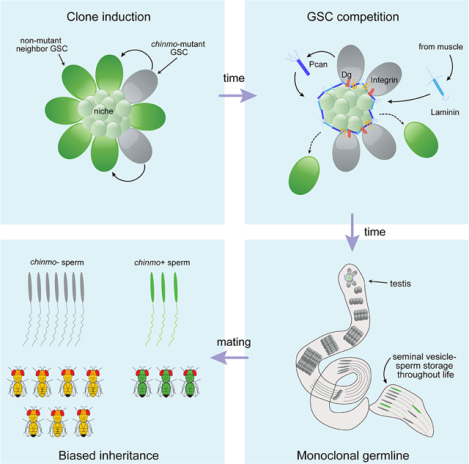
SUMMARY
Niches maintain a finite pool of stem cells via restricted space and short-range signals. Stem cells compete for limited niche resources, but the mechanisms regulating competition are poorly understood. Using the Drosophila testis model, we show that germline stem cells (GSCs) lacking the transcription factor Chinmo gain a competitive advantage for niche access. Surprisingly, chinmo−/− GSCs rely on a new mechanism of competition in which they secrete the extracellular matrix protein Perlecan to selectively evict non-mutant GSCs and then upregulate Perlecan-binding proteins to remain in the altered niche. Over time, the GSC pool can be entirely replaced with chinmo−/− cells. As a consequence, the mutant chinmo allele acts as a gene drive element: the majority of offspring inherit the allele despite the heterozygous genotype of the parent. Our results suggest the influence of GSC competition may extend beyond individual stem cell niche dynamics to population-level allelic drift and evolution.
Keywords: Drosophila, testis, germline stem cell, niche, competition, chinmo, Perlecan, Laminin, Dystroglycan, biased inheritance, aging
Graphical Abstract
eTOC Blurb
How stem cells compete for niche access remains largely unknown. Tseng et al. show that spermatogonial stem cells (SSCs) lacking the transcription factor Chinmo remodel the niche to their own benefit, causing the expulsion of non-mutant SSCs. This leads to a homozygous germline and biased inheritance of the chinmo-mutant allele.
INTRODUCTION
Stem cells reside in discrete microenvironments called niches that maintain a supply of undifferentiated stem cells via molecular signals. Because these signals are short-range and niche space is often limited, stem cells compete with one another for niche occupancy. In stem cell competition, “winners” remain in the niche and retain stem cell identity while “losers” exit and differentiate (Simons and Clevers, 2011). Under normal conditions, stem cells compete through a stochastic process: any given stem cell has an equal probability of remaining in the niche or being lost and replaced by its neighbor. In this homeostatic process, known as neutral competition, no one stem cell has a long-term competitive advantage over another (Klein and Simons, 2011). Competition can be biased if one stem cell gains an advantage over the others. Indeed, one advantaged stem cell and its progeny can hijack a niche to compose the entire stem cell pool, resulting in tissue monoclonality (Vermeulen et al., 2013, Amoyel et al., 2014, Issigonis et al., 2009, Snippert et al., 2014). However, the factors and mechanisms regulating stem cell competition – and germline stem cell competition in particular - remain largely mysterious.
The Drosophila testis is an ideal model system to study stem cell competition due to its well-characterized niche, genetic tractability, and conserved stem cell maintenance pathways (Fig. 1A and (Greenspan et al., 2015)). The testis is a coiled blind-ended tube, enveloped by a muscle sheath, responsible for spermatogenesis throughout adult male life (Hardy et al., 1979, Fuller, 1998). The niche includes a group of post-mitotic cells, which is anchored to the testis apex by integrins (Tanentzapf et al., 2007). The niche secretes self-renewal signals that support two distinct stem cell populations. GSCs divide continuously throughout life and ultimately produce sperm. Approximately 8 GSCs adhere to the niche through the adhesion molecule E-Cadherin (E-Cad) (Yamashita et al., 2003). The niche also supports somatic cyst stem cells (CySCs), which co-differentiate with GSCs into post-mitotic cysts that enclose developing spermatogonia (Hardy et al., 1979, Fabrizio et al., 2003).
Figure 1: chinmo−/− GSCs dominate the niche by evicting non-mutant GSCs.
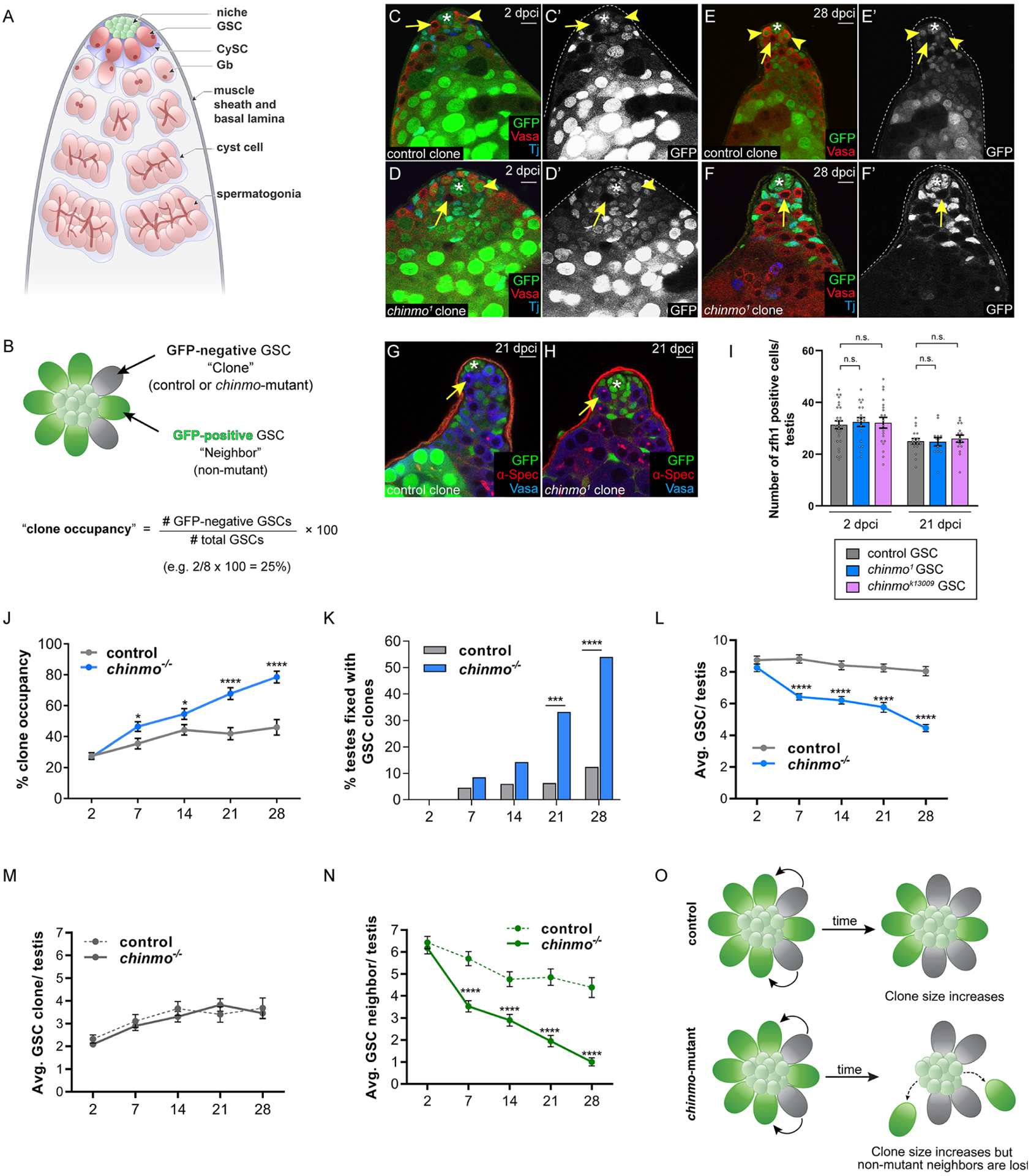
(A) The adult Drosophila testis. A GSC produces a gonialblast (Gb), which undergoes transita-mplifying divisions to produce spermatogonia that differentiate into sperm. CySCs divide to produce cyst cells, two of which envelope a Gb and its descendants.
(B) The clone occupancy assay. In a WT testis, 8 GSCs surround the niche. GSC clones (either control or chinmo−/−) lack GFP. The other GSCs (labeled “neighbors”) are not mutant and express GFP. Clone occupancy is measured by dividing the number of GSC clones by the total number of GSCs in that testis.
(C-F) Confocal images of testes with control (C,E, arrows) or chinmo−/− (D,F, arrows) GSC clones at 2 (C,D) or 28 (E,F) dpci. Clones lack GFP. Vasa (red) marks germ cells. Tj (blue) marks the nuclei of CySCs and early cyst cells. Arrowheads mark non-mutant GSC neighbors.
(G-H) Confocal images of testes with control (G, arrow) or chinmo−/− (H, arrow) clones at 21 dpci stained with αSpectrin (red) to mark the fusome. Clones lack GFP. Vasa (blue).
(I) Graph showing the number of Zfh1-positive CySCs in testes with control (gray), chinmo1 (blue) and chinmok13009 GSC clones (purple) at 2 and 28 dpci.
(J) Graph showing clone occupancy of control (gray) and chinmo−/− (blue) GSC clones at 2, 7, 14, 21, 28 dpci.
(K) Graph showing percent monoclonal testes within the GSC lineage at 2, 7, 12, 21 and 28 dpci when control (gray) or chinmo−/− (blue) GSC clones are present.
(L) Graph showing the average total number of GSCs in testes with control (gray) or chinmo−/− GSC clones (blue) at 2, 7, 14, 21 and 28 dpci.
(M) Graph showing the average number of control (dashed line) or chinmo−/− (solid line) GSC clones at 2, 7, 14, 21 and 28 dpci.
(N) Graph showing non-mutant GSC neighbors in testes with control (dashed line) or chinmo−/− (solid line) GSC clones at 2, 7, 14, 21 and 28 dpci.
(O) Model. Control GSC clones (gray cells, upper panel) and chinmo−/− GSC clones (gray cells, lower panel) are induced at the same low frequency. Over time, both types of clones expanded to a similar extent (gray cells). chinmo−/− clones cause the loss of non-mutant neighbor GSCs (green cells, lower panel) and this does not occur to the non-mutant neighbors (green cells, upper panel) of control GSC clones.
In C-F, G,H, an asterisk marks the niche.
Scale bar = 10 μM
In I,J,L,M,N, error bars represent SEM.
n.s. = not significant; * P ≤ 0.05; *** P ≤ 0.001; **** P ≤ 0.0001 as assessed by Student’s t-test (I,J,LM,N) or by χ2 test (K).
Stem cells in the Drosophila testis, as those in other tissues and organisms, compete for limited niche space and resources. CySCs have documented competitive behavior, and several mechanisms triggering biased competition in favor of single CySCs have been uncovered (Amoyel et al., 2016, Amoyel et al., 2014, Issigonis et al., 2009, Singh et al., 2010, Stine et al., 2014). Significantly less is known, however, about competition in GSCs. Regular GSC loss and replacement by neighbors has been observed (Wallenfang et al., 2006, Sheng and Matunis, 2011, Salzmann et al., 2013). While this behavior is suggestive of neutral competition, no underlying mechanisms have been elucidated. Biased competition has also never been observed in male GSCs, though single female GSCs harboring specific genetic mutations can outcompete their neighbors in the Drosophila ovary (Jin et al., 2008, Rhiner et al., 2009).
Here we report biased competition in male GSCs driven by mutations in the transcriptional repressor Chinmo (Zhu et al., 2006). Chinmo is a known regulator of somatic stem cell activity in the Drosophila ventral nerve cord and testis (Flaherty et al., 2010, Narbonne-Reveau et al., 2016). In the testis, Chinmo is required for the maintenance of male sexual identity. CySCs devoid of Chinmo undergo sex-reversal and transdifferentiate into feminized somatic stem cells (Grmai et al., 2018, Ma et al., 2014, Ma et al., 2016). Despite transcriptomic analyses of somatic stem cells lacking Chinmo, direct Chinmo target genes have not yet been identified (Grmai et al., 2021, Narbonne-Reveau et al., 2016). Here, we show that loss of chinmo in single GSCs confers a competitive advantage for niche access so that, over time, the GSC pool becomes “fixed” with chinmo−/− cells. Surprisingly, chinmo−/− GSCs do not rely on increased proliferation or canonical adhesion systems to gain their advantage. Instead, chinmo−/− GSCs ectopically secrete the extracellular matrix protein (ECM) Perlecan (Pcan) that creates a de novo ECM around the niche within the testis lumen. This resculpted niche selectively evicts non-mutant GSCs from the niche but retains chinmo−/− GSCs, which upregulate ECM-binding proteins. As a result this competition, the majority of adult offspring inherit the mutant chinmo allele despite the heterozygous genotype of the parent. These results provide the experimental evidence that GSC competition underlie “mitotic” drive, a proposed but not proven mechanism by which GSCs with a competition advantage transmit greater than the expected 50% Mendelian ratio (Otto and Hastings, 1998). Thus, GSC competition may not only be important for understanding the fundamental principles of stem cell dynamics but also have long-term implications for genetic drift and evolution (Hastings, 1989).
RESULTS
chinmo−/− GSCs evict non-mutant neighbor GSCs from the niche
Chinmo is expressed in male GSCs (Fig. S1A), but its role in these cells is unknown. To investigate this, we used mitotic recombination to induce single GFP-negative control and chinmo homozygous mutant (chinmo−/−) clones in adult flies and monitored their niche retention and lineages over time (Fig. 1B). We used a null allele (chinmo1), a hypomorphic allele (chinmok13009) and validated RNAi-depletion (chinmo-i) to generate chinmo-deficient GSCs clones (Fig. S1A–F). Control GSC clones and chinmo-deficient GSC clones were induced at the same frequency as assessed at 2 days post clone induction (dpci) (Fig. 1C,D). Both types of clones contained single, Vasa-positive cells at the niche that had dot fusomes characteristic of GSCs (Fig. 1G,H). Surprisingly, chinmo−/− GSCs composed a majority of the GSC pool by 28 dpci (Fig. 1F) compared to control clones (Fig. 1E). Prior work has shown that mutant CySCs with a competitive advantage prevail over both wild-type (WT) CySCs and WT GSCs for niche access. In contrast, chinmo−/− GSCs only outcompeted non-mutant GSCs while leaving the CySC population unaffected. We observed comparable numbers of CySCs expressing the stem cell marker Zfh1 in testes harboring either chinmo−/− or control GSC clones (Fig. 1-I, Table S1 #16). Thus, chinmo−/− GSCs provide a new model system for studying GSC-GSC specific stem cell competition.
We hypothesized that single GSCs lacking chinmo gain a competitive advantage over their non-mutant neighbors for niche access, so we quantified the percent of clones composing the GSC pool, which we termed “clone occupancy,” at 2, 7, 14, 21, and 28 dpci (Fig. 1B). At 2 dpci, both control and chinmo−/− GSC clones occupied the niche in equal proportions (Fig. 1M, Table S1 #37–46). By 28 dpci, however, chinmo−/− GSCs occupied a majority of the GSC pool (78.52 ± 3.77%, Table S1 #16), while control clones occupied significantly less (45.97 ± 5.00%, P < 2 × 10−5, Table S1 #11) (Fig. 1J). chinmo−/− GSCs occupied a majority of the GSC pool at 7, 14, and 21 dpci (Fig. 1J, Table S1 #13–15). Similar trends were observed at 14 dpci for the other chinmo alleles (Table S1 #76,80). Indeed, the probability of recovering testes with “fixed” niches - where a GSC clone composed the entire GSC pool - was significantly higher when GSC clones lacked chinmo (12.50% vs. 54.00% in control and chinmo−/− clones, respectively, at 28 dpci, P < 3 × 10−7) (Fig. 1K, Table S1 #17–26). These data indicate that loss of chinmo in single GSCs biases competition for niche access in their favor.
To determine whether chinmo−/− GSC clones expanded their niche presence by over-proliferation and/or by losing non-mutant neighbor GSCs, we quantified the average number of GSC clones and the average number of non-mutant neighbor GSCs over time in testes containing either control or chinmo−/− clones. Control and chinmo−/− clones themselves were present at nearly identical levels over time (3.68 ± 0.45 vs. 3.46 ± 0.25, control and chinmo−/− clones, respectively, by 28 dpci, P = 0.47) (Fig. 1M, Table S1 #37–46). By contrast, neighbor GSCs were precipitously lost from the niche in the presence of chinmo−/− but not control clones (4.38 ± 0.46 vs. 1.00 ± 0.18 neighbors of control or chinmo−/− clones, respectively, by 28 dpci, P = 7 × 10−7) (Fig. 1N, Table S1 # 47–56). As a result, niches harboring chinmo−/− clones experienced a net loss in total GSCs (8.05 ± 0.29 vs. 4.46 ± 0.22 in control and chinmo−/− clones, respectively, at 28 dpci, P = 4 × 10−21) (Fig. 1L, Table S1, #27–36). We obtained similar results with other chinmo alleles (Table S1, #98,102). These results suggest that chinmo−/− GSCs gain their competitive advantage not by increasing their numbers but rather by imperiling niche access of neighbor GSCs (Fig. 1O).
chinmo−/− GSCs ectopically secrete Pcan into the niche milieu
The competitive advantages of chinmo−/− GSCs could result from behaviors observed in “winning” cells in the Drosophila ovary or epithelia, including increased proliferation, increased E-Cad, or induced death of neighbors (Jin et al., 2008, Amoyel and Bach, 2014). However, there were no substantial differences in these parameters between chinmo−/− GSC clones and control GSC clones or non-mutant neighbors (Fig. S2A–C). Additionally, chinmo−/− GSC clones still divided asymmetrically as evidenced by the mother centrosome being located at the GSC-niche interface and the daughter centrosome 180° away (Fig. S2D–F). We considered the possibility that chinmo−/− GSCs had increased self-renewal signaling compared to neighbors. GSCs require BMP/Mad and JAK/STAT pathways for self-renewal and niche adhesion, respectively (Leatherman and Dinardo, 2010). However, there are no differences in phosphoMad or phospho-STAT between chinmo−/− clones and their non-mutant neighbors (Fig. S2G–H). We observed similar robust E-Cad levels at the GSC-niche interface in chinmo−/− clones and in their non-mutant neighbor GSCs (Fig. S2I), suggesting decreased E-Cad levels were not responsible for neighbor loss.
Electron micrograph of the stem cell niche at 28 dpci revealed a ring of proteinaceous material encircling the niche in testes with chinmo−/− GSCs but not those with control GSC clones (Fig. 2A,B). This material resembled and was contiguous with the ECM composing the muscle basal lamina (BL). We termed this structure the “moat” because it resembled the ditch around a castle. A similar “gap” between GSCs and the niche can be observed at 28 dpci in single confocal images of testes with chinmo−/− GSC clones (Fig. S3B) but not with control GSC clones (Fig. S3A). The majority of chinmo−/− GSCs were displaced significantly farther from the niche by 28 dpci than control clones (Fig. S3C, compare blue to gray bar). Moats were observed in a majority of testes with chinmo−/− clones but only rarely seen in testes with control clones (Fig. S3D). Based on these results, we speculate that chinmo−/− GSCs secrete ectopic ECM into the niche milieu.
Figure 2: chinmo−/− GSCs create a “moat” around the niche by secreting Pcan.
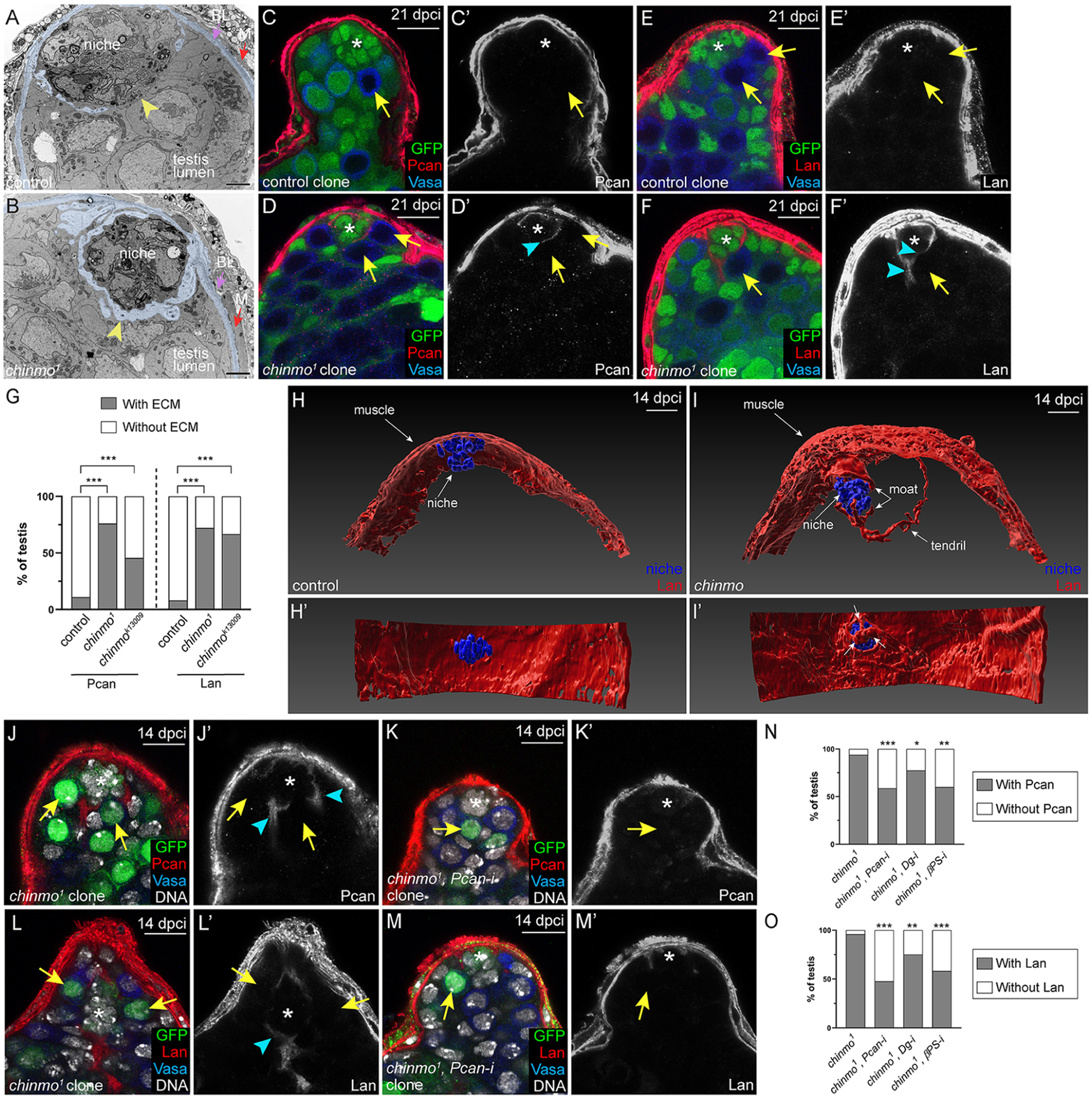
(A,B) TEMs of testes with control (A) or chinmo1 (B) GSC clones at 28 dpci. The micrographs are pseudocolored to show ECM-like material (light blue) in the muscle basal lamina (“BL”, purple arrowhead in A,B) or in the testis lumen (yellow arrowhead, B). Yellow arrowheads indicate GSC-niche interface. Magnification 5,600x. Scale bar is 2 μM.
(C-F) Pcan (C,D, red) and Lan (E,F, red) in testes with control (C,E arrows) chinmo−/− GSC (D,F, arrow) clones at 21 dpci. Arrowhead (D’,F’) indicates ectopic ECM.
(G) Graph quantifying the percentage testes with (gray portion of bar) or without (white portion of bar) ECM proteins Pcan or Lan surrounding the niche when control, chinmo1 or chinmok13009 clones are present.
(H,I) Imaris-generated view of a testis with control (H) or chinmo−/− (I) GSC clones (not shown) at 14 dpci. Niche cells (blue) are visible as a ball, and some niche cells adhere to Lan (red) present in the muscle (H,I). In I, some niche cells are partially obscured by a distended “hat” of Lan (red, upper arrow labeled “moat”) that is contiguous with Lan in the muscle. In I, there is ectopic Lan in the testis lumen, which form three “claws” on distal niche cells (lower arrow labeled “moat”). A “tendril” of ectopic Lan extends to the right. In H’,I’, the testis is rotated 90°. All niche cells are visible in H’, but in I’ several niche cells are covered by Lan (arrows).
(J-M) Expression of Pcan (red in J,K) and Lan (red in L,M) around the niche when chinmo−/− GSC clones (J,L) or chinmo−/− clones depleted for Pcan (K,M) are present. Arrowheads indicate ectopic ECM.
(N-O) Graph quantifying the percentage of testes with (gray portion of bar) or without (white portion of bar) Pcan (N) or Lan (O) in the moat when chinmo−/− clones (first bars), chinmo−/− clones depleted for Pcan (second bars), for Dg (third bars) or for βPS (fourth bars) are present. In A-F, clones lack GFP. In H-M, clones express GFP. In C-F, J-M, Vasa is blue and an asterisk marks the niche.
Scale bar = 10 μM
* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001 as assessed by χ2 test. See also Figs. S1, S2, S4, S7.
We examined expression of Pcan, a conserved secreted proteoglycan that cross-links the ECM, allows for cell-ECM adhesion, and facilities ligand diffusion (Broadie et al., 2011). In testes with control GSC clones, Pcan is found in the muscle BL and not within the testis lumen at 21 dpci (Fig. 2C,G). However, Pcan localized to the moat when chinmo−/− GSCs were present and this was observed as early as 7 dpci (Fig. 2D,G, Fig. S3E,F). We also examined expression of Laminin (Lan), a major component of BL. In testes with control GSC clones, Lan localized to the muscle BL and not the testis lumen (Fig. 2E,G). However, Lan localized to the moat when chinmo−/− GSCs were present as early as 7 dpci (Fig. 2F,G, Fig. S3G,H). Further, 3D reconstructions of confocal z-stacks revealed that Lan surrounded the niche in testes with chinmo−/− clones but not those with control clones (Fig. 3H,I, Movie S1). Other ECM proteins were expressed at low levels in testes and were not localized to the moat (Fig. S3M–T), indicating that the moat is specifically composed of Pcan and Lan.
Figure 3: chinmo−/− GSCs require Pcan to evict non-mutant neighbors.
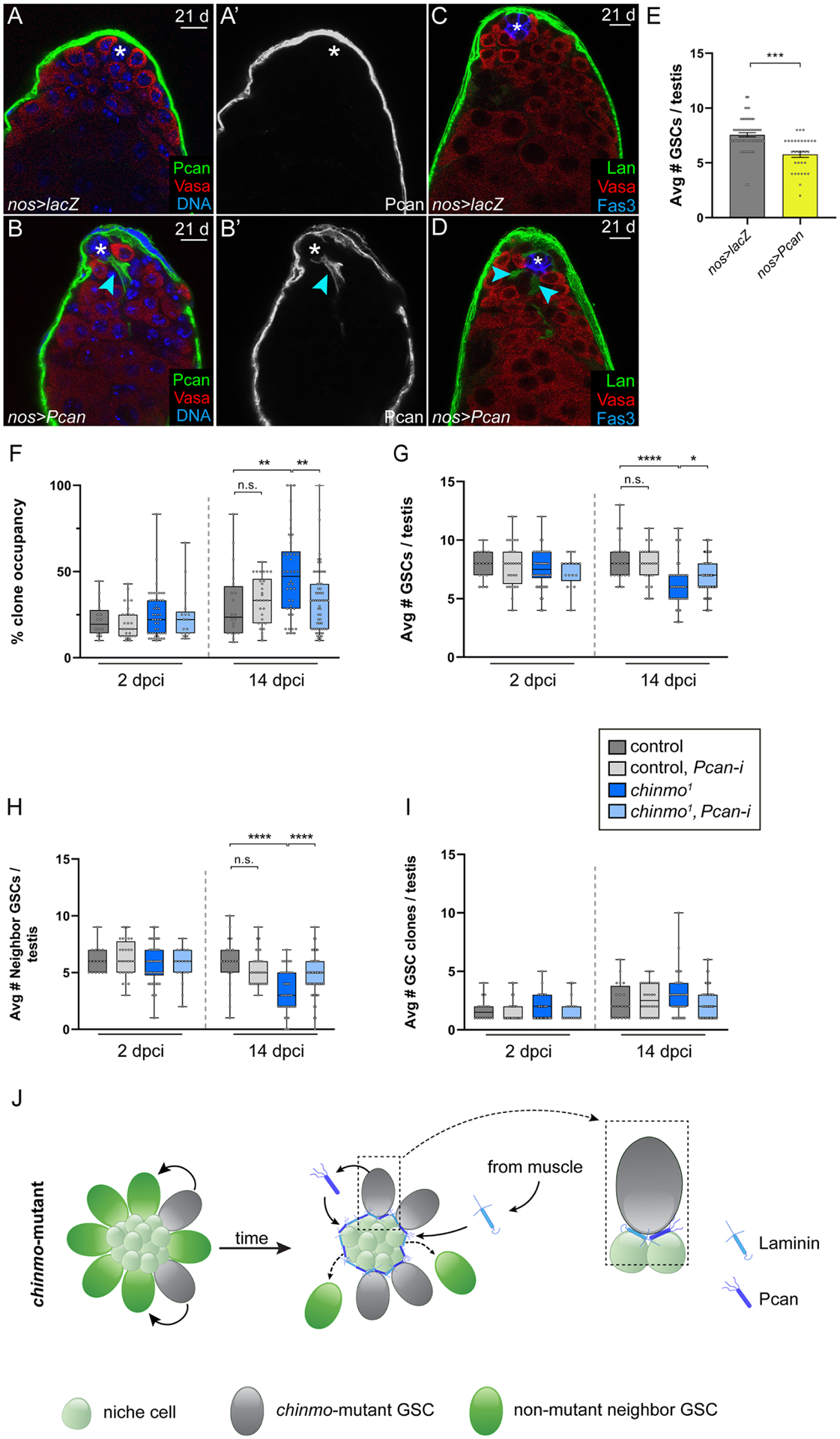
(A,D) Confocal images of nos>lacZ (A,C) or nos>Pcan (B,D) testes in which Pcan was mis-expressed in all GSCs for 21 days. Arrowheads (B,B’,D) indicate ectopic ECM. Pcan (green, A,B); Lan (green, C,D); Vasa (red) and DNA (ToPro, blue A,B); Fas3 (blue, C,D).
(E) Graph showing average number of GSCs in nos>lacZ (gray) or nos>Pcan (yellow) testes.
(F-I) Box and whisker plots showing clone occupancy (F), average total number of GSCs (G), average number of non-mutant GSC neighbors (H); average number of clones (I) in testis with control GSC clones (dark gray bars), with control GSC clones depleted for Pcan (light gray bars), with chinmo−/− GSC clones (dark blue bars) or chinmo−/− GSC clones depleted for Pcan (light blue bars) at 2 and 14 dpci.
(J) Model: chinmo−/− GSC clones (gray cells) secrete Pcan (dark blue symbol), which seeds the moat. Lan (light blue symbol) is recruited from the muscle BL. Boxed area at right illustrates a chinmo−/− GSC clone in contact with the moat. By contrast, non-mutant neighbor GSCs (green stem cells in middle cartoon) are lost from the niche. The smaller, light green cells are niche cells.
Scale bar = 10 μM
In F-I, error bars represent SEM.
n.s. = not significant; * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; **** P ≤ 0.0001, as assessed by Student’s t-test.
Pcan is necessary and sufficient for perturbing neighbor GSC niche occupancy
If ectopic Pcan underlies the competitive advantage of chinmo−/− GSCs, these should be true: (1) Lan accumulation in the moat should depend on Pcan secretion by chinmo−/− GSCs; (2) ectopic Pcan secretion should be sufficient to induce GSC loss in an otherwise WT testis; and (3) chinmo−/− GSCs should require Pcan for their competitive properties.
To test the first condition, we generated positive-marked chinmo−/− GSC clones and chinmo−/− GSC clones depleted for Pcan. In testes with chinmo−/− GSC clones, Pcan is found in the moat (Fig. 2J,N) but this was significantly reduced when these clones were depleted for Pcan (Fig. 2K,N). Lan in the moat also declined significantly in chinmo−/− clones depleted for Pcan (Fig. 2L,M,O). These results indicate that the moat is caused by ectopic Pcan secretion by chinmo−/− GSCs and suggest that Lan is recruited to the moat from the muscle BL. In support of this, Lan in the moat was significantly reduced when Lan was depleted from muscle (Fig. S3IK). HCR-FISH revealed that Pcan transcripts were significantly increased in chinmo−/− GSCs compared to control GSCs (Fig. S4A–F), suggesting that Pcan is Chinmo target in GSCs.
To test the second condition - whether ectopic Pcan can impede WT GSCs - we used the GSC driver nanos (nos) to mis-express Pcan in otherwise WT GSCs. We observed ectopic Pcan and Lan surrounding the niche in nos>Pcan testes, similar to the chinmo−/− GSC phenotype (Fig. 3A–D). As expected, GSC-secreted Pcan phenocopied the loss in total GSC number (Fig. 3E, Table S1 #57,58) observed in testes containing chinmo−/− GSC clones at same time point (i.e., 21 days) (Fig. 1L). Thus, ectopic Pcan secreted by GSCs is sufficient to alter the niche architecture and disadvantage WT GSC niche access.
To test the third condition - whether Pcan is necessary for chinmo−/− GSCs to expel non-mutant neighbor GSCs - we used the MARCM technique to deplete Pcan using two validated RNAi lines (Fig. S1K–N) in control or chinmo−/− GSC clones. Depletion of Pcan from chinmo−/− GSC clones robustly blocked the increase in chinmo−/− clone occupancy (48.21 ± 3.49% vs. 33.52 ± 2.50% in chinmo−/− and chinmo−/−, Pcan-RNAi clones, respectively, at 14 dpci, P < 0.0018) (Fig. 3F, Table S1 #68, 70). By contrast, clone occupancy of control clones was unaffected by Pcan depletion (Fig. 3F, Table S1 #60, 62). This reduction in competitive behavior is not because Pcan-depleted chinmo−/− GSCs were lost from the niche (Fig. 3I, Table S1 #134, 136), but rather because non-mutant neighbor GSCs were no longer preferentially evicted (3.37 ± 0.27 vs. 4.67 ± 0.23 WT neighbors of chinmo−/− and chinmo−/−, Pcan-RNAi clones, respectively, at 14 dpci, P < 0.0006) (Fig. 3H, Table S1, #112, 114). Consistent with the partial rescue of neighbor GSCs, the average total number of GSCs was also significantly increased by depleting Pcan in chinmo−/− GSCs (6.28 ± 0.26 vs. 6.97 ± 0.19 GSCs in testes with chinmo−/− and chinmo−/−, Pcan-RNAi clones, respectively, at 14 dpci, P < 0.044) (Fig. 3G, Table S1 #90, 92). A similar autonomous requirement for Pcan was observed in chinmok13009 clones (Table S1, #76, 78, 98, 100, 120, 122). By contrast, Lan depletion from chinmo−/− GSC clones using a validated RNAi line (Fig. S1H–J) did not significantly alter clone occupancy, the average number of GSCs, or the average number of neighbors (Table S1 #74, 96, 118). Thus, removal of Pcan but not Lan from chinmo−/− GSCs greatly impairs their competitive phenotype. Consistent with the model that Lan is recruited to the moat, depleting Lan from the muscle significantly reduced Lan in the moat and the clone occupancy of chinmo−/− GSCs (Fig. S3K,L, Table S1 #147, 148). We conclude that chinmo−/− GSCs non-autonomously compromise neighbor GSC access by secreting Pcan into the niche (Fig. 3J).
chinmo−/− GSCs remain in the altered niche by upregulating Dystroglycan (Dg)
We reasoned that chinmo−/− GSCs remain in the niche because they upregulate ECM-binding proteins. Dg is a transmembrane protein that interacts with Pcan via its extracellular domain and the actin cytoskeleton via its intercellular domain (Schneider et al., 2006). Dg is significantly increased at the GSC-niche interface in chinmo−/− GSCs but not in control GSC clones (Fig. 4A–C). Furthermore, HCR-FISH revealed that Dg transcripts were significantly increased in chinmo−/− GSCs compared to control GSCs (Fig. S4G–I), suggesting that Dg may be regulated by Chinmo in GSCs. To test this, we depleted Dg using a validated RNAi line (Fig. S1O–Q) from control and chinmo−/− clones. Depleting Dg from chinmo−/− clones significantly decreased their clone occupancy (Fig. 4D, Table S1 #154, 156), restored the average total number of GSCs (Fig. 4E, Table S1 #166, 168) and rescued non-mutant neighbors (Fig. 4F, Table S1 #178, 180). Dg knockdown in chinmo−/− clones – but not control clones - caused a slight but significant decrease in clone recovery at 14 dpci (Fig. 4G, Table S1 #190, 192). Similar results were observed with the chinmok13009 allele (Table S1 #158, 160, 170, 172, 182, 184, 194, 196). We conclude that Dg expression is an important mechanism used by chinmo−/− GSCs to remain in the remodeled niche (Fig. 4H).
Figure 4: chinmo−/− GSCs require Dg to remain in the altered niche.
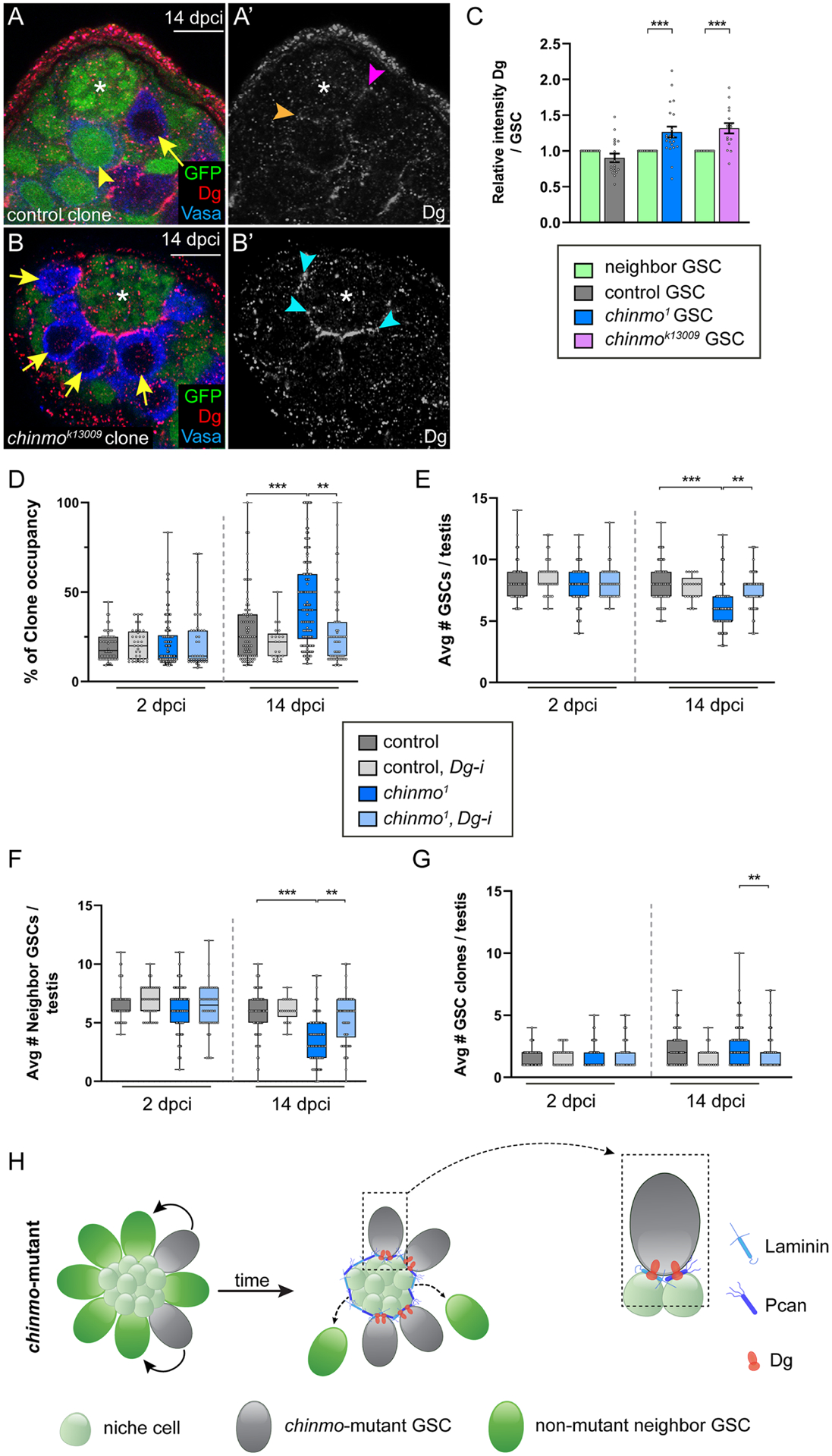
(A,B) Confocal images of Dg (red) in testes harboring control (A, arrow) or chinmok13009-mutant GSC clones (B, arrows) at 14 dpci. Orange arrowhead (A’), magenta arrowhead (A’) and blue arrowheads (B’) indicate Dg at the GSC-niche in a non-mutant GSC neighbor, a control clone and a chinmo−/− GSC, respectively. Clones lack GFP. Neighbors (yellow arrowhead) express GFP. Vasa (blue).
(C) Graph of relative Dg expression at the GSC-niche interface in control, chinmo1 or chinmok13009 GSC clones relative to that of neighbor GSCs in the same testis.
(D-G) Box and whisker plots showing clone occupancy (D), average total number of GSCs (E), average number of non-mutant GSC neighbors (F), and average numbers of GSC clones (G) in testes with control GSC clones (dark gray bars), with control GSC clones depleted for Dg (light gray bars), with chinmo−/− GSC clones (dark blue bars) or chinmo−/− GSC clones depleted for Dg (light blue bars) at 2 and 14 dpci.
(H) Model: chinmo−/− GSC clones (gray cells) have increased Dg (orange symbol) at the GSC-niche interface, allowing them to remain in the resculpted niche. Boxed area at right illustrates a chinmo−/− GSC clone remaining in contact with the moat through increased localized Dg expression. Non-mutant neighbor GSCs do not have increased Dg at the GSC-niche interface and cannot remain long-term in the niche.
Scale bar = 10 μM
In D-G, error bars represent SEM.
* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; **** P ≤ 0.0001, as assessed by Student’s t-test.
chinmo−/− GSCs remain in the altered niche via βPS integrin
We next examined integrins, which mediate cell-ECM interactions, in GSC competition. The integrin-associated protein Talin is significantly increased at the GSC-niche interface in chinmo−/− GSC clones compared to non-mutant GSC neighbors (Fig. S5B,C). By contrast, there is no significant change in Talin levels between control GSC clones compared to their neighbor GSCs (Fig. S5A,C). Only βPS - and not other integrin subunits - was increased at the GSC-niche interface in chinmo-deficient GSCs (Fig. S5D–P). HCR-FISH revealed that βPS transcripts were significantly increased in chinmo-depleted GSCs compared to control GSCs (Fig. S4J–L), suggesting that βPS may be regulated by Chinmo in GSCs.
To determine whether Talin or βPS was required for chinmo−/− clones to remain in the altered niche, we depleted either factor using validated RNAi lines (Fig. S1R–Y) in control or chinmo−/− clones and assessed parameters of competition. Knocking down Talin in chinmo−/− clones significantly decreased their clone occupancy (Fig. S6A, Table S1 #202, 204), restored the average total number of GSCs (Fig. S6B, Table S1 #210, 212) and rescued non-mutant neighbors (Fig. S6C, Table S1 #218, 220). Talin depletion from chinmo−/− clones - but not from control clones - reduced their niche residence (Fig. S6D, Table S1 #226, 228). Depleting βPS with either RNAi line in chinmo−/− clones significantly decreased clone occupancy (Fig. S6E, Table S1 #236, 238, 240), restored the average total number of GSCs (Fig. S6F, Table S1 #248, 250, 252) and rescued non-mutant neighbors (Fig. S6G, Table S1 #260, 262, 264). Control GSC clones depleted for βPS using RNA-i #1 could not be recovered at 2 dcpi (Fig. S6H, Table S1 #267), and control clone recovery was greatly reduced using the second βPS RNAi line (Table S1 #269). These data suggest that βPS is required for GSC maintenance. We could not test this hypothesis using mosaic analysis because mutation of the X-linked βPS gene is male lethal. However, in support of this model, there were significantly fewer GSCs at the niche when βPS or Talin were depleted from GSCs (Fig. S1Z). Importantly, chinmo−/− GSC clones depleted for βPS could remain in the altered niche (Fig. S6H, Table S1 #273, 274). These results indicate that increased integrin expression is used by chinmo−/− GSCs to remain in the resculpted niche (Fig. S6I). They also demonstrate that Dg and βPS integrin are non-redundant mechanisms employed by competitive GSCs for niche occupancy. Downregulation of ECM binding proteins in the chinmo−/− clone alters Pcan and Lan levels in the moat. In testes with chinmo−/− GSCs depleted for βPS or Dg, there is significantly less Pcan and Lan in the moat (Fig. 2N,O). These results suggest that (1) there is a feedback loop from ECM binding receptors promoting Pcan secretion or (2) ECM binding proteins in GSCs stabilize the moat.
Providing neighbors with more Dg rescues them from competition
We next tested whether non-mutant neighbor GSCs supplied with more Dg would adhere to the moat and be rescued from competition (Fig. 5A). To test this, we mis-expressed Dg in all GSCs using nos and then generated control or chinmo−/− GSC clones. We compared the number of non-mutant neighbor GSCs at 14 dpci when competition is robustly occurring (Fig. 5B). The moat generated by chinmo−/− GSCs (Fig. 5C,F) was still observed when Dg was mis-expressed in all GSCs (Fig. 5D–F). Despite the presence of the moat, non-mutant neighbor GSCs remained in the altered niche when they had increased Dg (Fig. 5D, yellow arrowheads, Fig. 5G, last bar, Table S2 #6, 8). Furthermore, the number of GSCs/testis is rescued when neighbors have increased Dg (Fig. 5H, Table S2 #14, 16).
Figure 5: Neighbors are rescued from competition when provide with ectopic Dg.
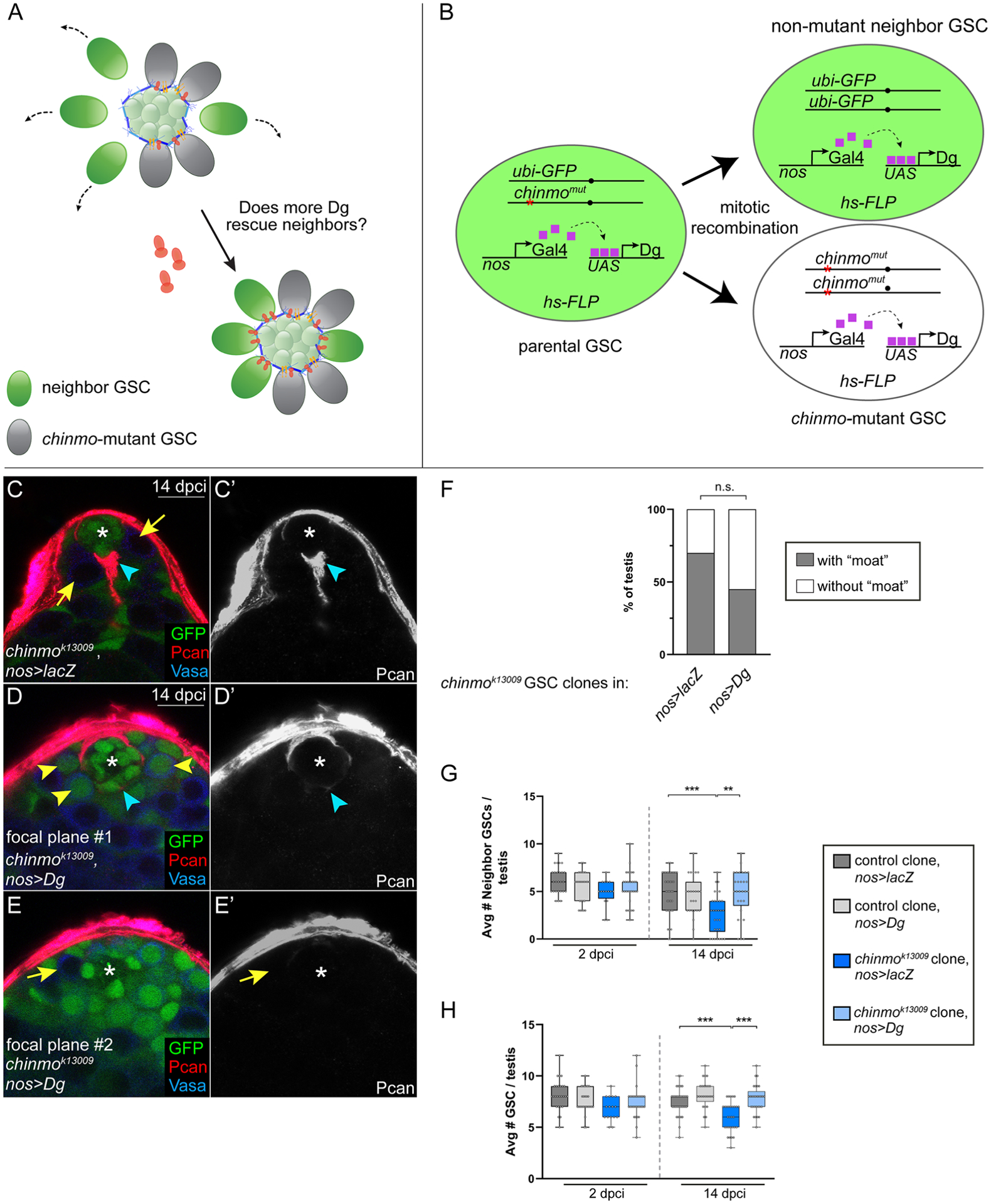
(A) Model: If non-mutant neighbor GSCs (green) are provided with increased Dg (dark orange), they should be able to remain in the niche despite the induction of chinmo−/− GSC clones (gray) and the formation of the moat (Pan, Lan, blue symbols). Yellow symbols represent integrins.
(B) In the “neighbor rescue” assay, chinmo−/− clones (abbreviated chinmomut) were generated in a background where all GSCs express UAS-Dg driven by the GSC driver nos-Gal4. We scored the number of non-mutant neighbors at 2 and 14 dpci (see G), average number of GSCs (see H) and presence a moat (see C,D,F).
(C-E) Pcan (red) in testes containing chinmok13009 GSC clones in nos>lacZ (C) or nos>Dg (D). The chinmo clone is not visible in D (“focal plan #1”) but is visible in E (arrow, “focal plane #2”). Clones lack GFP. Pcan (red). Vasa (blue).
(F) Graph showing percentage of testes with a “moat” when chinmok13009 GSC clones are generated in a nos>lacZ or nos>Dg background.
(G, H) Box and whisker plots showing average number of non-mutant GSC neighbors (G) or average number of GSCs (H) in testis with control GSC clones in nos>lacZ (dark gray bars), control GSC clones in nos>Dg (light gray bars), chinmok13009 GSC clones in nos>lacZ (dark blue bars), or chinmok13009 GSC clones in nos>Dg (light blue bars) at 2 and 14 dpci.
Scale bar = 10 μM
In G,H, error bars represent SEM.
n.s. = not significant; ** P ≤ 0.01; *** P ≤ 0.001 as assessed by Student’s t-test (G,H) and by χ2 test (F).
See also Table S2.
GSC competition causes biased inheritance
Since testes with chinmo−/− GSCs frequently have a homozygous mutant germline, the chinmo−/− allele should be inherited at a super-Mendelian frequency (i.e., greater > 50%). To assess this, we generated males which had a chinmo+ or chinmo− allele in trans to a ubi-GFP-labeled sister chromosome that could be scored in the next generation (Fig. 6A). We generated chinmo+/+ GSC clones or chinmo−/− GSC clones and aged the males until 21 dpci, at which time we mated single males with two virgin WT females for two days. At 23 dpci, we isolated the testes of the mated males and assessed germline clonality. Our model predicts that if competitive chinmo−/− GSCs cause mitotic drive, the chinmo− chromosome should be inherited by more than 50% of the F1 progeny while the GFP-positive sister chromosome should be passed on to less than 50%. By contrast, since chinmo+/+ GSCs should not have a competitive advantage, the chinmo+ chromosome should be inherited by 50% of the F1 progeny and the GFP-positive sister chromosome should be inherited by the other 50% (Fig. 6A).
Figure 6: GSC competition causes biased inheritance.
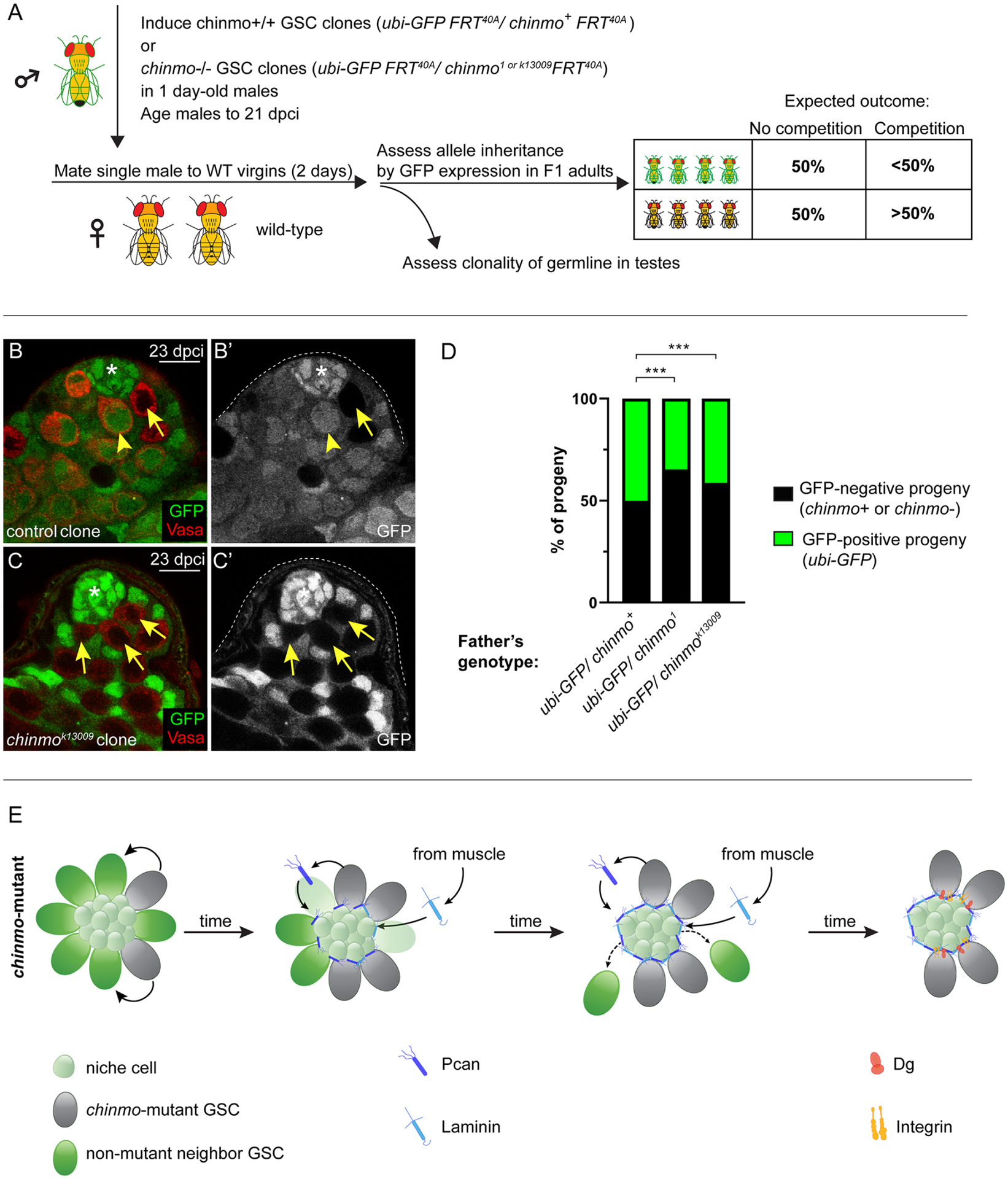
(A) Schematic of the “inheritance assay” - see STAR Methods for details. Box shows expected outcomes.
(B,C) Confocal images of a testis with a control (B,B’, arrow) or a chinmok13009 GSC clone (C,C’, arrows) at 23 dpci. Non-mutant neighbor GSCs are marked by an arrowhead. The only GFP-positive cells in C are somatic support cells. Clones lack GFP. Vasa is red.
(D) Graph showing inheritance of the chinmo chromosome (chinmo+, chinmo1 or chinmok13009 allele) (in black) or the ubi-GFP chromosome (in green).
(E) Model. chinmo−/− GSC clones (gray) secrete Pcan (dark blue), which causes the moat. Lan (light blue) is recruited to the moat from the muscle BL. chinmo−/− GSC clones increase Dg (orange) and βPS integrin (yellow) at the GSC-niche interface, allowing them to remain in the resculpted niche but non-mutant neighbor GSCs (green cells) do not and differentiate.
Scale bar = 10 μM
*** P ≤ 0.001 as assessed by χ2 test (D).
See also Table S3.
As expected, testes with chinmo+/+ clones contained both GFP-negative GSC clones and GFP-positive non-mutant neighbor GSCs (Fig. 6B). As predicted, testes with chinmo−/− GSC clones were frequently monoclonal or “fixed”, containing only GFP-negative chinmo−/− GSCs (Fig. 6C). For the allele inheritance assay, we followed the sister chromosome by inheritance of the GFP-positive transgene it harbors (Fig. 6D, green part of each bar) and the chinmo allele (either chinmo+ or chinmo−) by inheritance of the GFP-negative chromosome (Fig. 6D, black part of each bar). As predicted, 50% of the offspring of males with chinmo+/+ GSC clones inherited the chinmo+ allele and the other 50% inherited the GFP-positive sister chromosome (Fig. 6D, first bar, Table S3 #1, 2). Consistent with our model, 65% of offspring of males with chinmo1-mutant GSCs clones inherited the chinmo1 mutant allele and 35% inherited the GFP-positive sister chromosome (Fig. 6D, second bar, Table S3 #3, 4). We see a similar trend of biased inheritance with the chinmok13009 allele, (Fig. 6D, last bar, Table S3 #5, 6). These results indicate that GSC competition can cause biased inheritance.
Declining Chinmo levels in GSCs cause physiological aging of the testis niche
To assess whether the competition phenotypes could also be observed in physiological processes like aging and whether they also depended on reduced Chinmo expression, we examined testes from 2- (“young”) and 42-day-old (“aged”) males. Chinmo protein is expressed at a moderate level in GSCs in young testes (Fig. 7A,C) but is significantly decreased in GSCs in aged testes (Fig. 7B,C). A time course revealed that Chinmo is progressively and significantly decreased in GSCs during adulthood (Fig. 7C). The average total number of GSCs significantly declines during adulthood, and by 42 days there are on average 6.3 GSCs (Fig. 7D, Table S1 #283). This is similar to the decline in average total number of GSCs when chinmo−/− clones are induced in young males (Fig. 1L). A moat composed of Pcan and Lan is present in the lumen of aged but not young testes (Fig. 7E–K). Most testes from aged males had a moat, a significant increase compared to young males (Fig. 7I), that was phenotypically indistinguishable from the moat caused by competition (Fig. 2D,F). The moat is a valid age-related phenotype because it was observed two distinct genotypes, OregonR and nos>lacZ (see below). Additionally, GSCs in aged WT testes have increased Dg at the GSC-niche interface (Fig. 7M,N), similar to what we observe in chinmo−/− GSCs in younger males (Fig. 4B). Increased Dg was not observed in GSCs in young WT testes (Fig. 7L,N).
Figure 7: Age-related phenotypes of the testis stem cell niche are caused by declining Chinmo levels in GSCs.
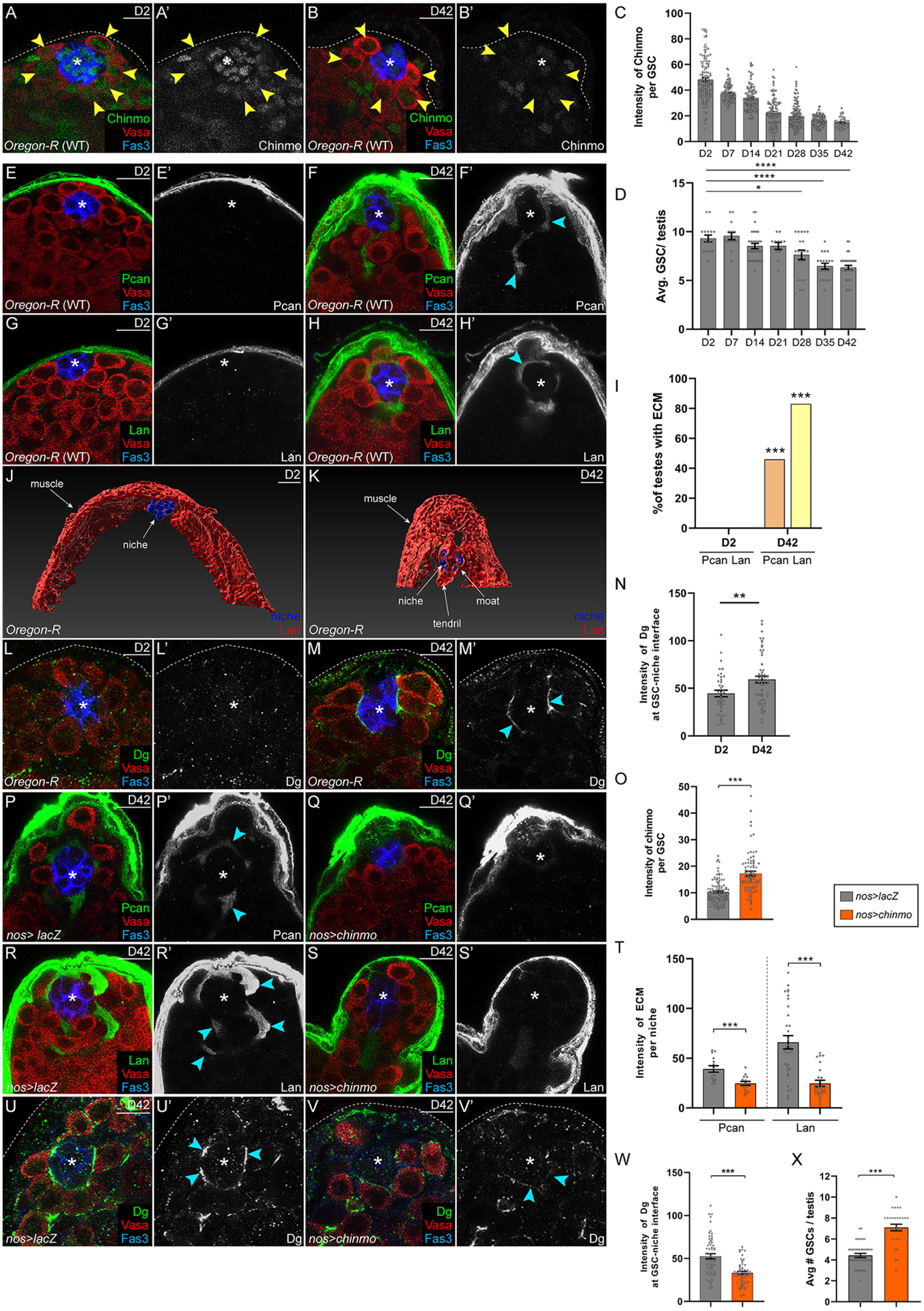
(A-D) Confocal images of young (2-day-old) (A) or aged (42-day-old) (B) WT testes. Chinmo (green). Arrowheads indicate GSCs in A,B. (C,D) Graphs showing Chinmo levels in GSCs (C) and average number of GSCs/testis (D) during aging.
(E-H) Confocal image of Pcan (green in E,F) and Lan (green in G,H) in young (E,G) and aged testes (F,H). (I) Graph showing percentage of testis with Pcan (orange) or Lan (yellow) surrounding the niche.
(J,K) Imaris-generated views of young (J) and aged (K) testes. Niche cells (blue) are visible as a ball (J,K). In a young testis, some niche cells interact with Lan (red) present in muscle BL (J). In an aged testis, some niche cells are partially obscured by ectopic Lan (red) in the testis lumen (K).
(L-N) Confocal image of young (L) and aged (M) testis stained with Dg (green). (N) Graph showing relative Dg levels at the GSC-niche interface in young or old testes.
(O) Graph showing Chinmo intensity in GSCs in nos>lacZ (gray) or nos>chinmo (orange).
(P-T) Confocal images of aged nos>lacZ (P,R) and aged nos>chinmo (Q,S) testes stained for Pcan (green in P,Q) or Lan (green in R,S). (T) Graph showing ECM intensity in aged nos>lacZ (gray) or aged nos>chinmo (orange) testes.
(U-X) Confocal images of aged nos>lacZ (U) or aged nos>chinmo (V) testes stained with Dg (green). Arrowheads indicate Dg at GSC-niche interface. (W) Graph of Dg intensity at the GSC-niche interface in aged nos>lacZ (gray) or in aged nos>chinmo (orange) testes.
(X) Graph showing number of GSCs in aged nos>lacZ (gray) or in aged nos>chinmo (orange) testes.
In A,B,E-H,L,M,P-S,U,V, Vasa is red and Fas3 is blue.
Scale bar = 10 μM
In C,D,N,O,T,W,X, error bars represent SEM.
* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; **** P ≤ 0.0001 as assessed by Student’s t-test (C,D,N,O,T,W,X) and by χ2 test (I).
See also Table S1.
To determine whether these aging phenotypes could be reserved by artificially maintaining Chinmo levels in GSCs throughout adulthood, we over-expressed Chinmo using nos (nos>chinmo) (Fig. 7O). As the control, we over-expressed a neutral protein lacZ using the same promoter (nos>lacZ). We found that all age-related phenotypes (the moat, Dg upregulation, and reduced GSC number) were significantly suppressed by maintaining moderate levels of Chinmo in GSCs throughout adulthood (Fig. 7O–X, Table S1 #284, 285). These data demonstrate that aging of the Drosophila testis stem cell niche is regulated by declining levels of Chinmo in GSCs. They also indicate that chinmo−/− GSCs in young males coopt an aging mechanism to remodel the niche to benefit themselves and to disadvantage non-mutant neighbors.
Chinmo represses Pcan expression in somatic stem cells
To test the possibility that Chinmo represses Pcan in other Drosophila stem cells to maintain tissue homeostasis, we examined the role of Pcan in male-to-female sex transformation of CySCs induced by loss of Chinmo. CySCs lacking chinmo transdifferentiate into ovarian, epithelial cells that express Fas3 and that generate an ectopic BL composed of Pcan (Fig. S7A–C”’, arrowheads, E). chinmo-mutant CySCs depleted for Pcan had significantly reduced Pcan deposition in the testis lumen (Fig. S7D–E), suggesting that the chinmo-deficient stem cells were the source of Pcan and that Chinmo normally represses Pcan in CySCs. Depleting Pcan from chinmo-deficient CySCs significantly reduced Fas3 expression, indicating that sex transformation was at least partially blocked (Fig. S7E). Thus, Chinmo represses Pcan expression in at least two distinct stem cell populations to maintain tissue function.
DISCUSSION
This work reveals an unexpected model of GSC competition that results in biased inheritance (Fig. 6E). These results provide mechanistic demonstration of the postulated “mitotic drive” by which germline stem cells with a competition advantage transmit competitive alleles at greater than the expected 50% Mendelian ratio (Otto and Hastings, 1998). Studies in plants, yeast, flies and mice have shown that selfish genetic elements can cause gene drive through various molecular mechanisms. These include “meiotic drivers” that coopt meiotic divisions or that kill viable gametes which do not inherit the selfish element (Bravo Nunez et al., 2018). “Mitotic drive” occurs earlier in the germline lineage, at the stem cell level, and is an understudied area. Prior work in the Drosophila ovary has shown that dedifferentiation-defective female GSCs “win” by upregulating E-Cad at the GSC-niche interface and gradually pushing WT GSCs out of the niche (Jin et al., 2008). Since differentiation-defective GSCs do not differentiate into gametes, this study could not examine biased allele inheritance. Female GSCs with 4x gene dose of Myc were reported to be “winners” but biased inheritance was not assessed (Rhiner et al., 2009).
Given the competitive advantage of GSCs lacking chinmo, why has evolution not selected for male GSCs with no chinmo expression? Since chinmo is an essential gene required for development (Zhu et al., 2006), loss of chinmo in GSCs might cause reduced chinmo expression in other tissues, likely reducing organismal fitness. Furthermore, chinmo-dependent competition is a progressive phenotype requiring at least two weeks of adulthood. If males with mosaic chinmo−/− clones in the germline were mated as young adults, both the chinmo+ and chinmo− allele would be passed on to offspring, and this would be sufficient to maintain chinmo+ allele in the population. Although the GSC pool in testes with chinmo−/− cells is often monoclonal, why is the chinmo− allele is not passed on to 100% of offspring. We maintain males as virgins until we dissect their testes or until mating. This means that both chinmo− GFP-negative and ubi-GFP-positive spermatids are stored in the seminal vesicle throughout the male’s lifetime and the single round of mating in our experiments allows for the transmission of both types of sperm.
We identified three phenotypes – competition, aging and transdifferentiation – that are dependent on ectopic Pcan expression in chinmo-deficient stem cells. This remarkable finding raises the important question of what factors regulate Chinmo expression in GSCs during adulthood and what genes are direct targets of Chinmo. We identified chinmo as a JAK/STAT target gene (Flaherty et al., 2010), and but STAT-deficient GSCs still express Chinmo protein (not shown), indicating that as-yet unidentified factors regulate Chinmo in GSCs and perhaps in other stem cells. Additionally, future molecular work will be needed to determine whether Pcan, Dg and βPS are direct Chinmo target genes in germline and somatic stem cells.
Our work raises the possibility that other mutant stem cells can “cheat” by resculpting their microenvironment and then ensuring their own retention in this remodeled milieu. Paternal age effect disorders (PAEs) encompass a broad spectrum of spontaneous congenital disorders and are thought to arise from rare selfish GSCs have that are positively selected and clonally expand (Goriely and Wilkie, 2012). While the current model of PAE postulates increased proliferation of mutant GSCs as the competitive mechanism, other selfish cellular behaviors such of the ones we have discovered could also be functioning in the mammalian testis. Cancer stem cells could utilize the mechanisms described in our study to colonize a tissue. Leukemic stem cells induce progressive remodeling of the bone marrow niche, and this altered niche favors the mutant stem cells while impairing normal hematopoietic stem cell residence and contributes to bone marrow fibrosis (Schepers et al., 2013, Sperling et al., 2017). In sum, our work raises the possibility that selfish stem cells across species cheat using the mechanisms we have discovered in competitive GSCs in the Drosophila testis.
Limitations of the Study
Although our study found significant increases in Pcan, Dg and βPS transcripts in GSCs deficient for Chinmo, our HCR FISH analyses of relative intensity are not strictly quantitative. As such, we cannot rule out that Chinmo affects other aspects of mRNA regulation, such as splicing. Because we have as-yet not obtained the transcriptome of chinmo−/− GSCs at sufficient resolution and have not been able to perform Chinmo ChIP-seq in GSCs, we cannot conclude that Chinmo directly represses these genes. It will be important in the future to determine Chinmo occupancy on chromatin in GSCs and in other stem cells. Surprisingly, the niche spaces vacated by non-mutant neighbor GSCs are occupied by CySCs and not by chinmo−/− GSCs. We do not understand why the CySCs predominate, and future studies employing ex vivo live-imaging will be important to gain insights into this process.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erika Bach (erika.bach@nyu.edu).
Materials availability
This study did not generate new unique reagents. All Drosophila stocks generated in this study are available from the Lead Contact without restriction.
Data and code availability
All data reported in this paper will be shared by the Lead Contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Lines and Maintenance
Drosophila melanogaster strains used in this study are listed in the Key Resources Table. Drosophila were reared on food made with these ingredients: 1800mL molasses (LabScientific, Catalog no. FLY-8008–16), 266 g agar (Mooragar, Catalog no. 41004), 1800 g cornmeal (LabScientific, Catalog no. FLY-8010–20), 744g Yeast (LabScientific, Catalog no. FLY-8040–20F), 47 L water, 56 g Tegosept (Sigma no. H3647–1KG), 560mL reagent alcohol (Fisher no. A962P4), and 190mL propionic acid (Fisher no. A258500). Flies were raised at 25°C except nos-Gal4 and mef2-Gal4 crosses, which were maintained at 18°C until eclosion, and the adult flies were transfer to 29°C.
We used the following Drosophila stocks: nos-Gal4-VP16 (Van Doren et al., 1998); tj-Gal4 (Kyoto #104055); UAS-lacZ (BDSC #3956); UAS-Pcan-i #1 (BDSC #33642) and #2 (BDSC #29440); Pcan-i on II (VDRC #24549); UAS-control-i (BDSC #61501); UAS-chinmo-i (BDSC #33638); UAS-LanB1-i (BDSC #42616); UAS-βPS-i #1(BDSC #33642) and #2 (BDSC #27735); UAS-talin-i (BDSC #32999); UAS-Dg-i (BDSC #34895); mef2-Gal4 (BDSC #50742); UAS-Dg (Deng et al., 2003); βPS-GFP (Klapholz et al., 2015); UAS-PcanRG (Cho et al., 2012); chinmo1 (Zhu et al., 2006); chinmok13009; (Kyoto #111100); tub-Gal80ts (McGuire et al., 2004); UAS-5’UTR-chinmo-3’UTR (Zhu et al., 2006). For a list of full genotypes by figure, see Table S4.
We used only adult Drosophila males in this study.
METHODS DETAILS
Drosophila Genetics and Clonal Analysis
Negatively-marked GSC clones were generated using the FLP/FRT technique after a single 1 hour heat shock at 37°C in 2-day old adult males (Xu and Rubin, 1993). Males were returned to 25°C until dissection at 2, 7, 14, 21, 23, 28 dpci.
Positively marked clones were generated by the MARCM technique after a single 1 hour heat shock at 37°C in 2-day old adult males (Lee and Luo, 1999). Males were returned to 25°C until dissection at 2, 14 dpci.
Lineage-wide mis-expression or depletion was achieved using the Gal4/UAS system (Brand and Perrimon, 1993). A Gal80ts transgene was used with nos-Gal4 to deplete Dg or Talin from adult GSCs in RNA-i efficiency experiments in Fig. S1O,P,T,U,W and X (McGuire et al., 2004). tj-Gal4 was used to deplete Chinmo from CySCs for 5 days of adulthood in Fig. S7A–D.
Immunofluorescence
Dissections and staining were carried out as previously described (Flaherty et al., 2010). Briefly, testes were dissected in 1x phosphate buffered saline (PBS), fixed for 15 minutes in 4% paraformaldehyde (PFA) in 1xPBS, washed for 1 hour at 25°C in 1xPBS with 0.5% Triton X-100, and blocked in PBTB (1xPBS 0.2% Triton X-100 and 1% bovine serum albumin) for 1 hour at 25°C. Primary antibodies were incubated overnight at 4°C except for pSTAT which was incubated overnight at 25°C. They were washed two times for 30 minutes in PBTB and incubated for 2 hours in secondary antibody in PBTB at 25°C and then washed two times for 30 minute in 1xPBS with 0.2% Triton X-100. They were mounted in Vectashield or Vectashield with DAPI (Vector Laboratories). For 5-ethynyl-20-deoxyuridine (EdU, Life Technologies) labeling, samples were incubated for 30 minutes before fixation in Ringer’s medium containing 10 μM EdU. Testes were fixed and processed normally for antibody labeling and then treated per manufacturer’s instructions. Confocal images were captured using Zeiss LSM 510 and LSM 700 microscopes with a 63x objective. Z-stacks for 2D and 3D images were captured on a Nikon W1 spinning disk confocal microscope with lasers at 405, 488, 561, 640 nm, narrow pass filters for emission, a SR HP Plan Apo 100X 1.35 Silicon Oil λS DIC lens, and an Andor 888 Live EMCCD camera.
Electron Microscopy
Prior to fixation, we dissected 10 testes of each genotype and visualized the extent of GFP-positivity (indicating non-mutant germ cells) under a GFP-dissecting microscope. Germ cells are the most numerous cell type in the testis, and it is relatively straightforward to gain a rough assessment of germline clonality in mosaic testes using GFP. Most of the testes with control clones contained approximately equal levels of GFP-negative and GFP-positive germ cells. Most of the testes with chinmo−/− clones contained few GFP-positive germ cells, indicating that the clones had outcompeted the non-mutant neighbors. All of the samples were processed for TEM, and we are presenting in Fig. 2A,B representative examples of each genotype.
Drosophila testes were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) with 1 mM CaCl2 for 2 hours. After fixation, they were treated with 1% osmium tetroxide for 1 hour, followed by block staining with 1% uranyl acetate aqueous solution overnight at 4°C. The samples were rinsed in water, dehydrated in graded series of ethanol, infiltrated with propylene oxide/EMbed 812 mixtures and embedded in EMbed 812 resin (Electron Microscopy Sciences, PA USA). 70 nm ultra-thin sections were cut and mounted on 200 mesh copper grids and stained with uranyl acetate and lead citrate. Imaging was performed by Talos120C transmission electron microscope (Thermo Fisher Scientific, Hillsboro, OR) and recorded using Gatan (4k × 4k) OneView Camera with software Digital Micrograph (Gatan Inc., Pleasanton, CA).
3D Image Rendering –
ImageJ 1.53 and Imaris 9.7 were used to generate 2D and 3D images in Fig. 2H,I, Fig. 7J,K and Movie S1.
Inheritance Assay
We induced control, chinmo1 or chinmok13009 GSC clones and aged the males for 21 days. Each male was mated singly to two OregonR (WT) virgins for 2 days. At 23 dpci, we dissected the testes of each mated male to determine the germline clonality. We scored the percentage of adult F1 offspring from the mated male for the inheritance of the chinmo+ allele (from control GSC clones) or the chinmo− allele (from chinmo1 or chinmok13009 GSC clones) by the lack of GFP expression under a Zeiss Stemi 11 GFP-dissecting scope. We scored inheritance in F1 offspring of the sister chromosome, which carries a ubi-GFP transgene, by the expression of GFP.
Hybridization Chain Reaction (HCR) Fluorescent in situ Hybridization (HCR-FISH)
We purchased from Molecular Instruments, Inc. the HCR probe set against Pcan, Dg and βPS mRNAs, the HCR amplifier, and the hybridization, wash, and amplification buffers. The protocol for immunostaining with HCR-FISH was adapted from (Zimmerman et al., 2013, Choi et al., 2018). Briefly, testes were fixed in 4% PFA in 0.1% Triton X-100 in 1xPBS-DEPC for 30 minutes at 25°C, washed with 0.5% Triton X-100 in 1xPBS-DEPC two times for 30 minutes at 25°C. Samples were blocked in 0.1% Triton X-100 in 1xPBS-DEPC with 50 μg/mL heparin and 250 μg/mL yeast tRNA (buffer hereafter called “PBTH”), and then they were then incubated with primary antibodies overnight at 4°C. The next day, the samples were washed twice in PBTH for 30 minutes. Samples were then incubated with fluorescently-labeled secondary antibodies in PBTH for 2 hours at 25°C. Samples were washed in PBTH twice for 30 minutes at 25°C. Samples were then dehydrated and rehydrated with a series of ethanol washes (25%, 50%, 75%, 100%) in 1xPBS-DEPC for 10 minutes at 25°C. Samples were treated for 7 minutes with 50 μg/mL Proteinase K, which was then inactivated by washing with 0.2% glycine twice in 1xPBS-DEPC for 5 minutes at 25°C. After Proteinase K treatment, the samples were fixed again in 4% PFA in 1xPBS-DEPC for 30 minutes at 25°C. The re-fixed samples were pre-hybridized in hybridization buffer provided by Molecular Instruments Inc. for 10 minutes at 25°C and then incubated with HCR probes overnight (12 – 16 hr) at 37°C. Samples were then washed 6 times 10 minutes at 37°C with wash buffer provided by Molecular Instruments Inc. and then twice for 5 min in 5xSCC at 25°C. Samples were incubated in amplification buffer provided by Molecular Instruments Inc. for 5 minutes at 25°C. The secondary reagents called “Hairpin h1 DNA” and “Hairpin h2 DNA” were prepared by heating each for 90 seconds at 95°C and cooling them at 25°C in a dark drawer for 30 minutes. Hairpin h1 DNA and Hairpin h2 DNA were mixed together at a 1:1 ratio and then added to the samples, which were then incubated in the dark environment overnight (16 hr) at 25°C. Samples were washed 6 times for 5 minutes with 5xSSC at 25°C and mounted in Vectashield plus DAPI for confocal analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Pcan/Lan in the moat
We captured confocal z-stacks (at 1 μM intervals) encompassing all niche cells (typically 16–20 slices) in testes with GSC clones and stained then with Pcan or Lan antibodies. The niche was counted as “with Pan/Lan” if the ECM protein was present next to at least one niche cell facing the testis lumen in any of the slices.
Quantification of βPS-GFP/Dg/Talin/E-Cad expression at the GSC-niche interface
We captured confocal z-stacks (at 1 μM intervals) encompassing all niche cells (typically 16–20 slices). We measured fluorescence intensity by ImageJ in single z slices at the area of maximal contact of a GSC with the niche. The background signal was measured in the nucleus of a gonialblast then subtracted from each GSC-niche measurement. In the mis-expression/knockdown experiments performed using the Gal4/UAS technique, each data point represents the intensity of βPS-GFP/Dg/Talin in one GSC. In the clonal analyses, the fluorescence intensity of Dg/Talin/E-Cad at the GSC-niche interface of the clone was normalized to that of a non-mutant neighbor GSC. In the clonal analyses, each data point represents one GSC.
Quantification of Chinmo protein and Pcan, Dg, βPS mRNAs in GSCs
We captured confocal z-stacks (at 1 μM intervals) encompassing all niche cells (typically 16–20 slices). We measured Chinmo (or HCR-probe) fluorescence intensity by ImageJ in a single z slice taken at the maximal width of the GSC. The background signal was measured in the nucleus of a muscle cell then subtracted from each measurement. Each data point represents one GSC.
Quantification of Pcan/Lan expression in the muscle basal lamina and in the testis lumen
We captured confocal z-stacks (at 1 μM intervals) encompassing the entire width of testes including the basal lamina and all niche cells (typically 20–25 slices). Measurements were performed using ImageJ on a single z section taken at the position where the niche attaches to the basal lamina. The background signal was measured in the nucleus of a niche cell then subtracted from each measurement. Each data point represents Pcan/Lan intensity in one testis.
Quantification of mis-oriented centrosomes in GSC clones
Centrosomes were labeled with a γ-tubulin antibody. GSCs with mis-oriented centrosomes were defined as having neither the mother nor the daughter centrosome located next to the niche as described in (Cheng et al., 2008). To measure the percentage of GSCs with mis-oriented centrosomes, we scanned each testis with GSC clones on a laser scanning confocal microscope. Z-sections were taken at 1 μM intervals (typically 16–20 slices in total/testis). We calculated the number of GFP-negative GSC clones and GFP-positive non-mutant neighbor GSCs with mis-oriented centrosomes (Fig. S2F).
Quantification of the GSC-niche distance
To measure the distance from GSCs to the niche (Fig. S3C), we used ImageJ to analyze images of testes captured on a laser scanning confocal microscope. We measured the distance from the GSC plasma membrane to the edge of the closest niche cell for each GSC in testes with control or chinmo−/− GSCs clones at 28 dpci.
Quantification of the number of testes with a moat
Testis with control or chinmo1 GSC clones were examined at 2, 7, 14, 21, 28 dpci for a gap (corresponding to the moat) between the GSCs and niche cells (Fig. S3D).
Statistical Analysis
Statistical analyses were performed using two-tailed Student’s t-tests except in Fig. 1K, which was performed using two-way Anova, and in Figs. 2G, 2N, 2O, 5F, 6D, 7I, S2F, S3K, and S7E, which were performed using χ2 tests; and Fig. S3D, which was performed by a Mann-Whitney test. Data were analyzed by GraphPad Prism and Microsoft Excel. Statistical significance was assumed by P < 0.05. Individual P values are indicated. Data are represented by the mean and standard error of mean (SEM), except Fig. S3D in which data are represented by the mean and standard deviation (SD).
Supplementary Material
Table S1 (excel file): CySC counts, clone occupancy, GSC number, number of neighbors GSC, and number of GSC clones in the analyzed genotypes (control and chinmo GSC clones), related to Figs. 1, 3, 4 and 7.
Movie S1 (mp4 file), related to Fig. 2
mp4 movie of Imaris-generated views of a testis with control GSC clones (left) and with chinmo-mutant GSC clones (right). Niche cells are shown by their blue nuclei and Lan (red) is highly expressed in the muscle basal lamina. The time point is 14 dpci. See also Fig. 2H,I.
(Left) Niche cells are visible as a ball. Some niche cells adhere to Lan present in the muscle basal lamina, thereby anchoring the niche to the apical tip of the testis.
(Right) Niche cells are visible as a ball. Some niche cells adhere to and are partially obscured by a distended “hat” of Lan (upper arrow labeled “moat”) that is contiguous with Lan in the muscle basal lamina. In addition, there is ectopic Lan in the testis lumen. Some of this ectopic Lan is present as three “claws” on distal niche cells (lower arrow labeled “moat”). An “tendril” of ectopic Lan extends from these three “claws” to the right side towards the muscle basal lamina.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP (1:1000) | Invitrogen | Cat# A6455 |
| Goat polyclonal anti-Vasa (1:200) | Santa Cruz | Cat# sc26877; RPID: AB_793877 |
| Rabbit polyclonal anti-Zfh1 (1:200) | K. White (University of Chicago, USA) | N/A |
| Guinea pig polyclonal anti-Traffic jam (Tj) (1:1000) | D. Godt (University of Toronto, Canada) | N/A |
| Rabbit polyclonal anti-Pcan (1:1000) | Baumgartner lab | N/A |
| Chicken polyclonal anti-Vasa (1:200) | P. Rangan (SUNY, Albany, USA) | N/A |
| Rat anti-Chinmo (1:200) | N. Sokol (Indiana University, USA) | N/A |
| Chicken polyclonal anti-GFP (1:1000) | Abcam | Cat# ab13970 RRID:AB_300798 |
| Guinea pig anti-Lan (1:1000) | T. Volk (Weizmann Institute, Israel) | N/A |
| Rabbit polyclonal anti-Dg (1:500) | Baumgartner lab | N/A |
| Mouse monoclonal anti-Talin carboxy terminus 534 amino acids (1:20) | Developmental Studies Hybridoma Bank (DSHB) | Cat# Talin E16B, RRID:AB_10683995 |
| Mouse monoclonal anti-Talin carboxy terminus 534 amino acids (1:20) | DSHB | Cat# Talin A22A, RRID:AB_10660289 |
| Mouse monoclonal anti-β-galactosidase (1:50) | DSHB | Cat# 40-1a RRID:AB_528100 |
| Mouse monoclonal anti-βPS integrin (1:20) | DSHB | Cat# cf.6g11 RRID:AB_528310 |
| Guinea pig anti-Myc (1:50) | G. Morata (CSIC-UAM, Spain) | N/A |
| Rabbit anti-cleaved Dcp-1 (1:500) | Cell Signaling | Cat# 9578 RRID:AB_2721060 |
| Mouse monoclonal anti-γ-tubulin (1:1000) | Sigma-Aldrich | Cat# T6557 RRID:AB_477584 |
| Rabbit polyclonal anti-phospho-Mad (1:1250) | E. Laufer (Columbia University, USA) | N/A |
| Rabbit polyclonal anti-pSTAT (1:50) | Bach lab | N/A |
| Rat monoclonal anti-E-Cad (1:5) | DSHB | Cat# DCAD2 RRID:AB_528120 |
| Rabbit polyclonal anti-ColIV (1:500) | B. Hudson (Vanderbilt University, USA) | N/A |
| Rabbit polyclonal anti-Testican (1:500) | Baumgartner lab | N/A |
| Rabbit polyclonal antiNidogen (1:400) | Baumgartner lab | N/A |
| Rabbit polyclonal anti-Sparc (1:200) | M. Ringuette (University of Toronto, Canada) | N/A |
| Mouse monoclonal anti-αPS1 integrin (1:100) | DSHB | Cat# dk.1a4 RRID:AB_528303 |
| Mouse monoclonal anti-αPS2 integrin (1:100) | DSHB | Cat# cf.2c7 RRID:AB_528304 |
| Rabbit polyclonal anti-αPS3 integrin (1:100) | S. Hayashi (RIKEN Center for Developmental Biology, Japan) | N/A |
| Rabbit polyclonal anti-αPS4 integrin (1:100) | M. Crozatier (Université de Toulouse, France) | N/A |
| Mouse monoclonal anti-βν integrin (1:200) | Y. Nakanishi (Kanazawa University, Japan) | N/A |
| Mouse monoclonal anti-Drosophila α-Spectrin (1:20) | DSHB | Cat# 3A9 RRID:AB_528473 |
| Cy3-AffiniPure Donkey Anti-Mouse IgG (1:400) | Jackson ImmunoResearch Labs | Cat# 715-165-150 RRID:AB_2340813 |
| Alexa Fluor 488-AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 711-545-152 RRID:AB_2313584 |
| Cy3-AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 711-165-152 RRID:AB_2307443 |
| Cy5-AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 711-175-152 RRID:AB_2340607 |
| Alexa Fluor 488-AffiniPure Donkey Anti-Rat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 712-545-150 RRID:AB_2340683 |
| Cy3-AffiniPure Donkey Anti-Rat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 712-165-150, RRID:AB_2340666 |
| Cy5-AffiniPure Donkey Anti-Rat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 712-175-150, RRID:AB_2340671 |
| Alexa Fluor 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 703-545-155, RRID:AB_2340375 |
| Cy3-AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 703-165-155, RRID:AB_2340363 |
| Cy5-AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 703-175-155, RRID:AB_2340365 |
| Cy3-AffiniPure Donkey Anti-Guinea Pig IgG (1:400) | Jackson ImmunoResearch Labs | Cat# 706-165-148, RRID:AB_2340460 |
| Cy5-AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 706-175-148, RRID:AB_2340462 |
| Cy3-AffiniPure Donkey Anti-Goat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 705-165-003, RRID:AB_2340411 |
| Alexa Fluor 647 AffiniPure Donkey Anti-Goat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 705-605-003, RRID:AB_2340436 |
| Chemicals, peptides, and recombinant proteins | ||
| VECTASHIELD Mounting Medium with DAPI | Vector Laboratories | Cat# H-1200, RRID:AB_2336790 |
| VECTASHIELD Mounting | Vector Laboratories | Cat# H-1000, RRID:AB_2336789 |
| Paraformaldehyde, 16% w/v aq. soln., methanol free (PFA) | Thermo Fisher Scientific | Cat# 43368-9L |
| Heparin | Sigma-Aldrich | Cat# H4784 |
| tRNA | Roche | Cat# 10109495001 |
| Protector RNase Inhibitor | Roche | Cat# 3335399001 |
| 20 × Saline Sodium Citrate (SSC) | Thermo Fisher | Cat# 15557-044 |
| Proteinase K | Thermo Fisher Scientific | Cat# EO0491 |
| Glycine | Thermo Fisher Scientific | Cat# BP381-500 |
| Diethyl pyrocarbonate (DEPC) | MilliporeSigma | Cat# D5758 |
| Click-iT EdU Imaging Kits | Thermo Fisher Scientific | Cat# C10340 |
| Molasses | Labscientific | Cat# FLY-8008-16 |
| Agar | Mooragar | Cat# 41004 |
| Cornmeal | LabScientific | Cat# FLY-8010-20 |
| Yeast | LabScientific | Cat# FLY-8040-20F |
| Tegosept | Sigma | Cat# H3647-1KG |
| Reagent alcohol | Fisher | Cat# A962P4 |
| Propionic acid | Fisher | Cat# A258500 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster; y, w, hs-flp122/Y; ubi-GFP, FRT40A | Bach lab | N/A |
| D. melanogaster; w; chinmo1,UAS-mCD8-GFP, FRT40A/CyO | T. Lee (Janelia Research Camp, USA) | N/A |
| D. melanogaster; w; P[ry[+t7.2]=neoFRT]40A; | Bach lab | N/A |
| D. melanogaster; y, w, hs-flp122, tub-Gal4, UAS-nls-GFP/Y; tub-Gal80, FRT40A/CyO | Bach lab | N/A |
| D. melanogaster; y1 v1; P[TRiP.GL01153]attP2 (Pcan-i #1) | BDSC | RRID:BDSC_42783 |
| D. melanogaster; y1 v1; P[TRiP.JF03376]attP2 (Pcan-i #2) | BDSC | RRID:BDSC_29440 |
| D. melanogaster; y, w; P[w+, nos-GAL4 VP16] on II | Bach lab | N/A |
| D. melanogaster; w1118; P[w[+mC]=UAS-lacZ.NZ]J312 Insertion on III | BDSC | RRID:BDSC_3956 |
| D. melanogaster; w; UAS-PcanRG Insertion on III | A. Kolodkin (Johns Hopkins School of Medicine, USA) | N/A |
| D. melanogaster; y1 sc* v1 sev21;P[TRiP.HMS01240]attP 2(Dg-i) | BDSC | RRID:BDSC_34895 |
| D. melanogaster; w; P[lacW]chinmok13009, FRT40A/CyO | Kyoto Stock Center | RRID:DGGR_111100 |
| D. melanogaster; y, w, βPS-GFP | N. Brown (University of Cambridge, UK) | N/A |
| D. melanogaster; y1 v1; P[TRiP.HMS00036]attP2/TM3, Sb1 (chinmo-i) | BDSC | RRID:BDSC_33638 |
| D. melanogaster; y1 v1; P[TRiP.HMS00043]attP2 (βPS-i #1) | BDSC | RRID:BDSC_33642 |
| D. melanogaster; y1 v1; P[TRiP.JF02819]attP2 (βPS-i #2) | BDSC | RRID:BDSC_27735 |
| D. melanogaster; w; UAS-Dg Insertion on III | W. Deng (Tulane University, USA) | N/A |
| D. melanogaster; y1 sc* v1 sev21; P[TRiP.HMS00799]attP2 (talin-i) | BDSC | RRID:BDSC_32999 |
| D. melanogaster; y1 sc* v1 sev21;P[TRiP.HMS02451]attP 2(Lan-i) | BDSC | RRID:BDSC_42616 |
| D.melanogaster; w*; P[Mef2-Gal4.247]3 | BDSC | RRID:BDSC_50742 |
| D. melanogaster; y1 sc* v1 sev21; P[TRiP.HMS00799]attP2 (talin-i) | BDSC | BDSC: 32999 FlyBase: FBst0032999 |
| D. melanogaster; w; +; UAS-5’UTR-chinmo-3’UTR | T. Lee (Janelia Research Camp, USA) | N/A |
| D. melanogaster; w*; P[UAS-mCherry.scramble.sponge]att P40; P[UAS mCherry.scramble.sponge]att P2 (control-i) | BDSC | RRID:BDSC_61507 |
| D. melanogaster; w1118; P[GD7744]v24549 (Pcan-i on II) | VDRC | RRID:SCR_24549 |
| D. melanogaster; w*; P[GawB]NP1624/CyO; (tj-Gal4) | Bach lab | RRID:DGGR_104055 |
| D. melanogaster; w 1118 ; P[w+mC=UAS-Dcr-2.D]10 (UAS-Dcr-2) | Bach lab | RRID:BDSC_24651 |
| D.melanogaster; w*; P[tubP-Gal80ts]2/TM2 | BDSC | RRID:BDSC_7071 |
| Oligonucleotide | ||
| HCR probe of Pcan | Molecular Instruments, Inc. | LOT: PRJ993 |
| HCR probe of Dg | Molecular Instruments, Inc. | LOT: PRJ994 |
| HCR probe of βPS | Molecular Instruments, Inc. | LOT: PRJ995 |
| Software and algorithms | ||
| ImageJ/Fiji | Fiji | http://fiji.sc/ |
| Photoshop/Illustrator | Adobe | https://www.adobe.com/products/ |
| Prism | GraphPad | https://www.graphpad.com |
| ZEN | Zeiss | https://www.zeiss.com/microscopy/us/products/microscopesoftware/zen.html |
| Excel | Microsoft | https://products.office.com/en-us/excel |
| Imaris | Oxford Instruments | https://imaris.oxinst.com/ |
Highlights.
chinmo-mutant GSCs secrete ECM proteins to remodel the niche, evicting WT GSCs.
chinmo-mutant GSCs remain in the altered niche by increasing ECM-binding proteins.
Inheritance of the chinmo-mutant allele is biased and occurs in >50% of F1 progeny.
Aged testes have a remodeled niche caused by declining levels of Chinmo in GSCs.
ACKNOWLEDGEMENTS
We thank NYU Langone Health DART Microscopy Laboratory, A. Liang, C. Petzold and K. Dancel-Manning for consultation and assistance with TEM work. The Microscopy Laboratory is partially supported by Laura and Isaac Perlmutter Cancer Center Support Grant NIH/NCI P30CA016087. We thank K. White, D. Godt, P. Rangan, N. Sokol, T. Volk, G. Morata, E. Laufer, B. Hudson, M. Ringuette, S. Hayashi, M. Crozatier, Y. Nakanishi for antibodies, and T. Lee, A. Kolodkin, N. Brown, Bloomington Stock Center (BDSC) and Kyoto Stock Center for fly stocks. The BDSC is supported by a grant from the Office of the Director of the NIH (P40OD018537). We are grateful to FlyBase, which is supported by a grant from the National Human Genome Research Institute at NIH (U41 HG000739).
Work is the Bach lab is supported by grants from the NIH (R03-HD090422; R01-GM085075). CYT was supported by a New York State Department of Health/NYSTEM institutional training grant (#C322560GG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no interests.
REFERENCES
- AMOYEL M, ANDERSON J, SUISSE A, GLASNER J & BACH EA 2016. Socs36E Controls Niche Competition by Repressing MAPK Signaling in the Drosophila Testis. PLoS Genet, 12, e1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMOYEL M & BACH EA 2014. Cell competition: how to eliminate your neighbours. Development, 141, 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMOYEL M, SIMONS BD & BACH EA 2014. Neutral competition of stem cells is skewed by proliferative changes downstream of Hh and Hpo. EMBO J, 33, 2295–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAND AH & PERRIMON N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–15. [DOI] [PubMed] [Google Scholar]
- BRAVO NUNEZ MA, NUCKOLLS NL & ZANDERS SE 2018. Genetic Villains: Killer Meiotic Drivers. Trends Genet, 34, 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROADIE K, BAUMGARTNER S & PROKOP A 2011. Extracellular matrix and its receptors in Drosophila neural development. Dev Neurobiol, 71, 1102–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENG J, TURKEL N, HEMATI N, FULLER MT, HUNT AJ & YAMASHITA YM 2008. Centrosome misorientation reduces stem cell division during ageing. Nature, 456, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHO JY, CHAK K, ANDREONE BJ, WOOLEY JR & KOLODKIN AL 2012. The extracellular matrix proteoglycan perlecan facilitates transmembrane semaphorin-mediated repulsive guidance. Genes Dev, 26, 2222–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI HMT, SCHWARZKOPF M, FORNACE ME, ACHARYA A, ARTAVANIS G, STEGMAIER J, CUNHA A & PIERCE NA 2018. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENG WM, SCHNEIDER M, FROCK R, CASTILLEJO-LOPEZ C, GAMAN EA, BAUMGARTNER S & RUOHOLA-BAKER H 2003. Dystroglycan is required for polarizing the epithelial cells and the oocyte in Drosophila. Development, 130, 173–84. [DOI] [PubMed] [Google Scholar]
- FABRIZIO JJ, BOYLE M & DINARDO S 2003. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol, 258, 117–28. [DOI] [PubMed] [Google Scholar]
- FLAHERTY MS, SALIS P, EVANS CJ, EKAS LA, MAROUF A, ZAVADIL J, BANERJEE U & BACH EA 2010. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell, 18, 556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FULLER MT 1998. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin Cell Dev Biol, 9, 433–44. [DOI] [PubMed] [Google Scholar]
- GORIELY A & WILKIE AO 2012. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet, 90, 175–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREENSPAN LJ, DE CUEVAS M & MATUNIS E 2015. Genetics of gonadal stem cell renewal. Annu Rev Cell Dev Biol, 31, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRMAI L, HARSH S, LU S, KORMAN A, DEB IB & BACH EA 2021. Transcriptomic analysis of feminizing somatic stem cells in the Drosophila testis reveals putative downstream effectors of the transcription factor Chinmo. G3-Genes Genomes Genetics, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRMAI L, HUDRY B, MIGUEL-ALIAGA I & BACH EA 2018. Chinmo prevents transformer alternative splicing to maintain male sex identity. PLoS Genet, 14, e1007203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDY RW, TOKUYASU KT, LINDSLEY DL & GARAVITO M 1979. The germinal proliferation center in the testis of Drosophila melanogaster. J Ultrastruct Res, 69, 180–90. [DOI] [PubMed] [Google Scholar]
- HASTINGS IM 1989. Potential germline competition in animals and its evolutionary implications. Genetics, 123, 191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISSIGONIS M, TULINA N, DE CUEVAS M, BRAWLEY C, SANDLER L & MATUNIS E 2009. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science, 326, 153–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIN Z, KIRILLY D, WENG C, KAWASE E, SONG X, SMITH S, SCHWARTZ J & XIE T 2008. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell, 2, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLAPHOLZ B, HERBERT SL, WELLMANN J, JOHNSON R, PARSONS M & BROWN NH 2015. Alternative mechanisms for talin to mediate integrin function. Curr Biol, 25, 847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN AM & SIMONS BD 2011. Universal patterns of stem cell fate in cycling adult tissues. Development, 138, 3103–11. [DOI] [PubMed] [Google Scholar]
- LEATHERMAN JL & DINARDO S 2010. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol, 12, 806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE T & LUO L 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron, 22, 451–61. [DOI] [PubMed] [Google Scholar]
- MA Q, DE CUEVAS M & MATUNIS EL 2016. Chinmo is sufficient to induce male fate in somatic cells of the adult Drosophila ovary. Development, 143, 754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA Q, WAWERSIK M & MATUNIS EL 2014. The Jak-STAT Target Chinmo Prevents Sex Transformation of Adult Stem Cells in the Drosophila Testis Niche. Dev Cell, 31, 474–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGUIRE SE, MAO Z & DAVIS RL 2004. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE, 2004, pl6. [DOI] [PubMed] [Google Scholar]
- NARBONNE-REVEAU K, LANET E, DILLARD C, FOPPOLO S, CHEN CH, PARRINELLO H, RIALLE S, SOKOL NS & MAURANGE C 2016. Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OTTO SP & HASTINGS IM 1998. Mutation and selection within the individual. Genetica, 102–103, 507–24. [PubMed] [Google Scholar]
- RHINER C, DIAZ B, PORTELA M, POYATOS JF, FERNANDEZ-RUIZ I, LOPEZ-GAY JM, GERLITZ O & MORENO E 2009. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development, 136, 995–1006. [DOI] [PubMed] [Google Scholar]
- SALZMANN V, INABA M, CHENG J & YAMASHITA YM 2013. Lineage tracing quantification reveals symmetric stem cell division in Drosophila male germline stem cells. Cell Mol Bioeng, 6, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHEPERS K, PIETRAS EM, REYNAUD D, FLACH J, BINNEWIES M, GARG T, WAGERS AJ, HSIAO EC & PASSEGUE E 2013. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell, 13, 285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHNEIDER M, KHALIL AA, POULTON J, CASTILLEJO-LOPEZ C, EGGER-ADAM D, WODARZ A, DENG WM & BAUMGARTNER S 2006. Perlecan and Dystroglycan act at the basal side of the Drosophila follicular epithelium to maintain epithelial organization. Development, 133, 3805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHENG XR & MATUNIS E 2011. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development, 138, 3367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMONS BD & CLEVERS H 2011. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell, 145, 851–62. [DOI] [PubMed] [Google Scholar]
- SINGH SR, ZHENG Z, WANG H, OH SW, CHEN X & HOU SX 2010. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J Cell Physiol, 223, 500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNIPPERT HJ, SCHEPERS AG, VAN ES JH, SIMONS BD & CLEVERS H 2014. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep, 15, 62–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPERLING AS, GIBSON CJ & EBERT BL 2017. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer, 17, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STINE RR, GREENSPAN LJ, RAMACHANDRAN KV & MATUNIS EL 2014. Coordinate regulation of stem cell competition by Slit-Robo and JAK-STAT signaling in the Drosophila testis. PLoS Genet, 10, e1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANENTZAPF G, DEVENPORT D, GODT D & BROWN NH 2007. Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol, 9, 1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DOREN M, WILLIAMSON AL & LEHMANN R 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol, 8, 243–6. [DOI] [PubMed] [Google Scholar]
- VERMEULEN L, MORRISSEY E, VAN DER HEIJDEN M, NICHOLSON AM, SOTTORIVA A, BUCZACKI S, KEMP R, TAVARE S & WINTON DJ 2013. Defining stem cell dynamics in models of intestinal tumor initiation. Science, 342, 995–8. [DOI] [PubMed] [Google Scholar]
- WALLENFANG MR, NAYAK R & DINARDO S 2006. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell, 5, 297–304. [DOI] [PubMed] [Google Scholar]
- XU T & RUBIN GM 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–37. [DOI] [PubMed] [Google Scholar]
- YAMASHITA YM, JONES DL & FULLER MT 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science, 301, 1547–50. [DOI] [PubMed] [Google Scholar]
- ZHU S, LIN S, KAO CF, AWASAKI T, CHIANG AS & LEE T 2006. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell, 127, 409–22. [DOI] [PubMed] [Google Scholar]
- ZIMMERMAN SG, PETERS NC, ALTARAS AE & BERG CA 2013. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat Protoc, 8, 2158–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (excel file): CySC counts, clone occupancy, GSC number, number of neighbors GSC, and number of GSC clones in the analyzed genotypes (control and chinmo GSC clones), related to Figs. 1, 3, 4 and 7.
Movie S1 (mp4 file), related to Fig. 2
mp4 movie of Imaris-generated views of a testis with control GSC clones (left) and with chinmo-mutant GSC clones (right). Niche cells are shown by their blue nuclei and Lan (red) is highly expressed in the muscle basal lamina. The time point is 14 dpci. See also Fig. 2H,I.
(Left) Niche cells are visible as a ball. Some niche cells adhere to Lan present in the muscle basal lamina, thereby anchoring the niche to the apical tip of the testis.
(Right) Niche cells are visible as a ball. Some niche cells adhere to and are partially obscured by a distended “hat” of Lan (upper arrow labeled “moat”) that is contiguous with Lan in the muscle basal lamina. In addition, there is ectopic Lan in the testis lumen. Some of this ectopic Lan is present as three “claws” on distal niche cells (lower arrow labeled “moat”). An “tendril” of ectopic Lan extends from these three “claws” to the right side towards the muscle basal lamina.
Data Availability Statement
All data reported in this paper will be shared by the Lead Contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


