Abstract
目的
分析miR-3682-3p在HCC中的表达及其与临床参数和预后的相关性。
方法
生物信息学分析miR-3682-3p在HCC中的表达以及与生存相关性; 实时荧光定量PCR和原位杂交分别检测miR-3682-3p(miR-3682)在18对HCC与癌旁新鲜肝组织以及90对石蜡包埋HCC及其癌旁组织中的表达差异。统计分析miR-3682-3p在HCC中的表达与患者临床参数和预后之间的关系。利用多因素回归分析探讨miR-3682-3p表达作为肝细胞癌预后独立因素的可能性。
结果
生物信息分析显示,miR- 3682-3p在HCC组织中高表达,且与HCC患者综合生存时间有统计学关联(χ2=8.793,P < 0.001)。实时荧光定量PCR分析18组配对的HCC和癌旁肝组织显示,miR-3682-3p在癌组织中表达明显上调(t=3.073,P=0.007)。原位杂交分析90组配对的HCC和癌旁组织中miR-3682-3p的表达,显示其在癌与癌旁组织的细胞浆中表达,并且在癌组织中表达上调(t=2.659,P=0.009)。miR-3682-3p表达的高低在美国癌症联合委员会(AJCC)第八版分期(χ2=4.272,P=0.039)、HBV表面抗原状态(χ2=5.143,P= 0.023)、复发(χ2=4.593,P=0.032)、肿瘤大小(χ2=4.580,P=0.032)和Edmondson-Steiner分级(χ2=4.068,P=0.044)方面差异存在统计学意义; Kaplan-Meier分析显示,miR-3682-3p表达越高,患者总生存时间(Log rank χ2=4.169,P=0.041)和无病生存时间(Log rank χ2=4.078,P=0.043)越短。多变量分析显示,miR-3682-3p表达是评估HCC患者预后独立因子。
结论
miR-3682-3p在HCC组织中表达上调,是促进HCC发病和预后不良的重要因子。
Keywords: 肝细胞癌, miR-3682-3p, 预后
Abstract
Objective
To investigate the expression of miR-3682-3p in hepatocellular carcinoma (HCC) and its correlation with clinical parameters and prognosis of HCC.
Methods
We conducted a bioinformatics analysis of the expression of miR-3682-3p in HCC and its correlation with the patients' survival, and examined its expression in 18 pairs of fresh and 90 pairs of paraffin-embedded HCC and adjacent tissues using real-time fluorescence quantitative PCR and in situ hybridization, respectively. The correlation of miR-3682-3p expression in HCC with the clinical parameters and prognosis of the patients was analyzed. Multivariate regression analysis was used to explore the possibility of miR-3682-3p expression as an independent prognostic factor of HCC.
Results
Bioinformatics analysis showed that miR-3682-3p was highly expressed in HCC and significantly correlated with the survival time of HCC patients (χ2=8.793, P < 0.001). The expression of miR-3682-3p was significantly up-regulated in fresh HCC tissues as compared with the adjacent liver tissues (t=3.073, P=0.007). In paraffin-embedded samples, in situ hybridization revealed positive miR-3682-3p expression in the cytoplasm of HCC and adjacent tissues, and its expression was signifcantly up-regulated in HCC tissues (t=2.659, P=0.009). The expression level of miR-3682-3p was significantly correlated with American Joint Commission on Cancer (AJCC; 8th edition) stage (χ2=4.272, P= 0.039), HBV surface antigen status (χ2=5.143, P=0.023), recurrence (χ2=4.593, P=0.032), tumor size (χ2=4.580, P=0.032) and Edmondson Steiner grade (χ2=4.068, P=0.044). Kaplan-Meier analysis showed that a higher expression of miR-3682-3p was associated with a shorter overall survival time (χ2=4.169, P=0.041) and disease-free survival time (χ2=4.078, P=0.043) of the patients. Multivariate analysis suggested that miR-3682-3p expression was an independent predictor of the prognosis of HCC patients.
Conclusion
MiR-3682-3p is up-regulated in HCC to serve as a significant factor that contributes to the occurrence and a poor prognosis of HCC.
Keywords: hepatocellular carcinoma, miR-3682-3p, prognostic factor
肝细胞癌(HCC)是一种异质性癌症,已有多种已知病因,如乙型肝炎病毒(HBV)或丙型肝炎病毒感染或黄曲霉毒素暴露,这些因素单独或联合长期作用于肝脏,可导致基因表达的慢性失衡[1-3],并最终促进了肝癌的发生和发展。MicroRNA(miRNA)参与包括HCC在内的多种肿瘤的发病机制[4-13]。MiR-3682-3p是一个与肿瘤发病相关的miRNA,研究发现miR-3682-3p在食管癌中高表达[14]。在HCC中,外泌体中miR-3682-3p通过改变Ras-MEK1/2-ERK1/2信号转导途径靶向血管生成素1(Ang-1),从而抑制血管生成,外泌体中miR- 3682-3p在HCC发病中可能发挥了候选肿瘤抑制因子[15]作用。然而,在另一项肝癌研究中,实时定量PCR的检测结果却显示,miR-3682-3p在HCC中高表达,其表达增加促进了HCC发病和不良预后[16],提示其在HCC中发挥了肿瘤促进作用。这两项研究结果相互矛盾,且相对较短的随访时间可能导致结论存在偏差[16]。因此需要对miR-3682-3p在HCC中的作用进行进一步验证。本研究对miR-3286-3p在HCC中的表达及其与临床参数和预后的关系进行了检测和分析,以明确miR-3286- 3p在HCC发病过程中的作用。
1. 资料和方法
1.1. 生物信息学分析
使用UALCAN(TCGA数据)网站(<a href="http://ualcan.path.uab.edu/" target="_blank">http://ualcan.path.uab.edu/</a>)确定miR-3682(miR-3682-3p)的表达及其与肝癌患者预后的相关性。
1.2. 临床样本
18配对肝细胞癌组织和癌旁组织来自贵州医科大学附属医院(液氮新鲜保存),其中男性12例,女性6例,发病年龄27~82岁,中位年龄53.5岁。90对石蜡包埋的HCC标本和其癌旁组织芯片[上海芯超生物技术有限公司(中国上海)]HLivH180Su15,病理类型均为肝细胞癌(<a href="http://www.superchip.com.cn/biology/tissue.html" target="_blank">http://www.superchip.com.cn/biology/tissue.html</a>),样本收集于2007~2008年,随访至2016年。所有病例均为手术切除,术前未经任何放化疗和其他治疗,并分别获得贵州医科大学附属医院和上海芯超生物技术有限公司伦理委员会的批准(2020025;SHY-JS-1901001)。
1.3. 原位杂交(ISH)
应用ISH对石蜡包埋的90对肝癌和癌旁组织芯片标本进行检测[17]。切片在二甲苯中脱蜡,并在不同乙醇浓度下进行梯度脱水处理。用蛋白酶K处理37 ℃、30 min,冲洗、固定切片,然后预混合2 h。将样品与BersinBio(中国广州)设计和合成的miRCURY miR-3682-3p地高辛标记探针杂交。冲洗载玻片,在室温下与与辣根过氧化物酶结合的抗地高辛抗体的Fab片段孵育1 h。添加3,3'-二氨基联苯胺(中国福州美新生物科技有限公司)观察信号,并按照文献[4]方法由两位经验丰富的病理医师进行双盲阅片,分别对染色强度和染色面积进行评分:阳性着色面积 < 5%为0分,5%~25%为1分,25%~ 50%为2分,51%~75%为3分,75%~100%为4分;染色强度:无色为0分,浅黄色为1分,棕黄色为2分,棕褐色为3分。两者计分的乘积为阳性等级:6分以下为低表达,6分及6分以上为高表达。
1.4. 统计分析
所有数据均使用SPSS 20.0进行分析。使用配对t检验分析基因差异表达;通过卡方检验评估基因表达与临床病理参数之间的相关性。Kaplan-Meier生存曲线用于分析基因表达与患者总生存率之间的关系。使用Cox比例风险回归模型进行单变量和多变量生存分析。P < 0.05为差异具有统计学意义。
2. 结果
2.1. 生物信息学分析miR-3682(miR-3682-3p)的表达与预后相关性
基于TCGA数据,使用UALCAN网站上的算法进行的分析显示,与邻近组织相比,miR-3682在HCC中明显高表达(t=4.149,P < 0.001,图 1A)。生存分析显示,miR-3682表达越高,患者的综合生存时间越短(χ2= 8.793,P < 0.001,图 1B)。
1.
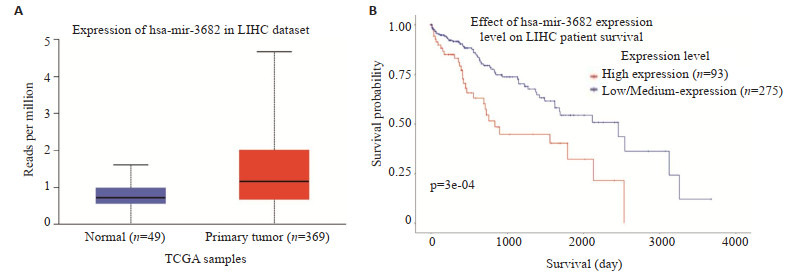
生物信息学分析miR-3682-3p表达水平与患者的预后的关系
Bioinformatics analysis of miR-3682-3p expression levels in relation to the patients' prognosis. A: MiR-3682 expression in HCC tissues and adjacent tissues based on data from TCGA database (P < 0.001). B: Relationship between miR-3682 expression and the patients' prognosis based on data from TCGA database.
2.2. RT-PCR检测miR-3682-3p在肝癌中的表达
为验证生物信息学结果,应用RT-PCR检测18对癌组织与它们匹配的癌旁组织中miR-3682-3p的表达。配对t检验显示,与18个非癌对照组织相比,miR-3682- 3p在18个肝癌组织中总体表达水平升高(t=3.073,P= 0.007,图 2)。
2.
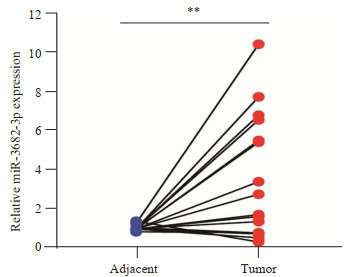
实时荧光定量PCR检测miR-3682-3p在18对配对新鲜肝癌组织和它们邻近癌旁组织中的表达水平(蓝点和红点分别代表癌旁组织和癌组织)
MiR-3682-3p mRNA expression level in 18 pairs of fresh HCC tissues (red dots) and adjacent tissues (blue dots) detected by real-time fluorescence quantitative PCR (**P < 0.01).
2.3. miR-3682-3p的原位杂交及亚细胞定位
原位杂交技术检测组织芯片中90例HCC和癌旁组织中miR-3682-3p的表达,并分析其亚细胞定位,结果显示,miR-3682-3p定位于HCC和癌旁组织的细胞浆中(图 3A)。与癌旁组织相比,miR-3682-3p在HCC中上调(t=2.659,P=0.009,图 3B)。
3.
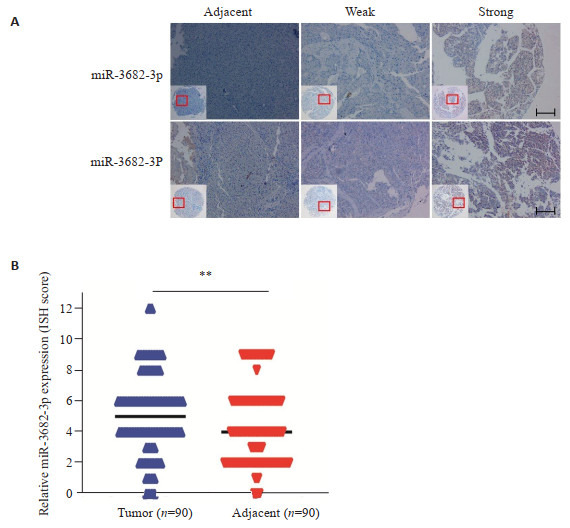
原位杂交法检测肝癌和癌旁组织中miR- 3682-3p的表达
Expression of miR-3682-3p detected by in situ hybridization in HCC and adjacent tissues. A: In situ hybridization of miR-3682-3p in HCC tissues and adjacent tissues (Scale bar: 50 μm). B: MiR-3682-3p is upregulated in HCC tissues compared with the adjacent tissues based on tissue microarrays (TMA) cell staining score (**P < 0.01).
2.4. miR-3682-3p的表达与肝癌临床进展的相关性
miR-3682-3p表达的高低在美国癌症联合委员会(AJCC)第八版分期(χ2=4.272,P=0.039)、HBV表面抗原状态(χ2=5.143,P=0.023)、复发(χ2=4.593,P=0.032)、肿瘤大小(χ2=4.580,P=0.032)和Edmondson-Steiner分级(χ2=4.068,P=0.044)方面差异存在统计学意义,但在年龄、性别、肿瘤数量、甲胎蛋白、总胆红素、丙氨酸转氨酶和谷氨酰转肽酶方面差异无统计学意义(P>0.05,表 1)。
1.
MiR-3682-3p表达与HCC患者临床参数之间的关系
Correlations between miR-3682-3p expression and clinicopathological features of HCC patients
| Characteristics | n | mir-3682-3p Expression | χ2 | P | |
| Low | High | ||||
| Age (year) | |||||
| ≤50 | 68 | 35 (51.47%) | 33 (48.53%) | 0.388 | 0.533 |
| > 50 | 22 | 13 (59.09%) | 9 (40.91%) | ||
| Gender | |||||
| Male | 80 | 42 (52.50%) | 38 (47.50%) | 0.013 | 0.911 |
| Female | 10 | 6 (60.00%) | 4 (40.00%) | ||
| AJCC Staging | |||||
| I | 67 | 40 (59.70%) | 27 (40.30%) | 4.272 | 0.039 |
| II-III | 23 | 8 (34.78%) | 15 (65.22%) | ||
| HBsAg | |||||
| Negative | 15 | 12 (80.00%) | 3 (20.00%) | 5.143 | 0.023 |
| Positive | 75 | 36 (48.00%) | 39 (52.00%) | ||
| Recurrence | |||||
| No | 43 | 28 (65.11%) | 15 (34.89%) | 4.593 | 0.032 |
| Yes | 47 | 20 (42.55%) | 27 (57.45%) | ||
| AFP (ng/L) | |||||
| > 400 | 33 | 17 (51.52%) | 16 (48.48%) | 0.069 | 0.792 |
| ≤400 | 57 | 31 (54.39%) | 26 (45.61%) | ||
| Total bilirubin (µmol/L) | |||||
| > 20 | 15 | 8 (53.33%) | 7 (46.67%) | 0 | 1 |
| ≤20 | 75 | 40 (53.33%) | 35 (46.67%) | ||
| ALT (U/L) | |||||
| > 45 | 32 | 14 (43.75%) | 18 (56.25%) | 1.832 | 0.176 |
| ≤45 | 58 | 34 (58.62%) | 24 (41.38%) | ||
| GGT (U/L) | |||||
| > 40 | 59 | 29 (49.15%) | 30 (50.85%) | 1.203 | 0.273 |
| ≤40 | 31 | 19 (61.29%) | 12 (38.71%) | ||
| Edmondson-Steiner | |||||
| I-II | 57 | 35 (61.40%) | 22 (38.60%) | 4.068 | 0.044 |
| III-IV | 33 | 13 (39.39%) | 20 (60.61%) | ||
| Tumor Number | |||||
| Single | 80 | 43 (53.75%) | 37 (46.25%) | 0 | 1 |
| Multiple | 10 | 5 (50.00%) | 5 (50.00%) | ||
| Tumor size (cm) | |||||
| > 5 | 19 | 6 (31.60%) | 13 (68.40%) | 4.58 | 0.032 |
| ≤5 | 71 | 42 (59.20%) | 29 (40.80%) | ||
2.5. miR-3682-3p表达与HCC患者不良预后的关系
在90例有预后信息的肝癌患者中,miR-3682-3p表达越高,患者总生存时间越短(χ2=4.169,P=0.041,图 4A),无病生存时间越短(χ2=4.078,P=0.043,图 4B)。单变量分析显示,患者生存率也与miR-3682-3p表达(HR=0.242[95%CI0.111-0.525],P < 0.001)、EdmondsonSteiner分级(HR=7.860[95%CI 3.479-3.497],P < 0.001)、GGT水平(HR=0.386[95%CI 0.159-0.937],P=0.035)、肿瘤大小(HR=0.169[95%CI 0.084-0.342],P < 0.001)和AJCC分期(HR=3.088[95%CI 1.535-6.212],P=0.002)有关。为了确定miR-3682-3p是否是HCC的独立预后因素,对HCC患者中miR-3682-3p的表达进行了多变量分析,并根据Edmondson-Steiner分级、肿瘤大小、GTT水平和AJCC分期进行调整。结果显示,miR-3682- 3p表达水平是影响肝癌预后的独立预后因素[HR=0.368(95%CI 0.161-0.842),P=0.018,表 2]。
4.
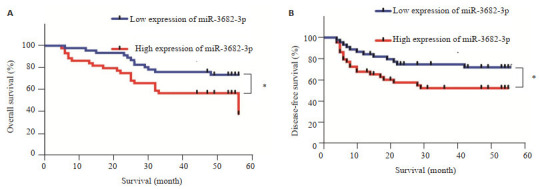
Kaplan-Meier生存曲线分析miR-3682-3p表达与肝癌患者总生存率和无病生存率的关系
Kaplan-Meier survival analysis of the correlation of miR-3682-3p expression with the overall survival and disease-free survival of HCC patients. A: Overall survival rate was markedly lower in patients with high miR-3682-3p expression level than in those with low miR-3682-3p expression (P=0.041, log-rank test). B: Disease-free survival rate was markedly lower in patients with high miR-3682-3p expression level than in those with low miR-3682-3p expression level (P=0.043, log-rank test).
2.
肝癌患者临床病理变量的单因素和多因素生存分析
Univariate and multivariate survival analysis of clinicopathological variables of HCC patients
| Characteristics | Overall survival | ||||||
| Univariate analysis | Multivariate analysis | ||||||
| HR | 95%CI | P | HR | 95%CI | P | ||
| miR-3682-3p expression | |||||||
| Low vs. High | 0.242 | 0.111-0.525 | < 0.001 | 0.368 | 0.161-0.842 | 0.018 | |
| Age (year) | |||||||
| ≤50 vs. > 50 | 0.771 | 0.356-1.667 | 0.508 | ||||
| Gender | |||||||
| Male vs. Female | 0.52 | 0.124-2.176 | 0.371 | ||||
| AJCC Staging | |||||||
| I vs. II-III | 3.088 | 1.535-6.212 | 0.002 | 1.678 | 0.803-3.507 | 0.168 | |
| HBsAg | |||||||
| Negative vs. Positive | 0.642 | 0.224-1.845 | 0.411 | ||||
| Recurrence | |||||||
| No vs. Yes | 0.054 | 0.264-1.086 | 0.083 | ||||
| AFP (ng/mL) | |||||||
| ≤400 vs. > 400 | 0.888 | 0.434-1.818 | 0.746 | ||||
| Total bilirubin (μmol/L) | |||||||
| ≤20 vs. > 20 | 1.113 | 0.428-2.893 | 0.826 | ||||
| ALT (U/L) | |||||||
| ≤45 vs. > 45 | 0.935 | 0.457-1.912 | 0.853 | ||||
| GGT (U/L) | |||||||
| ≤40 vs. > 40 | 0.386 | 0.159-0.937 | 0.035 | 0.489 | 0.190-1.259 | 0.138 | |
| Edmondson-Steiner grade | |||||||
| I-II vs. III-IV | 7.86 | 3.497-3.497 | < 0.001 | 5.269 | 1.985-13.990 | 0.001 | |
| Tumor number | |||||||
| Single vs. Multiple | 2.084 | 0.852-5.095 | 0.108 | ||||
| Tumor size (cm) | |||||||
| ≤5 vs. > 5 | 0.169 | 0.084-0.342 | < 0.001 | 0.368 | 0.161-0.842 | 0.633 | |
3. 讨论
许多miRNAs参与了HCC的发病机制[4-10],包括最近发现的miR-3682-3p。目前,关于miR-3682-3p在肿瘤中作用的研究仅限于食管癌和HCC,研究结论也存在矛盾[14-16],提示miR-3682-3p在肿瘤中可能发挥了相当复杂的作用。有研究对OncoMir数据库的数据进行分析,发现miR-3682-3p在HCC中表达显著增加,qPCR检测也获得了一致的结果[16],并且miR-3682-3p高表达与临床进展呈明显正相关,而与不良预后呈明显负相关,表明miR-3682-3p作为候选癌基因促进HCC的发病过程[16]。而有研究显示,HCC外泌体中的miR- 3682-3p能抑制Ras-MEK1/2-ERK1/2信号,降低Ang-1的生成,从而抑制血管生成,提示外泌体中miR-3682- 3p在HCC发病发挥了候选抑癌基因作用[15],该结论与Yao等[16]结果存在明显不同,提示miR-3682-3p在HCC发病中的复杂性,有必要进一步明确miR-3682-3p在HCC中作用。因此,本研究应用多种方法检测了miR- 3682-3p在HCC和肝组织样本中miR-3682-3p的表达,并重新分析了miR-3682-3p在HCC表达与临床参数及预后的关系。
本研究对TCGA数据进行分析的结果显示,miR- 3682-3p在肝癌组织中的表达明显升高,实时荧光定量检测也显示了同样的结果,这与部分报道相一致[16],支持了miR-3682-3p在OncoMir数据库中结果的可靠性。因既往研究的方法仅限于实时荧光定量PCR,本研究进一步应用原位杂交方法对miR-3682-3p的表达情况进行了验证,结果同样显示为miR-3682-3p在HCC中的表达明显升高。本研究明确回答了既往存在争议的问题,支持miR-3682-3p在HCC中呈高表达,为进一步评估miR-3682-3p表达在HCC中的意义提供了前提和依据。
HBV感染是HCC发病机制的关键因素[18-20],本研究发现miR-3682-3p高表达与HBV感染有关,HBV感染可能参与了对miR-3682-3p的诱导表达。此外,miR- 3682-3p的表达与AJCC分期、肿瘤复发、肿瘤大小以及Edmondson-Steiner分级等多个因素存在统计学关联,而肿瘤复发和生长等被认为与肿瘤干性增加有关[21-23],提示miR-3682-3p的表达可能促进对HCC肿瘤干性信号的调节,从而诱导HCC复发、细胞增殖和恶性进展。这些数据进一步支持miR-3682-3p作为刺激HCC发病的潜在癌基因的结论。
前期有研究报告了miR-3682-3p表达与肝癌患者3年生存时间的相关性[16]。本研究进行长期随访发现,miR-3682-3p表达越高,肝癌患者总生存时间越短。并且,高表达miR-3682-3p的HCC患者的无病生存时间明显低于低表达患者,表明miR-3682-3p高表达是促进HCC患者预后不良因素。本研究多变量分析显示,经调整了AJCC分期、肿瘤大小、Edmondson-Steiner分级和GGT水平,miR-3682-3p表达增加是肝癌患者预后不良的重要独立预测因子。
综上所述,本研究通过多种检测方法,证明了miR- 3682-3p在HCC组织中表达上调,是促进HCC发病和预后不良的重要因子,明确回答了这个既往存在争议的问题,为研究miR-3682-3p在肿瘤中的作用提供了新的证据。然而,关于miR-3682-3p在HCC中的作用机制仍未阐明,今后需要进一步开展相关机制方面的研究,为miR-3682-3p真正应用于肿瘤的临床治疗提供依据和方向。
Biography
刘绍华,副主任医师,E-mail: jxpxlsh20@163.com
Funding Statement
江西省科技厅自然科学基金(20192BAB205060); 东莞市社会科学发展计划(20195071524201)
Contributor Information
刘 绍华 (Shaohua LIU), Email: jxpxlsh20@163.com.
温 莹浩 (Yinghao WEN), Email: jxpxwyh@163.com.
韩 思源 (Siyuan HAN), Email: Hansiyuan2002@163.com.
References
- 1.Xie YH. Hepatitis B virus-associated hepatocellular carcinoma. http://link.springer.com/chapter/10.1007/978-981-10-5765-6_2. Adv Exp Med Biol. 2017;1018:11–21. doi: 10.1007/978-981-10-5765-6_2. [Xie YH. Hepatitis B virus-associated hepatocellular carcinoma[J]. Adv Exp Med Biol, 2017, 1018: 11-21.] [DOI] [PubMed] [Google Scholar]
- 2.Plissonnier ML, Herzog K, Levrero M, et al. Non-coding RNAs and hepatitis C virus-induced hepatocellular carcinoma. Viruses. 2018;10(11):591–9. doi: 10.3390/v10110591. [Plissonnier ML, Herzog K, Levrero M, et al. Non-coding RNAs and hepatitis C virus-induced hepatocellular carcinoma[J]. Viruses, 2018, 10(11): 591-9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kew MC. Aflatoxins as a cause of hepatocellular carcinoma. http://193.231.29.96/jgld/2013/3/13.pdf. J Gastrointestin Liver Dis. 2013;22(3):305–10. [Kew MC. Aflatoxins as a cause of hepatocellular carcinoma[J]. J Gastrointestin Liver Dis, 2013, 22(3): 305-10.] [PubMed] [Google Scholar]
- 4.Lin X, Zuo S, Luo R, et al. HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics. 2019;9(25):7583–98. doi: 10.7150/thno.37717. [Lin X, Zuo S, Luo R, et al. HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma[J]. Theranostics, 2019, 9(25): 7583-98.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma. Clin Chim Acta. 2020;500:10–9. doi: 10.1016/j.cca.2019.09.013. [Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma[J]. Clin Chim Acta, 2020, 500: 10-9.] [DOI] [PubMed] [Google Scholar]
- 6.Zheng YL, Li L, Jia YX, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9(3):796–810. doi: 10.7150/thno.28992. [Zheng YL, Li L, Jia YX, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway[J]. Theranostics, 2019, 9 (3): 796-810.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Z, Xia S, Liang Y, et al. Theranostics LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a transcription. Theranostics. 2020;10(19):8834–50. doi: 10.7150/thno.45158. [Lin Z, Xia S, Liang Y, et al. Theranostics LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a transcription[J]. Theranostics, 2020, 10(19): 8834-50.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Y, Guo J, Xiao P, et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/ MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310–22. doi: 10.1016/j.canlet.2019.11.001. [Ye Y, Guo J, Xiao P, et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/ MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma[J]. Cancer Lett, 2020, 469: 310-22.] [DOI] [PubMed] [Google Scholar]
- 9.Huang JL, Fu YP, Gan W, et al. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246- RORα-Wnt/β-Catenin axis. Cancer Lett. 2020;476:140–51. doi: 10.1016/j.canlet.2020.02.012. [Huang JL, Fu YP, Gan W, et al. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246- RORα-Wnt/β-Catenin axis[J]. Cancer Lett, 2020, 476: 140-51.] [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Feng J, Sun M, et al. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158–70. doi: 10.1016/j.canlet.2019.04.008. [Liu Y, Feng J, Sun M, et al. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma[J]. Cancer Lett, 2019, 454: 158-70.] [DOI] [PubMed] [Google Scholar]
- 11.Fu Q, Song X, Liu Z, et al. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/ras/c-myc feedback loop modulated by HDGF/DDX5/β-catenin complex in lung adenocarcinoma. Clin Cancer Res. 2017;23(20):6336–50. doi: 10.1158/1078-0432.CCR-16-2813. [Fu Q, Song X, Liu Z, et al. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/ras/c-myc feedback loop modulated by HDGF/DDX5/β-catenin complex in lung adenocarcinoma[J]. Clin Cancer Res, 2017, 23(20): 6336-50.] [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Liu X, Lin X, et al. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9. Signal Transduct Target Ther. 2019;4:48–56. doi: 10.1038/s41392-019-0084-3. [Li Y, Liu X, Lin X, et al. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9[J]. Signal Transduct Target Ther, 2019, 4: 48-56.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Jiang Q, Liu X, et al. Cinobufotalin powerfully reversed EBVmiR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain ⅡA/glycogen synthase 3β/β-catenin signaling pathway. EBioMedicine. 2019;48:386–404. doi: 10.1016/j.ebiom.2019.08.040. [Liu Y, Jiang Q, Liu X, et al. Cinobufotalin powerfully reversed EBVmiR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain ⅡA/glycogen synthase 3β/β-catenin signaling pathway[J]. EBioMedicine, 2019, 48: 386-404.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Xu L, Wang X, et al. A novel prognostic mRNA/miRNA signature for esophageal cancer and its immune landscape in cancer progression. Mol Oncol. 2021;15(4):1088–109. doi: 10.1002/1878-0261.12902. [Zhao Y, Xu L, Wang X, et al. A novel prognostic mRNA/miRNA signature for esophageal cancer and its immune landscape in cancer progression[J]. Mol Oncol, 2021, 15(4): 1088-109.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong SS, Dong DD, Yang ZF, et al. Exosomal miR-3682-3p suppresses angiogenesis by targeting ANGPT1 via the RAS-MEK1/ 2-ERK1/2 pathway in hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:633358–67. doi: 10.3389/fcell.2021.633358. [Dong SS, Dong DD, Yang ZF, et al. Exosomal miR-3682-3p suppresses angiogenesis by targeting ANGPT1 via the RAS-MEK1/ 2-ERK1/2 pathway in hepatocellular carcinoma[J]. Front Cell Dev Biol, 2021, 9: 633358-67.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao BW, Niu YS, Li YZ, et al. High-matrix-stiffness induces promotion of hepatocellular carcinoma proliferation and suppression of apoptosis via miR-3682-3p-PHLDA1-FAS pathway. J Cancer. 2020;11(21):6188–203. doi: 10.7150/jca.45998. [Yao BW, Niu YS, Li YZ, et al. High-matrix-stiffness induces promotion of hepatocellular carcinoma proliferation and suppression of apoptosis via miR-3682-3p-PHLDA1-FAS pathway[J]. J Cancer, 2020, 11(21): 6188-203.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang ZB, Chen WF, Xu Y, et al. MiR-4721, induced by EBV-miRBART22, targets GSK3β to enhance the tumorigenic capacity of npc through the WNT/β-catenin pathway. Mol Ther Nucleic Acids. 2020;22:557–71. doi: 10.1016/j.omtn.2020.09.021. [Tang ZB, Chen WF, Xu Y, et al. MiR-4721, induced by EBV-miRBART22, targets GSK3β to enhance the tumorigenic capacity of npc through the WNT/β-catenin pathway[J]. Mol Ther Nucleic Acids, 2020, 22: 557-71.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Li AM, Li YH, et al. Silencing MYH9 blocks HBx-induced GSK3β ubiquitination and degradation to inhibit tumor stemness in hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5(1):13–21. doi: 10.1038/s41392-020-0111-4. [Lin X, Li AM, Li YH, et al. Silencing MYH9 blocks HBx-induced GSK3β ubiquitination and degradation to inhibit tumor stemness in hepatocellular carcinoma[J]. Signal Transduct Target Ther, 2020, 5 (1): 13-21.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Gao Y, He Y, et al. HBV induced hepatocellular carcinoma and related potential immunotherapy. Pharmacol Res. 2020;159:104992–9. doi: 10.1016/j.phrs.2020.104992. [Jia L, Gao Y, He Y, et al. HBV induced hepatocellular carcinoma and related potential immunotherapy[J]. Pharmacol Res, 2020, 159: 104992-9.] [DOI] [PubMed] [Google Scholar]
- 20.Yao L, Zhou Y, Sui Z, et al. HBV-encoded miR-2 functions as an oncogene by downregulating TRIM35 but upregulating RAN in liver cancer cells. EBioMedicine. 2019;48:117–29. doi: 10.1016/j.ebiom.2019.09.012. [Yao L, Zhou Y, Sui Z, et al. HBV-encoded miR-2 functions as an oncogene by downregulating TRIM35 but upregulating RAN in liver cancer cells[J]. EBioMedicine, 2019, 48: 117-29.] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–95. doi: 10.1002/jcp.27740. [Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy[J]. J Cell Physiol, 2019, 234(6): 8381-95.] [DOI] [PubMed] [Google Scholar]
- 22.Liu YC, Yeh CT, Lin KH. Cancer stem cell functions in hepatocellular carcinoma and comprehensive therapeutic strategies. Cells. 2020;9(6):1331–40. doi: 10.3390/cells9061331. [Liu YC, Yeh CT, Lin KH. Cancer stem cell functions in hepatocellular carcinoma and comprehensive therapeutic strategies[J]. Cells, 2020, 9(6): 1331-40.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curry JM, Tuluc M, Whitaker-Menezes D, et al. Cancer metabolism, stemness and tumor recurrence: MCT1 and MCT4 are functional biomarkers of metabolic symbiosis in head and neck cancer. Cell Cycle. 2013;12(9):1371–84. doi: 10.4161/cc.24092. [Curry JM, Tuluc M, Whitaker-Menezes D, et al. Cancer metabolism, stemness and tumor recurrence: MCT1 and MCT4 are functional biomarkers of metabolic symbiosis in head and neck cancer[J]. Cell Cycle, 2013, 12(9): 1371-84] [DOI] [PMC free article] [PubMed] [Google Scholar]


