Abstract
Cryptosporidium present in 1,705 fecal samples from humans and 105 from livestock animals were analyzed by PCR-restriction fragment length polymorphism of the Cryptosporidium oocyst wall protein. Overall, genotype 1 (human exclusive type) was detected in 37.8% of the samples from humans, genotype 2 (broad host range) was detected in 61.5%, a third genotype designated genotype 3 (Cryptosporidium meleagridis) was detected in 0.3%, and both genotypes 1 and 2 were recovered from 0.4%. All samples from livestock yielded genotype 2. Among 469 patients infected during eight drinking water-related outbreaks, five outbreaks were predominantly due to genotype 1, and three were due to genotype 2. Fifty-four samples were collected from patients involved with five swimming pool-associated outbreaks: two outbreaks were due to genotype 1, one was due to genotype 2, and the remaining two involved both genotypes 1 and 2. Among 26 family outbreaks and 1 children's nursery outbreak (2 to 3 members per group), the same genotype was recovered from the different members of each outbreak: 13 were due to genotype 1, and 14 were due to genotype 2. In eighteen patients reporting contact with animals and/or farms, genotype 1 was recovered from one patient and genotype 2 was recovered from the remaining 17. Among the sporadic cases, there were distinct geographical and temporal variations in the distribution of the genotypes. The spring peak in cases was due to genotype 2. Genotype 1 was significantly more common in patients infected during the late-summer–autumn peak and in those with a history of foreign travel.
Cryptosporidium is increasingly recognized as a major cause of diarrheal disease worldwide (12). Infection occurs via the oral route, and the importance of water in disease transmission is well recognized (11, 12, 16, 19, 37). Despite this, the exact modes of transmission are often unclear, and the relative importance of foreign travel, consumption of foods, beverages, or water, and person-to-person transmission, as well as the role of infected animals in disease transmission, remains to be ascertained (11, 12, 19).
The genus Cryptosporidium is a group of coccidian protozoan parasites that infect a variety of mammals, birds, reptiles, and fish (20). Traditionally, the different species of this genus have been identified on the basis of host range, oocyst morphology, and site of infection. However, DNA sequence analysis indicates that this genus is complex and may contain more than 20 different species, and the traditional approaches do not provide a reliable method for the species identification (20). Molecular methods indicate that Cryptosporidium parvum is the major cause of cryptosporidiosis in humans and comprises at least two different genotypes (20) which may constitute separate species of parasite (18). However, other Cryptosporidium species have been associated with human infections: Cryptosporidium felis, Cryptosporidium meleagridis, and an as-yet-unnamed Cryptosporidium species designated the dog type in human immunodeficiency virus-positive individuals (21, 27) and C. meleagridis in the immunocompetent (25).
The two major types of C. parvum can be recognized by phenotypic and genotypic methods (5, 18, 20, 23, 26, 30, 31, 35): one type occurs exclusively in naturally infected humans and a single nonhuman primate (genotype 1, or human type), and a second occurs in livestock animals as well as humans (genotype 2, or calf type). Experimental animal infection of both calves and mice was successful with genotype 2, but not with genotype 1 (23). Recent data have shown that limited growth of a single genotype 1 strain occurs in an experimentally infected gnotobiotic piglet model (36). It has been suggested that these observations concerning the two genotypes of C. parvum reflect the epidemiology of two parasites with distinct and exclusive transmission cycles (20, 26). Since the host range of these two genotypes differs, the epidemiology of the disease in humans may also differ, although this has, to date, been incompletely investigated.
The Public Health Laboratory Service (PHLS) has been active in the investigation of the epidemiology of human cryptosporidiosis (9, 10, 17, 18). We previously reported a simple DNA extraction method applicable to whole feces and suitable for amplification of cryptosporidial DNA and presented data on the application of these techniques to 397 cases of cryptosporidiosis (18, 23). Here we further extend the results from this series of samples collected in the United Kingdom and report on the distributions of polymorphisms within the Cryptosporidium oocyst wall protein (COWP) gene amplified from 1,705 fecal samples from humans and 105 fecal samples from livestock animals. The distribution of genotypes from humans is analyzed with respect to patients associated with drinking water, swimming pool, and family outbreaks, and contacts with animals, together with seasonal and geographical distributions among the sporadic cases.
MATERIALS AND METHODS
Fecal samples and clinical information.
Whole feces were collected from humans with diarrhea and livestock animals (lambs and calves) in the United Kingdom where Cryptosporidium oocysts were recognized by conventional techniques (4). All samples were stored as whole feces at 4°C without preservatives. Epidemiological information (details of outbreaks, age, sex, recent foreign travel, geographical region, date of collection, etc.) was collected for all samples from humans.
National data on Cryptosporidium infections were collected through the PHLS surveillance network for laboratory-confirmed cases (34).
Oocyst disruption and DNA extraction.
Oocyst disruption and DNA purification were performed as described previously (18). The method was performed as follows. Approximately 200 μl of whole feces was added to 900 μl of 10 M guanidinium thiocyanate in 0.1 M Tris-HCl (pH 6.4) plus 0.2 M EDTA (pH 8.0), and 2% (wt/vol) Triton X-100, together with 0.3 g of 0.5-mm-diameter zirconia beads (Stratech Scientific, Luton, United Kingdom) plus 60 μl of isoamyl alcohol. The tube was shaken in a Mini-Beadbeater or a Beadbeater-8 (Stratech Scientific) for 2 or 1.5 min, respectively, at maximum speed, left at room temperature for 5 min, and centrifuged. Coarse activated silica suspension (100 μl; Severn Biotech, Kidderminster, United Kingdom) was added to the supernatant (6), and this was incubated at room temperature for 10 min with gentle agitation. The supernatant was discarded, and the pellet was washed: twice with 200 μl of 10 M guanidinium thiocyanate in 0.1 M Tris-HCl (pH 6.4), twice with 200 μl ice-cold 80% ethanol; and once with 200 μl of acetone. The pellet was then dried at 56°C for 5 min, and the DNA was eluted into 150 μl of water after vortex mixing and incubation at 56°C for 5 min. The supernatant (DNA sample) was recovered by centrifugation and either used directly for PCR amplification or stored at −20°C until further use.
For samples in which the PCR amplification (described below) was unsuccessful, a further DNA extraction with polyvinylpyrrolidone (PVP) was performed as described by Lawson et al. (15). This included the addition of 50 μl of the extracted DNA sample to 150 μl of PVP-TE (10% [wt/vol] PVP in Tris-EDTA [TE] buffer) and incubation at room temperature for 10 min. The DNA was then precipitated by the addition of 100 μl of 2 M ammonium acetate and 600 μl of isopropanol at −20°C for 30 min. DNA was recovered by centrifugation (11,000 × g for 10 min), dried, reconstituted in water, and used as described above.
PCR-RFLP analysis.
PCR amplification was performed in 25-μl total volumes and included 2.5 μl of extracted DNA. Positive (previously tested samples of known genotype) and negative (buffer only) controls were included in each batch of tests. PCR amplification of the COWP gene fragment using the primer pair cry-9 and cry-15 (Life Technologies, Ltd., Paisley, United Kingdom) was performed as described previously (30). Previous analysis with DNA extracted from a range of other intestinal pathogens (including other species of parasites) indicates that this assay is specific for Cryptosporidium (24, 33; S. Pedraza-Díaz et al., unpublished data). The PCR mixtures contained 2.5 μl of DNA sample; 1× PCR buffer (Life Technologies), 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphates (dNTPs), 30 pmol of each primer, and 1.25 U of Taq polymerase (Life Technologies, Ltd.). The PCR conditions used were 35 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 1 min, followed by 10 min at 72°C. Five-microliter aliquots of the PCR products were detected following electrophoresis in 1% agarose gels stained with either ethidium bromide or SYBR Green 1 nucleic acid stain (Cambridge Bioscience, Cambridge, United Kingdom) and recorded by UV transillumination and type 52 film (Polaroid, Ltd., St. Albans, Hertsfordshire, United Kingdom).
For restriction fragment length polymorphism (RFLP) analysis, the Cryptosporidium genotype was determined by restriction digestion of the COWP gene fragment using RsaI as described previously (30). Restriction fragments were resolved by electrophoresis in 3.2% typing-grade agarose gels and stained as described above. The fragments generated were 284, 129, 106, and 34 bp (C. parvum genotype 1); 413, 106, and 34 bp (C. parvum genotype 2); and 372, 147, and 34 bp (C. meleagridis; previously described as genotype 3 [23, 25]).
RESULTS
The COWP gene was amplified from 1,810 samples: 105 from livestock animals and 1,705 from humans (Table 1). Overall, genotype 1 was found in 37.8% of the human samples, genotype 2 was found in 61.5%, both genotypes 1 and 2 were detected in 0.4%, and a further unusual PCR-RFLP type (generating fragments at 372, 147, and 34 bp) designated as genotype 3 was found in 0.3%. A more complete analysis of the five samples in which genotype 3 was detected is published elsewhere (25), and these samples are not further considered here.
TABLE 1.
Cryptosporidium genotypes in 1,705 fecal samples from humans and 105 from livestock animals
| Type of case (yr) | No. of cases of COWP genotypea:
|
||||
|---|---|---|---|---|---|
| Total | 1 | 2 | 3 | 1 and 2 | |
| Human | 1,705 | 645 | 1,049 | 5 | 6 |
| Drinking water-associated outbreaks | 469 | 196 | 271 | 1 | 1 |
| Swimming pool-associated outbreaks | 54 | 38 | 13 | 0 | 3 |
| Sporadic | 1,182 | 411 | 765 | 4 | 2 |
| England (1995–1999) | 1,092 | 395 | 691 | 4 | 2 |
| Scotland (1998–1999) | 11 | 1 | 10 | 0 | 0 |
| N. Ireland (1998–1999) | 48 | 5 | 43 | 0 | 0 |
| Wales (1994–1998) | 31 | 10 | 21 | 0 | 0 |
| Livestock animals | 105 | 0 | 105 | 0 | 0 |
| Cows (natural infections) | 52 | 0 | 52 | 0 | 0 |
| Sheep (natural infections) | 16 | 0 | 16 | 0 | 0 |
| Experimental infections | 37 | 0 | 37 | 0 | 0 |
COWP, Cryptosporidium oocyst wall protein gene.
Genotyping results from 469 patients infected during eight drinking water-associated outbreaks were obtained, and this represents an analysis of 29% of the total number of microbiologically confirmed cases identified (range, 3.6 to 100%) in these outbreaks (Table 2). Genotype 1 was predominantly recovered from patients involved with outbreaks 1 to 5, albeit, only a small proportion of the total numbers of microbiologically confirmed cases were examined in outbreaks 1 (3.6%) and 2 (14.8%). Outbreaks 1 to 5 occurred throughout the year, and the water involved with these was derived from both boreholes and surface (river water) sources. In contrast, the remaining three outbreaks (outbreaks 6 to 8) were almost exclusively due to genotype 2, occurred during the spring, and involved contamination of surface (reservoir) water or a water tank used in a small private water supply (1).
TABLE 2.
Genetic analysis of Cryptosporidium from six drinking waterborne and swimming pool-waterborne outbreaks in England in 1994 to 1999
| Outbreak (period) | Total no. of microbiologically confirmed cases | Affected water source type | No. of patients (isolates) with COWP genotypea:
|
Reference(s) | ||||
|---|---|---|---|---|---|---|---|---|
| Total | 1 | 2 | 3 | 1 and 2 | ||||
| Drinking water | ||||||||
| 1 (August–December 1994) | 224 | Surface (river) | 8 | 8 | 0 | 0 | 0 | 7, 17 |
| 2 (August–September 1995) | 575 | Surface (river) | 85 | 84 | 0 | 0 | 1 | 7, 17 |
| 3 (January–March 1997) | 22 | Surface (river) | 11 | 11 | 0 | 0 | 0 | 7 |
| 4 (February–April 1997) | 345 | Borehole | 94 | 81 | 12 | 1 | 0 | 31 |
| 5 (December 1997–January 1998) | 34 | Borehole | 9 | 9 | 0 | 0 | 0 | 1 |
| 6 (April 1998) | 6 | Water tank | 6 | 0 | 6 | 0 | 0 | 2 |
| 7 (April 1998) | 62 | Surface (reservoir) | 24 | 0 | 24 | 0 | 0 | 2 |
| 8 (April–May 1999) | 347 | Surface (reservoir) | 232 | 3 | 229 | 0 | 0 | 2 |
| Swimming pool | ||||||||
| 9 (September–November 1998) | 11 | 3 | 3 | 0 | 0 | 0 | 2 | |
| 10 (September 1999) | 16 | 8 | 5 | 3 | 0 | 0 | 3 | |
| 11 (July 1999) | 11 | 9 | 0 | 9 | 0 | 0 | 3 | |
| 12 (October–November 1999) | 14 | 13b | 10 | 1 | 0 | 3 | 3 | |
| 13 (October–November 1999) | NKc | 20 | 20 | 0 | 0 | 0 | 3 | |
COWP, Cryptosporidium oocyst wall protein gene.
Sequential samples collected from one patient on two occasions. Genotype 1 alone and genotypes 1 and 2 together were recovered.
NK, not known.
Genotyping results were obtained from 52 patients collected during five swimming pool-associated outbreaks (Table 2). Two of these outbreaks (outbreaks 9 and 13) were due to genotype 1, one (outbreak 11) was due to genotype 2, and the remaining two (outbreaks 10 and 12) involved both genotypes 1 and 2.
Among the 1,705 samples from humans overall, there were 26 family groups (2 to 3 members per group) and one pair of patients in a children's nursery described as having contact (one woman 26 years of age and one male child 1 year of age). Two of the family groups were included in drinking waterborne outbreak 4 (Table 2), and two were included in the swimming pool-associated outbreaks 9 and 12 (Table 2). The remaining 21 groups all occurred among the sporadic cases diagnosed in England during 1999 and comprised 5 groups with a mother (29 to 38 years old) plus one or two children (6 months to 12 years) and 16 groups of 2 to 3 siblings (ages ranging from 1 to 34 years). In all instances, the same genotype of Cryptosporidium was recovered from members of the same family or contact group: 13 groups (26 individual cases) yielded genotype 1, and 14 groups (30 individual cases) yielded genotype 2.
Two sequential samples were tested from five patients; the times between collections varied from 10 min to 6 days. In four of these patients, the same genotype was recovered on both occasions: three patients yielded genotype 1, and one yielded genotype 2. Samples from the final patient (associated with swimming pool outbreak 12) yielded genotype 1 on the first occasion and 6 days later yielded both genotypes 1 and 2.
Genotyping results were obtained from 1,182 sporadic cases that occurred in the United Kingdom between 1994 and 1999 (Table 1). There were marked differences between the distributions of the genotypes in patients diagnosed in different countries in the United Kingdom: genotype 2 comprised 91 and 90% of the cases, respectively, in Scotland and Northern Ireland, but only 68 and 63% of the cases, respectively, in Wales and England. Because of the possibility of identifying regional and seasonal trends with the larger and more complete nature of the data set collected in England during 1998 to 1999, a more detailed analysis of cases in this country was performed.
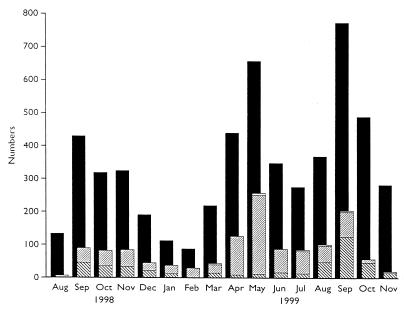
Genotyping results were obtained from 1,378 cases of cryptosporidiosis in humans occurring in England where specimens were collected between 19 August 1998 and 20 November 1999, and this includes both sporadic and outbreak-associated cases. During the same period, 5,447 laboratory-confirmed cases were reported to the PHLS Communicable Disease Surveillance Centre as part of routine surveillance for this disease. Monthly totals of the numbers of cases reported and genotypes are shown in Fig. 1. Since cases are reported anonymously, it is not possible to exactly match reported cases with those which had been genotyped; however, this is likely to represent a sample of approximately 25% of the reported cases during this period.
FIG. 1.
Monthly totals of cryptosporidiosis in humans in England in 1998 to 1999. ■, total number of microbiologically confirmed cases reported; ▧, genotype 1; ▨, genotype 2; □, other genotypes.
Among the genotyping results from cases in England occurring between 19 August 1998 and 20 November 1999, a single data set of 984 sporadic cases was established which excluded all cases associated with outbreaks, all cases without a date of collection, and all except one case per family group, or one per patient. Among these 984 cases, 340 (34.6%) were due to genotype 1, 638 (64.8%) were due to genotype 2, 4 (0.4%) were due to genotype 3, and 2 (0.2%) were due to both genotypes 1 and 2. The cases due to genotypes 1 plus 2 and genotype 3 are not further considered. Among the remaining 978 cases due to either genotype 1 or 2, 103 were associated with recent foreign travel (59 from European destinations, 22 from the Indian subcontinent, and 22 from other or unspecified countries). Within the 103 travel-associated cases, genotype 1 was recovered from 66 cases, and genotype 2 was recovered from 37 cases. The destinations were European (Cyprus, France, Greece, Malta, Portugal, Rumania, Spain, Switzerland, and Turkey), the Indian subcontinent (India, Nepal, and Pakistan), and other countries (Hong Kong, Kenya, Mexico, New Zealand, Nigeria, Tunisia, Singapore, South Africa, Tibet, United States, and Zimbabwe). Travel-associated cases were more often associated with the summer-autumn peak (Fig. 1 and 2) in cases, and genotype 1 was significantly associated with recent foreign travel irrespective of the destination (χ2 = 43.62, P < 0.0001000).
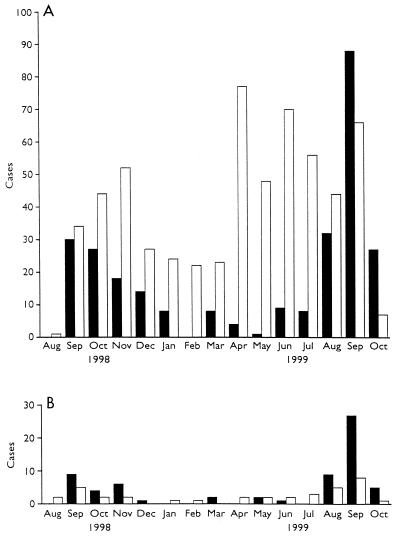
FIG. 2.
Monthly totals of cases of cryptosporidiosis in nontravelers (A) and travelers (B) in England in 1998 to 1999 due to genotype 1 (■) or genotype 2 (□).
Among the patients with sporadic cases, there were 17 who reported contact with farms or farm animals and 1 who had contact with a pet rabbit. Genotype 2 was recovered from all patients associated with farms and farm animals. Genotype 1 was recovered from the feces of the final patient reporting contact with a pet rabbit.
Overall, there was a marked seasonal pattern, with the spring peak in cases being almost exclusively due to genotype 2 and the late-summer–autumn peak due to both genotypes 1 and 2. The cases in patients with a recent history of foreign travel occurred predominantly in the late-summer–autumn peak and decreased earlier than those in patients for whom foreign travel was not recorded (Fig. 2 and 3).
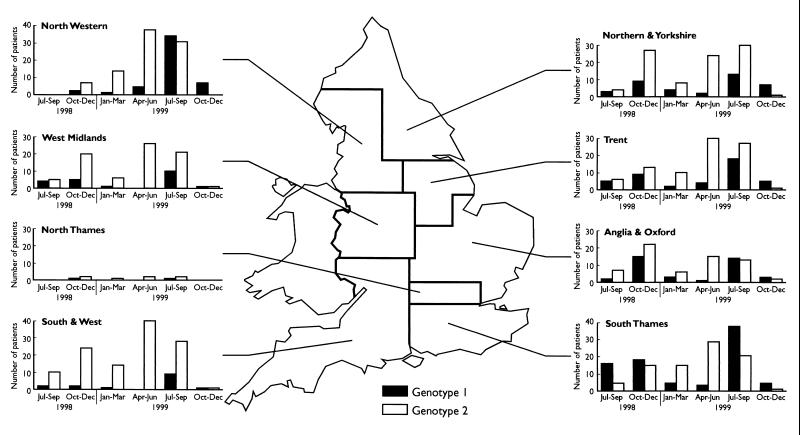
FIG. 3.
Distribution of three monthly totals of cases of cryptosporidiosis in humans in England in 1998 to 1999 by regional health authorities.
There was marked geographical variation in the distributions of genotypes, and these are shown in Fig. 3. For example, some regions showed a very high proportion of genotype 2 (e.g., 87% in the South and West and 79% in the West Midlands regions), and some showed a much higher proportion of genotype 1 (e.g., 50% in the South Thames region). The seasonal peaks in genotypes were present in all regions, although the start and duration of these showed some variation. For example, the genotype 1 peak was markedly present in July to September 1998 in the South Thames region, but was not evident in all other regions. There was also marked variation in the distribution of genotypes between the results from different towns within individual regions. The results from 11 different towns within the West Midlands and Trent regions are shown in Table 3. Laboratories in some towns (e.g., Birmingham, Nottingham, and Sheffield) showed a much higher proportion of cases due to genotype 1, while some towns (e.g., Lincoln and Shrewsbury) showed a much higher proportion of cases due to genotype 2.
TABLE 3.
Distribution in Trent and West Midlands by health authorities of C. parvum genotypes among 192 sporadic cases in individuals without a recent history of foreign travel
| Town | No. (%) of cases with COWP genotype:
|
|
|---|---|---|
| 1 | 2 | |
| Birmingham | 5 (50) | 5 (50) |
| Burton | 3 (18) | 14 (82) |
| Lincoln | 6 (15) | 35 (85) |
| Nottingham | 18 (37) | 31 (63) |
| Sheffield | 14 (48) | 15 (52) |
| Shrewsbury | 3 (16) | 16 (84) |
| Stoke on Trent | 10 (21) | 37 (79) |
| Other (4 laboratories) | 5 (26) | 14 (74) |
DISCUSSION
It is now well recognized that Cryptosporidium spp. do not multiply outside an infected host, and different species show marked host specificity (12, 20). In the early 1980s, however, human cryptosporidiosis was generally regarded as a zoonosis (29), and although this was not universally accepted (10), it is now clear that the epidemiology of human cryptosporidiosis is complex (11, 12, 19). For a disease such as cryptosporidiosis to be sustained in a community, there needs to be sufficient density of both susceptible and infected hosts and infectious particles (oocysts) available in the environment for transmission. Rational approaches to controlling cryptosporidiosis therefore require an understanding of both the host reservoirs and the routes of infection, together with host susceptibilities and survival of the pathogen in the environment. The purpose of this study was to further elucidate host reservoirs and routes of infection by the application of molecular markers for this parasite.
For human cryptosporidiosis, there are both multiple potential host reservoirs of infection and multiple routes of transmission (11, 12, 19), and although the consumption of contaminated water is generally regarded as important, infection also occurs via the consumption of contaminated foods or beverages, by recreational bathing, by person-to-person transmission, and by contact with infected animals (11, 12, 19). The advent of molecular biological methods has led to the identification of two major genotypes within C. parvum (the principal infectious agent for human cryptosporidiosis) with two different transmission cycles. C. parvum genotype 1 is exclusive to humans and a single nonhuman primate, and genotype 2 has a broader host range, including livestock and wild animals, as well as humans (15, 18, 20, 23, 26, 30, 31, 35). The lack of recombination between C. parvum genotypes 1 and 2 indicates that these are reproductively isolated populations (18, 20, 31) and further supports the supposition that these should be regarded as different species (18). Epidemiological studies have shown that genotype 1 was implicated in waterborne outbreaks (22, 26, 32), a food-borne outbreak associated with an infected food handler (28), person-to-person transmission in a day care center, and attendance of a water park (26). Genotype 2 was also associated with waterborne outbreaks (22, 26, 32), together with the consumption of apple juice contaminated with calf feces (26) and bovine contact (26). However, these epidemiological studies have been applied to samples from less than 80 patients overall, and for the waterborne outbreaks, comprise a small proportion of the total number of infected individuals. For example, among the estimated 400,000 cases associated with the 1993 waterborne outbreak in Milwaukee (16), genotyping data are available for five patients, all of which were genotype 1 (26, 32).
Within the large series of cryptosporidiosis cases presented here, all samples from livestock yielded genotype 2, and this is consistent with results from others (5, 20, 26, 30, 31, 35). Among samples collected from humans, 37.8% were due to genotype 1, 61.5% were due to genotype 2, 0.4% were due to both genotypes 1 and 2, and 0.3% were due to genotype 3. Further genetic analysis indicates that genotype 3 justifies its status as a Cryptosporidium species different from C. parvum, and the sequence of fragments of the 18S rRNA gene is identical to the published sequences of C. meleagridis (25). There are relatively few comparative data analyzing larger series from humans, although Sulaiman et al. (32) reported that out of 50 cases of infection in humans, 41 were due to genotype 1, and 9 were due to genotype 2. These results with respect to the proportions of the C. parvum genotypes 1 and 2 differed markedly from those described here for the United Kingdom; however, we further discuss marked seasonal and geographical differences in the cases studied here.
Data are presented here on eight drinking waterborne outbreaks, five of which were almost exclusively due to genotype 1, and three of which were due to genotype 2. Data from field epidemiological observations (1, 2) suggest that contamination of water supplies by sheep feces was involved with the three outbreaks due to genotype 2, and this is consistent with a zoonotic source of contamination. It may be of note that all outbreaks occurred in the spring, when lambing (together with outbreaks of cryptosporidiosis in sheep) occurs most commonly in the United Kingdom (9). In contrast, the likely source of C. parvum in the five drinking-waterborne outbreaks predominantly due to genotype 1 was by contamination with human sewage, and these occurred throughout the year. It may be of note that outbreaks 2 and 4 occurred after heavy rain (7, 17, 37), and the lack of treatment of sewage during overflow of storm drains may have been a contributing factor. Ong and colleagues (22) reported that genotype 1 was more likely to occur in people who were residents of urban areas and genotype 2 was more likely to occur in those from rural communities. This was certainly not true for the waterborne outbreaks described here. For example, both genotypes 1 and 2 were associated with outbreaks among predominantly rural communities (outbreaks 2 and 6) and predominantly urban communities (outbreaks 1, 3, 4, and 8). However, the source of water, together with the location and route of contamination by Cryptosporidium, is probably of prime importance. For example, waters derived from rural upland locations are more likely to be contaminated by C. parvum genotype 2 from livestock or wild animals, regardless of whether a rural or urban community is being served. Contamination by C. parvum at upland sites may occur via passage through birds. In support of this, retention of infectivity in orally dosed birds and the occurrence of C. parvum genotype 2 in the feces of wild birds have been demonstrated (13, 14). It is possible that C. parvum genotype 1 may be passively transferred to upland sites by birds feeding on sewage; however, this route of transmission has not, so far, been linked to human cryptosporidiosis in the United Kingdom. In contrast, if contamination of waters occurs at lowland sites (i.e., rivers, boreholes, etc.), this may be due to C. parvum genotype 1 or 2 from human sewage or to C. parvum genotype 2 from livestock or wild animals.
Among the five outbreaks associated with swimming pools, two were due to genotype 1, one was due to genotype 2, and the remaining two involved both genotypes 1 and 2. Both genotypes 1 and 2 were recovered both from different patients and simultaneously from the same patient in outbreak 12. Outbreaks in swimming pools may be associated with fecal contamination from a single infected individual (especially in toddler pools), resulting in a single genotype being recovered from the patients. However, outbreaks may also be due to more general problems, such as contamination with sewage, poor disinfection, or inadequate maintenance of filtration equipment (3), and these may be associated with genotypes 1 and 2, together with other pathogens. It may be of note that one of the outbreaks (outbreak 10) in which both genotypes were recovered from patients was also associated with giardiasis and with the detection of Giardia cysts in the swimming pool water (3).
Samples were analyzed here from 1 contact group within a nursery and 26 family groups. Among the sporadic cases, these are most likely to represent small intrafamily outbreaks where person-to-person secondary spread has occurred. The groups infected within drinking-waterborne and swimming pool-associated outbreaks may similarly represent secondary spread or may be coprimary cases resulting from common exposure to a single contaminated source. We previously presented preliminary data using a sodium dodecyl sulfate-polyacrylamide gel electrophoresis Western blotting technique to characterize a polymorphic cryptosporidial oocyst wall antigen isolated from feces (17). Results on the diversity of banding patterns suggested that the majority of cases within families infected during a single waterborne outbreak were infected as coprimary infections rather than as a result of secondary spread. Data presented in this report showed that the same genotype was detected from individuals within each family outbreak and from the sequential samples collected from four out of five individual patients. Genotype 1 was recovered from the remaining patient on the first sampling, and both genotypes 1 and 2 were recovered 6 days later. These results are encouraging, since they demonstrate the stability of results obtained with this polymorphic molecular marker. We are currently further investigating these groups (together with the sequential samples) by using more discriminatory molecular markers (8) than the COWP PCR-RFLP methods described in this report.
Marked differences were observed in the distribution of C. parvum genotypes between samples collected from different countries within the United Kingdom. However on consideration of the total numbers of reported cases, with the exception of samples collected from England, these are unlikely to be representative of all diagnosed cases. However, because of the much larger number of samples examined from England during 1998 to 1999, despite the fact that we are unable to exactly match these cases, we estimate that, for the first time, a national pattern of C. parvum genotypes has been established with a sample constituting approximately 25% of all cases diagnosed. The cases overall showed a previously recognized marked bimodal seasonal pattern (19), with one peak in the spring and the second in late summer to early autumn. The distribution of the genotypes also showed marked seasonal differences. The spring peak was almost exclusively due to genotype 2, and the late-summer–early-autumn peak was due to both genotypes 1 and 2. Although part of the spring peak is due to cases associated with waterborne outbreak 8 (genotype 2), which occurred in April to May 1999, genotype 2 was strongly associated with this peak in all other regions. Outbreaks of cryptosporidiosis in sheep in the United Kingdom show a marked spring peak, but those in cattle show both a spring and summer–early-autumn peak similar to those in the cases in humans (9). Although it is not possible yet to estimate the role of humans in the transmission of C. parvum genotype 2, livestock represent considerable potential for input of oocysts into the environment which coincides with the seasonal peaks in the distribution of genotype 2. Climatic features such as heavy rain and seasonal agricultural practices such as the spreading of excreta as fertilizer are also likely to be of importance.
We previously reported that patients with a recent history of foreign travel examined during the second half of 1998 were significantly more likely to be infected by genotype 1 (18). We report here that this was a consistent pattern over both 1998 and 1999. Further epidemiological investigations should be focused on elucidating risk factors for cryptosporidiosis as a result of foreign travel, particularly investigating sources in which there is potential contamination by human sewage (i.e., sea bathing in areas contaminated by sewage, swimming pools, drinking contaminated waters or eating contaminated foods, etc.). The remainder of the genotype 1 cases were also associated with a marked seasonal pattern similar to that obtained with the foreign travelers. Although there may be misclassification of some of the travelers as nontravelers (a very simple method of data capture was used here), this, together with reporting bias, is unlikely to explain the marked differences both within and between the regions. Some of the genotype 1 cases may be due to secondary person-to-person contact with a recent traveler. However, because of the marked geographical differences, we believe that the role of sewage contamination of drinking water sources (or of recreational water or food sources) from foreign travelers is likely to be important. We therefore propose the hypothesis that C. parvum genotype 1 in England usually results from either foreign travel, by direct (or indirect) contact with imported infections, or through the contamination of drinking water (or other sources) with sewage, and therefore is not primarily an endogenous disease. In contrast, because of the large potential reservoirs in livestock and wild animals (together with infected humans), C. parvum genotype 2 is primarily a disease endogenous to England. The source of infection and host range of the C. meleagridis (25) isolates remain to be elucidated.
ACKNOWLEDGMENTS
We thank colleagues in clinical microbiology laboratories for the donation of specimens and A. V. Swan (PHLS Statistics Unit) for statistical advice.
S.P.-D. is funded by Biomed grant PL 962557 from the European Commission and is jointly supervised by the Imperial College of Science and Technology and Medicine, London, United Kingdom.
REFERENCES
- 1.Anonymous. Surveillance of waterborne disease and water quality: January to June 1998. Public Health Lab Serv Commun Dis Rep. 1999;9:73–74. [PubMed] [Google Scholar]
- 2.Anonymous. Surveillance of waterborne disease and water quality. January to June 1999, and summary 1999. Public Health Lab Serv Commun Dis Rep. 1999;9:305–308. [Google Scholar]
- 3.Anonymous. Surveillance of waterborne disease and water quality: July to December 1999. Public Health Lab Serv Commun Dis Rep. 2000;10:65–68. [Google Scholar]
- 4.Arrowood M J. Diagnosis. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 43–64. [Google Scholar]
- 5.Awad-el-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox M T, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchier I. Cryptosporidium in water supplies: Third report of the group of experts. London, United Kingdom: Her Majesty's Stationery Office; 1998. [Google Scholar]
- 8.Cacció S, Homan W, Camilli R, Traldo G, Kortbeek T, Pozio E. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology. 2000;120:237–244. doi: 10.1017/s0031182099005508. [DOI] [PubMed] [Google Scholar]
- 9.Casemore D P. Epidemiological aspects of human cryptosporidiosis. Epidemiol Infect. 1990;104:1–28. doi: 10.1017/s0950268800054480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casemore D P, Jackson F B. Hypothesis: cryptosporidiosis in human beings is not primarily a zoonosis. J Infect. 1984;9:153–156. doi: 10.1016/s0163-4453(84)91117-4. [DOI] [PubMed] [Google Scholar]
- 11.Casemore D P, Wright S E, Coop R L. Cryptosporidiosis: human and animal epidemiology. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 65–92. [Google Scholar]
- 12.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 1–41. [Google Scholar]
- 13.Graczyk T K, Cranfield M R, Fayer R, Trout J, Goodale H J. Infectivity of Cryptosporidium parvum oocysts is retained upon intestinal passage through migratory water-fowl species (Canada goose, Branta canadensis) Trop Med Int Health. 1997;2:341–347. doi: 10.1111/j.1365-3156.1997.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graczyk T K, Fayer R, Trout J M, Lewis E J, Farley C A, Sulaiman I, Lal A A. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis) Appl Environ Microbiol. 1998;64:2736–2738. doi: 10.1128/aem.64.7.2736-2738.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson A J, Linton D, Stanley J, Owen R J. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 16.MacKenzie W R, Hoxie N J, Proctor M E, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 17.McLauchlin J, Casemore D P, Moran S, Patel S. The epidemiology of cryptosporidiosis: application of experimental sub-typing and antibody detection systems to the investigation of water-borne outbreaks. Folia Parasitol. 1998;45:83–92. [PubMed] [Google Scholar]
- 18.McLauchlin J, Pedraza-Diáz S, Amar-Hoetzeneder C, Nichols G L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meinhardt P L, Casemore D P, Miller K B. Epidemiological aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol Rev. 1996;18:118–136. doi: 10.1093/oxfordjournals.epirev.a017920. [DOI] [PubMed] [Google Scholar]
- 20.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C A. Variation within Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 21.Morgan U M, Weber R, Xiao L, Sulaiman I, Thompson R C A, Ndiritu W, Lal A, Moore A, Deplazes P. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–1183. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong C S L, Eisler D L, Goh S H, Tomblin J, Awad-el-Kariem F M, Beard C B, Xiao L, Sulaiman I, Lal A, Fyfe M, King A, Bowie W R, Isaac Renton J L. Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am J Med Hyg. 1999;61:63–69. doi: 10.4269/ajtmh.1999.61.63. [DOI] [PubMed] [Google Scholar]
- 23.Patel S, Pedraza-Díaz S, McLauchlin J, et al. The molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Commun Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 24.Patel S, Pedraza-Díaz S, McLauchlin J. The identification of Cryptosporidium species and Cryptosporidium parvum directly from whole faeces by analysis of a multiplex PCR of the 18S rRNA gene and by PCR/RFLP of the Cryptosporidium outer wall protein (COWP) gene. Int J Parasitol. 1999;29:1241–1247. doi: 10.1016/s0020-7519(99)00079-x. [DOI] [PubMed] [Google Scholar]
- 25.Pedraza-Díaz S, Amar C, McLauchlin J. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol Lett. 2000;189:189–194. doi: 10.1111/j.1574-6968.2000.tb09228.x. [DOI] [PubMed] [Google Scholar]
- 26.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphisms among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, et al. New Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiroz E S, Bern C, MacArthur J R, Xiao L, Fletcher M, Arrowood M J, Shay D K, Levy M E, Glass R, Lal A. An outbreak of cryptosporidiosis linked to a food handler. J Infect Dis. 2000;181:695–700. doi: 10.1086/315279. [DOI] [PubMed] [Google Scholar]
- 29.Schultz M G. Emerging zoonoses. N Engl J Med. 1983;308:1285–1286. doi: 10.1056/NEJM198305263082109. [DOI] [PubMed] [Google Scholar]
- 30.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 31.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human and animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suliaman I M, Xiao L, Lal A A. Evaluation of Cryptosporidium parvum genotyping techniques. Appl Environ Microbiol. 1999;65:4431–4435. doi: 10.1128/aem.65.10.4431-4435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall P G, de Louvois J, Gilbert R J, Rowe B. Food poisoning: notifications, laboratory reports, and outbreaks—where do the statistics come from and what do they mean? Public Health Lab Serv Commun Dis Rep Rev. 1996;7:R93–R100. [PubMed] [Google Scholar]
- 35.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:223–239. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 36.Widmer G, Akiyoshi D, Buckholt M A, Feng X, Rich S M, Deary K M, Bowman C A, Xu P, Wang Y, Wang X, Buck G A, Tzipori S. Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol Biochem Parasitol. 2000;108:187–197. doi: 10.1016/s0166-6851(00)00211-5. [DOI] [PubMed] [Google Scholar]
- 37.Willocks L, Crampin A, Milne L, et al. A large outbreak of cryptosporidiosis associated with a public water supply from a deep chalk borehole. Commun Dis Public Health. 1998;1:239–243. [PubMed] [Google Scholar]