Abstract
Objective
SARS‐CoV‐2 infection results in severe lung disease in up to 50% of hospitalised patients. The aetiopathogenesis in a subset of such patients, who continue to have progressive pulmonary disease following virus clearance, remains unexplored.
Methods
We investigated the role of NKG2C+/NKG2A− adaptive natural killer (ANK) cells, KLRC2 genotype and cytomegalovirus (CMV) reactivation in 22 such patients.
Results
The median duration of virus positivity was 23 days, and the median duration of hospitalisation was 48 days. The overall survival at 60 days in this group was 50%. Older age and comorbidities impacted survival negatively. CMV viraemia was documented in 11 patients, with a survival of 25% vs 80% in those without viraemia with viral load correlating with mortality. Both NK and T cells were markedly depressed in all patients at day 15. However, only persistently low ANK cells at 30 days along with an inversely high NKG2C−/NKG2A+ inhibitory NK cells significantly correlated with high CMV viraemia and mortality, irrespective of KLRC2 genotype. However, day 30 ANK cells were significantly lower in the KLRC2 deletion group. The release of IFN‐γ and perforin was severely compromised in all patients at day +15, with significant improvement in the survivors at day +30, but not in those with adverse outcome.
Conclusion
Patients with progressive lung disease even after negative SARS‐CoV‐2 status, with persistently reduced and functionally compromised ANK cells, are more likely to have CMV reactivation and an adverse outcome, independent of KLRC2 genotype.
Keywords: adaptive NK cells, CMV, COVID‐19, KLRC2, NKG2C
In a cohort of patients with severe COVID‐19 lung disease who continued to have severe illness even after clearance of virus, lack of recovery of NKG2C+ adaptive NK cells correlated with cytomegalovirus viral load and increased mortality.

Introduction
The recent pandemic caused by SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2) has been marked by morbidity and mortality because of lung disease in up to 50% of hospitalised patients. 1 Disease progression and adverse outcomes have been observed even after viral clearance in some patients. 2 This raises the question about the aetiopathogenesis of progressive disease and late death even after a decrease in virus load.
Natural killer (NK) cells have traditionally been considered as the front line of defence against viral pathogens. However, the canonical NK cells are not antigen‐specific and have a short lifespan. On the contrary, long‐lasting NK cells with virus‐specific memory following exposure to cytomegalovirus (CMV) in both mice and humans have gained attention in the last decade. 3 These cells, now commonly referred to as adaptive NK (ANK) cells, are characterised by clonal expansion, persistence and recall response, along with a strong expression of NKG2C, which is an activating receptor with HLA‐E as its ligand. NKG2A, which provides a strong inhibitory signal to NK cells and binds to the same ligand with a greater affinity, is not expressed on these cells, enabling them with interferon‐gamma (IFN‐γ) release potential and strong cytotoxicity. 4
At the early stages of COVID‐19 (coronavirus disease 2019 caused by SARS‐CoV‐2) pandemic, our group had hypothesised that ANK cells might have a significant impact on the severity and outcome of infection with SARS‐CoV‐2. 5 This was based on our observations on a strong correlation between an anti‐leukaemia and an antiviral effect of ANK cells derived from CMV‐seropositive donors. 6 , 7 , 8 Subsequently, a few studies have studied the NK cell profile in patients with COVID‐19 during the acute phase. 9 Deletion of KLRC2 gene, which encodes for NKG2C, has been described as a risk factor for severe disease. 10 However, a longitudinal study of ANK cells and their impact on late complications in those with prolonged COVID‐19 disease is lacking.
During the recent surge of COVID‐19 in India witnessed between April and June 2021, we prospectively evaluated non‐immunocompromised patients with severe lung disease surviving beyond 15 days from diagnosis, for KLRC2 genotype, ANK cells and CMV viraemia, with respect to their long‐term outcome.
Results
Twenty‐two patients with a median age of 56 years (range 29–78) were evaluated (Table 1). All had severe bilateral pulmonary involvement as per CT scan score 11 and received intensive respiratory support.
Table 1.
Patient characteristics and immunological parameters at day 30
| Alive (N = 11) | Dead (N = 11) | P‐value | |
|---|---|---|---|
| Age, median (range), years | 40 (29–73) | 67 (38–78) | 0.04 |
| Male/female | 8/3 | 5/6 | 0.4 |
| Duration of RT‐PCR positivity, median (range), days | 26 (10–48) | 20 (13–30) | 0.4 |
| Duration of hospitalisation, median (range), days | 49 (10–69) | 42 (32–57) | 0.13 |
| Comorbidities | 3 | 11 | 0.001 |
| KLRC2 gene deletion | 2 | 5 | 0.3 |
| Absolute lymphocyte count (cells µL−1), median (range) | 430 (30–854) | 485 (28–960) | 0.4 |
| Absolute lymphocyte count (% of total leucocytes), median (range) | 9.7 (4–33) | 10.2 (5–27) | 0.4 |
| CD3 (% of lymphocyte), median (range) | 31.3 (5.7–50.7) | 22 (12.3–70) | 0.94 |
| Natural killer cells (% of lymphocyte), median (range) | 9.3 (3–18) | 4 (2–15) | 0.06 |
CMV, cytomegalovirus; NK cells, natural killer cells; RT‐PCR, reverse transcription–polymerase chain reaction.
The median duration of RT‐PCR positivity was 23 days (range 10–48). There was no relationship between mortality and persistence of positive RT‐PCR. The median duration of illness (until discharge or death) was 47 days (range 15–84). Comorbidities were identified in 14 patients in the form of hypertension or diabetes mellitus or both.
Eleven patients died of progressive respiratory failure at a median of 42 days (range 32–57). The overall survival in this cohort at a median of 60 days was 50% (95% CI, 39.3–60.7). The median age was higher in those with mortality (median of 67 vs 40 years, P = 0.04), and all had comorbidities (P = 0.001) (Table 1).
CMV viraemia
All the patients were CMV‐seropositive. Only two patients had low‐level CMV viraemia at 15 days (< 1 × 103 copies mL−1). On day 30, 12 patients showed evidence of CMV viraemia. There was no relationship with the dose or duration of steroid therapy as this was similar for all patients in this study. Survival in patients with CMV viraemia was 25% (95% CI, 12.5–37.5) compared with 80% (95% CI, 67.4–92.6) in those without viraemia (P = 0.01). The median CMV viral load was 1 × 103 copies mL−1 (range 0.6 × 103 to 135 × 103). This was significantly higher for patients who died (median 22.4 × 103 copies mL−1 vs 0 copies mL−1 in survivors, P < 0.01; Figure 1).
Figure 1.
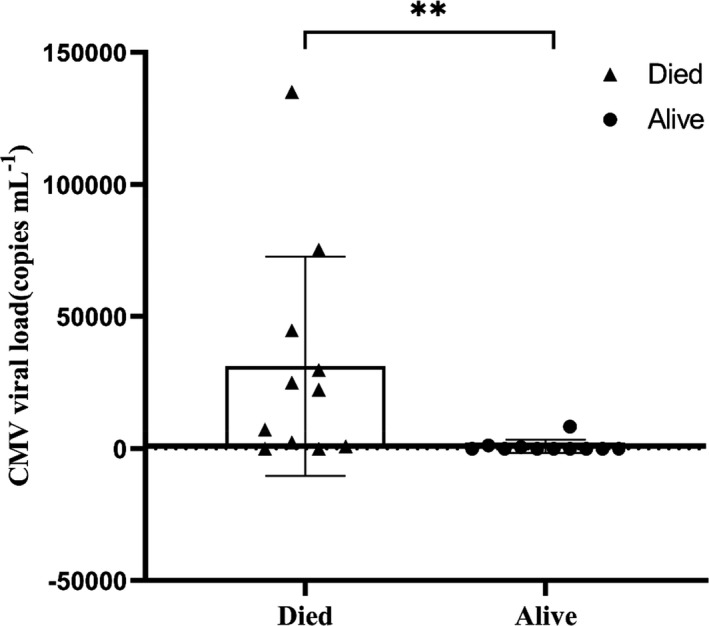
Cytomegalovirus (CMV) viral load in patients with severe SARS‐CoV‐2 lung disease. Scatter plot with bar showing difference in CMV viral load at day 30 in terms of patients alive (n = 11) compared with those who died (n = 11). ‘Died’ and ‘Alive’ are represented as shaded triangles (▲) and circles (●), respectively. **P < 0.01.
Kinetics of adaptive NK cells and cytokine release
Both absolute lymphocytes and CD3+ T cells were reduced in this cohort at 30 days (Table 1). However, there was no impact of these parameters on mortality. NK cells were reduced in the overall cohort but did not correlate with mortality. ANK cells were markedly reduced in all patients (< 5% of total NK cells) at day 15. However, at 30 days, this significantly improved in the survivors (14.1 ± 13%) and remained markedly reduced in those who died (2.2 ± 4.7%, P < 0.001; Figure 2). The inhibitory counterpart of ANK cells at 30 days, defined as NKG2C−/NKG2A+ cells, was significantly higher in those with mortality (83.1 ± 6.8% vs 62.3 ± 20.4%, P < 0.01; Figure 2). Those with lower ANK cells tended to have greater CMV reactivation (P = 0.07).
Figure 2.
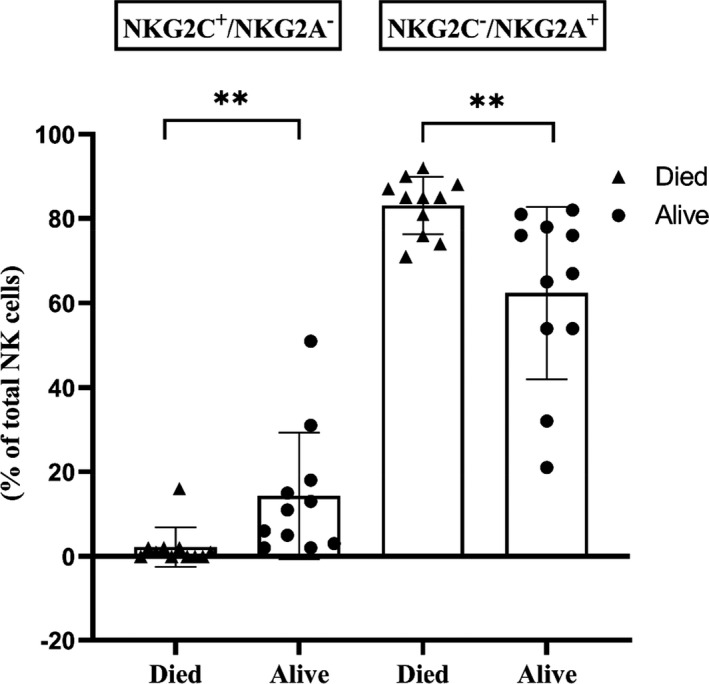
Natural killer (NK) cell subsets in patients with severe SARS‐CoV‐2 lung disease. Immunophenotyping of NK cell population was assessed by flow cytometric analysis. Scatter plot with bar showing NKG2C+/ NKG2A− adaptive NK cells and NKG2C−/NKG2A+ NK cells at day 30 in patients, in terms of patients alive (n = 11) compared with those who died (n = 11). ‘Died’ and ‘Alive’ are represented as shaded triangles (▲) and circles (●), respectively. ***P < 0.001 and **P < 0.01.
Cytokine release was evaluated at both days 15 and 30 on five patients each amongst survivors and non‐survivors. Both IFN‐γ release and perforin release were markedly impaired at day 15 in all 10 patients. However, this improved significantly in the survivors compared with those who died subsequently (Figure 3a–c, P < 0.01).
Figure 3.
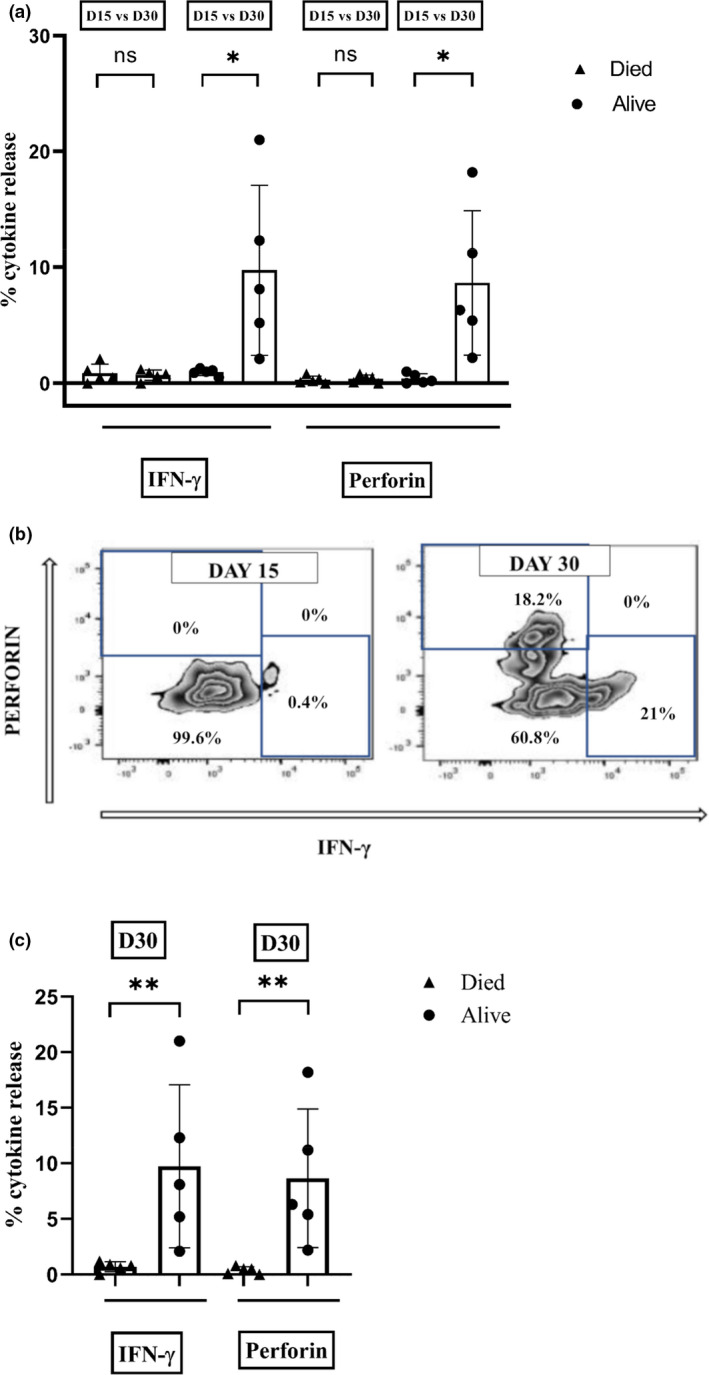
Intracellular cytokine release assessed by flow cytometry in patients with severe SARS‐CoV‐2 lung disease. Flow cytometric staining for intracellular cytokines IFN‐γ and perforin was performed on PBMC isolated from whole‐blood samples of patients at days 15 and 30 following the detection of SARS‐CoV‐2. (a) Scatter plot with bar showing difference in cytokine release (IFN‐γ and perforin) between day 15 and day 30 in those who died (n = 5) and those who were alive (n = 5). (b) Flow cytometric plot showing IFN‐γ and perforin release in a survivor (UPN‐6) at day 15 and day 30. (c) Scatter plot with bar showing difference in IFN‐γ and perforin release at day 30 in patients who were alive (n = 5) compared with those who died (n = 5). ‘Died’ and ‘Alive’ are represented as shaded triangles (▲) and circles (●), respectively. **P < 0.01, *P < 0.05 and ns, not significant.
KLRC2 deletion
Fifteen patients were KLRC2 wt+/wt+, and heterozygous KLRC2 deletion (wt+/del+) was detected in seven patients. KLRC2 del genotype tended to be associated with CMV viraemia (6/7 vs 6/15 without KLRC2 deletion, P = 0.07). ANK cells were significantly lower in the KLRC2 deletion group (2.2 ± 2.2%) than those with wild‐type genotype (10.96 ± 14.3%, P = 0.03; Figure 4). Even though 5 of 7 patients with KLRC2 deletion died, this was not statistically significant (P = 0.3; Table 1).
Figure 4.
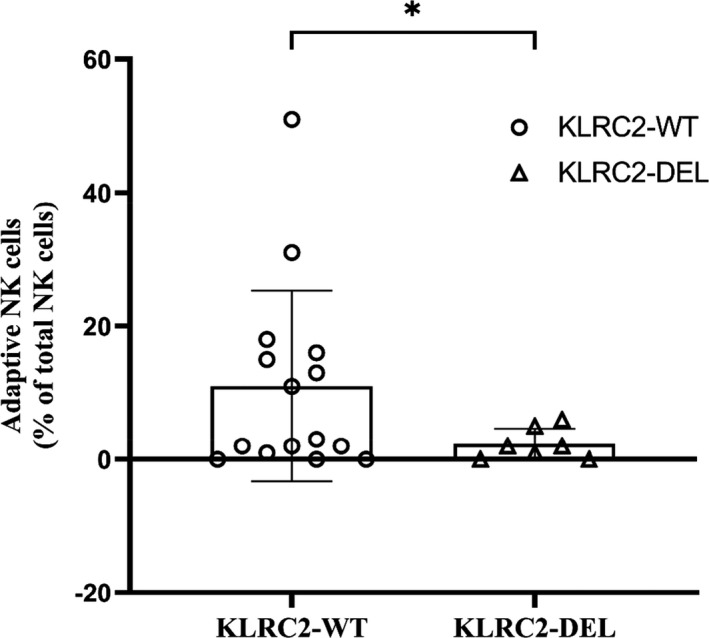
KLRC2 genotype and ANK cells. According to the KLRC2 genotyping, patients were categorised into KLRC2 wild type (WT) and deletion (DEL). Immunophenotyping of the adaptive NK (ANK) cell population was assessed by flow cytometry on PBMC isolated from the whole‐blood samples of patients. The scatter plot with bar shows the difference in ANK cells at day 30 in patients with KLRC2 genotype, WT (n = 15) vs DEL (n = 7). KLRC2 wild type and deletion are represented as unshaded circles (○) and triangles (▵), respectively. *P < 0.05.
Based on recursive partitioning to determine the optimal cut‐point for absolute counts of ANK cells at day 30, survival was 90% (95% CI, 80.5–99.5) in those with ANK > 2.5% vs 16.7% (95% CI, 5.9–27.5) in those with ANK < 2.5% (P < 0.0001; Figure 5).
Figure 5.
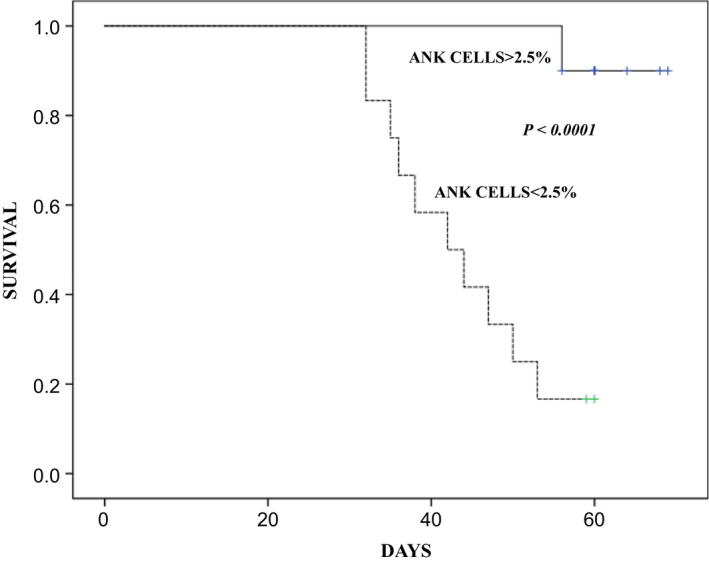
Survival in patients with severe SARS‐CoV‐2 lung disease with respect to adaptive NK cell recovery at day 30. A Kaplan–Meier plot showing survival in patients with adaptive NK (ANK) cells < 2.5% (n = 12) and > 2.5% (n = 10). The continuous line (—) denotes the cohort with ANK cells > 2.5% and the dashed line (‐‐) denotes the cohort with ANK cells < 2.5%. P < 0.0001.
Discussion
The immunology of COVID‐19 has been at the centre of the discourse through the pandemic, and NK cells have often been in the spotlight. 12 Most of these studies have focussed on the NK cell profile in the acute phase of the disease in patients with differing disease severity. In that context, the current study addresses a cohort of patients with severe lung disease, who survived the initial lung pathology beyond 30 days. Despite clearing the virus, over half the patients succumbed to progressive lung disease by 60 days. Our study focussed on the role of ANK cells in the aetiopathogenesis of this phenomenon, and the salutary findings in this small but unique cohort of patients highlight the role of ANK cells in CMV reactivation and subsequent adverse outcome.
While the contributory role of fungal pathogens in the late demise of patients with COVID‐19 has been well established, 13 little data exist on the role of CMV reactivation. 14 , 15 , 16 We found that majority of the patients had CMV reactivation, and this along with high CMV viraemia was associated with higher mortality. In the setting of an allogeneic haematopoietic cell transplantation, high‐dose steroids have been strongly correlated with CMV infection and disease. 17 It is important to note that although all patients received high‐dose steroids, there was no impact of the dose or duration of steroids on CMV reactivation. This brings to fore our endeavour to explore the impact of ANK cells on CMV infection and, more importantly, how and whether SARS‐CoV‐2 impacts the kinetics of ANK cells and vice versa.
Very few studies have longitudinally studied the kinetics of ANK cells in the context of prolonged and severe COVID‐19 disease. The studies so far have focussed almost exclusively on the acute phase of the disease with mixed reports. Several studies have emphasised on the high expression of NKG2A, a strong inhibitory counterpart of NKG2C, in those with severe disease. 18 On the one hand, an in vitro study demonstrated that spike protein‐1 (SP1) of SARS‐CoV‐2 increases the expression of both HLA‐E on lung epithelial cells and NKG2A on NK cells, impeding both NK cell activation and viral clearance. 19 On the other hand, a small study from Sweden suggested marked upregulation of ANK cells in the acute phase in those with severe COVID. 9 However, the implications remain unclear because of the lack of corroborative data on the outcome. Another study reported a correlation between KLR2C deletion genotype and disease severity without any data on NKG2C expression per se. 10
Our data showed a marked suppression of ANK cells in all patients in this cohort with severe disease at 15 days. There was a significant upregulation of NKG2A+ NK cells as well. Cytokine release data showed marked functional impairment as well. Yet, a subset of patients showed both phenotypic and functional recovery of ANK cells at 30 days, irrespective of clearance of SARS‐CoV‐2, which strongly correlated with a lack of CMV infection and a favorable outcome. On the contrary, those with persistently suppressed ANK cells, showing a high NKG2A phenotype, had CMV infection with a high level of viraemia and subsequent mortality. It is worth noting that the majority of the latter group were older with comorbidities. Some researchers have suggested age‐related CMV reactivation contributing to COVID‐19 morbidity. 20 , 21 The only other study, which has systematically examined the incidence of CMV reactivation in patients with severe COVID‐19 lung disease, detected pp65 antigenaemia in 6 of 26 critically ill patients with a higher incidence of both bacterial and fungal infections, and a higher mortality. There was no impact of age, corticosteroid use or other inflammatory parameters on CMV infection in this report. However, any immunological correlation of CMV reactivation in COVID‐19 disease has been lacking to date and our study highlighting the strong correlation between ANK cells and subsequent CMV reactivation might help understand this phenomenon better.
While age or comorbidities could have possibly impacted the ANK cell recovery, a direct role of SARS‐CoV‐2 in the downregulation of ANK cells cannot be discounted, given the in vitro findings demonstrating the same. 19 Either way, suppressed ANK population seemed to strongly correlate with CMV viraemia and the final outcome. In the absence of autopsy studies, it is difficult to ascertain the exact contribution of CMV infection to the lung pathology. It is possible that CMV reactivation in this context might be a surrogate for severe immune suppression, particularly that of ANK cells, rather than being a primary cause for mortality.
One might speculate as to whether the impact of ANK cells extends beyond its antiviral effect. In this context, it is worth noting that inversion of NKG2C and NKG2A expressions following EBV reactivation, in the context of allogeneic HCT, is strongly predisposed to chronic graft‐versus‐host disease (GVHD). 22 The same was demonstrated in the context of acute GVHD as well. 7 , 23 In contrast to conventional NK cells, which are suppressed by regulatory T cells (Tregs), ANK cells are inherently resistant to Treg‐mediated suppression and vice versa. 24 Thus, ANK cells are uniquely placed to augment antiviral cytotoxicity and downregulate exaggerated T‐cell reactivity at the same time.
Because of the small numbers in the cohort, no conclusive evidence could be drawn implicating KLRC2 genotype on the outcome, but the deletion phenotype was possibly contributory to the decremental ANK cells in the affected subset of patients. We have observed that KLRC2 wt+/del+ genotype does not necessarily correlate with compromised ANK cells in healthy adults and vice versa. Thus, the impact of KLRC2 deletion genotype is unlikely to be independent of the kinetics of ANK cells and a heterozygous state is not necessarily associated with impaired NKG2C phenotype, unlike that suggested in a previous study extrapolating KLRC2 deletion status with low NKG2C expression. 10 Our group is currently exploring the variable levels of NKG2C expression in those with KLRC2 heterozygous genotype.
Thus, our study suggests that prolonged severe COVID‐19 might have a detrimental impact on ANK cells, both phenotypically and functionally, which might persist in a subset of patients with higher age and comorbidities, contributing in turn to CMV infection and adverse outcome, even after viral clearance. Large prospective studies and further mechanistic explorations are needed to explore this further.
Methods
Patients with severe COVID‐19‐related lung disease diagnosed between 1 April and 15 May 2021, who survived for more than 15 days from the onset of the illness, were followed up with weekly RT‐PCR for SARS‐CoV‐2. Patients with malignant diseases and chronic renal failure were excluded. They were evaluated on days 15 and 30 of their illness for CMV reactivation and immunological parameters as listed below. The study was approved by the institutional ethics committee.
Treatment for COVID‐19 lung disease
Severe COVID‐19 was defined as per established criteria. 25 The severity of COVID‐19 lung disease was determined by a CT‐based semi‐quantitative score of pulmonary involvement as described previously. 11 All patients received intravenous methylprednisolone (2 mg kg−1) for 3 days, followed by tapering over the next 7 days, remdesivir, low molecular weight heparin and broad‐spectrum antibiotics. All required intensive care admission and invasive or non‐invasive ventilation support.
Real‐time reverse transcription–polymerase chain (RT‐PCR) for detection of SARS‐CoV‐2
All COVID tests were done by the TrueNat real‐time reverse transcription–polymerase chain reaction (RT‐PCR) test. Oropharyngeal or nasopharyngeal swabs were collected using a standard nylon‐flocked swab and inserted into the viral transport medium (VTM) provided by the same company (Molbio Diagnostics Pvt. Ltd., Goa, India). Samples were transported immediately to the laboratory maintaining proper temperature and processed as per the manufacturer’s guidelines (TrueNat Beta CoV chip‐based real‐time PCR test for betacoronavirus; Molbio Diagnostics Pvt. Ltd.). The target sequence for this assay is E gene of the Sarbeco virus and human RNase P (internal positive control). Confirmatory genes were RdRP gene and ORF1A gene.
CMV detection
Whole blood was collected from the patients on 15 and 30 days of SARS‐CoV‐2 positivity, for detection of cytomegalovirus DNA. DNA extraction was carried out from whole blood using a QIAamp® Mini Kit (Qiagen GmbH, Hilden, Germany). Cytomegalovirus (CMV) detection and quantitation were carried out using CMV R‐gene Kit (bioMerieux SA, Marcy‐l'Étoile, France) by q‐PCR on CFX96 Real‐time System (Bio‐Rad, Hercules, CA, USA). CMV viraemia was defined as the detection of greater than 5 × 102 virus copies mL−1.
Immunological monitoring
Peripheral blood mononuclear cells (PBMCs) were isolated from whole‐blood samples of the patients at days 15 and 30 following the detection of SARS‐CoV‐2, by density gradient centrifugations using HiSep™ LSM 1077 media (HiMedia Lab, Mumbai, India). For surface staining, 0.5 × 106 cells were washed with phosphate‐buffered saline (PBS) and stained with the following antibodies, which were used for phenotypic analysis: CD3 (APC‐H7, SK‐7), CD16 (PE‐Cy7, B73.1), CD56 (APC R700, NCAM16.2), CD57 (BV605, NK‐1) and NKG2A (PE‐Cy7, Z199) (BD Biosciences, Franklin Lakes, NJ, USA) and NKG2C (PE, REA205) (Miltenyi Biotec, Bergisch Gladbach, North Rhine‐Westphalia, Germany). Cells were then incubated for 30 min. Viability was assessed with 7‐AAD Viability Dye (Beckman Coulter, Brea, CA, USA). For intracellular staining, cells were stained using monoclonal antibodies for IFN‐γ (4S.B3) and perforin (Alexa 647, DG9) (BD Biosciences) after fixation and permeabilisation with appropriate buffer (BD Biosciences and eBiosciences, San Diego, CA, USA). Flow cytometry was performed using 10‐colour flow cytometry (BD FACSLyric), and the flow cytometric data were analysed using FlowJo software (v10.6.2; FlowJo). ANK cells were defined as CD56dimNKG2C+NKG2A−CD57+ subset of NK cells. The gating strategy is illustrated in Supplementary figure 1.
KLRC2 (NKG2C) genotyping by conventional PCR
KLRC2 gene encodes for NKG2C and is located in chromosome 12p13. DNA was isolated from the peripheral blood using a Qiagen QIAamp DNA Blood Mini Kit. PCR amplification was carried out with forward and reverse primer sequences as previously described. 26 PCR amplification was carried out in a 20 μL mixture, containing 1× PCR master mix, which has premixed Taq polymerase, dNTPs, PCR buffer (Thermo Fisher Scientific, Waltham, MA, USA), 1.65 pmol forward primers and reverse primers for KLRC2 deletion and wild‐type KLRC2 genes (Supplementary table 1), and 100 pg–1 μg genomic DNA. Amplification was performed using a T100 thermal cycler (Bio‐Rad). Cycling temperature profiles were adopted from a previous study 26 with minor modifications.
Briefly, the reaction mixture was subjected to one cycle of denaturation at 95°C for 10 min followed immediately by 29 cycles of 95°C for 20 s, 50°C for 30 s and 72°C for 40 s; and a final extension at 72°C for 10 min before cooling to 4°C. A non‐template control was included in each batch of PCRs. PCR products were identified by running the entire PCR product on a 2% agarose gel for 60 min at 70 Volts. The size of the amplicons was determined by comparison against the migration of a 100‐bp DNA ladder (GeneDireX, Inc., Taoyuan County, Taiwan). Agarose gels were visualised and documented using the Gel Doc XR+ Gel Documentation System (Bio‐Rad). An illustration of the PCR assay is shown in Supplementary figure 2.
The patients were categorised as KLRC2 wild‐type homozygous (wt+/wt+), KLRC2 deletion homozygous (del+/del+) and KLRC2 deletion heterozygous (del+/wt+).
Statistics
Total lymphocyte counts have been described, both in absolute numbers and in % of total leucocyte count in Table 1. Subsequent lymphocyte subsets have been represented as % of the parent population. Binary variables were compared between the groups using the chi‐squared test. The continuous variables were analysed using an independent‐sample t‐test considering Levene’s test for equality of variances and non‐parametric tests (the Mann–Whitney U‐test). Probabilities of survival were estimated using the Kaplan–Meier product‐limit method. A P‐value < 0.05 was considered to be significant. A recursive partitioning analysis was carried out using rpart package (https://cran.r‐project.org/web/pacskages/rpart/index.html) in R (https://cran.r‐project.org/) to generate optimal cut‐off for ANK cells at day 30 in terms of outcome.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Sarita Rani Jaiswal: Conceptualization; Formal analysis; Methodology; Supervision; Writing – original draft; Writing – review & editing. Jaganath Arunachalam: Formal analysis; Investigation; Methodology; Validation. Ashutosh Bhardwaj: Conceptualization. Ashraf Saifullah: Formal analysis. Rohit Lakhchaura: Validation. Mayank Soni: Investigation; Validation. Gitali Bhagawati: Investigation; Validation. Suparno Chakrabarti: Conceptualization; Formal analysis; Investigation; Methodology; Supervision; Writing – original draft; Writing – review & editing.
Supporting information
Supplementary figure 1
Supplementary figure 2
Supplementary table 1
Acknowledgments
We thank all the nursing and technical staff of the department who have assisted in carrying out this study. This study was supported by a grant from Indo‐US Science and Technology Forum (IUSSTF/VN‐COVID/049/2020). The research work and its publication was part funded by a grant from Narayana Health.
References
- 1. Tzotzos SJ, Fischer B, Fischer H, Zeitlinger M. Incidence of ARDS and outcomes in hospitalized patients with COVID‐19: a global literature survey. Crit Care 2020; 24: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borczuk AC. Pulmonary pathology of COVID‐19: a review of autopsy studies. Curr Opin Pulm Med 2021; 27: 184–192. [DOI] [PubMed] [Google Scholar]
- 3. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature 2009; 457: 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA‐E through mostly shared but partly distinct sets of HLA‐E residues. Eur J Immunol 2004; 34: 81–90. [DOI] [PubMed] [Google Scholar]
- 5. Jaiswal SR, Malhotra P, Mitra DK, Chakrabarti S. Focusing on a unique innate memory cell population of natural killer cells in the fight against COVID‐19: harnessing the ubiquity of cytomegalovirus exposure. Mediterr J Hematol Infect Dis 2020; 12: e2020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaiswal SR, Chakrabarti S. Natural killer cell‐based immunotherapy with CTLA4Ig‐primed donor lymphocytes following haploidentical transplantation. Immunotherapy 2019; 11: 1221–1230. [DOI] [PubMed] [Google Scholar]
- 7. Jaiswal SR, Chakraborty S, Lakhchaura R et al. Early and sustained expansion of adaptive natural killer cells following haploidentical transplantation and CTLA4Ig‐primed donor lymphocyte infusions dissociate graft‐versus‐leukemia and graft‐versus‐host effects. Transplant Cell Ther 2021; 27: 144–151. [DOI] [PubMed] [Google Scholar]
- 8. Jaiswal SR, Bhakuni P, Bhagawati G, Chakrabarti A, Chakrabarti S. CTLA4Ig‐based T‐cell costimulation blockade is associated with reduction of adenovirus viremia following post‐transplantation cyclophosphamide‐based haploidentical transplantation. Bone Marrow Transplant 2020; 55: 649–652. [DOI] [PubMed] [Google Scholar]
- 9. Maucourant C, Filipovic I, Ponzetta A et al. Natural killer cell immunotypes related to COVID‐19 disease severity. Sci Immunol 2020; 5: eabd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vietzen H, Zoufaly A, Traugott M, Aberle J, Aberle SW, Puchhammer‐Stockl E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA‐E variants are risk factors for severe COVID‐19. Genet Med 2021; 23: 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Francone M, Iafrate F, Masci GM et al. Chest CT score in COVID‐19 patients: correlation with disease severity and short‐term prognosis. Eur Radiol 2020; 30: 6808–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Market M, Angka L, Martel AB et al. Flattening the COVID‐19 curve with natural killer cell based immunotherapies. Front Immunol 2020; 11: 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh AK, Singh R, Joshi SR, Misra A. Mucormycosis in COVID‐19: a systematic review of cases reported worldwide and in India. Diabetes Metab Syndr 2021; 15: 102146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amiya S, Hirata H, Shiroyama T et al. Fatal cytomegalovirus pneumonia in a critically ill patient with COVID‐19. Respirol Case Rep 2021; 9: e00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niitsu T, Shiroyama T, Hirata H et al. Cytomegalovirus infection in critically ill patients with COVID‐19. J Infect 2021; 83: 496–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchi G, Vianello A, Crisafulli E et al. Cytomegalovirus‐induced gastrointestinal bleeding and pancreatitis complicating severe Covid‐19 pneumonia: a paradigmatic case. Mediterr J Hematol Infect Dis 2020; 12: e2020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suárez‐Lledó M, Martínez‐Cibrián N, Gutiérrez‐García G et al. Deleterious effect of steroids on cytomegalovirus infection rate after allogeneic stem cell transplantation depends on pretransplant cytomegalovirus serostatus of donors and recipients. Biol Blood Marrow Transplant 2018; 24: 2088–2093. [DOI] [PubMed] [Google Scholar]
- 18. Antonioli L, Fornai M, Pellegrini C, Blandizzi C. NKG2A and COVID‐19: another brick in the wall. Cell Mol Immunol 2020; 17: 672–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bortolotti D, Gentili V, Rizzo S, Rotola A, Rizzo R. SARS‐CoV‐2 spike 1 protein controls natural killer cell activation via the HLA‐E/NKG2A pathway. Cells 2020; 9: 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poloni C, Szyf M, Cheishvili D, Tsoukas CM. Are the healthy vulnerable? Cytomegalovirus seropositivity in healthy adults is associated with accelerated epigenetic age and immune‐dysregulation. J Infect Dis 2021: jiab365. 10.1093/infdis/jiab365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soderberg‐Naucler C. Does reactivation of cytomegalovirus contribute to severe COVID‐19 disease? Immun Ageing 2021; 18: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaiswal SR, Bhakuni P, Bhagwati G, Aiyar HM, Chakrabarti A, Chakrabarti S. Alterations in NKG2A and NKG2C subsets of natural killer cells following Epstein‐Barr virus reactivation in CTLA4Ig‐based haploidentical transplantation is associated with increased chronic graft‐versus‐host disease. Transplantation 2020; 104: e23–e30. [DOI] [PubMed] [Google Scholar]
- 23. Kordelas L, Steckel NK, Horn PA, Beelen DW, Rebmann V. The activating NKG2C receptor is significantly reduced in NK cells after allogeneic stem cell transplantation in patients with severe graft‐versus‐host disease. Int J Mol Sci 2016; 17: 1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarhan D, Miller JS. Assessing canonical and adaptive natural killer cell function in suppression assays in vitro . Methods Mol Biol 2019; 1913: 153–166. [DOI] [PubMed] [Google Scholar]
- 25. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323: 1239–1242. [DOI] [PubMed] [Google Scholar]
- 26. Miyashita R, Tsuchiya N, Hikami K et al. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int Immunol 2004; 16: 163–168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
Supplementary figure 2
Supplementary table 1


