Highlights
-
•
UCA1 acts as a binding partner of heterogeneous nuclear ribonucleoproteins (hnRNPs) I and L in bladder cancer.
-
•
UCA1 and hnRNP I/L have profound effects on metabolic reprogramming and proliferation in bladder cancer.
-
•
UCA1 specifically binds to and facilitates the combination of hnRNP I/L to the promoter of GPT2 and increases GPT2 expression.
-
•
UCA1-hnRNP I/L-GPT2 axis is critical for glutamine-driven anaplerosis in bladder cancer.
Keywords: LncRNA UCA1, HnRNP, Glutamine-driven anaplerosis, Metabolism, Bladder cancer
Abstract
Long noncoding RNA urothelial cancer associated 1 (UCA1), initially identified in bladder cancer, is associated with multiple cellular processes, including metabolic reprogramming. However, its characteristics in the anaplerosis context of bladder cancer (BLCA) remain elusive. We identified UCA1 as a binding partner of heterogeneous nuclear ribonucleoproteins (hnRNPs) I and L, RNA-binding proteins (RBPs) with no previously known role in metabolic reprogramming. UCA1 and hnRNP I/L profoundly affected glycolysis, TCA cycle, glutaminolysis, and proliferation of BLCA. Importantly, UCA1 specifically bound to and facilitated the combination of hnRNP I/L to the promoter of glutamic pyruvate transaminase 2 (GPT2), an enzyme transferring glutamate to α-ketoglutarate, resulting in upregulated expression of GPT2 and enhanced glutamine-derived carbons in the TCA cycle. We also systematically confirmed the influence of UCA1 and hnRNP I/L on metabolism and proliferation via glutamine-driven anaplerosis in BLCA. Our study revealed the critical role of UCA1-mediated mechanisms involved in glutamine-driven anaplerosis and provided novel evidence that lncRNA regulates metabolic reprogramming in tumor cells.
Introduction
Cancer cells reprogram their metabolism to fulfill anabolic and energetic requirements for survival and high proliferation. Enhanced uptake and utilization of glucose and glutamine is a prominent feature of cancer cells. Through aerobic glycolysis, or the Warburg effect, glucose is metabolized to lactate to generate energy rather than to enter the tricarboxylic acid (TCA) cycle [1], [2], [3], [4]. In addition to glycolysis, glutamine metabolism, or glutaminolysis, is another central route for anabolic metabolism and energy production. In cancer cells, the intermediates of the TCA cycle are also used as precursors for the synthesis of proteins, lipids, nucleic acids, and other biomolecules that are crucial to energy production and biosynthesis. The pool of intermediates in the TCA cycle must be replenished to maintain the TCA cycle function and balance the influx of metabolites, which is called anaplerosis [5,6].
Glutamine-driven anaplerosis is the most common way to fuel the TCA cycle in rapidly proliferative cells [7]. As the most abundant amino acid in the mammalian body, glutamine is an essential source that provides proliferating cells with carbon and nitrogen for biosynthesis [8], [9], [10]. Glutamine is introduced into the cell by glutamine transporters, such as solute carrier family 1, member 5 (SLC1A5) and solute carrier family 7, member 5 (SLC7A5). Through glutaminolysis, glutaminase (GLS) hydrolyzes glutamine to glutamate, which is then converted to α-Ketoglutarate (α-KG) with the aid of glutamate oxaloacetate transaminase (GOT) or glutamate pyruvate transaminase (GPT) or glutamate dehydrogenase (GLDH) [10], [11], [12]. Importantly, glutamine-derived α-KG serves as a critical anaplerotic substrate and carbon source for the TCA cycle in many cancers cells when TCA intermediates are used for biosynthetic precursors [13]. Glutaminolysis involved enzymes and glutamine transporters have been found dysregulated in cancers [14,15].
Long non-coding RNAs (lncRNAs), a group of endogenous RNAs longer than 200 nt and lack protein-coding ability, can affect genes involved in cancer metabolism [16,17]. LncRNA urothelial carcinoma associated 1 (UCA1), initially identified and highly upregulated in bladder cancer, is believed to function in tumorigenesis, development, as well as the metabolism of bladder cancer [16,18]. However, the exact mechanism by which UCA1 promotes metabolic reprogramming in bladder cancer cells remains unclear. For the tumorigenicity of UCA1, one mechanism worth mentioning is its capability of forming complexes with RNA-binding proteins (RBPs), including heterogeneous nuclear ribonucleoproteins (hnRNPs) which represent a large family of RBPs. HnRNPs assist in multiple cellular processes, such as alternative splicing, mRNA stabilization, and translation. Multiple hnRNPs play a crucial role in tumorigenesis, and their expression level is altered in various cancers [19,20]. HnRNP I and L are striking among the hnRNPs family. HnRNP I is involved in regulating metabolic shift from oxidative phosphorylation to anaerobic glycolysis in many cancers [21], [22], [23]. Huang et al. [24] reported hnRNP I can interact with 19 lncRNAs in breast cancer cells, among which UCA1 is on the top of the list in terms of enrichment. The interaction of UCA1 with hnRNP I suppresses the p27 and regulates breast cancer cell growth. The RNA-binding capacity of hnRNPs is mediated by RNA recognition motifs (RRMs). All four RRMs were found with high sequence similarity and shared between hnRNP I and L [19,25]. Therefore, it is likely that hnRNP I and L can interact with UCA1. But it is still unclear whether UCA1 binds to hnRNP I/L and how they play a part in bladder cancer metabolism.
In this study, we demonstrated that UCA1, hnRNP I/L, and GPT2 were markedly overexpressed in bladder cancer tissues and cell lines and profoundly impact bladder cancer metabolic reprogramming. Importantly, UCA1 bound specifically to hnRNP I/L and formed a functional UCA1-hnRNP I/L complex that upregulated GPT2 expression by binding to its promoter and promoted glutamine-driven TCA anaplerosis.
Materials and methods
Clinical samples and study approval
Tissues were collected from patients who underwent surgery at the First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, China. All patients were not treated with chemotherapy or radiotherapy before surgery. All samples were collected and used following the ethical policies of the institutional review board of the First Affiliated Hospital of Xi'an Jiaotong University. The clinical characteristics of bladder cancer patients are shown in Table 1.
Table 1.
Characteristics of bladder cancer patients.
| Characteristic | Patient frequency |
|---|---|
| Total | 43 |
| Gender | |
| Male | 35 (81.4%) |
| Female | 8 (18.6%) |
| Age | 66 (34–85) |
| <66 | 21 (48.8%) |
| ≥66 | 22 (51.2%) |
| Tumor stage | |
| T1 | 14 (32.6%) |
| T2-T4 | 29 (67.4%) |
| TNM stage | |
| I | 14 (32.6%) |
| II | 14 (32.6%) |
| III | 7 (16.3%) |
| IV | 8 (18.6%) |
| Grade (WHO2004) | |
| Low malignant potential | 8 (18.6%) |
| Low-grade | 19 (44.2%) |
| High-grade | 16 (37.2%) |
| Histopathological subtype | |
| Urothelial carcinoma | 43 (100%) |
Cell culture
Cell lines 5637 (HTB-9), T24 (HTB-4), UMUC2 (CRL-1748), and SV-HUC-1(CRL-9520) were purchased from ATCC and maintained in RPMI 1640 (containing 10% fetal bovine serum) unless otherwise stated. Stable cell lines with ectopic expression of UCA1, hnRNP I, and hnRNP L in UMUC2 cells, and cell lines with knockdown of UCA1, hnRNP I, hnRNP L, and GPT2 in 5637 cells were constructed and selected in our laboratory. All cell lines were verified by short tandem repeat analysis in the year 2019.
Lentivirus transduction
To establish 5637 cells stably expressing shRNAs against UCA1, hnRNP I, or hnRNP L, and UMUC2 cells ectopically expressing UCA1, hnRNP I, or hnRNP L, the pre-packaged lentivirus expressing pLKO.1-Puro-shRNA or pLV-EF1a-Puro-cDNA fragments of UCA1, hnRNP I, hnRNP L, and GPT2 were purchased from GenePharma (Shanghai, China) and used to infect cells following the manufacturer's instructions. In brief, cells were transduced with 10 MOI of lentivirus expressing shRNAs or genes and selected by 2 μg/ml puromycin for 2 days after infection. The transduction efficiency was validated by western blot analyses. Stably infected cells were maintained in 1 μg/ml puromycin. Cells infected with lentivirus containing non-sense shRNA or served as the control group.
Western blotting
Cells were lysed using RIPA buffer containing 1 mM PMSF and 1% complete protease inhibitor (Roche Applied Science; Indianapolis, IN, USA) to extract the proteins. The protein concentration was detected using BCA protein Assay Kit (Thermo Pierce). The proteins were subjected to SDS-PAGE and transferred to NC membrane (Pall Life Science; Port Washington, NY, USA). After blocking with 5% skim milk, NC membrane was incubated overnight at 4 °C with different primary antibodies: anti-hnRNP I (1:1000; Santa Cruz Biotechnology, Inc; CA, USA); anti-hnRNP L (1:1000; Santa Cruz Biotechnology, Inc; CA, USA); anti-GLS2 (1:1000; Santa Cruz Biotechnology, Inc; CA, USA); anti-GPT2 (1:1000; Abbacy, Cambridge, UK); anti-SLC1A5 (1:500; Santa Cruz Biotechnology, Inc; CA, USA); anti-SLC7A5 (1:500; Santa Cruz Biotechnology, Inc; CA, USA); anti-β actin (1:3000; CST; Beverly, MA, USA). After being washed, the membranes were incubated with HRP-conjugated goat-anti-mouse secondary antibodies (1:5000; Pierce; Rockford, IL, USA) for 1 hr at room temperature and visualized using Clarity™ Western ECL Substrate (Bio-Rad; Richmond, CA, USA).
Gene expression analysis
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) or RNAFast200 (Fastagen, Shanghai, China) according to the manufacturer's protocol. 1 μg of total RNA was reverse transcribed to cDNA using RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, Waltham, MA, USA). We performed qRT-PCR following the instructions of PrimeScript RT Master Mix Kit and SYBR Premix Ex Taq Kit (Takara, Clontech, Kyoto, Japan) with CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the thermal cycling conditions: 95 °C for 30 s, 40x (95 °C for 5 s, 60 °C for 30 s, 72 °C for 40 s with plate read), melt curve 65 to 95 °C, increment 0.5 °C, 5 s with plate read. The sequences of primers used for qPCR are listed in Table 2.
Table 2.
Oligonucleotides sequences (5′->3′).
| qRCR primers (5′->3′) | |||||
| UCA1 | Forward | CTCTCCATTGGGTTCACCATTC | |||
| Reverse | GCGGCAGGTCTTAAGAGATGAG | ||||
| hnRNP I | Forward | AATGACAAGAGCCGTGACTAC | |||
| Reverse | GGAAACCAGCTCCTGCATAC | ||||
| hnRNP L | Forward | TTGTGGCCCTGTCCAGAGAATT | |||
| Reverse | GTTTGTGTAGTCCCAAGTATCCTG | ||||
| SLC1A5 | Forward | GACCGTACGGAGTCGAGAAG | |||
| Reverse | GGGGGTTTCCTTCCTCAGTG | ||||
| SLC7A5 | Forward | GAAGGCACCAAACTGGATGT | |||
| Reverse | GAAGTAGGCCAGGTTGGTCA | ||||
| GLS2 | Forward | TGCCTATAGTGGCGATGTCTCA | |||
| Reverse | GTTCCATATCCATGGCTGACAA | ||||
| GPT2 | Forward | GGAGCTAGTGACGG CATTTCTACGA | |||
| GPT2 promotor | Reverse | CCCAGGGTTGATTATGCAGAGCA | |||
| Forward | CCTCCCCTGTCCCTACTGAT | ||||
| Reverse | ATGTCCATGCAGTCCCTTGC | ||||
| β-actin | Forward | TCCCTGGAGAAGAGCTACGA | |||
| Reverse | AGCACTGTGTTGGCGTACAG | ||||
| GAPDH | Forward | GTCGGAGTCAACGGATTTG | |||
| Reverse | TGGGTGGAATCATATTGGAA | ||||
| shRNA | |||||
| UCA1 | CCGGGTTAATCCAGGAGACAAAGACTCGAGTCTTTGTCTCCTGGATTAACTTTTTG | ||||
| hnRNP I | CCGGGCGTGAAGATCCTGTTCAATACTCGAGTATTGAACAGGATCTTCACGCTTTTTG | ||||
| hnRNP L | CCGGCCTCAACAACAACTTCATGTTCTCGAGAACATGAAGTTGTTGTTGAGGTTTTTG | ||||
| GPT2 | CCGGGACAACGTGTACTCTCCAGATCTCGAGATCTGGAGAGTACACGTTGTCTTTTTG | ||||
| Control | CCGGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAATTTTTG | ||||
| Anti-sense probes for selective retrieval of UCA1 by CHIRP | |||||
| Probe 1 | aagggttgtaggttgtttgt | ||||
| Probe 2 | cggcaggtcttaagagatga | ||||
| Probe 3 | gttttagacttttgacccag | ||||
| Probe 4 | atatgcgtgtactgttgtcc | ||||
| Probe 5 | ccaagccctctaacaacaaa | ||||
| Probe 6 | gcagatcctatgcagaagag | ||||
| Probe 7 | aatagtattccctgttgcta | ||||
| Probe 8 | aatgtaggtggcgatgagtt | ||||
Cell proliferation assay
Cells (3.5 × 103 per well) were seeded in triplicate in 96-well plates to measure cell proliferation by Cell Count Kit (7Sea, Shanghai, China). After 24, 48, 72, and 96 h, 10 μl CCK-8 was added to each well, and plates were shaken for 5 times, incubated for an additional 50 min, and measured by reading OD450 nm using an EnSpire Reader (PerkinElmer, USA). To further assess the effect of permeable dimethyl-αKG (DM-αKG), citrate, malate, and alanine, shUCA1, shhnRNP I, shhnRNP L, and shGPT2 cells (3.5 × 103 per well) were seeded in triplicate in 96-well plates and grown in 10% FBS-RPMI 1640 supplemented with 1 mM DM-αKG, 1 mM citrate, 1 mM malate, and 1 mM alanine respectively. After 24, 48, 72, and 96 h, cell proliferation was detected by CCK-8 assay.
Dual-luciferase reporter assay
Luciferase assays were performed using the Dual-Luciferase™ Reporter Assay System (Promega; Madison, WI, USA) according to the manufacturer's protocol. HnRNP I/L overexpressing cells and control cells were transfected with 0.1 μg reporter plasmids (pGL3-GLS2 promoter or pGL3-GPT2 promoter) and 0.01 μg internal control (Renilla plasmid) in 96-well plates utilizing X-treme GENE siRNA Transfection Reagent (Roche, Indianapolis, IN, USA). Cells were harvested and lysed for dual-luciferase reporter assay 48 h after transfection. Renilla luciferase plasmid served as normalization.
Immunohistochemistry
Immunohistochemical analysis for the protein expression was performed as previously described [26] using anti-hnRNP I (1:500; Santa Cruz Biotechnology, Inc; CA, USA) anti-hnRNP L (1:500; Santa Cruz Biotechnology, Inc; CA, USA) GPT2 (1:300; Abcam, Cambridge, MA, USA). Slides were incubated overnight at 4 °C, and immunostaining was performed with a SPlink Detection Kit and DAB (ZSBIO, Beijing, China). Stained slides were captured and imaged by microscopy. The image quantification was analyzed using ImageJ (1.52q) with the IHC Profiler plugin [27]. The staining intensity was graded as negative (0), low positive (1+), positive (2+), or high positive (3+). H score was calculated by the following formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)].
RNA pull-down
RNA pull-down assays were performed using Pierce™ Magnetic RNA-Protein Pull-Down Kit (#20,164, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacture's protocol. The RNA fragments of the entire UCA1 sequence, deleted form, or mutant form, were synthesized by GenePharma (Shanghai, China). Biotinylated RNA was bound to streptavidin magnetic beads and then incubated with cell lysates. RNA-binding protein complexes were washed and eluted. The retrieved samples were heated (10 min, 95 °C) and detected by western blot analysis.
Chromatin immunoprecipitation assay (ChIP)
ChIP experiments were performed using EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (#17–10,086, Merck Millipore, Billerica, MA, USA) according to the manufacturer's procedures. Cells were fixed in 1% formaldehyde for 10 min at room temperature to crosslink UCA1 to DNA. Nuclei were isolated with 500 μl nuclear lysis buffer supplemented with 2.5 μl protease inhibitor cocktail II. Chromatin DNA was sonicated (8 min total, AmpL 30%, pulse on 10 s, pulse off 30 s) and sheared to a length between 200 bp to 1000 bp. The sheared cross-linked chromatin was incubated and rotated with immunoprecipitating antibodies: anti-hnRNP I (1.2 μg/reaction; Santa Cruz Biotechnology, Inc; CA, USA)), anti-hnRNP L (1 μg/reaction; Santa Cruz Biotechnology, Inc; CA, USA), normal Mouse IgG (1 μg/reaction) or anti-RNA Polymerase (1 μg/reaction) overnight at 4 °C. Primers for ChIP-qPCR are listed in Table 2.
Chromatin isolation by RNA purification (ChIRP)
ChIRP was performed using EZ-Magna CHIRP™ RNA Interactome Kit (#17–10,495, Merck Millipore, Billerica, MA, USA) as previously described [28]. In brief, we designed 8 anti-sense oligo probes which enable affinity capture of UCA1-chromatin complex for selective retrieval of lncRNA UCA1 target by ChIRP at singlemoleculefish.com. Biotin-labeled oligo probes against UCA1 were synthesized by GenePharma (Shanghai, China). Eight oligo probes were diluted to 100 μM and combined as a pool. Cells were crosslinked in 1% formaldehyde at room temperature for 10 min. Cross-linked cell lysates were sonicated (15 min total, AmpL 30%, pulse on 10 s, pulse off 30 s) and centrifuge sonicated samples at 16,100 g for 10 min at 4 °C. Supernatants were aliquoted into 1 mL samples and hybridized with 100 pmol of the pool of biotinylated probes per mL. Retrieved DNA was quantified with SYBR Green PCR Kit (Takara, Clontech, Kyoto, Japan) to measure the enrichment of GPT2 promoter. The probes and primers sequences used in the ChIRP assay are listed in Table 2.
Metabolomics and metabolic flux analysis with mass spectrometry
Cells (1 × 106) were plated on 100 mm dishes in RPMI-based media for 6 hr. Withdrawn the medium from dishes, washed with PBS × 2, and changed to tracing media for an additional 24 h. The media were transferred to pre-chilled Eppendorf tubes. Supernatant samples were isolated after centrifugation at 1000 rpm for 5 min and frozen at −80 °C until analysis. Cells were transferred to pre-chilled Eppendorf tubes, washed with cold PBS × 2, pelleted by centrifugation, and flash frozen in liquid nitrogen. Before metabolite derivatization, media samples were dried under nitrogen gas and frozen at −80 °C. Cells were resuspended in 600 μl cold (−40 °C) 50% aqueous methanol, inserted in dry ice for 30 min, and thawed samples at 4 °C. Then added 400 μl chloroform, vortexed for 30 s before centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant was transferred to new Eppendorf tubes, dried, and stored at −80 °C. For xenografts, 800 µl of ice-cold chloroform-methanol-water (2:5:2, v/v/v) were added to the 40-mg tissue of each sample, and the mixture was homogenized in an ice-water bath. The samples were subsequently centrifuged at 14,000 g for 15 min at 4 °C, and the supernatant was collected. Following the addition of 600 μl cold (−40 °C) 50% aqueous methanol, the same steps for cell sample preparation were conducted.
Metabolomics profiling and metabolic flux analysis were performed on Shimadzu QP-2010 Ultra GC–MS with an injection temperature of 250 °C and injected volume of 1 µL. GC oven temperature started at 110 °C for 4 min, rising to 230 °C at 3 °C/min and to 280 °C at 20 °C /min with a final hold at this temperature for 2 min. GC flow rate with helium carrier gas was 50 cm/s. The GC column used was a 20 m x 0.25 mm x 0.25 mm Rxi-5 ms. GC–MS interface temperature was 300 °C and (electron impact) ion source temperature was set at 200 °C, with 70 V ionization voltages. The mass spectrometer was set to scan m/z range 50–800, with 1 kV detector. GC–MS data were analyzed to determine isotope labeling and quantities of metabolites. Metabolites with baseline separated peaks were quantified based on total ion count peak area, using standard curves generated from running standards in the same batch of samples. To determine 13C labeling, the mass distribution for known fragments of metabolites was extracted from the appropriate chromatographic peak. These fragments contained either the whole carbon skeleton of the metabolite, or lacked the alpha carboxyl carbon, or (for some amino acids) contained only the backbone minus the side-chain [29]. For each fragment, the retrieved data comprised mass intensities for the lightest isotopomer. These mass distributions were normalized and corrected for the natural abundance of heavy isotopes of the elements H, N, O, Si and C, using matrix-based probabilistic methods as described [30], and implemented in MATLAB. Labeling results were expressed as fractions of the particular compound containing isotopic labels from the particular precursor.
Xenograft assays in nude mice
All xenograft experiments have been approved by the Animal Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University. Twenty-five 5-weeks-old nude mice (female) were randomly divided into 5 groups (5 mice per group) and subcutaneously injected with 2 × 106 shCtrl, shUCA1, shhnRNP I, or shhnRNP L, 5637 cells. Cells were suspended in 100 μl PBS and kept on ice until injection. Xenograft size and mice weight were measured every 4 days until 30 days after injection. The tumor volume (V) was monitored by measuring the length (L) and width (W) with a caliper and calculated with the formula V = 0.5236 × (L × W2). Mice were sacrificed on the 30th day of the experiment, and the tumor samples were excised for analysis. The shRNA effects were validated by qRT-PCR or IHC and the levels of metabolites were determined by GC–MS analysis.
Statistical analysis
Data are presented as means ± SD and are representative of at least three independent experiments. Differences between two groups were analyzed with the unpaired/paired Student's t-test using GraphPad Prism 8 software. All reported differences are P < 0.05 unless otherwise stated. Details of each specific statistical analysis are indicated in the figure legends.
Results
High expression of UCA1 and hnRNP I/L in BLCA
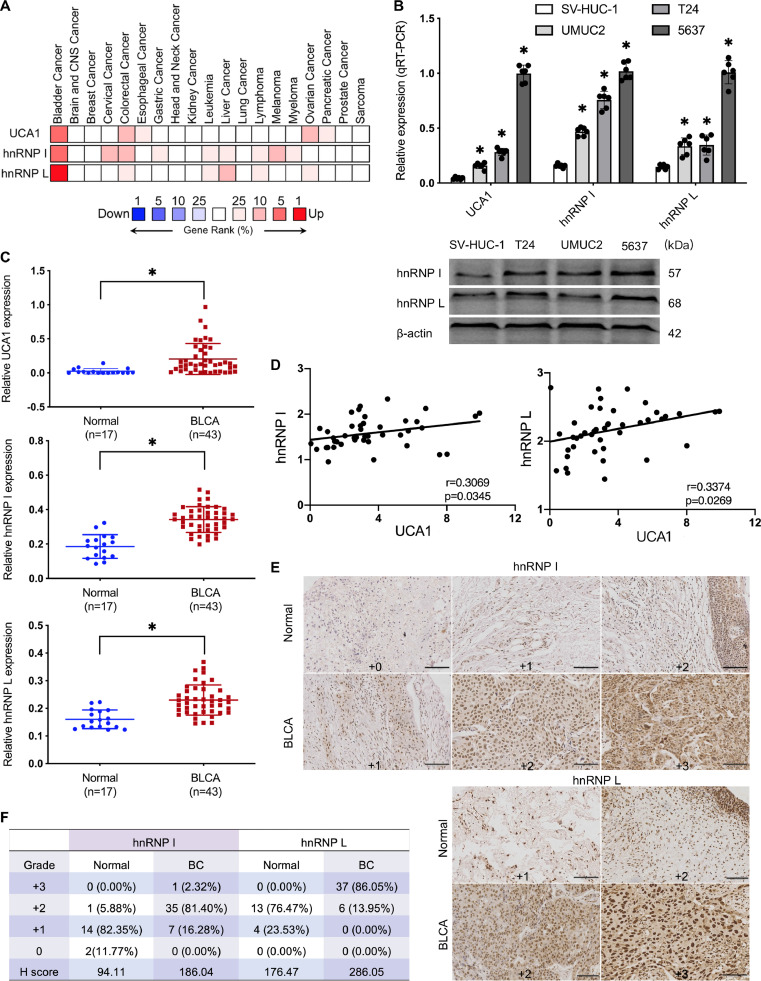
To investigate the expression profiles of UCA1 and hnRNP I/L in human cancers, we analyzed the available gene expression dataset, Oncomine, and compared the expression of UCA1 and hnRNP I/L across multi-cancer analyses. Rank for UCA1 and hnRNP I was in the top 5%, and hnRNP L was in the top 1%. Oncomine analysis showed that UCA1 and hnRNP I/L were more highly expressed genes in BLCA than other cancers (Fig. 1A). We further detected UCA1 and hnRNP I/L expression levels in a human uroepithelium cell line (SV-HUC-1) and three typical BLCA cells (UMUC2, T24, 5637) by qRT-PCR and western blot. We found hnRNP I/L expression was relatively higher in UCA1-upregulated BLCA cells (Fig. 1B). Additionally, we performed qRT-PCR assays and immunohistochemical staining (IHC) in clinical tissue samples. UCA1 and hnRNP I/L expression in BLCA tissues were higher than normal tumor-adjacent bladder tissues (Fig. 1C). Additionally, Pearson's correlations of UCA1 and hnRNP I/L showed a positive correlation between UCA1 and hnRNP I/L (Fig. 1D). IHC results further validated the upregulation of hnRNP I/L in BLCA (Fig. 1E and F).
Fig. 1.
Expression of UCA1 and hnRNP I/L in BLCA. A, High expression of UCA1 and hnRNP I/ L in BLCA. Oncomine analysis of UCA1 and hnRNP I/L across multi-cancer analyses (www.oncomine.org). The rank for a gene is the median rank for that gene across each of the analyses. P 〈 0.0001; Fold change 〉 2; 10% gene rank. B, High expression of UCA1 and hnRNP I/L in BLCA cells. One human uroepithelium cell and three BLCA cells were measured by qRT-PCR and western blot. n = 6 independent experiments. C to F, High expression of UCA1 and hnRNP I/L in BLCA tissues: qRT-PCR of UCA1 and hnRNP I/L (C) was performed using normal tumor-adjacent bladder tissues (n = 17) and BLCA tissues (n = 43), and Pearson's correlations of UCA1 and hnRNP I/L (D) were calculated; IHC of hnRNP I and hnRNP L (E) were performed using clinical tissues and H score (F) were determined. * P < 0.01. Scale bar=200 μm.
UCA1 interacts with hnRNP I/L
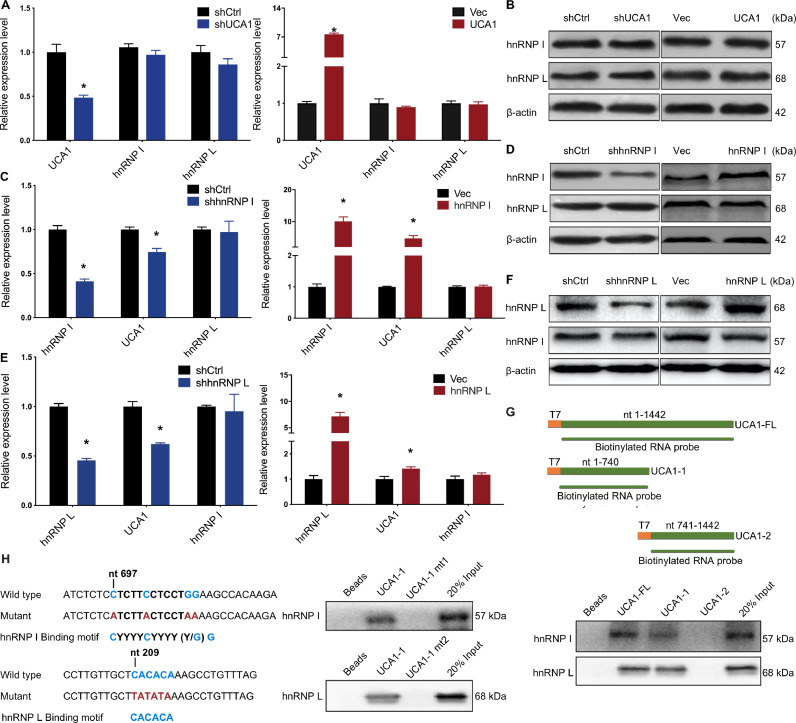
To further understand the relationship between UCA1 and hnRNP I/L, we first examined the effect of UCA1 knockdown and overexpression on hnRNP I/L. Results showed that hnRNP I/L is barely affected by UCA1 knockdown or overexpression (Fig. 2A and B). 5637 cells stably expressing shRNAs against hnRNP I showed the downregulation of UCA1. Conversely, ectopic expression of hnRNP I upregulated displayed the upregulation of UCA1 in UMUC2 cells (Fig. 2C). Similar results were observed for hnRNP L (Fig. 2E). Moreover, we examined whether the expression of hnRNP I or L could affect the other counterpart. 5637 cells stably expressing shhnRNP I showed no effect on the mRNA and protein expression of hnRNP L (Fig. 2C and D, left panel). HnRNP L expression was not affected by overexpression of hnRNP I in UMUC2 cells (Fig. 2C and D, right panel). Neither hnRNP I expression was affected by hnRNP L (Fig. 2E and F).
Fig. 2.
UCA1 is an hnRNP I/L binding partner. A, B, No effects of UCA1 on hnRNP I/L expression in BLCA cells. HnRNP I/L expression was analyzed in UCA1 knockdown and UCA1 overexpression BLCA cells by qRT-PCR (A) and western blot (B). C to F, Correlation between hnRNP I/L and UCA1 expression in BLCA cells. UCA1 expression was analyzed by qRT-PCR (C and E). The expression of hnRNP I/L was analyzed by qRT-PCR (C and E) and western blot (D and F). HnRNP I/L deletion decreases the expression of UCA1 in BLCA cells; Ectopic hnRNP I/L increases the expression of UCA1 in BLCA cells. The expression of hnRNP I or L do not affect the expression of the other counterpart. *P < 0.01. G, H, RNA pulldown assays validation of UCA1-hnRNP I/L binding regions in 5637 cells. Biotinylated full-length UCA1 (UCA1-FL) probe or truncated UCA1 probes (UCA1–1 and UCA1–2) were incubated with 5637 cell lysates. HnRNP I/L in samples pulled down by streptavidin beads were analyzed using western blot (G). Two forms of UCA1–1 mutated at the hnRNP I /L peak binding regions were described. HnRNP I/L in samples pulled down by wild type UCA1–1 and UCA1–1 mt probes labeled with biotin were analyzed by western blot (H).
HnRNP I and L are striking RBPs and have been reported to interact with lncRNAs. We then wonder whether UCA1 interacts with hnRNP I/L in BLCA. To verify this, we performed RNA pull down assays. Full-length UCA1 (1442 nt) and two truncated UCA1 (UCA1–1: 1–740 nt, UCA1–2: 741–1442 nt) were synthesized in vitro using T7 RNA polymerase and labeled with biotin. Those biotinylated RNA probes were used to pull down the 5637 cell lysates. Western blot analysis of hnRNP I/L in samples pulled down by biotinylated probes showed that the 5′ term of UCA1 was responsible for the binding with hnRNP I/L (Fig. 2G). Within the UCA1–1 region, the 697–708 nt sequence (5′-CTCTTCCTCCTGG-3′) is similar to hnRNP I binding motif CYYYYCYYY (Y/G) G, and the 209–214 nt sequence (5′-CACACA-3′) is similar to CA-enriched hnRNP L binding motif (Fig. 2H, left). Thus, we synthesized two mutant UCA1–1 probes (UCA1–1 mt1: mutant from 697 to 708 nt; UCA1–1 mt2: mutant 209–214 nt) to perform pull down assays. HnRNP I/L in samples pulled down by wild type UCA1–1 and mutant UCA1–1 probes were analyzed by western blot, and the results showed that the motifs were essential for binding hnRNP I and hnRNP L (Fig. 2H, right). These results confirmed that UCA1 could form an RNP complex with hnRNP I/L.
UCA1 and hnRNP I/L affect BLCA metabolic reprogramming
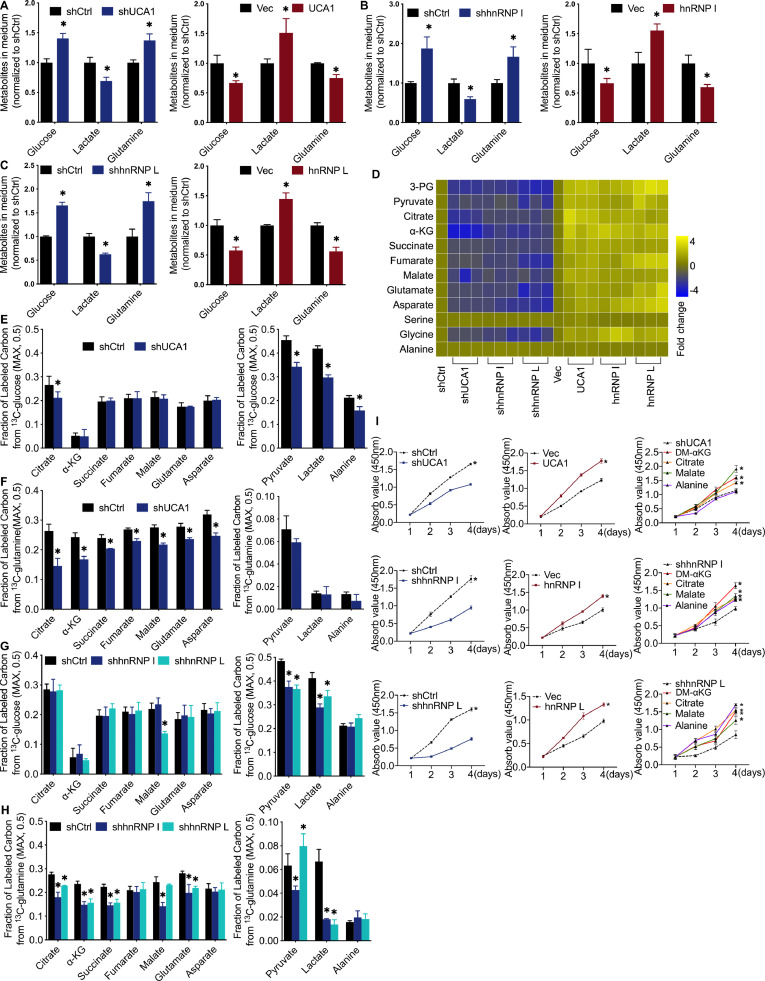
We were interested in investigating whether highly expressed UCA1 and hnRNP I/L may affect BLCA cell metabolism. Mass spectrometry (MS) analysis was utilized to measure multiple critical metabolites in the culture medium and cells. Increased glucose levels, decreased lactate levels, and increased glutamine levels were detected in the culture medium of UCA1 and hnRNP I/L knockdown 5637 cells (Fig. 3A–C, left). Moreover, decreased glucose levels, increased lactate levels, and decreased glutamine levels were detected in the medium of UCA1 and hnRNP I/L overexpression UMUC2 cells (Fig. 3A–C, right). This suggested that the consumption of glucose and glutamine and secretion of lactate were increased by upregulated UCA1 and hnRNP I/L. MS also measured twelve critical metabolites in UCA1 and hnRNP I/L knockdown cells. Among these were glycolysis intermediates (3-PG and pyruvate), TCA cycle intermediates (citrate, α-KG, succinate, fumarate, and malate), intermediates of glutamine catabolism (glutamate and aspartate), and glucogenic amino acid (glycine, alanine, and serine). Results showed that UCA1 and hnRNP I/L knockdown significantly reduced these intermediates except serine and alanine (Fig. 3D, left). In comparison, UCA1 and hnRNP I/L overexpression significantly increased these intermediates except serine and alanine (Fig. 3D, right).
Fig. 3.
The Effect of UCA1, hnRNP I, and hnRNP L on cell metabolism in vitro. A to C, Glucose uptake, lactate secretion, and glutamine uptake decreased by UCA1 or hnRNP I/L knockdown in BLCA cells. Cells were grown in the medium for 24 h, and levels of glucose, lactate, and glutamine in the medium were analyzed by MS and normalized to shCtrl group. D, Cellular metabolites levels examined by MS and normalized to shCtrl group (n = 3 independent experiments). E to H, Fraction of labeled carbon in metabolic intermediates tested by 13C glucose and 13C-glutamine tracing experiments. I, DM-αKG, citrate, or malate rescues 5637 cells from suppressed cell proliferation caused by shUCA1, shhnRNP I, and shhnRNP L. Cells were grown in medium with the presence of the indicated nutrient. CCK8 assay was performed to determine the cell proliferation at indicated time points. n = 3 independent samples. *P < 0.01 compared with shCtrl group.
To further explore the different metabolic responses to shUCA1, shhnRNP I, and shhnRNP L, we performed stable-isotope tracing experiments with glucose or glutamine labeled with 13C (at all carbon atoms). Results showed that UCA1 knockdown did not affect the fraction of 13C-glucose-derived carbon of α-KG, succinate, fumarate, malate, glutamate, and asparate (Fig. 3E, left) and the fraction of 13C-glutamine derived carbon of pyruvate, lactate, and alanine (Fig. 3F, right), but reduced the fraction of 13C-glutamine derived carbon of citrate, α-KG, succinate, fumarate, malate, glutamate, and asparate (Fig. 3F, left) and the fraction 13C-glucose-derived carbon of citrate, pyruvate, lactate, and alanine (Fig. 3E, right). HnRNP I knockdown did not affect the fraction of 13C-glucose-derived carbon of citrate, α-KG, succinate, fumarate, malate, glutamate, asparate, and alanine (Fig. 3G) and the fraction of 13C-glutamine derived carbon of fumarate, asparate, and alanine (Fig. 3H). However, it reduced the fraction of 13C-glutamine derived carbon of citrate, α-KG, succinate, malate, glutamate, pyruvate, and lactate (Fig. 3H) and the fraction of 13C-glucose-derived carbon of pyruvate and lactate (Fig. 3G). HnRNP L knockdown had no effect on the fraction of 13C-glucose-derived carbon of citrate, α-KG, succinate, fumarate, glutamate, asparate, and alanine (Fig. 3G) and the fraction of 13C-glutamine derived carbon of fumarate, malate, asparate, and alanine (Fig. 3H), reduced the fraction of 13C-glutamine derived carbon of citrate, α-KG, succinate, glutamate, and lactate (Fig. 3H) and the fraction of 13C-glucose-derived carbon of malate, pyruvate, and lactate (Fig. 3G), and increased the fraction of 13C-glutamine derived carbon of pyruvate (Fig. 3H).
Besides, we examined the effect of UCA1 and hnRNP I/L on cell proliferation. CCK8 assays revealed that UCA1 and hnRNP I/L downregulation significantly slowed cell proliferation (Fig. 3I, left), whereas upregulation of UCA1 and hnRNP I/L significantly accelerated cell proliferation (Fig. 3I, middle). Moreover, the addition of DM-αKG, citrate, and malate (TCA cycle intermediates) in culture media could rescue the restrained proliferation induced by knockdown of UCA1, hnRNP I, and hnRNP L (Fig. 3I, right). These results suggest that UCA1 and hnRNP I/L significantly impact cell metabolic reprogramming, especially in enhancing the fraction of glutamine-derived carbons of TCA cycle intermediates.
UCA1 regulates GPT2 expression by interacting with hnRNP I/L
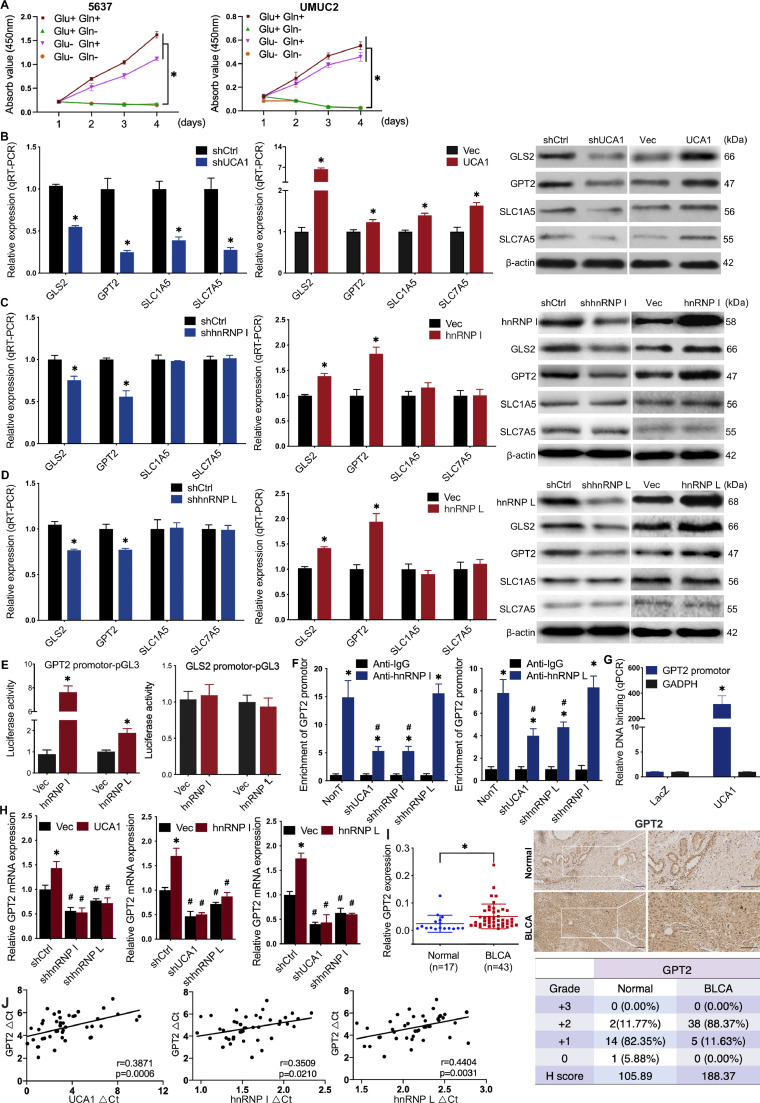
Glucose and glutamine are the two most essential fuels for cancer cell proliferation and growth. The 5637 and UMUC2 cells were able to proliferate in complete or glucose-free RPMI 1640 medium (10% FBS), whereas unable to proliferate in glutamine-free or both glutamine-free and glucose-free RPMI 1640 medium (10% FBS), suggesting that these BLCA cells required glutamine more than glucose for proliferation (Fig. 4A). Then we were interested in investigating how UCA1 and hnRNP I/L reprogram the glutamine metabolism of BLCA cells. First, we evaluated whether UCA1 and hnRNP I/L regulated important genes involved in the glutaminolysis of 5637 and UMUC2 cells using qRT-PCR (Fig. 4B–D, right). Next, western blot was performed to assess the relationship between the changed genes and UCA1 or hnRNP I/L (Fig. 4B–D, right). The results showed that SLC1A5 and SLC7A5 were positively related to UCA1 but not affected by hnRNP I/L. GLS2 and GPT2 were positively related to both UCA1 and hnRNP I/L. Also, dual-luciferase reporter assay showed that upregulated hnRNP I or L significantly increased the luciferase activity of GPT2 promoter-pGL3 reporter vector but did not affect the luciferase activity of GLS2 promoter-pGL3 reporter vector (Fig. 4E), indicating that the overexpressed hnRNP I or L increased the promoter activity of GPT2.
Fig. 4.
UCA1 regulates GPT2 expression by interacting with hnRNP I/L A, Glutamine more than glucose was required for the proliferation of BLCA cells. B to D, Screening of glutaminolysis associated enzymes: mRNA expression levels in UCA1 or hnRNP I/L knockdown and UCA1 or hnRNP I/L overexpressed BLCA cells were analyzed by qRT-PCR (left and middle). Protein levels were detected by western blot (right). E, Activation of the GPT2 promoter-pGL3 reporter by hnRNP I/L. Luciferase activity under the control of the GPT2 or GLS2 promoter was normalized to constitutively expressed renilla luciferase in 5637 cells transfected with control vector or hnRNP I/L plasmid. F, ChIP-qRCR detection of hnRNP I/L binding to the GPT2 promoter. Anti-IgG was used as the negative control. The fold enrichment of GPT2 promoter sequence in hnRNP I/L was normalized to IgG ChIP. *P < 0.01, compared with Anti-IgG; #P < 0.01 compared with NonT. G, CHIRP-qRCR detection of UCA1 binding to GPT2 promoter. UCA1 targeted probes and negative LacZ probes were used for ChIRP assay. The fold enrichment of GPT2 promoter or GADPH sequence in UCA1 CHIRP was normalized to LacZ CHIRP. H, Overexpression of UCA1 cannot rescue the reduction of GPT2 caused by hnRNP I/L knockdown. Overexpression of hnRNP I cannot rescue the reduction of GPT2 caused by hnRNP L knockdown either. I to L High expression of GPT2 in BLCA tissues: qRT-PCR of GPT2 (I) was performed using normal tumor-adjacent bladder tissues (n = 17) and BLCA tissues (n = 43), and Pearson's correlations of GPT2 (J) were calculated; IHC of GPT2 (K) was performed using clinical tissues, and H score (L) was determined. * P < 0.01. Scale bar = 200 μm.
Because UCA1 and hnRNP I/L could form RNP complex, and analysis of GPT2 promoter found hnRNP I/L binding sequences, we hypothesized that UCA1 and hnRNP I/L might regulate GPT2 gene expression by forming a complex at the promoter of GPT2 gene. ChIP assays revealed that hnRNP I/L bound to the promoter of GPT2 gene. And knockdown of UCA1 or hnRNP I/L decreased the binding of hnRNP I/L to GPT2 promoter region (Fig. 4F). We also confirmed by CHIRP assays that UCA1 bound to the GPT2 promoter (Fig. 4G). Overexpression of UCA1 did not rescue the reduction of GPT2 expression caused by hnRNP I/L knockdown (Fig. 4H), which further confirmed the close collaboration of UCA1 and hnRNP I/L to regulate the transcriptional activity of GPT2 promoter. As a supplement, we verified the significant upregulation and positive correlation between GPT2 and UCA1 or hnRNP I/L in clinical BLCA tissues (Fig. 4I–L).
We next further tested whether hnRNP I and L influenced each other on the regulation of GPT2. The results showed that the enrichment of GPT2 promoter in hnRNP I or L was barely affected by hnRNP L or hnRNP I knockdown (Fig. 4F); overexpression of hnRNP I or L cannot rescue the downregulated GPT2 expression in shhnRNP L or I cells (Fig. 4H). Together with the previous results that the expression of hnRNP I or L cannot affect the other counterpart, we validated that there was no interaction between hnRNP I and L as for regulating GPT2 expression.
The role of GPT2 in glutamine-driven anaplerosis
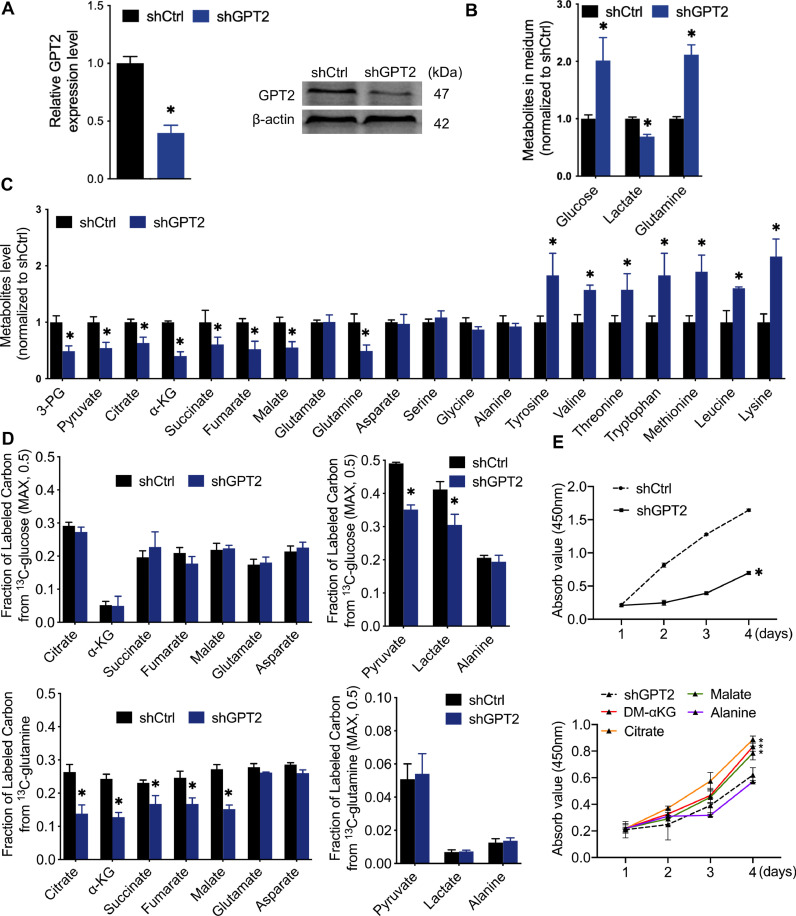
We then investigated the role of GPT2 in BLCA metabolism reprogramming. We generated 5637 cells that stably expressed shRNAs against GPT2 and validated the downregulated GPT2 expression (Fig. 5A). Critical metabolites in culture medium and cells were detected by MS analysis. Also, 13C-glucose and 13C-glutamine tracing with MS were used to observe differential fractions of labeled carbon in the critical intermediates of glutamine metabolism and TCA cycle. In the culture medium of GPT2-knockdown 5637 cells, we observed increased glucose levels, decreased lactate levels, and increased glutamine levels (Fig. 5B), which meant the GPT2 knockdown suppressed the consumption of glucose and glutamine and the secretion of lactate. GPT2-knockdown 5637 cells exhibited significantly decreased levels of several critical metabolites, including glycolysis intermediates (3-PG and pyruvate), TCA cycle intermediates (citrate, α-KG, succinate, fumarate, and malate), but did not affect intermediates of glutamine catabolism (glutamate and aspartate) and glucogenic amino acid (glycine, alanine, and serine) (Fig. 5C). Surprisingly, there was no increase in cellular glutamate levels accompanied the decrease in a-KG levels. To investigate if GPT2 silencing caused decreased precursor or increased efflux of intracellular glutamate, we detected the levels of cellular glutamine and essential amino acids (EAAs). Decreased cellular glutamine and increased cellular EAAs, including tyrosine, valine, threonine, tryptophan, methionine, leucine, and lysine, were observed in GPT2-knockdown 5637 cells (Fig. 5C). Furthermore, 13C-tracing experiments showed that GPT2 knockdown did not affect glucose contribution to TCA cycle intermediates and glutamine contribution to glycolysis intermediates, but suppressed glutamine contribution to TCA cycle intermediates and glucose contribution to glycolysis intermediates (Fig. 5D). GPT2-knockdown 5637 cells showed the inhibition to supply glutamine-derived metabolites for the TCA cycle or to maintain glycolysis. Insistently, suppressed 5637 cell proliferation induced by GPT2 knockdown (Fig. 5E, up) could be rescued by TCA cycle intermediates (DM-αKG, citrate, and malate) (Fig. 5E, down). Together these findings suggest that GPT2 is crucial for glutamine-driven anaplerosis that fueled the TCA cycle function in highly proliferative 5637 cells.
Fig. 5.
The metabolic characteristics of GPT2 in glutamine-driven anaplerosis A, Validation of the shGPT2 effect by qRT-PCR and Western blot detection of GPT2 in 5637 cells. B and C, Silencing GPT2 decreased glucose uptake, lactate secretion, and glutamine uptake and increased essential amino acids uptake in 5637 cells. Cells were grown in the medium for 24 h, and metabolites levels in culture medium and cells were analyzed by MS and normalized to shCtrl group. D, Fraction of labeled carbon in metabolic intermediates tested by 13C glucose and 13C-glutamine tracing experiments. E, DM-αKG, citrate, or malate rescues 5637 cells from suppressed cell proliferation caused by shGPT2. Cells were grown in medium with the presence of the indicated nutrient. CCK8 assay was performed to determine the cell proliferation at indicated time points. n = 3 independent samples. * P < 0.01 compared with shCtrl group.
Knockdown of UCA1, hnRNP I, hnRNP L, and GPT2 inhibits the growth of BLCA xenografts
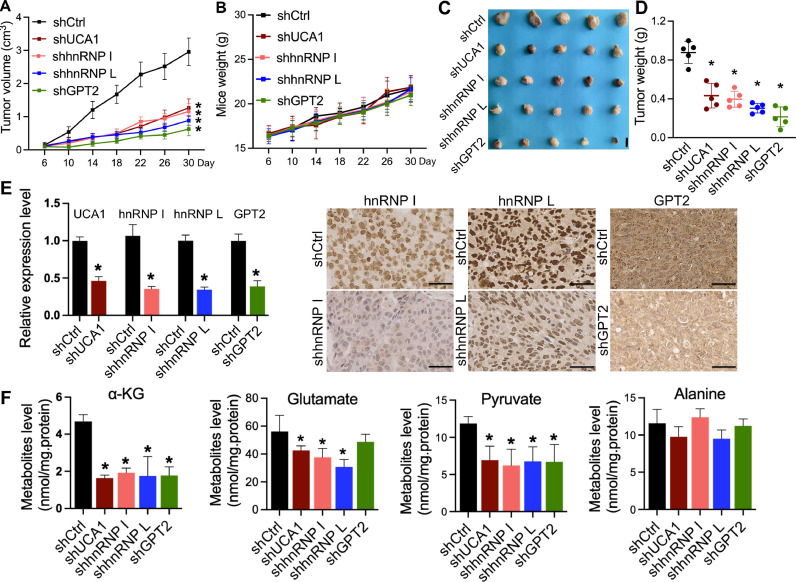
To further determine the tumor-promoting function of UCA1, hnRNP I, hnRNP L, and GPT2 in BLCA in vivo, we injected 2 × 106 shCtrl, shUCA1, shhnRNP I, shhnRNP L, and shGPT2 cells into the nude mice for creating the xenograft model. The tumors grow much faster in the shCtrl group. Knockdown of UCA1, hnRNP I, hnRNP L, and GPT2 dramatically suppressed tumor growth (Fig. 6A, C, and D), but did not affect the weight of mice (Fig. 6B). To further validate the shRNA effects in xenografts, the expression level of UCA1, hnRNP I, hnRNP L, and GPT2 were detected by qRT-PCR or IHC (Fig. 6E). We next tested the function of UCA1, hnRNP I, hnRNP L, and GPT2 in the glutamine-driven anaplerosis in xenografts. The critical step of glutamine-driven anaplerosis is the production of α-KG from glutamate transamination. Therefore, we detected the level of α-KG, glutamate, pyruvate, and alanine involved in this transamination by GC–MS. As shown in Fig. 6F, UCA1, hnRNP I, and hnRNP L knockdown reduced amounts of α-KG, glutamate, and pyruvate but did not affect alanine level; GPT2 depletion reduced amounts of α-KG and pyruvate but did not alter glutamate and alanine levels. These results obtained from mouse xenograft assay are consistent in vitro results and further validate that UCA1, hnRNP I, hnRNP L, and GPT2 functionally promote tumor growth and are involved in the glutamine-driven anaplerosis in BLCA.
Fig. 6.
Knockdown of UCA1, hnRNP I, hnRNP L, or GPT2 inhibits the growth of BLCA xenografts.
A to D, Two million cells were injected subcutaneously into nude mice. Tumor volumes (A) and mice weights (B) were measured every 4 days. Tumor weights (C and D) were measured after xenografts excision at the end of the experiment. Tumor growth in the shCtrl group was substantially faster than in other groups. No significant differences were observed on mice weights between the shCtrl and the other groups. Scale bar = 1 cm. E, The qRT-PCR and IHC staining of UCA1, hnRNP I, hnRNP L, and GPT2 in xenograft samples. F, Detection of the metabolites related to the produce of α-KG from glutamate transamination in BLCA xenografts. Xenografts were harvested, and the indicated metabolites were measured by GC–MS. n = 5 tumors for each group. * p < 0.01 compared with shCtrl group. Scale bar=50 μm.
Discussion
Here, we report a previously uncharacterized mechanism by which UCA1, combining with hnRNP I/L, significantly impacts cell metabolic reprogramming, especially in replenishing TCA cycle intermediates by upregulating glutamine anaplerosis. We observed that UCA1 and hnRNP I/L were markedly upregulated and positively correlated in bladder cancer. They could form a ribonucleoprotein complex and affect bladder cancer cell metabolism and proliferation via glutamine metabolism. Significantly, the UCA1-hnRNP I/L complex was involved in the glutamine-driven TCA anaplerosis of bladder cancer by binding with the GPT2 promoter. These observations complement our previous studies linked UCA1 to dysregulated cancer metabolism, reveal the mechanism of UCA1 involved in glutamine-driven TCA cycle anaplerosis, and provide a novel experimental basis of lncRNA regulating metabolic reprogramming in tumor cells.
The regulatory roles of LncRNAs in dysregulated cancer metabolism exert through diverse mechanisms, particularly in regulating glucose metabolism, glutamine metabolism, and mitochondrial function [17,31,32]. As for UCA1, several mechanisms of regulating metabolism in bladder cancer have been demonstrated. It stimulates glycolysis indirectly through activating the mTOR-STAT3 pathway and inhibiting microRNA-143 expression to upregulate the expression of hexokinase 2 [33]. UCA1 promotes glutaminolysis by interfering with the negative regulation of GLS2 mRNA by microRNA-16 to upregulate GLS2 expression and inhibiting ROS production to protect cells from oxidative toxicity in bladder cancer [34,35]. UCA1 acts as a competing endogenous RNA to inhibit microRNA-195, resulting in elevated ARL2 expression, which is essential for mitochondrial activity in bladder cancer [26]. Herein, our results show that the downregulation of UCA1 plays metabolic inhibition roles, including glucose and glutamine metabolism, suggesting that UCA1 gets potential clinical translation for bladder cancer via the regulation of cancer metabolism. We also observed a similar negative effect on the metabolic profile with the downregulation of hnRNP I/L (Fig 3). Given the RNA stabilization function of hnRNP I/L [19,20,36,37] and their combination with UCA1 (Fig 2), it is likely that hnRNP I/L may affect metabolism at least partially through increasing the UCA1 stability, which explains the similar metabolic profiles.
Glutamine-driven anaplerosis, another step beyond the Warburg effect, is required for oncogene-induced tumorigenesis and metabolic reprogramming in many cancer cells [5,[38], [39], [40]]. Despite the increasing evidence implicating the vital role of UCA1 in cancer metabolism, it has been poorly characterized in the anaplerosis context of bladder cancer. As pointed out earlier, glutamine can function in anaplerotic reactions through glutaminolysis. Metabolic enzymes are often found to be targeted by lncRNAs directly or indirectly in cancers. In this study, we observed that bladder cancer cells required glutamine, rather than glucose, for survival. And glutaminolytic enzymes GLS2 and GPT2 expression were upregulated by UCA1, hnRNP I, and hnRNP L. Besides, glutamine transporter SLC1A5 and SLC7A5 were upregulated by UCA1, not hnRNP I/L, and the underlying mechanism needs further investigation. Significantly, UCA1 and hnRNP I/L form a functional RNP complex, which could activate the expression of GPT2 by binding to its promoter region. In consistent with these results, stable-isotope tracing experimental data show a profound suppression of glutamine-driven carbons contributing to the TCA cycle by knockdown of UCA1, hnRNP I, hnRNP L, and GPT2. Therefore, our findings uncover a novel mechanism for UCA1 in glutamine-driven anaplerosis and complement previous studies linking UCA1 to bladder cancer glutamine metabolism [34].
The role of UCA1 in the interaction of hnRNP I/L with GPT2 promoter is an interesting paradigm. HnRNP I/L are known for their post-transcriptional regulation activities that control mRNA splicing, stability, and translation [19,41]. However, recent studies have indicated that lncRNAs interact with hnRNP I or L to function in transcriptional regulation. Linc1992 interacts with hnRNP L and binds to regulate TNFα expression [42]. LncLGR forms a functional complex with hnRNP L to facilitate the recruitment of hnRNP L to the glucokinase promoter in fasted mice [43]. Our study indicated the role of UCA1, hnRNP I/L, and GPT2 in metabolic reprogramming, especially in glutamine-driven anaplerosis. And our data do not rule out the possibility that UCA1 or hnRNP I/L may regulate GPT2 expression by other unidentified pathways or cellular factors. Intriguingly, the impact of UCA1 and hnRNP I/L on glutamine-driven anaplerosis strongly suggests that they may also contribute to other metabolic processes. And their mechanism in regulating cancer metabolic reprogramming will emerge in the future.
Mitochondrial GPT2 converts glutamate to α-KG. In our study, silencing of GPT2 causes a significant decrease in α-KG levels, but showed no effect on glutamate levels in vitro and in vivo. Knockdown of GPT2 decreased glutamine uptake and increased cellular EAAs levels. Intracellular EAAs uptake is accompanied by the simultaneous efflux of glutamine out of cells, which is regulated by the bidirectional transporter SLC7A5/SLC3A2. These results indicated glutamine, the major source of glutamate, is significantly reduced by silencing of GPT2, and partially explained why increased glutamate levels did not accompany with the decrease in α-KG levels. As GPT2 is crucial for the glutamine-driven anaplerosis and the BLCA cell proliferation, the inhibition of GPT2 function may be a promising adjuvant strategy to conventional treatments. For example, aminooxyacetate (AOA), a compound of aminotransferase inhibitor, is able to block the conversion of glutamate to a-KG, which results in tumor inhibitory effect in a xenograft model of PIK3CA mutant colorectal cancer [44]. It is conceivable that a GPT2-specific inhibitor could be more potent have a favorable therapeutic index.
Together, our data show that UCA1 and hnRNP I/L exhibit substantial effects on bladder cancer metabolic reprogramming and reveal the unexpected mechanism of UCA1 forming a functional UCA1-hnRNP I/L complex that upregulates GPT2 expression to promote glutamine-driven TCA anaplerosis. This work provides new insights into the pivotal role of glutamine addiction and glutamine-driven anaplerosis in bladder cancer, implying a potentially therapeutic value of targeting these pathways for bladder cancer treatment.
CRediT authorship contribution statement
Hua Zhao: Conceptualization, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. Wenjing Wu: Conceptualization, Formal analysis, Methodology, Writing – review & editing. Xu Li: Supervision, Resources, Supervision, Writing – review & editing. Wei Chen: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81772735).
References
- 1.Devic S. Warburg Effect - a consequence or the cause of carcinogenesis? J. Cancer. 2016;7:817–822. doi: 10.7150/jca.14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Y., Wang D., Xu T., Liu P., Cao Y., Wang Y., Yang X., Xu X., Wang X., Niu H. Bladder cancer cells re-educate TAMs through lactate shuttling in the microfluidic cancer microenvironment. Oncotarget. 2015;6:39196–39210. doi: 10.18632/oncotarget.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seyfried T.N., Flores R.E., Poff A.M., D'Agostino D.P. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghaffari P., Mardinoglu A., Nielsen J. Cancer metabolism: a modeling perspective. Front. Physiol. 2015;6:1–9. doi: 10.3389/fphys.2015.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa-Ruiz E., Diaz-Ruiz R. Anaplerosis in cancer: another step beyond the warburg effect. Am. J. Mol. Biol. 2012;02:291–303. doi: 10.4236/ajmb.2012.24031. [DOI] [Google Scholar]
- 6.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 7.Cluntun A.A., Lukey M.J., Cerione R.A., Locasale J.W. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 2017;3:169–180. doi: 10.1016/j.trecan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina M.A. Glutamine and cancer. J. Nutr. 2001;131:2539S–2542S. doi: 10.1093/jn/131.9.2539S. [DOI] [PubMed] [Google Scholar]
- 9.Chambers J.W., Maguire T.G., Alwine J.C. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 2010;84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J.B., Bardeesy N., Asara J.M., Haigis M.C., DePinho R.A., Cantley L.C., Kimmelman A.C. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otter S.J., Chatterjee J., Stewart A.J., Michael A. The role of biomarkers for the prediction of response to checkpoint immunotherapy and the rationale for the use of checkpoint immunotherapy in cervical cancer. Clin. Oncol. 2019;31:834–843. doi: 10.1016/j.clon.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Li T., Le A. Glutamine metabolism in cancer. Adv. Exp. Med. Biol. 2018;1063:13–32. doi: 10.1007/978-3-319-77736-8_2. [DOI] [PubMed] [Google Scholar]
- 13.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y., Lin S.H., Wang Y., Chin Y.E., Kang L., Mi J. Glutamic pyruvate transaminase GPT2 promotes tumorigenesis of breast cancer cells by activating sonic hedgehog signaling. Theranostics. 2017;7:3021–3033. doi: 10.7150/thno.18992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sreedhar A., Zhao Y. Dysregulated metabolic enzymes and metabolic reprogramming in cancer cells. Biomed. Rep. 2018;8:3–10. doi: 10.3892/br.2017.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W., Zhou Q., Wang C.Q., Zhu L., Bi C., Zhang S., Wang X., Jin H. LncRNAs regulate metabolism in cancer. Int. J. Biol. Sci. 2020;16:1194–1206. doi: 10.7150/ijbs.40769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan C., Tang Y., Wang J., Xiong F., Guo C., Wang Y., Zhang S., Gong Z., Wei F., Yang L., He Y., Zhou M., Li X., Li G., Xiong W., Zeng Z. Role of long non-coding RNAs in glucose metabolism in cancer. Mol. Cancer. 2017;16:1–11. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Li X., Xie X., Zhao L., Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung L., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu W., lun Zhou B., juan Rong L., Ye L., juan Xu H., Zhou Y., jun Yan X., dong Liu W., Zhu B., Wang L., jun Jiang X., ping Ren C. Roles of PTBP1 in alternative splicing, glycolysis, and oncogensis. J. Zhejiang Univ. Sci. B. 2020;21:122–136. doi: 10.1631/jzus.B1900422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minami K., Taniguchi K., Sugito N., Kuranaga Y., Inamoto T., Takahara K., Takai T., Yoshikawa Y., Kiyama S., Akao Y., Azuma H. MiR-145 negatively regulates Warburg effect by silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget. 2017;8:33064–33077. doi: 10.18632/oncotarget.16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugiyama T., Taniguchi K., Matsuhashi N., Tajirika T., Futamura M., Takai T., Akao Y., Yoshida K. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107:1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M., Mo Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 26.Li H.J., Sun X.M., Li Z.K., Yin Q.W., Pang H., Pan J.J., Li X., Chen W. LncRNA UCA1 promotes mitochondrial function of bladder cancer via the miR-195/ARL2 signaling pathway. Cell. Physiol. Biochem. 2017;43:2548–2561. doi: 10.1159/000484507. [DOI] [PubMed] [Google Scholar]
- 27.Varghese F., Bukhari A.B., Malhotra R., De A. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9:e96801. doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu C., Quinn J., Chang H.Y. Chromatin isolation by RNA purification (ChIRP) J. Vis. Exp. 2012:e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanchen A., Fuhrer T., Sauer U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 2007;358:177–197. doi: 10.1007/978-1-59745-244-1_11. [DOI] [PubMed] [Google Scholar]
- 30.van Winden W.A., Wittmann C., Heinzle E., Heijnen J.J. Correcting mass isotopomer distributions for naturally occurring isotopes. Biotechnol. Bioeng. 2002;80:477–479. doi: 10.1002/bit.10393. [DOI] [PubMed] [Google Scholar]
- 31.Lu W., Cao F., Wang S., Sheng X., Ma J. LncRNAs: the regulator of glucose and lipid metabolism in tumor cells. Front. Oncol. 2019;9:1–11. doi: 10.3389/fonc.2019.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z.D., Zhuang L., Gan B. Long non-coding RNAs in cancer metabolism. BioEssays. 2016;38:991–996. doi: 10.1002/bies.201600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Li X., Wu S., Xue M., Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–955. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H.J., Li X., Pang H., Pan J.J., Xie X.J., Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 2015;45:1055–1063. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]
- 35.D'Autréaux B., Toledano M.B. ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 36.Arake De Tacca L.M., Pulos-Holmes M.C., Floor S.N., Cate J.H.D. PTBP1 mRNA isoforms and regulation of their translation. RNA. 2019;25:1324–1336. doi: 10.1261/rna.070193.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hui J., Reither G., Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q., Kirk K., Shurubor Y.I., Zhao D., Arreguin A.J., Shahi I., Valsecchi F., Primiano G., Calder E.L., Carelli V., Denton T.T., Beal M.F., Gross S.S., Manfredi G., D'Aurelio M. Rewiring of glutamine metabolism is a bioenergetic adaptation of human cells with mitochondrial DNA mutations. Cell Metab. 2018;27:1007–1025. doi: 10.1016/j.cmet.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Zhao X., Chen V., Feng Y., Wang L., Croniger C., Conlon R.A., Markowitz S., Fearon E., Puchowicz M., Brunengraber H., Hao Y., Wang Z. Colorectal cancers utilize glutamine as an anaplerotic substrate of the TCA cycle in vivo. Sci. Rep. 2019;9:19180. doi: 10.1038/s41598-019-55718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishore S., Luber S., Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genom. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Chao T.C., Chang K.Y., Lin N., Patil V.S., Shimizu C., Head S.R., Burns J.C., Rana T.M. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan X., Li P., Cangelosi A., Yang L., Cao H. A Long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep. 2016;14:1867–1875. doi: 10.1016/j.celrep.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng X., Hao Y., Wang Z. Targeting glutamine metabolism in PIK3CA mutant colorectal cancers. Genes Dis. 2016;3:241–243. doi: 10.1016/j.gendis.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]