Abstract
HIV-1 infection-induced cGAS–STING–TBK1–IRF3 signaling activates innate immunity to produce type I interferon (IFN). The HIV-1 nonstructural protein viral infectivity factor (Vif) is essential in HIV-1 replication, as it degrades the host restriction factor APOBEC3G. However, whether and how it regulates the host immune response remains to be determined. In this study, we found that Vif inhibited the production of type I IFN to promote immune evasion. HIV-1 infection induced the activation of the host tyrosine kinase FRK, which subsequently phosphorylated the immunoreceptor tyrosine-based inhibitory motif (ITIM) of Vif and enhanced the interaction between Vif and the cellular tyrosine phosphatase SHP-1 to inhibit type I IFN. Mechanistically, the association of Vif with SHP-1 facilitated SHP-1 recruitment to STING and inhibited the K63-linked ubiquitination of STING at Lys337 by dephosphorylating STING at Tyr162. However, the FRK inhibitor D-65495 counteracted the phosphorylation of Vif to block the immune evasion of HIV-1 and antagonize infection. These findings reveal a previously unknown mechanism through which HIV-1 evades antiviral immunity via the ITIM-containing protein to inhibit the posttranslational modification of STING. These results provide a molecular basis for the development of new therapeutic strategies to treat HIV-1 infection.
Keywords: Vif, cGAS–STING, FRK, Immune evasion
Subject terms: Signal transduction, Immune evasion
Introduction
Human immunodeficiency virus (HIV)-1 infection activates the antiviral innate immune response and triggers the type I interferon (IFN) response [1–4]. HIV-1 RNA and cDNA are sensed by Toll-like receptors (TLRs) and cytosolic sensors, respectively. The single-stranded RNA of HIV-1 may be sensed by TLR7 and TLR8 in endosomes. This results in signal transduction that leads to the translocation of IFN regulatory factor 7 (IRF7) or nuclear factor-kappa B (NF-κB) to the nucleus and upregulates the transcription of type I IFNs and proinflammatory cytokines [5, 6]. The Y-form cDNA of HIV-1, generated by RNA reverse transcription, is recognized by IFN-γ-inducible protein 16 (IFI16) and cyclic GMP–AMP synthase (cGAS) [7–11]. cGAS specifically detects cDNA and responds by producing cyclic GMP–AMP (cGAMP). Both IFI16 and cGAMP activate stimulator of interferon genes (STING) to recruit and activate TANK-binding kinase 1 (TBK1), which phosphorylates IRF3 and induces the production of type I IFNs and proinflammatory cytokines [12–16].
Type I IFNs, including both IFN-α and IFN-β, are well-characterized innate antiviral proteins contributing to the fight against HIV-1 infection. Upon recognition of HIV-1 by TLRs or cytosolic DNA sensors, type I IFNs activate the JAK/signal transducer and activator of transcription signaling pathway and induce the expression of IFN-stimulated genes (ISGs). ISGs contribute to the inhibition of viral infection and indirectly activate both innate and adaptive antiviral immune responses [17]. In addition, type I IFNs enhance the antigen presentation and chemokine production of antigen-presenting cells, which promote the antibody production of B cells and amplify T cell effector function [18]. Therefore, the modulation of the type I IFN response prior to or during infection is an important step for developing novel therapeutic strategies to prevent HIV infection.
Although immune cells protect the host by producing type I IFNs and pro-inflammatory cytokines [19, 20], HIV-1 uses several unique strategies to evade host restriction and achieve successful infection. For example, the HIV-1 capsid protein recruits host cyclophilin A and polyadenylation-specific factor 6 to facilitate infection [21–23]. Similarly, the viral protein U (Vpu) of HIV-1 promotes the production of virions by interacting with and antagonizing tetherin, a host factor that specifically inhibits the release of virions [24]. Apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) exhibits cytidine deaminase activity and restricts HIV-1 replication by converting cytosines to uracils in newly synthesized viral cDNA. However, the HIV-1 regulatory protein viral infectivity factor (Vif) not only binds to APOBEC3G mRNA to inhibit its translation but also induces APOBEC3G ubiquitination and degradation through the actions of the cullin 5–Skp1–Cullin–F-box complex and core-binding factor-β [25–27]. In addition, the deoxynucleoside triphosphate (dNTP) triphosphohydrolase SAMHD1 assists HIV-1 in suppressing the production of IFNs, ISGs, and costimulatory molecules in myeloid cells in a cGAS-STING pathway-dependent manner [28, 29]. Furthermore, HIV-1 evades the antiviral innate immune response by inhibiting mitochondrial antiviral signaling protein (MAVS) after sensing abortive HIV-1 RNA via the host helicase DEAD-box polypeptide 3 [30]. HIV-1 also negatively regulates the type I IFN response by exploiting the nucleotide-binding domain and leucine-rich repeat-containing protein X1 to inhibit the interaction between STING and TBK1 [31]. However, it is unclear which HIV-1 protein specifically mediates immune evasion, and the detailed mechanisms involved in the signaling cascade need to be elucidated.
In this study, we discovered that the HIV regulatory protein Vif inhibited the production of type I IFNs during HIV-1 infection through its interaction with SHP-1 in an ITIM-dependent manner. We demonstrated that Vif enhanced the association of SHP-1 with STING and inhibited its K63-linked ubiquitination at Lys337 via Tyr162 dephosphorylation. Furthermore, the FRK inhibitor D-65495 counteracted the phosphorylation of Vif, restored the antiviral innate immune response, and antagonized HIV-1 infection.
Methods and materials
Reagents and plasmids
The following antibodies were used: anti-TBK1 (3504), anti-Myc (2276, 2278), anti-phospho-TBK1 (5483), anti-phospho-IRF3 (29047), anti-phospho-p65 (3033), anti-phospho-p38 (9215), anti-phospho-Erk1/2 (9101), rabbit anti-phospho-Tyr (8954), fluorescent Alexa 488-conjugated anti-mouse IgG (4408), fluorescent Alexa 555-conjugated anti-mouse IgG (4409), fluorescent Alexa 594-conjugated anti-rabbit IgG (8889), and fluorescent Alexa 647-conjugated anti-rabbit IgG (4414), all from Cell Signaling Technology; anti-Vif (ab66643) and anti-SHP-1 (ab55356), both from Abcam; anti-Flag M2 Affinity Gel (A2220), anti-HA (H9658), anti-HA (H6908), and anti-Flag antibodies (F7425), all from Sigma-Aldrich; and anti-GFP (CW0087), anti-GST (CW0085M) and anti-His (CW0083M), all from Cwbio. Polyclonal anti-phospho-STING Y162 was generated via immunization of rabbits with the peptide AWS-pY-YIGYLRL (GeneCreate Biotech). Protein G Sepharose 4 Fast Flow was purchased from GE Healthcare. The expression constructs for Flag-RIG-I, Flag-N-RIG-I, Flag-TBK1, Flag-IRF3, and HA-Ubs were obtained from Dr. B. Ge (Tongji University, Shanghai, China). HA–Vif was a gift from Dr. Y. Zheng (Michigan State University, Michigan); Flag-STING was obtained from Dr. B. Sun (Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China); cDNAs encoding SRC, BLK, FRK, FYN, LYN, and LCK were obtained from Dr. J. Han (Xiamen University, Fujian, China); and VSV-G-pseudotyped NL4.3-Δenv was obtained from Dr. D. Sauter (Ulm University, Meierhofstrasse, Germany). Site-directed point mutagenesis of STING was performed using a KOD Plus Mutagenesis kit (SMK101, TOYOBO) according to the manufacturer’s protocols.
Mice
Mice heterozygous for SHP-1 deficiency (“motheaten” C57BL/6J Ptpn6me-v/+/J mice; 000811; Jackson Laboratories) and Tmem173−/− mice (obtained from Z. Jiang, Peking University, Beijing, China) were bred under specific pathogen-free conditions at the Shanghai Research Center for Biomodel Organisms. Three-week-old homozygous Ptpn6me-v/me-v mice and their wild-type littermates (control mice) and 6-week-old Tmem173−/− and wild-type control mice were utilized in the experiments. All animal studies were approved by the Institutional Animal Care and Use Committee of Fudan University.
Cell culture
Mouse peritoneal macrophages, MEFs, TZM-bl, and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Gibco) and 100 U/mL penicillin and streptomycin (HyClone). HeLa, THP-1, and Jurkat cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) heat-inactivated FBS (Gibco) and 100 U/mL penicillin and streptomycin (HyClone); MDMs were cultured in IMDM (Life Technologies) supplemented with 10% (v/v) heat-inactivated FBS (Gibco) and 100 U/mL penicillin and streptomycin (HyClone).
Isolation of mouse peritoneal macrophages, MEFs, and human MDMs
For peritoneal macrophages, 3-week-old homozygous Ptpn6me-v/me-v mice and their wild-type littermates were intraperitoneally injected with 1 mL of 4% Brucella Broth. Three days later, peritoneal lavage fluid was collected from the mice, and the cells were washed three times with phosphate-buffered saline (PBS). Peritoneal macrophages were cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin and streptomycin. For MEFs, Tmem173−/− or wild-type control mice were sacrificed at Day 13.5 of gestation via cervical dislocation. The embryos were separated from the placenta and the embryonic sac under aseptic conditions. After washing with PBS, the embryos were finely minced using a sterile razor blade. A total of 5 mL of 0.05% trypsin with 0.02% ethylenediaminetetraacetic acid (EDTA) was added, and the cells were dissociated by thorough pipetting before incubation at 4 °C overnight. The digested samples were centrifuged at a low speed (300 × g) at 4 °C for 5 min, and the cell pellets were suspended and cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin and streptomycin for 24 h. The cells were continuously cultured for passage or frozen when they reached 90–95% confluence. For MDMs, peripheral blood mononuclear cells (PBMCs) were isolated from healthy adult blood using CD14 microbeads (130-091-153; Miltenyi Biotec) according to the manufacturer’s protocol. Information on the volunteer blood donors is given in Table S1. The PBMCs were cultured for 6 days in endotoxin-free IMDM (Life Technologies) containing 10% FBS to obtain MDMs.
HIV-1ΔVif and HIV-1 Vif(Y147F) strains
HIV-1ΔVif and HIV-1 Vif(Y147F) were produced via site-directed mutagenesis based on the VSV-G-pseudotyped NL4.3-Δenv virus. For HIV-1ΔVif, two stop codons were inserted into the open reading frame of Vif using specific primers. For HIV-1 Vif(Y147F), the codon TAC encoding tyrosine at 147 was mutated to TTT, encoding phenylalanine. KOD FX polymerase (KFX-101, TOYOBO) was utilized for PCR amplification. Briefly, a 50-μL PCR solution containing 25 μL of 2× PCR buffer for KOD FX, 10 μL of dNTPs, 2.5 μL of primers, 1 μL of KOD FX polymerase, 200 ng of template plasmid, and ddH2O was used for the following PCR program: 94 °C for 2 min, 25 cycles of 98 °C for 10 s followed by 68 °C for 13 min, and 68 °C for 10 min. After digestion with DpnI at 37 °C for 1 h, the PCR products were transformed into DH5α for positive clone screening. The valid mutant vectors were identified by sequencing.
Virus infection
For HIV-1 infection, VSV-G-pseudotyped NL4.3-Δenv viruses were produced in HEK293T cells. After filtering through a 0.22-μm mesh, the viral supernatants were concentrated using 100K Amicon Ultra-15 filter tubes (Millipore). The virus concentrations were quantified by p24 ELISA (PerkinElmer Life Sciences) and titrated in TZM-bl cells using a luciferase assay system (E1500, Promega) according to the manufacturer’s protocol. MDMs and THP-1 cells (1 × 106 cells) were cultured without serum for 12 h and infected with HIV-1 pseudoviruses (MOI = 1) for the indicated times. For HSV-1 infection, viruses were collected from the supernatants of infected Vero cells. THP-1 cells (1 × 106 cells) were incubated for 12 h without serum and infected with HSV-1 (MOI = 5) for the indicated times.
RNA-seq library preparation, sequencing, and data processing
Human MDMs were treated for 24 h with medium or infected with HIV-1 or HIV-1ΔVif and lysed with RNAiso Plus (Takara) according to the manufacturer’s protocol. The lysates were sent to Personalbio for cDNA library construction and sequenced using an Illumina NovaSeq 6000. The RNA-seq reads were first trimmed to remove poly-A sequences, and unqualified reads were eliminated using Cutadapt. The remaining reads were then aligned to the Homo sapiens reference genome GRCh38 with HISAT2. The counts were summarized at the gene level using HTSeq. The gene expression values were computed from fragments per kilobase per million fragment (FPKM) values generated by addition of a pseudocount of 1 and log2-transformation of the results. These FPKM values were used to draw a heatmap using the pheatmap R package. Paired differential gene expression analyses were conducted using the DEseq R package with the criteria of a fold-change >2 and a p value < 0.05. Volcano plots representing differential gene expression were drawn using the ggplot2 R package. GO and KEGG enrichment analyses of differentially expressed genes were conducted using topGO and KAAS, respectively. The terms or pathways involved in the host antiviral immune response with a fold-change >2 and a p value < 0.05 were used to identify GO term enrichment or KEGG pathway enrichment. PPI analysis of the differential genes was conducted using the STRING database, and a PPI network model was drawn using Cytoscape.
Real-time quantitative PCR and HIV-1 Tat-Rev mRNA expression detection
Cells were incubated for 12 h without serum and infected with viruses for the indicated time. Total RNA was isolated using RNAiso Plus (9109; Takara) according to the manufacturer’s protocol. RNA (1 µg) was reverse-transcribed using a PrimeScript™ RT Reagent Kit (RR037; Takara) to generate cDNA. A LightCycler (LC480; Roche) and a SYBR RT–PCR kit (QPK-212; Toyobo) were utilized for quantitative real-time RT–PCR analysis. Gene amplification was analyzed using the ΔΔCt method. Gene expression was normalized to that of GAPDH. The primers used to amplify the human genes, mouse genes, and HIV-1 Tat-Rev mRNA are listed in Table S2.
ELISA
Cells were incubated for 12 h without serum and infected with viruses for the indicated time. A VerikineTM Human IFN-β ELISA Kit (41410-1; PBL Assay Science) and a Human TNF ELISA Kit (430204; BioLegend) were used to measure immunoreactive IFN-β and TNF in supernatants from MDMs, respectively.
Pervanadate treatment, immunoprecipitation, and Western blot analysis
For immunoprecipitation, HEK293T cells were transfected with the indicated plasmids. After 48 h, the cells were treated with pervanadate (0.1 mM sodium orthovanadate and 10 mM H2O2) for 30 min at 37 °C and washed three times with ice-cold PBS. The cells were lysed in lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, and 1 mM EDTA (pH 8.0)) supplemented with a protease inhibitor cocktail (04693159001; Roche), 1 mM PMSF, 1 mM Na3VO4, and 1 mM NaF. After 30 min on ice, the lysates were centrifuged for 15 min at 13,200 rpm and 4 °C to remove debris. The cell lysates were incubated with anti-Flag M2 Affinity Gel at 4 °C overnight. For immunoprecipitation of endogenous protein, peritoneal macrophages or THP-1 cells were lysed, and the lysates were incubated with specific antibodies and Protein G Sepharose 4 Fast Flow at 4 °C overnight. The Sepharose samples were centrifuged and washed three times with ice-cold PBST (1% Triton X-100 in PBS). For immunoblotting, the precipitates or cell lysates were boiled in 1× SDS loading buffer at 100 °C for 10 min and then analyzed via immunoblot assay.
GST pulldown
His or GST fusion proteins were expressed in E. coli BL-21(DE3) (Tiangen Biotech) or TKB1 cells (E. coli with a plasmid-encoded inducible gene encoding tyrosine kinase) (StrateGene) according to the manufacturer’s protocol. The purified His-fusion proteins or lysates from THP-1 cells were incubated with GST-fusion proteins conjugated to glutathione beads at 4 °C for 4 h. After centrifugation and washing three times with ice-cold PBS, the beads were incubated in 1× SDS loading buffer at 100 °C for 10 min and then analyzed via immunoblot assay.
In vitro kinase assay
GST-fusion proteins were expressed in E. coli BL-21(DE3) (Tiangen Biotech) and purified with GST Bestarose 4FF (Bestchrom) according to the manufacturer’s protocol. HEK293T cells were transfected with Myc-FRK or control vectors. After 48 h, the cells were lysed as described above. The cell lysates were incubated with Protein G Sepharose 4 Fast Flow (GE Healthcare) and a mouse anti-Myc tag antibody at 4 °C overnight. After washing three times with ice-cold PBST (1% Triton X-100 in PBS), the Sepharose samples were incubated with GST-fusion proteins in kinase reaction buffer (5 mM Tris–HCl (pH 7.5), 2.5 mM beta-glycerophosphate, 1 mM EGTA, 1 mM Na3VO4, 4 mM MgCl2, 100 nM ATP, and 1 mM DTT) at 30 °C for 30 min and evaluated via Western blot analysis.
Cell staining and confocal microscopy
HeLa cells were transfected with the appropriate vectors for 48 h, infected with HIV-1 or HIV-1ΔVif pseudoviruses for the times indicated, and fixed with 4% formaldehyde for 20 min at room temperature. The cells were permeabilized for 30 min in PBS containing 0.3% Triton X-100 and blocked at 4 °C for 1 h in blocking buffer (1% BSA in PBS). The cells were then incubated with primary antibodies at 4 °C overnight and secondary antibodies at room temperature for 1 h. After staining with DAPI, images were taken using a Leica TCS SP5 confocal laser microscopy system.
Transfection and RNA knockdown by adenoviral vectors
HEK293T cells were transiently transfected with polyethylenimine (23966-2; Polysciences) according to the manufacturer’s protocol. HeLa cells were transfected with Lipofectamine 2000 (11668; Invitrogen), and MEFs were transfected with Lipofectamine 3000 (L3000; Invitrogen). For SHP-1 knockdown, HEK293T cells were transfected with rADs with 2 μg of human α-synuclein and 4 μg of shSHP-1 or control vectors using LipofiterTM (Hanbio). For FRK knockdown with rADs, HEK293T cells were transfected with 2 μg of shFRK or control vectors and 4 μg of pBHGlox(delta)E1/3Cre using Nanofusion (Biomedicine Biotech). When most cells had detached from the substratum, the cells and supernatants were collected and subjected to three freeze–thaw cycles in liquid nitrogen at 37 °C. The virus was amplified twice by infecting fresh HEK293T cells with virus-containing supernatants, and virions were isolated by ultracentrifugation using CsCl. The viral titer was determined via TCID50 assay in HEK293T cells. For rAD infection, MDMs or THP-1 cells were incubated with rAD carrying shNC, shSHP-1, or shFRK for 8 h and cultured with fresh RPMI-1640 containing 10% FBS for 48 h. SHP-1-specific shRNA (5ʹ-AATTCGCGGCTGACATTGAGAACCGAGTGTTTGTGCTTAACACTCGGTTCTCAATGTCAGCCGTTTTTTG-3ʹ) and control shRNA (5ʹ-GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTC-3ʹ) was used to knock down SHP-1 in MDMs and THP-1 cells. FRK-specific shRNA (5ʹ-GGAGGCACAGATAATGAAGAACCTAACTCGAGTTAGGTTCTTCATTATCTGTGCCTCTTTTTG-3ʹ) and control shRNA (5ʹ-GTGGCCACTACTTTGTG GCTTTGTTTCTCGAGAAACAAAGCCACAAAGTAGTGGCCATTTTTG-3ʹ) were used for the FRK knockdown experiments.
Adenovirus-mediated overexpression
The rADs carrying various proteins, including Nef, Rev, Tat, Vif, Vpr, Vpu, and Vif(Y147F), were packaged as described previously. For infection, THP-1 cells were incubated with rAD carrying a mock insert or the indicated genes for 8 h and cultured with fresh RPMI-1640 containing 10% FBS for 48 h.
Dual-luciferase reporter assay
HEK293T cells were transiently transfected with pRL-IFN-β–Luc or pRL-ISRE–Luc, pRL-TK, and other vectors as indicated for 24 h. A Dual-Luciferase Reporter Assay System (RG028; Beyotime) was used to detect luciferase activity according to the manufacturer’s protocol.
Native PAGE
A dimerization assay was performed as previously described (Song et al., 2016). Briefly, THP-1 cells (1 × 106 cells) were cultured without serum for 12 h and infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F) pseudoviruses (MOI = 1). THP-1 cells were harvested with 50 μL of ice-cold lysis buffer (50 mM Tris (pH 7.4); 150 mM NaCl; 1% Triton X-100; 1 mM EDTA (pH 8.0); and protease inhibitor cocktail, 1 mM PMSF, 1 mM Na3VO4, and 1 mM NaF). After centrifugation at 13,000 × g for 10 min, the supernatants were quantified and diluted with 2× Native PAGE sample buffer (125 mM Tris–HCl (pH 6.8), 30% glycerol, and 0.1% bromophenol blue). Then, 30 μg of total protein was applied to a prerun 7.5% native polyacrylamide gel. After electrophoresis, the proteins were transferred onto a PVDF membrane for immunoblot analysis.
Protein structure analysis with Phyre2
The structure of Vif (residues 139-164) was analyzed using Phyre2 tools at the following website: http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index.
Statistical analysis
Statistical analysis of the RNA-seq data was performed as described previously. All data are expressed as the mean ± SEM. Two-tailed unpaired Student’s t tests were employed to compare the corresponding groups as indicated. GraphPad Prism 7.0 software was used for data analysis and presentation. P values ≤ 0.05 were considered to indicate statistical significance.
Results
Vif suppresses the antiviral innate immune response
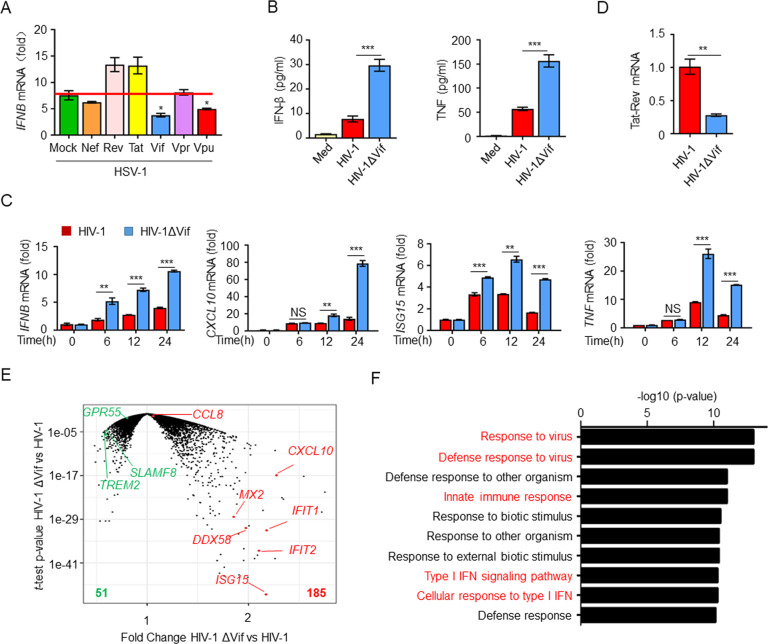
To elucidate the roles of HIV-1 proteins in the antiviral innate immune response during infection, we transfected various HIV-1 regulatory proteins into THP-1 cells and then infected the cells with herpes simplex virus 1 (HSV-1). The results indicated that overexpression of Vif significantly inhibited the HSV-1-induced expression of IFNB mRNA (Fig. 1A). HIV-1 infection promotes the activation of antiviral signaling pathways and thereby induces the production of type I IFNs; chemokines; ISGs; and proinflammatory cytokines, including IFN-β, CXCL10, ISG15, and tumor necrosis factor (TNF) [32]. To determine whether Vif suppresses these mediators during HIV-1 infection, we infected monocyte-derived macrophages (MDMs) with pseudotyped strains of HIV-1 and glycoproteins of vesicular stomatitis virus (VSV-G)-pseudotyped NL4.3-Δenv and its deletion mutant (HIV-1ΔVif). In these strains, the envelope protein of HIV-1 was replaced with the glycoprotein of vesicular stomatitis virus (VSV-G), which limits HIV-1 to a single round of replication in HEK293T cells and facilitates the infection of a large variety of human cell types [7, 31, 33]. As expected, we found that the deletion of Vif induced significantly elevated levels of IFN-β and TNF in MDMs (Fig. 1B). Accordingly, the expression of IFNB, CXCL10, ISG15, and TNF mRNA in HIV-1ΔVif-infected MDMs and IFNB, IFNG, IL2, and CD69 mRNA in HIV-1ΔVif-infected Jurkat cells were also higher than that in cells infected with the HIV-1 strain (Fig. 1C and Supplementary Fig. S1). Moreover, the viral replication level, which was represented by the expression of HIV-1 Tat-Rev mRNA in HIV-1ΔVif-infected cells, was significantly lower than that in cells infected with HIV-1 (Fig. 1D) [30]. These findings indicate that HIV-1 Vif inhibits cytokine production.
Fig. 1.
Vif suppresses the antiviral immune response. A IFNB mRNA levels in THP-1 cells expressing the HIV-1 regulatory proteins Nef, Rev, Tat, Vif, Vpr, and Vpu and infected with HSV-1 for 12 h (n = 3). B ELISA of IFN-β and TNF in the supernatants of MDMs infected with HIV-1 or HIV-1ΔVif for 24 h (n = 3). C IFNB, CXCL10, ISG15, and TNF mRNA levels in MDMs infected with HIV-1 or HIV-1ΔVif (n = 3). D Tat-Rev mRNA expression in MDMs infected with HIV-1 or HIV-1ΔVif for 24 h (n = 3). E Volcano plots (fold-change vs. t test P value) comparing the gene expression in MDMs infected with HIV-1 or HIV-1ΔVif. F GO enrichment analysis of differentially expressed genes. The data are representative of at least three independent experiments. The data are the means ± SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed unpaired Student’s t test)
To explore the regulatory role of Vif and its impact on overall host antiviral immunity, we conducted an RNA-seq analysis of human MDMs infected with HIV-1 or HIV-1ΔVif. Compared with HIV-1 infection, infection of MDMs with HIV-1ΔVif resulted in 236 differentially expressed genes (185 upregulated and 51 downregulated; Fig. 1E and Supplementary Fig. S2A). Protein–protein interaction (PPI) analysis based on the differential gene expression profile indicated a significant interaction network among the host antiviral genes, including ISG15, CXCL10, MX2, STAT1, TRIM22, IFIT-family genes, and OAS-family genes (Supplementary Fig. S2B). Consistently, a functional classification analysis of the upregulated genes revealed enrichment of positive regulatory host defense responses to virus infection (Supplementary Fig. S2C). Gene Ontology (GO) analyses revealed that the deletion of Vif resulted in upregulation of the expression of multiple genes related to the type I IFN signaling pathway, the cellular response to type I IFN, defense responses to viruses, and the innate immune response (Fig. 1F and Supplementary Fig. S2D). Similarly, a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated the enrichment of genes associated with antiviral immune signaling pathways, including the NOD-like receptor signaling pathway, cytosolic DNA-sensing pathway, RIG-I-like receptor signaling pathway, and chemokine signaling pathways (Supplementary Fig. S2E, F). Given the pivotal role of Vif in the regulation of the host antiviral transcriptional program, we conclude that Vif is important to HIV-1 infection, a finding consistent with the results of previous experiments.
Vif interacts with SHP-1 to suppress cytokine production
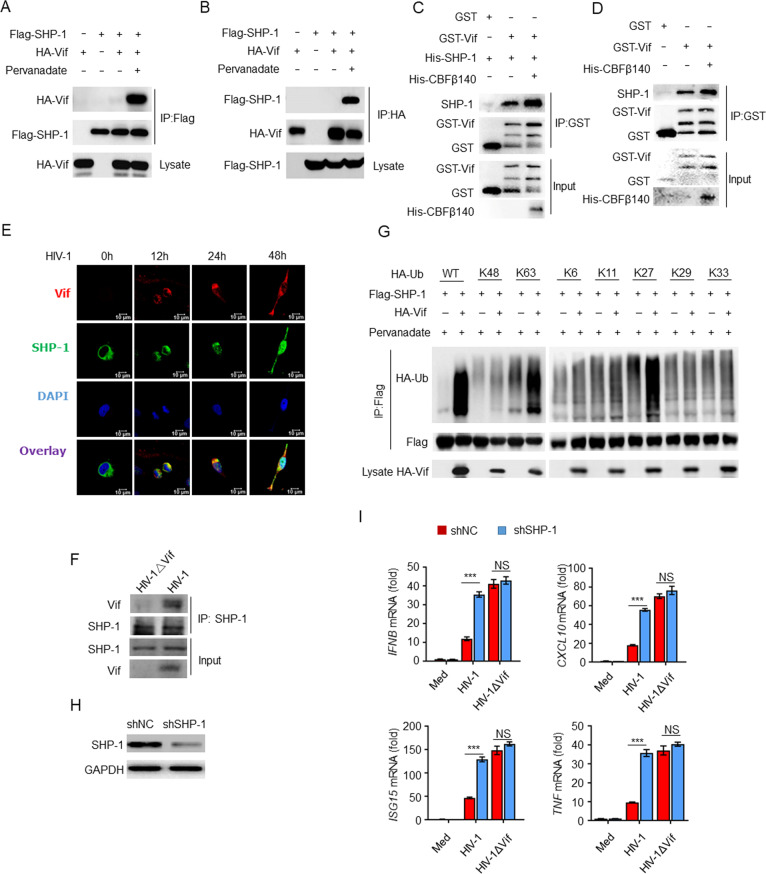
To explore the molecular mechanism through which Vif mediates immune inhibition, we reviewed the amino acid sequence of Vif and identified a host cellular ITIM (145LQYLAL150) motif. Host ITIMs are critical negative regulatory elements in immune cells that can be phosphorylated and recruit SHPs to inhibit the immune response [34]. To determine whether HIV-1 Vif interacts with SHP-1, we conducted forward and reverse coimmunoprecipitation assays (Co-IPs) and discovered that Flag–SHP-1 and HA–Vif interacted with each other in the presence of the phosphatase inhibitor pervanadate [35] (Fig. 2A, B). Furthermore, a phosphatase-defective SHP-1 mutant also interacted with Vif (Supplementary Fig. S3A). In addition, an in vitro glutathione S-transferase (GST) pulldown assay demonstrated that recombinant GST–Vif purified from TKB1 cells (Escherichia coli with a plasmid-encoded inducible gene for tyrosine kinase) was directly bound to purified recombinant His–SHP-1 or endogenous SHP-1 from THP-1 cells, while coexpression of CBFβ increased the binding (Fig. 2C, D). Consistent with these findings, Vif colocalized with SHP-1 following HIV-1 infection, as determined via immunofluorescence confocal microscopy (Fig. 2E). We also infected THP-1 cells with either HIV-1 or HIV-1ΔVif and found that Vif interacted with endogenous SHP-1 (Fig. 2F). To map the binding regions of both Vif and SHP-1, we prepared deletion mutants to determine whether these mutants lost their ability to interact with one another. Vif fragments containing amino acid residues 1–99 or 1-144 were unable to bind to SHP-1. However, SHP-1 bound to the regions of Vif corresponding to amino acid residues 1-150 or 100–192 (Supplementary Fig. S3B, C), which include the aforementioned ITIM (145LQYLAL150) element. These results indicate that ITIM phosphorylation is essential to the interaction with SHP-1. Similarly, SHP-1 fragments containing amino acid residues 1–242 were capable of interacting with Vif, whereas SHP-1 amino acid residues 243–595 exhibited only weak binding (Supplementary Fig. S3D, E). In addition, we found that Vif promoted the K27- and K63-linked ubiquitination of SHP-1, which are both essential to SHP-1 activation (Fig. 2G) [36]. These results indicate that Vif interacts with SHP-1 via the ITIM and promotes SHP-1 activation.
Fig. 2.
Vif interacts with SHP-1 to inhibit the immune response. A, B Immunoassay of HEK293T cell lysates expressing various vectors and treated with pervanadate. C Direct binding of GST–Vif with His–SHP-1. D The binding of GST–Vif with endogenous SHP-1 from THP-1 cells. E Confocal imaging of HeLa cells transfected with Flag–SHP-1 and infected with HIV-1. F Endogenous interaction of Vif and SHP-1 in HIV-1-infected THP-1 cells. G Immunoassay of lysates from HEK293T cells expressing various vectors. H Impact of SHP-1 shRNA in MDMs. I IFNB, CXCL10, ISG15, and TNF mRNA levels in MDMs transfected with SHP-1 or control shRNA and infected with HIV-1 or HIV-1ΔVif for 24 h (n = 3). The data are representative of at least three independent experiments. The data are the means ± SEMs. ***P < 0.001 (two-tailed unpaired Student’s t test)
As Vif was capable of interacting with SHP-1, we investigated whether SHP-1 mediated the inhibition of cytokine production by Vif. We found that transfection of MDMs with SHP-1 short hairpin (sh)RNA resulted in the specific inhibition of gene expression (Fig. 2H). SHP-1 knockdown caused the expression of IFNB, CXCL10, ISG15, and TNF mRNA (Fig. 2I) to be elevated in response to HIV-1 infection. In contrast, no significant differences were observed in cytokine production in MDMs infected with HIV-1ΔVif, regardless of SHP-1 expression (Fig. 2I). Taken together, these results suggest that the inhibition of cytokine production by Vif is mediated by SHP-1.
SHP-1 interacts with STING to inhibit K63-linked ubiquitination of STING
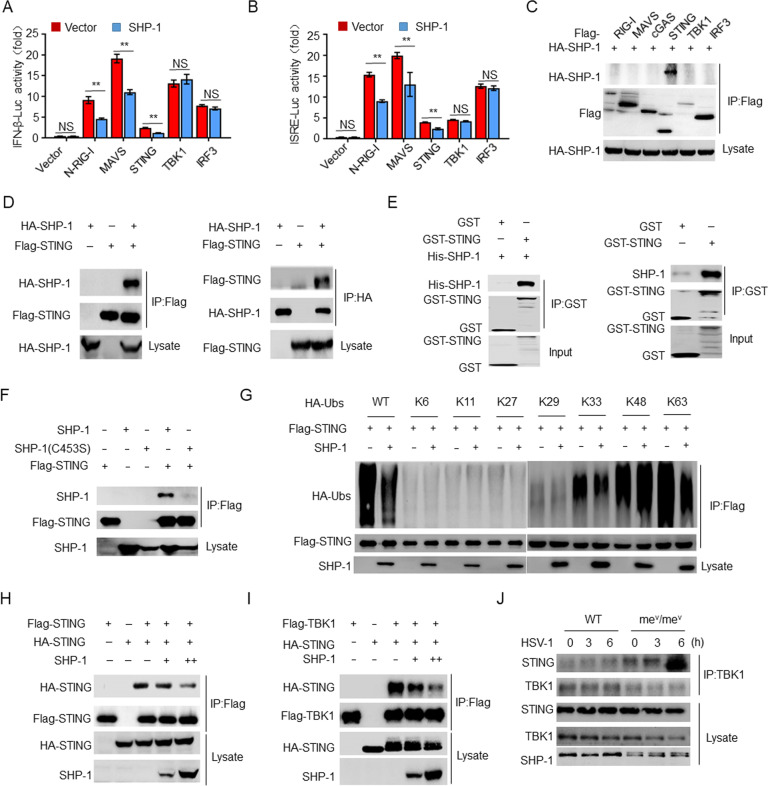
Our finding that the inhibition of antiviral immunity by Vif is mediated by SHP-1 raises the issue of the molecular mechanism through which SHP-1 exerts its effects during HIV-1 infection. A luciferase assay demonstrated that SHP-1 inhibited N-RIG-I-, MAVS-, and STING-induced IFN-β and ISRE activation but had no effect on the activation pathways induced by TBK1 and IRF3. This indicates that SHP-1 acts upstream of TBK1-IRF3 (Fig. 3A, B). Co-IP-based screening revealed that SHP-1 was capable of interacting with STING (Fig. 3C), which was confirmed in a forward and reverse Co-IP experiment (Fig. 3D). A GST pulldown assay revealed that STING directly bound to both His–SHP-1 and endogenous SHP-1 from peritoneal macrophages (Fig. 3E). However, the phosphatase-defective SHP-1 mutant was unable to interact with STING (Fig. 3F). Furthermore, STING colocalized with SHP-1, as revealed by immunofluorescence confocal microscopy (Supplementary Fig. S4A). To map the binding regions, we generated STING and SHP-1 deletion mutants and found that amino acid residues 1–137 of STING and amino acid residues 243–595 of SHP-1 are essential for their interaction (Supplementary Fig. S4B–E).
Fig. 3.
SHP-1 interacts with STING to inhibit the activation of STING. Luciferase assay of IFN-β (A) and ISRE (B) activation in HEK293T cells expressing various vectors (n = 3). C, D Immunoassay of lysates of HEK293T cells expressing various vectors. E Direct binding of GST-STING to His–SHP-1 (left) or to endogenous SHP-1 from peritoneal macrophages (right). F–I Immunoassay of lysates of HEK293T cells expressing various vectors. J Immunoassay of lysates of peritoneal macrophages from WT or mev/mev mice infected with HSV-1. The data are representative of at least three independent experiments. The data are the means ± SEMs. **P < 0.01 (two-tailed unpaired Student’s t test)
K63-linked ubiquitination of STING is required for TBK1 recruitment and type I IFN induction [37, 38]. To determine whether SHP-1 affects the ubiquitination of STING, we overexpressed SHP-1 in HEK293T cells and found that SHP-1 inhibited both the total and K63-linked ubiquitination of STING but had no effect on other types of ubiquitination (Fig. 3G and Supplementary Fig. S4F), whereas SHP-1(C453S) had no inhibitory activity (Supplementary Fig. S4G). Furthermore, SHP-1 inhibited the oligomerization of STING and the interaction between STING and TBK1 in a dose-dependent manner (Fig. 3H, I). In contrast, STING bound to TBK1 more efficiently in SHP-1-deficient peritoneal macrophages (marked as mev/mev) than in peritoneal macrophages from wild-type mice (Fig. 3J).
SHP-1 inhibits K63-linked ubiquitination of STING at Lys337 through dephosphorylation at Tyr162
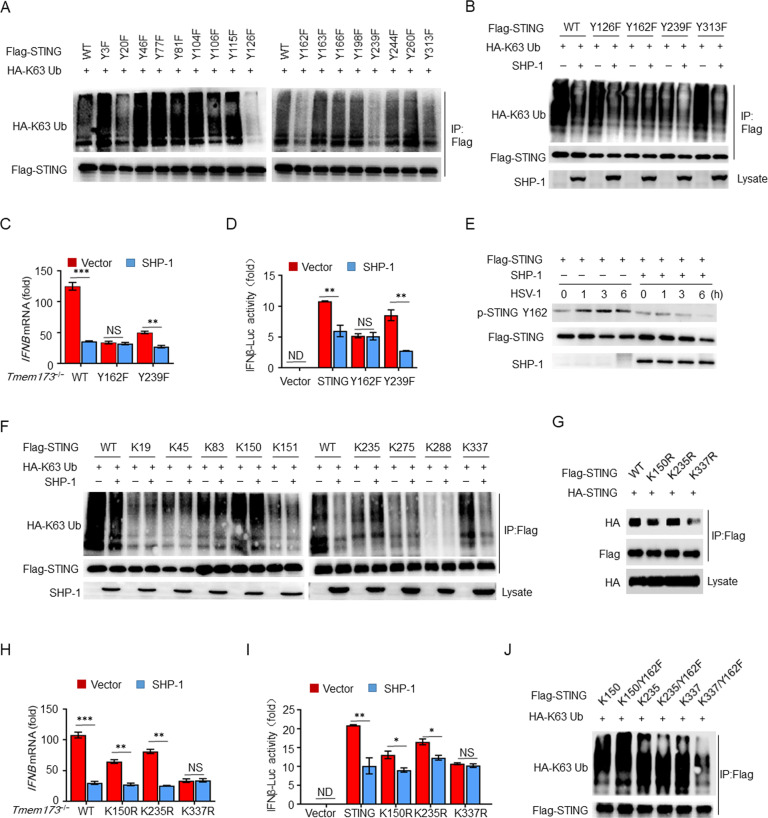
Next, we investigated why and how SHP-1, a protein tyrosine phosphatase, could inhibit the K63-linked ubiquitination of STING. We hypothesized that SHP-1 may counteract tyrosine phosphorylation of STING and thereby inhibit its ubiquitination. We generated STING mutants in which each of the 17 tyrosine residues was replaced with a phenylalanine (Y→F) residue. We found that the specific Y126F, Y162F, Y239F, and Y313F STING mutations caused reductions in K63-linked ubiquitination (Fig. 4A). Overexpression of SHP-1 inhibited the K63-linked ubiquitination of STING(WT), STING(Y126F), STING(Y239F), and STING(Y313F) but had a minimal effect on STING(Y162F) (Fig. 4B). Similarly, SHP-1 had no effect on HSV-1-stimulated IFNB in STING-deficient Tmem173−/− mouse embryonic fibroblasts (MEFs) transfected with STING(Y162F) or IFN-β luciferase activity in HEK293T cells expressing STING(Y162F) (Fig. 4C, D). Next, we generated a polyclonal antibody against pTyr162-STING and demonstrated that SHP-1 inhibited the HSV-1-induced phosphorylation of STING at Tyr162 (Fig. 4E).
Fig. 4.
SHP-1 inhibits K63-linked ubiquitination of STING at Lys337 by dephosphorylating STING at Tyr162. A, B Immunoassay of HEK293T cell lysates expressing various vectors. C IFNB mRNA levels in STING-deficient Tmem173−/− MEFs transfected with WT, Y162F, or Y239F mutants of STING together with control or SHP-1 and then infected with HSV-1 for 12 h (n = 3). D Luciferase assay of IFN-β activation in HEK293T cells expressing various vectors (n = 3). E Immunoassay of lysates of transfected HEK293T cells infected with HSV-1. F, G Immunoassay of lysates of HEK293T cells expressing various vectors. H IFNB mRNA levels in Tmem173−/− MEFs transfected with WT, K150R, K235R, or K337R mutants of STING together with control or SHP-1 and infected with HSV-1 for 12 h (n = 3). I Luciferase assay of IFN-β activation in HEK293T cells expressing various vectors (n = 3). J Immunoassay of lysates from HEK293T cells expressing various vectors. The data are representative of at least three independent experiments. The data are the means ± SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed unpaired Student’s t test)
To further confirm this mechanism, we generated STING mutants in which all of the lysine residues (except for those in the transmembrane domain) were replaced with arginine residues (K→R) with one exception [39, 40]. Most of the mutants exhibited nearly a complete loss of K63-linked ubiquitination, except for those with K83, K150, K235, and K337 mutations (each containing only one lysine) (Fig. 4F). Furthermore, overexpression of SHP-1 inhibited the ubiquitination of K337 but had no impact on the ubiquitination of K83, K150, or K235 (Fig. 4F). Interestingly, the single K337R mutation (in which only K337 was mutated) blocked the oligomerization of STING (Fig. 4G). SHP-1 was unable to suppress HSV-1-stimulated IFNB expression in Tmem173−/− MEFs transfected with K337R, nor did it have any effect on IFN-β luciferase activity in HEK293T cells expressing K337R (Fig. 4H, I). These results indicate that SHP-1 inhibits the K63-linked ubiquitination of STING specifically at Lys337.
To determine the relationship between Tyr162 and Lys337, Tyr162 was replaced with phenylalanine (Y→F) in the K150, K235, and K337 mutants. The results indicated that Y162F conversion dramatically reduced K63-linked ubiquitination at K337 but had no effect on K150 or K235 (Fig. 4J). These results indicate that SHP-1 inhibits the K63-linked ubiquitination of STING at Lys337 by dephosphorylating STING at Tyr162.
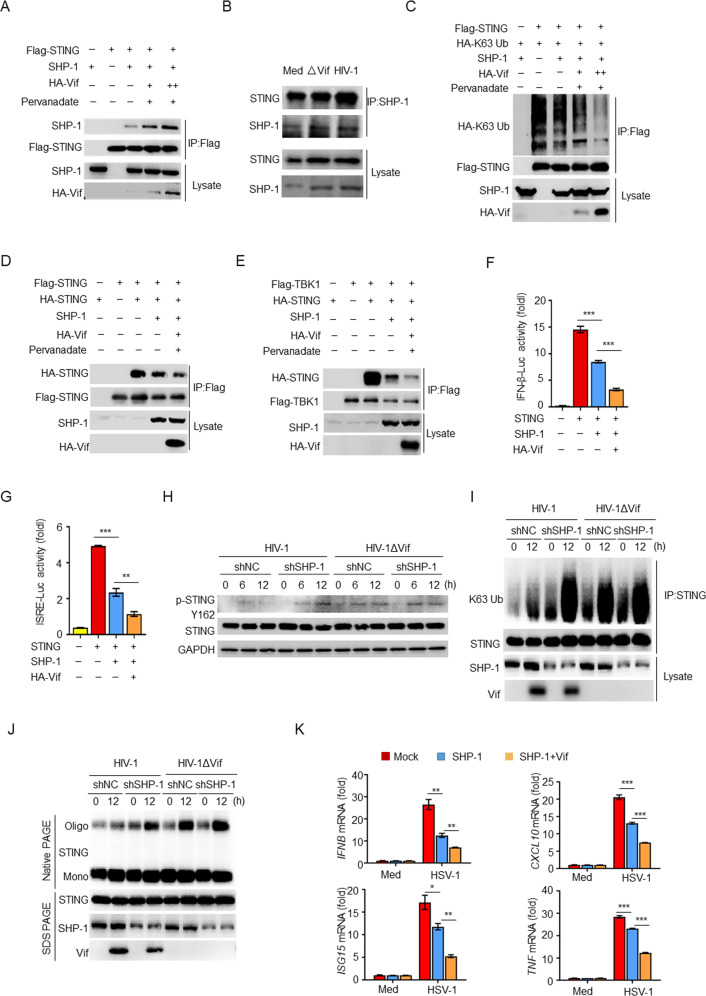
Vif facilitates the inhibitory effect of SHP-1 on STING activation
We subsequently investigated the regulatory relationship of the Vif-SHP-1-STING complex. We found that Vif not only promoted the recruitment of SHP-1 to STING in HEK293T cells (Fig. 5A) but also enhanced the interaction between endogenous SHP-1 and STING in HIV-1-infected THP-1 cells (Fig. 5B). In addition, overexpression of Vif enhanced SHP-1-mediated inhibition of STING phosphorylation (Supplementary Fig. S5A) and total and K63-linked ubiquitination (Supplementary Fig. S5B and Fig. 5C) but had no effect on K48-linked ubiquitination (Supplementary Fig. S5C). Similarly, Vif facilitated SHP-1-mediated inhibition of STING oligomerization and the interaction between STING and TBK1 (Fig. 5D, E), resulting in further inhibition of STING-induced IFN-β and ISRE luciferase activity (Fig. 5F, G). Conversely, knockdown of SHP-1 with shRNA resulted in increased Tyr162 phosphorylation (Fig. 5H), K63-linked ubiquitination (Fig. 5I), and oligomerization (Fig. 5J) of STING in response to HIV-1 infection. In contrast, we observed no significant differences in THP-1 cells infected with HIV-1ΔVif, irrespective of whether SHP-1 was knocked down. Furthermore, overexpression of Vif significantly decreased IFNB, CXCL10, ISG15, and TNF expression upon SHP-1 inhibition (Fig. 5K). These results indicate that Vif facilitates SHP-1-mediated inhibition.
Fig. 5.
Vif facilitates SHP-1-mediated inhibition of STING. A Immunoassay of lysates of HEK293T cells expressing various vectors. B Endogenous interaction of SHP-1 and STING in THP-1 cells infected with HIV-1 or HIV-1ΔVif. C–E Immunoassay of lysates from HEK293T cells expressing various vectors. Luciferase assays of IFN-β (F) and ISRE (G) activation in HEK293T cells expressing the indicated vectors (n = 3). H Immunoassay of lysates of MDMs transfected with SHP-1 or control shRNA and infected with HIV-1 or HIV-1ΔVif. I Immunoassay of lysates of THP-1 cells transfected with SHP-1 or control shRNA and infected with HIV-1 or HIV-1ΔVif. J Immunoblot (native PAGE) demonstrating STING dimerization in THP-1 cells transfected with SHP-1 or control shRNA and infected with HIV-1 or HIV-1ΔVif. K IFNB, CXCL10, ISG15, and TNF mRNA levels in THP-1 cells transfected with the indicated vectors and infected with HSV-1 for 12 h (n = 3). The data are representative of at least three independent experiments. The data are the means ± SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed unpaired Student’s t test)
ITIM phosphorylation is required for immune inhibition
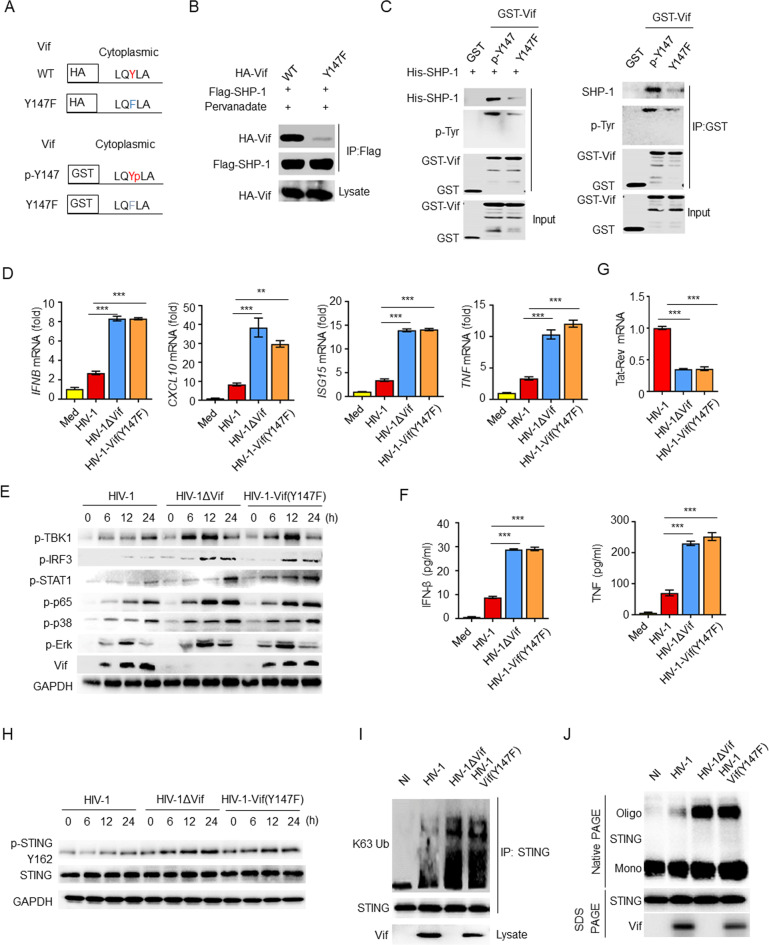
To determine whether the tyrosine phosphorylation of the ITIM is required for the interaction between Vif and SHP-1, we generated HA–Vif or GST–Vif mutants, in which the Tyr147 residue of the ITIM was replaced with a phenylalanine residue (Fig. 6A). Using a Co-IP assay, we observed that the ITIM mutation abolished the interaction between Vif and SHP-1 (Fig. 6B). Furthermore, we found that mutant Vif(Y147F) was neither phosphorylated nor capable of interacting with His–SHP-1 or endogenous SHP-1 using THP-1 cells in GST pulldown assays (Fig. 6C).
Fig. 6.
ITIM phosphorylation is required for the Vif–SHP-1 interaction and cytokine inhibition. A ITIM mutants of HA–Vif and GST–Vif. B Immunoassay of lysates of HEK293T cells expressing various vectors. C Direct binding of GST–Vif or its ITIM mutant to His–SHP-1 (left) or endogenous SHP-1 from THP-1 cells (right). D IFNB, CXCL10, ISG15, and TNF mRNA levels in MDMs infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F) for 24 h (n = 3). E Immunoblot of lysates of MDMs infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F). F ELISA of IFN-β and TNF in supernatants from (D) (n = 3). G Tat-Rev mRNA expression in MDMs infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F) for 24 h (n = 3). H Immunoblot of lysates of THP-1 cells infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F). I Immunoassay of lysates of THP-1 cells infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F). J Immunoblot (native PAGE) demonstrating STING dimerization in THP-1 cells infected with HIV-1, HIV-1ΔVif, or HIV-1-Vif(Y147F). The data are representative of at least three independent experiments. The data are the means ± SEMs. ***P < 0.001 (two-tailed unpaired Student’s t test)
Although Vif(Y147F) was detected in HIV-1 Vif(Y147F)-infected cells, it lost the ability to inhibit cytokine production and signal transduction in MDMs (Fig. 6D, E). Similarly, increased cytokine production was observed in HIV-1 Vif(Y147F)-infected THP-1 cells compared with HIV-1-infected cells (Supplementary Fig. S6A). Accordingly, the IFN-β and TNF protein levels were also higher in HIV-1 Vif(Y147F)-infected MDMs than in MDMs infected with HIV-1 (Fig. 6F). Moreover, viral replication in HIV-1 Vif(Y147F)-infected cells was lower than that in cells infected with HIV-1 (Fig. 6G). Mechanistically, compared with WT Vif, the ITIM mutant of Vif was not able to inhibit phosphorylation at Tyr162 (Fig. 6H), K63-linked ubiquitination (Fig. 6I), or oligomerization (Fig. 6J) of STING. Furthermore, overexpression of Vif(Y147F) in THP-1 cells resulted in the inability to inhibit HSV-1-induced expression of IFNB, CXCL10, ISG15, and TNF mRNA (Supplementary Fig. S6B, C). The viral load of HSV-1 was decreased to baseline in Vif(Y147F)-overexpressing THP-1 cells (Supplementary Fig. S6D). Taken together, the findings indicate that ITIM phosphorylation of Vif is essential for Vif–SHP-1 interaction and cytokine inhibition.
FRK is responsible for phosphorylating the ITIM of Vif during HIV-1 infection
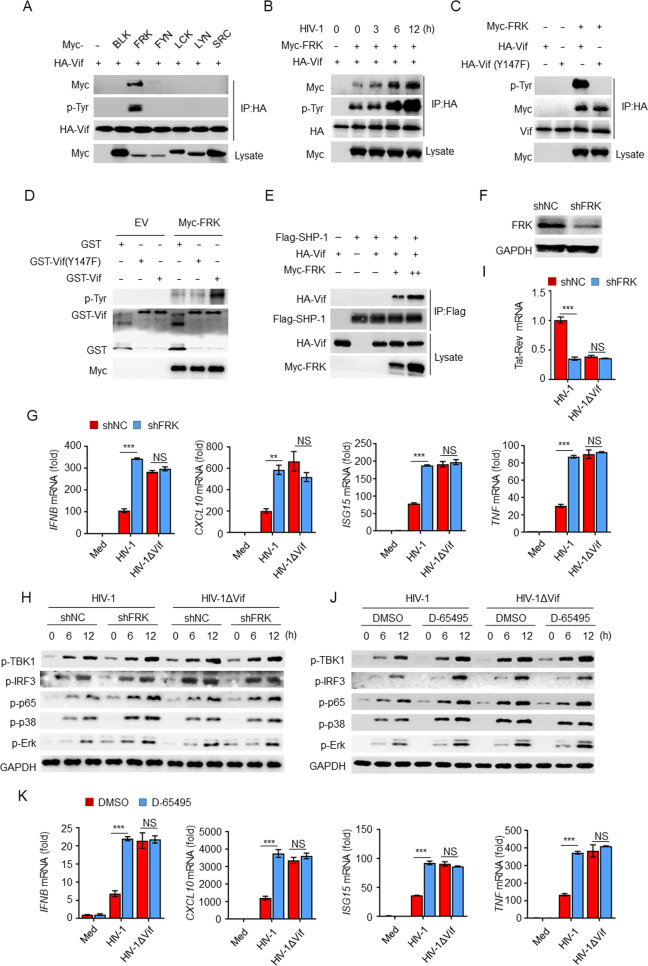
The tyrosine in the ITIM is normally phosphorylated by SRC-family kinases. To determine whether the ITIM of Vif was phosphorylated by a host tyrosine kinase, we transfected HEK293T cells with Vif in conjunction with six SRC-family tyrosine kinases, namely, BLK, FRK, FYN, LCK, LYN, and SRC, and performed a Co-IP experiment. The results indicated that FRK was the only kinase that interacted with Vif and promoted its tyrosine phosphorylation (Fig. 7A). In addition, HIV-1 infection enhanced the interaction between Vif and FRK as well as the tyrosine phosphorylation of Vif (Fig. 7B). In contrast, FRK interacted with Vif(Y147F), but no phosphorylation was observed (Fig. 7C, D). Furthermore, FRK facilitated the recruitment of Vif to SHP-1 in a dose-dependent manner in the absence of pervanadate (Fig. 7E). These results indicate that FRK is responsible for phosphorylating the ITIM of Vif.
Fig. 7.
FRK phosphorylates the ITIM of Vif to suppress cytokine production. A Immunoassay of lysates of HEK293T cells expressing various vectors. B Immunoassay of lysates of HEK293T cells transfected with the indicated vectors and infected with HIV-1. C Immunoassay of lysates of HEK293T cells. D Immunoblot of the in vitro kinase assay performed with purified GST, GST–Vif(Y147F), or GST–Vif and Myc-FRK. E Immunoassay of lysates of HEK293T cells expressing various vectors. F Impact of FRK shRNA in THP-1 cells. G IFNB, CXCL10, ISG15, and TNF mRNA levels in THP-1 cells transfected with FRK or control shRNA and infected with HIV-1 or HIV-1ΔVif (n = 3). H Immunoblot of lysates from THP-1 cells treated as in G. I Tat-Rev mRNA expression in THP-1 cells transfected with FRK or control shRNA and infected with HIV-1 or HIV-1ΔVif for 24 h (n = 3). J Immunoblot of lysates of THP-1 cells treated with D-65495 or DMSO and infected with HIV-1 or HIV-1ΔVif for the indicated times. K IFNB, CXCL10, ISG15, and TNF mRNA levels in THP-1 cells treated with D-65495 or DMSO and infected with HIV-1 or HIV-1ΔVif (n = 3). The data are representative of at least three independent experiments. The data are the means ± SEMs. **P < 0.01 and ***P < 0.001 (two-tailed unpaired Student’s t test)
We next investigated whether FRK mediates Vif inhibition. We transfected THP-1 cells with shRNA and found that FRK-specific shRNA decreased the expression of FRK (Fig. 7F). The knockdown of FRK resulted in significantly increased expression of IFNB, CXCL10, ISG15, and TNF mRNA (Fig. 7G); elevated phosphorylation of TBK1, IRF3, p65, p38, and Erk (Fig. 7H); and decreased viral replication in response to HIV-1 infection (Fig. 7I). In contrast, there were no significant differences in THP-1 cells infected with HIV-1ΔVif, regardless of whether FRK was knocked down.
As FRK signaling can be inhibited by D-65495 [41], we hypothesized that Vif-mediated inhibition may be blocked by D-65495. Our results indicated that treatment with D-65495 resulted in significantly elevated phosphorylation of IRF3, TBK1, p65, p38, and Erk (Fig. 7J), as well as IFNB, CXCL10, ISG15, and TNF (Fig. 7K), in HIV-1-infected THP-1 cells, whereas no differences were observed in HIV-1ΔVif-infected cells, regardless of the D-65495 treatment. These results suggest that FRK is a critical kinase that mediates the inhibition of cytokine production by Vif during HIV-1 infection.
Discussion
HIV-1 activates antiviral innate immune responses through its interactions with PRRs, including TLRs and DNA sensors, to induce the production of type I IFNs and proinflammatory cytokines. To the best of our knowledge, the role of Vif has not been fully elucidated in this process. In the present study, we found that Vif is phosphorylated by host FRK and then interacts with SHP-1 in an ITIM-dependent manner. Activated SHP-1 inhibits the K63-linked ubiquitination of STING at Lys337 by dephosphorylating STING at Tyr162, which subsequently inhibits STING oligomerization, TBK1 recruitment, and type I IFN production. Collectively, this is an effective means by which HIV-1 evades the antiviral innate immune response and attains symbiosis (Fig. S7).
During HIV-1 infection, the ITIM of Vif may be continuously phosphorylated by activated FRK to recruit SHP-1, which causes subsequent inhibition of STING activation. The FRK inhibitor D-65495 blocks Vif-mediated immune evasion and promotes the activation of IFN-β signaling. We have further demonstrated that the specific inhibition of FRK abolishes Vif-mediated inhibition of IFN-β and cytokine production, which facilitates virus clearance and further supports the identification of FRK as a critical kinase for inhibition of IFN-β and cytokine production by Vif.
As a rapidly emerging focus for understanding virus-induced innate immune signaling pathways, the cGAS–STING pathway is essential for the host immune response triggered by microbial DNAs, including the Y-form cDNA of HIV-1. Numerous DNA sensors, such as cGAS and IFI16, initiate signaling pathways that ultimately converge on STING, indicating the central position of STING in the antiviral immune response. However, the regulatory mechanisms underlying STING-mediated signaling have not been fully elucidated. By screening the 17 tyrosine residues and 9 lysine residues of STING, we identified Tyr162 phosphorylation and Lys337 ubiquitination as essential elements underlying STING activation. Moreover, we found that mutation of Tyr162 dramatically decreased the K63-linked ubiquitination of STING at Lys337. Our results indicate that Vif may recruit SHP-1 to inhibit the K63-linked ubiquitination of STING at Lys337 by dephosphorylating Tyr162, which ultimately inhibits IFN-β production. Other viruses, such as HSV-1, also induced the phosphorylation of STING at Tyr162, which was inhibited by SHP-1 (Fig. 4E). This suggests that this regulatory mechanism is a common strategy employed by viruses to interfere with the host immune response.
APOBEC3G potently inhibits HIV-1 replication by deaminating cytidine in viral DNA intermediates [42, 43]. However, Vif can degrade APOBEC3G and thereby promote viral replication in nonpermissive cells, including CD4+ T cells and MDMs [25, 26]. In contrast, permissive cells, such as HEK293T cells, do not express APOBEC3G. Thus, Vif has no effect on the replication and infectivity of HIV-1 in these cells [44]. In the present study, HIV-1 or HIV-1ΔVif pseudoviruses were collected at equivalent concentrations from the supernatants of APOBEC3G-deficient HEK293T cells transfected with VSV-G-pseudotyped NL4.3-Δenv vector (an env-deficient single-round clone) and pCL-VSVG. HIV-1 or HIV-1ΔVif single-round pseudoviruses do not proliferate in MDMs or THP-1 cells due to the absence of the Env gene. Thus, any impact of Vif on HIV-1 replication via APOBEC3G was ruled out in our study.
Although HIV primarily infects CD4+ T cells, it can also infect macrophages and dendritic cells without triggering a strong innate immune response [7, 45]. The absence of a rigorous innate immune response to HIV-1 in dendritic cells is a major factor that limits a productive T-cell response [46]. Furthermore, HIV-1 exploits numerous negative regulators, such as SHP-1, to restrict the host innate immune response. In most cases, SHP-1 inhibits the activation of NF-κB and MAP kinases via direct dephosphorylation [47, 48]. In our study, we found that SHP-1 inhibited type I IFN production, which was dominantly induced by cGAS–STING during HIV-1 infection, as cDNA is more effective in eliciting type I IFN production than RNA in the context of HIV-1 infection [7]. The interplay between the NF-κB and type I IFN signaling pathways coordinately modulates antiviral innate immune responses [49]. During HIV-1 infection, the interaction between the glycoprotein gp41 and TAK1 results in the phosphorylation of IκB and the translocation of NF-κB to the cell nucleus, which ultimately leads to the production of proinflammatory cytokines [50]. In our experiments, we found that HIV-1 Vif inhibited both the NF-κB and type I IFN signaling pathways, which modulated HIV-1 replication and mediated immune evasion.
The ITIM 145LQYLAL150 element overlaps with Vif’s BC-box motif (residues 144–155), which interacts with the E3 ubiquitin ligase components EloginB/EloginC with high affinity to form the Vif–CBF-b–CUL5–ELOB–ELOC complex. We are curious whether the interaction of Vif and SHP-1 affects this complex assembly. Previous publications have reported that Vif residues V142, L145, L148, A149, A152, and L153 in the BC box create a hydrophobic face that interacts with EloC [51, 52]. Interestingly, the polar residue Y147 is located on the opposite side of the hydrophobic face, which makes it possible that SHP-1 binds with Vif [53] (Fig. S8A, B). Theoretically, increased polarity of Y147 due to phosphorylation is beneficial for hydrophobic face formation. Indeed, our data demonstrated that SHP-1 significantly enhanced the binding of Vif to EloginB and EloginC, while EloginB/EloginC promoted the interaction between Vif and SHP-1 in the presence of pervanadate (Fig. S8C-F). In contrast, the ITIM mutation Y147F decreased the ability of the ITIM to bind with EloginB and EloginC (Fig. S8G, H). We speculate that phosphorylation of Y147 is critical for Vif-dependent APOBEC3G degradation via recruitment of SHP-1. Consequently, Vif recruits SHP-1 not only to inhibit STING activation but also to enhance Vif-dependent APOBEC3G degradation during HIV-1 infection in APOBEC3G + cells. The synergistic effect of Vif greatly increases HIV-1 infectivity. On the other hand, SHP-1-mediated inhibition of Vif in the cGAS-STING pathway is effective for immune inhibition in the absence of APOBEC3G.
In summary, we have demonstrated that Vif recruits SHP-1 and results in the decreased production of type I IFNs through its interactions with STING. In addition, SHP-1-mediated dephosphorylation at Tyr162 results in the inhibition of K63-linked ubiquitination of STING at Lys337. Overall, our results reveal a completely novel mechanism whereby an ITIM-containing HIV-1-encoded protein inhibits the posttranslational modification of STING. Moreover, our finding that STING phosphorylation regulates STING ubiquitination may represent a common strategy employed by viruses to modulate type I IFNs. Finally, we discovered that FRK phosphorylates the ITIM of Vif and thereby promotes its interactions with SHP-1. In contrast, the FRK inhibitor D-65495 counteracts Vif-mediated immune evasion. Our study sheds light on a previously undiscovered mechanism of immune evasion by HIV-1 and raises the possibility of developing therapeutic strategies for HIV-1 infection based on Vif.
Supplementary information
Acknowledgements
This work was supported by grants from the Program of Shanghai Academic Research Leader (21XD1402900), the Natural Science Foundation of Shanghai (21ZR1481400), the National Natural Science Foundation of China (31972900), the National Youth Talent Support Program (Ten Thousand Talent Program), the National Key Research and Development Program of China (2018YFC1705505), and the National Megaproject on Key Infectious Diseases (2017ZX10202102). We thank Dr. Y. Zheng (Michigan State University, Michigan) for HA-Vif, Dr. B. Sun (Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China) for Flag-STING, Dr. D. Sauter (Ulm University, Meyerhofstrasse, Germany) for pBR322-HIV-1-M-NL4-3-IRES-eGFP env STOP plasmid (pseudotyping is required for infection), and Dr. J. Han (Xiamen University, Xiamen, China) for cDNAs encoding SRC-family kinases.
Author contributions
JX, QZ, and DY conceived the project and designed the experiments. YW and DY wrote the manuscript. YW, GQ, LZ, and ZZ performed most of the experiments and analyzed the data. YL, WH, XZ, YZ, and TX assisted with the experiments and provided technical help. HZ, XY, XY, and XZ provided comments and assisted with manuscript preparation.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yu Wang, Gui Qian, Lingyan Zhu, Zhuo Zhao.
Contributor Information
Jianqing Xu, Email: xujianqing@shphc.org.cn.
Quanming Zou, Email: qmzou2007@163.com.
Dapeng Yan, Email: dapengyan@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00802-9.
References
- 1.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10:1081–8. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 2.Gringhuis SI, van der Vlist M, van den Berg LM, den Dunnen J, Litjens M, Geijtenbeek TB. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat Immunol. 2010;11:419–26. doi: 10.1038/ni.1858. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsen MR, Olagnier D, Hiscott J. Innate immune sensing of HIV-1 infection. Curr Opin HIV AIDS. 2015;10:96–102. doi: 10.1097/COH.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 4.Soper A, Kimura I, Nagaoka S, Konno Y, Yamamoto K, Koyanagi Y, et al. Type I interferon responses by HIV-1 infection: association with disease progression and control. Front Immunol. 2017;8:1823. doi: 10.3389/fimmu.2017.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 6.Cohen KW, Dugast AS, Alter G, McElrath MJ, Stamatatos L. HIV-1 single-stranded RNA induces CXCL13 secretion in human monocytes via TLR7 activation and plasmacytoid dendritic cell-derived type I IFN. J Immunol. 2015;194:2769–75. doi: 10.4049/jimmunol.1400952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110:E4571–80. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–42. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–32. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol. 2015;16:1025–33. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, et al. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell. 2018;175:488–501 e22. doi: 10.1016/j.cell.2018.08.062. [DOI] [PubMed] [Google Scholar]
- 15.Hotter D, Bosso M, Jønsson KL, Krapp C, Stürzel CM, Das A, et al. IFI16 targets the transcription factor Sp1 to suppress HIV-1 transcription and latency reactivation. Cell Host Microbe. 2019;25:858–872 e13. doi: 10.1016/j.chom.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567:394–8. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivro A, Su RC, Plummer FA, Ball TB. Interferon responses in HIV infection: from protection to disease. Aids Rev. 2014;16:43–51. [PubMed] [Google Scholar]
- 18.Scagnolari C, Antonelli G. Type I interferon and HIV: subtle balance between antiviral activity, immunopathogenesis and the microbiome. Cytokine Growth Factor Rev. 2018;40:19–31. doi: 10.1016/j.cytogfr.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsikis PD, Mueller YM, Villinger F. The cytokine network of acute HIV infection: a promising target for vaccines and therapy to reduce viral set-point? PLoS Pathog. 2011;7:e1002055. doi: 10.1371/journal.ppat.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9:1138–43. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 22.Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, KewalRamani VN, et al. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 2012;8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–5. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 26.Mercenne G, Bernacchi S, Richer D, Bec G, Henriet S, Paillart JC, et al. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–46. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2011;481:376–79. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 28.Ayinde D, Casartelli N, Schwartz O. Restricting HIV the SAMHD1 way: through nucleotide starvation. Nat Rev Microbiol. 2012;10:675–80. doi: 10.1038/nrmicro2862. [DOI] [PubMed] [Google Scholar]
- 29.Maelfait J, Bridgeman A, Benlahrech A, Cursi C, Rehwinkel J. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 2016;16:1492–1501. doi: 10.1016/j.celrep.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gringhuis SI, Hertoghs N, Kaptein TM, Zijlstra-Willems EM, Sarrami-Forooshani R, Sprokholt JK, et al. HIV-1 blocks the signaling adaptor MAVS to evade antiviral host defense after sensing of abortive HIV-1 RNA by the host helicase DDX3. Nat Immunol. 2017;18:225–35. doi: 10.1038/ni.3647. [DOI] [PubMed] [Google Scholar]
- 31.Guo H, König R, Deng M, Riess M, Mo J, Zhang L, et al. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA viruses. Cell Host Microbe. 2016;19:515–28. doi: 10.1016/j.chom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol. 2015;13:403–13. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–13. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 35.Yan D, Wang X, Luo L, Cao X, Ge B. Inhibition of TLR signaling by a bacterial protein containing immunoreceptor tyrosine-based inhibitory motifs. Nat Immunol. 2012;13:1063–71. doi: 10.1038/ni.2417. [DOI] [PubMed] [Google Scholar]
- 36.Aki D, Li H, Zhang W, Zheng M, Elly C, Lee JH, et al. The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR. Nat Immunol. 2018;19:766–75. doi: 10.1038/s41590-018-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–76. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–55. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18:214–24. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 41.Welsh M, Welsh C, Ekman M, Dixelius J, Hägerkvist R, Annerén C, et al. The tyrosine kinase FRK/RAK participates in cytokine-induced islet cell cytotoxicity. Biochem J. 2004;382:261–68. doi: 10.1042/BJ20040285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 43.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 44.Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 45.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–22. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luban J. Innate immune sensing of HIV-1 by dendritic cells. Cell Host Microbe. 2012;12:408–18. doi: 10.1016/j.chom.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson DJ, Pao LI, Dhanji S, Murakami K, Ohashi PS, Neel BG. Shp1 regulates T cell homeostasis by limiting IL-4 signals. J Exp Med. 2013;210:1419–31. doi: 10.1084/jem.20122239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nandan D, Lo R, Reiner NE. Activation of phosphotyrosine phosphatase activity attenuates mitogen-activated protein kinase signaling and inhibits c-FOS and nitric oxide synthase expression in macrophages infected with Leishmania donovani. Infect Immun. 1999;67:4055–63. doi: 10.1128/iai.67.8.4055-4063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 50.Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS, et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc Natl Acad Sci USA. 2018;115:E3798–807. doi: 10.1073/pnas.1801213115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, et al. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol. 2008;82:8656–63. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, et al. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–33. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 53.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.