Abstract
The covalently closed circular DNA (cccDNA) of HBV plays a crucial role in viral persistence and is also a risk factor for developing HBV-induced diseases, including liver fibrosis. Stimulator of interferon genes (STING), a master regulator of DNA-mediated innate immune activation, is a potential therapeutic target for viral infection and virus-related diseases. In this study, agonist-induced STING signaling activation in macrophages was revealed to inhibit cccDNA-mediated transcription and HBV replication via epigenetic modification in hepatocytes. Notably, STING activation could efficiently attenuate the severity of liver injury and fibrosis in a chronic recombinant cccDNA (rcccDNA) mouse model, which is a proven suitable research platform for HBV-induced fibrosis. Mechanistically, STING-activated autophagic flux could suppress macrophage inflammasome activation, leading to the amelioration of liver injury and HBV-induced fibrosis. Overall, the activation of STING signaling could inhibit HBV replication through epigenetic suppression of cccDNA and alleviate HBV-induced liver fibrosis through the suppression of macrophage inflammasome activation by activating autophagic flux in a chronic HBV mouse model. This study suggests that targeting the STING signaling pathway may be an important therapeutic strategy to protect against persistent HBV replication and HBV-induced fibrosis.
Keywords: STING activation, Epigenetic suppression of HBV cccDNA, HBV-induced liver fibrosis, Inflammasome activation, Autophagic flux
Subject terms: Viral infection, Inflammasome, Macroautophagy
Introduction
HBV infection remains an important public health problem worldwide. Globally, ~260 million people are chronically infected with HBV, and nearly one million people die of HBV-related diseases, such as liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC), annually [1, 2]. Liver fibrosis is a key factor in the prognosis of HBV-induced liver diseases. The removal of fibrotic response-causing agents may aid in the regression of fibrosis; otherwise, HBV-associated liver fibrosis progresses toward excessive scarring and organ failure, such as in liver cirrhosis, and eventually progresses to HCC [3, 4]. However, a lack of reliable and convenient animal models has long been a bottleneck for studying the pathogenesis of HBV-induced liver fibrosis.
Nuclear-localized covalently closed circular DNA (cccDNA) plays a pivotal role in the HBV life cycle. Because of its high stability and long-term presence in the nucleus, cccDNA is the key to persistent HBV infection and the main obstacle for a cure for chronic hepatitis B (CHB) [5, 6]. In addition to its crucial role in HBV infection, cccDNA is also closely associated with the occurrence and development of HBV-induced liver diseases, including liver fibrosis [7, 8]. Inflammatory mediators, especially IL-1β, released during viral infection may play more important roles in the development of liver fibrosis than the virus itself [9–13]. Of note, the inflammatory response may often lead to a positive feedback loop in the progression of fibrosis [14, 15]. Therefore, strategies that target both the virus and the virus-induced inflammatory response may be of great value in treating HBV-induced fibrosis. Recently, Li et al. [16]. reported a recombinant cccDNA (rcccDNA) mouse model that supported long-term HBV persistence, developed significant signs of liver fibrosis, and provided a suitable platform to design strategies against CHB and HBV-induced liver fibrosis. Evidence indicates that the regulation of innate immune signaling may play a crucial role in controlling viral replication and virus-related diseases [17–20]. However, whether HBV can efficiently induce the activation of innate immune responses is somewhat controversial [21], and it is now well accepted that innate immune activation can effectively inhibit HBV replication [22, 23]. Recent evidence demonstrates that stimulator of interferon genes (STING) is a key adapter protein in DNA-induced innate immune activation [24, 25]. When cyclic GMP-AMP synthase (cGAS) senses dsDNA in the cytoplasm, it catalyzes ATP and GTP formation into the second messenger molecule cGAMP, which then binds to STING and activates TBK1/IRF3 and IKK/NF-κB signaling, thereby promoting the production of type I interferon (IFN-I) and proinflammatory cytokines, such as TNF-α and IL-6 [19, 26]. In addition to serving as the adapter protein for cGAS, STING is also a signaling hub for other DNA sensors, such as IFI16 and DDX41, and induces the production of IFN-α/β and proinflammatory cytokines [27, 28]. Of interest, recent studies have shown that STING has a typical LC3-interacting region, which can activate cellular autophagy by direct interactions with LC3 independent of TBK1 activation or interferon induction [29, 30], and evidence indicates that macrophage autophagy is crucial for alleviating liver fibrosis by inhibiting inflammatory responses [31, 32].
Given its importance in the activation of innate immunity and autophagy, STING is emerging as an important therapeutic target for a variety of diseases, including cancer, inflammatory diseases, and viral infections [33, 34]. It has been reported agonist-induced STING signaling activation promotes antitumor immunity by triggering IFN-I production in the tumor microenvironment [35], and may also contribute to the priming of CD8+ T cells against immunogenic tumors, including HCC [20, 36]. Increasing evidence indicates that STING signaling may also play a pivotal role in controlling a variety of viral diseases. Lio et al. [37] reported that STING was required for triggering the first phase of IFN-I production and curbing cytomegalovirus infection. Reinert et al. [38] reported that STING deficiency rendered mice susceptible to herpes simplex encephalitis after peripheral infection in the eye. Ding et al. [39] reported that HCV NS4B could block the interaction between STING and TBK1 to evade the host IFN response. In terms of HBV infection, Guo et al. [40] showed that the expression level of STING was low in hepatocytes but high in macrophages, and the STING agonist DMXAA induced an intense cytokine response in macrophages to effectively inhibit HBV replication in hepatocytes. The researchers also showed that activation of the STING signaling pathway in hepatocytes by cGAMP or a relatively high concentration of DMXAA also contributed to the inhibition of HBV [41]. Their data showed that HBV infection did not impair the expression of STING or the STING-activated immune response, and HBV even appeared to upregulate the expression level of STING. However, data from Liu et al. [42] revealed that HBV polymerase could directly interact with STING and subsequently disrupt its K63-linked ubiquitination to suppress the production of IFN-β. Additionally, the expression level of STING in hepatocytes is somewhat controversial. Thomsen et al. [43]. and Yu et al. [44] reported that human or murine hepatocytes lacked STING expression and thus failed to produce IFN-I in response to foreign DNA or HBV infection; in contrast, resident liver macrophages, which are known as Kupffer cells (KCs), express a level of STING and efficiently respond to cytosolic DNA. However, Guo et al. [41] and Zhu et al. [45] reported that although STING expression was undetected in Huh7 cells, HepG2 cells or HepG2-derived cell lines did express STING and thus, to some extent, sense HBV DNA. However, some reports have revealed that human hepatomas, including HepG2 cells, are not ideal for studying innate immune responses because innate immune signaling in these cells is impaired [46]. In the present study, we focused on the potential inhibitory effect of STING activation on HBV cccDNA and HBV-induced liver fibrosis.
Our data showed that activation of STING signaling in macrophages could efficiently inhibit cccDNA-driven transcription and HBV replication in hepatocytes through epigenetic suppression. Notably, our investigation further indicated that STING activation could efficiently attenuate the severity of liver injury and HBV-induced liver fibrosis by autophagic inhibition of macrophage inflammasome activation. These data suggested that, apart from inhibiting HBV replication, STING activation might attenuate liver fibrosis by suppressing inflammasome activation.
Results
STING activation inhibits cccDNA-driven transcription and HBV replication in vitro
To investigate the potential effect of STING activation on HBV cccDNA, we constructed a rcccDNA cell model by cotransfecting the prcccDNA and pCMV-Cre plasmids (prcccDNA/Cre) into AML12 cells, which is a murine hepatoma cell line, and the rcccDNA cell model was then directly treated with the STING agonist DMXAA (Fig. 1a). Direct DMXAA stimulation did not significantly affect HBV transcription or replication in this rcccDNA cell model (Fig. 1b–d and Supplementary Fig. 1a, b), as demonstrated by the small alterations in the levels of HBsAg, HBV pgRNA, intracellular core particle-associated HBV-DNA and rcccDNA. Furthermore, we treated murine macrophages (RAW264.7 cells) with DMXAA for 12 h, transferred the culture supernatants onto prcccDNA/Cre-transfected AML12 cells, and incubated these cells for another 48 h (Fig. 1e). Our data showed that the culture supernatants of DMXAA-treated RAW264.7 cells significantly downregulated the levels of HBsAg, pgRNA, and HBV DNA (Fig. 1f, g and Supplementary Fig. 1c, d) in prcccDNA/Cre-transfected AML12 cells, while the level of rcccDNA was not affected (Fig. 1h), suggesting that STING signaling activation by DMXAA in macrophages could suppress rcccDNA-driven transcription and HBV replication without affecting the level of rcccDNA itself in hepatocytes. We also examined the effect of the STING agonist cGAMP on HBV replication in this rcccDNA cell model. Similar to the effect of DMXAA, cGAMP-induced STING signaling activation in RAW264.7 macrophages efficiently suppressed rcccDNA-mediated transcription and replication without affecting cccDNA levels in AML12 cells (Supplementary Fig. 2a, b). Furthermore, we determined the effect of STING activation on HBV in another well-established cccDNA cell model by transfecting AML12 cells with linear HBV monomers, as described by Pollicino et al. (Fig. 1i) [47]. Consistent with the rcccDNA data, DMXAA treatment significantly inhibited cccDNA transcription and HBV replication but had no impact on the level of cccDNA itself (Fig. 1j).
Fig. 1.
STING signaling pathway activation inhibited HBV gene expression and replication in an rcccDNA cell model. a Schematic illustration of the direct effect of DMXAA on prcccDNA/Cre-transfected AML12 cells. Briefly, AML12 cells were transfected with prcccDNA and pCMV-Cre plasmids. Forty-eight hours posttransfection, the cells were then treated with increasing doses of DMXAA (20 and 40 μg/mL) for another 48 h. b Cells were treated as shown in a, and the levels of HBsAg in the culture supernatants, pgRNA in the transfected cells, and intracellular HBV DNA were analyzed by ELISA, qRT–PCR and qPCR, respectively. c Cells were treated as shown in a, and the levels of intracellular HBV DNA were analyzed by Southern blotting. RC, relaxed circular DNA; RI, replicative intermediate. d Cells were treated as shown in a, rcccDNA was extracted by Hirt’s procedure as described in the Materials and Methods and measured by qPCR. e Schematic illustration of the indirect effect of DMXAA on prcccDNA/Cre-transfected AML12 cells. Briefly, RAW264.7 cells were treated with increasing doses of DMXAA (10 and 20 μg/mL). Twelve hours later, the culture medium of DMXAA-treated RAW264.7 cells was collected and used to treat the prcccDNA/Cre-transfected AML12 cells for another 48 h. Cells were treated as shown in e, and the levels of HBsAg, pgRNA, and HBV-DNA (f, g) and rcccDNA (h) were determined as shown in b–d. i, j The effect of the STING agonist DMXAA on cccDNA-driven transcription and HBV replication in a linear HBV monomer-transfected cccDNA cell model. k AML12 or RAW264.7 cells were treated with different doses of DMXAA (20 and 40 μg/mL for AML12 cells; 10 and 20 μg/mL for RAW264.7 cells). 4 h later, cell lysates were collected and subjected to Western blotting using antibodies against phosphorylated IRF3 (p-IRF3), total IRF3, phosphorylated TBK1 (p-TBK1), total TBK1, IκBα, phosphorylated p65 (p-p65), total p65 and GAPDH. l, m AML12 or RAW264.7 cells were treated as shown in k, and the mRNA levels of IFN-β (l) and TNF-α (m) were quantified by qRT–PCR (normalized to GAPDH). n The protein levels of STING in RAW264.7, AML12 or prcccDNA/Cre-transfected AML12 cells were determined by Western blotting using antibodies against STING and GAPDH. o–q The effect of STING knockdown in RAW264.7 cells on DMXAA-mediated innate immune activation and inhibition of rcccDNA transcription and HBV replication. Primary human hepatocytes (PHHs) or human KCs (hKCs) were treated with the STING agonist cGAMP (5 μg/mL) for 6 h, the levels of STING and the phosphorylation of IRF3 and p65 were determined by Western blotting (r), and the induction of IFN-β and TNF-α was determined by qRT–PCR (normalized to GAPDH) (s, t). u hKCs were treated with 2 μg/mL cGAMP for 12 h, and the culture supernatants of hKCs were then collected and used to treat HBV-infected PHHs. Forty-eight hours posttreatment, the levels of secreted HBsAg, pgRNA, intracellular HBV DNA and cccDNA were determined by ELISA, qRT–PCR and qPCR, respectively. The data are shown as the means ± SD and were compared by unpaired Student’s t test or one-way analysis of variance (ANOVA). *P < 0.05, **P < 0.01, ***P < 0.001; NS, no significance
To explore why STING activation only inhibited cccDNA transcription and HBV replication in an indirect way, as shown in Fig. 1e–j, we examined the effect of DMXAA on innate immune signaling in AML12 cells and RAW264.7 cells. After DMXAA treatment, IFN-I and, to a lesser extent, NF-κB signaling in RAW264.7 cells but not in AML12 cells was significantly activated, as revealed by the increased levels of p-IRF3, p-TBK1, and p-p65 and the downregulation of IκBα (Fig. 1k and Supplementary Fig. 1e), as well as by the increased mRNA levels of IFN-β and TNF-α (Fig. 1l, m and Supplementary Fig. 1f, g). Of note, our data showed that the expression level of STING in AML12 cells and in prcccDNA/Cre-transfected AML12 cells was barely detectable, which was consistent with the lack of an effect of direct DMXAA treatment on AML12 cells in terms of HBV inhibition and innate immune activation (Fig. 1n). STING was knocked down in RAW264.7 cells, as shown in Fig. 1o, and DMXAA-mediated innate immune activation (Fig. 1p) and its inhibitory effect on rcccDNA transcription and HBV replication (Fig. 1q) were significantly downregulated. Furthermore, we examined the effect of STING activation on HBV replication in human cells. Consistent with data obtained from mouse cells, STING was mainly expressed in human Kupffer cells (hKCs) but not in primary human hepatocytes (PHHs) (Fig. 1r). Accordingly, treatment with cGAMP significantly activated innate immune signaling in hKCs but not in PHHs (Fig. 1r–t). STING signaling activation by cGAMP in hKCs significantly inhibited HBV transcription and replication without affecting the level of HBV cccDNA (Fig. 1u). It has been reported that APOBEC3 cytidine deaminases can cause the degradation of HBV cccDNA [48]. However, our data revealed that STING signaling activation failed to upregulate human APOBEC3A (hAPOBEC3A) or murine APOBEC3 (mAPOBEC3) (Supplementary Fig. 3a, b), which was consistent with the inability of STING activation to cause cccDNA degradation.
Taken together, these data indicated that STING signaling activation in macrophages could significantly inhibit rcccDNA-mediated transcription and HBV replication in hepatocytes.
STING activation inhibits HBV gene expression and replication in a chronic rcccDNA mouse model
To further investigate the effect of STING activation on cccDNA, we established a chronic rcccDNA mouse model by injecting 1.5 × 109 PFU of Ad-rcccDNA into Alb-Cre Tg mice via the tail vein (Fig. 2a), as described previously [17], and then examined the effect of STING signaling pathway activation on rcccDNA-driven transcription and HBV replication in this mouse model. Our results showed that STING signaling activation by DMXAA significantly downregulated HBsAg and HBV DNA in mouse sera (Fig. 2b, c and Supplementary Fig. 4a–d) and HBcAg-positive liver cells (Fig. 2d), pgRNA (Fig. 2e) and significantly downregulated HBV DNA in mouse livers (Fig. 2f and Supplementary Fig. 4e) but had little effect on the level of rcccDNA itself (Fig. 2g). We also evaluated the effect of DMXAA treatment on innate immune signaling in rcccDNA mice. As expected, DMXAA treatment efficiently activated IFN-I signaling and, to a lesser extent, NF-κB signaling in this HBV mouse model (Fig. 2h–k and Supplementary Fig. 4f-h). To determine which cell type was responsible for STING-mediated innate activation, we isolated primary murine hepatocytes (PMHs) and murine KCs from DMXAA-treated or untreated rcccDNA mice and then examined the innate immune responses in PMHs and KCs. Consistent with the in vitro data, STING was significantly expressed in KCs but not in PMHs, and treatment with the STING agonist DMXAA significantly activated IFN-I and NF-κB signaling in KCs but not in PMHs, as revealed by the upregulation of p-IRF3, p-TBK1, and p-p65 and the downregulation of IкBα (Fig. 2l), as well as by the increased mRNA levels of IFN-β and TNF-α (Fig. 2m, n). Taken together, these data suggested that DMXAA treatment could activate the STING signaling pathway in liver KCs and then suppress rcccDNA-mediated transcription and HBV replication in mice.
Fig. 2.
STING signaling pathway activation inhibited HBV gene expression and replication in a chronic rcccDNA mouse model. a Schematic illustration of the effect of STING activation on HBV transcription and replication in a chronic rcccDNA mouse model. Briefly, Alb-Cre Tg mice were injected with Ad-rcccDNA (1.5 × 109 PFU) by the tail vein. On Day 45 postinjection, rcccDNA mice were treated with 10 mg/kg DMXAA or vehicle by intraperitoneal injection for the indicated times. On Day 15 after the initial DMXAA treatment, the effect of STING activation on HBV replication and the innate immune response was examined. b, c Serum levels of HBsAg and HBV DNA in DMXAA-treated or untreated mice were determined by ELISA and qPCR, respectively (n = 10/group). d The levels of HBcAg in the liver tissues of DMXAA-treated or untreated mice were determined by immunohistochemistry. e The levels of pgRNA in the liver tissues of DMXAA-treated or untreated mice were analyzed by qRT–PCR (n = 10/group). f Intracellular HBV DNA in the liver tissues of DMXAA-treated or untreated mice was determined by Southern blotting. g The level of rcccDNA in the liver tissues of DMXAA-treated or untreated mice was determined by qPCR (n = 6/group). h, i Serum levels of IFN-β or TNF-α were determined by ELISA (n = 6/group). j, k The mRNA levels of IFN-β or TNF-α in the livers of DMXAA-treated or untreated mice were determined by qRT–PCR (normalized to GAPDH) (n = 6/group). l PMHs or KCs were isolated from DMXAA-treated or untreated mice and analyzed by Western blotting using antibodies against p-IRF3, IRF3, p-TBK1, TBK1, IκBα, p-p65, p65, STING, and GAPDH. m, n The mRNA levels of IFN-β or TNF-α in PMHs or KCs were quantified by qRT–PCR (normalized to GAPDH) (n = 6/group). The data are shown as the means ± SD and were compared by unpaired Student’s t test. ***P < 0.001; ****P < 0.0001; NS, no significance
STING activation leads to functional suppression of cccDNA through epigenetic modification
The above data showed that agonist-mediated STING activation significantly inhibited cccDNA-mediated transcription and HBV replication but had little effect on the level of cccDNA itself, indicating that STING signaling activation mainly led to the suppression of cccDNA function. Evidence from our group and others indicates that epigenetic modification plays an important role in the functional silencing of cccDNA [49–51]. We thus explored the effect of STING activation on the epigenetic regulation of HBV cccDNA. ChIP assays showed that STING activation by DMXAA or cGAMP in RAW264.7 cells significantly increased the recruitment of the heterochromatin markers H3K9me3 and H3K27me3 onto rcccDNA but decreased that of the euchromatin markers AcH3 and H3K4me3 onto rcccDNA in prcccDNA/Cre-transfected AML12 cells (Fig. 3a, b and Supplementary Fig. 5a). We further investigated the effect of STING activation on the epigenetic modification of cccDNA in linear HBV monomer-transfected AML12 cells, and similar data as those from the rcccDNA cell model were obtained (Fig. 3c, d and Supplementary Fig. 5b). Furthermore, we explored the role of STING signaling in the epigenetic regulation of rcccDNA in vivo. Consistent with the in vitro data, STING signaling activation by DMXAA caused heterochromatinization of rcccDNA in the liver tissues of rcccDNA mice, as revealed by increased recruitment of the repressive markers H3K9me3 and H3K27me3 and decreased recruitment of the active markers AcH3 and H3K4me3 onto rcccDNA mini chromosomes (Fig. 3e).
Fig. 3.
The STING signaling pathway participates in the epigenetic regulation of HBV cccDNA. RAW264.7 cells were treated with increasing doses of DMXAA (10 and 20 μg/mL). Twelve hours later, the culture medium of DMXAA-treated RAW264.7 cells was used to treat prcccDNA/Cre-transfected AML12 cells (a) or linear HBV monomer-transfected AML12 cells (c) for another 48 hours. AML12 cells were then harvested and used for ChIP assays with the indicated antibodies. RAW264.7 cells were treated with increasing doses of cGAMP (5 and 10 μg/mL). Twelve hours later, the culture medium of cGAMP-treated RAW264.7 cells was used to treat prcccDNA/Cre-transfected AML12 cells (b) or linear HBV monomer-transfected AML12 cells (d) for another 48 h. AML12 cells were then harvested and used for ChIP assays with the indicated antibodies. e rcccDNA mice were treated with or without DMXAA, and the livers were harvested and used for ChIP assays with the indicated antibodies (n = 3/group). The data are shown as the means ± SD and were compared by unpaired Student’s t test or one-way analysis of variance (ANOVA). *P < 0.05, **P < 0.01; NS, no significance
Studies have shown that the chromosome 5/6 complex (Smc5/6) may play an important role in the epigenetic silencing of cccDNA [52, 53]. Our previous study also showed that IFN-α-inducible IFI16 could bind to cccDNA and lead to its heterochromatinization [51]. We were thus interested in the effect of STING activation on the expression of these epigenetic modulators. Agonist-induced STING signaling activation in macrophages failed to upregulate the expression of SMC5 or SMC6 in hepatocytes but significantly increased hepatic expression of IFI16 or its murine homolog (IFI204) (Supplementary Fig. 6a, b). The role of IFI16/IFI204 in STING agonist-mediated epigenetic suppression of cccDNA merits further investigation.
STING activation attenuates HBV-induced liver fibrosis in a chronic rcccDNA mouse model
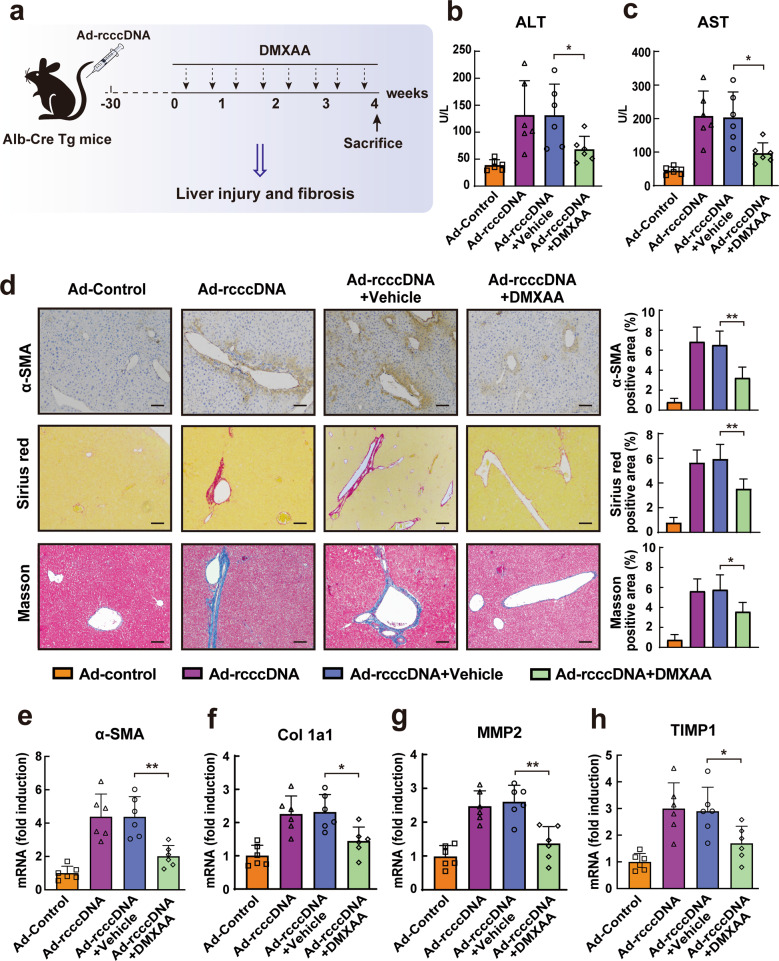
Apart from the inhibitory effect of STING activation on viral replication, STING signaling has also been proposed as a potential therapeutic target for various virus-associated diseases. Of interest, the chronic rcccDNA mouse model used in the present study has been proposed to be a valuable model for investigating HBV-induced fibrosis; thus [16], we evaluated the possible effect of STING activation on HBV-induced liver fibrosis, as shown in Fig. 4a. We first examined the serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), which are significantly associated with the progression of liver fibrosis [3, 4]. Consistent with a previous investigation [16], the serum levels ALT (Fig. 4b) and AST (Fig. 4c) in rcccDNA mice were significantly higher than those in control mice, while activation of STING by DMXAA treatment efficiently decreased the serum levels ALT and AST in rcccDNA mice (Fig. 4b, c). Furthermore, the immunohistochemical staining results showed that, compared with that of control mice, the level of alpha-smooth muscle actin (α-SMA), a marker of activated hepatic stellate cells (HSCs), was elevated in the liver tissue of rcccDNA mice, while treatment with DMXAA inhibited this effect (Fig. 4d). Sirius red and Masson’s trichrome staining also demonstrated that DMXAA treatment could efficiently reduce collagen deposition in the portal areas of rcccDNA mouse livers (Fig. 4d). In line with the histological results, the qRT–PCR data showed that treatment with DMXAA could significantly downregulate the expression levels of fibrosis-related genes, including α-SMA, alpha-1 type 1 collagen (Col 1a1), matrix metallopeptidase 2 (MMP2), and tissue inhibitor of metalloproteinases-1 (TIMP1), in the liver tissue of this chronic rcccDNA mouse model (Fig. 4e–h).
Fig. 4.
STING signaling activation alleviated liver injury and fibrosis in rcccDNA mice. a Schematic illustration of the effect of STING activation on HBV-induced fibrosis in a chronic rcccDNA mouse model. Briefly, rcccDNA mice were treated with 10 mg/kg DMXAA or vehicle by intraperitoneal injection twice per week for 4 weeks. On the day after the last DMXAA treatment, the effect of STING activation on liver injury and HBV-induced fibrosis was examined (n = 6/group). b, c The levels of ALT or AST in mouse sera were determined by ALT or AST ELISA kits. d The levels of α-SMA in liver sections were determined by immunohistochemical staining, and liver collagen deposition was assessed by Sirius red staining and Masson’s trichrome staining. Right panel: The percentages of α-SMA-, Sirius red- and Masson-positive areas were measured in 5 random fields from each animal. 10 mg/kg data are represented as the means ± SEM and were compared by unpaired Student’s t test. *P < 0.05, **P < 0.01. Scale bar: 100 μm. e–h The levels of α-SMA, Col 1a1, MMP2, and TIMP1 in livers were determined by qRT–PCR (normalized to GAPDH). Except for the data in d, all data are shown as the means ± SD and were compared by unpaired Student’s t test. *P < 0.05, **P < 0.01
Taken together, these results indicated that STING activation could inhibit the severity of liver injury and the progression of HBV-induced liver fibrosis in this rcccDNA mouse model.
STING activation suppresses macrophage inflammasome activation, which is crucial for the progression of HBV-induced liver fibrosis
Increasing evidence indicates that inflammasome/IL-1β signaling plays a critical role in the development of HBV-induced liver fibrosis [4, 15]. Of note, inflammatory mediators, including IL-1β, can often trigger a positive amplification circuit of inflammatory responses in fibrosis-related diseases [14, 15], which may be hard to eradicate even after viral inhibition. We, therefore, investigated the effect of STING signaling activation on inflammasome activation in rcccDNA mice. Our data showed that, compared with Ad-control, Ad-rcccDNA administration significantly increased the level of IL-1β in mouse sera (Fig. 5a) and led to the upregulation of caspase-1 activity in liver tissue, as demonstrated by the upregulation of caspase-1 p20 (Fig. 5b). Furthermore, we blocked IL-1β activity using a neutralizing anti-IL-1β antibody (Fig. 5c) and found that blockade of IL-1β activity significantly attenuated liver injury (Fig. 5d, e) and liver fibrosis (Fig. 5f), confirming the role of inflammasome activation in these processes. Because DMXAA significantly ameliorated liver injury and fibrosis in rcccDNA mice, we examined the effect of DMXAA on inflammasome activation in rcccDNA mice. DMXAA treatment significantly decreased the levels of IL-1β (Fig. 5g) and IL-18 (Fig. 5h) in mouse sera and caspase-1 p20 activity in the liver tissue of rcccDNA mice (Fig. 5i). Furthermore, we investigated which cell type was responsible for DMXAA-mediated inhibition of inflammasome/IL-1β activation in rcccDNA mice. As shown in Fig. 5j and k, Ad-rcccDNA significantly upregulated the level of caspase-1 p20 and the secretion of IL-1β in KCs but not in PMHs; however, this process could be significantly attenuated by treatment with DMXAA, indicating that, similar to its effect on HBV replication, DMXAA exerts its inhibitory effect on inflammasome activation by targeting KCs but not PMHs. We also examined the effect of DMXAA treatment on inflammasome activation in KCs treated with LPS plus ATP (Supplementary Fig. 7a). DMXAA alone did not have a significant effect on inflammasome activation, but it efficiently suppressed LPS/ATP-mediated inflammasome activation (Supplementary Fig. 7b, c). Furthermore, we downregulated STING expression in KCs by siRNA and then examined the effect of DMXAA on inflammasome activation induced by LPS plus ATP (Fig. 5l). DMXAA significantly inhibited inflammasome activation induced by LPS/ATP, as expected; however, once STING was knocked down in KCs, DMXAA-mediated inhibition of inflammasome activation was significantly reversed (Fig. 5m, n).
Fig. 5.
STING signaling pathway activation suppressed inflammasome activation in macrophages. a Serum levels of IL-1β from Ad-control or Ad-rcccDNA-transduced mice were determined by ELISA at the indicated time points (n = 6/group). b The levels of caspase-1 p20 in the liver tissues of Ad-control or Ad-rcccDNA-transduced mice were determined by Western blotting. Right panel: The relative level of caspase-1 p20 was determined by densitometric analysis, and the value of Ad-control mice was set as 1.0 (n = 6/group). Anti-IL-1β or isotype control antibodies (25 mg/mL) were intraperitoneally administered twice per week for 4 weeks to the rcccDNA chronic mouse model (n = 5/group): c Serum levels IL-1β were measured by ELISA. d, e The levels of ALT or AST in mouse sera were determined by ALT or AST ELISA kits. f Liver fibrosis was evaluated by determining the α-SMA-, Sirius red- and Masson-positive areas. Right panel: The percentages of α-SMA-, Sirius red- and Masson-positive areas were measured in 5 random fields from each animal. The data are represented as the means ± SEM and were compared by unpaired Student’s t test. **P < 0.01. Scale bar: 100 μm. Serum levels IL-1β (g) and IL-18 (h) in rcccDNA mice treated with or without DMXAA were determined by ELISA (n = 6/group). i The levels of caspase-1-p20 in the liver tissues of rcccDNA mice treated with or without DMXAA were determined by Western blotting and quantified as shown in b (n = 6/group). j The levels of caspase-1-p20 in PMHs and KCs from rcccDNA mice treated with or without DMXAA were determined by Western blotting and quantified as shown in b (n = 3/group). k The levels of IL-1β secreted by PMHs and KCs from rcccDNA mice treated with or without DMXAA were determined by ELISA (n = 3/group). l Schematic illustration of the effect of siRNA-mediated STING knockdown on DMXAA-mediated inhibition of inflammasome activation. m, n Cells were treated as shown in l, and the protein levels of IL-1β in culture supernatants and the levels of caspase-1 p20 in cell lysates were determined by ELISA and Western blotting, respectively. Except for the data in f, all data are shown as the means ± SD and were compared by unpaired Student’s t test or one-way analysis of variance (ANOVA). *P < 0.05, **P < 0.01
Taken together, these data indicated that STING activation induced by DMXAA could inhibit inflammasome activation both in vitro and in rcccDNA mice.
STING activation suppresses macrophage inflammasome activation by enhancing autophagic flux
Accumulating evidence shows that autophagy plays an important role in the regulation of inflammasome activation in macrophages [54–56] and contributes to protection against liver fibrosis [57, 58], while recent evidence demonstrates the crucial role of STING in the activation of autophagy; [29, 30] thus, we examined whether STING signaling activation could inhibit inflammasome activation by activating autophagic flux in macrophages. Our data showed that the activation of STING by DMXAA indeed upregulated the level of LC3II, a well-established marker of autophagy induction (Fig. 6a), in liver tissues. Furthermore, our data showed that DMXAA activated autophagic flux in KCs but not in PMHs in rcccDNA mice, as demonstrated by the upregulation of LC3II and the downregulation of p62 protein, which is widely used as an indicator of autophagic flux (Fig. 6b). We also determined the effect of DMXAA on autophagy in AML12 and RAW264.7 cells. DMXAA significantly upregulated the level of LC3II and downregulated the level of p62 in RAW264.7 cells but not in AML12 cells, which was consistent with the role of STING in DMXAA-mediated autophagic flux (Supplementary Fig. 8a–c). Furthermore, our data revealed that siRNA-mediated knockdown of STING in KCs significantly inhibited DMXAA-mediated autophagic flux (Fig. 6c, Supplementary Fig. 9). To investigate the effect of DMXAA-mediated autophagic flux on inflammasome activation, we treated KCs with leupeptin, a well-established inhibitor of autophagic flux, as described previously (Fig. 6d) [59]. Once autophagic flux in KCs was blocked by leupeptin treatment (Fig. 6e), the inhibitory effect of DMXAA on inflammasome activation in LPS/ATP-treated KCs was significantly reversed, as demonstrated by the upregulation of caspase-1 p20 and secretion of IL-1β (Fig. 6f, g). Furthermore, our data showed that, consistent with the data obtained from mouse cells, STING signaling activation by cGAMP efficiently activated autophagic flux in human KCs (Fig. 6h), and STING activation suppressed inflammasome activation induced by LPS/ATP in human KCs, which could be significantly attenuated by blocking autophagic flux with leupeptin (Fig. 6i, j). Taken together, these data indicated that STING signaling activation could suppress macrophage inflammasome activation by enhancing autophagic flux.
Fig. 6.
STING signaling pathway activation suppressed inflammasome activation by inducing autophagic flux. a The levels of LC3 in the liver tissues of rcccDNA mice treated with or without DMXAA were examined by Western blotting. Lower panel: The relative level of LC3II was determined by densitometric analysis, and the value of Ad-control mice was set as 1.0. b The levels of LC3 and p62 in PMHs or KCs from rcccDNA mice treated with or without DMXAA were determined by Western blotting. Right panel: The relative levels of LC3II or p62 were determined by densitometric analysis, and the value of vehicle-treated cells was set as 1.0. c The levels of LC3 and p62 in KCs with or without STING knockdown were determined by Western blotting and quantified as shown in b. d Schematic illustration of the effect of blocking DMXAA-mediated autophagic flux using leupeptin (200 μg/mL) on inflammasome activation induced by LPS (500 ng/mL) plus ATP (5 nM) in KCs. e KCs were treated as shown in d, and the levels of LC3 and p62 were determined by Western blotting. The relative level of p62 was determined by densitometric analysis, and the value of control cells was set as 1.0. f, g KCs were treated as shown in d, and the levels of caspase 1-p20 and secreted IL-1β were determined by Western blotting and ELISA, respectively. h Human KCs (hKCs) were treated with the STING agonist cGAMP (5 μg/mL) for 6 h, and the levels of LC3 and p62 were determined by Western blotting. i LPS plus ATP-stimulated hKCs were treated with the STING agonist cGAMP in the presence or absence of leupeptin, and the levels of caspase 1-p20, LC3II, and p62 were examined by Western blotting. j Human KCs were treated as described in i, and secreted IL-1β was measured by ELISA. The data are shown as the means ± SD and were compared by unpaired Student’s t test. *P < 0.05,**P < 0.01
It has been reported that interleukin-1 receptor antagonist (IL-1ra) plays an important role in suppressing inflammasome-dependent inflammation and contributes to TLR-mediated alleviation of liver injury and fibrosis [60–62]. We therefore examined the effect of STING activation on the expression level of IL-1ra in macrophages. However, our data showed that DMXAA treatment failed to significantly upregulate the expression level of IL-1ra in macrophages (Supplementary Fig. 10a, b).
STING activation attenuates the severity of HBV-induced liver fibrosis through autophagy-mediated inflammasome suppression
The above data showed that the blockade of autophagic flux by leupeptin significantly reversed the inhibitory effect of DMXAA on inflammasome activation in vitro. We further investigated whether blocking autophagic flux could attenuate macrophage inflammasome activation and thus reverse the DMXAA-mediated amelioration of HBV-induced liver fibrosis in rcccDNA mice (Fig. 7a). Our results showed that after leupeptin treatment, DMXAA-mediated autophagic flux in rcccDNA mice was efficiently inhibited, as demonstrated by the upregulation of p62 protein expression (Fig. 7b). Notably, blocking autophagic flux with leupeptin significantly reversed DMXAA-mediated inhibition of inflammasome activation, as demonstrated by the upregulation of caspase-1-p20 in liver tissues (Fig. 7c) and upregulated IL-1β secretion in sera (Fig. 7d). Consistent with its effect on inflammasome activation, blocking autophagic flux with leupeptin significantly decreased DMXAA-mediated inhibition of liver injury, as demonstrated by the upregulation of ALT and AST (Fig. 7e, f), and reversed DMXAA-mediated inhibition of liver fibrosis, as demonstrated by immunohistochemical staining of α-SMA, Sirius red staining and Masson’s trichrome staining (Fig. 7g). qRT–PCR also showed that leupeptin-mediated blockade of autophagic flux significantly reversed DMXAA-mediated inhibition of α-SMA, Col 1a1, MMP2, and TIMP1 mRNA levels in rcccDNA mice (Fig. 7h–k).
Fig. 7.
STING activation attenuated HBV-induced liver fibrosis via autophagy-mediated suppression of inflammasomes. a Schematic illustration of the effect of blocking STING-mediated autophagic flux on inflammasome activation and liver fibrosis in rcccDNA mice. Briefly, rcccDNA mice were treated with DMXAA (10 mg/kg) with or without leupeptin (40 mg/kg) twice per week for 4 weeks. 5 h after the last DMXAA plus leupeptin treatment, the effect of DMXAA on autophagic flux, inflammasome activation, liver injury, and fibrosis was determined (n = 6/group). b The effect of DMXAA on the levels of LC3 and p62 in KCs in the presence or absence of leupeptin was determined by Western blotting. Right panel: The relative level of p62 was determined by densitometric analysis, and the value of control mice was set as 1.0. c The effect of DMXAA on the levels of caspase-1 p20 in KCs in the presence or absence of leupeptin was determined by Western blotting. Right panel: The relative level of caspase-1 p20 was determined by densitometric analysis, and the value of control mice was set as 1.0. d The effect of DMXAA on the serum levels IL-1β in rcccDNA mice in the presence or absence of leupeptin was determined by ELISA. e, f The effect of DMXAA on the levels of ALT and AST in rcccDNA mouse sera in the presence or absence of leupeptin. g The effect of DMXAA on liver fibrosis in rcccDNA mice in the presence or absence of leupeptin was evaluated by immunohistochemical staining for α-SMA, Sirius red staining, and Masson’s trichrome staining. Right panel: The percentages of α-SMA-, Sirius red- and Masson-positive areas were measured in five random fields from each animal. The data are represented as the means ± SEM and were compared by unpaired Student’s t test. **P < 0.01. Scale bar: 100 μm. h–k The effect of DMXAA on the mRNA expression of α-SMA, Col 1a1, MMP2, and TIMP1 in the liver tissue of rcccDNA mice in the presence or absence of leupeptin was determined by qRT–PCR (normalized to GAPDH). Except for the data in g, all data are shown as the means ± SD and were compared by unpaired Student’s t test. *P < 0.05, **P < 0.01
Taken together, our data indicated that STING activation could attenuate HBV-induced liver fibrosis via autophagy-mediated suppression of inflammasomes.
Discussion
STING plays a central role in innate immune activation, especially in DNA-mediated innate signaling. Recent evidence demonstrates that STING signaling is an important therapeutic target for diverse diseases, including viral infection and virus-related diseases.
In the present study, we showed that STING activation significantly inhibited cccDNA-driven transcription and HBV replication but had little effect on the level of cccDNA itself, indicating that STING activation mainly led to the functional silencing of cccDNA. It has been reported that IFN-α can lead to the degradation of cccDNA by upregulating APOBEC3 cytidine deaminases [48]. However, our data showed that STING signaling activation failed to upregulate hAPOBEC3A or mAPOBEC3, which was consistent with the lack of effect of STING activation on cccDNA itself. Similar to our findings, although TLR7 activation by GS-9620 could inhibit HBV replication via IFN-α induction, TLR7 activation also failed to upregulate the expression level of APOBEC3A and lead to the degradation of cccDNA itself [63]. We propose that the activation of STING or TLR7 signaling may not be able to stimulate the production of IFN-α to a level that is required for the upregulation of APOBEC3A, as treatment with a high dose of IFN-α may cause severe side effects [64]. Since evidence indicates that IFN-α may cooperate with other cytokines for viral inhibition [65], it is suggested that STING-induced production of IFN-α and other cytokines (such as TNF-α and IL-6) might achieve synergistic effects and suppress HBV replication without causing significant side effects.
Although it is appealing to eliminate cccDNA completely from hepatocytes, evidence indicates that epigenetic silencing of cccDNA may be a more realistic and practical way to treat HBV infection [49]. In the present study, our data revealed that STING signaling activation could cause heterochromatinization of cccDNA, as demonstrated by the concomitant decrease in the active chromatin markers AcH3 and H3K4me3 and the increase in the repressive chromatin markers H3K9me3 and H3K27me3 on HBV cccDNA. Our previous study showed that the innate DNA sensor IFI16 could bind to cccDNA and lead to its heterochromatinization [50]. Other studies reported that the chromosome 5/6 complex (Smc5/6) may play an important role in the epigenetic silencing of cccDNA [52, 53]. We thus examined whether STING-mediated suppression of cccDNA involved these epigenetic modifiers. Our data showed that STING signaling activation by an agonist failed to upregulate the expression levels of SMC5 or SMC6; however, STING activation could significantly upregulate the expression level of IFI16 and its mouse homolog (IFI204) in hepatocytes. A detailed study of the role of IFI16/IFI204 in STING-mediated epigenetic suppression of cccDNA is beyond the scope of this report, but this topic deserves further investigation. IFNα has been reported to inhibit HBV transcription and replication through the epigenetic suppression of cccDNA [66]. As one of the most important innate immune adapters, STING activation stimulates the production of various antiviral cytokines, including type I/III IFNs, TNF-α, and IL-6 [24, 25]. Type I IFNs surely play a major role in STING agonist-mediated anti-cccDNA activity. Our data also showed that upon STING activation, the production of IFN-β was far greater than that of TNF-α. Considering the possible cooperation and synergism of antiviral processes, STING agonizts may achieve a similar inhibitory effect on cccDNA transcription with fewer side effects.
In addition to its central role in persistent HBV infection, cccDNA is also closely related to HBV-induced liver diseases, including liver fibrosis. HBV-induced liver fibrosis is a process of chronic mild to moderate liver injury accompanied by scarring within the liver; however, investigating its pathogenic mechanisms has been significantly challenging, as there is a lack of suitable laboratory models. Recently, Li et al. [16] reported that the Ad-rcccDNA mouse model closely resembled clinical chronic viral hepatitis with progressive liver pathology and developed signs of liver fibrosis, indicating that this model might be a suitable platform to design strategies against HBV-induced liver fibrosis. Our data confirmed that there was indeed significant liver injury and the occurrence of liver fibrosis in this rcccDNA mouse model; however, STING signaling activation by DMXAA could efficiently ameliorate the severity of liver injury and fibrosis. Although HBV alone can cause liver fibrosis directly [67], a growing number of studies have shown that virus-induced inflammatory responses, especially the activation of macrophage inflammasomes, may play a more crucial role in the development of liver fibrosis [10]. Importantly, the activation of inflammasome/IL-1β signaling may enhance the recruitment and activation of other immune cells to amplify inflammatory responses [68]. Our data showed that the inflammasome was indeed significantly activated in rcccDNA mice and contributed to the progression of liver fibrosis, while STING signaling activation by DMXAA significantly inhibited inflammasome activation. Furthermore, KCs but not PMHs were identified to be responsible for rcccDNA-induced inflammasome activation and the effect of STING on inflammasomes, which is consistent with the report that macrophages throughout the body, including liver resident KCs, are the principal sources of proinflammatory cytokines [9, 69]. We attempted to use STING-knockout (KO) mice to explore the effect of STING activation on the progression of HBV-induced fibrosis, and a chronic rcccDNA mouse model was established by injecting Ad-rcccDNA into Alb-Cre transgenic mice. Additionally, as we and others demonstrated that STING might be important for the inhibition of HBV, STING deletion might lead to excessive viral replication, which might result in the blockade of immune tolerance and be unfavorable for establishing a persistent HBV mouse model [70, 71]. However, we further investigated the effect of STING activation on inflammasome activation in STING siRNA-transfected KCs and found that STING knockdown significantly reversed DMXAA-mediated inhibition of inflammasome activation in KCs. In contrast to our findings, Yu et al. [72] reported that HBV suppressed rather than activated inflammasomes in the livers of HBV mice. The exact reason for the discrepancy between their and our data remains unclear. This difference may be because the study by Yu et al. [72] used the adeno-associated virus (AAV) vector-mediated HBV model, while the rcccDNA model used in the present study was established by injecting an adenovirus (Ad)-harbored HBV construct into Alb-Cre mice. It is now evident that Ad may induce stronger inflammatory responses than AAV [73]. Consistently, in a recent study, AAV-rcccDNA was injected into Alb-Cre mice to establish a novel rcccDNA mouse model that had been proven to exhibit longer-term maintenance of rcccDNA and antigenemia but elicited weaker immune responses than rcccDNA mice derived from Ad-rcccDNA [74]. Reports indicate that Ad infection may contribute to caspase-1-dependent IL-1β secretion by activating the NLRP3 inflammasome [75, 76], although the effect of Ad on immune activation and subsequent liver injury is dose dependent [77]. In terms of HBV-associated liver fibrosis in clinical patients, Ad may not be the “natural” pathogenic factor for liver fibrosis. However, Ad may mimic some “injurious stimuli”, which can synergize with HBV to break the threshold of inflammasome activation and thus contribute to the development of liver fibrosis in CHB patients.
There is another intriguing issue: STING activation has been reported to activate the NLRP3 inflammasome by translocating STING to the lysosome, leading to the rupture of lysosomes and the initiation of potassium efflux [78]. However, recent evidence demonstrates that STING activation does not lead to the translocation of STING to the lysosome but to the endoplasmic reticulum-Golgi intermediate compartment, resulting in the induction of autophagy [29]. It has also been reported that in NASH (nonalcoholic steatohepatitis), the expression of STING is increased in liver macrophages, contributing to the development of NASH fibrosis [44, 79], which appears to contradict our findings. This discrepancy is not fully understood and may be due to several reasons. (1) Although the outcomes of NASH fibrosis and HBV-induced fibrosis may be similar, their etiologies may differ significantly; for example, the inflammatory cytokines released from inflamed adipose tissues and the ROS derived from dysregulated hepatic lipid and cholesterol metabolism may contribute greatly to the progression of NASH fibrosis. HBV-induced liver fibrosis may be caused by virus-induced inflammatory responses, and HBV per se can activate HSCs, leading to liver fibrosis [80]. (2) The expression level of STING was reported to be increased in the liver tissue of NASH patients; however, viral infection might lead to the functional suppression of STING or a decrease in STING expression for the purpose of viral evasion [42, 81]. (3) Activation of STING or other innate immune signaling pathways to different extents may lead to different outcomes. Our data also showed that treatment with DMXAA at a concentration greater than 40 mg/kg/d led to significant hepatotoxicity. (4) In addition to its role in NF-κB activation and its subsequent contribution to the progression of NASH fibrosis, STING signaling may play a more crucial role in IFN-I production and autophagy induction, both of which are involved in the alleviation of liver fibrosis [31, 60, 61, 82]. It has been reported that activation of type 1 IFN signaling by TLR7 or TLR9 induces anti-inflammatory responses and contributes to the alleviation of liver damage or fibrosis, which may be mediated by the induction of interleukin-1 receptor antagonist (IL-1ra), a potent suppressor of inflammasome activity [60, 61]. Similarly, a recent report indicated that activation of TLR5 signaling by flagellin could ameliorate CCL4- or BDL-induced liver fibrosis by inducing IFN-β-modulated IL-1ra [62]. Of note, it has also been reported that the overactivation of TLR5 signaling by a high dose of flagellin causes inflammatory responses and leads to acute liver injury [83]. Taken together, these results suggest that the role of STING and other innate immune signaling pathways in liver injury and fibrosis may depend on the nature, extent, and duration of the stimulus.
Autophagy, as a homeostatic process, is involved in diverse pathophysiological processes [84]. In addition to its central role in IFN induction, STING was revealed to play a critical role in autophagy regulation in recent studies [29, 30]. Accumulating evidence demonstrates that macrophage autophagy may play an important role in protecting against inflammasome/IL-1β-mediated hepatotoxic injury. For example, Saitoh et al. [55] reported that Atg16L1-deficient macrophages had enhanced production of IL-1β and IL-18 but not IL-6 or TNFα upon LPS stimulation, and Lodder et al. [32] reported that macrophages from ATG5-/- mice exhibited impaired autophagy and proinflammatory properties, and macrophage autophagy was demonstrated to protect against liver fibrosis in mice. We were interested in the effect of STING activation on autophagy induction in rcccDNA mice. Our data showed that STING activation significantly enhanced autophagic flux in KCs but had no effect on PMHs, as expected. Notably, further results revealed that blocking STING-mediated autophagic flux in macrophages with leupeptin could significantly enhance inflammasome activation in KCs and thus reverse the STING-mediated attenuation of liver fibrosis in rcccDNA mice. Considering the important role of IFN-I-modulated IL-1ra in controlling inflammasome activation and thus contributing to the alleviation of liver fibrosis [60–62], we also evaluated the effect of STING activation on IL-1ra expression in macrophages. However, our data showed that DMXAA treatment did not significantly affect the expression level of IL-1ra in murine macrophages, further suggesting the crucial role of autophagic flux in the STING-mediated alleviation of HBV-induced liver fibrosis.
In summary, our data showed that STING signaling activation could lead to the functional suppression of cccDNA via epigenetic modification. Of note, STING activation could efficiently attenuate the severity of liver injury and HBV-induced hepatic fibrosis. Mechanistically, apart from inhibiting HBV, STING activation could alleviate the progression of HBV-induced liver fibrosis through the suppression of macrophage inflammasome activation by inducing autophagic flux (Fig. 8). These data suggest that the STING signaling pathway may be a novel therapeutic target to protect against chronic hepatitis B and HBV-induced fibrosis.
Fig. 8.
Schematic illustration of the inhibitory effect of STING activation on HBV cccDNA and HBV-induced liver fibrosis. STING signaling activation in macrophages leads to the functional suppression of cccDNA in hepatocytes via epigenetic modification. In addition to inhibiting HBV replication, STING activation ameliorates HBV-induced liver fibrosis by suppressing the activation of macrophage inflammasomes by stimulating autophagic flux
Material and methods
Cell culture and transfection
AML12 murine hepatocytes (cat# CRL-2254) and RAW264.7 murine macrophages (cat# TIB-71, passage number: 10–15) were obtained from ATCC Laboratories. The cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 100 μg/mL penicillin, and 100 μg/mL streptomycin in an incubator with 5% CO2 at 37 °C. PHHs and hKCs were prepared from liver resections as described in detail by Kegel et al. [85]. Human liver resections were obtained from the Department of Liver Surgery and Transplantation of Zhongshan Hospital (Fudan University, Shanghai, China). All patients provided written informed consent. The study conformed to the ethical guidelines of the 1975 Declaration of Helsinki. Hepatocytes were seeded into collagen-I-coated culture plates in Williams medium supplemented with 5% FCS, 100 μg/mL penicillin, 100 μg/mL streptomycin, 5 μg/mL insulin, 5 × 10−5 M hydrocortisone, and 2% DMSO. For HBV infection experiments, PHHs were infected with HBV at 100 vge/cell as described previously [51]. Seven days after HBV infection, PHHs were used for experiments. Kupffer cells were maintained in culture plates in RPMI 1640 supplemented with 10% FCS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM L-glutamine. The precursor plasmid prcccDNA and pCMV-Cre (encoding Cre recombinase) were kind gifts from Dr. Deng [71]. In prcccDNA, a loxP-chimeric intron was engineered into a monomeric HBV genome, with the single loxP site replaced with two directly repeated loxP segments flanking a prokaryotic plasmid backbone, which can be excised using Cre/loxP-mediated DNA recombination into a 3.3-kb rcccDNA in the nuclei of hepatocytes. rcccDNA carries a chimeric intron, which can be used to specifically identify rcccDNA. However, the chimeric intron will be spliced from RNA transcripts during RNA processing without interrupting the HBV life cycle. SiRNA specific for STING (5′-CGAAAUAACUGCCGCCUCA-3′) was obtained from Dharmacon (catalog# M-024333-00). Plasmids and siRNAs were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Animal experiments
Albumin (Alb)-Cre Tg mice (males, aged 6–8 weeks) expressing Cre recombinase under the albumin promoter were bred and maintained under specific pathogen-free conditions. The mice were injected with 1.5 × 109 plaque-forming units (PFUs) of recombinant virus in 200 μL of phosphate-buffered saline (PBS) via the tail vein to establish an rcccDNA chronic mouse model as described previously [17]. Three to seven days after injection, the successful establishment of the rcccDNA mouse model was confirmed by measuring serum HBsAg. To isolate PMHs and KCs, mouse livers were perfused through the portal vein with Hank’s balanced salt solution (HBSS) and HBSS containing 0.025% collagenase IV. The digested liver was then dissected out thoroughly in collagenase buffer. Then, the suspension was centrifuged at 50 × g for 3 min. For PMH isolation, the collected pellet was separated by a 40% Percoll solution, centrifuged at 450 × g for 10 min and then washed and plated in DMEM containing antibiotics and 10% FBS. For KC isolation, the nonparenchymal cell fraction was washed twice with buffer, and cells were centrifuged on a density cushion of Percoll at 1000 × g for 15 min. Cells were then collected from the interface between the two density cushions, further stained with PE-conjugated anti-F4/80 antibody coupled with anti-PE microbeads and isolated using a MACS system (Miltenyi Biotec). The purity of KCs was more than 90%, as determined by flow cytometry (Supplementary Fig. 11). Animal experiments were performed in accordance with the Guide for the Care and Use of Medical Laboratory Animals (Ministry of Health, China) and approved by the Animal Ethics Committee of Fudan University (permit number: 20180302-035).
Quantitative reverse transcription PCR (qRT–PCR)
Total RNA was extracted from cells or liver tissues using TRIzol reagent according to the manufacturer’s protocol and reverse-transcribed into cDNA. qRT–PCR analysis of cDNA targets was performed using a Roche LightCycler 480 II system and SYBR green (Takara, Dalian, China). GAPDH served as an endogenous control. The primers used in this study are listed in Supplementary Table 1. To examine HBV pgRNA, the PrimeScript™ RT reagent Kit with genomic DNA (gDNA) Eraser (Takara, Dalian, China) was used, in which gDNA Eraser was used to remove contaminating transfected HBV DNA.
Extraction and analysis of intracellular core particle-associated HBV DNA
The method for the extraction and quantification of intracellular core particle-associated HBV DNA in cell or rcccDNA mouse livers was described previously [51, 86]. For Southern blotting, HBV DNA was hybridized with a digoxigenin-labeled full-length HBV probe synthesized with a DIG probe synthesis kit (Roche, Mannheim, Germany). The primers used in this study are shown in Supplementary Table 1.
Quantification of HBsAg and cytokines by ELISA
The levels of HBsAg in cell culture supernatants or mouse sera were quantified by a commercial ELISA kit (Kehua Biotech, Shanghai, China) according to the manufacturer’s instructions. The levels of IFN-β, TNF-α, IL-1β, and IL-18 in cell culture supernatants or mouse sera were analyzed using DuoSet ELISA (eBioscience. USA) according to the manufacturers’ instructions.
Western blotting
Western blotting was performed as described previously [51, 86]. Briefly, cells or liver tissues were lysed on ice in lysis buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 0.1% SDS) supplemented with a protease inhibitor cocktail and PMSF. Total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. Then, the membranes were incubated with the indicated primary antibodies and visualized with peroxidase-conjugated secondary antibodies using SuperSignalTM West Femto Maximum Sensitivity Substrate (Thermo Scientific). The antibodies used in this study are shown in Supplementary Table 2.
Immunohistochemical (IHC) staining, Sirius red staining, and Masson’s trichrome staining
Liver tissues from mice were fixed in 4% paraformaldehyde and embedded in paraffin wax For IHC analysis of HBcAg or α-SMA, liver sections were routinely deparaffinized and rehydrated; after being treated with 3% hydrogen peroxide and blocked with 5% bovine serum albumin, the sections were then incubated sequentially with antibodies against HBcAg or α-SMA (Supplementary Table 2), biotin-labeled secondary antibodies and avidin-biotin complex. The peroxidase stain was developed with 3,3′-diaminobenzidine solution and counterstained with hematoxylin. Sirius red and Masson’s trichrome staining of liver tissues was performed by Servicebio (Shanghai, China). Microscopy images were acquired using a Nikon Eclipse Ts2 microscope and analyzed using Image Pro-plus software.
Extraction and quantification of HBV cccDNA or rcccDNA
HBV cccDNA or rcccDNA was isolated from cells and liver tissues using a modified Hirt extraction procedure [87]. Liver tissues were homogenized on ice in prechilled PBS using a tissue homogenizer. After being filtered by a cell strainer (BD), the liver homogenate was centrifuged at 500 × g for 5 min. The pelleted cells were lysed in lysis buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.2% NP-40 and 150 mM NaCl). The lysate was then centrifuged at 12,000 × g for 30 min at 4 °C. Then, the pellet was dissolved in nuclear lysis buffer (6% SDS, 0.1 M NaCl) and neutralized with 3 M potassium acetate (pH 4.8). The protein-free DNA was further extracted with phenol and chloroform and then precipitated with ethanol. The extracted DNA was further treated for 1 h at 37 °C with Plasmid-Safe DNase (Epicenter Biotechnologies, Madison, WI) and then HBV cccDNA or rcccDNA was analyzed by quantitative PCR (qPCR). The absence of contaminating genomic DNA was further confirmed by negative PCR results for β-globin. The primers used in this study are shown in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP) assay
AML12 cells were treated with 1% formaldehyde for 10 min to generate DNA-protein crosslinks. Liver tissues were homogenized in lysis buffer (5 mM PIPES, 85 mM KCl, 0.5% NP40) and incubated for 10 min at 4 °C. After centrifugation, the culture supernatants were removed, and the pelleted nuclei were fixed in 1% formaldehyde for 30 min at 4 °C. Then, the isolated crosslinked nuclei were sonicated to generate chromatin fragments of 200–300 bp and immunoprecipitated with antibodies, as shown in Supplemental Table 2. After reverse crosslinking, HBV cccDNA or rcccDNA was extracted and further treated with plasmid-safe DNase and then analyzed by real-time quantitative PCR (qPCR) with the corresponding primers, as shown in Supplementary Table 1.
Statistical analysis
The data are expressed as the mean ± SD unless otherwise stated. Statistical significance was determined using a two-tailed, unpaired Student’s t test (for two sample comparisons) or one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison correction data (for multiple comparisons). All statistical analyses were performed using GraphPad Prism software version 8.0. P values < 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (C31872731, C32070910, C31470839) and Zhengyi Scholar Foundation of School of Basic Medical Sciences, Fudan University (S25-01).
Author contributions
BG and JBC conceived and supervised the study. BG, JBC, YQL, MJH, and ZYW participated in the study design and analyzed the data. YQL, MJH, ZYW, ZYD, ZWG ZTW, RJG, and THC performed the experiments. BG, YQL, and MJH wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yuqi Li, Minjing He, Ziyu Wang.
Contributor Information
Jiabin Cai, Email: cai.jiabin@zs-hospital.sh.cn.
Bo Gao, Email: gaobofd@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00801-w.
References
- 1.Seto WK, Lo YR, Pawlotsky JM, Yuen MF. Chronic hepatitis B virus infection. Lancet. 2018;392:2313–24. doi: 10.1016/S0140-6736(18)31865-8. [DOI] [PubMed] [Google Scholar]
- 2.Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–63. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 3.Parola M, Pinzani M. Liver fibrosis: pathophysiology, pathogenetic targets and clinical issues. Mol Asp Med. 2019;65:37–55. doi: 10.1016/j.mam.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. Nat Rev Immunol. 2014;14:181–94. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 5.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–84. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed M, Wang F, Levin A, Le C, Eltayebi Y, Houghton M, et al. Targeting the Achilles heel of the hepatitis B virus: a review of current treatments against covalently closed circular DNA. Drug Discov Today. 2015;20:548–61. doi: 10.1016/j.drudis.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Lenci I, Marcuccilli F, Tisone G, Di Paolo D, Tariciotti L, Ciotti M, et al. Total and covalently closed circular DNA detection in liver tissue of long-term survivors transplanted for HBV-related cirrhosis. Dig Liver Dis. 2010;42:578–84. doi: 10.1016/j.dld.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Cheng PN, Liu WC, Tsai HW, Wu IC, Chang TT, Young KC. Association of intrahepatic cccDNA reduction with the improvement of liver histology in chronic hepatitis B patients receiving oral antiviral agents. J Med Virol. 2011;83:602–7. doi: 10.1002/jmv.22014. [DOI] [PubMed] [Google Scholar]
- 9.Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66:1300–12. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Altamirano-Barrera A, Barranco-Fragoso B, Mendez-Sanchez N. Management strategies for liver fibrosis. Ann Hepatol. 2017;16:48–56. doi: 10.5604/16652681.1226814. [DOI] [PubMed] [Google Scholar]
- 11.Tao Y, Wang N, Qiu T, Sun X. The role of autophagy and NLRP3 inflammasome in liver fibrosis. Biomed Res Int. 2020;2020:7269150. doi: 10.1155/2020/7269150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50:778–95. doi: 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbier L, Ferhat M, Salamé E, Robin A, Herbelin A, Gombert JM, et al. Interleukin-1 family cytokines: keystones in liver inflammatory diseases. Front Immunol. 2019;10:2014. doi: 10.3389/fimmu.2019.02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karhadkar TR, Chen W, Gomer RH. Attenuated pulmonary fibrosis in sialidase-3 knockout (Neu3(-/-)) mice. Am J Physiol Lung Cell Mol Physiol. 2020;318:L165–L179. doi: 10.1152/ajplung.00275.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G, Sun L, Kato T, Okuda K, Martino MB, Abzhanova A, et al. IL-1beta dominates the promucin secretory cytokine profile in cystic fibrosis. J Clin Invest. 2019;129:4433–50. doi: 10.1172/JCI125669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Zhu Y, Shao D, Chang H, Zhang X, Zhou D, et al. Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology. 2018;67:56–70. doi: 10.1002/hep.29406. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14:315–28. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altfeld M, Gale MJ. Innate immunity against HIV-1 infection. Nat Immunol. 2015;16:554–62. doi: 10.1038/ni.3157. [DOI] [PubMed] [Google Scholar]
- 19.Hu MM, Shu HB. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu Rev Immunol. 2020;38:79–98. doi: 10.1146/annurev-immunol-070119-115052. [DOI] [PubMed] [Google Scholar]
- 20.Thomsen MK, Skouboe MK, Boularan C, Vernejoul F, Lioux T, Leknes SL, et al. The cGAS-STING pathway is a therapeutic target in a preclinical model of hepatocellular carcinoma. Oncogene. 2020;39:1652–64. doi: 10.1038/s41388-019-1108-8. [DOI] [PubMed] [Google Scholar]
- 21.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–80. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol. 2005;79:7269–72. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maini MK, Gehring AJ. The role of innate immunity in the immunopathology and treatment of HBV infection. J Hepatol. 2016;64:S60–S70. doi: 10.1016/j.jhep.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Cui X, Zhang R, Cen S, Zhou J. STING modulators: predictive significance in drug discovery. Eur J Med Chem. 2019;182:111591. doi: 10.1016/j.ejmech.2019.111591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–65. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–6. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Wu H, Wang C, Li Y, Tian H, Siraj S, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019;26:1735–49. doi: 10.1038/s41418-018-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, Bae J, Choi CY, Choi SP, Kang HS, Jo EK, et al. Autophagy induced by AXL receptor tyrosine kinase alleviates acute liver injury via inhibition of NLRP3 inflammasome activation in mice. Autophagy. 2016;12:2326–43. doi: 10.1080/15548627.2016.1235124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodder J, Denaës T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, et al. Macrophage autophagy protects against liver fibrosis in mice. Autophagy. 2015;11:1280–92. doi: 10.1080/15548627.2015.1058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, Gallant CW, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun. 2020;11:2739. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao L, Qi J, Zhao Q, Wu QN, Wei DL, Wei XL, et al. Targeting the STING pathway in tumor-associated macrophages regulates innate immune sensing of gastric cancer cells. Theranostics. 2020;10:498–515. doi: 10.7150/thno.37745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–30. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–42. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lio CW, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, et al. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J Virol. 2016;90:7789–97. doi: 10.1128/JVI.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinert LS, Lopušná K, Winther H, Sun C, Thomsen MK, Nandakumar R, et al. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun. 2016;7:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Guo F, Han Y, Zhao X, Wang J, Liu F, Xu C, et al. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob Agents Chemother. 2015;59:1273–81. doi: 10.1128/AAC.04321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo F, Tang L, Shu S, Sehgal M, Sheraz M, Liu B, et al. Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother. 2017;61:e00771–17. doi: 10.1128/AAC.00771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Li J, Chen J, Li Y, Wang W, Du X, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol. 2015;89:2287–300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen MK, Nandakumar R, Stadler D, Malo A, Valls RM, Wang F, et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology. 2016;64:746–59. doi: 10.1002/hep.28685. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Liu Y, An W, Song J, Zhang Y, Zhao X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J Clin Invest. 2019;129:546–55. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Q, Hu H, Liu H, Shen H, Yan Z, Gao L. A synthetic STING agonist inhibits the replication of human parainfluenza virus 3 and rhinovirus 16 through distinct mechanisms. Antivir Res. 2020;183:104933. doi: 10.1016/j.antiviral.2020.104933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melén K, Julkunen I. Impaired antiviral response in human hepatoma cells. Virology. 1999;263:364–75. doi: 10.1006/viro.1999.9983. [DOI] [PubMed] [Google Scholar]
- 47.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–37. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–8. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology. 2017;66:2066–77. doi: 10.1002/hep.29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, et al. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093–102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 51.Yang Y, Zhao X, Wang Z, Shu W, Li L, Li Y, et al. Nuclear sensor interferon-inducible protein 16 inhibits the function of hepatitis B virus covalently closed circular DNA by integrating innate immune activation and epigenetic suppression. Hepatology. 2020;71:1154–69. doi: 10.1002/hep.30897. [DOI] [PubMed] [Google Scholar]
- 52.Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–9. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 53.Tsai K, Cullen BR. Epigenetic and epitranscriptomic regulation of viral replication. Nat Rev Microbiol. 2020;18:559–70. doi: 10.1038/s41579-020-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giegerich AK, Kuchler L, Sha LK, Knape T, Heide H, Wittig I, et al. Autophagy-dependent PELI3 degradation inhibits proinflammatory IL1B expression. Autophagy. 2014;10:1937–52. doi: 10.4161/auto.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Li Z, Feng D, Zu G, Li Y, Zhao Y, et al. Autophagy induction ameliorates inflammatory responses in intestinal ischemia-reperfusion through inhibiting NLRP3 inflammasome activation. Shock. 2019;52:387–95. doi: 10.1097/SHK.0000000000001259. [DOI] [PubMed] [Google Scholar]
- 57.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 58.Jia D, Wang YY, Wang P, Huang Y, Liang DY, Wang D, et al. SVIP alleviates CCl4-induced liver fibrosis via activating autophagy and protecting hepatocytes. Cell Death Dis. 2019;10:71. doi: 10.1038/s41419-019-1311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7:629–42. doi: 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrasek J, Dolganiuc A, Csak T, Kurt-Jones EA, Szabo G. Type I interferons protect from Toll-like receptor 9-associated liver injury and regulate IL-1 receptor antagonist in mice. Gastroenterology. 2011;140:697–708. doi: 10.1053/j.gastro.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roh YS, Park S, Kim JW, Lim CW, Seki E, Kim B. Toll-like receptor 7-mediated type I interferon signaling prevents cholestasis- and hepatotoxin-induced liver fibrosis. Hepatology. 2014;60:237–49. doi: 10.1002/hep.26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Z, Kim JW, Qi J, Eo SK, Lim CW, Kim B. Toll-like receptor 5 signaling ameliorates liver fibrosis by inducing interferon beta-modulated IL-1 receptor antagonist in mice. Am J Pathol. 2020;190:614–29. doi: 10.1016/j.ajpath.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 63.Niu C, Li L, Daffis S, Lucifora J, Bonnin M, Maadadi S, et al. Toll-like receptor 7 agonist GS-9620 induces prolonged inhibition of HBV via a type I interferon-dependent mechanism. J Hepatol. 2018;68:922–31. doi: 10.1016/j.jhep.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 64.Wong DKH. Effect of alpha-interferon treatment in patients with hepatitis B antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–23. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 65.Fink K, Martin L, Mukawera E, Chartier S, De Deken X, Brochiero E, et al. IFNbeta/TNFalpha synergism induces a non-canonical STAT2/IRF9-dependent pathway triggering a novel DUOX2 NADPH oxidase-mediated airway antiviral response. Cell Res. 2013;23:673–90. doi: 10.1038/cr.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, et al. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Investig. 2012;122:529–37. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, Zhu ST, You H, Cong M, Liu TH, Wang BE, et al. Hepatitis B virus infects hepatic stellate cells and affects their proliferation and expression of collagen type I. Chin Med J. 2009;122:1455–61. [PubMed] [Google Scholar]
- 68.Kubes P, Mehal WZ. Sterile Inflammation in the Liver. Gastroenterology. 2012;143:1158–72. doi: 10.1053/j.gastro.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 69.Song K, Kwon H, Han C, Chen W, Zhang J, Ma W, et al. Yes-associated protein in kupffer cells enhances the production of proinflammatory cytokines and promotes the development of nonalcoholic steatohepatitis. Hepatology. 2020;72:72–87. doi: 10.1002/hep.30990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang LR, Gäbel YA, Graf S, Arzberger S, Kurts C, Heikenwalder M, et al. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology. 2012;142:1447–50. doi: 10.1053/j.gastro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 71.Qi Z, Li G, Hu H, Yang C, Zhang X, Leng Q, et al. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88:8045–56. doi: 10.1128/JVI.01024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu X, Lan P, Hou X, Han Q, Lu N, Li T, et al. HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1beta production via suppressing the NF-kappaB pathway and ROS production. J Hepatol. 2017;66:693–702. doi: 10.1016/j.jhep.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 73.Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76:4580–90. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu M, Wang C, Shi B, Fang Z, Qin B, Zhou X, et al. A novel recombinant cccDNA-based mouse model with long term maintenance of rcccDNA and antigenemia. Antivir Res. 2020;180:104826. doi: 10.1016/j.antiviral.2020.104826. [DOI] [PubMed] [Google Scholar]
- 75.Liu Q, Muruve DA. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 2003;10:935–40. doi: 10.1038/sj.gt.3302036. [DOI] [PubMed] [Google Scholar]
- 76.McGuire KA, Barlan AU, Griffin TM, Wiethoff CM. Adenovirus type 5 rupture of lysosomes leads to cathepsin B-dependent mitochondrial stress and production of reactive oxygen species. J Virol. 2011;85:10806–13. doi: 10.1128/JVI.00675-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muruve DA, Barnes MJ, Stillman IE, Libermann TA. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum Gene Ther. 1999;10:965–76. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- 78.Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell. 2017;171:1110–24. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo X, Li H, Ma L, Zhou J, Guo X, Woo SL, et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology. 2018;155:1971–84. doi: 10.1053/j.gastro.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rockey DC. Liver fibrosis reversion after suppression of hepatitis B virus. Clin Liver Dis. 2016;20:667–79. doi: 10.1016/j.cld.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yi G, Wen Y, Shu C, Han Q, Konan KV, Li P, et al. Hepatitis C virus NS4B can suppress STING accumulation to evade innate immune responses. J Virol. 2016;90:254–65. doi: 10.1128/JVI.01720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanabe J, Izawa A, Takemi N, Miyauchi Y, Torii Y, Tsuchiyama H, et al. Interferon-beta reduces the mouse liver fibrosis induced by repeated administration of concanavalin A via the direct and indirect effects. Immunology. 2007;122:562–70. doi: 10.1111/j.1365-2567.2007.02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiao Y, Liu F, Yang J, Zhong M, Zhang E, Li Y, et al. Over-activation of TLR5 signaling by high-dose flagellin induces liver injury in mice. Cell Mol Immunol. 2015;12:729–42. doi: 10.1038/cmi.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 85.Kegel V, Daniela D, Pfeiffer E, Zeilinger K, Seehofer D, Damm G, et al. Protocol for isolation of primary human hepatocytes and corresponding major populations of non-parenchymal liver cells. J Vis Exp. 2016: e53069. [DOI] [PMC free article] [PubMed]
- 86.Zhong L, Shu W, Dai W, Gao B, Xiong S. Reactive oxygen species-mediated c-Jun NH2-terminal kinase activation contributes to hepatitis B virus X protein-induced autophagy via regulation of the Beclin-1/Bcl-2 interaction. J. Virol. 2017;91:e00001–17. doi: 10.1128/JVI.00001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–9. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.