Abstract
A nucleic acid sequence-based amplification (NASBA) assay for qualitative detection of human cytomegalovirus (CMV) pp67 mRNA was evaluated in a multicenter study. Negative results were obtained for all specimens from 50 CMV-seronegative and 50 CMV-seropositive low-risk whole-blood donors. No interference with CMV mRNA amplification was observed in the testing of 288 specimens containing various potential interfering substances, nonspecifically reacting substances (including mRNA from other herpesviruses), and three anticoagulants. A total of 95% (50 of 51) of CMV-positive (cell culture- and antigenemia immunofluorescence [AG-IFA]-positive) clinical specimens were positive by the NASBA assay. Results from different operators over multiple testing days were consistent for each of four panel members containing different concentrations of CMV mRNA, indicating the reproducibility of the assay. The estimated 95% reliable upper detection limit of the assay was 600 mRNA copies; the lower limit of detection was less than 25 mRNA copies. The clinical utility of the assay was evaluated with longitudinally collected specimens from solid-organ transplant patients (n = 21). A total of 98% (81 of 83) of the specimens from CMV-negative patients were negative by the NASBA assay, while 90% (10 of 11) of patient specimens that were positive by cell culture or AG-IFA were positive by the NASBA assay. Positive NASBA assay results were obtained earlier than AG-IFA or cell culture results for 55% of the patients and at the same time for the remainder of the patients (45%). The overall agreement between the NASBA assay and current reference tests was 86% when active CMV infection was present. These studies indicate that the CMV pp67 mRNA NASBA assay has reproducible and sensitive performance characteristics that should enable more rapid diagnosis of CMV infection.
Primary infection with cytomegalovirus (CMV), a beta-herpesvirus with high prevalence (40 to 80%) in developed countries (10, 17), is usually asymptomatic and results in latent lifetime infections in individuals with normal immune function (11). In individuals with compromised immune systems, such as AIDS patients (5, 6) and transplant recipients (solid organ or bone marrow) (12), a high rate of morbidity and mortality can occur upon reactivation of CMV infection.
Early and accurate diagnosis of active CMV infection is important for optimal clinical efficacy of treatment with antiviral drugs, such as ganciclovir, cidofovir, or foscarnet (13). Current standard methods for the detection of active CMV infection use either tube or shell vial cell culture (9) as a measure of viremia or an immunofluorescence assay (IFA) as a test for the presence of pp65 antigen (AG-IFA) (16). While each approach has an overall benefit for the diagnosis of CMV infection, the inherent characteristics of these methods tend to preclude a rapid or objective test result. For example, conventional tube cell culture of CMV can require up to 2 weeks before a definitive determination of virus replication can be reached.
Application of nucleic acid amplification techniques for the diagnosis of CMV has been described primarily for the detection of viral DNA with PCR. In general, comparative studies with PCR and other tests used for the detection of CMV (i.e., culture or AG-IFA) indicate an increased sensitivity with the molecular assay (for an example, see reference 15). However, not all studies of this type present clearly superior results obtained with PCR compared to those obtained with other tests for CMV diagnosis (e.g., AG-IFA) (19). Further, qualitative PCR is limited in the determination of active CMV infection, because the technique can amplify latent viral DNA. This problem may be lessened somewhat with the use of quantitative PCR, as recently described for transplant patients (14, 18, 21).
Since the presence of mRNA in the host cell can be considered a fundamental indicator of viral replication, detection of this analyte would provide a marker for active CMV infection. Using this approach, recent studies have demonstrated the feasibility of using either CMV pp67 mRNA, which is translated to a phosphorylated matrix protein expressed late in the viral replication cycle, or immediate-early-protein mRNA in renal-allograft recipients (1, 2).
This study evaluated the analytical performance of a nucleic acid sequence-based amplification assay for the qualitative detection of CMV pp67 mRNA and its clinical applicability for a population of solid-organ transplant patients.
MATERIALS AND METHODS
Nucleic acid sequence-based amplification assay.
The NucliSens CMV pp67 assay (Organon Teknika Corporation [OTC], Durham, N.C.), which was used according to the manufacturer's directions (1), incorporates a stringent nucleic acid isolation technique that releases RNA present in the specimen with detergent and guanidine thiocyanate, with subsequent capture on silica (3). CMV-specific mRNA primers and enzymes (avian myeloblastosis virus reverse transcriptase, RNase H, and T7 RNA polymerase) were used for isothermal nucleic acid amplification at 41°C (20). Detection of amplified nucleic acids was based on electrochemiluminescence (ECL) resulting from the excitation of ruthenium-labeled probes specific for the wild-type (WT) CMV mRNA and for the System Control RNA, which functions as an internal process control for the assay. ECL emission is proportional to the amount of amplified nucleic acid products. Data analysis software associated with the NucliSens system was used to calculate a qualitative negative or positive result using a cutoff of 200 ECL units.
Specimens.
Anticoagulated whole-blood (0.1-ml volume) specimens were used for analysis with the NucliSens CMV pp67 assay. Clinical specimens for the analytical performance characterization studies were collected from subjects at North Shore University Hospital (Manhasset, N.Y.), Massachusetts General Hospital (Boston), BioClinical Partners (Franklin, Mass.), and Sacramento Medical Foundation (Sacramento, Calif.) with institutional review board approval and informed consent. Basic demographic information (age and gender) and medical information were obtained for each subject at the time of enrollment by chart review. For longitudinally collected specimens, medical information was obtained at each subsequent specimen collection. Clinical data for the transplant patients studied longitudinally at the Mayo Clinic (Rochester, Minn.) cohort (described below) were obtained prospectively during the first 4 months posttransplantation. The medical information obtained from each subject was associated specifically with the following parameters, including conditions relating to CMV infection or disease: temperature, leukocyte count, platelet count, renal function, retinitis, pneumonia, and gastrointestinal disease. Results from reference laboratory test methods (i.e., AG-IFA, shell vial cell culture, and tube cell culture) were obtained from the laboratory of each participating institution. For this study, CMV disease is defined as a positive diagnostic test result accompanied by symptoms, and CMV infection is defined as a positive diagnostic test result without accompanying symptoms. Diagnosis criteria for CMV infection and disease were applied for the present study at the Mayo Clinic, as previously described (18).
For specificity studies, specimens were obtained from healthy volunteer whole-blood donors who were either CMV seronegative or CMV seropositive, as confirmed by a qualitative solid-phase red-cell adherence test system (CAPTURE-CMV; IMMUCOR, Inc., Norcross, Ga.).
For interference studies, specimens were obtained from subjects with defined medical conditions or elevated physiological analytes but without overt symptoms of illness. Each specimen was tested in the presence and absence of a CMV-specific mRNA spike (described below). Matched whole-blood specimens with and without anticoagulant were collected from individuals with autoimmune diseases (antinuclear antibody, systemic lupus erythematosus, and rheumatoid factor). Serum was used for IFA to confirm the autoimmune condition for each individual, and the anticoagulated whole-blood specimen from each subject was tested with the NucliSens CMV pp67 assay. Icteric specimens (bilirubin range, 1.6 to 3.8 mg/dl) were obtained from patients with liver metastasis. Lipemic specimens (triglyceride range, 243 to 2,400 mg/dl) were obtained from volunteer whole-blood donors.
The potential for nonspecific amplification of the mRNAs of other viruses by the NucliSens CMV pp67 assay was evaluated by testing clinical specimens with and without a CMV-specific mRNA spike (see below). Clinical specimens were obtained from individuals infected with human immunodeficiency virus type 1 (HIV-1) and from others infected with human T-cell lymphotropic virus (HTLV). Cell cultures infected with different strains of Epstein-Barr virus were obtained from OTC and the American Type Culture Collection (Manassas, Va.). Varicella-zoster virus, herpes simplex virus (HSV) type 1, and HSV type 2 clinical isolates were propagated in susceptible human cells until the development of cytopathic effect, at which time infection was confirmed with IFA. The infected cells were harvested and stored at −70°C until used. The number of cells in each virus-infected culture was determined by hemocytometer counting, and each culture was added to EDTA-anticoagulated whole blood from individual volunteer blood donors at a standardized concentration of 106 cells/ml.
Matched specimens from 10 volunteer whole-blood donors were collected into EDTA (K3), sodium heparin, and acid citrate dextrose solution B Vacutainer brand tubes (Becton Dickinson, Franklin Lakes, N.J.) and tested with the NucliSens CMV pp67 assay for possible anticoagulant interference effects.
Using a panel of specimens containing known quantities of CMV mRNA, the reproducibility of the NucliSens CMV pp67 assay's performance was evaluated at three different laboratories with three kit lots over multiple days.
CMV mRNA spike.
For reproducibility and specificity studies, an RNA spike was synthesized using standard techniques to obtain an exact sequence of 1,125 nucleotides corresponding to authentic CMV pp67 mRNA. Following purification with an anion-exchange column (Qiagen, Leusden, The Netherlands), the RNA concentration was determined by measuring the absorbance at 260 nm and then diluted to yield an estimated number of input copies for subsequent experimentation. For the reproducibility study, the mRNA spike ranged from 2,500 to 25 copies/assay input. For specificity (interference and nonspecific amplification) studies, 4 × 103 copies of the CMV mRNA spike were added to each specimen prior to the isolation procedure used with the assay.
Methods.
The manner in which specimens were tested was dependent upon the type of specimen and the experimental design. Whereas clinical specimens were tested directly following collection, specimens for analytical performance characterization studies and specificity (interference and nonspecifically reacting substances) studies were tested with and without the CMV mRNA spike using the NucliSens CMV pp67 assay. Each mRNA spiked specimen was tested at a final volume of 100 μl (a 5-μl spike containing 4 × 103 copies and 95 μl of specimen).
Specimens from subjects suspected to have CMV infection and from posttransplantation subjects were inoculated into standard tube and shell vial fibroblast cultures using standard techniques (9). For this study, each of these cell culture-based procedures was considered an independent reference test. Inoculated cell cultures were monitored for cytopathic effect, and CMV infectivity was confirmed with IFA using CMV-specific monoclonal antibodies. AG-IFA, using 5 × 104 cells processed from each clinical specimen, was performed according to the manufacturers' directions (which do not require quantitation of cells positive for antigenemia) (CMV Brite [BioTest Diagnostics Corp., Denville, N.J.] or Light Diagnostics [Chemicon International, Inc., Temecula, Calif.]). Specimens with discordant results between the NucliSens CMV pp67 assay and any of the other tests were reevaluated with the NucliSens CMV pp67 assay.
Clinical utility in a solid-organ transplant population.
At the Mayo Clinic, a longitudinal study of 21 solid-organ transplant (19 liver and 2 heart) recipients was conducted from December 1997 to April 1999. Each specimen (140 total), collected at sequential time intervals posttransplantation, was evaluated with the NucliSens CMV pp67 assay and three conventional reference laboratory procedures (tube and shell vial cell culture and AG-IFA). Clinical specimens were stored at −70°C prior to analysis with the NucliSens CMV pp67 assay. The results obtained from specimens collected in conjunction with this prospective study were analyzed independently of the results from clinical specimens obtained for the analytical characterization of the NucliSens CMV pp67 assay.
Statistical analysis.
Amplification of the NucliSens CMV pp67 System Control RNA was used to assess the effects of interferents on assay performance, as the use of the mRNA spike precluded using the WT CMV mRNA amplification level for this comparison. ECL results were transformed to log10 values for calculations. For comparison of NucliSens CMV pp67 assay results with standard reference test (cell culture and AG-IFA) results, the kappa statistic was constructed to estimate agreement beyond chance.
RESULTS
Analytical characterization.
Specimens from all CMV-seronegative (n = 50) and -seropositive (n = 50) volunteer blood donors were negative upon testing with the NucliSens CMV pp67 assay. The specimens from the CMV-seropositive volunteer blood donors were also AG-IFA negative. Based on these data, the exact 95% binomial confidence limits for the specificity of the assay in these populations were 92.9 and 100%.
For 100 specimens from five different classes of interfering substances, all expected results, either positive in the presence of the RNA spike or negative in its absence, were obtained with the NucliSens CMV pp67 assay (Table 1). The presence of authentic viral mRNAs in whole-blood specimens from HIV-1-infected individuals and HTLV-infected individuals, as well as in cultured cells infected with non-CMV herpesviruses, did not interfere with obtaining the expected positive NucliSens CMV pp67 results in the presence of the CMV mRNA spike or negative results in the absence of the spike (Table 1). System Control RNA amplification levels were similar for the spiked and unspiked specimens in each of the experimental groups. For matched specimens collected in each of three different anticoagulants and tested unspiked and spiked with CMV mRNA, each unspiked anticoagulated whole-blood specimen was NucliSens CMV pp67 negative, while each CMV mRNA-spiked specimen was positive. Amplification levels of the NucliSens CMV pp67 System Control RNA were similar for each of the nonspiked (mean = 5.6702) and spiked (mean = 5.4893) anticoagulated whole-blood specimens. The mean amplification level for the CMV mRNA-spiked anticoagulated whole-blood specimens was 5.5257 (range, 5.4910 to 5.5487).
TABLE 1.
Summary of NucliSens CMV pp67 assay results with specimens containing potentially interfering substances and nonspecifically reacting substancesa
| Specimen group | Unspiked
|
Spiked
|
|||
|---|---|---|---|---|---|
| No. negative/no. tested | System Control RNA (log of the mean ECL reading) | No. positive/no. tested | System Control RNA (log of the mean ECL reading) | WT (log of the mean ECL reading) | |
| HIV-1 | 10/10 | 5.6695 | 10/10 | 5.4059 | 5.3754 |
| HTLVb | 9/9 | 5.6510 | 9/9 | 5.5278 | 5.2713 |
| HSV type 1 | 10/10 | 5.6902 | 10/10 | 5.5343 | 5.4515 |
| HSV type 2 | 10/10 | 5.7199 | 10/10 | 5.4907 | 5.4323 |
| Epstein-Barr virusc | 10/10 | 5.7317 | 10/10 | 5.5073 | 5.4416 |
| Varicella-zoster virus | 10/10 | 5.7585 | 10/10 | 5.5223 | 5.4196 |
| Icteric | 10/10 | 5.6737 | 10/10 | 5.5002 | 5.4001 |
| Lipemic | 10/10 | 5.6971 | 10/10 | 5.4867 | 5.3007 |
| Antinuclear antibody | 10/10 | 5.6509 | 10/10 | 5.4111 | 5.3761 |
| Systemic lupus erythematosus | 10/10 | 5.7632 | 10/10 | 6.5622 | 5.5068 |
| Rheumatoid factor | 10/10 | 5.7140 | 10/10 | 5.4819 | 5.4225 |
| Total | 109/109 | 5.7017 | 109/109 | 5.5845 | 5.3998 |
Each specimen was tested with and without a CMV mRNA spike.
HTLV-1, n = 3; HTLV-2, n = 6. HTLV was typed by Western blotting (Cambridge Biotech Diagnostics, Worcester, Mass.).
Cells from OTC: Raji, P3HR1, and NC37. Cells from the American Type Culture Collection: Daudi, EB1, Ramos, Jiyoye, EB3, CA46, and EB2.
Using a panel of specimens containing different concentrations of CMV mRNA, reproducible NucliSens CMV pp67 results were obtained at three laboratories regarding the number of positive and negative results for each panel member tested (Table 2). The rate of CMV mRNA detection with these specimens was directly proportional to the analyte concentration. The NucliSens CMV pp67 assay 95% reliable upper detection limit was estimated by interpolation of these results to be 600 mRNA copies, and the lower limit of detection was estimated to be less than 25 mRNA copies. All the panel specimens without the CMV mRNA spike were negative.
TABLE 2.
Reproducibility of the NucliSens CMV pp67 assay at three different laboratories
| No. of input CMV mRNA copies | % (no. positive) at site:
|
% of total (no. positive) | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 2,500 | 100 (48) | 100 (48) | 100 (48) | 100 (144) |
| 250 | 77 (37) | 79 (38) | 85 (41) | 81 (116) |
| 25 | 27 (13) | 19 (9) | 21 (10) | 22 (32) |
| 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Three different lots of the NucliSens CMV pp67 assay were tested in equivalent numbers at each site, each over a 4-day period. A total of 144 samples were tested.
Of 51 clinical specimens positive for CMV by either conventional tube or shell vial cell culture, AG-IFA, or both, 43 (84%) were positive by the NucliSens CMV pp67 assay. Forty-one percent (14 of 34) of the specimens were from subjects with CMV disease (11 HIV-1-infected individuals, 10 with retinitis and 1 with pneumonia; 1 recipient of an allogeneic bone-marrow transplant; 1 renal-transplant recipient; and 1 individual with Wegener's disease). Ninety-seven percent (34 of 35) of the specimens from HIV-1 patients and transplant recipients that were both cell culture and AG-IFA positive were NucliSens CMV pp67 positive. The one specimen in this group with NucliSens CMV pp67-negative results was from a bone marrow transplant recipient with no observable CMV disease but with positive shell vial cell culture and AG-IFA (16 positive cells) results.
Eighty percent (4 of 5) of the cell culture-positive and antigenemia-negative specimens were NucliSens CMV pp67 positive.
Forty-five percent (5 of 11) of the AG-IFA-positive and cell culture-negative specimens were NucliSens CMV pp67 positive. One of these five NucliSens CMV pp67-positive specimens was from a renal-transplant patient with CMV disease, while the other four specimens were from either transplant (one liver and one cardiac) recipients or HIV-1-infected individuals without disease symptoms. The remaining six specimens, with positive AG-IFA results only, were from patients without any manifestations of CMV disease (two renal-transplant recipients, two liver transplant recipients, and two HIV-1-infected individuals). In this group of 11 specimens, the mean number of AG-IFA-positive cells in those specimens that were NucliSens CMV pp67 positive was 88 (range, 221 to 3) and that in NucliSens CMV pp67-negative specimens was 2 (range, 3 to 1).
The agreement of NucliSens CMV pp67 results with those from the culture-positive clinical specimens was 95% (38 of 40), and the agreement with those from the AG-IFA-positive clinical specimens was 85% (39 of 46).
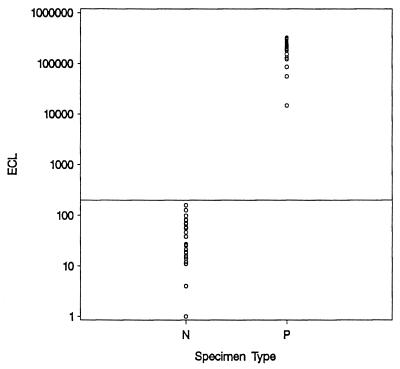
Analysis of the NucliSens CMV pp67 ECL values obtained known consensus negative specimens and, with known consensus positive specimens (cell culture and antigenemia positive) as described above, indicated that the assay cutoff is properly positioned, with a distinct separation of the two subpopulations (Fig. 1).
FIG. 1.
Distribution of the NucliSens CMV pp67 ECL signals for CMV-negative (N) and CMV-positive (P) specimens. The horizontal line at 200 ECL units represents the cutoff for the assay.
Clinical utility.
Of 83 total specimens from 11 liver transplant subjects with no evidence of active CMV infection, as determined by negative results with the three standard reference tests (tube cell culture, shell vial cell culture, and AG-IFA), 81 (98%) were NucliSens CMV pp67 negative. The two specimens with discordant results were from one CMV-seronegative recipient of a liver transplant from a CMV-seropositive donor. At the times the two specimens with positive NucliSens CMV pp67 results were collected, symptoms were present (fever and leukopenia) that were suggestive of an underlying CMV disease state for this patient. These two specimens were NucliSens CMV pp67 positive upon repeat testing.
Of 57 specimens from the 10 patients (8 liver and 2 heart transplant recipients) with at least one positive reference test, 28 (49%) were NucliSens CMV pp67 positive, whereas 18 (32%) were positive by at least one of the three standard reference tests. Seventeen specimens were NucliSens CMV pp67 positive and were negative by each of the three standard reference tests, 6 specimens were NucliSens CMV pp67 positive and had one reference test result that was positive, and 4 specimens were NucliSens CMV pp67 positive and had two reference test results that were positive. Complete agreement among the three reference tests and the NucliSens CMV pp67 assay was observed for one specimen. In this subpopulation, the overall agreement between the NucliSens CMV pp67 assay and any one of the standard reference tests was 86% (121 of 140 individual test observations). However, better agreement with NucliSens CMV pp67 results was found with culture-based methods for CMV detection than with AG-IFA (Table 3).
TABLE 3.
Summary of result agreement between the NucliSens CMV pp67 assay and standard reference tests with specimens from solid-organ transplant patientsa
| Test | No. of specimens with CMV reference assay result/NucliSens CMV pp67 test result:
|
Total concordance | Agreement (kappa) | |||
|---|---|---|---|---|---|---|
| +/+ | −/− | +/− | −/+ | |||
| Tube cell culture | 2 | 110 | 0 | 28 | 0.80 | 0.09 |
| Shell vial cell culture | 11 | 110 | 1 | 18 | 0.86 | 0.47 |
| Antigenemiab | 4 | 111 | 0 | 26 | 0.81 | 0.15 |
A total of 140 samples were tested.
Antigenemia was determined with pp65 IFA.
For 90% (9 of 10) of the patients, the NucliSens CMV pp67 assay results were concordant with those obtained with the reference tests used to detect the presence of active CMV infection. For the one patient in this group with NucliSens CMV pp67 results discordant with the reference test results, a positive shell vial result was reported only for the seventh specimen in the collection series of eight. Symptoms consistent with active CMV disease were observed at the time of collection for the two preceding specimens (the fifth and sixth of the series), but none of the reference tests was reported positive at these times. For this subject, the NucliSens CMV pp67 results were negative for specimens from all eight time points, with both initial and repeat testing for the specimen at time point 7.
A variable temporal relationship between the NucliSens CMV pp67 results and those from the reference tests with the longitudinally collected specimens from this subpopulation of nine subjects was observed. A positive NucliSens CMV pp67 result was obtained before a positive result by any one of the reference tests for five of the nine subjects (55%) (mean = 8 days; range, 5 to 14 days) and at the same time as any of the reference tests in the remaining four subjects (45%). This observation accounts for the increase in the number of positive results obtained with the NucliSens CMV pp67 assay compared to the standard reference assays described above. In no instance was the NucliSens CMV pp67 assay reported initially positive after any of the standard reference assays were reported positive for a given subject.
In this group of nine subjects with at least one standard reference test reported with a positive result, symptoms consistent with active CMV disease were observed in five. For each of these subjects, the NucliSens CMV pp67 results were positive either at the same time as (n = 2) or before (n = 3) a positive result was obtained by any one of the standard reference tests. These results suggest that the NucliSens CMV pp67 assay has clinical sensitivity at least as good as those of the standard reference tests for the detection of active CMV infection used for the present study with solid-organ transplant patients.
DISCUSSION
The study results described herein indicated that the NucliSens CMV pp67 assay is highly specific. No false-positive or false-negative results were obtained with specimens either from a low-risk blood donor population or from numerous classes of specimens containing potentially interfering and nonspecifically reacting substances. Based on these studies, the analytical specificity of the assay is estimated at 100% (95% binomial intervals at 92.5 to 100%). Significantly, none of the specimens from CMV-seropositive whole-blood donors were NucliSens CMV pp67 positive, which was the same result obtained with a CMV pp65 antigenemia test. Thus, the NucliSens CMV pp67 assay can discriminate between latent infection, as shown with this group of subjects, and active infection, as shown with the specimens from other subjects confirmed to be CMV positive by both antigenemia testing and cell culture. These results indicate that the negative predictive value of the assay will be beneficial for the accurate assessment of active CMV infection.
Studies with numerous specimens containing potentially interfering substances and nonspecifically reacting substances indicated that the presence of these biologic entities did not adversely affect the NucliSens CMV pp67 assay performance. The effective removal of potentially interfering substances during CMV mRNA isolation can be attributed to the use of Boom methodology (3), which has been previously shown to effectively remove interferents with known inhibitory effects in PCR (22). The isolation of the target analyte free of interferents also contributes to the high level of specificity observed with the NucliSens CMV pp67 assay.
Results from studies characterizing the sensitivity of the NucliSens CMV pp67 assay indicated a lower detection limit of less than 25 CMV mRNA copies. However, the rate of CMV mRNA detection at concentrations lower than 25 copies would be expected to be less than the 22% positive results seen with specimens at this concentration level in the reproducibility study. As cells infected with CMV and undergoing active replication would be expected to have this concentration of mRNA molecules (4, 7), the assay detection limits are expected to provide sufficient sensitivity to detect an active infection in the early stages of viral pathogenesis. However, with some clinical populations (8), other assays for CMV may be reported as positive earlier than the NucliSens CMV pp67 assay. In these cases, either the amount of infectious virus detected by shell vial cell culture or the number of pp65 antigen-positive cells detected by IFA appears to be quite low. This further suggests a relationship between the detectability of active CMV infection by the NucliSens CMV pp67 assay and that by other diagnostic tests which is dependent on the precise stage of CMV infection and the presence of enough CMV-infected cells in the 0.1-ml whole-blood sample to detect the pp67 mRNA. Further study is necessary to determine the stoichiometric relationship between the concentration of mRNA in clinical specimens relative to the number of infected blood cells and the capability of detection by the NucliSens CMV pp67 assay.
The present study compared currently used non-PCR assays for detection of CMV with the NucliSens CMV pp67 assay. The non-PCR assays for CMV detection are based on detection either of antigen present in the sample with IFA or of infectious virions with cell culture techniques. In contrast, the NucliSens CMV pp67 assay amplifies the CMV mRNA present in the specimen to a level greater than that originally present. This type of approach affords some advantage compared to either the static IFA assay or the cell culture techniques, which can require an extended period of time postinoculation to reach an outcome. The NucliSens CMV pp67 assay is advantageous regarding time to result compared to the other assays, because a final reportable result can be obtained in approximately 5 h for 10 specimens. Results obtained by the NucliSens CMV pp67 assay were in excellent agreement with those obtained by the standard tests when both were reported as positive, as seen both in single clinical specimens (95%) and in transplant subjects (90%). The NucliSens CMV pp67 assay tended to be in better agreement overall in cases where the only positive standard test result was by cell culture (93%) than in cases where the only positive result was by antigen detection (78%). However, the results for specimens sequentially collected from clinical transplantation subjects suggest that other factors can influence which test is reported as positive first, although in each case studied anti-CMV therapy was initiated immediately after the first positive result with a reference test. Consequently, the direct comparison of the results for the different tests used for CMV diagnosis in this study is difficult. Each assay used for the diagnosis of CMV infection has unique characteristics, with a specific target that make comparison of the different methods complicated. However, the time to positivity in individuals with active CMV infection has important consequences for the start of antiviral treatment before the patient progresses to clinical disease. In this study, the NucliSens CMV pp67 assay was shown to be better than or equivalent to the currently used assays with comparative testing of clinical specimens from transplant patients. Therefore, it is possible to consider the NucliSens CMV pp67 assay as one that may have clinical utility as an indication to effect preemptive antiviral therapy in transplant recipients. The results presented also suggest that the NucliSens CMV pp67 assay can be used in conjunction with other laboratory assays for the detection of CMV. Based on the results obtained in the present study, a clinical algorithm utilizing the NucliSens CMV pp67 assay for early detection and shell vial cell culture for subsequent confirmation of CMV viremia could be one approach. The goal of fully understanding the clinical status of an individual patient with the ultimate benefit of providing the clinician useful information for management of patient therapeutic treatment regimens may be achieved ultimately by using tests that detect different markers of CMV infection. Further study is needed to ascertain which treatment decision to make based on positive results reported by the NucliSens CMV pp67 assay.
In conclusion, the results from these studies indicate that the NucliSens CMV pp67 assay reliably detects active CMV infection. As demonstrated for specimens from solid-organ transplant recipients and confirmed by standard reference laboratory tests for CMV, the assay has clinical utility for the detection of active CMV infection.
ACKNOWLEDGMENTS
OTC, the manufacturer of the NucliSens CMV pp67 assay, funded this study. C. C. Ginocchio was funded in part by the Jane and Dayton Brown and Dayton Brown, Jr., Molecular Diagnostics Laboratory.
REFERENCES
- 1.Blok M J, Goossens V J, Vanherle S J V, Top B, Tacken N, Middeldorp J M, Christiaans M H L, van Hooff J P, Bruggeman C A. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J Clin Microbiol. 1998;36:1341–1346. doi: 10.1128/jcm.36.5.1341-1346.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blok M J, Christiaans M H L, Goossens V J, van Hooff J P, Sillekens P, Middeldorp J M, Bruggeman C A. Early detection of human cytomegalovirus infection after kidney transplantation by nucleic acid sequence based amplification. Transplantation. 1999;67:1274–1277. doi: 10.1097/00007890-199905150-00013. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Depto A S, Stenberg R M. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drew W L. Cytomegalovirus infection in patients with AIDS. Clin Infect Dis. 1992;14:608–615. doi: 10.1093/clinids/14.2.608-a. [DOI] [PubMed] [Google Scholar]
- 6.Drew W L, Mintz L, Miner R C, Sands M, Ketterer B. Prevalence of cytomegalovirus infection in homosexual men. J Infect Dis. 1981;143:188–192. doi: 10.1093/infdis/143.2.188. [DOI] [PubMed] [Google Scholar]
- 7.Geballe A P, Leach F S, Mocarski E S. Regulation of cytomegalovirus late gene expression: γ genes are controlled by posttranscriptional events. J Virol. 1986;57:864–874. doi: 10.1128/jvi.57.3.864-874.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerna G, Baldanti F, Middeldorp J M, Furione M, Zavattoni M, Lilleri D, Revello M G. Clinical significance of expression of human cytomegalovirus pp67 late transcript in heart, lung, and bone marrow transplant recipients as determined by nucleic acid sequence-based amplification. J Clin Microbiol. 1999;37:902–911. doi: 10.1128/jcm.37.4.902-911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gleaves C A, Smith T F, Shuster E A, Pearson G R. Comparison of standard tube and shell vial cell culture techniques for the detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 1985;21:217–221. doi: 10.1128/jcm.21.2.217-221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gold E, Nankervis G A. Cytomegalovirus. In: Evans A S, editor. Viral infections of humans: epidemiology and control. New York, N.Y: Plenum Press; 1976. pp. 143–161. [Google Scholar]
- 11.Hanshaw J V. Cytomegalovirus. In: Remingon J S, Klein J O, editors. Infectious diseases of the fetus and newborn infant. W. B. Philadelphia, Pa: Saunders; 1990. pp. 242–281. [Google Scholar]
- 12.Hibberd P L, Snydman D R. Cytomegalovirus infection in organ transplant recipients. Infect Dis Clin N Am. 1995;9:863–877. [PubMed] [Google Scholar]
- 13.Ho M. Cytomegalovirus infection and indirect sequelae in the immunocompromised transplant patient. Transplant Proc. 1991;23:2–7. [PubMed] [Google Scholar]
- 14.Humar A, Gregson D, Caliendo A M, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation. 1999;68:1305–1311. doi: 10.1097/00007890-199911150-00015. [DOI] [PubMed] [Google Scholar]
- 15.Matsunaga T, Sakamaki S, Ishigaki S, Kohda K, Takeda M, Katoh J, Kuroda H, Hirayama Y, Kusakabe T, Akiyama T, Kuga T, Niitsu Y, Masaoka T, Sagawa T, Matsumoto Y. Use of PCR serum in diagnosing and monitoring CMV reactivation in bone marrow transplant recipients. Int J Hematol. 1999;69:105–111. [PubMed] [Google Scholar]
- 16.Revello M G, Percivalle E, Di Matteo A, Morini F, Gerna G. Nuclear expression of the lower matrix protein of human cytomegalovirus in peripheral blood leukocytes of immunocompromised viraemic patients. J Gen Virol. 1992;73:437–442. doi: 10.1099/0022-1317-73-2-437. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds D W, Stagno S, Hosty T S, Tiller M, Alford C A., Jr Maternal cytomegalovirus excretion and perinatal infection. N Engl J Med. 1973;289:1–5. doi: 10.1056/NEJM197307052890101. [DOI] [PubMed] [Google Scholar]
- 18.Sia I G, Wilson J A, Espy M J, Paya C V, Smith T F. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J Clin Microbiol. 2000;38:600–606. doi: 10.1128/jcm.38.2.600-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanabe K, Tokumoto T, Ishikawa N, Koyama I, Takahashi K, Fuchinoue S, Kawai T, Koga S, Yagisawa T, Toma H, Ota K, Nakajiima H. Comparative study of cytomegalovirus (CMV) antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation. 1997;64:1721–1725. doi: 10.1097/00007890-199712270-00016. [DOI] [PubMed] [Google Scholar]
- 20.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescence (ECL) labeled probes. J Virol Methods. 1994;49:157–168. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 21.Weinberg A, Hodges T N, Li S, Cai G, Zamora M R. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol. 2000;38:768–772. doi: 10.1128/jcm.38.2.768-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witt D J, Kemper M. Techniques for the evaluation of nucleic acid amplification technology performance with specimens containing interfering substances: efficacy of Boom methodology for extraction of HIV-1 RNA. J Virol Methods. 1999;79:97–111. doi: 10.1016/s0166-0934(99)00011-7. [DOI] [PubMed] [Google Scholar]