Summary
Huntington's disease (HD) is an autosomal dominant disorder with progressive motor dysfunction and cognitive decline. The disease is caused by a CAG repeat expansion in the IT15 gene, which elongates a polyglutamine stretch of the HD protein, Huntingtin. No therapeutic treatments are available, and new pharmacological targets are needed. Retrotransposons are transposable elements (TEs) that represent 40% and 30% of the human and Drosophila genomes and replicate through an RNA intermediate. Mounting evidence suggests that mammalian TEs are active during neurogenesis and may be involved in diseases of the nervous system. Here we show that TE expression and mobilization are increased in a Drosophila melanogaster HD model. By inhibiting TE mobilization with Reverse Transcriptase inhibitors, polyQ-dependent eye neurodegeneration and genome instability in larval brains are rescued and fly lifespan is increased. These results suggest that TE activation may be involved in polyQ-induced neurotoxicity and a potential pharmacological target.
Subject areas: Biological sciences, Molecular biology, Neuroscience
Graphical abstract

Highlights
-
•
Transposable elements: new pathogenic players in Huntington's disease
-
•
polyQ-Htt expression leads to widespread heterochromatin relaxation
-
•
Reverse transcriptase inhibitors rescue HD-induced neurodegeneration
Biological sciences; Molecular biology; Neuroscience
Introduction
Huntington's disease (HD) (OMIM #143100) is a late-onset, autosomal dominant disorder characterized by progressive motor dysfunction, cognitive decline, and psychiatric disturbances, which lead to death approximately 15–20 years after the initial symptoms (Folstein, 1989). The most prominent pathological manifestation of the disease is a selective and gradual loss of medium-sized spiny neurons of the striatum, although, in the late stage of the disease, pathological alterations occur in additional brain regions (Vonsattel and DiFiglia, 1998). The disease is caused by a CAG repeat expansion in the IT15 gene, which elongates a stretch of polyglutamine (polyQ) at the amino-terminal end of the HD protein, Huntingtin (Htt) (The Huntington’s Disease Collaborative Research Group, 1993). The number of CAG repeats varies between 6 and 35 units on normal chromosomes, whereas on HD chromosomes the repeat is expanded above the pathological threshold of 36 CAGs and can range as high as 150 or more. Htt is ubiquitously expressed, and it is mainly, but not exclusively, localized to the cytoplasm where it may associate to organelles (DiFiglia et al., 1995). Htt interacts with numerous proteins implicated in processes as diverse as gene transcription, RNA splicing, intracellular transport, signal transduction, and metabolism.
The HD mutation, by virtue of the expanded glutamine tract, confers a novel toxic property to the mutant protein. A gain-of-function mechanism is indeed supported by genetic and experimental data (Duyao et al., 1995; Nasir et al., 1995). In HD target neurons, the trigger event driven by mutant Huntingtin is likely to occur many years before the appearance of the first signs of neurodegeneration. The identification of genetic modifiers of the age of onset has established the role of somatic mosaicism of the CAG trinucleotide repeats in neural genomes (Gusella et al., 2014; Lee et al., 2019). Mutant Huntingtin triggers extensive epigenetic-chromatin deregulation as shown by the analysis of DNA methylation and histone (Bassi et al., 2017; Seong et al., 2010). The disease cascade that leads to neuronal cell death involves several pathways including defects in the proteasome apparatus, glutamate-mediated excitotoxicity, mitochondrial dysfunction, and neuroinflammation (Ross and Tabrizi, 2011).
Our understanding of the molecular basis of HD pathogenesis remains incomplete and a treatment for this devastating disease is still not available. It is therefore important to identify previously unnoticed pathways that may be altered in HD and potential target of therapeutic treatments.
Drosophila melanogaster is an excellent choice for modeling HD pathology and disease mechanisms. It has a relatively simple central nervous system with an architecture that separates specialized functions such as vision, olfaction, learning, and memory similarly to mammalian nervous systems (Armstrong et al., 1995). Moreover, the majority of pathological features of HD can be recapitulated in transgenic fly models, including dominant gain-of-function neurotoxicity, nuclear inclusion formation, progressive neurodegeneration, behavioral abnormalities, and early death (Chan et al., 2002).
In the last few years, correlations between transposon activation and neurological diseases have been observed (Douville et al., 2011; Fukuda et al., 2021; Guo et al., 2018; Kaneko et al., 2011; Li et al., 2012, 2015; McConnell et al., 2017; Perron et al., 1997; Prudencio et al., 2017; Sun et al., 2018; Tam et al., 2019; Thomas et al., 2017). Transposable elements (TEs) are mobile genetic elements that constitute a large fraction of eukaryotic genomes (Belancio et al., 2008). Retrotransposons (RTEs) replicate through an RNA intermediate by taking advantage of reverse transcriptase (RT) activity and represent approximately 40% and 30% of the human and Drosophila genomes, respectively. During the co-evolution of TEs with their host genomes, organisms have evolved efficient mechanisms to prevent and regulate TE activation and mobilization including methylation and Piwi-interacting RNAs (piRNAs) expression (Ozata et al., 2019). Mounting evidence shows that long interspersed nuclear element 1 (LINE-1) sequences are normally active during neurogenesis in both rodent and human tissues, and somatic mobilization of TEs has been observed in different human brain regions giving rise to mosaicism (Bodea et al., 2018). The extent and the functional role of TE mobilization in brain physiology remain unclear (Cappucci et al., 2018; Evrony et al., 2016; Upton et al., 2015). LINE-1 mobilization has also been associated to deletion of genomic fragments in human brains (Erwin et al., 2016; Rodriguez-Martin et al., 2020). Emerging evidence suggests an association between unregulated activation of TEs and diseases of the nervous system (Jacob-Hirsch et al., 2018). Pathological TE activation has been observed in, among others, Rett syndrome (Muotri et al., 2010), ataxia telangiectasia (Coufal et al., 2011), amyotrophic lateral sclerosis (ALS) (Krug et al., 2017; Li et al., 2012), Alzheimer disease (AD) (Guo et al., 2018; Sun et al., 2018), and Parkinson disease (Blaudin de Thé et al., 2018).
In a Drosophila model of ALS with the ectopic expression of human TAR DNA-binding protein 43 (TDP-43), a de-repression of both LINE and LTR families of TEs was observed (Krug et al., 2017). The degenerative fly phenotypes were rescued by pharmacologically inhibiting RT activity or by interfering with the expression of the endogenous retrovirus (ERV) gypsy (Krug et al., 2017). Gyspy, together with copia and Het-A RNAs, was also found induced in a fly model of AD overexpressing wild-type or mutant Tau (Guo et al., 2018). Interestingly, chromatin decondensation and decreased expression of piwi/piRNAs led to TE activation in another fly model of tauopathy where inhibition of RT activity was able to rescue the neurodegenerative phenotype (Sun et al., 2018).
These data suggest that resurrection of TEs could be causally involved in neurodegeneration and recapitulated in fly models of disease.
By taking advantage of a well-established fly HD model (Romero et al., 2008), here we show that TE activation may also be involved in HD pathogenesis, and its inhibition may represent a new therapeutic strategy.
Results
TE expression is induced in HD fly model
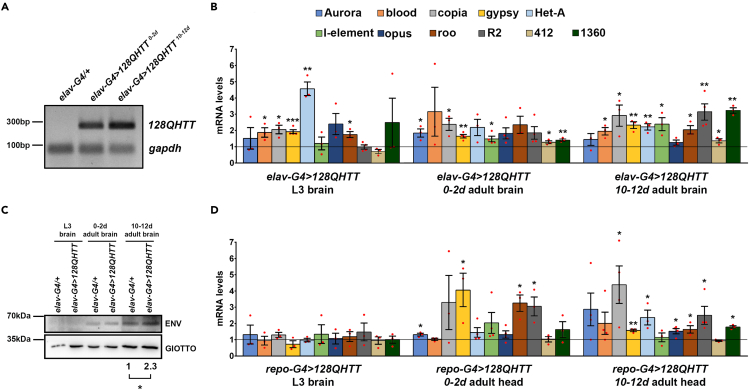
In this study, we take advantage of the Drosophila HD experimental model generated by Romero and collaborators (Romero et al., 2008). It is based on the expression of full-length human Huntingtin (Htt) protein with 128 glutamines (128QHttFL, pathogenic HD construct) under the control of the UAS-Gal4 system (Brand and Perrimon, 1993). In this bipartite system, the expression of the HD pathogenic construct placed downstream of UAS sequences depends on the spatial and temporal expression of the yeast transcriptional activator Gal4. In order to induce a pan-neuronal expression of the pathogenic HD construct, we crossed flies carrying the UAS-128QHttFL transgene (hereafter referred to as 128QHtt) to flies elav-Gal4 driving the expression of the pathogenic construct in every post mitotic neuronal cell, starting from very early stages of development (Robinow and White, 1988). We first checked by semi-quantitative RT-PCR the correct expression of 128QHtt mRNA in Drosophila HD heads at two different time points (0–2 and 10–12 days) (Figure 1A); we then analyzed different transposon transcripts in larval and adult brain comparing transposon RNA levels from HD flies (elav-Gal4>128QHtt) with those from controls (elav-Gal4/+) (Figure 1B). The time points for this analysis (0–2 and 10–12 days) were chosen taking into account the dynamic of survival curves, indicating that HD flies begin to die shortly (after 10 days of age) and have a dramatically reduced lifespan compared with control flies (elav-Gal4/+) (see Figure S1A). We initially profiled 11 Drosophila TEs, including 1 DNA transposon (1360), 7 LTR retrotransposons (Aurora, blood, copia, gypsy, opus, roo, and 412), and 3 non-LTR retrotransposons (Het-A, I-element, and R2).
Figure 1.
Neuronal and glial expression of 128QHtt results in transposable element derepression
(A) Semiquantitative RT-PCR analysis to assess the induction levels of the HD transgenic construct by elav-Gal4. cDNA was prepared from total RNA purified from HD (elav-G4>128QHtt) and control (elav-G4/+) head tissues. The constitutive gapdh was examined as an endogenous control.
(B) qRT-PCR analysis of transposable element expression in larval and adult brains of flies expressing 128QHtt in neurons (elav-G4>128QHtt); adult brains were analyzed at both young (0–2 days) and aged (10–12 days) time points; transcript levels were normalized to rp49 and displayed as fold change relative to flies carrying the elav-Gal4 driver with no 128QHtt transgene (elav-G4/+).
(C) Western blot assay of gypsy envelope protein (ENV) expression in HD larval and adult brains. GIOTTO protein was used as a loading control. Result was expressed as means for at least three independent biological replicates (∗p < 0.05; one-sample t test).
(D) qRT-PCR analysis of TE expression in larval brains and adult heads isolated from 0- to 2-day-old and 10- to 12-day-old flies expressing 128QHtt with the pan-glial repo-Gal4 driver (repo-Gal4>128QHtt). Transcript levels were normalized to gapdh and displayed as fold change relative to flies carrying the repo-Gal4 driver with no128QHtt transgene (repo-G4/+). (B and D) Bar graph represents the mean ± SEM from at least three independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; unpaired t tests). Red dots indicate individual data points. The black horizontal line indicates the fold change control value, set to 1. See also Figures S1–S3.
We found that different classes of TE transcripts were significantly upregulated in HD neurons at both larval and adult stages (Figure 1B). The (LINE)-like Het-A was the most induced class at the larval stage, whereas several classes presented similar induction at 0–2 and 10–12 days of adult brain. Interestingly, the increase of TE transcripts at 10–12 days correlated with age-dependent neurodegeneration because at this stage an extensive neuronal loss was evident, as demonstrated by the strong decrease of the neuronal marker Elav (see Figure S1B). In addition, consistently with the expression of gypsy RNA, an endogenous insect retrovirus similar to human ERVs, a significant age-dependent increase of the gypsy ENV (envelope) protein levels was observed in adult brains expressing pan-neuronally 128QHtt (Figure 1C). In order to provide further evidence that TE activation was due to expression of the polyQ-expanded 128QHtt, we examined the effect on TE deregulation of a wild-type nonexpanded transgenic construct containing 16 glutamines (16QHtt) (Romero et al., 2008). As shown in Figure S2A, the absence of significant differences between elav-Gal4/+ and elav-Gal4>16QHtt demonstrates that TE activation shown in Figure 1B is the direct effect of the expression of pathogenic polyQ-expanded 128QHtt transgene. We also observed that pan-neuronal expression of 16QHtt did not affect the median lifespan of elav-Gal4>16QHtt with respect to control elav-Gal4/+ (see Figure S2B).
Because glial cell lineage expression of human mutant Htt (hHtt103Q) was previously shown to induce developmental and late-onset neuronal pathologies in Drosophila model (Tamura et al., 2009), we expressed the pathogenic HD protein in glial cells by using the pan-glial driver repo-Gal4 (Awasaki et al., 2008) and tested TE RNA induction. Repo-Gal4>128QHtt raised at 29°C, showed severe locomotive defects and very early death; over 70% of the repo-Gal4>HD flies died within 4–5 days following eclosion, and the survivors died more gradually over the ensuing 8–10 days. Aiming to analyze TE expression in repo-Gal4>128QHtt adult heads, we raised repo-Gal4>128QHtt flies at 25°C to partially suppress Gal4 activity and increase their survival rate (as summarized in Figure S3A). Pan-glial expression of mutant Htt induced a significant decrease of Repo protein levels, especially at 10–12 days posteclosion (see Figure S3B), suggesting that the reduced expression of Repo could be important for the effect of 128QHtt on lifespan shortening in repo-Gal4>128QHtt flies. By qRT-PCR we analyzed RNA samples from larval brains and whole heads of repo-Gal4>128QHtt flies collected at both 0–2 and 10–12 days posteclosion. As shown in Figure 1D, although no significant increase in repo-Gal4>128QHtt larval brains was observed, several TE classes were upregulated in repo-Gal4>128QHtt adult heads with a different pattern of expression when compared with elav-Gal4>128QHtt.
Loss of histone H3 trimethylation at Lys9 (H3K9me3) and global heterochromatin relaxation contribute to TE silencing release in HD
To investigate the molecular mechanisms by which HD might contribute to transposon activation, we analyzed, by chromatin immunoprecipitation coupled to quantitative PCR (ChIP-qPCR), the levels of H3K9me3 on promoters and coding regions of upregulated TEs. Trimethylation on lysine 9 of histone H3 is a specific epigenetic mark of heterochromatic domains, and it is essential for heterochromatin formation and maintenance (Allshire and Madhani, 2018). As shown in Figure 2A, we found a drastic reduction of H3K9me3 occupancy in adult heads isolated from 10- to 12-day-old elav-Gal4>HD flies for almost all sequences of TE analyzed, especially on their promoters.
Figure 2.
Global heterochromatin relaxation mediates HD-induced TE activation
(A) Transposon DNA sequences show decreased H3K9me3 levels in HD. The relative fold enrichment, normalized to the heterochromatic F22 control region, was calculated by dividing the amount of immunoprecipitated DNA from elav-G4>128QHtt heads by that from the control (elav-G4/+). Bars represent mean ± SEM of three independent experiments performed in duplicate (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01; unpaired t tests). The gray area represents the control value set to 1.
(B) HP1 and Su(var)3–9 are genetic modifiers of HD eye phenotype. GMR-driven HP1 overexpression (GMR-Gal4>HP1OE;128QHtt) or GMR driven Su(var)3–9 overexpression (GMR-Gal4>Su(var)3–9OE;128QHtt) strongly prevents HD-induced eye neurodegeneration. GMR-G4/+ was used as control. The lower graph represents the phenotypic scores of each indicated genotype. The phenotypic scores are concordant with the visual assessment of the eye phenotypes. The number of images used for this analysis ranged from 7 to 17 (∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA with Tukey's post hoc test).
(C) Survival curves of control (elav-Gal4/+) and HD flies (elav-Gal4>128QHtt) overexpressing HP1 or Su(var)3–9. The log rank test with Bonferroni correction for multiple comparisons indicates a significant difference between all survival curves (∗∗∗p < 0.001), except for elav-G4/+ vs elav-G4>Su(var)3–9OE;128QHtt lifespan curves (p = 1).
(D) Mutant Htt suppresses BL2 PEV. Histochemical staining for β-galactosidase activity in larval brains isolated from HD (elav-G4/Tp(3;Y)BL2; UAS-128QHtt, right panel) and control (elav-G4/Tp(3;Y)BL2, left panel) male larvae. Scale bar indicates 100 μm. Scatter plot indicates the quantitative analysis of X-gal staining intensities. Data are the means ± SD. The dots indicate individual data points (∗∗∗p < 0.01; unpaired t tests). See also Figures S3, S4, and S6.
The overall reduction in H3K9me3 levels on TE sequences in HD could result from a global heterochromatin decondensation, a phenomenon recently reported in Drosophila models of tauopathy (Frost et al., 2014; Napoletano et al., 2021; Sun et al., 2018). To verify whether constitutive heterochromatin widespread relaxation represents a distinctive feature also in the HD model, we analyzed the effects of perturbing the expression of HP1 and Su(var)3–9, two key positive regulators of heterochromatin formation and maintenance (Casale et al., 2021; Eissenberg et al., 1990; Ebert et al., 2004; James and Elgin, 1986; Piacentini and Pimpinelli, 2010; Schotta et al., 2002).
To this purpose, we took advantage of the ability of 128QHtt to elicit photoreceptors' degeneration upon its expression in the developing eye using the GMR-Gal4 driver (Ellis et al., 1993; Romero et al., 2008). The examination of the external eye morphology using bright-field microscopy revealed that overexpression of HP1 or Su(var)3–9 in HD genetic background induced a significant recovery of pigmentation and structure of HD eye (Figure 2B). To quantify the degree of severity of the eye phenotypes, we used Flynotyper (Iyer et al., 2016), a computational method that calculates a phenotypic score based on the disorganization or altered symmetry of the ommatidial arrangement.
In addition, consistent with the previous results, we observed an increase in median lifespan by 116.72% in elav-Gal4>Su(var)3–9OE;128QHtt flies and by 29.71% in elav-Gal4>HP1OE;128QHtt flies compared with elav-Gal4>128QHtt flies (the median lifespan was 55.59 ± 2.28 days for elav-Gal4>Su(var)3–9OE;128QHtt flies and 33.27 ± 0.81 days for elav-Gal4>HP1OE;128QHtt flies versus 25.65 ± 0.43 days for elav-Gal4>128QHtt flies) (Figure 2C). In accord with previous studies (Wood et al., 2016), the pan-neuronal overexpression of HP1 and Su(var)3–9 resulted in increased lifespan (13.09% and 18.57%, respectively) also in elav-Gal4/+ flies, suggesting that heterochromatin levels and integrity in neuronal cells are important to promote longevity even under normal physiological conditions (the median lifespan was 70.14 ± 1.22 days for elav-Gal4>Su(var)3–9OE and 73.54 ± 1.48 for elav-Gal4>HP1OE flies versus 62.02 ± 0.85 days for elav-Gal4/+ flies).
To account for the possible titration of Gal4 protein in the presence of two UAS transgenes and subsequent reduction in expression of the pathogenic construct 128QHtt, we carried out a new quantitative assessment of eye phenotypes and lifespan experiments by adding a second unrelated UAS transgene (UAS-mCD8:GFP) in the HD genetic background. As shown in Figure S4, we found no significant differences in eye phenotypes and longevity between elav-Gal4>128QHtt and elav-Gal4>mCD8:GFP;128QHtt, confirming that the obtained results are not due to titration of Gal4 protein. Moreover, to exclude the possibility that HP1 or Su(var)3–9 overexpression might affect 128QHtt expression, we also evaluated the expression profile of the pathogenic construct 128QHtt in adult heads overexpressing these pro-heterochromatin genes. As shown in Figure S5, we did not find any significant difference in 128QHtt transcript levels between elav-Gal4> 128QHtt and both elav-Gal4>Su(var)3–9OE;128QHtt and elav-Gal4>HP1OE;128QHtt flies.
These results suggest that changes in heterochromatin structure play a central role in HD pathogenesis.
In Drosophila, a widely accepted genetic tool in understanding the dynamic of heterochromatin state is the position-effect variegation (PEV), a heterochromatin-induced gene silencing that occurs when an euchromatic gene, juxtaposed with heterochromatin by chromosomal rearrangements or translocations, is transcriptionally silenced in some cells but active in others, producing a characteristic mosaic phenotype (Wakimoto, 1998). Genes or transgenes that impact heterochromatin structure and organization act as dose-dependent modifiers of PEV-based gene silencing (Elgin and Reuter, 2013). For instance, loss of dHtt and HP1 mutations have been previously shown to suppress PEV (Eissenberg et al., 1990; Dietz et al., 2015), leading to reduced silencing of variegating genes, whereas HP1 overexpression resulted in an increased heterochromatic gene silencing (Eissenberg et al., 1992). To assay whether 128QHtt expression can induce heterochromatin decondensation, thus suppressing PEV, we used the Tp(3;Y)BL2, a chromosome rearrangement carrying the Hsp70-lacZ inducible variegating transgene into Y pericentromeric heterochromatin. This can be used to detect PEV modifications in larval tissues upon staining for β-galactosidase activity (Lu et al., 1996). To analyze PEV in larval brain tissues, we crossed elav-G4 females to Tp(3;Y)BL2 or Tp(3;Y)BL2; UAS-128Htt males. We then compared elav-Gal4/Tp(3;Y)BL2 and elav-Gal4/Tp(3;Y)BL2; UAS-128QHtt/+ F1 males for LacZ expression in larval brains (Figure 2D). X-Gal staining revealed that the expression of 128QHtt strongly suppressed the position-effect variegation in the Tp(3;Y)BL2 line (Figure 2D).
We then asked whether HP1 or Su(var)3–9 overexpression rescues both HD eye phenotype and reduced lifespan through restoration of heterochromatin organization. To this end, we tested the sensitivity of the BL2 reporter to HP1 or Su(var)3–9 overexpression in the HD genetic background. To produce the appropriate genotypes for BL2 variegation analysis, another pan-neuronal driver, nSyb-Gal4 (Riabinina et al., 2015), was used to drive the expression of transgenic constructs. We crossed Tp(3;Y)BL2; nSyb-Gal4 males to four different female genotypes: wild-type (Oregon-R), UAS-128Htt, UAS-Su(var)3–9;UAS-128Htt, and UAS-HP1;UAS-128Htt. In the F1 offspring, all genotypes were compared for LacZ expression in larval brains. As shown in Figure S6, an increase in the dose of Su(var)3–9 or HP1 in HD genetic background enhanced heterochromatin-induced silencing of the BL2 transgene, suggesting that overactivation of Su(var)3–9 or HP1 in HD genetic background leads to heterochromatin reorganization.
In summary, evidence in larval and adult brains and in the eye strongly suggests that mutant Htt induces TE activation through a widespread heterochromatin decondensation.
RT inhibitors rescue HD eye phenotypes and HD-induced genome instability
To assess the functional role of TE mobilization in poly-Q induced neurotoxicity, we adopted a pharmacological approach to block the transposition of RTEs with RT inhibitors and monitored their effects on the HD eye phenotype. Flies expressing GMR-Gal4>128QHtt were fed over the entire developmental period on standard cornmeal agar yeast medium supplemented with 1 mg/mL Lamivudine (3TC) or 5 mg/mL Zidovudine (AZT), two well-known RT inhibitors (Goic et al., 2013; Krug et al., 2017; Sun et al., 2018; Wood et al., 2016) and therefore blockers of RTE transposition. As shown in Figure 3A, 3TC and AZT treatments strongly rescued the altered HD eye phenotype in 100% of HD flies carrying only one copy of the HD mutagenic construct (GMR-Gal4/+; UAS-128QHtt/+) (AZT, n = 60; 3TC, n = 55). In addition, AZT treatment resulted more effective because, unlike 3TC, it was able to rescue the HD eye phenotype also in 56% of flies carrying two copies of the HD mutagenic construct (GMR-Gal4/+; UAS-128QHtt/UAS-128QHtt) (AZT, n = 64; 3TC, n = 46). AZT was therefore used in all subsequent experiments.
Figure 3.
Treatment with reverse transcriptase inhibitors rescues HD eye phenotype and prevents HD-induced genome instability
(A) 3TC and AZT treatments significantly improve the altered HD eye-phenotype. Transgenic flies expressing GMR-Gal4>128QHtt and treated with 3TC at 1 mg/mL or AZT at 5 mg/mL for the entire development period show a significant improvement of the polyQ-induced neurodegenerative phenotype relative to untreated flies. GMR-G4/+ was used as control. The right graph indicates the phenotypic scores of each genotype. The phenotypic scores are concordant with the visual assessment of the eye phenotypes. The number of images used for this analysis ranged from 5 to 6 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA with Tukey's post hoc test).
(B) Mitotic chromosomes from third instar larval HD brains stained with DAPI. HD chromosomes show a higher degree of chromatin decondensation and structural rearrangements (see arrows for examples) when compared with the control. Scale bar indicates 5 μm.
(C) Quantification of chromosomal abnormalities observed in HD larval brains (∗∗p < 0.01, ∗∗∗p < 0.001; Chi-square test with Bonferroni correction).
(D) Confocal microscopy images showing immunofluorescence against γH2Av on HD (elav-Gal4>128QHtt) and control (elav-Gal4/+) larval brains. Images were captured at 63X magnification. Scale bar indicates 10 μm.
(E) Bar graph represents mitotic index (percentage of cells in mitosis per optical field, at 40X magnification) observed in the brains of HD and control larvae treated with or not with AZT (∗∗∗p < 0.001; one-way ANOVA with Tukey's post hoc test). See also Figure S7.
PolyQ-expansion mutations in HD have been recently shown to lead to chromosomal instability with a molecular mechanism that remains unclear (Ruzo et al., 2018). It is well known that transposition events could represent a potential cause of genomic instability (Ayarpadikannan and Kim, 2014; Belgnaoui et al., 2006; Gasior et al., 2006). Therefore, in order to determine whether HD-induced TE activation can impact genomic stability, we analyzed metaphase chromosomes obtained from elav-Gal4>128QHtt larval brains and control ones (elav-Gal4/+), showing that HD metaphases present abnormal chromosome configurations such as breakages, fusions, chromosomal rearrangements, and a higher degree of chromatin decondensation (Figure 3B). Interestingly, the frequencies of chromosome abnormalities observed in HD larval brain were significantly decreased after AZT pharmacological treatment (Figure 3C), supporting that TE activation participates in HD-induced genomic instability, as previously shown for TE in other biological contexts (Ayarpadikannan and Kim, 2014). We also analyzed the frequencies of chromosomal aberrations in HD larval brains overexpressing HP1 or Su(var)3–9. As shown in Figure S7 and consistently with the data presented in Figure 2, we found that pan-neuronal overexpression of HP1 or Su(var)3–9 induced a significant decrease in the frequency of HD-dependent chromosomal aberrations.
As a confirmation of the presence of genomic damage, immunofluorescence analysis was performed on HD larval brains using an antibody against the histone variant γH2Av, an early and specific marker of DNA damage. Confocal microscopy images clearly showed that HD larval brains accumulate γH2Av-positive foci, confirming HD-induced genomic instability (Figure 3D). This was consistent with a drastic reduction in mitotic index, indicating that DNA damage may also affect mitotic entry or, more in general, cell-cycle progression. Notably, as reported in Figure 3E, the mitotic index in AZT-treated HD larvae was significantly higher than in untreated HD larvae. The mechanistic details of the role of TE activation in the establishment of abnormal chromosome configurations remain unclear and need further studies.
All together, these results strongly suggest a functional role of RTEs mobilization in HD pathogenesis.
RT inhibitors extend lifespan of HD flies
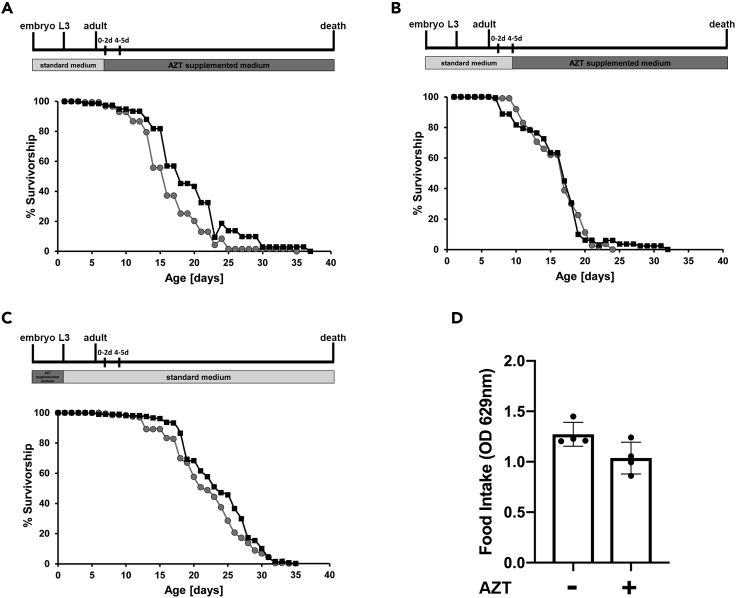
In order to study a systemic effect of the AZT inhibitor, we performed lifespan assays for elav-Gal4>128QHtt flies treated (or not) with AZT. We crossed parental lines on normal medium and transferred the HD offspring on AZT supplemented medium, starting the treatment at 0–2 or 4–5 days after eclosion (Figures 4A and 4B).
Figure 4.
AZT treatment is more effective in rescuing median lifespan only when it is administered in the larval and early adult stage
(A and B) Survival curves of elav-Gal4>128QHtt flies untreated (gray circles) or treated with AZT (black squares) starting at 0–2 (A) or 4–5 days (B) following eclosion. The survival curves shown in (A) are significantly different (∗∗∗p < 0.001; log rank test). In (B) the survival difference is not statistically significant (p = 0.68; log rank test).
(C) Survival curves of elav-Gal4>128QHtt flies untreated (gray circles) or treated with AZT (black squares) from eggs hatch to postfeeding larval stage. The log rank test showed that the two curves were significantly different (∗∗p < 0.01; log rank test). The schemes at the top represent an outline of experimental strategy. Lifespan experiment was conducted with at least 150–200 flies per condition.
(D) AZT supplementation does not affect solid food intake in elav-Gal4>128QHtt HD flies. Food intake was quantified by spectrophotometric absorbance measurements (OD 629 nm) (p = 0.06; unpaired t tests). See also Figures S8, S10, and S11.
The log rank analysis of lifespan curves showed that starting the AZT treatment at 0–2 days after eclosion significantly increased the median lifespan by 15.36% (the median lifespan was 15.43 ± 0.31 days for untreated flies versus 17.80 ± 0.38 days for AZT treated flies) (Figure 4A). On the contrary, starting the AZT treatment at 4–5 days after eclosion did not ameliorate the shortened lifespan of HD flies (the median lifespan was 16.53 ± 0.28 days for untreated flies versus 16.27 ± 0.32 days for AZT treated flies) (Figure 4B). These results strongly suggest that AZT treatment is effective only starting from early stages of HD pathogenesis, whereas it has no effect at late stages of the disease. To extend these results, we crossed the parental lines directly on AZT-medium letting the HD offspring develop on AZT-supplemented medium until postfeeding larval stage, when larvae stop feeding and commit to metamorphosis. After eclosion, HD flies were transferred to vials containing normal food medium, and the survivorship was determined. Also in this case, we found a significant rescue effect in median lifespan (7.8%) between treated and untreated HD flies (the median lifespan was 22 ± 0.39 days for untreated flies versus 23.72 ± 0.37 days for AZT treated flies) (Figure 4C), suggesting that AZT treatment was more effective in rescuing median lifespan only when it is administered in the larval and early adult stage. Caloric restriction (CR) pathway is the most robust nongenetic interventions known to increase longevity in Drosophila and other organisms (see Green et al., [2021] for a review). To rule out the possibility that AZT supplementation in the medium could reduce food consumption, thus extending lifespan due to caloric restriction, we measured feeding rates using the blue food dye (1% FD&C Blue#1) mixed with either ethanol or AZT. Quantification of food consumption using spectrophotometric analysis (OD 629 nm) demonstrated that AZT (at the concentration used in lifespan analyses) does not significantly alter feeding behavior of elav-Gal4>128QHtt flies as compared with the same vehicle-treated genotype (Figure 4D).
As a further experimental control we also examined the effect of AZT on control elav-Gal4/+ flies. Unexpectedly, AZT negatively affected longevity of control flies for each treatment condition (see Figure S8), suggesting that AZT could induce toxicity in normal condition and at the concentration used in fly food. This result is consistent with previous studies demonstrating that antiretroviral drugs, although effective in reverse transcriptase inhibition, could have genotoxic effects (Chen-Chen et al., 2018; Guimarães et al., 2008).
Quantification of TE content by analysis of WGS data and validation by TaqMan CNV assays
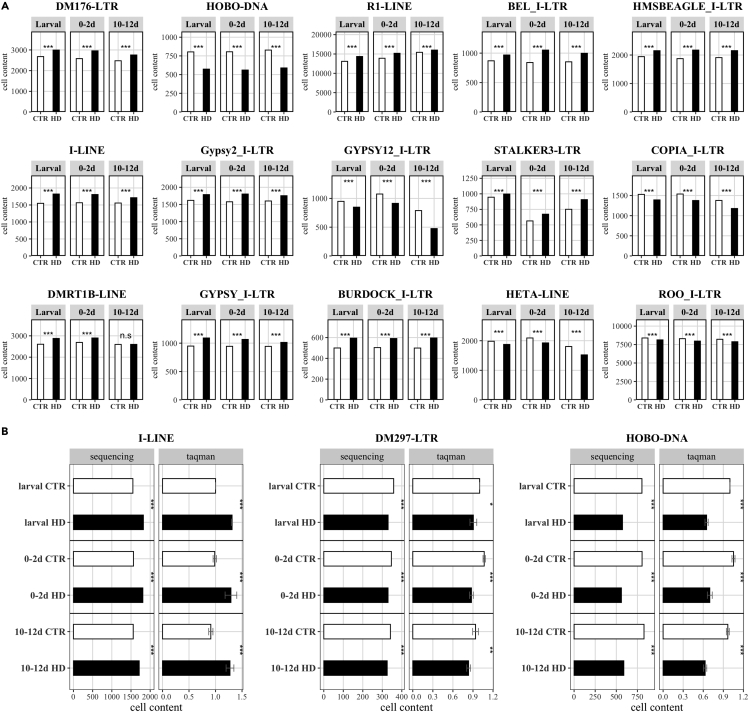
To better investigate the role played by TEs in the genome of HD fly models, we performed whole genome sequencing (WGS) on 1 μg of DNA from offspring control (elav-Gal4/+, offspr. CTR) and HD brain tissues (elav-Gal4>128QHtt, offspr. HD) at three different developmental time points (larvae, young, and aged adults). Given the size of the Drosophila melanogaster genome, about 60 individuals were sequenced. The 6 DNA samples were sequenced using paired-end (PE) Illumina technology. Raw reads were assessed for quality by using FastQC (Andrews, 2010), and the 125 bp PE reads were mapped to the Drosophila genome (UCSC dm6 version [Casper et al., 2017]) using bwa mem (Li and Durbin, 2009) with default parameters. On average, >90% of the reads of the 6 samples mapped properly. The coverage calculation highlighted an average value of about 50×. We therefore used a bioinformatic strategy to quantify the overall TE content of our samples. DNA-seq reads mapped on each TE consensus sequence were counted, and the normalized values of the coverages were interpreted as the sample content for each TE. By analyzing the coverage proportions of 189 TE classes, we highlighted 100 TEs showing a read coverage significantly different between CTR and HD samples at the pathogenic stage (0–2 days stage) (FDR < 0.001) (see Table S1). Almost two-thirds of them (65%) showed a higher content in HD samples. Figure 5A presents data on the top 15 elements according to their statistical significance at 0–2 days stage. Among them, 7 LTR retrotransposons (DM176, BEL_I, HMSBEAGLE_I, Gypsy2_I, STALKER-3, GYPSY_I, and BURDOCK_I) and 3 LINE retrotransposons (R1, I, and DMRT1B) displayed a higher proportion of mapped reads in HD than CTR sample, whereas the DNA element HOBO, 3 LTRs (GYPSY12_I, COPIA_I, and ROO_I), and the LINE HET-A showed an opposite trend.
Figure 5.
TE copy number according to sequencing analyses and TaqMan CNV assay
(A) Fifteen top significant TEs at the 0–2 days stage. Cell content of each TE has been calculated at larval, 0–2 days, and 10–12 days for both CTR and HD samples. The top15 significant TEs at 0–2 days stage (the pathogenetic one) are here represented. All the top15 TEs are classified as retrotransposons (LINE and LTR elements) except for HOBO, a DNA transposon. Of the 15 reported TEs, ten showed a higher cell TE content in HD than CTR 0–2 days sample (DM176, R1, BEL_I, HMSBEAGLE_I, I-element, Gypsy2_I, STALKER-3, DMRT1B, GYPSY_I, and BURDOCK_I). The cell TE content reported on y axis has been calculated counting the reads mapping against each TE RepBase consensus and normalizing on the total number of sequenced reads and multiplied by 1,000,000 (∗∗∗ FDR< 0.001, two proportions Z-test with Benjamin-Hochberg FDR correction).
(B) Validation of the TE copy number by TaqMan CNV assay. The I-element (retrotransposon) assay fully confirmed the sequencing results highlighting higher content of I-element in HD samples than CTR samples, in all the 3 time points (left). TaqMan CNV assay and sequencing analyses showed concordant significant results for DM297 element (middle) (retrotransposon). TaqMan CNV assay displayed a lower CNV of HOBO (DNA transposon) in HD than control samples in all the 3 time points confirming sequencing results (right). Sequencing panel: cell content indicated as number of reads mapped on the TE consensus normalized on the total number of sequenced reads multiplied by 1,000,000. Two proportions Z-test with Benjamin-Hochberg FDR correction. TaqMan panel: cell content reported as ddCt using as reference gene DMRT1C and as calibrator offspring larval CTR sample. Data reported as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; two-way ANOVA with Bonferroni post hoc test). Offspring CTR samples are depicted in white, whereas offspring HD are in black. See also Figures S9, S12 and Table S1.
The I-element is the LINE element displaying the highest ratio between mapped reads in HD and CTR samples at the pathogenetic stage (0–2 days). GYPSY_I displayed the highest number of mapped reads in HD than CTR samples, in all the 3 analyzed time points.
To validate WGS data, we carried out a series of copy number variation (CNV) analysis by TaqMan quantitative PCR (qPCR) of the very same genomic DNA samples. Although CNV TaqMan assays are available for some TE classes in human and mouse genomes, a comprehensive toolbox is still lacking in Drosophila. Firstly, a properly designed TaqMan assay requires a normalizer. To this purpose we identified a specific, invariant DMRT1C fragment sequence that was stable in all samples, and it presented a number of copies comparable with TEs (using the 2–ΔΔCt method for both relative and absolute quantification, as described in STAR methods and reported in supplementary information) (Livak and Schmittgen, 2001). Then, we designed and optimized CNV TaqMan assays for one representative class of each type of the following TEs: LINEs (I-element), LTRs (DM297), and DNA transposons (HOBO). As shown in Figures 5B and S9, TaqMan CNV assays confirmed the differential genomic TE content as measured by bioinformatic analysis of WGS data. When actin was used as invariant control in selective assays, results were comparable (representative experiments are shown in Figures S9C and S9D).
Preliminary evaluation of AZT treatment effects on TEs tissue content
We have previously shown that AZT treatment of HD flies rescued the eye phenotype and the genomic instability observed in larval brains and increased lifespan when administered at 0–2 days. Therefore, we investigated whether AZT affects the brain content of selected TE classes during the same time window when it exerts its systemic protective effects. As previously reported, we crossed parental lines on normal medium and transferred the HD offspring on AZT supplemented medium, starting the treatment at 0–2 or 4–5 days following eclosion. Ten- to twelve-day-old flies were then harvested and brain tissue isolated. For each time point (0–2 days: dark gray and 4–5 days: stripped bars), 3 adult brain gDNAs (HD untreated, HD treated with ethanol (solvent), and HD treated with AZT) were analyzed and compared with gDNA from a healthy control with similar age (10–12 days old) (white). By taking advantage of TaqMan CNV assays on arbitrarily selected, representative TE classes, we monitored the tissues content of LINE I-element, LTR DM297, and DNA transposon Hobo. As shown in Figures S10 and S11, AZT treatment had no effects on tissue content of I-element. This result suggests that the I-element activity might have started before AZT treatment at 0–2 days. In addition, AZT treatment had no effects on the tissue content of the Hobo element. This result was expected, as Hobo is a DNA transposon whose transposition mechanism does not exploit the reverse transcriptase enzyme and therefore was not affected by the AZT treatment. On the contrary, AZT decreased DM297 in a statistically significant manner when administered at 0–2 days, suggesting that AZT treatment may have impaired DM297 mobilization in this time window (see Figures S10 and S11).
Discussion
Growing evidence suggests a detrimental effect of TEs activity in neurodegenerative disorders (Chang and Dubnau, 2019; Douville et al., 2011; Fukuda et al., 2021; Guo et al., 2018; Kaneko et al., 2011; Krug et al., 2017; Li et al., 2012, 2015; Liu et al., 2019; Perron et al., 1997; Prudencio et al., 2017; Romano et al., 2020; Sun et al., 2018; Thomas et al., 2017; Tam et al., 2019). Taking advantage of a fly model, this study proposes a pivotal role played by TEs in HD pathogenesis. We provided in vivo functional evidence that pan-neuronal expression of 128Q mutant Htt in Drosophila induces a widespread deregulation of TE silencing and the activation of their expression. We found that different families of TE transcripts were significantly upregulated in larval, young (0–2 days old), and aged (10–12 days old) HD brains. Moreover, the increase of TE transcripts at 10–12 days correlated with age-dependent HD neurodegeneration. Interestingly, we found that TE upregulation in response to 128Q mutant Htt expression is not a specific feature of postmitotic neurons. Indeed, the expression of the pathogenic Htt protein in glial cell lineage by using the pan-glial driver repo-Gal4 induced similar altered levels of TE transcripts.
On the basis of previous studies documenting a global constitutive heterochromatin relaxation in neurodegenerative disorders (Frost et al., 2014) and identifying constitutive heterochromatin as an epigenetic silencing mechanism for TE (Fedoroff, 2012; Sun et al., 2018), we investigated the underlying molecular mechanisms of HD-induced TE activation and demonstrated that the loss of TE silencing in 10- to 12-day-old HD fly brains is supported by a significant reduction of H3K9me3 occupancy in their promoters and coding regions. Moreover, PEV assay on larval brains expressing HD pathogenic construct confirmed that the overall reduction in H3K9me3 levels on TE sequences results from a global heterochromatin relaxation that impairs its ability to silence the variegating reporter transgene Hsp70-lacZ into Y pericentromeric heterochromatin (Lu et al., 1996). This is consistent with the observation that HP1 and Su(var)3–9 overexpression increase lifespan in short-lived HD flies and rescue the severe neurodegenerative phenotype induced by eye-specific expressions of the 128Q mutant Htt. In order to discriminate if the release of transposon silencing in HD is a cause or a consequence of neurodegeneration, we adopted a pharmacological approach to block the transposition of RTEs with RT inhibitors and demonstrated that RT inhibitors treatments rescued (1) the HD eye phenotype, (2) the genomic instability observed in HD larval brains, and (3) the lifespan in HD flies treated from larvae and 0–2 days adult stage. These results suggest that the HD phenotype may depend on RT activity and that retrotransposition at 0–2 days stage disease actively contributes to neurotoxicity. In addition, given the phenotypic rescues highlighted by the RT inhibitor treatments, it is likely that DNA transposons, whose transposition is not mediated by RT, do not contribute to neurotoxicity.
In the second part of our study, we explored the genomic complexity of TE mobilization by WGS and TaqMan qPCR. To this purpose we sequenced gDNA from HD brain tissues (elav-Gal4>128QHtt, offspr. HD) and offspring control (elav-Gal4/+, offspr. CTR) at 3 different developmental time points (larvae, young, and aged adults). A bioinformatic approach was developed to measure and compare the genomic content for each TE class in HD and control flies along lifespan. To this end, we assessed the normalized number of reads that map on the consensus sequence for each of the 189 TE classes in the Drosophila melanogaster genome, identifying 100 TE classes in which sequence content was different between HD and CTR flies at 0–2 days, the time when systemic RT inhibitory treatment was increasing lifespan. The differential genomic content for three of them was validated with TaqMan qPCR showing a very good correspondence between the two experimental approaches. The list of TE classes with a differential tissue content presents candidates for being molecular effectors of RT-dependent phenotypes. Among them, in a proof-of-concept experiment, the variation in LTR DM297 copy number at 0–2 days of HD brains, was blocked in the presence of AZT. Interestingly, a minor but substantial fraction of TEs showed a decrease in HD samples.
In general, a lack of correlation between TE expression and CNVs was evident, as previously reported in other biological systems (Jachowicz et al., 2017; Wang et al., 2018). This is probably due to multiple regulatory layers that define the quantity of TE-encoded ribonucleoparticles available for retrotransposition in specific cell types at a given time. Increasing evidence shows that a substantial amount of TE RNA is nuclear retained (Liu et al., 2020), where they may function as regulatory long noncoding RNAs (Percharde et al., 2018). Further studies are therefore needed to define the correlation between mRNA, protein expression, and mobilization rate for each TE class in HD flies.
Interestingly, GYPSY_I showed both a strong increase of RNA expression levels and the highest number of mapped reads in HD than CTR samples, in all the 3 analyzed time points. GYPSY_I element was previously described (Krug et al., 2017) to be overexpressed and contributes to neurodegeneration in a Drosophila ALS model (Krug et al., 2017). Furthermore, gypsy expression was found induced in a fly model of tauopathy (Guo et al., 2018). Our results suggest that GYPSY_I may be involved in three different fly models of neurodegenerative diseases such as ALS, AD, and HD.
In summary, the genomic analysis of TE content in HD fly brains lays down the foundations for the identification of the TE classes causally involved in the establishment of RT-dependent HD phenotypes and for new strategies of pharmacological intervention.
Limitations of the study
In this study we investigate the molecular mechanisms by which TEs contribute to cellular toxicity in Huntington's disease (HD). We provide evidences that TE expression and mobilization are increased in a Drosophila HD model. Moreover, we show that by preventing TE mobilization with reverse transcriptase (RT) inhibitors, we can mitigate polyQ-dependent neurodegeneration, reduce genomic instability, and increase the survival of HD flies. The results presented in this paper were obtained from experiments performed on a Drosophila transgenic line expressing the full-length mutant human Huntingtin under the control of UAS sequences. Although Drosophila is an excellent and well-established genetic model system, it could provide an incomplete modeling of HD. Aside from this general consideration, a limitation of our study may be represented by some experimental roadblocks concerning the measurements of differential genomic content of TEs. First and foremost, data from sequencing coverage and qPCR experiments may depend on differential access to selective regions of the genome caused by chromatin condensation, and this may lead to apparent changes in TE content. The lack of increase in copy number of a class of TE in the presence of RT-inhibitors, as for DM297, is indeed an experimental proof that data unveil RT-dependent processes. However, together with mobilization, RT activity may also lead to the synthesis of extrachromosomal cDNA from TE RNAs (De Cecco et al., 2019). The unequivocal identification and validation of single integration sites is therefore necessary to assess the extent of retrotransposition events and their contribution to differential copy number variation. However, this approach is limited by sequencing “bulk” DNA from 60 individuals and from samples composed by a highly heterogeneous pool of different cell types. Given the depth of sequencing and the heterogeneity of the sample, somatic, rare integration events are under the limits of detection. The decrease in TE genomic content requires further attention. Again, data can be the result of differential access to selective regions of the genome due to changes in heterochromatin content. In addition, loss of TE DNA in bulk samples may result from alternative or concurring mechanisms. Genomic deletions may be caused by TE mobilization (Rodriguez-Martin et al., 2020) and TE-mediated recombination (Startek et al., 2015), whereas surviving cells, although unlikely, may have been selected for their TE content. Further studies are needed to address these important points.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 555 conjugated goat anti-rabbit | Thermo Fisher Scientific | Cat: #A21428; RRID: AB_2535849 |
| HRP-conjugated goat anti-mouse IgG secondary antibody | Thermo Fisher Scientific | Cat: #31430; RRID: AB_228307 |
| HRP-conjugated goat anti-rabbit IgG secondary antibody | Thermo Fisher Scientific | Cat: #31460; RRID: AB_228341 |
| Mouse monoclonal anti-Elav 9F8A9 | Developmental Studies Hybridoma Bank | Cat: #ELAV 9F8A9; RRID: AB_2314364 |
| Mouse monoclonal anti-Gypsy envelope | Song et al., 1994 | RRID: AB_2569690 |
| Mouse monoclonal anti-Histone H3K9me3 | Active Motif | Cat: #61013; RRID: AB_2687870 |
| Mouse monoclonal anti-Repo 8D12 | Developmental Studies Hybridoma Bank | Cat: #8D12; RRID: AB_528448 |
| Rabbit polyclonal anti-Giotto | Giansanti et al., 2006 | RRID: AB_2892585 |
| Rabbit polyclonal anti-γH2AV | Rockland Immunochemicals | Cat: #600401914; RRID: AB_828383 |
| Chemicals, peptides, and recombinant proteins | ||
| Blue Dye N.1 | Sigma-Aldrich | Cat: #861146; CAS: 3844-45-9 |
| DAPI | Sigma-Aldrich | Cat: #D9542; CAS: 28718-90-3 |
| Lamivudine | Sigma-Aldrich | Cat: #PHR-1365; CAS: 134678-17-4 |
| TOTO3-Iodide | Invitrogen | Cat: #T3604; CAS: 166196-17-4 |
| Zidovudine | Sigma-Aldrich | Cat: #PHR-1292; CAS: 30516-87-1 |
| X-Gal | Sigma-Aldrich | Cat: #B4252; CAS: 7240-90-6 |
| Critical commercial assays | ||
| iQ multiplex powermix | Bio-Rad | Cat: #1725849 |
| NucleoSpin™ Gel and PCR clean-up Kit | Macherey-Nagel | Cat: #740609.50 |
| Platinum Taq DNA polymerase | Invitrogen | Cat: #10966018 |
| QIAquick Gel Extraction Kit | Qiagen | Cat: #28706X4 |
| Quant-iT PicoGreen™ dsDNA Assay Kit | Thermo Fisher Scientific | Cat: #P7589 |
| QuantiFast SYBR green PCR Kit | Qiagen | Cat: #204057 |
| SuperScript III Reverse Transcriptase | Invitrogen | Cat: #18080093 |
| TaKaRa Ex Taq DNA Polymerase | Takara | Cat: #RR001C |
| Deposited data | ||
| Analysed data: see Table S1 | This paper | N/A |
| Raw data: See European Nucleotide archive under accession number PRJEB41409 | This paper | https://www.ebi.ac.uk/ena/browser/view/PRJEB41409 |
| Drosophila reference genome (dm6 version) | UCSC | http://genome.ucsc.edu/ |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Overexpression of human Htt with 128Q under UAS control: w[1,118]; P{w[+mC]=UAS-HTT.128Q.FL}f27b | Bloomington Drosophila Stock Center | RRID: BDSC_33808 FlyBase: FBst0033808 |
| D. melanogaster: Overexpression of human Htt with 16Q under UAS control: w[1,118]; P{w[+mC]=UAS-HTT.16Q.FL}F24/CyO | Bloomington Drosophila Stock Center | RRID: BDSC_33810 FlyBase: FBst0033810 |
| D. melanogaster: Carries a rearrangement causing position effect variegation of the w[+] transgene marker: w[∗]/Dp(3;Y)BL2, P{w[enh]=HS-lacZ.scs}65E | Bloomington Drosophila Stock Center | RRID: BDSC_57371 FlyBase: FBst0057371 |
| D. melanogaster: Expresses membrane-localized GFP under UAS control: y1 w∗; P{w+mC=UAS-mCD8::GFP.L}LL5, P{UAS-mCD8::GFP.L}2 | Bloomington Drosophila Stock Center | RRID: BDSC_5137 FlyBase: FBst0005137 |
| D. melanogaster: Expresses GAL4 in neurons under elav control: P{w[+mW.hs]=GawB}elav[C155] | Bloomington Drosophila Stock Center | RRID: BDSC_458 FlyBase: FBst0000458 |
| D. melanogaster: Expresses GAL4 in glia: w[1,118]; P{w[+m∗]=GAL4}repo/TM3, Sb[1] | Bloomington Drosophila Stock Center | RRID: BDSC_7415 FlyBase: FBst0007415 |
| D. melanogaster: Expresses GAL4 in the eye under GMR control: w[∗]; P{w[+mC]=GAL4-ninaE.GMR}12 | Bloomington Drosophila Stock Center | RRID: BDSC_1104 FlyBase: FBst0001104 |
| D. melanogaster: Expresses GAL4 pan-neuronally under nSyb control and CD8-tagged GFP under the control of UAS: w[1,118]; Pin[1]/CyO; P{y[+t7.7] w[+mC]=nSyb-GAL4.P}attP2, P{w[+mC]=UAS-mCD8::GFP.L}LL6 | Bloomington Drosophila Stock Center | RRID: BDSC_51944 FlyBase: FBst0051944 |
| D. melanogaster: Overexpression of HP1a under UAS control: w[1,118]; UAS-Hp1/CyO | Our lab | N/A |
| D. melanogaster: Overexpression of Su(var)3–9-EGFP under UAS control | G. Reuter Lab (Schotta and Reuter, 2000) | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR: see Table S2 | This paper; Jin et al., 2013; Preall et al., 2012; Padeken et al., 2013; Klenov et al., 2011; Lu and Clark, 2010 | N/A |
| Primers for ChIP-qPCR: see Table S3 | This paper; Papp and Müller, 2006; Wang and Elgin, 2011 | N/A |
| Primers and probes for CNV assay: see Table S4 | This paper | N/A |
| Primers for semi-quantitative PCR: 128QHtt F: CACCGACCAAAGAAAGAACTTTCA | Branco et al., 2008 | N/A |
| Primers for semi-quantitative PCR: 128QHtt R: TTTAATTTCCTTATAGAGCTCGAGCTGTAA | Branco et al., 2008 | N/A |
| Primers for semi-quantitative PCR: gapdh F: CAGCCCCGACATGAAGGT | Vanden Broeck et al., 2013 | N/A |
| Primers for semi-quantitative PCR: gapdh R: CGATCTCGAAGTTGTCATTGATG | Vanden Broeck et al., 2013 | N/A |
| Software and algorithms | ||
| Adobe Photoshop CS6 | Adobe | https://www.adobe.com/products/photoshop.html |
| Bowtie 1.2 version | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| BWA Aligner | Li and Durbin, 2009 | https://emea.illumina.com/products/by-type/informatics-products/basespace-sequence-hub/apps/bwa-aligner.html |
| FastQC v0.11.7 version | BaseSpace Labs | https://emea.illumina.com/products/by-type/informatics-products/basespace-sequence-hub/apps/fastqc.html |
| Flynotyper | Iyer et al., 2016 | http://flynotyper.sourceforge.net/ |
| GraphPad Prism 6.00 version | La Jolla California USA | www.graphpad.com |
| Mobile Element Locator Tool (MELT) 2.1.4 version | Gardner et al., 2017 | https://melt.igs.umaryland.edu/ |
| Primer3 | Rozen and Skaletsky, 1999 | https://primer3.org/ |
| Samtools view 1.3.1 version | Li et al., 2009 | http://www.htslib.org/ |
| ZEN Software | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
Resource availability
Lead contact
Further information and requests for resources, material and reagents should be direct to and will be fulfilled by the lead contact, Lucia Piacentini (lucia.piacentini@uniroma1.it).
Materials availability
All Drosophila lines and reagents generated in this study are available from the lead contact.
Experimental models and subject details
Drosophila strains
The Drosophila stocks used in this study were raised at 24°C with 12:12 h light/dark cycle on standard cornmeal-sucrose-yeast-agar medium. All crosses were performed at 24°C unless otherwise noted. The Ore-R stock and balancer stocks, used to balance inserts on the X, second, and third chromosomes, respectively, have been kept in our laboratory for many years. To generate UAS-HP1 transgenic flies, the coding sequence for HP1 was amplified by PCR using a cDNA as template with specific primers, HP1 F (CCGGTACCATATGGGCAAGAAAATCGACAAC) and HP1 R (GTGCGGCCGCTGTTTAATCTTCATTATCAGAG) that introduced restriction enzyme sites KpnI-NotI for cloning into pUASp vector. Germline transformation into w1118 embryos was carried out by BestGene Inc. Sources of fly stocks are provided in the key resource table.
Methods details
NRTI food preparation
Zidovudine (AZT; PHR-1292 Sigma-Aldrich) and Lamivudine (3TC; PHR-1365 Sigma-Aldrich) were dissolved in 95% ethanol and water, respectively. NRTI solutions were added in standard medium (cornmeal-sucrose-yeast-agar) to give a final concentration of 5 mg/mL for AZT and 1 mg/mL for 3TC.
Food intake assay
Four groups of eight 3-day-old mated flies (four males and four females) were transferred onto fresh food medium containing 1% (w/v) blue food dye (Blue Dye no. 1, Merck) and supplemented with 5 mg/mL AZT (or solvent alone). After 3 h, flies were homogenized in 100 μL of PBS1x and the absorbance of the liquid sample was then measured at 629 nm (Thermo Scientific Multiskan GO Microplate Spectrophotometer).
Mitotic chromosome preparations
Cytological preparations of mitotic chromosomes from Drosophila larval brain were obtained according to Pimpinelli et al. (2000) (Pimpinelli et al., 2000) and stained with DAPI (4,6-diamidino-2-phenilindole, 0.01 mg/mL) to visualize DNA.
The slides were mounted in antifading medium (23,3 mg/mL of DABCO (1,4-Diazobicyclo-(2,2,2) octane) in 90% glycerol – 10% 1X PBS) and all images were acquired on Ellipse Epifluorescence microscope (E1000 Nikon) equipped with a CCD camera (Coolsnap). Images were analyzed and further processed using Adobe Photoshop CS6.
Immunofluorescent staining of Drosophila larval brains
The immunofluorescent staining of larval brains was performed according to Wu and Luo (2006) (Wu and Luo, 2006). The primary antibody used was rabbit anti-γH2AV (1:50, 600-401-914 Rockland Immunochemicals). Fluorescent-labelled secondary antibody raised in goat was Alexa Fluor 555 conjugated anti-rabbit (1:300, A21428 Thermo Fisher Scientific). The brains were stained with TOTO-3 Iodide (1 μM) to visualize DNA and mounted in antifading medium. Confocal observations were performed using a Leica DMIRE (Leica Microsystems, Hiedelberg, Germany) and a Zeiss LSM 780 (Zeiss, Berlin, Germany). Images were analyzed and further processed using Zen Software and Adobe Photoshop CS6.
Drosophila eye imaging
Age- and genotype-specific flies were ether-anesthetized, and eye external images were taken with a Nikon camera D5000 mounted onto a stereoscopic microscope. Images were further processed using Adobe Photoshop CS6. The Drosophila eye defects were quantified using Flynotyper (Iyer et al., 2016), a software that detects alterations in eye morphology and that provides a Phenotypic Score as a quantitative measure of the severity of the ommatidial arrangement defects.
Lifespan analysis
100–200 flies were collected for each experimental group (30 flies per vial). Every 2–3 days, flies were passed into new vials and dead flies were counted. The survival rate was calculated by the percentage of total flies surviving.
Total RNA extraction and qRT-PCR
RNA samples from larval and adult brains were isolated using Qiazol reagent (Qiagen), according to the manufacturer's instructions. 5 μg of total RNA was reverse transcribed using oligo dT and SuperScript Reverse Transcriptase III (Invitrogen) according to the manufacturer's protocol. The qPCR reactions were carried out with QuantiFast SYBR Green PCR Kit (Qiagen) according to manufacturer's protocol. Relative abundance of the different transcripts was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001) using rp49 or gapdh transcript as controls. qRT-PCR experiments were performed in three independent biological replicates each with three technical replicates. All used primers are listed in Table S2.
Semi-quantitative PCR
About 100 ng of reverse transcribed total RNA purified from fly heads, was added to the following reaction mix: 1X PCR Buffer, primers (0.2 μM), MgCl2 (1.5 mM), dNTPs (0.2 mM each), and 2U/rxn of Platinum™ Taq DNA Polymerase (Invitrogen). The thermal profile was: 94°C for 3 min for the initial denaturation, followed by 28 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 30 s and a final extension at 72°C for 7 min. PCR products were analyzed by agarose gel electrophoresis. The oligonucleotides used as primers are listed in the key resource table.
Western blot analyses
Western blot was carried out as previously described in Cappucci et al. (2019) (Cappucci et al., 2019). To obtain a total protein extract, Drosophila brain or head samples were homogenized in Sample Buffer 1X (50 mM Tris-HCl pH 6.8, 100 mM dithiothreitol, 2% sodium dodecyl sulfate, 2.5% glycerol, 0.1% bromophenol blue) and heated at 85°C for 8 min to complete the protein denaturation.
Protein extracts were fractionated by 10% SDS-PAGE and electroblotted onto Immobilion-P polyvinyl difluoride membranes (Bio-rad) in CAPS-based transfer buffer (10 mM CAPS pH 11, 10% methanol) in a semi-dry transfer apparatus (Amersham Biosciences). The membranes were blocked with 5% non-fat dry milk in Tris-buffered saline with Tween 20 (TBST) buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween 20) and incubated with the following antibodies diluted in TBST: mouse anti-ENV 8E7 (1:500 (Song et al., 1994), kindly provided by J. Gall), rabbit anti-GIOTTO (1:10,000 (Giansanti et al., 2006)), mouse anti-elav (1:500, 9F8A9 DSHB) and mouse anti-repo (1:500, 8D12 DSHB). Proteins of interest were detected with HRP-conjugated goat anti-mouse or anti-rabbit IgG antibody and visualized with the ECL Western blotting substrate (GE Healthcare), according to the provided protocol. The chemiluminescence detection was performed on the ChemiDoc XRS + System (Bio-rad) and analyzed using the included ImageLab software.
Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation was performed according to Casale et al. (2019) (Casale et al., 2019) with some modifications. Approximately 250 μL of 10- to 12-days-old fly heads were homogenized in 3 mL of NEB buffer (10 mM HEPES-Na pH 8, 10 mM NaCl, 0.1 mM EGTA Na pH 8, 0.5 mM EDTA-Na pH 8, 1 mM DTT, 0.5% NP-40, 0.5 mM Spermidine, 0.15 mM Spermine, 1× EDTA-free Complete Protease Inhibitors) with a Polytron homogenizer (Kinematica Swizerland) with a PT300 tip five times (2 min and 1 min on ice) at 3,500 rpm and five times at 4,000 rpm. The homogenate was transferred to a pre-chilled glass dounce (Wheaton) and 20 full strokes were applied with a tight pestle. Free nuclei were filtered on a 70 μM strainer and then centrifuged at 6,000 × g for 10 min at 4°C. The nuclei-containing pellets were resuspended in 1 mL of NEB + 0.25 M sucrose and centrifuged at 20,000 × g for 20 min on sucrose gradient (0.65 mL of 1.6 M sucrose in NEB, 0.35 mL of 0.8 M sucrose in NEB). The pellet was resuspended in 1 mL of NEB and formaldehyde to a final concentration of 1%. Nuclei were cross-linked for 10 min at room temperature and quenched by adding 1/10 vol of 1.375 M glycine. The nuclei were collected by centrifugation at 6,000 × g for 5 min. Nuclei were washed in PBS 1X and then twice in 1 mL of NEB and resuspended in 1.5 mL of Lysis Buffer (15 mM HEPES-Na pH 7.6, 140 mM NaCl, 0.5 mM EGTA, 1 mM EDTA pH 8, 1% Triton X-100, 0.5 mM DTT, 0.1% Na-Deoxycholate, 0.1% SDS, 0.5% N-lauroylsarcosine and 1× EDTA-free Complete Protease Inhibitors). Nuclei were sonicated using a Hielscher Ultrasonic Processor UP100H (100 W, 30 kHz) thirty times for 30s and 30s on ice. Sonicated nuclei were centrifuged at 15,000 × g for 10 min at 4°C. The majority of sonicated chromatin was 300–500 base pairs (bp) in length. To allow the immunoprecipitation, 3 μg of H3K9me3 monoclonal antibody (61,013 Active Motif) were incubated in the presence of dynabeads protein G (Invitrogen) for 3 h at room temperature on a rotating wheel, then chromatin extract was added, and incubation was continued overnight at 4°C on a rotating wheel. The supernatants were discarded, and samples were washed twice in Lysis Buffer (each wash 15 min at 4°C) and twice in TE Buffer (1 mM EDTA, 10 mM Tris HCl pH 8). Chromatin was eluted from beads in two steps; first in 100 μL of Elution Buffer 1 (10 mM EDTA, 1% SDS, 50 mM Tris HCl pH 8) at 65°C for 15 min, followed by centrifugation and recovery of the supernatant. Beads material was re-extracted in 100 μL of TE + 0.67% SDS. The combined eluate (200 μL) was incubated overnight at 65°C to revert the cross-link and treated by 50 μg/mL RNaseA for 30 min at 37°C and by 500 μg/mL Proteinase K (Invitrogen) for 3 h at 65°C. Samples were phenol–chloroform extracted, and ethanol precipitated. DNA was resuspended in 15 μL of water and candidate genes were amplified through qPCR. The DNA oligonucleotide primers needed for ChIP-qPCR are listed in Table S3.
Histochemical localization of β-galactosidase in PEV analysis
The induction and localization of β-galactosidase on larval brains was performed according to Lu et al. (1996) (Lu et al., 1996). To induce β-galactosidase expression, larvae were heat-shocked at 37°C for 45 min followed by 1 h of recovery at room temperature. Brains were dissected in 1X PBS, fixed in 4% formaldehyde for 20 min, washed in 1X PBS and incubated in 0.2% X-gal (5-bromo-4-chloro-3-indolyl-p-D-galactopyranoside) assay buffer (3.1 mM potassium ferricyanide, 3.1 mM potassium ferrocyanide, 10 mM PB (pH 7.2), 0.15 M NaCl, 1 mM MgCl2) for 16 h. Brains were immersed in 1X PBS and the images were acquired through a stereomicroscope equipped with a Nikon D5000 camera and further processed using Adobe Photoshop CS6. X-Gal staining intensities were analyzed using ImageJ software. All RGB images were converted in 8-bit gray scale and then inverted with the command Edit > Invert. An outline was drawn around each brain, area and intensities measured, along with adjacent background reading. Intensity values were then calculated following the total corrected cell fluorescence (TCCF) method as described in McCloy et al. (2014) (McCloy et al., 2014).
Isolation of genomic DNA
Brain tissues were homogenized in 200 μL of extraction buffer (120 mM Tris-HCl pH 8, 60 mM EDTA pH 8, 80 mM NaCl, 160 mM sucrose, 0.5% v/v SDS, 200 μg/mL RNase DNase free) and incubated at 65°C for 60 min. After cooling at room temperature for a few minutes, 28 μL 8M K-acetate was added. After 30 min on ice, the samples were spun at 10,000 rpm for 15 min at 4°C; DNA was precipitated by adding 0.5 volume of isopropanol to the supernatant, leaving for 10 min at room temperature, and spinning again for 10 min. The pellet was washed with 70% ethanol, dried and redissolved in 60 μL H2O. 1 μg of gDNA was sequenced by high-throughput Illumina technology.
DNA sequencing and raw reads quality controls
1 μg of DNA was extracted from every fly sample. Sample1, sample3 and sample5 were obtained from homogenized brain tissue from offspring control flies (elavG4/+; +/TM6) at larval, young (0–2 days old) and aged (10–12 days old) stages while sample2, sample4 and sample6 were obtained from offspring HD flies (elavG4/+; 128QHtt/+) at the 3 different time points used for control samples. DNA was sequenced by Institute of Applied Genomics (IGA, Udine, Italy) obtaining 125 bp paired-end (PE) reads. The quality of the raw reads was tested using FastQC (version v0.11.7).
In silico TE tissue content quantification
In order to in silico quantify TE cell content in the 6 sequenced samples, DNA-seq reads of every sample were mapped on each of the Repbase (Bao et al., 2015) Drosophila melanogaster TE consensus sequences (with exception for SAT and Unknown TEs) using bwa (Li and Durbin, 2009) with default parameters (bwa version 0.7.15-r1140). Reads mapping on each TE consensus sequence were then counted using samtools view (-F 4 and -c parameters, version 1.3.1) (Li et al., 2009). Two proportions Z-test (prop.test function in R) was used at larval, 0–2d and 10–12d stages to highlight significant differences in the proportion of reads mapping on TEs in CTR and HD samples with respect to the total number of sequenced reads. p value scores were then adjusted applying false discovery rate (FDR) correction (Benjamini & Hochberg) and the set of significant TEs was defined selecting TEs with FDR < 0.001 at 0–2 d stage.
Evaluation of the in silico TE tissue content quantification
The computational method used to produce data in Figure 5 is the result of a bioinformatics pipeline exploiting publicly available tools. To calculate the TEs cell content, WGS reads were mapped to the Drosophila TE consensus and counted. To test whether this method provides proper estimations of the TEs cell content, we generated WGS reads in silico from the Drosophila (dm6) reference genome at a coverage similar to that of our WGS data (∼50×) with a uniform distribution throughout the genome. Next, our pipeline has been run on these simulated reads measuring the content of each TE consensus sequence annotated in RepBase for Drosophila. Considering that reads were generated in silico from the Drosophila reference genome, the TEs cell content estimation for each TE should correlate with its genomic occupancy in the reference genome. To test this, Pearson correlation coefficients between estimated TE cell content and TE genomic occupancy were calculated showing strong positive and significant correlation (R2 = 0.9, p < 2.2e-16) suggesting a correct estimation of the TEs content computed by our pipeline (Figure S12A). To further investigate the proper functioning of our strategy, WGS reads were additionally simulated from a custom Drosophila reference genome where the consensus sequences of three TEs were added (GYPSY12_I from LTR class, HOBO from DNA transposon class and I-element from LINE class). TE cell content of each TE was then estimated in the reads deriving from both the canonical Drosophila reference genome and from the genome where the three additional TE consensus were inserted. Results show that our strategy was capable to identify the increase for the three TEs in the modified genome with respect to the reference, while all other TEs remained unaltered (Figures S12B and S12C). Altogether, the in silico tests performed indicate that our computational strategy may properly estimate the TE cell content.
DNA quantification for primers validation and CNV assays
In order to discriminate small variations in TE contents, a precise quantification of genomic DNA is required. The first DNA dilution to 2 ng/μL was spectrophotometrically measured using NanoDROP (Thermo Scientific, Thermo Fisher Inc, USA). Further dilutions in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) to a final concentration of 100 pg/μL were performed using QuantiT PicoGreenR dsDNA kit (Invitrogen, Thermo Fisher Inc, California, USA), following manufacturer's instructions. Standard curve ranging from 20 pg/mL to 640 pg/mL, DNA samples and blank were measured in duplicate using EnSpire Multimode Plate Reader (PerkinElmer Inc, Waltham, USA). By plotting the measured fluorescence versus DNA, concentrations were calculated through a standard curve plot. Final DNA concentrations ranging from 90 to 110 pg/μL were accepted for the PCR analysis.
Primers and probes design and PCR amplification
Following standard experimental protocols for validation assay, primers and probes were designed on the candidate TE consensus sequences using the online tool Primer3 (Rozen and Skaletsky, 1999) (https://primer3.org/), as listed in Table S4. Primers and probes specificity was tested using an in silico amplification custom bioinformatics pipeline. Briefly, for each candidate TE, primers and probe were mapped using bowtie (version 1.2) (Langmead et al., 2009) against the dm6 fly genome allowing 0 mismatches in the last 7 nucleotides and up to 3 in the remaining part. Then only the in silico amplicons i) recognized by primers on opposite strand, ii) with the probe mapped in the between and iii) with a length comprised between 50 and 2,000 nt, were selected. To test their specificity, the genomic coordinates of the selected in silico amplicons were intersected (intersectBed, -wo, -bed, -F 0.99 parameters (Quinlan and Hall, 2010) with the coordinates of TEs annotated in the fly dm6 genome. The same custom pipeline was then re-run mapping primers and probes against the TE consensus sequences, in order to select primers and probe that recognize a unique TE sequence. Primers and probes were finally synthesized by Sigma-Aldrich. All primers were tested on 100 ng of genomic DNA in order to validate experimentally their specificity. PCR amplifications were performed using 5 units/μL TaKaRa ExTaq Polymerase, 10x TaKaRa Buffer (both ClonTech), dNTPs (2.5 μM) and primers (10 μM each). PCR conditions are as follows: a denaturation step at 94°C for 3 min, followed by 40 cycles of amplification at 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, ending with a final extension at 72°C for 5 min. The amplified products together with a negative control, such as non-templates, were separated on a 1.5% agarose gel and visualized with Midori staining. Amplicons were extracted from gel following manufacturer's instructions (QIAquick Gel Extraction Kit, Qiagen) and confirmed through Sanger sequencing (Eurofins Genomics). For the DM297 LTR and Hobo DNA elements primer and probe have been designed against the 3′ portion of the consensus sequence while for I-element a design on the consensus 5′ has been performed.
Invariant gene design for TaqMan CNV assay
In order to evaluate the number of copies of repetitive elements, a proper invariant gene should be a multi-copied genomic element. A TaqMan CNV assay requires an invariant gene resulting stable in all the samples analyzed and with a number of copies similar to the analyzed elements. To our knowledge, all the previous genomic studies performed with a TaqMan qPCR assay to estimate copy number variations of TEs in fruit flies, were performed using single-copy genes as invariant gene, such as actin, gapdh, rp49 (rpl32) (Guo et al., 2018; Krug et al., 2017). However, in our experimental conditions (100 pg DNA) and following Real Time qPCR technical protocols, single copy gene as actin amplified within 35 cycles threshold (Ct), resulting not comparable to the TEs (on average 26 Ct) in the final 2–ΔΔCt method for the relative quantification (Livak and Schmittgen, 2001). To overcome this technical problem, we scanned the fly genome to find a stable multi-copied element. We thus took advantage of the IS analysis carried out with MELT on our WGS data considering that ancient, fixed and inactive TEs could represent candidate invariant sequences. 56 TE classes showed no novel ISs in all the 6 fly samples. Given that we are selecting TEs with no novel ISs, the balancer-associated TE ISs over-estimation in CTR samples should not affect this analysis. We then selected TEs with at least 30 annotated genomic copies and with a consensus sequence longer than 400 bp, shortening the list to 5 TEs. Among these, DMRT1C LINE element was the only element displaying no full-length copies fixed in the fly genome. The DMRT1C consensus sequence is, indeed, 5,443 bp long, whereas the length of the 224 DMRT1C fixed fragments in the fly genome is comprised between 30 and 3,000 bp. Additionally, none of the DMRT1C 224 copies retained a full-length reverse transcriptase functional domain. Together, these observations strongly suggest that all the 224 DMRT1C genomic copies are likely truly truncated and inactive. DMRT1C copy number in silico predicted, was further confirmed by PCR when compared to actin allowed the co-amplification with the other TEs under study at very low concentrations (100 pg), in optimal technical conditions (31 Ct). The DMRT1C was thus validated as the best candidate invariant element and used in TaqMan assays as normalizer (data not shown).
Taqman based copy number variation (CNV) assay
TaqMan-based Copy Number Variation (CNV) assay was performed to detect TE content in DNA in Drosophila samples in all timing and life conditions, as previously described. All custom TaqMan probes sequences are listed above and were designed following standard protocol and the vendor's custom assay design service manual (Thermo Fisher Scientific). Probes for all TE assays were conjugated to the fluorophore label MGB 6-FAM, whereas the invariant genes Actin or DMRT1C probes were conjugated with MGB-VIC. Actin assay Act5C (assay ID Dm02361909_s1) was acquired from Thermo Fisher Scientific.
A 20 μL reaction volume containing 10 μL of 2X iQ Multiplex Powermix (Biorad), 0.60 μL of primers (10 μM), 0.2 μL of probes (10 μM) and 1 μL DNA (100 pg for DMRT1C; 1 ng for Actin) was used for qPCR amplification. PCR conditions are as follows: 95°C for 20 s, followed by 40 cycles of amplification at 95°C for 10 s and 59°C for 30 s.
Standard curves were performed for each couple of target and invariant gene on genomic DNA ranging from 32 pg to 2 ng. The slope of linear regression to standard curve was nearly −3.32 for all assays in this study, which means that primers and multiplexing efficiency were acceptable.
The experiments were carried out using CFX96 Real-Time PCR detection system (Biorad), according to standard protocol. Three replicas of qPCR reactions were performed in duplicates. Standardization was performed considering a control sample as calibrator and an independent inter-run calibrator for each plate. Data obtained from the co-amplifications of the target DNA sequence and the internal invariable control were analyzed using the 2–ΔΔCt method for the relative quantification (Livak and Schmittgen, 2001).
In addition to the 2–ΔΔCt method for the relative quantification, we carried out TE quantification using standard dilution in qPCR. To this purpose, I-element, DM297, HOBO, DMRT1C and Actin were amplified through endpoint PCR using gDNA from control sample of head fly. Program and primers were described above. Amplicons were run on agarose gel (1.8%) and extracted with a purification kit (NucleoSpin Gel and PCR Clean Up by Macherey-Nagel). DNAs were quantified at Nanodrop and then diluted in serial dilutions for the standard quantification in qPCR. Number of copies was calculated with the formula: number of copies = (amount ∗ 6.022 × 1023)/(length ∗ 1 × 109 ∗ 650) using dsDNA copy number calculator (University of Rhode island: https://cels.uri.edu/gsc/cndna.html).
Quantitative PCR was the performed using TaqMan assays and protocols described above. Standard curves for TE amplicons are shown in Table S5, calculated directly from the graphs reported in the Figure S13 by linear regression analysis. Results for quantitative method are reported in supplemental information.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism version 6.00 (GraphPad Software, La Jolla, CA, USA, www.graphpad.com). For all statistics, a p value ≤ 0.05 was considered statistically significant. qRT-PCR, ChIP-qPCR, X-Gal intensity and food intake statistical analysis was performed by the Unpaired t test or one-way ANOVA test (followed by Tukey's or Dunnett's multiple comparison tests). Western blots were analyzed by One-sample t test. Lifespan data were analyzed by log rank test with Bonferroni adjustment. Statistical significance of chromosomal abnormalities was determined by Chi-Square test with Bonferroni correction. Mitotic index and Phenotypic Score (Flynotyper) statistical significance was determined by one-way ANOVA followed by Tukey's post-hoc test. For the TaqMan CNV assay, to compare all study groups and experimental conditions, two-Way ANOVA with Bonferroni post hoc test were used. Statistical significance for all the results obtained from the bioinformatics analyses (number of variations with/without chr3 and TE cell content) was determined by using the two proportions Z-test (prop. test function in R). Calculated p values were then corrected using Benjamini & Hochberg FDR correction. FDR < 0.05 were considered as statistically significant.
All statistical tests performed in this manuscript, with correspondent significance levels, dispersion and precision measures, are further specified in figure legends.
Acknowledgments
We are grateful to Sergio Pimpinelli for critically reading the manuscript. We thank the Bloomington Stock Center, VDRC Stock Center, and the Developmental Studies of Hybridoma Bank for fly strains and antibodies. We would like to thank Gianluca Cestra, Joseph Gall, Victor Corces, Marta Marzullo, and Gunter Reuter for providing antibodies, Su(var)3–9 primers, and fly stocks. We sincerely appreciate Silvana Caristi for assistance with confocal image acquisition. We are indebted to Ms Eva Ferri, Fabrizio Torri, and Alessandra Sanna (IIT [Istituto Italiano di Tecnologia]), and Cristina Leonesi (SISSA) for administrative support. This study has been supported by the Fondazione Terzo Pilastro Internazionale (L.P.), the research project grant from Sapienza University of Rome (L.P.), the IIT and SISSA intramural grants (S.G. and R.S.).
Author contributions
A.M.C., F.L., and U.C. performed the molecular and genetic experiments, contributed to the analysis of the results and to the design of the research; F.A. performed the bioinformatic analysis of WGS data and developed TaqMan CNV PCR assays; S.F. developed and carried out the TaqMan CNV PCR experiments; G.S. performed some bioinformatic analysis of WGS data; F.P. conceived and supervised the study; R.S. conceived and supervised the bioinformatic analysis and the TaqMan CNV PCR assays, wrote the manuscript; S.G. and L.P. wrote the manuscript, conceived, and supervised the entire study. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103702.
Contributor Information
Stefano Gustincich, Email: stefano.gustincich@iit.it.
Lucia Piacentini, Email: lucia.piacentini@uniroma1.it.
Supplemental information
Data and code availability
-
•
Whole genome sequencing (WGS) data from control and HD flies at larval stage, 0–2 d- and 10–12 d-aged have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB41409 (https://www.ebi.ac.uk/ena/browser/view/PRJEB41409).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Allshire R.C., Madhani H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. Babraham Bioinformatic; 2010. FASTQC. A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Armstrong J.D., Kaiser K., Müller A., Fischbach K.F., Merchant N., Strausfeld N.J. Flybrain, an on-line atlas and database of the Drosophila nervous system. Neuron. 1995;15:17–20. doi: 10.1016/0896-6273(95)90059-4. [DOI] [PubMed] [Google Scholar]
- Awasaki T., Lai S.-L., Ito K., Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayarpadikannan S., Kim H.-S. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12:98–104. doi: 10.5808/GI.2014.12.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kojima K.K., Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi S., Tripathi T., Monziani A., Di Leva F., Biagioli M. In: Neuroepigenomics in Aging and Disease. Delgado-Morales R., editor. Springer International Publishing; 2017. Epigenetics of Huntington’s disease; pp. 277–299. [DOI] [Google Scholar]
- Belancio V.P., Hedges D.J., Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Belgnaoui S.M., Gosden R.G., Semmes O.J., Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaudin de Thé F.X., Rekaik H., Peze Heidsieck E., Massiani Beaudoin O., Joshi R.L., Fuchs J., Prochiantz A. Engrailed homeoprotein blocks degeneration in adult dopaminergic neurons through LINE-1 repression. EMBO J. 2018;37:e97374. doi: 10.15252/embj.201797374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodea G.O., McKelvey E.G.Z., Faulkner G.J. Retrotransposon-induced mosaicism in the neural genome. Open Biol. 2018;8:180074. doi: 10.1098/rsob.180074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco J., Al-Ramahi I., Ukani L., Pérez A.M., Fernandez-Funez P., Rincón-Limas D., Botas J. Comparative analysis of genetic modifiers in Drosophila points to common and distinct mechanisms of pathogenesis among polyglutamine diseases. Hum. Mol. Genet. 2008;17:376–390. doi: 10.1093/hmg/ddm315. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cappucci U., Torromino G., Casale A.M., Camon J., Capitano F., Berloco M., Mele A., Pimpinelli S., Rinaldi A., Piacentini L. Stress-induced strain and brain region-specific activation of LINE-1 transposons in adult mice. Stress. 2018;21:575–579. doi: 10.1080/10253890.2018.1485647. [DOI] [PubMed] [Google Scholar]
- Cappucci U., Noro F., Casale A.M., Fanti L., Berloco M., Alagia A.A., Grassi L., Le Pera L., Piacentini L., Pimpinelli S. The Hsp70 chaperone is a major player in stress-induced transposable element activation. Proc. Natl. Acad. Sci. U S A. 2019;116:17943–17950. doi: 10.1073/pnas.1903936116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casale A.M., Cappucci U., Fanti L., Piacentini L. Heterochromatin protein 1 (HP1) is intrinsically required for post-transcriptional regulation of Drosophila Germline Stem Cell (GSC) maintenance. Sci. Rep. 2019;9:4372. doi: 10.1038/s41598-019-40152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casale A.M., Cappucci U., Piacentini L. Unravelling HP1 functions: post-transcriptional regulation of stem cell fate. Chromosoma. 2021;130:103–111. doi: 10.1007/s00412-021-00760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D., et al. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2017;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.Y., Warrick J.M., Andriola I., Merry D., Bonini N.M. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum. Mol. Genet. 2002;11:2895–2904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- Chang Y.-H., Dubnau J. The gypsy endogenous retrovirus drives non-cell-autonomous propagation in a Drosophila TDP-43 model of neurodegeneration. Curr. Biol. 2019;29:3135–3152.e4. doi: 10.1016/j.cub.2019.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Chen L., de Jesus Silva Carvalho C., de Moraes Filho A.V., Véras J.H., Cardoso C.G., Bailão E., Spanó M.A., Cunha K.S. Toxicity and genotoxicity induced by abacavir antiretroviral medication alone or in combination with zidovudine and/or lamivudine in Drosophila melanogaster. Hum. Exp. Toxicol. 2018;38:446–454. doi: 10.1177/0960327118818248. [DOI] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C.N., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., Gage F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U S A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D., et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.N., Di Stefano L., Maher R.C., Zhu H., Macdonald M.E., Gusella J.F., Walker J.A. The Drosophila Huntington’s disease gene ortholog dhtt influences chromatin regulation during development. Hum. Mol. Genet. 2015;24:330–345. doi: 10.1093/hmg/ddu446. [DOI] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J.P., Carraway R., Reeves S.A., et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- Douville R., Liu J., Rothstein J., Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Ebert A., Schotta G., Lein S., Kubicek S., Krauss V., Jenuwein T., Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., James T.C., Foster-Hartnett D.M., Hartnett T., Ngan V., Elgin S.C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., Morris G.D., Reuter G., Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S.C.R., Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M.C., O’Neill E.M., Rubin G.M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Erwin J.A., Paquola A.C.M., Singer T., Gallina I., Novotny M., Quayle C., Bedrosian T.A., Alves F.I.A., Butcher C.R., Herdy J.R., et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 2016;19:1583–1591. doi: 10.1038/nn.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony G.D., Lee E., Park P.J., Walsh C.A. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016;5:e12966. doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N.V. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Folstein S.E. Johns Hopkins University Press; 1989. Huntington’s Disease: A Disorder of Families, the Johns Hopkins Series in Contemporary Medicine and Public Health. [Google Scholar]
- Frost B., Hemberg M., Lewis J., Feany M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., Varshney A., Fowler B.J., Wang S., Narendran S., Ambati K., Yasuma T., Magagnoli J., Leung H., Hirahara S., et al. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2022751118. e2022751118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner E.J., Lam V.K., Harris D.N., Chuang N.T., Scott E.C., Pittard W.S., Mills R.E., 1000 Genomes Project Consortium. Devine S.E. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27(11):1916–1929. doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M.G., Bonaccorsi S., Kurek R., Farkas R.M., Dimitri P., Fuller M.T., Gatti M. The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 2006;16(2):195–201. doi: 10.1016/j.cub.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Goic B., Vodovar N., Mondotte J.A., Monot C., Frangeul L., Blanc H., Gausson V., Vera-Otarola J., Cristofari G., Saleh M.-C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- Green C.L., Lamming D.W., Fontana L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2021 doi: 10.1038/s41580-021-00411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães N.N., de Castro Pereira K., de Andrade H.H.R., Lehmann M., Silva Cunha K. Comparative analysis of genetic toxicity of AZT and ddI antiretrovirals in somatic cells of Drosophila melanogaster. Environ. Mol. Mutagen. 2008;49:312–317. doi: 10.1002/em.20389. [DOI] [PubMed] [Google Scholar]
- Guo C., Jeong H.H., Hsieh Y.C., Klein H.U., Bennett D.A., De Jager P.L., Liu Z., Shulman J.M. Tau activates transposable elements in Alzheimer’s disease. Cell Rep. 2018;23:2874–2880. doi: 10.1016/j.celrep.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella J.F., MacDonald M.E., Lee J.-M. Genetic modifiers of Huntington’s disease. Mov. Disord. 2014;29:1359–1365. doi: 10.1002/mds.26001. [DOI] [PubMed] [Google Scholar]
- Iyer J., Wang Q., Le T., Pizzo L., Grönke S., Ambegaokar S.S., Imai Y., Srivastava A., Troisí B.L., Mardon G., et al. Quantitative assessment of eye phenotypes for functional genetic studies using Drosophila melanogaster. G3. 2016;6:1427–1437. doi: 10.1534/g3.116.027060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz J.W., Bing X., Pontabry J., Bošković A., Rando O.J., Torres-Padilla M.-E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017;49:1502–1510. doi: 10.1038/ng.3945. [DOI] [PubMed] [Google Scholar]
- Jacob-Hirsch J., Eyal E., Knisbacher B.A., Roth J., Cesarkas K., Dor C., Farage-Barhom S., Kunik V., Simon A.J., Gal M., et al. Whole-genome sequencing reveals principles of brain retrotransposition in neurodevelopmental disorders. Cell Res. 2018;28:187–203. doi: 10.1038/cr.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C., Elgin S.C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986;6:3862–3872. doi: 10.1128/MCB.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Flynt A.S., Lai E.C. Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr. Biol. 2013;23:1442–1448. doi: 10.1016/j.cub.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov M.S., Sokolova O.A., Yakushev E.Y., Stolyarenko A.D., Mikhaleva E.A., Lavrov S.A., Gvozdev V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. U S A. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug L., Chatterjee N., Borges-Monroy R., Hearn S., Liao W.-W., Morrill K., Prazak L., Rozhkov N., Theodorou D., Hammell M., Dubnau J. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 2017;13:e1006635. doi: 10.1371/journal.pgen.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-M., Correia K., Loupe J., Kim K.-H., Barker D., Hong E.P., Chao M.J., Long J.D., Lucente D., Vonsattel J.P.G., et al. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell. 2019;178:887–900.e14. doi: 10.1016/J.CELL.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Jin Y., Prazak L., Hammell M., Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Lee M.-H., Henderson L., Tyagi R., Bachani M., Steiner J., Campanac E., Hoffman D.A., von Geldern G., Johnson K., et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015;7:307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E.Y., Russ J., Cali C.P., Phan J.M., Amlie-Wolf A., Lee E.B. Loss of nuclear TDP-43 is associated with decondensation of LINE retrotransposons. Cell Rep. 2019;27:1409–1421.e6. doi: 10.1016/j.celrep.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Dou X., Chen C., Chen C., Liu C., Xu M.M., Zhao S., Shen B., Gao Y., Han D., He C. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J., Clark A.G. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–227. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.Y., Bishop C.P., Eissenberg J.C. Developmental timing and tissue specificity of heterochromatin-mediated silencing. EMBO J. 1996;15:1323–1332. [PMC free article] [PubMed] [Google Scholar]
- McCloy R.A., Rogers S., Caldon C.E., Lorca T., Castro A., Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M.J., Moran J.V., Abyzov A., Akbarian S., Bae T., Cortes-Ciriano I., Erwin J.A., Fasching L., Flasch D.A., Freed D., et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: the Brain Somatic Mosaicism Network. Science. 2017;356:eaal1641. doi: 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A.R., Marchetto M.C.N., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoletano F., Ferrari Bravo G., Voto I.A.P., Santin A., Celora L., Campaner E., Dezi C., Bertossi A., Valentino E., Santorsola M., et al. The prolyl-isomerase PIN1 is essential for nuclear Lamin-B structure and function and protects heterochromatin under mechanical stress. Cell Rep. 2021;36:109694. doi: 10.1016/j.celrep.2021.109694. [DOI] [PubMed] [Google Scholar]
- Nasir J., Floresco S.B., Kusky J.R.O., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Ozata D.M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Padeken J., Mendiburo M.J., Chlamydas S., Schwarz H.-J., Kremmer E., Heun P. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol. Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Papp B., Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C.-J., Yin Y., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405.e19. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron H., Garson J.A., Bedin F., Beseme F., Paranhos-Baccala G., Komurian-Pradel F., Mallet F., Tuke P.W., Voisset C., Blond J.L., et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc. Natl. Acad. Sci. U S A. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Pimpinelli S. Positive regulation of euchromatic gene expression by HP1. Fly (Austin) 2010 doi: 10.4161/fly.4.4.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S., Bonaccorsi S., Fanti L., Gatti M. In: Drosophila Protocols. Sullivan W., Ashburner M., Hawley S., editors. Cold Spring Harbor Laboratory Press; 2000. Preparation and analysis of drosophila mitotic chromosomes; pp. 1–24. [Google Scholar]
- Preall J.B., Czech B., Guzzardo P.M., Muerdter F., Hannon G.J. Shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M., Gonzales P.K., Cook C.N., Gendron T.F., Daughrity L.M., Song Y., Ebbert M.T.W., van Blitterswijk M., Zhang Y.-J., Jansen-West K., et al. Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum. Mol. Genet. 2017;26:3421–3431. doi: 10.1093/hmg/ddx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M.N., Luo L., Potter C.J. Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods. 2015;12:219–222. doi: 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S., White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev. Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-X. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin B., Alvarez E.G., Baez-Ortega A., Zamora J., Supek F., Demeulemeester J., Santamarina M., Ju Y.S., Temes J., Garcia-Souto D., et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020;52:306–319. doi: 10.1038/s41588-019-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G., Klima R., Feiguin F. TDP-43 prevents retrotransposon activation in the Drosophila motor system through regulation of Dicer-2 activity. BMC Biol. 2020;18:82. doi: 10.1186/s12915-020-00816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero E., Cha G.H., Verstreken P., Ly C.V., Hughes R.E., Bellen H.J., Botas J. Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length Huntingtin accumulating in the cytoplasm. Neuron. 2008;57:27–40. doi: 10.1016/j.neuron.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., Tabrizi S.J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. In: Misener S., Krawetz S.A., editors. Humana Press; 1999. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. (BT - Bioinformatics Methods and Protocols). [DOI] [Google Scholar]
- Ruzo A., Croft G.F., Metzger J.J., Galgoczi S., Gerber L.J., Pellegrini C., Wang H., Fenner M., Tse S., Marks A., et al. Chromosomal instability during neurogenesis in Huntington’s disease. Development. 2018;145:dev156844. doi: 10.1242/dev.156844. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Reuter G. Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol Gen Genet MGG. 2000 doi: 10.1007/pl00008659. [DOI] [PubMed] [Google Scholar]
- Seong I.S., Woda J.M., Song J.-J., Lloret A., Abeyrathne P.D., Woo C.J., Gregory G., Lee J.-M., Wheeler V.C., Walz T., et al. Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 2010;19:573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.U., Gerasimova T., Kurkulos M., Boeke J.D., Corces V.G. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Startek M., Szafranski P., Gambin T., Campbell I.M., Hixson P., Shaw C.A., Stankiewicz P., Gambin A. Genome-wide analyses of LINE–LINE-mediated nonallelic homologous recombination. Nucleic Acids Res. 2015;43:2188–2198. doi: 10.1093/nar/gku1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Samimi H., Gamez M., Zare H., Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018;21:1038–1048. doi: 10.1038/s41593-018-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam O.H., Ostrow L.W., Gale Hammell M. Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob. DNA. 2019;10:32. doi: 10.1186/s13100-019-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Sone M., Yamashita M., Wanker E.E., Okazawa H. Glial cell lineage expression of mutant ataxin-1 and Huntingtin induces developmental and late-onset neuronal pathologies in Drosophila models. PLoS One. 2009;4:e4262. doi: 10.1371/journal.pone.0004262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- Thomas C.A., Tejwani L., Trujillo C.A., Negraes P.D., Herai R.H., Mesci P., Macia A., Crow Y.J., Muotri A.R. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell. 2017;21:319–331.e8. doi: 10.1016/j.stem.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton K.R., Gerhardt D.J., Jesuadian J.S., Richardson S.R., Sánchez-Luque F.J., Bodea G.O., Ewing A.D., Salvador-Palomeque C., van der Knaap M.S., Brennan P.M., et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Broeck L., Naval-sánchez M., Adachi Y., Diaper D., Dourlen P., Chapuis J., Kleinberger G., Gistelinck M., van Broeckhoven C., Lambert J.-C., et al. TDP-43 loss-of-function causes neuronal loss due to defective steroid receptor-mediated gene program switching in Drosophila. Cell Rep. 2013;3:160–172. doi: 10.1016/j.celrep.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wakimoto B.T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wang S.H., Elgin S.C.R. Drosophila piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. U S A. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dou K., Moon S., Tan F.J., Zhang Z.Z. Hijacking oogenesis enables massive propagation of LINE and retroviral transposons. Cell. 2018;174:1082–1094. doi: 10.1016/j.cell.2018.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.G., Jones B.C., Jiang N., Chang C., Hosier S., Wickremesinghe P., Garcia M., Hartnett D.A., Burhenn L., Neretti N., Helfand S.L. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. U S A. 2016;113:11277–11282. doi: 10.1073/pnas.1604621113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.S., Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 2006;1:2110. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Whole genome sequencing (WGS) data from control and HD flies at larval stage, 0–2 d- and 10–12 d-aged have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB41409 (https://www.ebi.ac.uk/ena/browser/view/PRJEB41409).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.