Abstract
Imatinib is an ATP-competitive inhibitor of Bcr-Abl kinase and the first drug approved for chronic myelogenous leukemia (CML) treatment. Here we show that imatinib binds to a secondary, allosteric site located in the myristoyl pocket of Abl to function as an activator of the kinase activity. Abl transitions between an assembled, inhibited state and an extended, activated state. The equilibrium is regulated by the conformation of the αI helix, which is located nearby the allosteric pocket. Imatinib binding to the allosteric pocket elicits an αI helix conformation that is not compatible with the assembled state, thereby promoting the extended state and stimulating the kinase activity. Although in wild-type Abl the catalytic pocket has a much higher affinity for imatinib than the allosteric pocket does, the two binding affinities are comparable in Abl variants carrying imatinib-resistant mutations in the catalytic site. A previously isolated imatinib-resistant mutation in patients appears to be mediating its function by increasing the affinity of imatinib for the allosteric pocket, providing a hitherto unknown mechanism of drug resistance. Our results highlight the benefit of combining imatinib with allosteric inhibitors to maximize their inhibitory effect on Bcr-Abl.
Keywords: Drug resistance, allosteric regulation, allosteric activator, Bcr-Abl, NMR
Graphical Abstract
Introduction
Because of its stringent specificity for the Bcr-Abl oncoprotein and its favorable safety profile, imatinib (Gleevec) was introduced as a first-line drug treatment for chronic myeloid leukemia (CML) .1,2 Imatinib recognizes specifically a low-populated inactive conformational state of the Abl kinase domain3 and binds with high affinity (dissociation constant, Kd ~10 nM) to its ATP site thereby blocking catalytic activity.4 Despite imatinib’s remarkable success in improving survival rates,5 acquired drug resistance due to amino acid substitutions in patients is a major complication.6–8 The most severe of these substitutions occurs in the so-called gatekeeper residue (Thr334; Abl1b numbering) that is mutated to an Ile. The T334I substitution reduces drastically the affinity of imatinib for Abl because of steric clashes and the loss of a H-bond.4 Among the clinically available Abl kinase inhibitors only ponatinib is effective against T334I;9,10 however, its management is complicated by adverse cardiovascular effects.11,12 An alternative strategy to improve therapy for patients carrying such mutations has been to increase the dose of imatinib administration.1,2 However, it has been shown that a high imatinib dose is not associated with better long-term clinical outcomes, despite improved early responses.13–15
In an effort to overcome resistance to ATP-competitive inhibitors, a series of highly selective allosteric inhibitors (e.g. GNF2, GNF5 and asciminib) have been designed that target a deep pocket located in the C-lobe of Abl.16–18 In the downregulated form of Abl, the pocket is occupied by the myristoyl moiety of the post-translationally modified N-terminus of full-length Abl (Figure 1A). Binding of the myristoyl group into the pocket elicits a bent conformation to the αI helix that creates a docking site for the SH2 domain, thereby stabilizing an assembled, autoinhibited form of Abl19 (Figure 1A). It was recently reported that the assembled state stabilizes an inactive conformation in the kinase domain thus providing the structural basis for the allosteric inhibition.3 In Bcr-Abl the N-terminus of Abl is deleted and thus the fused kinase cannot be autoinhibited. Allosteric inhibitors function by mimicking the binding effect of the myristoyl group, which elicits the bent conformation to the αI helix. Allosteric inhibitors shift the population drastically towards the assembled, autoinhibited conformation of Abl thereby suppressing its activity.3,20 Asciminib remains active against Bcr-Abl-T334I, although IC50 and GI50 in both biochemical and cellular assays are higher than against the wild type kinase,18 likely because the T334I substitution stabilizes the active conformational state in the kinase domain.3 By contrast, allosteric activators bind to the myristoyl pocket and prevent the αI helix from adopting the bent conformation, thereby promoting an extended and active structure of Abl.21,22 Combinatorial therapy of asciminib with nilotinib has been shown to lead to complete tumor suppression in mice without re-growth even after the treatment is discontinued.18 Combinatorial treatment of asciminib with ponatinib shows activity against T334I-inclusive compound mutants and represses tumor growth in mice.23
Figure 1.
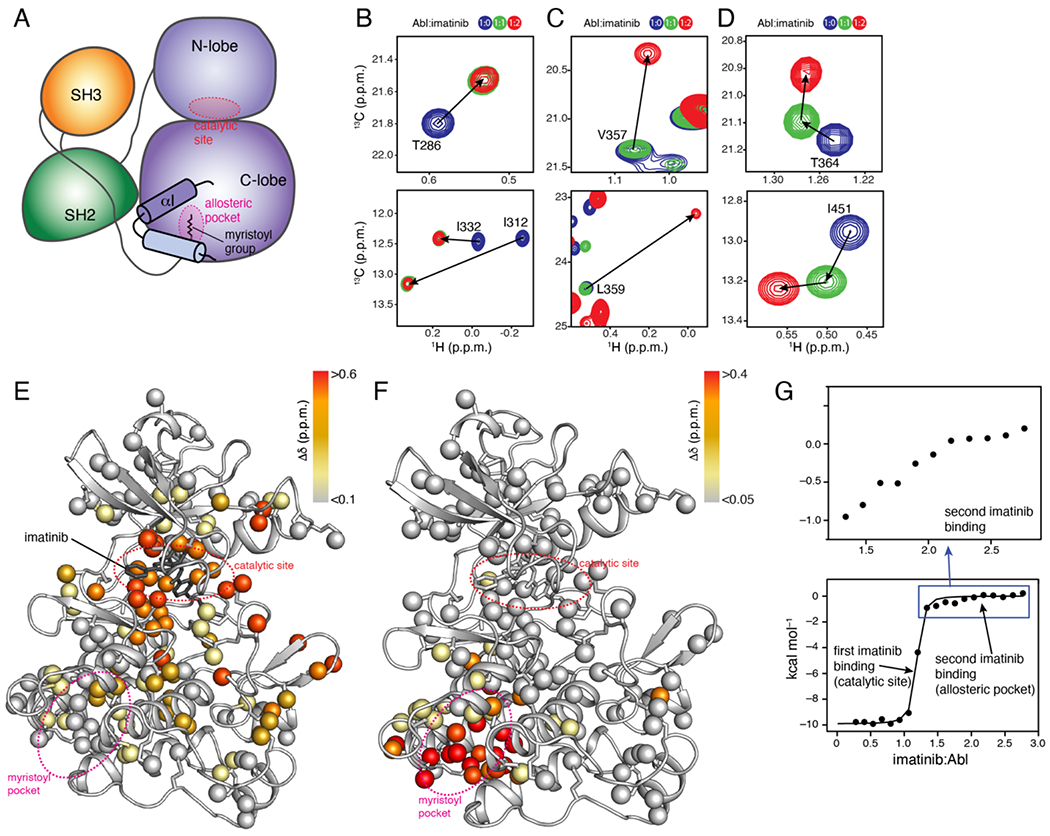
Imatinib binds to the allosteric pocket of Abl. (A) Schematic of the assembled state of Abl highlighting the kink in the αI-helix induced by the docking of the myristic moiety into the pocket. (B-D) Overlaid 1H-13C methyl HMQC NMR spectra of the Abl kinase domain (blue) bound to one (green) and two (red) equivalents of imatinib. Analysis of the NMR data revealed three classes of Abl residues with respect to their response to imatinib binding: those affected by the first imatinib equivalent, but not the second (panel B), those affected by the second imatinib equivalent but not the first (panel C), and those affected by both the first and second imatinib equivalent (panel D). (E,F) Chemical shift perturbation (Δδ) elicited by the first (panel E) and second (panel F) equivalent of imatinib mapped onto the structure of Abl (PDB 1IEP). (G) ITC traces of the titration of imatinib to Abl.
We have used NMR spectroscopy to better understand at the atomic level how imatinib interacts with Abl and imatinib-resistant Abl variants. Our structural and biochemical data show that imatinib binds to the allosteric pocket, albeit with a lower affinity than to the catalytic site and promotes the extended/active conformation of Abl thereby enhances the kinase activity.
Results
Imatinib-Abl interaction involves two binding processes. NMR-monitored stepwise titration of imatinib to isotopically labeled wild-type Abl kinase domain (hereafter Abl) revealed two distinct binding processes (Figure 1(B–D) and Figure S1(A)). The first binding process is slow on the NMR chemical shift time scale and occurs when imatinib is titrated to Abl up to an equimolar amount (Figure 1B–D, green peaks). Analysis of the chemical shift perturbation (CSP) data shows that imatinib binds to the catalytic site (Figure 1E), in agreement with the crystallographic4 and previous NMR data.24,25 The second binding process is also slow on the NMR chemical shift time scale and CSP data indicate that imatinib binds into the myristoyl pocket (Figure 1F). The NMR data show that the two binding processes are distinct, and binding takes place in a sequential manner (Figure 1D): binding to the myristoyl pocket occurs only after the catalytic site is filled with imatinib (Figure 1D). The presence of an inhibitor in the catalytic site does not appear to affect imatinib binding to the myristoyl pocket (Figure S1(B,C)). We also tested five additional ATP-competitive inhibitors of Abl (dasatinib, nilotinib, bosutinib, PD173955, and ponatinib). Ponatinib binds to the allosteric pocket, albeit weakly, whereas the other inhibitors show no binding (Figure S2).
The Abl-imatinib isothermal titration calorimetry (ITC) binding isotherm is dominated by the high affinity of the inhibitor for the catalytic site (Figure 1G and Table S2). However, a closer look at the isotherm shows the presence of a second, weaker binding process (Figure 1G). Indeed, ITC data of imatinib titrated to a sample of Abl wherein the catalytic site is already filled with imatinib reveals that a second imatinib molecule binds to the myristoyl pocket with a Kd of ~10 μM (Figure S1(D) and Table S2). Thus, the affinity of imatinib for the catalytic site is three-orders-of-magnitude higher than for the myristoyl pocket, explaining the purely sequential binding mechanism of imatinib to Abl observed by NMR (Figure 1(B–D)). Imatinib binds to the myristoyl pocket with a two-fold lower affinity than the myristic peptide (in trans), and with substantially lower affinity than the allosteric inhibitors GNF5 (Kd ~1 μM) (Figure S3(A) and Table S2) and asciminib (Kd ~1 nM15). Imatinib binds to the myristoyl pocket with a 10-100 fold higher association rate (kon ~107 M−1 s−1) than to the catalytic site25,26 but dissociates much faster (koff ~102 s−1), as measured by relaxation NMR (Figure S4(A)).
Crystal structure of Abl with imatinib bound to the allosteric pocket.
To gain insight into how imatinib interacts with the Abl myristoyl pocket, we determined the crystal structure of their complex (Table S1). A truncated Abl fragment (residues 248-518) with half of the αI helix deleted yielded high-quality crystals, whereas Abl fragments encompassing the longer (residues 248-522) or the whole αI helix (residues 248-534) yielded low quality crystals. To ensure that the allosteric pocket is saturated with imatinib, we blocked the catalytic site by adding dasatinib. The crystal structure of Abl–imatiniballost (denoting that imatinib is bound to the allosteric pocket) is shown in Figure 2. Imatinib binds to the allosteric pocket with the methyl group (C20) attached to the middle phenyl-ring inserted deeply into the pocket (Figure 2(B,C)). The same position is occupied by the N-terminal methyl group of the myristic moiety (Figure 2(D)), the CF3 group of GNF2 (Figure 2(E)), and the CClF2 group of asciminib (Figure 2(F)). The overall structure is similar to the one seen for imatinib binding to the Abl-related gene kinase.27 The part of the imatinib molecule encompassing the second phenyl and the piperazine rings has a disposition similar to the one seen in the bound myristic group, GNF2, and asciminib ligands (Figure 2(D–F)). The part of imatinib encompassing the pyrimidine and pyridine rings stretches towards the αI helix (Figure 2(B,C)). The interaction is dominated by hydrophobic contacts with only two hydrogen bonds formed between imatinib and the backbone of Ala356 and Gln517 (Figure 2(C)). The most important Abl residue for imatinib binding appears to be Leu359 as it shows the largest chemical shift change upon binding to imatinib (Figure S1(A)) and its substitution by Ala results in no detectable complex formation (Figure S5 and Table S2). Leu360, L448, L490 and M515 also appear to contribute significantly to binding (Figure S1(A),Figure S5 and Table S2). Substitution of Ala356 and Ala452 by Val, a bulkier residue, abrogates binding (Figure S5 and Table S2). The total surface area buried when imatinib is bound to the allosteric pocket amounts of ~880 Å2. This is similar or even larger than the area buried when other ligands bind to the allosteric pocket (Figure S6). The myristic moiety, for example, buries a much smaller amount of surface (~600 Å2) but binds with higher affinity than imatinib, indicating the lack of intimate contacts in the imatinib complex.
Figure 2.
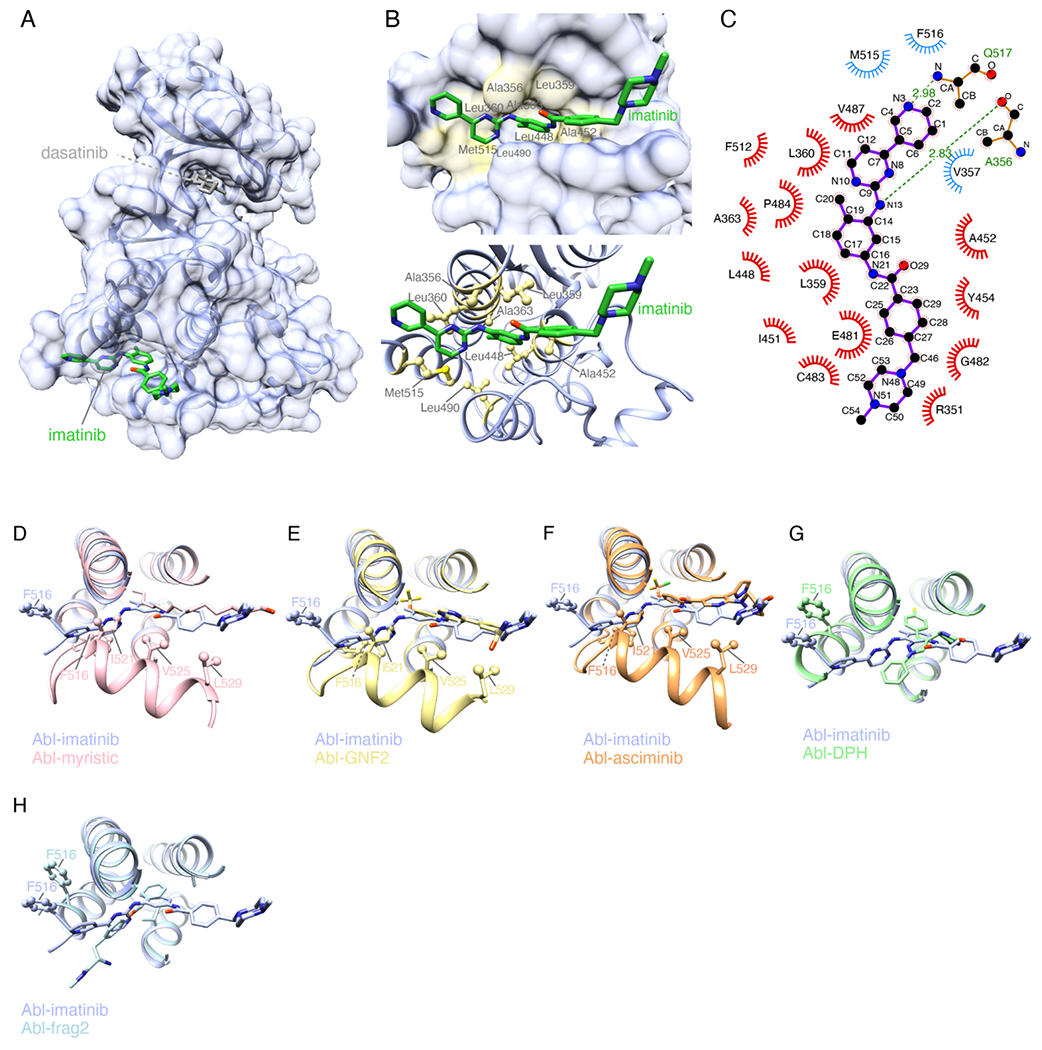
Structural basis for the binding of imatinib to the allosteric pocket. (A) Crystal structure of Abl in complex with imatinib bound to its allosteric pocket (Abl-imatiniballost). The catalytic site is bound by dasatinib. (B) Zoomed view of the allosteric pocket highlighting residues interacting with imatinib. (C) Schematic showing the inter-molecular contacts between imatinib and Abl generated by Ligplot+. (D-H) Superposition of the structure of Abl-imatiniballost with the structure of Abl bound to: myristic moiety (panel D, PDB 2fO0), GNF2 (panel E, PDB 3K5V), asciminib (panel F, PDB 5MO4), DPH (panel G, PDB 3PYY) and “frag2” (panel H, PDB 3MSS).
Superposition of the structures of the Abl–imatiniballost complex and Abl in complex with any of the ligands (myristic moiety, GNF2, and asciminib) known to induce the kink to the αI helix shows that imatinib binding is not compatible with the kinked αI helix conformation (Figure 2(D–F)). The pyridine and pyrimidine rings of imatinib sterically clash with residues Phe516 and Ile521 in the kinked αI helix conformation (Figure 2(D–F)) thereby preventing the αI helix to make the turn at Ser519. Similar steric clashes were seen in the crystal structures of the allosteric activator DPH19 and a compound referred to as frag221 bound to the Abl allosteric pocket (Figure 2(G,H)), and imatinib bound to the Abl2 allosteric site.27
The αI helix intrinsically populates an extended and a bent conformation. In the autoinhibited form of Abl19 the myristoyl group inserts into the allosteric pocket to induce a kink in the αI helix (Figure 2(D) and Figure 3(A)). The kink breaks the helix at Phe516, with residues Phe516-Ser519 forming a turn, followed by another helical region up to residue Leu529. This structural effect is also elicited by allosteric inhibitors, such as GNF216–18 (Figure 3(A)). Although the αI helix is central to the regulation of Abl activity, its preferred intrinsic conformation when the allosteric pocket is vacant is not known. In most crystal structures of Abl in the absence of a ligand in the allosteric pocket the αI helix ends at Phe516 (for example, PDB IDs 2GQG, 1FPU, 1IEP, and 3QRI) but in other structures it extends in a straight conformation by an additional 3-turns-of-helix up to residue Leu529 (for example, PDB IDs 2F4J, 3KF4, and 3OXZ). To find out the preferred conformation of the αI helix in solution, we studied by NMR the structural and dynamic properties of unliganded Abl. The NMR data are consistent with the αI helix adopting two distinct conformational states. In one of them αI adopts a straight helix throughout residue Ser522, as clearly demonstrated by the observation of characteristic NH-NH NOEs (Figure 3(B)). In the second conformation, αI likely adopts a conformation similar to the one observed in the complexes with the myristic moiety or GNF2 (Figure 3(A)), as indicated by characteristic methyl-methyl NOEs (e.g. between Ile521 and Leu360, Figure 3(C)). The intensity of the NOE between Ile521 and Leu360 is much stronger in the complex of Abl with the allosteric inhibitor GNF2, indicating a lower population of the bent conformation in unliganded Abl (Figure 3(C)). This observation is further supported by the lower order parameters (S2axis) of the α helical segment Ile521-Leu529 indicating that is more dynamic in the unliganded Abl than in its complex with GNF2 (Figure 3(D)). The sharp bend induced by the myristoyl group and allosteric inhibitors such as GNF2 also manifests in the characteristic NMR chemical shift perturbation that αI residues experience (Figure 3(E) and Figure S7(A,B)). Thus, the αI helix in Abl intrinsically adopts both an extended and a bent conformation, and both are significantly populated.
Figure 3.
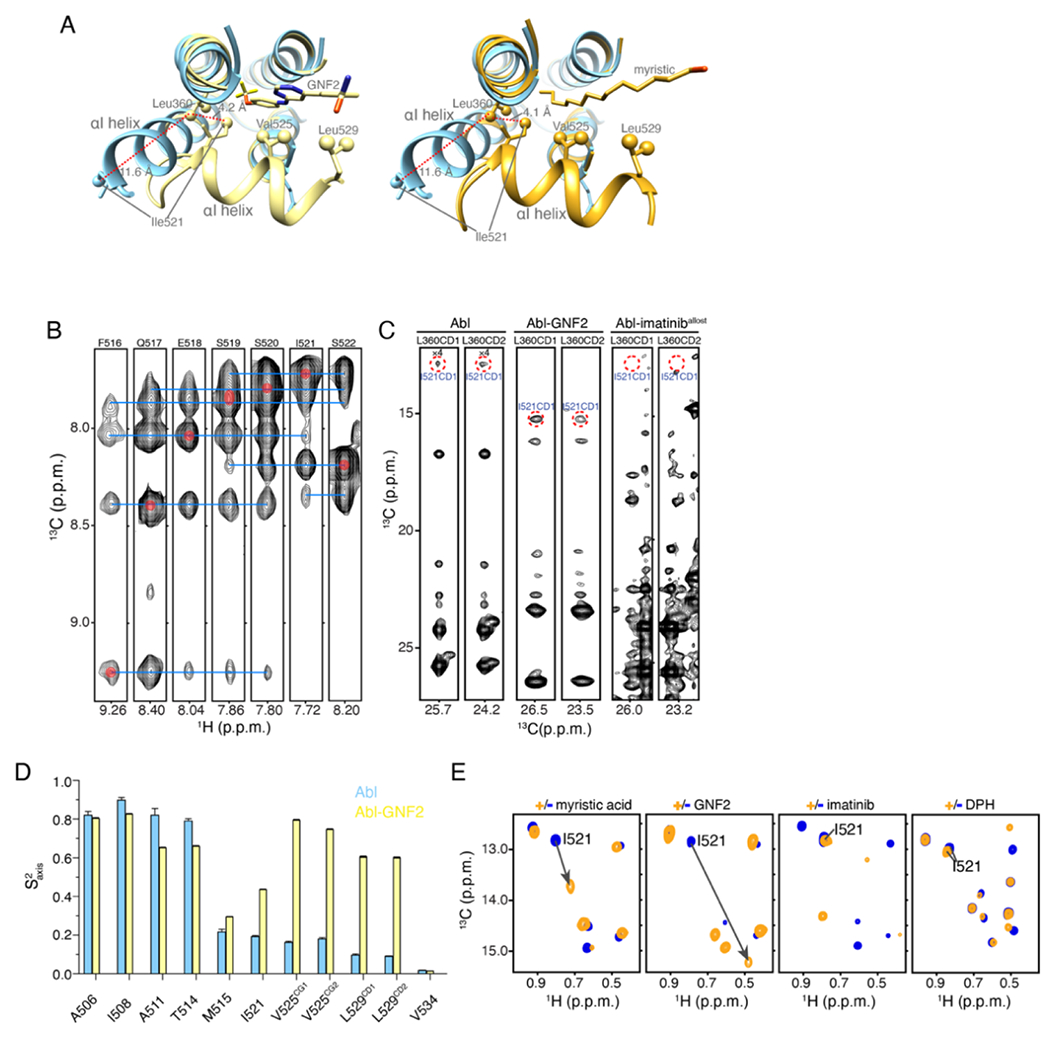
αI helix bending in Abl. (A) The structure of Abl in complex with PD173955 (blue, PDB 1M52), wherein the αI helix is not kinked, is superimposed onto the structure of Abl in complex with the allosteric inhibitor GNF2 (yellow, PDB 3K5V), which induces a kink in the αI helix (left panel) and superimposed on the structure of Abl in complex with the myristic moiety (orange, PDB 2FO0), which induces a kink in the αI-helix (right panel). (B) Strips from 3D 15N-edited 1H-1H NOESY spectra showing HN-HN NOEs that are characteristic of a helix formation. (C) Strips from 3D 13C-edited 1H-1H NOESY spectra showing the characteristic NOE between the methyl group of Leu360 and Ile521. These two residues are nearby in space in the bent conformation of the I-helix but remote in the extended αI helix conformation (panel A). (D) Methyl order parameters (S2axis) of the αI helix methyl-bearing residues in the unliganded Abl and in complex with GNF2. (E) 1H-13C methyl HMQC spectra of Abl in complex with the myristic peptide, GNF2, imatinib, and DPH. The resonance of Ile521, whose chemical shift is characteristic of the conformation of the αI helix is shown. The myristic peptide and GNF2 induce the kink to the αI helix, whereas imatinib and DPH prevent the αI helix from bending.
Notably, binding of imatinib to the myristoyl pocket prevents the αI helix from forming the bent conformation, a finding supported by the quench of the NOE between Ile521 and Leu360 (Figure 3(C)) and chemical shift analysis (Figure 3(E) and Figure S7(A,B)). Ile521, Val525 and Leu529 are located in the αI helix and are directly involved in the interaction with the myristic moiety or allosteric inhibitors as indicated by the observed major chemical shift perturbation (Figure 3(E) and Figure S7(A,B)). In comparison, these residues are much less affected by imatinib or DPH (Figure 3(E)), suggesting that the αI helix upon imatinib binding adopts a similar conformation to that bound to allosteric activator DPH.
Imatinib allosterically promotes the extended/activated conformation.
Given that the bent conformation of the αI helix is required for Abl to adopt the assembled, autoinhibited conformational state, binding of imatinib to the myristoyl pocket could in fact destabilize the autoinhibited state thereby stimulating Abl activity. To test this intriguing hypothesis, we sought to assess the effect of imatinib on the conformational ensemble of an Abl fragment that encompasses the SH3-SH2 module and the kinase domain, which, for simplicity, we refer to as the full-length Abl kinase (AblFK). Abl interconverts rapidly between two distinct conformational states: the assembled/autoinhibited state, wherein the kinase activity is suppressed, and the extended/activated state (Figure 4(A)) that is the most active form of Abl and has been associated with increased leukemogenic activity28. We recently showed that NMR can be used to measure the populations of the inhibited vs. activated state of Abl17. Specifically, the resonance of Met263, located at the interface between the N lobe and the SH2, provides a sensitive and quantitative probe of the populations of the two states20 (Figure 4(A,B)).
Figure 4.
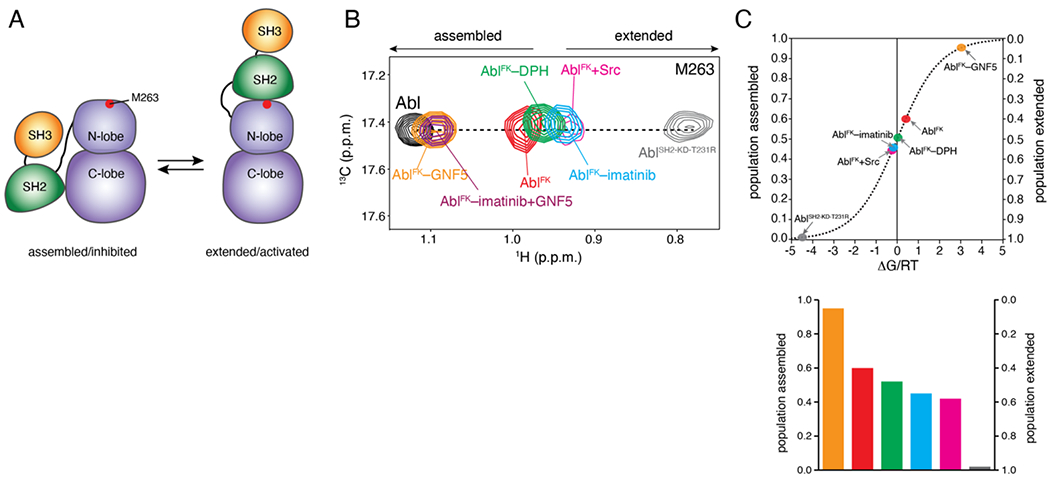
Allosteric activation of Abl by imatinib. (A) Schematic showing the transition of Abl between the assembled and the extended conformational states. The position of M263 is shown as a red circle. (B) Overlaid 1H-13C methyl HMQC NMR spectra of the indicated Abl variants, showing the M263 residue. M263 is located at the interface between the SH2 and the kinase domain and provides the most sensitive probe for determining the populations of the two states in the variants, as shown previously [17]. (C) Populations of the assembled and extended states for Abl variants, determined by NMR from the spectra shown in panel B. The populations are plotted as a function of the associated free energy, ΔG/RT, where R is the gas constant, T is the temperature, and ΔG is given as GE-GA. A 0.6 kcal mol−1 change in ΔG corresponds to a change by 1 unit in ΔG/RT at room temperature. As variants approach the free-energy degeneracy (ΔG/RT=0), small changes in energy result in substantial changes in the populations.
The largest population of the extended state is achieved by the AblSH2-KD-T231R variant (AblSH2-KD-T231R), which also exhibits the highest kinase activity20 (Figure 4(B,C)). On the other hand, the largest population of the assembled state is reached by the addition of the allosteric inhibitor GNF5, which induces the bent conformation of the αI helix thereby promoting the assembled state (Figure 4(B,C)). AblFK prefers the inhibited state (~60% population). Interestingly, binding of imatinib to the allosteric pocket shifts the equilibrium in favor of the activated state (Figure 4(B,C)). This finding suggests that imatinib acts as an allosteric activator of Abl by promoting the activated state of Abl. Addition of GNF5 displaces imatinib from the myristoyl pocket and shifts the population almost fully towards the inhibited state (Figure 4(B)). In fact, imatinib appears to be a more potent allosteric activator than DPH (Figure 4(C)), a small molecule that was found to bind to the allosteric pocket and to increase markedly the kinase activity of Abl by preventing the kinked conformation of αI helix22. Abl phosphorylation by Src kinase is known to activate Abl29 and we showed previously that the underlying mechanism involves a shift of the population towards the activated state20. Of note, our data now demonstrate that allosteric activation of Abl by imatinib is as efficient as Src-mediated phosphorylation, with the two processes inducing a very similar population of the activated state (Figure 4(C)).
The allosteric pocket competes with the ATP-binding site for imatinib. The affinity of imatinib for the myristoyl pocket (Kd ~10 μM) is much weaker than its affinity for the catalytic site (Kd ~10 nM). The so-called gatekeeper mutation (T334I) reduces the affinity of imatinib for the catalytic site by several orders of magnitude (Kd ~4 μM) and combined with other imatinib-resistant mutations that often occur in patients (e.g. G269E and Y272H) they further weaken imatinib binding to the catalytic site (Kd ~40 μM) (Figure S8). Notably, stepwise titration of imatinib to AblT334I/G269E shows that the allosteric pocket becomes saturated with imatinib before the catalytic site (Figure 5(A)). This produces a small but significant population (~10%) of AblT334I/G269E wherein only the allosteric pocket is bound with imatinib whereas the catalytic site is vacant. This population will be associated with higher kinase activity because of the promotion of the activated state by the allosteric effect of imatinib (Figure 4(C)).
Figure 5.
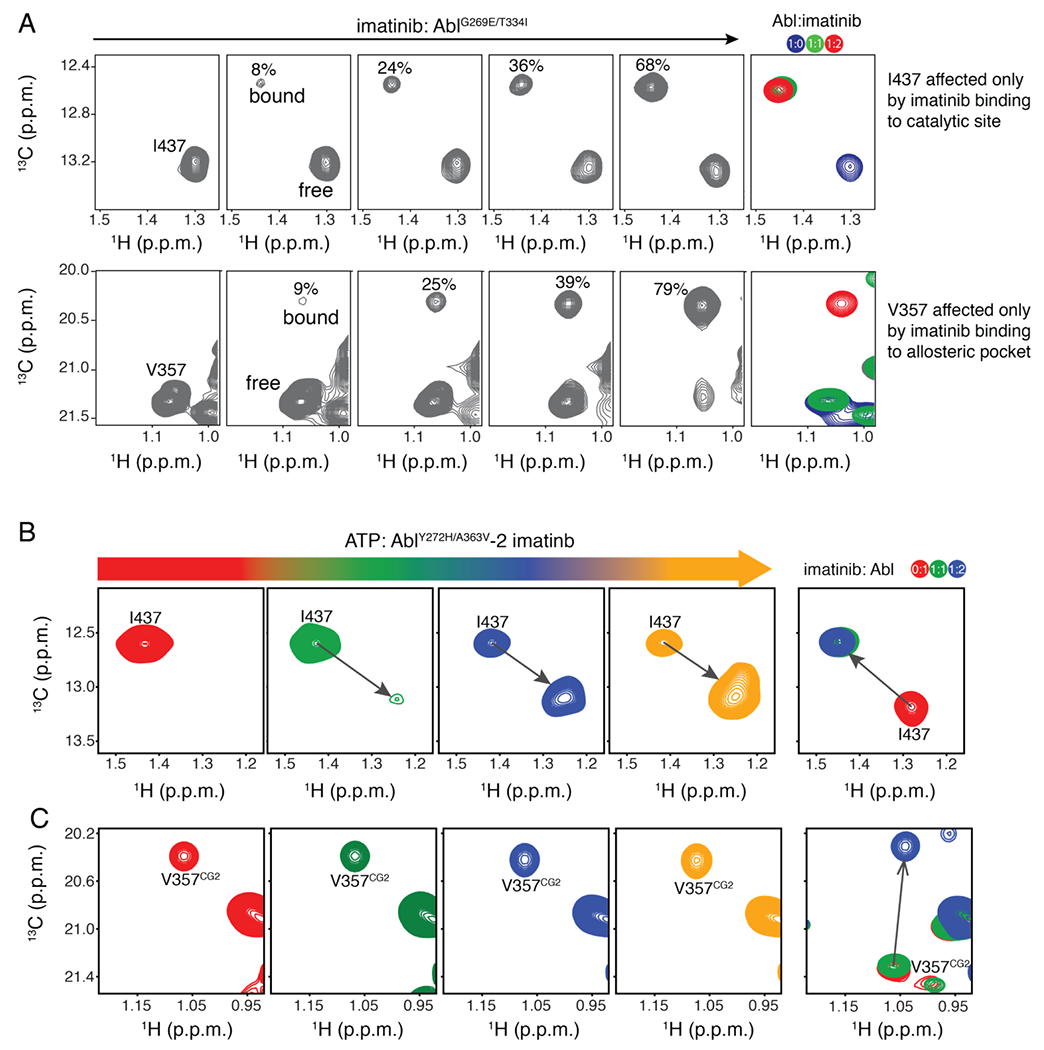
Competition of the allosteric and catalytic sites for imatinib. (A) Stepwise titration of imatinib to the imatinib-resistance AblG269E/T334I variant monitored by NMR. (B,C) Stepwise titration of ATP to the AblY272H/A363V variant occupied with imatinib at both the catalytic and allosteric sites monitored by NMR.
A363V, an imatinib-resistance mutation that is seen in patients30,31, is located inside the myristoyl pocket (Figure 2(B)). Interestingly, whereas the A363V substitution has no effect on the affinity of imatinib for the catalytic site (Kd ~10 nM; Figure S8) and on the equilibration between extended and assembled conformations (Figure S9), the substitution increases the affinity of imatinib for the myristoyl pocket by a factor of two (Figure S5). Therefore, it appears that this mutation exerts its function by enhancing the “activating” effect of the allosteric pocket-binding imatinib molecule and may serve as a hitherto unknown mechanism of developing resistance to imatinib.
Standard treatment of CML by daily imatinib dose of 400 mg yields an average steady-state plasma concentration of ~3.4 μM.32,33 Under these conditions, a substantial fraction of Bcr-Abl is expected to have imatinib bound to its allosteric pocket. The problem can become even more serious when higher doses of imatinib are administered, as is the case in patients who have developed imatinib resistance.2
The problem is further compounded by the competition of ATP and imatinib for the catalytic site in Abl carrying imatinib-resistance mutations. Stepwise titration of ATP to AblY272H/A363V saturated by imatinib at both the catalytic site and the allosteric pocket clearly shows that ATP outcompetes imatinib for the catalytic site (Figure 5(B)). By contrast, there is no competition between ATP and imatinib for the allosteric pocket since ATP does not bind to it (Figure 5(C)). Imatinib binds to the allosteric site about 1000-fold weaker than to the catalytic site. Given that imatinib needs to compete with high-concentration of ATP at the physiological condition for the catalytic site, the apparent difference of binding affinity between the two sites is much smaller than 1000-fold. For imatinib-resistant mutants, the occupancy of imatinib at the allosteric site can be comparable to or even bigger than that at the catalytic site. Hence imatinib binding to the allosteric pocket magnifies the resistance of these mutants to imatinib. In other words, imatinib-resistant mutants appear to be more severe than they are if imatinib doesn’t bind to the allosteric site.
Imatinib binding to myristoyl pocket activates Abl.
To assess the effect of imatinib binding to the allosteric pocket on the kinase activity, we measured the phosphorylation activity of Abl, AblY272H/L359A, and AblY272H/A363V at a concentration of 2 mM ATP (Figure 6A). We selected L359A and A363V because the first mutant decreases whereas the latter mutant increases the affinity of imatinib for the allosteric pocket (Figure S5). As expected, imatinib inhibits the catalytic activity of wild-type Abl (Figure 6A). Because the Y272H mutation increases markedly the IC50 of imatinib for the catalytic pocket,34 the drug has a weaker inhibiting effect (Figure 6A.). Interestingly, the kinase activity of AblY272H/A363V is significantly higher than the activity of AblY272H/L359A in the presence of imatinib (Figure 6A), indicating that imatinib binding to the allosteric pocket may activate Abl. The kinase activity of AblY272H/A363V in the presence of both imatinib and GNF5 is strongly suppressed (Figure 6A) due to the much stronger binding of GNF5 into the allosteric pocket (Figure S3(A)). To evaluate the effect of imatinib binding to the allosteric pocket on the kinase activity with less interference of imatinib binding to the ATP-site, we further compared the phosphorylation activity of AblY272H/L359A AblY272H/A363V in the presence of a higher concentration (20 mM) of ATP (Figure 6B). Importantly, imatinib binding to AblY272H/A363V significantly increases the catalytic activity of Abl (Figure 6B), further confirming that imatinib binding to the allosteric pocket can activate Abl. These findings suggest that designing an imatinib analog that doesn’t interact with the allosteric pocket could improve its overall inhibitory effect.
Figure 6.
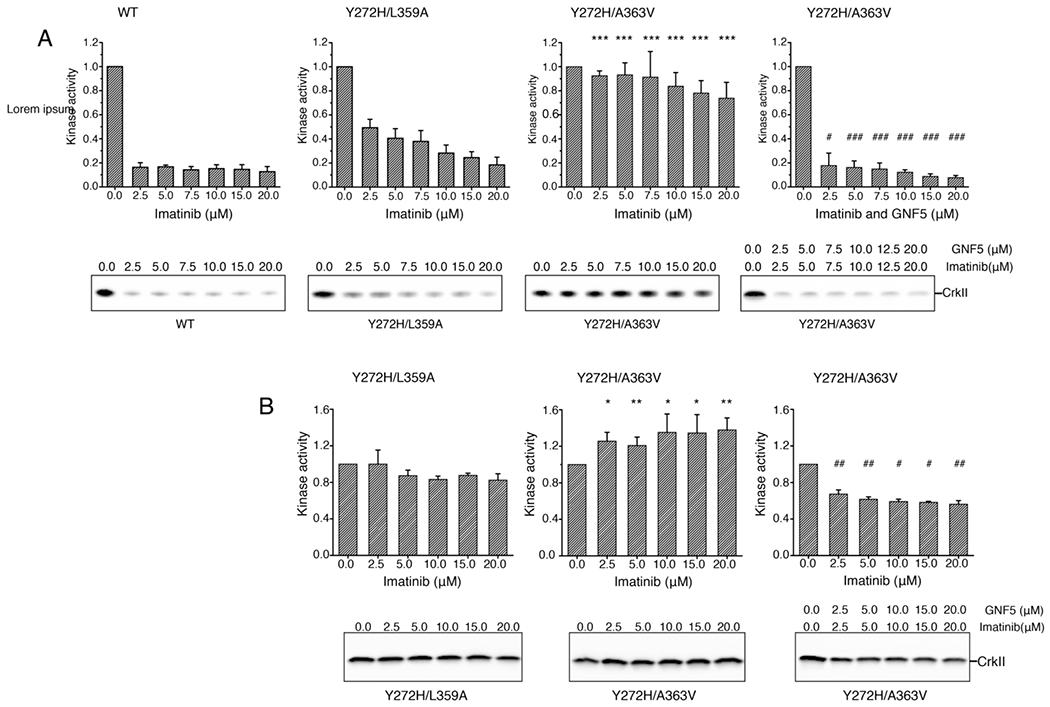
Effect of imatinib on kinase activity of AblFK and variants measured by monitoring the phosphorylation of CrkII at low (A) and high (B) concentration of ATP. Quantification of the kinase activity and representative gels are shown from at least three measurements except Y272H/A363V in the presence of both imatinib and GNF5 in panel (A) which were measured two times. The significance of differences of kinase activity between Abl variants was estimated using two-sample t-test assuming unequal variances. The kinase activity of Y272H/A363V was hypothesized to be greater than that of Y272H/L359A in the presence of imatinib, and the kinase activity of Y272H/A363V in the presence of both imatinib and GNF5 was hypothesized to be lower than that in the presence of only imatinib. *: P<0.05 vs Y272H/L359A, **: P<0.01 vs Y272H/L359A, ***: P<0.001 vs Y272H/L359A; #: P<0.05 vs Y272H/A363V, **: P<0.01 vs Y272H/A363V, ***: P<0.001 vs Y272H/A363V. The differences are significant.
An NMR method to differentiate Abl allosteric inhibitors from activators.
Small molecules that bind to the allosteric pocket can either act as inhibitors or activators, depending on whether they stabilize the bent or the extended helical form of αI helix.21 The chemical shifts of residues Ile521, Val525 and Leu529 are sensitive to the conformation of the αI helix and they are affected in a distinct way by the binding of an activator or an inhibitor (Figure 3E and Figure S7). Thus, monitoring the methyl chemical shifts of these residues presents a convenient way to screen the binding of small molecules to the allosteric pocket and assess their inhibitory or activating effect (Figure 7). Among these allosteric binders, the ones that induce major chemical shift changes to any of these three residues function as allosteric inhibitors, and those leading to no or minor chemical shift perturbations can be identified as activators (Figure 7).
Figure 7.
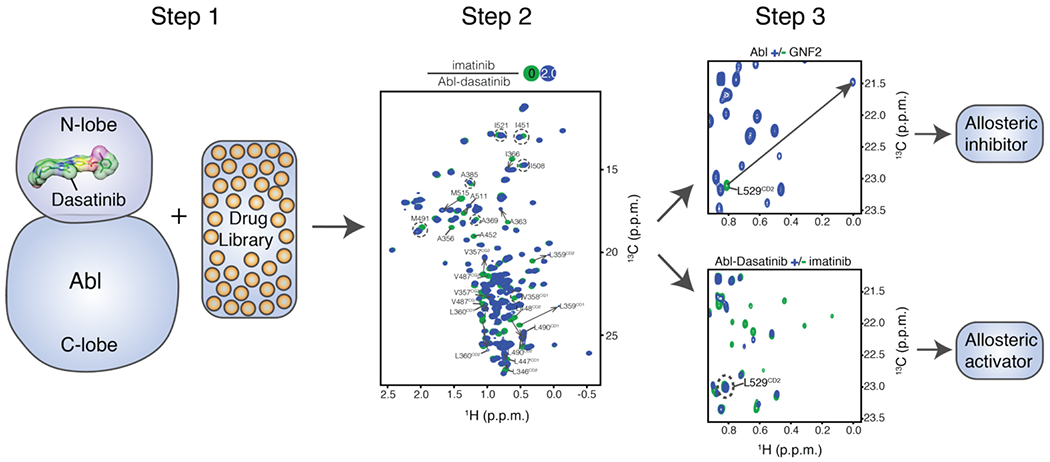
An NMR method to differentiate allosteric inhibitors from activators. The ATP site of [U-2H; Ala-13CH3; Met-13CH3; Ile-δ1-13CH3; Leu, Val-13CH3/13CD3]-labelled Abl kinase domain is occupied by ATP-competitive inhibitors such as dasatinib or nilotinib which bind to the ATP-site with much higher affinity than that of imatinib. Small molecule binders of the myristoyl pocket can be identified based on the chemical shift perturbations. Among these allosteric binders, the ones that induced major chemical shift changes to any one of residues Ile521, Val525 and Leu529 serve as allosteric inhibitors, and those causing no or minor chemical shift perturbations can be identified as activators.
DISCUSSION
The unexpected allosteric activating function of imatinib can exert a dual negative effect on inhibition of Abl kinase. First, the allosteric pocket competes with the catalytic site for imatinib, thereby decreasing the available amount of imatinib; second, the extended Abl conformation promoted by imatinib binding to the allosteric pocket has an increased kinase activity. Although imatinib binds to the catalytic site with much higher affinity than to the allosteric pocket, the two sites have comparable affinities for imatinib in Bcr-Abl variants occurring in patients that develop resistance to imatinib. Given the competition of imatinib and the cellular ATP for the catalytic site, the activating effect of imatinib binding to the allosteric pocket is even more amplified. Moreover, promotion of the extended Abl conformation by imatinib binding to the allosteric pocket shifts the population of the kinase domain from the inactive state to the active one.3 The active state is not compatible with imatinib binding to the catalytic site, which further reduces its affinity.3 Our findings are in line with clinical results showing that high imatinib doses do not provide improved long-term outcomes, despite better early responses.13–15 It is of interest that previously isolated imatinib-resistant mutations located in the allosteric pocket (e.g. A363V) appear to be mediating their effect by increasing the affinity of imatinib for the allosteric pocket. It should be noted that Bcr-Abl’s oncogenicity may be mediated, in addition to its stimulated catalytic activity, by the interaction of its SH2 and SH3 domains with a number of signaling proteins.35 Such an activating mechanism is independent of the catalytic activity of the kinase and is stimulated by the transition of Bcr-Abl from the assembled to the extended state. Our data highlight the benefit of combining competitive with allosteric inhibitors18 to maximize their inhibitory effect on Bcr-Abl.
It was previously reported that imatinib promotes the Abl extended/active state by binding to the catalytic pocket36,37. The authors argue that the extended/active state is stabilized by the closed activation loop elicited by imatinib binding to the catalytic pocket, and the assembled/inactive state is promoted by the open activation loop. This is not in agreement with recent structural findings on inhibitor-free Abl that clearly showed the closed activation loop being favored in the assembled state of Abl.3 In our previous work we did not find any evidence that imatinib binding to the catalytic pocket shifts the equilibrium towards the extended/activated state.3,20 It is possible that the authors, unbeknown to them, captured the effect of imatinib binding to the allosteric pocket. It is a common practice in NMR experiments to add excess ligand to ensure saturation. Given the sample concentration (150-200 μM) used in their NMR experiment, it is likely that in their experimental setup the allosteric pocket is bound by imatinib. Unfortunately, the authors do not mention the Abl:imatinib ratio used in their experiments and thus we cannot conclude if the allosteric pocket is indeed occupied by imatinib.
Materials and methods
Protein production.
The coding sequences of human Abl and its variants were cloned into the pET16b vector for expression as maltose-binding-protein (MBP)-His6 fusion proteins with a tobacco etch virus (TEV) protease cleavage site at the N-terminus. Abl mutants were generated using QuikChange site-directed mutagenesis (Agilent) and their sequences were confirmed by DNA sequencing. Abl constructs were expressed and purified as described previously.3, 20,38 Unlabeled proteins were grown at 37 °C in Luria-Bertani (LB) broth. When the A600nm reached ~0.7-0.8, protein expression was induced at 16 °C by adding 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and the cells were harvested ~48 hours thereafter. Cell pellets were resuspended in 50 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl, 5 mM β-mercaptoethanol and lysed by sonication followed by centrifugation at 48,000 x g for 30 min. Proteins were purified by Ni-NTA agarose resin (GE Healthcare) followed by incubation with TEV protease at 4 °C overnight The cleaved proteins were passed through Ni-NTA agarose resin again to remove protease and His tag followed by size exclusion chromatography purification using Superdex 75 16/60 or 200 16/60 columns (GE Healthcare) pre-equilibrated with 25 mM sodium phosphate buffer (pH 7.1) containing 75 mM NaCl, 2.0 mM β-mercaptoethanol. Protein concentration was determined spectrophotometrically at 280 nm using the corresponding extinction coefficient. CrkII was cloned into the pET42a vector, and expression and purification were performed as previously described.38,39.
Inhibitors, myristic peptide and ATP.
All small molecule inhibitors and activators used in this study were bought from Selleck Chemicals, except DPH which was bought from Sigma-Aldrich (SML0202). The myristic peptide (Mry-GQQPGKVLGDQRRPSL) was synthesized by GeneScript. ATP was bought from Sigma-Aldrich (A7699).
ITC experiments.
ITC experiments were performed on MicroCal iTC200 or Auto-iTC200 calorimeters (Malvern Instruments Inc.). ITC titrations for each protein-drug pair were typically performed at 25 °C in 25 mM sodium phosphate buffer (pH 7.1) including 75 mM NaCl, and 2.0 mM tris (2-carboxyethyl)phosphine (TCEP). Proteins were purified by size exclusion chromatography using corresponding ITC buffers before use. Concentrations of protein and small molecules were measured spectrophotometrically and by weight, respectively. Proteins or protein-drug complexes were placed in the cell, while the drugs or myristic peptide were in the syringe with concentrations of 7 to 100 μM and 70 to 2000 μM respectively. Binding curves were fitted using Origin 7.0 (OriginLab Corporation).
Isotope labeling for NMR studies.
Isotopically labeled samples for NMR studies were produced by growing the cells in M9 minimal medium. U-[2H,13C,15N]-labeled samples were generated for the backbone assignment by supplementing the growing medium with 15NH4Cl (1g/L), 2H7, 13C6-glucose (2g/L) in 99.9% 2H2O (Cambridge Isotope Laboratories or Isotec). The methyl-protonated samples in an otherwise deuterated background were prepared as described40–42 by adding 50 mg/L α-ketobutyric acid, 90 mg/L α-ketoisovaleric acid, 50 mg/L of 13CH3-Met, 50 mg/L 2H2, 13CH3-Ala, and 50 mg/L 2H2,13CH3-Thr to the cultures one hour before IPTG induction. The NMR samples of apo Abl kinase domain for backbone and methyl assignments were typically prepared in 25 mM sodium phosphate buffer (pH 7.1) including 75 mM NaCl, 2.0 mM β-mercaptoethanol, in 8% 2H2O. [U-2H; Ala-13CH3; Met-13CH3; Ile-δ1-13CH3; Thr-13CH3; Leu,Val-13CH3/12CD3]-labeled Abl samples were titrated with increasing amount of inhibitors and/or the myristic peptide. NMR titration experiments were run at 10 °C in 25 mM sodium phosphate buffer (pH 7.1), 75 mM NaCl, 2.0 mM β-mercaptoethanol, and 100% 2H2O with typical protein concentration of 0.05-0.1 mM.
NMR Spectroscopy.
NMR experiments were acquired on Bruker 850, 800, 700 and 600 MHz spectrometers equipped with cryogenic probes at 10 °C, 20 °C or 25 °C. All NMR data were processed using NMRPipe43 and analyzed using NMRFAM-Sparky44. Backbone 1H, 15N, and 13C resonance assignment was achieved using 3D HNCACB, CBCA(CO)NH, HNCA, and HNCO experiments. Sidechain methyls were assigned by analyzing 3D-13C,15N-NOESY-HMQC and 13C-HMQC-NOESY-HMQC spectra with a mixing time of 300 ms.45,46
Crystallization, data collection, structure determination and model quality.
Protein-inhibitor co-crystals were grown by the sitting-drop vapor diffusion method at 4 °C. Prior to setup, dasatinib and imatinib were incubated with Abl248-518 at a 1:1:1 molar ratio, with each at a concentration of 450 μM. The 500 μl well solution contained 0.36 M Ammonium dihydrogen phosphate. The drop contained 2 μl of well solution and 3 μl of protein-inhibitor mixture (10 mM HEPES, pH 7.1, 75 mM NaCl and 3 mM BME). For cryo-preservation, crystals were soaked in crystallization buffer containing 30 % glycerol (v/v) prior to flash-cooling in liquid nitrogen. The cryo-solution also contained 5.7 mM of imatinib to ensure good occupancy of the ligand.
Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) Sector 22-BM beamline at 1 Å. Data were integrated and scaled using XDS47 and Aimless48 respectively, to 2.2 Å. The structure was solved by molecular replacement with Phaser49 using Abl kinase domain (PDB 2GQG) as the search model with ligands removed. Iterative rounds of model building and refinement were performed with COOT50 and Refmac51, respectively. Data collection and refinement statistics are summarized in Table S1. The final model is of high quality, with three Abl248-518 protomers in the I 222 cell. All protomers form a 1:1:1 complex with Dasatinib and Imatinib. Representative electron density is provided in Figure S10. Due to insufficient electron density, the N-lobe of protomer C is largely modeled as poly-alanine. Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 7N9G.
CEST experiment.
13CHD2-CEST experiments52,53 for measuring kinetics of imatinib binding to the myristoyl pocket were performed on a Bruker 850 MHz spectrometer on a 0.3 mM sample of [U-2H; Leu,Val-13CHD2/13CHD2]-labeled Abl filled with dasatinib in the catalytic site and in the presence of ~15% imatinib at 10 °C. 114 spectra in a pseudo-3D mode were recorded with a mixing time (Tmix) of 450 ms and a weak B1 radiofrequency field of 14 Hz was applied to 17.9-27.5ppm in an 18 Hz step. A recycle time of 2.1 s was used. CEST profiles were made by plotting the intensity ratios with and without the Tmix period. CEST data were fit to a two-state exchange model using program Chemex (https://github.com/gbouvignies/chemex) to get kex and the population of Abl in the free and complex with imatinib. Errors in peak intensity for Chemex input were measured from the background noise of spectra. CEST was used to determine the kinetics of imatinib binding to the myristoyl pocket. The exchange rate constant kex is given by kex=kon[imatinib]+koff, where [imatinib] is the concentration of the unbound imatinib. The kon and koff values were calculated based on kd measured from ITC and kex and population of the complex were obtained from fitting of CEST.
Ps-ns Dynamics of the αI helix.
We performed 1H triple-quantum coherence transfer experiment54,55 to measure the squared order parameter of the sidechain methyl groups (S2axis) of the αI-helix on apo Abl and in complex with GNF2. The experiments were run at 10 °C in 25 mM sodium phosphate buffer (pH 7.1), 75 mM NaCl, 2.0 mM β-mercaptoethanol and 100% 2H2O with protein concentration of about 0.2-0.25 mM. S2axis values were obtained by measuring the cross-correlated relaxation rate (η) between pairs of 1H-1H vectors in methyls using the following equations:54–55
| (1) |
Where RF2,H and RS2,H are the fast and slow relaxation rates of each of the single-quantum 1H transitions respectively, τc is the global tumbling time of protein molecule, μ0 is the vacuum permittivity constant, γH is the gyromagnetic ratio of proton, rHH is the distance between pairs of methyl protons (1.813 Å), P2(x)=(1/2)(3x2-1), and θaxis,HH (90°) is the angle between the methyl 3-fold axis and a vector connecting a pair of methyl 1H nuclei. The time dependencies of peak intensity in the measurements of cross-correlated relaxation rates were monitored with relaxation delays of 1, 1.7, 2.5, 3.5, 4.5, 6, 8, 10, 12, and 14 ms for each pair of allowed single-quantum and forbidden triple-quantum datasets. Values of η were calculated by fitting ratios of peak intensities measured in pairs recorded as a function of relaxation time (T) to the following equation:
| (2) |
Where Ia and Ib are peak intensities in triple-quantum and single-quantum spectra respectively, T is parametrically varied delays, and value of δ depends on the 1H spin density around the methyl group. τc value of 45 ns was used to calculate S2axis so that all S2axis values are within 0-1. In order to reduce the proton density, two methyl protonated samples were generated: one was [U-2H; Leu,Val-13CH3/12CD3] labeled and the second was [U-2H; Ala-13CH3; Met-13CH3; Ile-δ1-13CH3, and Thr-13CH3] labeled.
ATP titration.
Increasing amount of ATP and MgCl2 was added to 80 M [U-2H; Ala-13CH3; Met-13CH3; Ile-δ1-13CH3; Thr-13CH3; Leu,Val-13CH3/12CD3]-labeled AblY272H/A363V bound to imatinib at both ATP-binding and myristoyl sites. NMR titration experiments were acquired at 10° in 10 mM Tris-HCl buffer (pH 7.4) containing 75 mM KCl, 2.0 mM β-mercaptoethanol, and 8% 2H2O. The stoichiometry of ATP: MgCl2 was kept at ~1:2 during the titration.
Kinase assays.
The kinase assays were conducted in 50 mM Tris (pH 7.5), 5 mM MgCl2, 100 mM KCl, and 3.0 mM BME at room temperature with CrkII as substrate. 10 μM CrkII was incubated with 0.2 μM Abl and mutants in the presence of Imatinib of different concentration in the reaction buffer, with the addition of ATP to a final concentration of 2 or 20 mM to initiate the reaction. Reactions were stopped at 60 sec by adding SDS-containing loading buffer. Proteins were resolved in a 4-20% SDS-PAGE gel (Bio-Rad Laboratories) and probed by western blot with antibody to CrkII pTyr221 (3491S, Cell Signaling Technology). All blots were loaded with SuperSignalTM West Pico Plus Chemiluminescent Substrate (34577, Thermo Scientific), visualized using an Amersham Imager 600 (GE Life Sciences) and quantified with ImageJ. Activity of Abl proteins on CrkII is normalized as percentage of activity at 0 μM imatinib for each time point.
Statistical analysis of kinase assasy.
The significance of kinase activity difference between Abl variants was estimated using two-sample one-tailed t-test assuming unequal variances. The kinase activity of Y272H/A363V was hypothesized to be greater than that of Y272H/L359A in the presence of imatinib, and the kinase activity of Y272H/A363V in the presence of both imatinib and GNF5 was hypothesized to be lower than that in the presence of only imatinib.
Supplementary Material
Highlights.
Imatinib binds to the allosteric pocket of Abl kinase
Interaction of imatinib with the allosteric site promotes the Abl extended/activated conformation
Binding of imatinib to the allosteric pocket activates Abl
Imatinib binding to the allosteric pocket enhances drug resistance of catalytic site mutations
Acknowledgments
We thank S. Vaithiyalingam for help with some of the ITC experiments. This work was supported by U.S. National Institutes of Health grant R35 GM122462 (C.G.K.) and ALSAC. NMR spectra were acquired at the St. Jude Biomolecular NMR Spectroscopy Center. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions (see www.ser-cat.org/members.html), and equipment grants (S10_RR25528 and S10_RR028976) from the National Institutes of Health. We thank the St. Jude Structural Biology X-ray Center for crystallography support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
PDB ID: 7N9G
CRediT author statement
Tao Xie: Conceptualization, Methodology, Investigation, Writing-Original Draft, Visualization. Tamjeed Saleh: Methodology, Investigation. Paolo Rossi: Methodology, Investigation: Darcie Miller: Methodology, Investigation. Charalampos Kalodimos: Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Rosti G, Castagnetti F, Gugliotta G, Baccarani M, (2017). Tyrosine kinase inhibitors in chronic myeloid leukaemia: which, when, for whom? Nat. Rev. Clin. Oncol, 14, 141–54. [DOI] [PubMed] [Google Scholar]
- 2.Braun TP, Eide CA, Druker BJ, (2020). Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell, 37, 530–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie T, Saleh T, Rossi P, Kalodimos CG, (2020). Conformational states dynamically populated by a kinase determine its function. Science, 370, eabc2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, et al. , (2002). Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res, 62, 4236–43. [PubMed] [Google Scholar]
- 5.Deininger M, Buchdunger E, Druker BJ, (2005). The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood, 105,2640–53. [DOI] [PubMed] [Google Scholar]
- 6.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. , (2001). Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science, 293, 876–80. [DOI] [PubMed] [Google Scholar]
- 7.Soverini S, De Benedittis C, Papayannidis C, Paolini S, Venturi C, Iacobucci I, et al. , (2014). Drug resistance and BCR-ABL kinase domain mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia from the imatinib to the second-generation tyrosine kinase inhibitor era: The main changes are in the type of mutations, but not in the frequency of mutation involvement. Cancer, 120,1002–9. [DOI] [PubMed] [Google Scholar]
- 8.Talati C, Pinilla-Ibarz J, (2018). Resistance in chronic myeloid leukemia: definitions and novel therapeutic agents. Curr. Opin. Hematol, 25:154–61. [DOI] [PubMed] [Google Scholar]
- 9.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. , (2009). AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer cell, 16, 401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pemovska T, Johnson E, Kontro M, Repasky GA, Chen J, Wells P, et al. ,(2015). Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature, 519,102–5. [DOI] [PubMed] [Google Scholar]
- 11.Breccia M, Pregno P, Spallarossa P, Arboscello E, Ciceri F, Giorgi M, et al. , (2017). Identification, prevention and management of cardiovascular risk in chronic myeloid leukaemia patients candidate to ponatinib: an expert opinion. Ann. Hematol, 96, 549–58. [DOI] [PubMed] [Google Scholar]
- 12.Anagnostou T, Litzow MR, (2018). Spotlight on ponatinib in the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia: patient selection and perspectives. Blood Lymphat Cancer, 8, 1–9. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baccarani M, Druker BJ, Branford S, Kim DW, Pane F, Mongay L, et al. , (2014). Long-term response to imatinib is not affected by the initial dose in patients with Philadelphia chromosome-positive chronic myeloid leukemia in chronic phase: final update from the Tyrosine Kinase Inhibitor Optimization and Selectivity (TOPS) study. Int. J. Hematol, 99, 616–24 [DOI] [PubMed] [Google Scholar]
- 14.Deininger MW, Kopecky KJ, Radich JP, Kamel-Reid S, Stock W, Paietta E, et al. , (2014). Imatinib 800 mg daily induces deeper molecular responses than imatinib 400 mg daily: results of SWOG S0325, an intergroup randomized PHASE II trial in newly diagnosed chronic phase chronic myeloid leukaemia. Br. J. Haematol, 164, 223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hehlmann R, Lauseker M, Saussele S, Pfirrmann M, Krause S, Kolb HJ, et al. , (2017). Assessment of imatinib as first-line treatment of chronic myeloid leukemia: 10-year survival results of the randomized CML study IV and impact of non-CML determinants. Leukemia, 31, 2398–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adrián FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, et al. , (2006). Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat. Chem. Biol, 2, 95–102. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. , (2010). Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature, 463, 501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, et al. , (2017). The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature, 543, 733–7. [DOI] [PubMed] [Google Scholar]
- 19.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, et al. , (2003). Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell, 112, 859–71. [DOI] [PubMed] [Google Scholar]
- 20.Saleh T, Rossi P, Kalodimos CG, (2017). Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol, 24, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahnke W, Grotzfeld RM, Pellé X, Strauss A, Fendrich G, Cowan-Jacob SW, et al. , (2010). Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay. J. Am. Chem. Soc, 132, 7043–8. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Campobasso N, Biju MP, Fisher K, Pan XQ, Cottom J, et al. , (2011). Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chem. Biol, 18, 177–86. [DOI] [PubMed] [Google Scholar]
- 23.Eide CA, Zabriskie MS, Savage Stevens SL, Antelope O, Vellore NA, Than H, et al. , (2019). Combining the Allosteric Inhibitor Asciminib with Ponatinib Suppresses Emergence of and Restores Efficacy against Highly Resistant BCR-ABL1 Mutants. Cancer Cell, 36:431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Grzesiek S, et al. , (2008). Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J. Bio. Chem, 283, 18292–302. [DOI] [PubMed] [Google Scholar]
- 25.Agafonov RV, Wilson C, Otten R, Buosi V, Kern D, (2014). Energetic dissection of Gleevec’s selectivity toward human tyrosine kinases. Nat. Struct. Mol. Biol, 21, 848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan Y, Seeliger MA, Eastwood MP, Frank F, Xu H, Jensen MØ, et al. , (2009). A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc. Natl. Acad. Sci. U.S.A, 106,139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salah E, Ugochukwu E, Barr AJ, von Delft F, Knapp S, Elkins JM, (2011). Crystal structures of ABL-related gene (ABL2) in complex with imatinib, tozasertib (VX-680), and a type I inhibitor of the triazole carbothioamide class. J. Med. Chem, 54, 2359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grebien F, Hantschel O, Wojcik J, Kaupe I, Kovacic B, Wyrzucki AM, et al. , (2011). Targeting the SH2-Kinase Interface in Bcr-Abl Inhibits Leukemogenesis. Cell,147, 306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM, (1999). cAbl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes. Dev 13, 2400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barouch-Bentov R, Sauer K, (2011). Mechanisms of drug resistance in kinases. Expert Opin. Investig. Drugs., 20, 153–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soverini S, De Benedittis C, Mancini M, Martinelli G, (2016). Present and future of molecular monitoring in chronic myeloid leukaemia. Br. J. Haematol, 173,337–49. [DOI] [PubMed] [Google Scholar]
- 32.Peng B, Hayes M, Resta D, Racine-Poon A, Druker BJ, Talpaz M, et al. , (2004). Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J. Clin. Oncol 22, 935–42. [DOI] [PubMed] [Google Scholar]
- 33.Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, et al. , (2014). BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell, 26, 428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, et al. , (2005). In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res, 65, 4500–5. [DOI] [PubMed] [Google Scholar]
- 35.Ichim CV, (2014). Kinase-independent mechanisms of resistance of leukemia stem cells to tyrosine kinase inhibitors. Stem Cells Transl. Med, 3, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skrora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S, (2013). NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc. Natl. Acad. Sci. U.S.A, E4437–E4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonti R, Hertel-Hering I, Lamontanara AJ, Hantschel O, Grzesiek S, (2018). ATP site ligands determine the assembly state of the Abelson kinase regulatory core via the activation loop conformation. J. Am. Chem. Soc, 140, 1863–1869. [DOI] [PubMed] [Google Scholar]
- 38.Saleh T, Jankowski W, Sriram G, Rossi P, Shah S, Lee KB, et al. , (2016). Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Nat. Chem. Biol, 12, 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG, (2011). Structural basis for regulation of the Crk signaling protein by a proline switch. Nat. Chem. Biol, 7, 51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monneau YR, Ishida Y, Rossi P, Saio T, Tzeng SR, Inouye M, et al. , (2016). Exploiting E. coli auxotrophs for leucine, valine, and threonine specific methyl labeling of large proteins for NMR applications. J. Biomol. NMR, 65, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Rossi P, Kalodimos CG, (2019). Structural basis for client recognition and activity of Hsp40 chaperones. Science, 365, 1313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi P, Monneau YR, Xia Y, Ishida Y, Kalodimos CG, (2019). Toolkit for NMR Studies of Methyl-Labeled Proteins. Meth. Enzymol, 614,107–42. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A, (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–93. [DOI] [PubMed] [Google Scholar]
- 44.Lee W, Tonelli M, Markley JL, (2015). NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics, 31, 1325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossi P, Xia Y, Khanra N, Veglia G, Kalodimos CG, (2016). 15N and 13C- SOFAST-HMQC editing enhances 3D-NOESY sensitivity in highly deuterated, selectively [1H,13C]-labeled proteins. J. Biomol. NMR, 66, 259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monneau YR, Rossi P, Bhaumik A, Huang C, Jiang Y, Saleh T, et al. , (2017). Automatic methyl assignment in large proteins by the MAGIC algorithm. J. Biomol. NMR, 69, 215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W, (2010). Integration, scaling, space-group assignment and post-refinement. Acta. Crystallogr. D. Biol. Crystallogr, 66, 133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans PR, Murshudov GN, (2013). How good are my data and what is the resolution? Acta. Crystallogr. D. Biol. Crystallogr, 69, 1204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, (2007). Phaser crystallographic software. J. Appl. Crystallogr, 40, 658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K, (2010). Features and development of Coot Acta. Crystallogr. D. Biol. Crystallogr, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ, (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D. Biol. Crystallogr, 53, 240–55. [DOI] [PubMed] [Google Scholar]
- 52.Vallurupalli P, Bouvignies G, Kay LE, (2012). Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc, 134, 8148–61. [DOI] [PubMed] [Google Scholar]
- 53.Rennella E, Huang R, Velyvis A, Kay LE, (2015). (13)CHD2-CEST NMR spectroscopy provides an avenue for studies of conformational exchange in high molecular weight proteins. J. Biomol. NMR 63, 187–99. [DOI] [PubMed] [Google Scholar]
- 54.Tugarinov V, Sprangers R, Kay LE, (2007). Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J. Am. Chem. Soc, 129, 1743–50. [DOI] [PubMed] [Google Scholar]
- 55.Sun H, Kay LE, Tugarinov V, (2011). An optimized relaxation-based coherence transfer NMR experiment for the measurement of side-chain order in methyl-protonated, highly deuterated proteins. J. Phys. Chem. B, 115, 14878–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


