressive because of their overall high read counts, although their expression changed only slightly between 18 and 28 h of separated coculture. Highest expressed was the general amino acid control transactivator/cross-pathway control protein A gene cpcA (gcn4, CC1G_05406) with a bZIP domain that acts as a repressor of protein synthesis while it induces TFs of stress-response genes under various stress conditions [56, 57]. The other highly expressed TF contains a leucine zipper domain and shares 68.7% sequence similarity with Skn7 of S. cerevisiae. Skn7 is an oxidative stress-responsive transcription factor shown in other fungi to contribute to the expression of detoxification genes (CC1G_08281 [58, 59]). As verified further by qRT-PCR (Fig. 3B), transcription of Skn7 was upregulated from 0 h to 36 h in separated cocultivation of C. cinerea with Gongronella sp. w5, and peaked at 24 h, indicating a function in oxidative stress response and regulation of expression of detoxification proteins. Moreover, Skn7 showed no increase at the transcription level after NAC (N-acetyl-l-cysteine) as ROS scavenger was added into the culture broth at 0 h of cocultivation, further supporting a role in stress response (Fig. 3C).
ROS is involved in the synthesis of laccase during C. cinerea-Gongronella sp. w5 interspecific interaction.
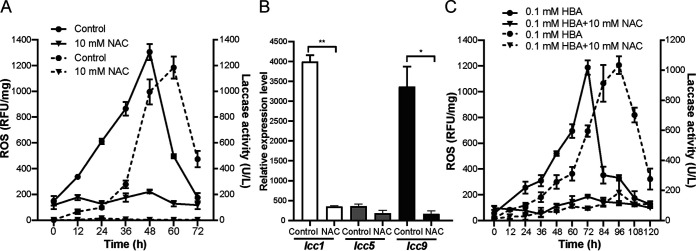
Based on the transcription data, we hypothesized that the oxidative stress in C. cinerea derived from antagonistic compounds secreted by Gongronella sp. w5 is a signal for induction of laccase production in C. cinerea. Thus, the intracellular ROS concentration in C. cinerea and the laccase activity during the separated coculture were analyzed to verify this hypothesis. As shown in Fig. 4A, the ROS level in C. cinerea increased rapidly after the start of fungal cocultivation, with a peak at 48 h and a sharp decline afterwards. Notably, the laccase and ROS curves shared the same growth trends, with a lag-phase in between with which the growth curve of laccase activity followed that of the intracellular ROS concentration. The laccase activity curve thus peaked 12 h after the highest ROS level reached and fell sharply with a delay of also 12 h (Fig. 4A). ROS scavenger NAC added to the coculture system at 0 h further supported the above hypothesis. NAC prevented an increase in intracellular ROS concentration (Fig. 4A) and, as tested for both lcc9, lcc5 and lcc1 transcription (Fig. 4B) as well as total enzyme activity levels (Fig. 4A), the expression of laccase in C. cinerea under coculture conditions.
FIG 4.
ROS was involved in the synthesis of laccase during C. cinerea-Gongronella sp. w5 interspecific interaction in separated coculture. (A, B) ROS increased in C. cinerea separated cocultured with Gongronella sp. w5 in SAHX with sucrose (A) and added ROS scavenger NAC decreased laccase activity (A) and transcripts (B) of C. cinerea. Control represented separated cocultures without NAC. (C) NAC decreased laccase activity in monoculture of C. cinerea in SAHX medium using fructose as carbon source with 0.1 mM HBA added. Solid lines and dotted lines represent ROS values and laccase activities, respectively. The data were analyzed using Student's t test (*, P < 0.05; **, P < 0.01). Data show mean ± SD, n = 3. RFU, relative fluorescence units.
Previously, HBA as produced by Gongronella sp. w5 was evidenced as a principle to induce C. cinerea laccase activity in cocultures [25]. In order to rule out an indirect influence of NAC added in coculture due to a possible changed behavior of Gongronella sp. w5 and to link the transmission of the ROS signal in C. cinerea with induction of laccase production, HBA at 0.1 mM concentration was employed in C. cinerea SAHX (sucrose dl-asparagine XH medium) single species-cultures with fructose as the carbon source. The addition of HBA continuously increased the intracellular ROS level in the first 72 h of culture incubation. Consistent with the results of the cocultures and with a time lag compared to ROS, it also increased the laccase activity of C. cinerea, which peaked at 96 h of single-species cultivation. The decrease of laccase activity at 108 h followed the drastic decrease in the ROS level at 84 h. However, in treatments with 10 mM NAC, HBA had an inducing effect neither on ROS generation nor on the laccase production (Fig. 4C), suggesting that HBA was not a direct inducer of laccase activity.
Laccase expression in C. cinerea lcc9 silencing strains.
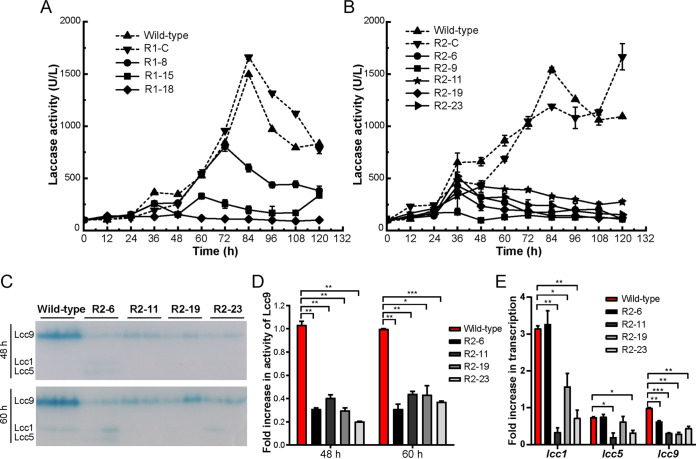
Laccase lcc9 silencing strains were constructed based on two antisense constructs, pYSK-lcc9-antisense-1 and pYSK-lcc9-antisense-2 [60], to further evaluate the role of lcc9 during interspecific fungal interaction. Three strains of pYSK-lcc9-antisense-1 (R1-7, R1-15, and R1-18 as positive strains, plus R1-C transformed with pCcAde8 as a control strain) and five strains of pYSK-lcc9-antisense-2 (R2-6, R2-9, R2-11, R2-19, and R2-23 as positive strains, plus R2-C as a pCcAde8 control strain) were randomly chosen from transformations for laccase activity determination in liquid cocultivation with Gongronella sp. w5. As shown in Fig. 5A and B, the laccase activities of the two control strains R1-C and R2-C were similarly increasing to that of the wild-type strain, but the eight lcc9 silencing strains in comparison showed much lower laccase activities over the whole time of cocultivation (< 500 U/liter) (Fig. 5A and B). Separated cocultivation experiments provided similar tendencies of laccase activities for the clones (not shown) and further suggested that the fragment lcc9 antisense 2 performed better than the antisense 1 on lcc9 silencing. As a result, silencing strains R2-6, R2-11, R2-19, and R2-23 were chosen for further experiments.
FIG 5.
Lcc9 was downregulated in lcc9 silencing strains. (A, B) The laccase activity of four transformants of pYSK-lcc9 antisense 1 (A) and six transformants of pYSK-lcc9 antisense 2 (B) tested in separated coculture with Gongronella sp. w5 was found to be decreased, using ABTS as the substrate. R1-7, R1-15, R1-18, R2-6, R2-9, R2-11, R2-19, and R2-23 were positive lcc9 silencing strains represented by solid lines. pCcAde8 transformants R1-C and R2-C used as control strains are represented by dotted lines, as well as wild-type strains. (C) Native-PAGE of laccase activities of C. cinerea wild-type and of four C. cinerea lcc9 silencing strains from 48 h and 60 h separated coculture with Gongronella sp. w5 indicates downregulation in the lcc9 silencing strains. Equal amounts of culture supernatants (10 μl) were loaded onto the gels and activity-stained after gel-electrophoresis. (D) Quantification of lcc9 activity from the separated cocultures deduced from gels as shown in panel C. (E) lcc1, lcc5, and lcc9 laccase transcripts were shown by qRT-PCR analysis to be downregulated in the four lcc9 silencing strains. The lcc9 transcription level of wild-type strain at 36 h was set as the baseline. The data were analyzed using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Data show mean ± SD, n = 3.
Native-PAGE of culture samples taken from 48 h and 60 h coculturing showed the Lcc9 activity of the four silencing strains was at both time points significantly suppressed, accompanied by a partial reduction of Lcc1 and Lcc5 expression in several clones at the same time (Fig. 5C and D, P < 0.05, P < 0.01 or P < 0.001). qRT-PCR analysis, in which the lcc9 transcription level of the wild-type strain at 36 h was set as the baseline, showed that lcc9 transcripts were substantially downregulated in the four silencing strains compared to the wild-type at 36 h in separated coculture with Gongronella sp. w5 (Fig. 5E, P < 0.01 or P < 0.001). Among them, R2-19 had the highest downregulation observed (75%).
Laccase is involved in cellular oxidative stress resistance of C. cinerea during coculture with Gongronella sp. w5.
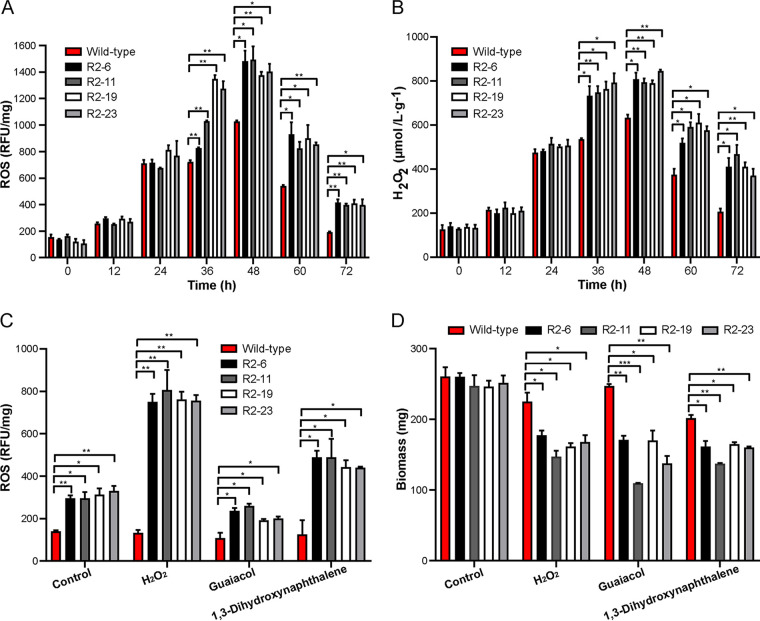
To analyze whether the Lcc9 could protect C. cinerea against ROS, the intracellular ROS and H2O2 concentrations of the wild-type strain and four lcc9 silencing clones were tested and compared during separated cocultivation with Gongronella sp. w5. The four silencing strains always showed 1.5–2 times higher concentrations of intracellular ROS and H2O2 than the wild-type strain during the entire cocultivation from 36 h to 72 h (Fig. 6A and B). Furthermore, the lcc9 silencing strains also showed higher sensitivity than the wild-type strain to oxidative stresses caused by H2O2, guaiacol, or 1,3-dihydroxynaphthalene added at 36 h of cultivation to the cocultures. Within 36 h of further incubation with H2O2 and 1,3-dihydroxynaphthalene, ROS levels in the lcc9 silencing strains increased above those of untreated control cultures (Fig. 6C). Their biomass was much less than that for the wild-type strain (Fig. 6D), although they all showed no apparent difference in single-species cultures of C. cinerea with the addition of oxidative stressors (not further shown).
FIG 6.
Laccase was involved in cellular oxidative stress resistance of C. cinerea during interspecific interaction with Gongronella sp. w5. C. cinerea exhibited higher ROS level (A) and H2O2 content (B) in lcc9 silencing strains compared to the wild-type strain after separated coculturing for 48 h in SAHX with sucrose with Gongronella sp. w5 sealed in sterile dialysis tubes. (C, D) The lcc9 silencing strains showed higher ROS level (C) and less biomass (D) than the wild-type strain in separated cocultures when exposed to 100 mM H2O2, 2 mM guaiacol, and 0.5 mM 3, 5-dihydroxynaphthalene. Chemicals were added at 36 h of separated cocultivation and cocultures were incubated for further 36 h. The data were analyzed using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Data show mean ± SD, n = 3.
Lcc9 is involved in cell growth and cell morphology of C. cinerea during coculture with Gongronella sp. w5.
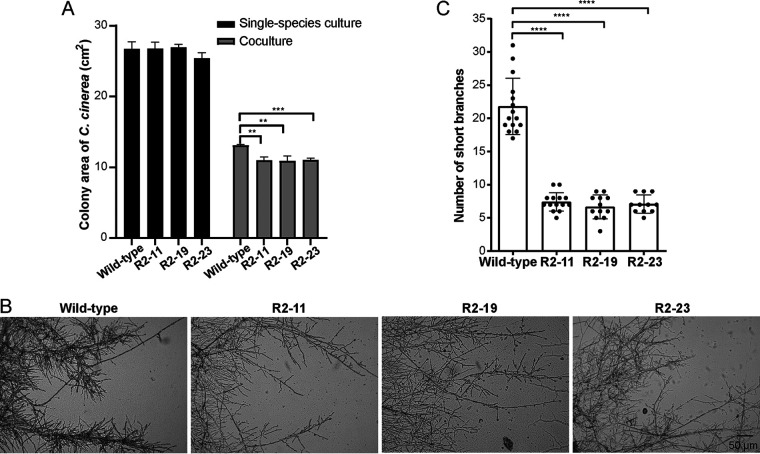
The enhanced lcc9 production may be necessary for C. cinerea to confront stress and survive during coculture with Gongronella sp. w5. Thus, we compared the growth rate of the C. cinerea wild-type strain and three lcc9 silencing strains R2-11, R2-19, and R2-23. All strains in single-species cultures showed no difference in speed of mycelial growth on SAHX-fructose agar plates. However, when cocultured with Gongronella sp. w5, the expansion rate of the three lcc9 silencing strains was much reduced (Fig. 7A and Fig. S1).
FIG 7.
Lcc9 was involved in cell growth and cell morphology of C. cinerea in coculture with Gongronella sp. w5 on solid medium. (A) Quantification of the colony area of the C. cinerea wild-type strain and three lcc9 silencing strains under single-species culture or coculture with Gongronella sp. w5. (B) Mycelial growth fronts with short hyphal branches in the interspecific region with Gongronella sp. w5 of the C. cinerea wild-type strain and three lcc9 silencing strains were cultured on microscope slides with SAHX medium with sucrose and observed by light microscopy. (C) Quantification of the numbers of short branches from 15 different randomly chosen views of the mycelia as shown in panel B. The data were analyzed using Student’s t- test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Data show mean ± SD, n = 3. Scale bar, 50 μm.
The mycelial morphologies of the C. cinerea wild-type strain and the lcc9 silencing strains as cocultured with Gongronella sp. w5 on slides with the SAHX-sucrose agar medium are shown in Fig. 7B. C. cinerea mycelial extension fronts in the interspecific interaction zone were observed in regions still separated from Gongronella sp. w5 when the two fungi with hyphae had just reached each other at some places. Light microscopy results suggested that although lcc9 silencing had no effect on the formation of an aerial mycelial mat by C. cinerea, it significantly reduced the mycelial density and the formation of hyphal side branches compared to the hyperbranching wild-type (Fig. 7B and C). Indeed, the hyphal branching patterns of the three lcc9 silencing clones appeared more normal, comparable to the hyphal growth of the wild-type when growing unstressed on fungal media (Fig. S2). Moreover, after incubation for 7 days on SAHX-sucrose agar plates, there were many more mitotic oidia distributed in liquid droplets in the aerial mycelium of the C. cinerea wild-type strain than in the aerial mycelium of the three lcc9 silencing clones (Fig. S2 and Table S3).
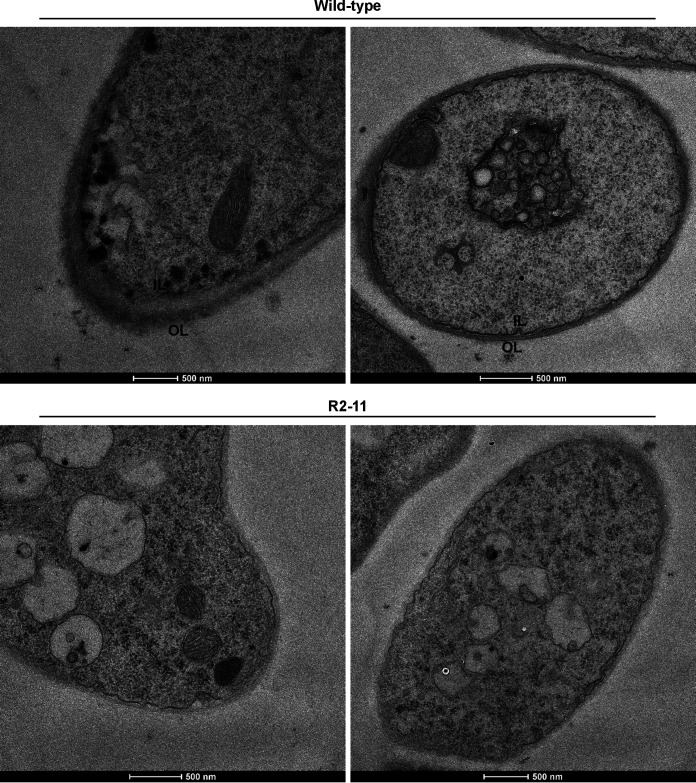
TEM observation results of C. cinerea hyphae collected from growth zones at the confrontation side with Gongronella sp. w5 showed that the hyphal cell walls of the wild-type strain comprised an inner layer (IL) and an outer layer (OL). However, the hyphal cell walls of the three lcc9 silencing strains were thinner and did not have two clear layers, as randomly presented in Fig. 8 for R2-11. Thus, the development of cell wall was also affected during interactions with Gongronella sp. w5 after lcc9 silencing. Furthermore, our results demonstrated that the vacuoles in the wild-type strain were large and few in most cells, whereas the silencing strains R2-11, R2-19, and R2-23 had a number of small vacuoles (Fig. 8 and data not shown).
FIG 8.
Lcc9 silencing affected cell walls of C. cinerea during coculture with Gongronella sp. w5. The hyphal cell walls with inner (IL) and outer layer (OL) and fungal vacuoles of the wild-type strain and lcc9 silencing strain R2-11 are shown by transmission electron microscopy. Scale bar, 500 nm.
DISCUSSION
Most fungi will come across competitive or antagonistic organisms in their natural population and communities and will be subject to attack, defenses, and possibly defeat [3, 61]. Activating silent gene clusters for the production of antimicrobial secondary metabolites is among the promising strategies adopted in this warfare [62–64] and also induction of expression of various types of combative and protective enzymes, including laccase [3, 7, 10, 11, 24, 61, 65]. In some cases, however, the increase in laccase production is simply due to the co-production between the two species; only some laccase isoenzyme genes could be activated in specific combinatorial cultivations [66, 67]. It is still unknown what role the upregulated laccases and those triggered newly in expression play during the fungal coculture. In this study, comparative transcriptomic analysis of C. cinerea and gene silencing of laccase lcc9 were performed to reveal the function of enhanced-expressed laccase during the fungal C. cinerea-Gongronella sp. w5 interaction.
Specific utilization of fructose as carbohydrate substrate and energy supply.
Our previous data showed that Gongronella sp. w5 in cocultivation with C. cinerea hydrolyzed almost all of the sucrose in the medium into glucose and fructose within 24 h and provided C. cinerea with mainly the fructose and traces of glucose [25]. Compared to the no-growth of C. cinerea in monocultures with sucrose as the sole carbon source [25, 68], C. cinerea grew fast in coculture in the first 48 h by using released fructose spurned by Gongronella sp. w5 [25]. Consistent with these results, the transcriptome results in this study showed that functions in the catabolic primary metabolisms, including glycolysis/gluconeogenesis, pentose phosphate pathway, citric acid cycle, and oxidative phosphorylation pathway for energy supply, were mostly upregulated (Fig. 2 and Table S1-2). The increased pentose phosphate pathway could also supply sufficient NADPH for the fast growth of C. cinerea in coculture. Meanwhile, catabolic pathways for energy generation involved in fatty acid and several amino acid metabolisms were also upregulated, together with adaptations in expression of potential TFs for nitrogen and amino acid regulation (Table S1-2).
In paired cultures of P. ostreatus with either Dichomitus squalens or Trametes versicolor and of D. squalens with T. versicolor, with glucose used as sole carbon and energy source, expression of the oxidative phosphorylation pathway is upregulated in P. ostreatus and D. squalens, and in addition, the expression of the TCA cycle in P. ostreatus [46, 69]. Expression of functions in glycolysis/gluconeogenesis is in addition enhanced in Pleurotus eryngii var. ferulae in dual cultures with Rhodotorula mucilaginosa in a medium with a more complex mixture of carbon sources (glucose, corn powder, wheat bran) [70], similar to what was observed here in C. cinerea in separated coculture with Gongronella sp. w5 (Fig. 2 and Table S1-2). Direct competition between two fungi for the same nutrients is responsible for the increases in the TCA cycle and the oxidative phosphorylation pathway [46, 69, 70]. Different preferences for supplied carbon sources between P. eryngii var. ferulae and R. mucilaginosa and specific utilization of fructose provided by Gongronella sp. w5 for C. cinerea might have also activated the upstream glycolysis pathway. Noteworthy is that all these examples of fungal cocultures had in common that specific laccases are selectively overexpressed in the basidiomycetes analyzed by transcriptomics [25, 46, 69, 70]. Moreover, pathways for toxic metabolite biodegradation are also induced in all species studied [46, 69, 70] (Table S1-3). ROS generation, oxidative stress responses, and protective responses are observed during the interspecies confrontations in P. ostreatus, D. squalens, and C. cinerea [46, 69] (Table S1-3) but not in P. eryngii var. ferulae in the confrontation with R. mucilaginosa that provided the antioxidant β-carotene [70].
ROS and laccase induction in interspecies interactions.
Phenolic compounds are often effective inducers for laccase production. β-Carotene released by red basidiomycetous yeasts into the medium is confirmed to be the inducer of laccase activity in fungal coculture systems including P. eryngii var. ferulae [65, 70]. This phenomenon is also exemplified by inducing laccase production with phenolic compounds, such as o-toluidine, in single-species cultures of various basidiomycetes [71–74]. Based on our previous data [25] and that presented in this study (Table S2), we summarize that HBA and likely also other secondary organic compounds produced by Gongronella sp. w5 were the main inducers for the lcc9 expression in C. cinerea (Fig. 2 and Fig. S3).
The detailed strategy of how antagonistic compounds induce the expression of laccase during fungal cocultivations is still under investigation. However, it is well known that extracellular agents and toxic compounds can destroy the intracellular redox balance, resulting in increased intracellular ROS concentrations [61, 75]. ROS is reported to play a central role in the molecular cross talk that occurs during interactions between fungi and organisms in their environment, including plants and other microbes, leading to interspecific recognition and eventually the activation of fungal defense reactions [43]. Several studies have reported that oxidative stress increases during fungal cocultivation [6, 11, 46]. Our data supported that compounds from Gongronella sp. w5 raised the ROS concentration in C. cinerea cells and suggested that C. cinerea encountered oxidative stress during cocultivation (Fig. 4, Fig. S3).
Transcriptional regulation.
Unfortunately, little knowledge has yet been gained on relationships between increased oxidative stress and laccase production during fungal cocultivations. Interestingly in this study, ROS was speculated to be a signal to induce lcc9 expression at the early stage of cocultivation (Fig. 4), and the signal might be transmitted into the nucleus by the oxidative stress response regulator Skn7 (Fig. 3, Fig. S3, and Table S1-5). S. cerevisiae Skn7 is activated following several phosphorelays, responsible for sensing upstream signals and controlling the expression of downstream antioxidative proteins [58, 59]. Deleting skn7 in the yeast increases sensitivity to a wide variety of oxidizing agents [76] and decreases expression of detoxification genes such as trx2, gpx2, ctt1, sod1, or sod2 [59]. Another study showed that Skn7 can interact with heat shock factor Hsf1 in vivo, may bind to the same sites as Hsf1, and is required to induce heat shock genes by oxidative stress [58]. In the human pathogen Candida glabrata or Candida albicans, a significant role of skn7 in regulating antioxidant genes such as trx2, trr1, tsa1, and cta1 was demonstrated [77], and many other ROS-detoxifying enzymes are regulated by skn7 directly or indirectly [78]. Similarly, the expression levels of two genes encoding for Zn-superoxide dismutase and glutathione reductase for antioxidant defense are lower in skn7 deleted mutants in Aspergillus flavus [79], catalase B expression in A. nidulans requires the oxidative stress Skn7 protein SrrA [80], and deletion of FoSkn7 in Fusarium oxysporum f. sp. cubense negatively affects expression of many genes encoding for ROS-detoxifying enzymes and heat shock proteins [81].
Lcc1 and Lcc5 are the main laccases secreted by C. cinerea monokaryons in liquid single-species cultures [23], while Lcc9 is the main laccase expressed during coculture with Gongronella sp. w5 (data from reference 24 and this study). In addition, with the qRT- PCR data indicating that lcc1 was also induced strongly with lcc9 here (Fig. 3A), increased laccase activity in coculture might be produced as a sum effect of both upregulated genes. In phylogenetic analysis, the 17 laccase isozymes of C. cinerea branch into four subgroups, with laccases Lcc5 and Lcc9 grouped in a same branch in the same subgroup as Lcc1 [22]. This reinforced the fact that although the amino acid sequences of these three laccase isozymes are very similar, lcc9 might be transcribed differently from lcc5 and might specifically be expressed to deal with ROS. Similarly, upregulated lcc1 might also contribute to ROS detoxification. In contrast to the promoter of lcc1 and lcc5, one motif TCTAGA was found in the promoter region of lcc9 (position −180 to −175), which is confirmed in C. albicans by CHIP-Seq as a binding motif of Skn7 [78], suggesting possible regulation of lcc9 in C. cinerea by Skn7 when expressed under ROS.
Previously, Kombrink et al. (2019) reported transcriptome studies of C. cinerea homokaryon AmutBmut challenged by the bacteria Bacillus subtilis and Escherichia coli, with three laccase genes being overexpressed, which are lcc9, lcc10, and lcc14 (JGI AmutBmut IDs 488706, 502564, and 208308) [82]. The activating principle is secreted by the bacteria into the medium and is, as here, the presence of Gongronella sp. w5, also inducing expression of the antibacterial lagopodin gene cluster with genes cox1, cop6, and cox2 [36, 82]. Of these genes in C. cinerea, other than lcc9, the promoters of lcc10 (position −284 to −279) and cop6 (position −389 to −384) contained a TCTAGA sequence. The lacc2 gene of P. eryngii var. ferulae (JGI ID 1521536) induced by the antioxidant β-carotene in cultivation with R. mucilaginosa in the absence of ROS stimulation, in contrast, has no such motif in its promoter, but it possesses a stress STRE element CCCCT (position −249 to −245) [70] to which in S. cerevisiae the zinc finger transcription factors Msn2 and Msn4 bind [83]. P. ostreatus has the same STRE element in its lacc2 promoter (JGI ID 1067328) but none in the lacc10 promoter (JGI ID 1089723), and both have no TCTAGA sequence, while both were overexpressed in the confrontation with D. squalens [69]. Despite oxidative stress, also neither of the laccase genes induced by fungal confrontations in D. squalens (Dslcc11, JGI ID 110823; Dslcc5, JGI ID 169869) contain a TCTAGA sequence but Dslcc5 has a STRE element in its promoter (position −39 to −35). Finally, STRE elements are also found in the promoter of gene cox1 (position −59 to −63) in C. cinerea as well as in the promoters of genes clp1 (positions −143 to −138 and −98 to −102) and CC1G_04209 (positions −211 to −207 and −205 to −201) in the coprinoferrin siderophore gene cluster, whereas the promoter of the upregulated cpl2 gene contains a TCTAGA sequence (−138 to −133). In S. cerevisiae, Skn7 with some overlap in gene regulatory function may bind as Hsf1 to a heat shock element (HSE) AGAAnnTTC [58], but none of the basidiomycete genes discussed here had such sequence in their promoters. Summing up, a clear picture on stress regulation in expression of laccase genes in confrontations of different fungal species and usage of potential transcription factor binding sites is not evolving.
By the increased expression of skn7 in interspecies interaction and a target site for Skn7 in the promoter region of lcc9, the oxidative stress factor Skn7 in C. cinerea is likely to directly participate in specific lcc9 activation while this might not be the same case for the overexpressed laccase genes in fungal confrontations of other species. A recent literature overview on fungal skn7 deletion mutants of various species indicates that the Skn7 transcription factor variably effects tolerance to oxidative and sometimes also osmotic stresses, cell wall integrity, hyphal growth, and asexual sporulation of different fungal species [84]. Many of these phenotypes reoccurred here in C. cinerea in interspecific interactions with Gongronella sp. w5 and in the lcc9 silencing clones. Suggested from the results of the combined HBA-NAC treatment of C. cinerea single species cultures (Fig. 4C), compound HBA as an inducer of gene lcc9 expression might act indirectly on the promoter in C. cinerea in a complex regulation network involving oxidative stress. Two potential benzoate-response elements were reported for Aspergillus niger, i.e., TAGGTAGCC and TAGTCA [85, 86]. These were neither found in the promoter of gene lcc9 nor in the promoter of gene skn7.
Lcc9 and ROS in fungal growth and morphology.
Lcc9 was expressed and secreted to the extracellular environment to relieve the oxidative stress caused by secondary compounds from Gongronella sp. w5 and to then also lower the intracellular ROS level here (Fig. 5 and 6 and Fig. S3). Actually, maintenance of ROS detoxification is paramount for fungi. Several intracellular and extracellular defense mechanisms have been evolved to scavenge intracellular ROS [27]. Among them, laccases are seen as a resistant approach of some ascomycetes and basidiomycetes to organic compounds. In fact, gene deletion in the ascomycete P. anserina indicates constitutive laccases to play a protective role against aromatic compounds and H2O2 [17]. Expression of a heterologous laccase in Pichia pastoris increases the resistance of the yeast to H2O2-mediated oxidative stress by stimulating the activity of the glutathione-based antioxidative system (including glutathione, glutathione peroxidase, γ-glutamylcysteine synthetase, and glutathione reductase) [16]. In addition, fungal vacuoles are acidic compartments containing abundant hydrolases and lipases, and are involved in storage, osmoregulation, homeostasis, and detoxification [87]. According to the fact that endogenous ROS is associated with vacuole fragmentation in S. cerevisiae [88, 89], lcc9 silencing leading to vacuole fragmentation might be attributed to the increased ROS level in C. cinerea (Fig. 8).
Several studies have suggested a clear link between laccases and fungal morphology and growth. For example, deletion of laccase-like fungal pigment multicopper oxidase genes in the human pathogenic A. fumigatus, the insect pathogenic Metarhizium anisopliae, and the dung fungus P. anserina all affect the pigment synthesis [17, 90, 91]. Laccases in Cryptococcus neoformans and L. edodes are associated with tension, thickness, and strength of cell walls [20, 92]. Moreover, it has been reported that laccase overexpression affects mycelial growth in T. versicolor [93], P. ostreatus [94–96], H. marmoreus [21], and Trichoderma viride [97]. Our research on C. cinerea suggested that Lcc9 facilitated reasonable fungal growth during cocultivation with Gongronella sp. w5 and under extracellular ROS stress (Fig. 6 and 7, and Fig. S1), provoked hyperbranching with bunches of multiple short side branches on parental hyphae in the wild-type when stressed by Gongronella sp. w5, helped in cellular morphology by maintaining vacuole and cell wall structures, and supported constitutive asexual sporulation (Fig. 7 and 8, Fig. S2 and S3).
Because laccase lcc9 by its pattern of activation only functions in coculture, the opposing phenotypic effects seen in wild-type and lcc9 silencing strains might be related to ROS regulation. In fact, the ability of ROS to influence cell morphology, disturb cell wall integrity, and affect cell growth is demonstrated in C. albicans [98]. For growth in P. anserina, the assimilation of complex plant biomass needs the production of external H2O2, and it has been shown that catalases are required in the protection of the fungal cells during this process [99]. Our work revealed that ROS was produced excessively in C. cinerea in the interspecific interaction with Gongronella sp. w5. After silencing lcc9, the levels of ROS and H2O2 were gradually increased (Fig. 6), then the cellular accumulation of ROS will have caused oxidative damage with harm to the growth and development of C. cinerea mycelium.
MATERIALS AND METHODS
Fungi and culture media.
C. cinerea Okayama 7 (#130; A43, B43, ade8) (ATCC No. MYA-4618) and Gongronella sp. w5 (China Center for Type Culture Collection No. AF2012004) were maintained on YMG agar (yeast malt glucose; per liter, 4 g yeast extract, 10 g malt extract, 4 g glucose, and 15 g agar) or PDA (potato dextrose agar; per liter, filtrate of 200 g boiled potato, 20 g glucose, and 15 g agar) plates at 4°C according to Pan et al. [24]. All culturing of fungi was done at 37°C.
Classical mixed coculture, separated coculture, and chemical applications to cultures.
Classical mixed coculture and separated coculture were performed according to Hu et al. [25] in SAHX (sucrose dl-asparagine XH; per liter, 15.0 g sucrose, 1.5 g dl-asparagine, 1.0 g KH2PO4, 0.5 g MgSO4·7H2O, 0.1 g Na2HPO4·5H2O, 10.0 g CaCl2, 1.0 mg FeSO4·7H2O, 28.0 mg adenine, 2.0 mg CuSO4·5H2O, and 50.0 μg vitamin B1) medium using sucrose as the carbon source. In all classical mixed coculture experiments in this study, C. cinerea was always first inoculated and cultivated for 36 h before addition of homogenized Gongronella sp. w5 mycelium (2.5%, vol/vol), and the time zero h of cocultivation refers to the start of coculture or the 36 h of initial cultivation. For separated coculture, Gongronella sp. w5 were sealed in dialysis tubes (molecular mass cutoff of 50 kDa), which were immersed in flasks to avoid direct mycelial contact of the two species. When p-hydroxybenzoic acid (HBA, Sigma, St. Louis, MO, USA) was employed to induce laccase expression in single-species culture of C. cinerea, it was added at the final concentration of 0.1 mM (solved in ethanol) at 0 h to liquid shaken SAHX cultures of C. cinerea (5% inoculum, vol/vol) that had been precultured for 36 h with 7.5 g/L fructose instead of sucrose as the carbon source. When N-acetyl-l-cysteine (NAC, Sigma, St. Louis, MO, USA) was used to remove the reactive oxygen species (ROS) in C. cinerea in separated coculture with Gongronella sp. w5 or when cultured alone with HBA induction, 10 mM final concentration was added at 0 h of cocultivation or initial cultivation, respectively. Cultures were incubated for another 4–5 d and laccase activity and the intracellular ROS levels were regularly assayed every 12 h. For stress tests with chemicals, 100 mM H2O2 (Sangon Biotech, Shanghai, China), 2 mM guaiacol (Sigma, St. Louis, MO, USA), or 0.5 mM 1,3-dihydroxynaphthalene (Sangon Biotech, Shanghai, China), which were secreted by Gongronella sp. w5 (Table S2) and effective in stress induction in C. cinerea in preliminary experiments, were added to separated cocultures of C. cinerea and Gongronella sp. w5 at 36 h of cocultivation, and the intracellular ROS levels and biomasses of C. cinerea were detected within 36 h of further cocultivation, respectively. The experiments were performed three times, always with triplicate cultures per test case.
Measurement of C. cinerea biomass from cocultures.
After removing Gongronella sp. w5 from the dialysis bags, the total liquid cultures of C. cinerea in the flasks were filtered through Whatman No. 1 filter paper (GE Healthcare, Hangzhou, China). The C. cinerea mycelia were washed two times with distilled water, transferred into pre-weighed weighing bottles, and for measurements dried in an oven at 105°C to a constant weight.
C. cinerea transcriptome sequencing.
For comparative transcriptome analysis, wild-type C. cinerea mycelia were harvested from the culture liquid after 18 h and 28 h in separated cocultivation with Gongronella sp. w5, respectively. The mycelia of C. cinerea were homogenized using glass beads in a freezing grinding instrument JXFSTPRP-CL (Jingxin Co., Ltd., Shanghai, China), and the total RNA of C. cinerea was extracted using the TRIzol reagent (Thermo Fisher, Catalogue No. 15596026) according to the manufacturer’s protocol. The integrity and quantity of the isolated RNA were assessed on a NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA, USA). RNA library preparation was carried out by poly(A)-selection of mRNA, cDNA synthesis, adenylation of 3′ ends, adapter ligation, DNA fragment enrichment, and purification. Library quality check was performed using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent, CA, USA). The library was then sequenced on NovaSeq 6000 platform (Illumina) by Shanghai Personal Biotechnology Co. Ltd. Transcriptome sequencing was performed for three independent samples per time point.
Differentially expressed gene (DEG) calculation and analysis.
Raw sequencing data were filtered by deleting the sequences containing connectors and low quality reads using Cutadapt (v1.15) [100]. The clean reads were then mapped to the reference genes of C. cinerea Okayama 7 (GenBank, accession No. AACS00000000) [101] using HISAT2 (v2.0.5) [102]. HTSeq (v0.9.1) statistics was used to compare the Read Count values on each gene as the original expression of this gene, and then FPKM (Fragments Per Kilobase per Million) was used to standardize the expression [103]. Then, gene expression differences were analyzed by DESeq (v1.30.0) with |log2FoldChange| > 1 and false discovery rate (FDR) < 0.05 [104]. In a few cases, DEGs with a threshold of |log2FoldChange| > 0.75 or < −0.75 were considered. At the same time, R language Pheatmap (1.0.8) software package was used to perform bi-directional clustering analysis of all different genes of samples. Heatmap was gated according to the expression level of the same gene in different samples and the expression patterns of different genes in the same sample with Euclidean method to calculate the distance and Complete Linkage method to cluster. Go enrichment analysis was performed with topGo (v3.2), and the P value was calculated by hypergeometric distribution method. GO terms with a P value ≤ 0.05 were considered significantly enriched. ClusterProfiler (v3.4.4) software was used to carry out the enrichment analysis of the KEGG pathway of differential genes, focusing on the significant enrichment pathway with P value <0.05.
Quantitative reverse transcription-PCR (qRT-PCR) analysis.
To confirm results from the transcriptome analysis, 15 genes were randomly selected for qRT-PCR verification using a SYBR green kit (TaKaRa, Dalian, China) on a Roche LightCycler 96 real-time PCR system (Roche, Basel, Switzerland) according to the manufacturer’s instructions (primers for qRT-PCR analysis are shown in Table 1). Three biological replicates and three technical replicates per sample were included for each cDNA template. Then, the relative expression levels were calculated using the 2-ΔΔCt method [105]. In order to normalize the qRT-PCR data, five genes, including β-actin, capzb (F-actin capping protein beta subunit), α-tub (tubulin), cyt-c (cytochrome c), and ubc (ubiquitin) were chosen for their stable expression levels across different time points. They were classified and screened according to their suitability as internal standards using the NormFinder (https://www.moma.dk/normfinder-software/) and BestKeeper (https://www.gene-quantification.de/bestkeeper.html) algorithms [106, 107]. The most stable transcribed gene was chosen as endogenous reference gene.
TABLE 1.
Primers used in this studya
| Name | Sequence (5′–3′) | Purpose |
|---|---|---|
| lcc9-antisense1-fwd | CTCCCATCTACACACAACAAGCTTATCGCCCGGGATTCTCATAGTTGTAAGTGCTGC | Cloning of lcc9 antisense fragment 1 |
| lcc9- antisense 1-rev | CACTGGCCCTCTGGTCAACTATAATATTATAGATGGGCCTTGGACCTGCCG | Cloning of lcc9 antisense fragment 1 |
| lcc9- antisense 2-fwd | CTCCCATCTACACACAACAAGCTTATCGCCCGGACCACTTCCTCCTGGGGCA | Cloning of lcc9 antisense fragment 2 |
| lcc9- antisense 2-rev | CACTGGCCCTCTGGTCAACTATAATATTATCTCTCATGGTCGACGAAATCCAGATC | Cloning of lcc9 antisense fragment 2 |
| PF | ACATCCACCATCTCCGTTTTCTCCCAT | PCR of co-transformants |
| PR | TGACTATAGCAGCCTCCTACCACTG | PCR of co-transformants |
| qRT-13048-F | CTCTGGAGTTATGGTAGGAATGGGC | qRT-PCR of β-actin |
| qRT-13048-R | GATGCCATGTTCGATGGGGTACTTG | qRT-PCR of β-actin |
| qRT-00146-F | CCGCCAACAACTATGCCCGTGGTCA | qRT-PCR of α-tubulin |
| qRT-00146-R | TCCTCCACCGAAGGAGTGGAAGACG | qRT-PCR of α-tubulin |
| qRT-10735-F | TTCGCTCCTGGTGACGCTTCTA | qRT-PCR of cyt-c |
| qRT-10735-R | GCCGAAGATGCCGAAGAGGTTG | qRT-PCR of cyt-c |
| qRT-11833-F | AAGGCGAAGATTCAGGACAAGGAAGGTA | qRT-PCR of ubc |
| qRT-11833-R | CGAAGGCGGAGAACCAAGTGAAGAGTA | qRT-PCR of ubc |
| qRT-07869-F | GAAGAAGACTCTCACTCCGTCAACG | qRT-PCR of capzb |
| qRT-07869-R | CTGTGTGGTGAGCTGGAGCATGA | qRT-PCR of capzb |
| qRT-lcc9-F | ATGTCCAGGAAACTTTTCTCTCTCG | qRT-PCR of lcc9 |
| qRT-lcc9-R | ATGTTCGAGACCGTCATGGTACT | qRT-PCR of lcc9 |
| qRT-07201-F | AGCGCAAGCTGACCCCTTGGT | qRT-PCR of sga1 |
| qRT-07201-R | ACAAGTCCAGCCGCTGCCCC | qRT-PCR of sga1 |
| qRT-05652-F | CAAGGATCGACGGAGGGCGCA | qRT-PCR of tkl |
| qRT-05652-R | TCCACTTCCGAGCCGCCACC | qRT-PCR of tkl |
| qRT-02494-F | AGCAAGGTGGATGTCCACGAGTCC | qRT-PCR of vwfa |
| qRT-02494-R | GGCCACTGCCAGCGACTTGGT | qRT-PCR of vwfa |
| qRT-06452-F | TACGCTGGAATGGACGCCGGA | qRT-PCR of exg |
| qRT-06452-R | TGCCGCGGGCCCAAAAGTTCTTC | qRT-PCR of exg |
| qRT-03564-F | CACCTTCGCTTGTTCTACGGATCCA | qRT-PCR of cyp98A3 |
| qRT-03564-R | TCAACCCAGAAGCGTCCGGGAATAC | qRT-PCR of cyp98A3 |
| qRT-05291-F | CTAAGCAGCATAATGTGAAAGCCCT | qRT-PCR of Chi |
| qRT-05291-R | GGAAATTCCCAGTCAAAGTCGATACCA | qRT-PCR of Chi |
| qRT-11387-F | ATCGAGATGATCGAGCATGTCGGC | qRT-PCR of cfa |
| qRT-11387-R | TGATGACAGCGGCGGCCTTTTG | qRT-PCR of cfa |
| qRT-08324-F | TGCAGAGGAAGAGCGACCTCGGA | qRT-PCR of samB |
| qRT-08324-R | CCAGATGCTGAGAGGGAAACGGAGG | qRT-PCR of samB |
| qRT-09241-F | CCGGATTTGAAGACGATCAAGAGCTT | qRT-PCR of psd |
| qRT-09241-R | GAGAAGGTACATGGGAACGCCGATAG | qRT-PCR of psd |
| qRT-09422-F | TTATCGCATACTCGCCCCTCGGA | qRT-PCR of akr |
| qRT-09422-R | AAGGCATCGACGATTGCCATGTTG | qRT-PCR of akr |
| qRT-14829-F | TGCTGGAGAGCAAGGTTGTGTCTCA | qRT-PCR of gst |
| qRT-14829-R | AAGGGAATGGTGTAGTGGGGCGAGC | qRT-PCR of gst |
| qRT-05005-F | GCCATCCGCTCCAGTGCCTTCAAT | qRT-PCR of cgl-II |
| qRT-05005-R | GCGGATGCTGATGTGGAGGAGGATG | qRT-PCR of cgl-II |
| qRT-02499-F | CAGTCAGAGAGTTGCCTATACTCTCA | qRT-PCR of tdo |
| qRT-02499-R | GGATGATGGTGGCAGAGAGTGAATG | qRT-PCR of tdo |
| qRT-07229-F | CATCTTAGCTGGCTCGGCTTCTTC | qRT-PCR of nrt |
| qRT-07229-R | ACGATGTTGGAGTTGGCGACTTCG | qRT-PCR of nrt |
| qRT-lcc1-F | ATGTTCAAGAACCTCCTCTCGT | qRT-PCR of lcc1 |
| qRT-lcc1-R | ACGTTCGCGTTGGTGAGGGTCAT | qRT-PCR of lcc1 |
| qRT-lcc5-F | ATGTCGTTTGCTTGGAAAGCAT | qRT-PCR of lcc5 |
| qRT-lcc5-R | TGGTCATAGTATCTTGGTTGCCAAT | qRT-PCR of lcc5 |
| qRT-08281-F | TGTACAAGATGCTCGAAGACCCA | qRT-PCR of skn7 |
| qRT-08281-R | TTCATGTCCTTTACGACGAAACAGTC | qRT-PCR of skn7 |
Five candidate reference genes chosen were β-actin, capzb (F-actin capping protein beta subunit), α-tub (tubulin), cyt-c (cytochrome c), and ubc (ubiquitin). Fifteen randomly chosen genes for qRT-PCR to validate gene expression data in the RNA-seq experiment were lcc9 (laccase 9), sga1 (glucoamylase), tkl (TKL/TKL-ccin protein kinase), vwfa (VWFA domain protein), exg (glucan 1, 3-beta-glucosidase), cyp98A3 (cytochrome P450 98A3), Chi (endochitinase), cfa (cyclopropane-fatty-acyl-phospholipid synthase), samB (MYND-type Zinc finger protein), psd (phosphatidylserine decarboxylase), akr (aldo/keto reductase), gst (glutathione S-transferase), cgl-II (galectin-2), tdo (tryptophan 2,3-dioxygenase), and nrt (nitrate transporter).
Further, qRT-PCR analysis for lcc1, lcc5, lcc9, and skn7 transcription was also done from separated cocultures at the indicated times using the same protocol as described above with the primers listed in Table 1.
Intracellular ROS levels and H2O2 assay.
Intracellular ROS levels were detected using the fluorogenic probe DCFH-DA (2, 7-dichlorodihydrofluorescein diacetate) [108] according to the manufactural instructions (Beyotime Biotech, Shanghai, China). C. cinerea cells in liquid cultures were harvested at certain time points, washed twice with PBS (phosphate-buffered saline), resuspended in 10 μM DCFH-DA in PBS, and incubated at 37°C for 20 min in the dark. After PBS washing, C. cinerea hyphae were ground to a fine powder under liquid nitrogen and mixed with 1 ml cold PBS buffer. A total of 200 μl supernatant was measured with excitation and emission wavelengths of 488 nm and 525 nm, respectively, on a SpectraMax M5 instrument (Molecular Devices, San Jose, CA, USA). All measurements were performed in triplicate.
Intracellular H2O2 concentration measurements were analyzed in 96-well plates (Thermo Fisher, Waltham, MA) using an H2O2 detection kit (Beyotime Biotech, Shanghai, China) in which Fe2+ could be oxidized, formed purple product with xylenol orange and be detectable on a SpectraMax M5 instrument at wavelengths of 560 nm. C. cinerea cells were harvested, washed twice with PBS, ground to a fine powder in liquid nitrogen, and mixed with 1 ml cold PBS buffer. After that, 50 μl supernatant was mixed with 100 μl H2O2 detection reagent and incubated for 30 min at room temperature before testing. The concentrations of H2O2 in the samples were calculated according to a standard curve obtained by testing the absorbance of different dilutions of H2O2.
Laccase assay.
Aliquots of culture broth from classical mixed coculture, separated coculture, and single-species culture systems were withdrawn every 12 h. Laccase activity was determined with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonate) (ABTS) (0.5 mM) as the substrate according to Bourbonnais and Paice [109]. One activity unit (U) was defined as the amount of laccase required for oxidizing 1 μmol of ABTS per minute. Reactions with heat-treated enzyme samples (5 min, 100°C) were used as control.
Activity staining of laccases was performed in 100 mM citrate-phosphate buffer (pH 4.0) after polyacrylamide gel electrophoresis (PAGE) using ABTS as the substrate, according to Pan et al. [24].
Data availability.
Transcriptomic data are publicly available in National Center for Biotechnology Information (NCBI) under BioProject number PRJNA755756. The data sets generated/analyzed during the present study are available and included either within this article or in the supplemental material. The DNA sequence of the 18S rRNA gene for molecular identification of Gongronella sp. w5 was previously deposited in the GenBank database (accession no. EU253966) [110]. In addition, the GenBank accession numbers of the genome sequence of C. cinerea Okayama 7 [101] and Gongronella sp. w5 [111] were AACS00000000 and GCA_001650995.1, respectively.
ACKNOWLEDGMENTS
This work was supported by the Chinese National Natural Science Foundation grant (No. 31800051, No. 31870068, and No. 31870098), the Science Fund for Distinguished Young Scholars of Anhui Province (No. 2008085J12), and the Doctoral Research Start-Up Funding of Anhui University (Y040418162).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Zemin Fang, Email: zemin_fang@ahu.edu.cn.
Ursula Kües, Email: ukuees@gwdg.de.
Irina S. Druzhinina, Nanjing Agricultural University
REFERENCES
- 1.Boddy L. 2000. Interspecific combative interactions between wood decaying basidiomycetes. FEMS Microbiol Ecol 31:185–194. 10.1111/j.1574-6941.2000.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 2.Arfi Y, Levasseur A, Record E. 2013. Differential gene expression in Pycnoporus coccineus during interspecific mycelial interactions with different competitors. Appl Environ Microbiol 79:6626–6636. 10.1128/AEM.02316-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiscox J, Boddy L. 2017. Armed and dangerous—chemical warfare in wood decay communities. Fungal Biol Rev 31:169–184. 10.1016/j.fbr.2017.07.001. [DOI] [Google Scholar]
- 4.Chi Y, Hatakka A, Maijala P. 2007. Can co-culturing of two white-rot fungi increase lignin degradation and the production of lignin-degrading enzymes? Int Biodeterior Biodegrad 59:32–39. 10.1016/j.ibiod.2006.06.025. [DOI] [Google Scholar]
- 5.Bader J, Mast-Gerlach E, Popović MK, Bajpai R, Stahl U. 2010. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol 109:371–387. 10.1111/j.1365-2672.2009.04659.x. [DOI] [PubMed] [Google Scholar]
- 6.Silar P. 2005. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol Res 109:137–149. 10.1017/s0953756204002230. [DOI] [PubMed] [Google Scholar]
- 7.Baldrian P. 2004. Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol Ecol 50:245–253. 10.1016/j.femsec.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Flores C, Vidal C, Trejo-Hernández MR, Galindo E, Serrano-Carreón L. 2009. Selection of Trichoderma strains capable of increasing laccase production by Pleurotus ostreatus and Agaricus bisporus in dual cultures. J Appl Microbiol 106:249–257. 10.1111/j.1365-2672.2008.03998.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Yu G, Li P, Gu Y, Li J, Liu G, Yao J. 2009. Overproduction of Trametes versicolor laccase by making glucose starvation using yeast. Enzym Microb Technol 45:146–149. [Google Scholar]
- 10.Kannaiyan R, Mahinpey N, Kostenko V, Martinuzzi RJ. 2015. Nutrient media optimization for simultaneous enhancement of the laccase and peroxidases production by coculture of Dichomitus squalens and Ceriporiopsis subvermispora. Biotechnol Appl Biochem 62:173–185. 10.1002/bab.1263. [DOI] [PubMed] [Google Scholar]
- 11.Du W, Sun C, Wang J, Xie W, Wang B, Liu X, Zhang Y, Fan Y. 2017. Conditions and regulation of mixed culture to promote Shiraia bambusicola and Phoma sp. BZJ6 for laccase production. Sci Rep 7:17801. 10.1038/s41598-017-17895-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U. 2006. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273:2308–2326. 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 13.Kües U, Rühl M. 2011. Multiple multi-copper oxidase gene families in basidiomycetes—what for? Curr Genomics 12:72–94. 10.2174/138920211795564377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundell TK, Mäkelä MR, Hildén K. 2010. Lignin-modifying enzymes in filamentous basidiomycetes—ecological, functional and phylogenetic review. J Basic Microbiol 50:5–20. 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- 15.Janusz G, Pawlik A, Świderska-Burek U, Polak J, Sulej J, Jarosz-Wilkołazka A, Paszczyński A. 2020. Laccase properties, physiological functions, and evolution. Int J Mol Sci 21:966. 10.3390/ijms21030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Fan F, Zhuo R, Ma F, Gong Y, Wan X, Jiang M, Zhang X. 2012. Expression of the laccase gene from a white rot fungus in Pichia pastoris can enhance the resistance of this yeast to H2O2-mediated oxidative stress by stimulating the glutathione based antioxidative system. Appl Environ Microbiol 78:5845–5854. 10.1128/AEM.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie N, Chapeland-Leclerc F, Silar P, Ruprich-Robert G. 2014. Systematic gene deletions evidences that laccases are involved in several stages of wood degradation in the filamentous fungus Podospora anserina. Environ Microbiol 16:141–161. 10.1111/1462-2920.12253. [DOI] [PubMed] [Google Scholar]
- 18.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol 181:6469–6477. 10.1128/JB.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakade K, Watanabe H, Sakamoto Y, Sato T. 2011. Gene silencing of the Lentinula edodes lcc1 gene by expression of a homologous inverted repeat sequence. Microbiol Res 166:484–493. 10.1016/j.micres.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto Y, Nakade K, Sato S, Yoshimi A, Sasaki K, Konno N, Abe K. 2018. Cell wall structure of secreted laccase-silenced strain in Lentinula edodes. Fungal Biol 122:1192–1200. 10.1016/j.funbio.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Chen H, Chen M, Ren A, Huang J, Wang H, Zhao M, Feng Z. 2015. Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus. Microbiol Res 179:54–63. 10.1016/j.micres.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Kilaru S, Hoegger PJ, Kües U. 2006. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet 50:45–60. 10.1007/s00294-006-0074-1. [DOI] [PubMed] [Google Scholar]
- 23.Rühl M, Majcherczyk A, Kües U. 2013. Lcc1 and Lcc5 are the main laccases secreted in liquid cultures of Coprinopsis cinerea strains. Antonie Van Leeuwenhoek 103:1029–1039. 10.1007/s10482-013-9883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan K, Zhao N, Yin Q, Zhang T, Xu X, Fang W, Hong Y, Fang Z, Xiao Y. 2014. Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour Technol 162:45–52. 10.1016/j.biortech.2014.03.116. [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Zhang Y, Xu Y, Sun Q, Liu J, Fang W, Xiao Y, Kües U, Fang Z. 2019. Gongronella sp. w5 elevates Coprinopsis cinerea laccase production by carbon source syntrophism and secondary metabolite induction. Appl Microbiol Biotechnol 103:411–425. 10.1007/s00253-018-9469-4. [DOI] [PubMed] [Google Scholar]
- 26.Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15. 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 27.Harms H, Schlosser D, Wick LY. 2011. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9:177–192. 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender JL. 2014. Metabolite induction via microorganism coculture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv 32:1180–1204. 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Sjaarda CP, Abubaker KS, Castle AJ. 2015. Induction of lcc2 expression and activity by Agaricus bisporus provides defence against Trichoderma aggressivum toxic extracts. Microb Biotechnol 8:918–929. 10.1111/1751-7915.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agger S, Lopez-Gallego F, Schmidt-Dannert C. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72:1181–1195. 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, Jaeger GD, Mely Y, Goossens A, Werck-Reichhart D. 2012. Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell 24:4465–4482. 10.1105/tpc.112.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi X, Yu X, Xu D, Fang H, Dong K, Li W, Liang C. 2017. Identification and analysis of CYP450 genes from transcriptome of Lonicera japonica and expression analysis of chlorogenic acid biosynthesis related CYP450s. Peer J 5:e3781. 10.7717/peerj.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelosi L, Ducluzeau AL, Loiseau L, Barras F, Schneider D, Junier I, Pierrel F. 2016. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 1:e00091-16. 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasconcelos MRS, Vieira GAL, Otero IVR, Bonugli-Santos RC, Rodrigues MVN, Rehder VLG, Ferro M, Boaventura S, Bacci M, Jr, Sette LD. 2019. Pyrene degradation by marine-derived ascomycete: process optimization, toxicity, and metabolic analyses. Environ Sci Pollut Res Int 26:12412–12424. 10.1007/s11356-019-04518-2. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Farradá G, Manzano-León AM, Rineau F, Ramos Leal M, Thijs S, Jambon I, Put J, Czech J, Rivera GG, Carleer R, Vangronsveld J. 2019. Biodegradation of polycyclic aromatic hydrocarbons by native Ganoderma sp. strains: identification of metabolites and proposed degradation pathways. Appl Microbiol Biotechnol 103:7203–7215. 10.1007/s00253-019-09968-9. [DOI] [PubMed] [Google Scholar]
- 36.Stöckli M, Morinaka BI, Lackner G, Kombrink A, Sieber R, Margot C, Stanley CE, deMello AJ, Piel J, Künzler M. 2019. Bacteria-induced production of the antibacterial sesquiterpene lagopodin B in Coprinopsis cinerea. Mol Microbiol 112:605–619. 10.1111/mmi.14277. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Gallego F, Schmidt-Dannert C. 2010. Multi-enzymatic synthesis. Curr Opin Chem Biol 14:174–183. 10.1016/j.cbpa.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 38.Masuya T, Tsunematsu Y, Hirayama Y, Sato M, Noguchi H, Nakazawa T, Watanabe K. 2019. Biosynthesis of lagopodins in mushroom involves a complex network of oxidation reactions. Org Biomol Chem 17:234–239. 10.1039/c8ob02814a. [DOI] [PubMed] [Google Scholar]
- 39.Tsunematsu Y. 2021. Genomics-directed activation of cryptic natural product pathways deciphers codes for biosynthesis and molecular function. J Nat Med 75:261–274. 10.1007/s11418-020-01466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sass H, Ansari SR, Dietl AM, Déziel E, Haas H, Stevens DA. 2019. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS One 14:e0216085. 10.1371/journal.pone.0216085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mügge V, Heine T, Baraibar AG, van Berkel WJH, Paul CE, Tischler D. 2020. Flavin-dependent N-hydroxylating enzymes: distribution and application. Appl Microbiol Biotechnol 104:6481–6499. 10.1007/s00253-020-10705-.w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Autréaux B, Toledano MB. 2007. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Bio 8:813–824. 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 43.Nanda AK, Andrio E, Marino D, Pauly N, Dunand C. 2010. Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52:195–204. 10.1111/j.1744-7909.2010.00933.x. [DOI] [PubMed] [Google Scholar]
- 44.Montibus M, Pinson-Gadais L, Richard-Forget F, Barreau C, Ponts N. 2015. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit Rev Microbiol 41:295–308. 10.3109/1040841X.2013.829416. [DOI] [PubMed] [Google Scholar]
- 45.Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci USA 101:6564–6569. 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z, Li N, He B, Igarashi Y, Luo F. 2019. Transcriptome analysis of differential gene expression in Dichomitus squalens during interspecific mycelial interactions and the potential link with laccase induction. J Microbiol 57:127–137. 10.1007/s12275-019-8398-y. [DOI] [PubMed] [Google Scholar]
- 47.Breitenbach M, Weber M, Rinnerthaler M, Karl T, Breitenbach-Koller L. 2015. Oxidative stress in fungi: its function in signal transduction, interaction with plant hosts, and lignocellulose degradation. Biomolecules 5:318–342. 10.3390/biom5020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bultman KM, Kowalski CH, Cramer RA. 2017. Aspergillus fumigatus virulence through the lens of transcription factors. Med Mycol 55:24–38. 10.1093/mmy/myw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krüger M, Fischer R. 1998. Integrity of a Zn finger-like domain in SamB is crucial for morphogenesis in ascomycetous fungi. EMBO J 17:204–214. 10.1093/emboj/17.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Převorovský M, Oravcová M, Tvarůžková J, Zach R, Folk P, Půta F, Bähler J. 2015. Fission yeast CSL transcription factors: mapping their target genes and biological roles. PLoS One 10:e0137820. 10.1371/journal.pone.0137820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim J, Heo J, Ju H, Shin JW, Kim Y, Lee S, Yu HY, Ryu CM, Yun H, Song S, Hong KS, Chung HM, Kim HR, Roe JS, Choi K, Kim IG, Jeong EM, Shin DM. 2020. Glutathione dynamics determine the therapeutic efficacy of mesenchymal stem cells for graft-versus-host disease via CREB1-NRF2 pathway. Sci Adv 6:eaba1334. 10.1126/sciadv.aba1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Songdech P, Ruchala J, Semkiv MV, Jensen LT, Sibirny A, Ratanakhanokchai K, Soontorngun N. 2020. Overexpression of transcription factor ZNF1 of glycolysis improves bioethanol productivity under high glucose concentration and enhances acetic acid tolerance of Saccharomyces cerevisiae. Biotechnol J 15:e1900492. 10.1002/biot.201900492. [DOI] [PubMed] [Google Scholar]
- 53.Rai R, Tate JJ, Georis I, Dubois E, Cooper TG. 2014. Constitutive and nitrogen catabolite repression-sensitive production of Gat1 isoforms. J Biol Chem 289:2918–2933. 10.1074/jbc.M113.516740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song X, Wang Y, Wang P, Pu G, Zou X. 2020. GATA-type transcriptional factor Gat1 regulates nitrogen uptake and polymalic acid biosynthesis in polyextremotolerant fungus Aureobasidium pullulans. Environ Microbiol 22:229–242. 10.1111/1462-2920.14841. [DOI] [PubMed] [Google Scholar]
- 55.Jordá T, Puig S. 2020. Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes (Basel) 11:795. 10.3390/genes11070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450. 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 57.Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, Zavolan M. 2017. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun 8:457. 10.1038/s41467-017-00539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell 11:2335–2347. 10.1091/mbc.11.7.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fassler JS, West AH. 2011. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot Cell 10:156–167. 10.1128/EC.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dörnte B, Peng C, Fang Z, Kamran A, Yulzivar C, Kües U. 2020. Selection markers for transformation of the sequenced reference monokaryon Okayama 7/#130 and homokaryon AmutBmut of Coprinopsis cinerea. Fungal Biol Biotechnol 7:15. 10.1186/s40694-020-00105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crowe JD, Olsson S. 2001. Induction of laccase activity in Rhizoctonia solani by antagonistic Pseudomonas fluorescens strains and a range of chemical treatments. Appl Environ Microbiol 67:2088–2094. 10.1128/AEM.67.5.2088-2094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brakhage AA, Schroeckh V. 2011. Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol 48:15–22. 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Schroeckh V, Brakhage AA. 2015. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 6:299. 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Hindra, Elliot MA. 2019. Unlocking the trove of metabolic treasures: activating silent biosynthetic gene clusters in bacteria and fungi. Curr Opin Microbiol 51:9–15. 10.1016/j.mib.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Guo C, Zhao L, Wang F, Lu J, Ding Z, Shi G. 2017. β-Carotene from yeasts enhances laccase production of Pleurotus eryngii var. ferulae in coculture Front Microbiol 8:1101. 10.3389/fmicb.2017.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mali T, Kuuskeri J, Shah F, Lundell TK. 2017. Interactions affect hyphal growth and enzyme profiles in combinations of coniferous wood-decaying fungi of Agaricomycetes. PLoS One 12:e0185171. 10.1371/journal.pone.0185171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mali T, Mäki M, Hellén H, Heinonsalo J, Bäck J, Lundell T. 2019. Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns. FEMS Microbiol Ecol 95:fiz135. 10.1093/femsec/fiz135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore D. 1969. Sources of carbon and energy used by Coprinus lagopus sensu Buller. J Gen Microbiol 58:49–56. 10.1099/00221287-58-1-49. [DOI] [PubMed] [Google Scholar]
- 69.Zhong Z, Li L, Chang P, Xie H, Zhang H, Igarashi Y, Li N, Luo F. 2017. Differential gene expression profiling analysis in Pleurotus ostreatus during interspecific antagonistic interactions with Dichomitus squalens and Trametes versicolor. Fungal Biol 121:1025–1036. 10.1016/j.funbio.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Q, Zhao L, Li Y, Wang F, Li S, Shi G, Ding Z. 2020. Comparative transcriptomics and transcriptional regulation analysis of enhanced laccase production induced by coculture of Pleurotus eryngii var. ferulae with Rhodotorula mucilaginosa. Appl Microbiol Biotechnol 104:241–255. 10.1007/s00253-019-10228-z. [DOI] [PubMed] [Google Scholar]
- 71.Xiao Y, Tu X, Wang J, Zhang M, Cheng Q, Zeng W, Shi Y. 2003. Purification, molecular characterization and reactivity with aromatic compounds of a laccase from basidiomycete Trametes sp. strain AH28-2. Appl Microbiol Biotechnol 60:700–707. 10.1007/s00253-002-1169-3. [DOI] [PubMed] [Google Scholar]
- 72.Xiao Y, Hong Y, Li J, Hang J, Tong P, Fang W, Zhou C. 2006. Cloning of novel laccase isozyme genes from Trametes sp. AH28-2 and analyses of their differential expression. Appl Microbiol Biotechnol 71:493–501. 10.1007/s00253-005-0188-2. [DOI] [PubMed] [Google Scholar]
- 73.Janusz G, Rogalski J, Szczodrak J. 2007. Increased production of laccase by Cerrena unicolor in submerged liquid cultures. World J Microbiol Biotechnol 23:1459–1464. 10.1007/s11274-007-9390-y. [DOI] [Google Scholar]
- 74.Yang J, Wang G, Ng TB, Lin J, Ye X. 2016. Laccase production and differential transcription of laccase genes in Cerrena sp. in response to metal ions, aromatic compounds, and nutrients. Front Microbiol 6:1558. 10.3389/fmicb.2015.01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marschall R, Schumacher J, Siegmund U, Tudzynski P. 2016. Chasing stress signals—exposure to extracellular stimuli differentially affects the redox state of cell compartments in the wild type and signaling mutants of Botrytis cinerea. Fungal Genet Biol 90:12–22. 10.1016/j.fgb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J 16:1035–1044. 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saijo T, Miyazaki T, Izumikawa K, Mihara T, Takazono T, Kosai K, Imamura Y, Seki M, Kakeya H, Yamamoto Y, Yanagihara K, Kohno S. 2010. Skn7p is involved in oxidative stress response and virulence of Candida glabrata. Mycopathologia 169:81–90. 10.1007/s11046-009-9233-5. [DOI] [PubMed] [Google Scholar]
- 78.Basso V, Znaidi S, Lagage V, Cabral V, Schoenherr F, Leibundgut-Landmann S, d'Enfert C, Bachellier-Bassi S. 2017. The two-component response regulator Skn7 belongs to a network of transcription factors regulating morphogenesis in Candida albicans and independently limits morphogenesis-induced ROS accumulation. Mol Microbiol 106:157–182. 10.1111/mmi.13758. [DOI] [PubMed] [Google Scholar]
- 79.Zhang F, Xu G, Geng L, Lu X, Yang K, Yuan J, Nie X, Zhuang Z, Wang S. 2016. The stress response regulator AflSkn7 influences morphological development, stress response, and pathogenicity in the fungus Aspergillus flavus. Toxins 8:202. 10.3390/toxins8070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vargas-Pérez I, Sánchez O, Kawasaki L, Georgellis D, Aguirre J. 2007. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot Cell 6:1570–1583. 10.1128/EC.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qi X, Liu L, Wang J. 2019. Stress response regulator FoSkn7 participates in the pathogenicity of Fusariumoxysporum f. sp. cubense race 4 by conferring resistance to exogenous oxidative stress. J Gen Plant Pathol 85:382–394. 10.1007/s10327-019-00858-6. [DOI] [Google Scholar]
- 82.Kombrink A, Tayyrov A, Essig A, Stöckli M, Micheller S, Hintze J, van Heuvel Y, Dürig N, Lin CW, Kallio PT, Aebi M, Künzler M. 2019. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J 13:588–602. 10.1038/s41396-018-0293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moskvina E, Schüller C, Maurer CT, Mager WH, Ruis H. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14:1041–1050. . [DOI] [PubMed] [Google Scholar]
- 84.Zhou G, Ying S, Hu Y, Fang X, Feng M, Wang J. 2018. Roles of three HFS domain-containing proteins in mediating heat-shock protein genes and sustaining asexual cycle, stress tolerance, and virulence in Beauveria bassiana. Front Microbiol 9:1677. 10.3389/fmicb.2018.01677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van den Brink JM, Punt PJ, van Gorcom RF, van den Hondel CA. 2000. Regulation of expression of the Aspergillus niger benzoate para-hydroxylase cytochrome P450 system. Mol Gen Genet 263:601–609. 10.1007/s004380051207. [DOI] [PubMed] [Google Scholar]
- 86.Antunes MS, Hodges TK, Carpita NC. 2016. A benzoate-activated promoter from Aspergillus niger and regulation of its activity. Appl Microbiol Biotechnol 100:5479–5489. 10.1007/s00253-016-7373-3. [DOI] [PubMed] [Google Scholar]
- 87.Richards A, Gow NA, Veses V. 2012. Identification of vacuole defects in fungi. J Microbiol Methods 91:155–163. 10.1016/j.mimet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 88.Corson LB, Folmer J, Strain JJ, Culotta VC, Cleveland DW. 1999. Oxidative stress and iron are implicated in fragmenting vacuoles of Saccharomyces cerevisiae lacking Cu, Zn-superoxide dismutase. J Biol Chem 274:27590–27596. 10.1074/jbc.274.39.27590. [DOI] [PubMed] [Google Scholar]
- 89.Pujol-Carrion N, Petkova MI, Serrano L, de la Torre-Ruiz MA. 2013. The MAP kinase Slt2 is involved in vacuolar function and actin remodeling in Saccharomyces cerevisiae mutants affected by endogenous oxidative stress. Appl Environ Microbiol 79:6459–6471. 10.1128/AEM.01692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugareva V, Härtl A, Brock M, Hübner K, Rohde M, Heinekamp T, Brakhage AA. 2006. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch Microbiol 186:345–355. 10.1007/s00203-006-0144-2. [DOI] [PubMed] [Google Scholar]
- 91.Fang W, Fernandes EK, Roberts DW, Bidochka MJ, St Leger RJ. 2010. A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet Biol 47:602–607. 10.1016/j.fgb.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 92.Zhu X, Williamson PR. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res 5:1–10. 10.1016/j.femsyr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 93.Thiruchelvam AT, Ramsay JA. 2007. Growth and laccase production kinetics of Trametes versicolor in a stirred tank reactor. Appl Microbiol Biotechnol 74:547–554. 10.1007/s00253-006-0695-9. [DOI] [PubMed] [Google Scholar]
- 94.Tlecuitl-Beristain S, Sánchez C, Loera O, Robson GD, Díaz-Godínez G. 2008. Laccases of Pleurotus ostreatus observed at different phases of its growth in submerged fermentation: production of a novel laccase isoform. Mycol Res 112:1080–1084. 10.1016/j.mycres.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Das N, Sengupta S, Mukherjee M. 1997. Importance of laccase in vegetative growth of Pleurotus florida. Appl Environ Microbiol 63:4120–4122. 10.1128/aem.63.10.4120-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das N, Chakraborty TK, Mukherjee M. 2001. Purification and characterization of a growth-regulating laccase from Pleurotus florida. J Basic Microbiol 41:261–267. . [DOI] [PubMed] [Google Scholar]
- 97.Divya L, Sadasivan C. 2016. Trichoderma viride laccase plays a crucial role in defense mechanism against antagonistic organisms. Front Microbiol 7:741. 10.3389/fmicb.2016.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu Q, Zhang B, Li J, Zhang B, Wang H, Li M. 2016. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radic Biol Med 99:572–583. 10.1016/j.freeradbiomed.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 99.Grimm C, Böhl L, Osiewacz HD. 2015. Overexpression of Pa_1_10620 encoding a mitochondrial Podospora anserina protein with homology to superoxide dismutases and ribosomal proteins leads to lifespan extension. Curr Genet 61:73–86. 10.1007/s00294-014-0446-x. [DOI] [PubMed] [Google Scholar]
- 100.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 101.Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, Deng J, Dietrich FS, Fargo DC, Farman ML, Gathman AC, Goldberg J, Guigó R, Hoegger PJ, Hooker JB, Huggins A, James TY, Kamada T, Kilaru S, Kodira C, Kües U, Kupfer D, Kwan HS, Lomsadze A, Li W, Lilly WW, Ma LJ, Mackey AJ, Manning G, Martin F, Muraguchi H, Natvig DO, Palmerini H, Ramesh MA, Rehmeyer CJ, Roe BA, Shenoy N, Stanke M, Ter-Hovhannisyan V, Tunlid A, Velagapudi R, Vision TJ, Zeng Q, Zolan ME, Pukkila PJ. 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA 107:11889–11894. 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sirén J, Välimäki N, Mäkinen V. 2014. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans Comput Biol Bioinform 11:375–388. 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- 103.Anders S, Pyl PT, Huber W. 2015. HTSeq―a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:1–12. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 106.Llanos A, François JM, Parrou JL. 2015. Tracking the best reference genes for RT-qPCR data normalization in filamentous fungi. BMC Genomics 16:71. 10.1186/s12864-015-1224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pathan EK, Ghormade V, Deshpande MV. 2017. Selection of reference genes for quantitative real-time RT-PCR assays in different morphological forms of dimorphic zygomycetous fungus Benjaminiella poitrasii. PLoS One 12:e0179454. 10.1371/journal.pone.017945.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Narasaiah KV, Sashidhar RB, Subramanyam C. 2006. Biochemical analysis of oxidative stress in the production of aflatoxin and its precursor intermediates. Mycopathologia 162:179–189. 10.1007/s11046-006-0052-7. [DOI] [PubMed] [Google Scholar]
- 109.Bourbonnais R, Paice MG. 1990. Oxidation of nonphenolic substrates: an expanded role for laccase in lignin biodegradation. FEBS Lett 267:99–102. 10.1016/0014-5793(90)80298-W. [DOI] [PubMed] [Google Scholar]
- 110.Wei F, Hong Y, Liu J, Yuan J, Fang W, Peng H, Xiao Y. 2010. Gongronella sp induces overproduction of laccase in Panus rudis. J Basic Microbiol 50:98–103. 10.1002/jobm.200900155. [DOI] [PubMed] [Google Scholar]
- 111.Dong Y, Sun Q, Zhang Y, Wang X, Liu P, Xiao Y, Fang Z. 2018. Complete genome of Gongronella sp. w5 provides insight into its relationship with plant. J Biotechnol 286:1–4. 10.1016/j.jbiotec.2018.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Table S1 legend, Tables S2 and S3, Fig. S1 to S4. Download AEM.01760-21-s0001.pdf, PDF file, 0.7 MB (734.1KB, pdf)
Table S1. Download AEM.01760-21-s0002.xlsx, XLSX file, 0.1 MB (130.4KB, xlsx)
Data Availability Statement
Transcriptomic data are publicly available in National Center for Biotechnology Information (NCBI) under BioProject number PRJNA755756. The data sets generated/analyzed during the present study are available and included either within this article or in the supplemental material. The DNA sequence of the 18S rRNA gene for molecular identification of Gongronella sp. w5 was previously deposited in the GenBank database (accession no. EU253966) [110]. In addition, the GenBank accession numbers of the genome sequence of C. cinerea Okayama 7 [101] and Gongronella sp. w5 [111] were AACS00000000 and GCA_001650995.1, respectively.