Abstract
目的
探讨硒代半胱氨酸tRNA特异性真核延伸因子(EEFSEC)对人前列腺癌细胞增殖、迁移和侵袭的影响。
方法
采用qRT-PCR法检测人正常前列腺细胞系RWPE1和人前列腺癌细胞系22Rv1、LNcap、Vcap、PC-3细胞中EEFSEC mRNA的表达;收集前列腺癌患者的癌组织和癌旁组织,采用Western blot法检测前列腺癌组织中EEFSEC的蛋白表达情况。慢病毒感染22Rv1细胞,Western blot检测敲低效率;XTT实验检测各组细胞的增殖能力;划痕实验和Transwell实验用来检测细胞迁移能力;Transwell试验检测细胞侵袭能力;流式细胞术检测细胞周期;qRT-PCR检测敲低EEFSEC后周期相关基因的mRNA水平的变化。
结果
与对照组相比,EEFSEC在前列腺癌中高表达(P < 0.05);且EEFSEC高表达导致前列腺癌患者不良预后;感染敲低EEFSEC的慢病毒后,可明显降低22Rv1细胞中EEFSEC的蛋白表达水平;与对照组相比,敲低EEFSEC可显著抑制22Rv1细胞的增殖(P < 0.001),迁移(P < 0.001)和侵袭能力(P < 0.001);敲低EEFSEC细胞周期主要阻滞在G0/G1期,qRT-PCR实验显示敲低EEFSEC可明显下调C-myc和CCNB1的表达,上调p15的表达。
结论
敲低EEFSEC可能通过降低C-myc表达来抑制前列腺癌细胞22Rv1的增殖、迁移和侵袭能力。
Keywords: 前列腺癌, EEFSEC, 增殖, 迁移, 侵袭
Abstract
Objective
To investigate the role of selenocysteine-tRNA specific eukaryotic elongation factor (EEFSEC) in regulating the proliferation, migration, and invasion of human prostate cancer 22Rv1 cells.
Methods
We detected EEFSEC mRNA expression levels in human normal prostate cell line RWPE1 and human prostate cancer cell lines 22Rv1, LNCaP, Vcap and PC-3 using qRT-PCR and EEFSEC protein expression in surgical specimens of prostate cancer and adjacent tissues using Western blotting. 22Rv1 cells were infected with a lentiviral vector carrying EEFSEC shRNA or a control lentivirus and the interference efficiency was determined using Western blotting. XTT assay was used to assess the changes in the viability of the infected cells, and Transwell chamber assay was used to examine the changes in cell migration and invasion. The effect of EEFSEC knockdown on cell cycle progression was determined with flow cytometry and by detecting the expressions of cell cycle proteins using qRT-PCR.
Results
EEFSEC was significantly upregulated in prostate cancer cells (P < 0.05), and a high expression of EEFSEC was associated with a poor prognosis of the patients with prostate cancer. In 22Rv1 cells, EEFSEC knockdown significantly suppressed the proliferation (P < 0.001), migration (P < 0.001) and invasion (P < 0.001) of the cells, resulted in cell cycle arrest in G0/G1 phase, obviously inhibited the expression of C-myc and CCNB1, and significantly increased the expression of p15.
Conclusion
EEFSEC knockdown can inhibit the proliferation, migration, and invasion of prostate cancer cells in vitro possibly by down-regulating the expression of C-myc.
Keywords: prostate cancer, EEFSEC, proliferation, migration, invasion
前列腺癌是仅次于肺癌的全球男性易患癌症中的第二大癌症。全球新增男性癌症人数的13.5%为前列腺癌,前列腺癌死亡率占男性因癌症死亡总人数的6.7%[1]。目前,在美洲、北欧、西欧、澳大利亚、新西兰和撒哈拉以南的非洲大部分地区,前列腺癌成为当地男性最易患的癌症。尽管前列腺癌在部分西方国家死亡率逐渐下降,但是在世界其他大部分地方其死亡率逐渐上升,这归因于医疗水平的差距。现如今,前列腺癌的发病机制仍然不明确,许多因素可能导致前列腺癌的发生。
硒对于哺乳动物来说是一种必须微量元素,以硒代半胱氨酸(Sec)的形式在人体内存在。有关硒预防前列腺癌的综述指出,补充硒能够降低前列腺癌的发生风险[2]。研究发现脚趾甲里硒水平与晚期前列腺癌风险的显著降低相关[3]。适量补充硒可以预防某些癌症,但是硒的过度摄入可能会促进肿瘤的发生,硒与肿瘤的关系表现为U型分布[4]。通过对前列腺癌患者进行基因型分析发现硒相关转运蛋白发生突变会影响发生前列腺癌的概率[5]。硒代半胱氨酸的翻译是由UGA密码子来介导。UGA通常作为终止密码子来行使其功能,因此硒蛋白的翻译需要硒代半胱氨酸插入序列(SECIS)与UGA引导翻译。该引导过程由丝氨酸(Ser)与Sec特异性tRNA结合形成Ser-tRNASec启动翻译,在硒代半胱氨酸tRNA特异性真核延伸因子(EEFSEC)的介导下向核糖体传递,参与硒蛋白的合成[6]。
人EEFSEC由四个结构域(D1-D4)组成一个似圣杯的结构,结构域D1-D3形成类似于一般翻译延伸因子Tu(EF-Tu)的杯子。结构域D4是杯子的基部,在许多与核酸相互作用的蛋白质中被发现。由两条β-链组成的连接区代表连接杯和脚的圣杯茎。EF-Tu样结构域带有GTPase位点和Sec结合口袋[7]。人类的EEFSEC基因定位于第3号染色体。EEFSEC蛋白由596个氨基酸残基组成。EEFSEC具有核定位信号序列,其编码的蛋白可定位到细胞核中,并在胞浆胞核之间进行穿梭[8]。通过基因组研究发现,EEFSEC与早产风险相关[9]。EEFSEC作为真核延伸因子的一员,其在前列腺癌中的作用,至今还未见报道。
本研究通过构建敲低EEFSEC的前列腺癌细胞稳定株、XTT、Transwell等方法来研究EFFSEC在前列腺癌中的功能,为探究前列腺癌的发病机制奠定基础。
1. 材料和方法
1.1. 细胞和主要试剂
实验细胞系22Rv1、LNcap、PC-3、293T、RWPE1、Vcap(ATCC);RPMI 1640基础培养基(Lot No. RNBJ9906)(Sigama);F-12 Kaighn′s Modification培养基(HyClone);SYBR Select Master Mix和Lipofectamine 2000转染试剂、DMEM高糖培养基、SFM培养基、胎牛血清、HRP标记的山羊抗兔IgG、ECL化学发光液、PCR产物回收试剂盒(Lot No.00912090)和逆转录相关试剂(Lot No.4368814)(ThermoFisher Scientific);XTT实验试剂盒(Roche);Western blot所用一抗EFFSEC、GAPDH和HRP标记的山羊抗鼠IgG(Abclonal);Transwell小室(Corning);RNA纯化取试剂盒(TIANGEN);病毒衣壳质粒pMD2.G、psPAX2和慢病毒载体PLKO.1(Addgene);慢病毒寡核苷酸序列EEFSEC shRNA1和EEFSEC shRNA2由上海生工生物工程股份有限公司合成,EEFSEC shRNA1正义链为CCGGCGGCAAGTTCAAGATCCATCTCGAGATGT GGATCTTGAACTTGCCGTTTTTTG,EEFSEC shRNA1的反义链为CAAAAAACGGCAAGTTCAAGATC CACATCTCGAGATGTGGATCTTGAACTGCCGCC GG。EEFSEC shRNA2的正义链为CCGGCAGATT TCCATCCCAACGAGACTCGAGTCTCGTTGGGAT GGAAATCTGTTTTTTG,EEFSEC shRNA2的反义链为CAAAAAACAGATTTCCATCCCAACGAGACTC GAGTCTCGTTGGGATGGAAATCTGCCGG。
1.2. 标本来源
山东第一医科大学附属成武医院收集癌旁组织及前列腺癌组织,所有患者手术前均没有接受过靶向治疗、内分泌等治疗或放化疗。所有患者均签署知情同意书,实验方案由山东第一医科大学附属成武医院伦理委员会审核批准,伦理编号(2021-030)。所有样本离体后迅速放入液氮中保存,用于后续检测EEFSEC的表达。
1.3. 细胞培养
22Rv1细胞和LNcap细胞使用RPMI 1640培养基,PC-3细胞使用F-12K培养基,RWPE1细胞使用SFM培养基,Vcap细胞和293T细胞使用DMEM培养基,所有培养基添加1%青链霉素,所有细胞均置于37 ℃、5% CO2条件下常规培养。每隔3~4 d用胰酶消化传代1次,利用对数生长期的细胞进行后续实验。
1.4. shRNA设计与合成
从NCBI中检索人的EEFSEC基因序列(NM_ 021937.5),将序列号输入在线软件BLOCK-iT<sup>TM</sup> RNAi Designer(<a href="https://rnaidesigner.thermofisher.com/rnaiexress/Design.do" target="_blank">https//rnaidesigner.thermofisher.com/rnaiexress/Design.do</a>)寻找靶点。
1.5. 慢病毒制备
将293T细胞用胰酶消化3 min,然后重悬细胞,将其接种至六孔板,在细胞培养箱中培养24 h。然后利用脂质体Lipofectamine 2000将构建好的EEFSEC shRNA1、EEFSEC shRNA2、包膜质粒pMD2.G和包装质粒psPAX2共转染细胞,24 h后更换成正常的含有10%血清的低糖DMEM培养基。转染48 h后收集含慢病毒的培养基3次,每次间隔24 h。病毒经0.45 μm的滤膜过滤后分装并冻存至-80 ℃。
1.6. 干涉EEFSEC的22Rv1稳定株的构建
22Rv1细胞经胰酶消化3 min,然后重悬细胞,将其接种至六孔板,在细胞培养箱中培养24 h。弃除培养基,然后加入500 μL新鲜的RPMI 1640完全培养基,每孔分别加入500 μL的EEFSEC干涉慢病毒(EEFSEC shRNA)以及对照组慢病毒(Control shRNA)原液,再加入3 μL的4 mg/mL的聚凝胺,在细胞培养箱孵育2 h后,各孔再补充1 mL新鲜的RMPI 1640培养基。感染24 h后将细胞进行换液。22Rv1细胞感染干涉病毒48 h后用1 μg/mL的嘌呤霉素对其进行筛选,连续筛选3 d,当阴性对照细胞全部死亡后,成功构建干涉EEFSEC的22Rv1细胞稳定株。
1.7. XTT实验
将敲低EEFSEC及对照组的22Rv1细胞株分别接种至96孔板(2×103/孔)中,每一组均设置5个重复孔。将细胞置于37 ℃、5% CO2条件下分别培养0、2、4、6 d,每孔加入10 μL XTT混合溶液,随后放置培养箱培养12 h,最后使用超微量分光光度计检测在450 nm波长下每孔细胞的光密度值A450 nm。
1.8. 划痕实验
将敲低EEFSEC及对照组的22Rv1细胞株分别接种至24孔板(1×105/孔)中,每组均设置3个重复孔,培养细胞使其汇合度接近100%,用小枪头在24孔板内画1条均匀的直线,PBS清洗掉悬浮的细胞,补充完全培养基。将细胞置于37 ℃、5% CO2条件下培养,分别于0、48 h在同一位置用倒置显微镜观察拍照,计算相对划痕愈合率,相对划痕愈合率=实验组(0 h划痕面积-48 h划痕面积)/对照组(0 h划痕面积-48 h划痕面积)。
1.9. Transwell小室实验
Transwell小室实验包含迁移和侵袭两种实验,迁移实验把处理好的细胞悬液直接转移至小室上层;侵袭实验需在小室的上层先加入100 μL的基质胶,待其凝固,并进行后续实验。利用胰酶将对照组和干涉组细胞进行消化,不含血清培养基重悬细胞,在不含或包含基质胶小室上层分别添加105和2×105细胞,下室均加入700 μL的完全培养基。迁移实验24 h后取出小室,侵袭实验48 h后取出小室,利用PBS冲洗2次洗去杂质,3.7%多聚甲醛固定,吉姆萨染色,并用PBS清洗,随后用棉签将各小室的上层细胞小心擦拭掉,最后在显微镜下进行拍照并计数。
1.10. 实时荧光定量PCR(qRT-PCR)
使用RNA纯化试剂盒(TIANGEN)分别提取22Rv1、LNcap、PC-3、RWPE1、Vcap、Control shRNA组、EEFSEC shRNA1和EEFSEC shRNA2组的RNA,取2 μg总RNA逆转录为cDNA,利用qRT-PCR方法检测EEFSEC的RNA表达水平,以β-actin作为内参。qRT-PCR引物均上海生工生物工程股份有限公司合成。qRT-PCR引物序列见表 1。
1.
qRT-PCR引物序列
Sequences of qRT-PCR primer
| Name of primer | Sequences (5′ to 3′) |
| β-actin-RT-F | AGAAAATCTGGCACCACACC |
| β-actin-RT-R | AGAGGCGTACAGGGATAGCA |
| EEFSEC-RT-F | CATCCCAACGAFACCCCT |
| EEFSEC-RT-R | GGAAACATCTGCATGGACTTCA |
| p15-RT-F | GGGACTAGTGGAGAAGGTGC |
| p15-RT-R | CCATCATCATGACCTGGATCG |
| p53-RT-F | ACCTATGGAAACTACTTCCTGAAA |
| p53-RT-R | GGAGCTTCATCTGGACCTGG |
| CCNB1-RT-F | AATAAGGCGAAGATCAACATGGC |
| CCNB1-RT-R | TTTGTTACCAATGTCCCCAAGAG |
| CCND1-RT-F | CAATGACCCCGCACGATTTC |
| CCND1-RT-R | CATGGAGGGCGGATTGGAA |
| CDKN1A-RT-F | AAACGGCGGCAGACCAGC |
| CDKN1A-RT-R | GCTTCCTGTGGGCGGATTAG |
| C-myc-RT-F | GTCAAGAGGCGAACACACAAC |
| C-myc-RT-R | TTGGACGACAGGATGTATGC |
1.11. Western blot
提取前列腺癌组织和不同细胞系总蛋白,检测蛋白浓度进行SDS-PAGE电泳、蛋白质转印、封闭、一抗4 ℃孵育过夜、洗膜、二抗孵育、洗膜、暗室显影。一抗EEFSEC(1∶500)、GAPDH(1∶4000)、二抗IgG(1∶5000)进行配制。
1.12. 细胞周期检测
收集对数生长期的细胞制备成细胞悬液接种至6孔板中,细胞培养箱培养48 h,用胰酶消化离心,收集全部细胞,用4 ℃预冷PBS洗涤细胞,并用70%酒精对细胞进行固定并4 ℃过夜。细胞离心去上清,PBS重悬细胞,细胞悬液加入RNase和PI染色孵育30 min。300目尼龙膜过滤,流式细胞仪上机检测。
1.13. 统计学分析
每组实验独立重复3次,用GraphPad Prism8.0软件对实验结果进行统计学分析。数据来自人类癌症转移数据库(HCMDB)和高通量基因表达数据库(GEO)。组间比较采用t检验,以P < 0.05为差异具有统计学意义。
2. 结果
2.1. EEFSEC在前列腺癌组织中的表达
实时定量PCR结果显示,与正常人的前列腺上皮细胞系RWPE1相比,前列腺癌细胞系22Rv1、LNcap、Vcap、PC-3中EEFSEC表达水平明显上调(P < 0.05,图 1A)。通过HCMDB查询得知,与正常对照组相比,EEFSEC在前列腺癌组织中显著高表达(P < 0.001,图 1B)。通过分析GEO数据库中前列腺癌转移数据的数据集GSE35988和GSE21032得知,前列腺癌转移组织和局限性前列腺癌中EEFSEC的mRNA表达水平差异有统计学意义(P < 0.01,图 1C、D)。
1.
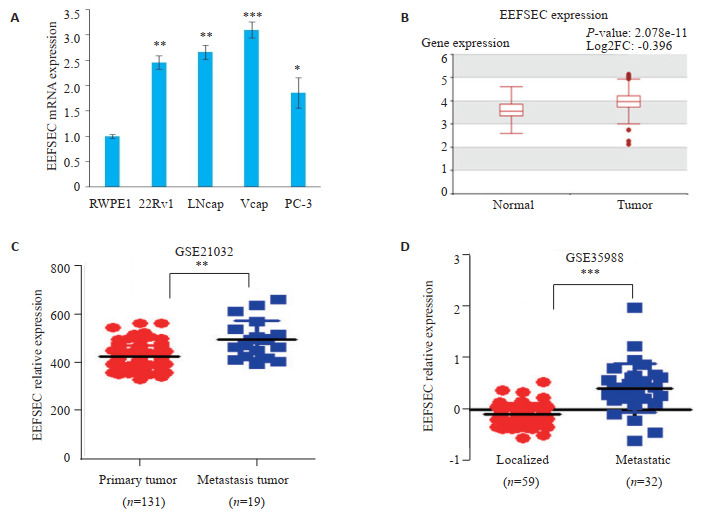
EEFSEC在前列腺癌组织中的表达
Expression of EEFSEC in prostate cancer tissues and adjacent tissues from TCGA and GEO databases. A: Expression of EEFSEC mRNA in RWPE1, 22Rv1, LNcap, Vcap and PC-3 cells detected by qRT-PCR; *P < 0.05, **P < 0.01, ***P < 0.001 vs RWPE1. B: Expression of EEFSEC in normal tissue and prostate tumor (TCGA); C: Expression of EEFSEC in primary tumor and metastasis tumor (GSE21032); D: Expression of EEFSEC in localized tumor and metastasis tumor (GSE35988).
2.2. EEFSEC在病人样本中的蛋白表达情况
Western blot实验结果显示,与癌旁组织相比,EEFSEC在前列腺癌组织中的蛋白表达明显增加(图 2)。
2.
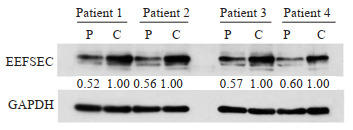
EEFSEC在前列腺癌(P)和癌旁组织(C)中的蛋白表达情况
Protein expression of EEFSEC in prostate cancer(C) and adjacent tissues (P).
2.3. EEFSEC高表达导致前列腺癌患者不良预后
基于TCGA数据库,在GEPIA2网站分析显示EEFSEC高表达导致了前列腺癌患者无病生存期有所降低(P=0.55,图 3)。
3.
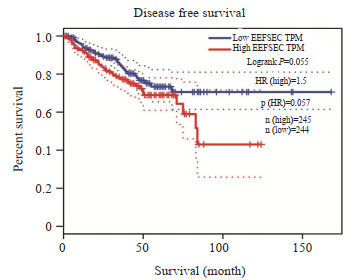
EEFSEC高表达导致前列腺癌患者预后不良
High expression of EEFSEC is associated with a poor prognosis in patients with prostate cancer.
2.4. 干涉EEFSEC稳定株的构建
Western blot实验结果显示,与Control shRNA组相比,干涉EEFSEC的22Rv1细胞EEFSEC表达量下降(P < 0.01,图 4)。
4.
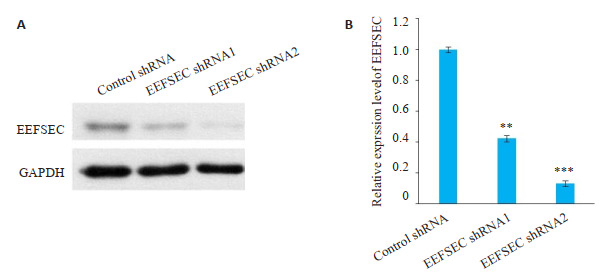
EEFSEC在22Rv1细胞中干涉成功
EEFSEC is successfully knocked down in 22Rv1 cells. A: EEFSEC knockdown results in significantly reduced expression of EEFSEC in 22Rv1 cells; B: Relative expression of EEFSEC protein. **P < 0.01, ***P < 0.001 vs control shRNA.
2.5. 干涉EEFSEC影响前列腺癌细胞22Rv1的增殖
XTT实验结果显示,与Control shRNA组相比,干涉EEFSEC明显抑制22Rv1细胞的增殖,Control shRNA组与EEFSEC shRNA1组和EEFSEC shRNA2组增殖能力差异有统计学意义(P < 0.001,图 5)。
5.
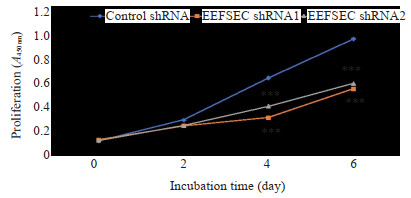
XTT实验检测EEFSEC基因敲低对22Rv1细胞增殖的影响
Effect of EEFSEC knockdown on proliferation of 22Rv1 cells assessed using XTT assay; ***P < 0.001 vs control shRNA.
2.6. 干涉EEFSEC影响前列腺癌细胞22Rv1的迁移
划痕实验显示,干涉EEFSEC表达后,与Control shRNA组相比,EEFSEC shRNA1组和EEFSEC shRNA2组的22Rv1细胞的迁移距离低于对照组(P < 0.05,图 6A、B)。Transwell迁移实验中,与Control shRNA组相比,EEFSEC shRNA1组和EEFSEC shRNA2组的22Rv1细胞穿膜细胞数明显减少(图 6C),两组间进行比较发现差异有统计学意义(P < 0.001,图 6D)。
6.
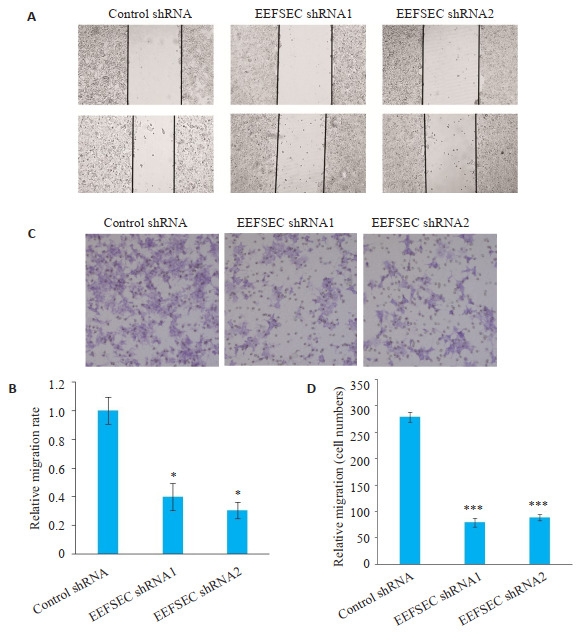
划痕和Transwell实验检测EEFSEC基因敲低对22Rv1细胞迁移能力的影响
Effect of EEFSEC knockdown on migration ability of 22Rv1 cells. A: Wound healing assay; B: Relative migration rate; *P < 0.05 vs control shRNA; C: Transwell assay (Original magnification: ×200); D: Migrated cells. ***P < 0.001 vs control shRNA.
2.7. 干涉EEFSEC影响前列腺癌细胞22Rv1的侵袭
Transwell侵袭实验显示,与Control shRNA组相比,EEFSEC shRNA1组和EEFSEC shRNA2组的22Rv1细胞穿膜细胞数明显减少(图 7A),两组间进行比较发现差异存在统计学差异(P < 0.001,图 7B)。
7.
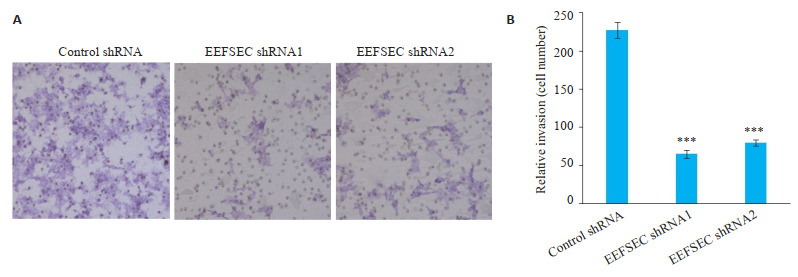
Transwell实验检测EEFSEC基因敲低对22Rv1细胞侵袭能力的影响
Effect of EEFSEC gene silencing on invasion ability of 22Rv1 cells. A: Transwell assay (×200); B: Migrated cells.***P < 0.001 vs control shRNA.
2.8. 干涉EEFSEC影响前列腺癌细胞22Rv1的周期
细胞周期实验显示,与Control shRNA组相比,EEFSEC shRNA组的细胞被阻滞于G0/G1期(图 8A),两组间进行比较发现存在显著的统计学差异(P < 0.01,图 8B)。另外对细胞周期蛋白进行mRNA表达水平进行检测,发现敲低EEFSEC显著上调p15,显著下调CCNB1和C-myc(P < 0.05,图 8C)。
8.
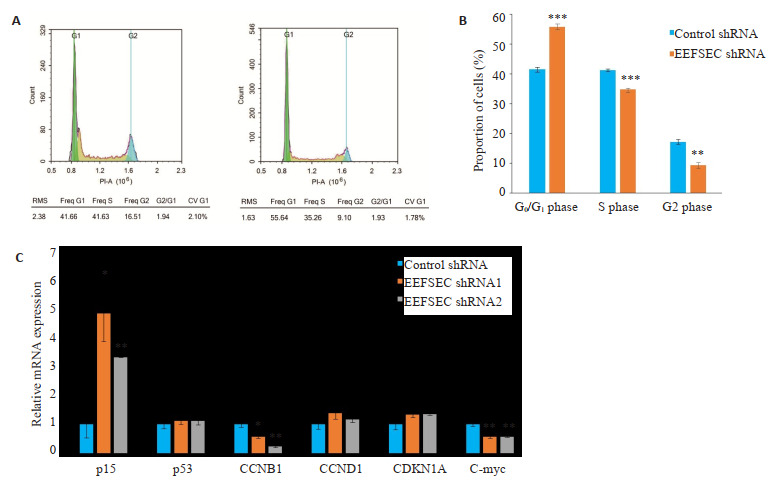
EEFSEC基因敲低对22Rv1细胞周期的影响
Effect of EEFSEC gene silencing on cell cycle of 22Rv1 cells. A: Flow cytometry; B: Proportion of cells in each phase determined by flow cytometry; **P < 0.01, ***P < 0.001 vs control shRNA; C: The mRNA expression of cell cycle related genes detected by qRT-PCR. *P < 0.05, **P < 0.01 vs control shRNA.
3. 讨论
前列腺癌作为最常见的男性生殖系统恶性肿瘤,严重地影响男性的生命健康。前列腺癌治疗虽然取得了一定的进展,但前列腺癌的治疗仍然是一个重要的医学问题,其中包括对良性疾病的过度治疗和对转移性前列腺癌的治疗方法不足[10]。许多因素会影响前列腺癌的发生,其中种族和遗传因素是重要原因[1]。因此对前列腺癌的发生、发展以及转移机制研究至关重要。
延伸因子(EF)是在mRNA翻译过程中,可促进多肽链延伸的蛋白质因子。研究表明真核生物延伸因子1(EEF1)和真核生物延伸因子2(EEF2)两者共同构成了真核生物的延伸因子,并在翻译过程中发挥作用[11]。真核生物延伸因子1-α(EEF1A)和真核生物延伸因子1-β(EEF1B)的复合物一起构成EEF1复合物。
EEF1A是G蛋白家族的一个成员,也是构成真核生物延伸因子的四个亚单位之一[12, 13]。目前已知有两个EEF1A亚型在人类中表达,分别是EEF1A1和EEF1A2[14]。这两个亚型在体外翻译延伸活性相似,但它们对GDP和GTP的结合亲和力存在差异[14]。Sandbaken等[15]发现EEF1A1的突变直接影响了酿酒酵母中氨基酸的错掺和氨基酸错读的频率。Scaggiante等[16]发现EEF1A2可能是前列腺细胞转化的标记或癌症进展的标志。其它研究报道发现EEF1A2与乳腺癌[17, 18]、卵巢癌[19, 20]、肝癌[21, 22]和胰腺癌[23, 24]的发生发展具有紧密的联系。
EEFSEC属于真核生物延伸因子家族的一员,其在肿瘤细胞中的作用至今未见报道。我们利用实时定量PCR方法发现,与人的正常前列腺上皮细胞系RWPE1相比,EEFSEC的mRNA在前列腺癌细胞系22Rv1、LNcap、Vcap、PC-3中水平明显上调。通过HCMDB查询得知,与正常对照组相比,EEFSEC在前列腺癌组织中显著高表达(P < 0.001)。通过分析GEO数据库中前列腺癌转移数据的数据集GSE35988和GSE21032得知,EEFSEC的mRNA表达水平在前列腺癌转移组织和局限性前列腺癌中存在显著差异(P < 0.01)。随后我们收集了4例前列腺癌患者的前列腺癌及癌旁组织,Western blot结果显示,与癌旁组织相比,EEFSEC在前列腺癌组织中的蛋白水平明显增加。以上结果提示,EEFSEC在前列腺癌中发挥一定的作用。随后利用慢病毒感染的方式干涉前列腺癌细胞系22Rv1中EEFSEC的表达,利用抗生素筛选出敲低EEFSEC稳定细胞株。并通过Western blot验证EEFSEC在蛋白水平的干涉效果。利用XTT实验、划痕实验和Transwell实验发现干涉EEFSEC的22Rv1细胞的增殖、迁移和侵袭能力均受到明显抑制。
C-myc基因位于人类8q24染色体上,并编码C-myc蛋白,其作为转录因子在调控许多细胞行为中起着关键作用[25]。C-myc在许多肿瘤样本中高表达[26],C-myc的扩增可作为前列腺癌发生发展的潜在标志物[27]。C-myc的异常表达与病人前列腺肿瘤发展和死亡率显著相关;C-myc拷贝数的增加还与病人的预后不良相关[28]。在小鼠前列腺和正常人前列腺上皮细胞中过表达C-myc会导致前列腺癌发生转化,并具有侵袭性表型[29]。以上研究表明C-myc的过表达会导致前列腺癌细胞的恶化。研究报道,C-myc在癌症中可调控许多细胞周期蛋白的表达,例如p15[30]、CDKN1A[31]、CCNB1[32]。Taewan Kim等人发现在前列腺癌中,敲低C-myc的靶向基因MYCLo-1后发现前列腺癌细胞增殖变慢,p15表达增加,使细胞积累在G0/G1期[30]。有报道发现在DU-145和PC-3中敲低C-myc明显降低CCNB1的表达,同时阻断C-myc的转录共激活剂,使细胞阻滞在G0/ G1期,更加验证了本实验结论[33]。通过qRT-PCR实验发现,在干涉EEFSEC的22Rv1细胞系的C-myc的表达显著下降,提示EEFSEC可能通过调节的C-myc表达,进而影响p15和CCNB1的表达,使细胞阻滞在G0/G1期,从而抑制22Rv1细胞的增殖、迁移和侵袭能力,而EEFSEC如何调控C-myc的表达仍需进一步研究。
综上所述,本研究发现EEFSEC在前列腺癌中高表达,干涉22Rv1细胞EEFSEC可下调C-myc的表达,进而调节细胞周期相关基因的表达,从而影响前列腺癌细胞22Rv1的增殖、迁移和侵袭。本研究探索了前列腺癌的发病机制并为后续前列腺癌的治疗提供了基础。
Biography
徐彬兵,硕士研究生,E-mail: xubingdyx@163.com
Funding Statement
国家自然科学基金(81972417);陕西省自然科学基金(2020JM-292,2020JQ-430);中央高校基本科研业务费(GK201902002,GK201903061);大学生创新创业训练计划项目(S202110718036,S202010718306,S202010718091)
Supported by National Natural Science Foundation of China (81972417)
Contributor Information
徐 彬兵 (Binbing XU), Email: xubingdyx@163.com.
高 平 (Ping GAO), Email: ping.gao@snnu.edu.cn.
石 光耀 (Guangyao SHI), Email: lylengshan@126.com.
董 小明 (Xiaoming DONG), Email: xiaomingdong@snnu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer J Clin, 2018, 68(6): 394-424.] [DOI] [PubMed] [Google Scholar]
- 2.Etminan M, FitzGerald JM, Gleave M, et al. Intake of selenium in the prevention of prostate cancer: a systematic review and meta-analysis. Cancer Causes Control. 2005;16(9):1125–31. doi: 10.1007/s10552-005-0334-2. [Etminan M, FitzGerald JM, Gleave M, et al. Intake of selenium in the prevention of prostate cancer: a systematic review and meta-analysis[J]. Cancer Causes Control, 2005, 16(9): 1125-31.] [DOI] [PubMed] [Google Scholar]
- 3.Geybels MS, Verhage BAJ, van Schooten FJ, et al. Advanced prostate cancer risk in relation to toenail selenium levels. J Natl Cancer Inst. 2013;105(18):1394–401. doi: 10.1093/jnci/djt186. [Geybels MS, Verhage BAJ, van Schooten FJ, et al. Advanced prostate cancer risk in relation to toenail selenium levels[J]. J Natl Cancer Inst, 2013, 105(18): 1394-401.] [DOI] [PubMed] [Google Scholar]
- 4.Rayman MP. Selenium and human health. Lancet. 2012;379(9822):1256–68. doi: 10.1016/S0140-6736(11)61452-9. [Rayman MP. Selenium and human health[J]. Lancet, 2012, 379 (9822): 1256-68.] [DOI] [PubMed] [Google Scholar]
- 5.Chan JM, Darke AK, Penney KL, et al. Selenium- or vitamin E-related gene variants, interaction with supplementation, and risk of high-grade prostate cancer in SELECT. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1050–8. doi: 10.1158/1055-9965.EPI-16-0104. [Chan JM, Darke AK, Penney KL, et al. Selenium- or vitamin E-related gene variants, interaction with supplementation, and risk of high-grade prostate cancer in SELECT[J]. Cancer Epidemiol Biomarkers Prev, 2016, 25(7): 1050-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serrão VHB, Silva IR, da Silva MTA, et al. The unique tRNASec and its role in selenocysteine biosynthesis. http://www.researchgate.net/profile/Vitor_Serrao/publication/325732038_The_unique_tRNASec_and_its_role_in_selenocysteine_biosynthesis/links/5b2178f1458515270fc6da1e/The-unique-tRNASec-and-its-role-in-selenocysteine-biosynthesis.pdf. Amino Acids. 2018;50(9):1145–67. doi: 10.1007/s00726-018-2595-6. [Serrão VHB, Silva IR, da Silva MTA, et al. The unique tRNASec and its role in selenocysteine biosynthesis[J]. Amino Acids, 2018, 50(9): 1145-67.] [DOI] [PubMed] [Google Scholar]
- 7.Simonović M, Puppala AK. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins. Biochim Biophys Acta Gen Subj. 2018;1862(11):2463–72. doi: 10.1016/j.bbagen.2018.03.018. [Simonović M, Puppala AK. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins[J]. Biochim Biophys Acta Gen Subj, 2018, 1862 (11): 2463-72.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubey A, Copeland PR. The selenocysteine-specific elongation factor contains unique sequences that are required for both nuclear export and selenocysteine incorporation. PLoS One. 2016;11(11):e0165642. doi: 10.1371/journal.pone.0165642. [Dubey A, Copeland PR. The selenocysteine-specific elongation factor contains unique sequences that are required for both nuclear export and selenocysteine incorporation[J]. PLoS One, 2016, 11 (11): e0165642.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallman M, Haapalainen A, Huusko JM, et al. Spontaneous premature birth as a target of genomic research. Pediatr Res. 2019;85(4):422–31. doi: 10.1038/s41390-018-0180-z. [Hallman M, Haapalainen A, Huusko JM, et al. Spontaneous premature birth as a target of genomic research[J]. Pediatr Res, 2019, 85(4): 422-31.] [DOI] [PubMed] [Google Scholar]
- 10.Wang GC, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. http://www.onacademic.com/detail/journal_1000040457287810_526b.html. Genes Dev. 2018;32(17/18):1105–40. doi: 10.1101/gad.315739.118. [Wang GC, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer[J]. Genes Dev, 2018, 32(17/18): 1105-40.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.何 天琦, 卢 松岩, 朱 东韦, et al. 真核翻译延伸因子eEF1A功能研究进展. https://www.cnki.com.cn/Article/CJFDTOTAL-ZGXQ202003019.htm. 中国预防兽医学报. 2020;42(3):315–9. [何天琦, 卢松岩, 朱东韦, 等. 真核翻译延伸因子eEF1A功能研究进展[J]. 中国预防兽医学报, 2020, 42(3): 315-9.] [Google Scholar]
- 12.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269(22):5360–8. doi: 10.1046/j.1432-1033.2002.03290.x. [Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells[J]. Eur J Biochem, 2002, 269(22): 5360-8.] [DOI] [PubMed] [Google Scholar]
- 13.Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci Biotechnol Biochem. 2002;66(1):1–21. doi: 10.1271/bbb.66.1. [Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization[J]. Biosci Biotechnol Biochem, 2002, 66(1): 1-21.] [DOI] [PubMed] [Google Scholar]
- 14.Lee MH, Surh YJ. eEF1A2 as a putative oncogene. Ann N Y Acad Sci. 2009;1171:87–93. doi: 10.1111/j.1749-6632.2009.04909.x. [Lee MH, Surh YJ. eEF1A2 as a putative oncogene[J]. Ann N Y Acad Sci, 2009, 1171: 87-93.] [DOI] [PubMed] [Google Scholar]
- 15.Sandbaken MG, Culbertson MR. Mutations in elongation factor EF-1 alpha affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics. 1988;120(4):923–34. doi: 10.1093/genetics/120.4.923. [Sandbaken MG, Culbertson MR. Mutations in elongation factor EF-1 alpha affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae[J]. Genetics, 1988, 120(4): 923-34.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaggiante B, Dapas B, Bonin S, et al. Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: the potential of EEF1A2 as a hallmark for prostate transformation and progression. Br J Cancer. 2012;106(1):166–73. doi: 10.1038/bjc.2011.500. [Scaggiante B, Dapas B, Bonin S, et al. Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: the potential of EEF1A2 as a hallmark for prostate transformation and progression[J]. Br J Cancer, 2012, 106(1): 166-73.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlinson VAL, Newbery HJ, Wray NR, et al. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer. 2005;5:113. doi: 10.1186/1471-2407-5-113. [Tomlinson VAL, Newbery HJ, Wray NR, et al. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours[J]. BMC Cancer, 2005, 5: 113.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinestrosa MC, Dickersin K, Klein P, et al. Shaping the future of biomarker research in breast cancer to ensure clinical relevance. Nat Rev Cancer. 2007;7(4):309–15. doi: 10.1038/nrc2113. [Hinestrosa MC, Dickersin K, Klein P, et al. Shaping the future of biomarker research in breast cancer to ensure clinical relevance[J]. Nat Rev Cancer, 2007, 7(4): 309-15.] [DOI] [PubMed] [Google Scholar]
- 19.Kim A, Ueda Y, Naka T, et al. Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res. 2012;31:14. doi: 10.1186/1756-9966-31-14. [Kim A, Ueda Y, Naka T, et al. Therapeutic strategies in epithelial ovarian cancer[J]. J Exp Clin Cancer Res, 2012, 31: 14.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinke DE, Kalloger SE, Francetic T, et al. The prognostic significance of elongation factor eEF1A2 in ovarian cancer. Gynecol Oncol. 2008;108(3):561–8. doi: 10.1016/j.ygyno.2007.11.019. [Pinke DE, Kalloger SE, Francetic T, et al. The prognostic significance of elongation factor eEF1A2 in ovarian cancer[J]. Gynecol Oncol, 2008, 108(3): 561-8.] [DOI] [PubMed] [Google Scholar]
- 21.Schlaeger C, Longerich T, Schiller C, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47(2):511–20. doi: 10.1002/hep.22033. [Schlaeger C, Longerich T, Schiller C, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis[J]. Hepatology, 2008, 47(2): 511-20.] [DOI] [PubMed] [Google Scholar]
- 22.Grassi G, Scaggiante B, Farra R, et al. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Biochimie. 2007;89(12):1544–52. doi: 10.1016/j.biochi.2007.07.007. [Grassi G, Scaggiante B, Farra R, et al. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade[J]. Biochimie, 2007, 89(12): 1544-52.] [DOI] [PubMed] [Google Scholar]
- 23.Duanmin H, Chao X, Qi Z. eEF1A2 protein expression correlates with lymph node metastasis and decreased survival in pancreatic ductal adenocarcinoma. http://www.ncbi.nlm.nih.gov/pubmed/23165190. Hepatogastroenterology. 2013;60(124):870–5. doi: 10.5754/hge12869. [Duanmin H, Chao X, Qi Z. eEF1A2 protein expression correlates with lymph node metastasis and decreased survival in pancreatic ductal adenocarcinoma[J]. Hepatogastroenterology, 2013, 60(124): 870-5.] [DOI] [PubMed] [Google Scholar]
- 24.Xu C, Hu DM, Zhu Q. eEF1A2 promotes cell migration, invasion and metastasis in pancreatic cancer by upregulating MMP-9 expression through Akt activation. Clin Exp Metastasis. 2013;30(7):933–44. doi: 10.1007/s10585-013-9593-6. [Xu C, Hu DM, Zhu Q. eEF1A2 promotes cell migration, invasion and metastasis in pancreatic cancer by upregulating MMP-9 expression through Akt activation[J]. Clin Exp Metastasis, 2013, 30 (7): 933-44.] [DOI] [PubMed] [Google Scholar]
- 25.Gurel B, Iwata T, Koh CM, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–67. doi: 10.1038/modpathol.2008.111. [Gurel B, Iwata T, Koh CM, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis[J]. Mod Pathol, 2008, 21(9): 1156-67.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesbit C E, Tersak J M, Prochownik E V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–16. doi: 10.1038/sj.onc.1202746. [Nesbit C E, Tersak J M, Prochownik E V. MYC oncogenes and human neoplastic disease[J]. Oncogene, 1999, 18(19): 3004-16.] [DOI] [PubMed] [Google Scholar]
- 27.Jenkins RB, Qian J, Lieber MM, et al. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. http://www.researchgate.net/profile/Daniel_Lerda3/publication/307656257_Evaluation_of_Myc_Gene_Amplification_in_Prostate_Cancer_Using_a_Dual_Color_Chromogenic_in-Situ_Hybridization_Dual_CISH_Assay/links/57e0280f08aec6ce9f28dddd.pdf. Cancer Res. 1997;57(3):524–31. [Jenkins RB, Qian J, Lieber MM, et al. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization[J]. Cancer Res, 1997, 57(3): 524-31.] [PubMed] [Google Scholar]
- 28.Sato K, Qian JQ, Slezak JM, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst. 1999;91(18):1574–80. doi: 10.1093/jnci/91.18.1574. [Sato K, Qian JQ, Slezak JM, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma[J]. J Natl Cancer Inst, 1999, 91(18): 1574-80.] [DOI] [PubMed] [Google Scholar]
- 29.Chuan YC, Iglesias-Gato D, Fernandez-Perez L, et al. Ezrin mediates c-Myc actions in prostate cancer cell invasion. Oncogene. 2010;29(10):1531–42. doi: 10.1038/onc.2009.442. [Chuan YC, Iglesias-Gato D, Fernandez-Perez L, et al. Ezrin mediates c-Myc actions in prostate cancer cell invasion[J]. Oncogene, 2010, 29(10): 1531-42.] [DOI] [PubMed] [Google Scholar]
- 30.Kim T, Jeon YJ, Cui R, et al. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. http://search.ebscohost.com/login.aspx?direct=true&db=aph&AN=107783883&site=ehost-live. J Natl Cancer Inst. 2015;107(4):dju505. doi: 10.1093/jnci/dju505. [Kim T, Jeon YJ, Cui R, et al. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis[J]. J Natl Cancer Inst, 2015, 107(4): dju505.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Cetinkaya C, Munoz-Alonso MJ, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22(3):351–60. doi: 10.1038/sj.onc.1206145. [Wu S, Cetinkaya C, Munoz-Alonso MJ, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter[J]. Oncogene, 2003, 22(3): 351-60.] [DOI] [PubMed] [Google Scholar]
- 32.Yin XY, Grove L, Datta NS, et al. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 overexpression. http://hwmaint.cancerres.aacrjournals.org/cgi/reprint/61/17/6487.pdf. Cancer Res. 2001;61(17):6487–93. [Yin XY, Grove L, Datta NS, et al. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 overexpression[J]. Cancer Res, 2001, 61(17): 6487-93.] [PubMed] [Google Scholar]
- 33.Zhou Q, Chen X, He H, et al. WD repeat domain 5 promotes chemoresistance and Programmed Death-Ligand 1 expression in prostate cancer. Theranostics. 2021;11(10):4809–24. doi: 10.7150/thno.55814. [Zhou Q, Chen X, He H, et al. WD repeat domain 5 promotes chemoresistance and Programmed Death-Ligand 1 expression in prostate cancer[J]. Theranostics, 2021, 11(10): 4809-24.] [DOI] [PMC free article] [PubMed] [Google Scholar]


