Key Points
CEBPA mutation in the bZIP domain is associated with favorable prognosis in de novo AML, even if it was detected as CEBPAsm.
Visual Abstract
Abstract
Mutations of CCAAT/enhancer–binding protein alpha (CEBPAmu) are found in 10% to 15% of de novo acute myeloid leukemia (AML) cases. Double-mutated CEBPA (CEBPAdm) is associated with a favorable prognosis; however, single-mutated CEBPA (CEBPAsm) does not seem to improve prognosis. We investigated CEBPAmu for prognosis in 1028 patients with AML, registered in the Multi-center Collaborative Program for Gene Sequencing of Japanese AML. It was found that CEBPAmu in the basic leucine zipper domain (bZIP) was strongly associated with a favorable prognosis, but CEBPAmu out of the bZIP domain was not. The presence of CEBPAmu in bZIP was a strong indicator of a higher chance of achieving complete remission (P < .001), better overall survival (OS; P < .001) and a lower risk of relapse (P < .001). The prognostic significance of CEBPAmu in bZIP was also observed in the subgroup with CEBPAsm (all patients: OS, P = .008; the cumulative incidence of relapse, P = .063; patients aged ≤70 years and with intermediate-risk karyotype: OS, P = .008; cumulative incidence of relapse, P = .026). Multivariate analysis of 744 patients aged ≤70 years showed that CEBPAmu in bZIP was the most potent predictor of OS (hazard ratio, 0.3287; P < .001). CEBPAdm was validated as a cofounding factor, which was overlapping with CEBPAmu in bZIP. In summary, these findings indicate that CEBPAmu in bZIP is a potent marker for AML prognosis. It holds potential in the refinement of treatment stratification and the development of targeted therapeutic approaches in CEBPA-mutated AML.
Introduction
Genetic abnormalities are potent prognostic markers for acute myeloid leukemia (AML). Recently, mutated genes in AML have been assessed in addition to the conventional chromosomal analysis, which dates back to the 1990s. Mutated genes have been affirmed as important prognostic markers, and hence widespread screening for genetic mutations has been initiated in clinical practice.1-3 CCAAT/enhancer–binding protein alpha (CEBPA) is one of the most important AML prognostic genes, and its mutation (CEBPAmu) is detected in 10% to 15% of de novo AML cases. One-third of CEBPAmu are single-mutated CEBPA (CEBPAsm), a heterozygous monoallelic mutation, and two-thirds are double-mutated CEBPA (CEBPAdm), usually biallelic N- and C-terminal mutations. Of these mutations, CEBPAdm is frequently detected in the intermediate-risk karyotype group, making it a favorable prognostic factor. However, CEBPAsm has a poorer prognosis than CEBPAdm; thus, the usefulness of CEBPAsm as a prognostic factor has not been clarified.4-9 The mechanisms underlying this biological paradox, wherein the group with biallelic mutations is associated with a better prognosis than the group with monoallelic mutations, has yet to be explained.
CEBPA messenger RNA has 2 translation initiation sites and 2 isoforms: a full-length p42 (42 kDa) and a p30 (30 kDa) with weak transcriptional activity and lacking an N-terminal transactivation domain-1.8 The structure common to both isoforms is the basic leucine zipper domain (bZIP domain), located on the C-terminal side. This bZIP is a structure found in many transcription factors and has an important role in protein dimerization. This part enables DNA binding to the major groove of the DNA molecule. Interestingly, mutation in both the C-terminal and N-terminal of the bZIP domain accounts for the majority of the mutations. Both types of mutations have different functions and augment the development of AML.10 Conversely, one-third of CEBPAsm are mutations in the bZIP domain, and the remaining two-thirds are mutations out of the bZIP domain. However, investigations regarding the effect of the location of CEBPAsm on prognosis are rare.
In this study, we retrospectively analyzed the clinical features of AML with CEBPAmu based on clinical sequencing data conducted at a multicenter joint study in Japan, and we investigated the effect of the location of CEBPAmu on prognosis.
Materials and methods
Study population
This study was reviewed and approved by the Human Subjects Institutional Review Board (project approval number 29-07-783) of the Nippon Medical School (Tokyo, Japan), and informed consent was obtained in accordance with the Declaration of Helsinki from all participants. All patients in this analysis were enrolled and selected by the Multi-center Collaborative Program for Gene Sequencing of Japanese AML conducted by Nippon Medical School. Briefly, Japanese residents aged ≥16 years, who had had de novo AML since 2001, were enrolled in this observational study after obtaining their consent. The following test results of the patients with AML since 2009 were provided by the investigators to physicians within 1 month: FLT3 internal tandem duplication (FLT3-ITD) (polymerase chain reaction assay from 2009, fragment analysis from 2018), nucleophosmin1 (NPM1) exon12 (from 2009), CEBPA (from 2009), and DNA methyltransferase 3A (DNMT3A) R882 (from 2017). These data were used for decision-making in clinical practice. Baseline clinical, laboratory, and treatment data were abstracted by using a standard protocol. Treatment response data were extracted from medical records; the response was solely based on the treating physician’s documentation.
Patient samples
This analysis included patients with de novo AML (excluding the FAB-M3 subtype) who were enrolled in the Multi-center Collaborative Program for Gene Sequencing of Japanese AML from 2001 to 2019. Patient samples were collected after diagnosis, and genomic DNA extraction was conducted in patients with ≥20% blasts in bone marrow or peripheral blood. For genomic DNA extraction, mononuclear cells from bone marrow or peripheral blood were isolated by density gradient centrifugation using lymphocyte separation medium (Organon Teknika Corp., Durham, NC). Genomic DNA of mononuclear cells was extracted with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The clinical sequencing for FLT3-ITD, NPM1 exon12, CEBPA, and DNMT3A R882 were conducted within 1 month, whereas target-captured sequencing for the AML gene panel using cryopreserved samples was performed later according to the procedures stated in the "Mutational analysis" section.
Screening for cytogenetic mutations
G-band analysis was performed on bone marrow samples obtained from patients at initial presentation. When obtaining bone marrow samples was difficult, peripheral blood was used. For patients suspected of being M2 (myeloblastic with differentiation), M3 (promyelocytic), or M4eo (myelomonocytic with eosinophilia) based on the French-American-British classification, fluorescence in situ hybridization analysis was used to additionally search for Runt-related transcription factor (RUNX1-RUNX1T1), promyelocytic leukemia/retinoic acid receptor alpha (PML-RARA), and core-binding factor b-myosin heavy chain 11 (CBFB-MYH11) mutations. The cytogenetic prognosis was then classified in accordance with the system recommended by the 2017 European Leukemia Net classification (ELN 2017).
Mutational analysis
For the screening of NPM1 and CEBPA mutations, Sanger sequencing of exon12 of the NPM1 gene and the entire exon of CEBPA gene was performed.11 We defined mutations of the bZIP domain as mutations whose starting positions were in the bZIP coding region. FLT3-ITD was screened by polymerase chain reaction assay, and quantitative fragment analysis of FLT3-ITD was conducted for calculating the FLT3-ITD allelic ratio (ITD-AR).12 For this study, high-AR and low-AR were defined as >0.5 and <0.5 of ITD-AR, respectively. DNMT3A R822 mutations were detected by using i-densy (ARKRAY, Inc., Kyoto, Japan), a fully automated single-nucleotide polymorphism genotyping system based on the quenching probe method.13 The detected mutations were confirmed by Sanger sequencing.
Target-captured sequencing for the AML gene panel
An oligonucleotide library construction and template preparation were generated by Ion Chef (Thermo Fisher Scientific, Waltham, MA) using order-made probes designed against the genes of the AML gene panel (supplemental Table 1). The library was sequenced with a next-generation sequencer, Ion Proton (Thermo Fisher Scientific). With respect to detected mutations, the National Center for Biotechnology Information and the Catalogue Of Somatic Mutations In Cancer databases were used to search for polymorphisms and cancer-related mutations. For newly identified mutations, we searched for genetic polymorphisms using Sanger sequencing with remission-stage samples.
Statistical analysis
The primary end point was overall survival (OS). Cumulative incidence of relapse (CIR) for patients who had achieved complete remission (CR) was calculated from the time interval between the date of CR to the date of relapse. The χ2 test and Fisher’s exact test were used to test the association between categorical variables and the presence and absence of mutations. The nonparametric Mann-Whitney U test was used to determine the statistical significance of differences in median values. All statistical tests were two-sided. The Kaplan-Meier method and log-rank test were used to analyze OS and CIR. Patients were alive when censored at the last follow-up. The Pearson coefficient was used to score the correlations among concurrent mutations. With respect to the prognostic factors, multivariate analysis was conducted with the Cox proportional hazards model. A backward and forward stepwise procedure selection model with the Akaike information criterion was used to extract independent events.
Statistical analyses were performed by using GraphPad Prism version 9.00 for Windows (GraphPad Software, La Jolla, CA) and IBM SPSS Statistics version 21.0 for Windows (IBM SPSS Statistics, IBM Corporation, Armonk, NY). Power calculations for sample size estimation were performed by using GraphPad StatMate version 2.00 for Windows.
Results
Patient background
Of the 1414 patients enrolled in the study, 377 were excluded either because their samples did not satisfy our sample criteria or their clinical information was not available. Moreover, 9 patients were excluded because they were treated with FLT3 inhibitors at the induction phase. The remaining 1028 patients were included in the analysis. The follow-up information of 41 patients was missing, and they were censored at time 0 for the OS analysis.
Supplemental Table 2 details the background of the 1028 patients enrolled in this study. Of these patients, 864 were ≤70 years old, and the median observation period was 686.1 days. Chromosomal classification according to ELN 2017 yielded 146 cases with favorable-risk, 647 cases with intermediate-risk, and 160 cases with adverse-risk karyotype; 73 cases had unknown karyotypes due to reasons such as no cell division. In accordance with previous reports, OS prognosis stratification was possible with chromosome analysis in this cohort as well (OS for all patients, P < .001; OS for patients aged ≤70 years, P < .001) (supplemental Figure 1). The 7 + 3 induction regimen, which consists of 7 days of standard-dose cytarabine (100-200 mg/m2 continuous infusion) and 3 days of an anthracycline antibiotic infusion (idarubicin 12 mg/m2 or daunorubicin 60-90 mg/m2), was used as a first induction therapy in 80.8% of the cases. As a postremission treatment, hematopoietic stem cell transplantation was performed in 151 patients during the first remission phase. The donor sources were auto peripheral blood stem cell (n = 5), HLA-matched related donor (n = 37), HLA-matched unrelated donor (n = 73), cord blood (n = 32), and 1-haploidentical donor (n = 4).
Distribution and frequency of CEBPA mutations
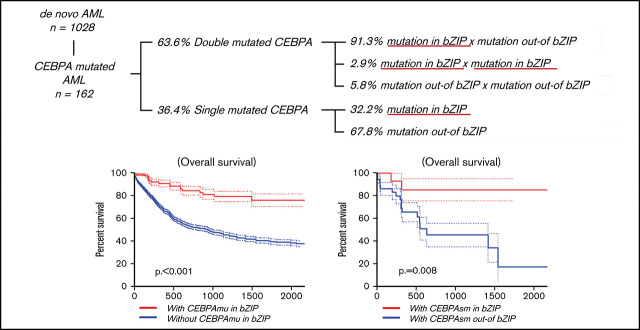
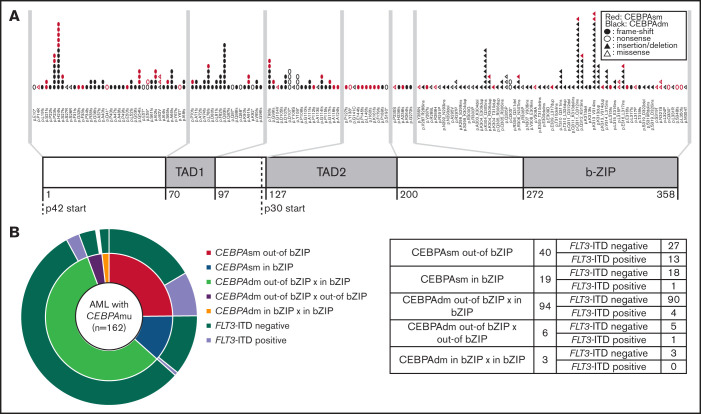
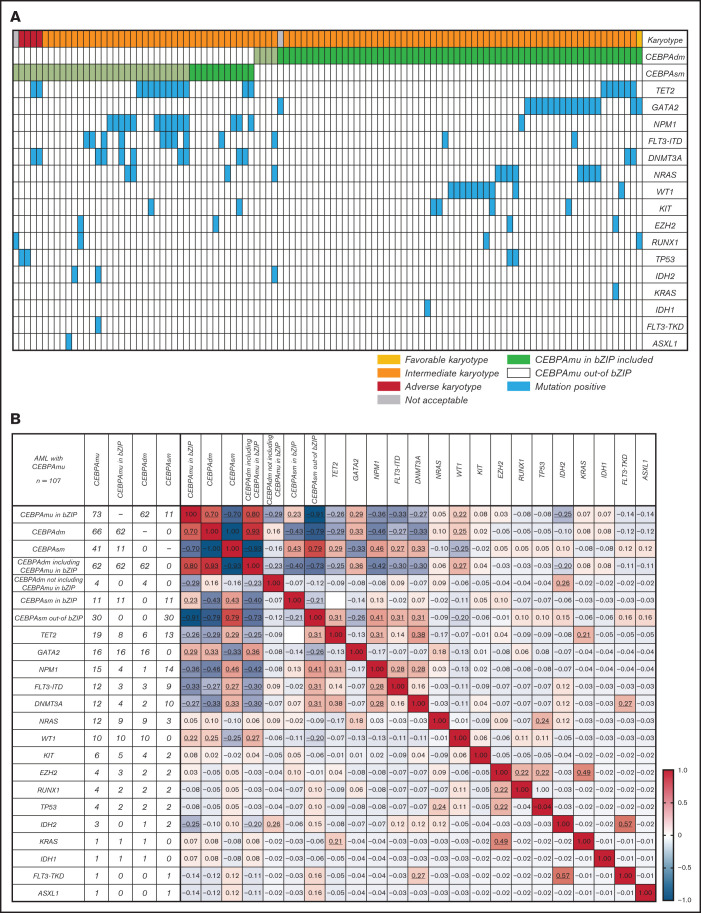
The distribution and frequency of all CEBPAmu are shown in Figure 1A-B, respectively. There were 59 patients with CEBPAsm and 103 patients with CEBPAdm among the total 1028 patients. Of the patients with CEBPAdm, 91.3% (94 of 103) had a combination of mutations in the bZIP domain (CEBPAmu in bZIP) and mutations out of the bZIP domain (ie, CEBPAmu out-of bZIP), 2.9% (3 of 103) of the patients had 2 CEBPAmu in bZIP, and 5.8% (6 of 103) of the patients had 2 CEBPAmu out-of bZIP. Furthermore, 32.2% (19 of 59) of the patients with CEBPAsm had mutations in the bZIP domain (CEBPAsm in bZIP), and 67.8% (40 of 59) of patients had mutations out-of the bZIP domain (CEBPAsm out-of bZIP). In total, 116 of all the patients had CEBPAmu in bZIP, irrespective of AML with CEBPAdm or CEBPAsm. AML patients with CEBPAdm had long OS and CIR, but CEBPAsm was not a significant prognostic marker (OS for all patients, P < .001; CIR for all patients, P = .015; OS for patients aged ≤70 years and with intermediate-risk karyotype, P < .001; CIR for patient aged ≤70 years and with intermediate-risk karyotype, P = .009) (supplemental Figure 2).
Figure 1.
Summary of CEBPA mutations in the primary cohort. (A) Distribution of CEBPA mutations. (B) Overlapping pattern of CEBPA mutations
Clinical significance of AML with CEBPAmu in bZIP
Supplemental Table 2 describes the background of patients with AML with and without CEBPAmu in bZIP. Almost all cases of AML with CEBPAmu in bZIP were classified into the intermediate-risk chromosomal classification. The NPM1 mutation was detected in 27.8% (286 of 1028) of all AML cases and 5.2% (6 of 116) of patients with AML with CEBPAmu in bZIP. FLT3-ITD was detected in 21.0% (216 of 1028) of all AML cases and 4.3% (5 of 116) of patients with AML with CEBPAmu in bZIP. The incidence of both these mutations was significantly lower than AML without CEBPAmu in bZIP (P < .001). Based on these findings, the NPM1 mutation and FLT3-ITD were considered far less frequent in AML with CEBPAmu in bZIP.
The rate of achieving CR in the induction phase was calculated only for patients who underwent remission induction therapy. The CR rate for all the AML cases was 67.2% for all age groups and 72.0% for patients aged ≤70 years. The CR rate for AML with CEBPAmu in bZIP was 90.2% for all age groups and 92.7% for patients aged ≤70 years; these values were significantly higher than AML without CEBPAmu in bZIP (all age groups, P < .001; patients aged ≤70 years, P < .001) (supplemental Table 2).
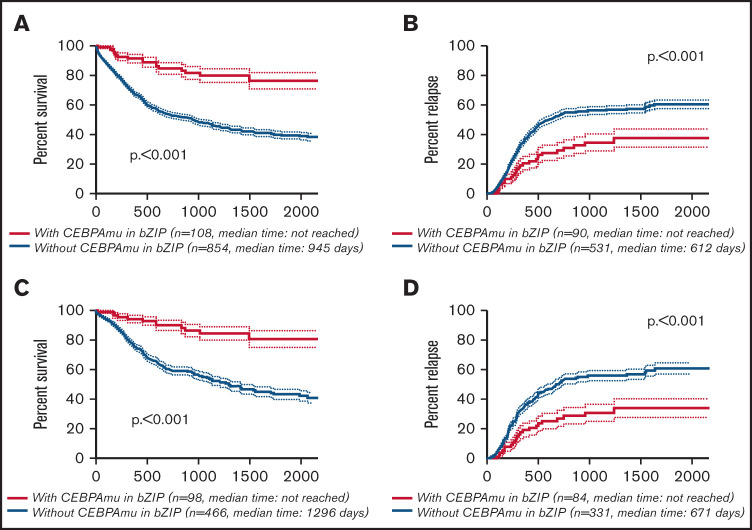
The Kaplan-Meier curve showing OS and CIR of AML with CEBPAmu in bZIP and AML without CEBPAmu in bZIP is shown in Figure 2. AML with CEBPAmu in bZIP had significantly longer OS and was associated with lower CIR than AML without CEBPAmu in bZIP (OS for all patients, P < .001; CIR for all patients, P < .001; OS for patients aged ≤70 years and with intermediate-risk karyotype, P < .001; CIR for patients aged ≤70 years and with intermediate-risk karyotype, P < .001).
Figure 2.
Kaplan-Meier survival curves for OS and CIR comparing patients without CEBPAmu in bZIP and patients with CEBPAmu in bZIP. Analyses were conducted for 962 of 1028 patients who were followed up. Kaplan-Meier curves were stratified according to whether patients with AML have the CEBPA mutation in the bZIP domain: with CEBPAmu in bZIP (red), without CEBPAmu in bZIP (blue). Kaplan-Meier curve of OS for all patients (A), Kaplan-Meier curve of CIR for all patients (B), Kaplan-Meier curve of OS for patients aged ≤70 years and with intermediate-risk karyotype (C), and Kaplan-Meier curve of CIR for patients aged ≤70 years and with intermediate-risk karyotype (D).
Differences between CEBPAsm in bZIP and CEBPAsm out-of bZIP
The effect of the position of CEBPAsm on AML prognosis was analyzed to rule out the possibility of confounding bias caused by the inclusion of CEBPAmu in bZIP in the majority of AML cases with CEBPAdm, given that CEBPAmu in bZIP has a favorable prognosis.
As stated earlier, 67.8% (40 of 59) of patients with AML with CEBPAsm had CEBPAsm out-of bZIP, and 32.2% (19/59) had CEBPAsm in bZIP. Supplemental Table 3 describes the characteristics of patients with AML with CEBPAsm out-of bZIP and in bZIP. As shown, FLT3-ITD mutations were detected in 32.5% (13 of 40) of patients in the CEBPAsm out-of bZIP group, which was higher than the 5.3% (1 of 19) of patients in the CEBPAsm in bZIP group (P = .021). NPM1 mutations were detected in 35.0% (14 of 40) of patients in the CEBPAsm out-of bZIP group and 26.3% (5 of 19) of patients in the CEBPAsm in bZIP group. There was no significant difference in the frequency of NPM1 mutations between the CEBPAsm out-of bZIP group and the CEBPAsm in bZIP group.
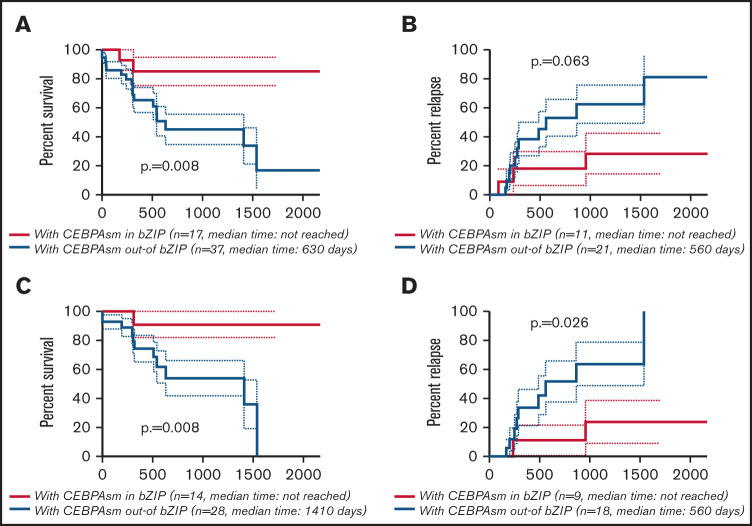
The CR rate was higher in AML with CEBPAsm in bZIP, but this difference was not significant (AML with CEBPAsm in bZIP, 80.0%; AML with CEBPAsm out-of bZIP, 57.5%; P = .132) (supplemental Table 3). The Kaplan-Meier curves showing OS and CIR of AML with CEBPAsm in bZIP and out-of bZIP are shown in Figure 3A-B (all patients with CEBPAsm) and Figure 3C-D (patients with CEBPAsm, aged ≤70 years and with intermediate-risk karyotype), respectively. AML with CEBPAsm in bZIP had a significantly longer OS than AML with CEBPAsm out-of bZIP. The median OS of patients with CEBPAsm was as follows: AML with CEBPAsm in bZIP, not reached; AML with CEBPAsm out-of bZIP, 630 days (P = .008). The median time to relapse in patients with CEBPAsm was as follows: AML with CEBPAsm in bZIP, not reached; AML with CEBPAsm out-of bZIP, 560 days (P = .063). In the group of patients aged ≤70 years and with intermediate-risk karyotype, AML with CEBPAsm in bZIP had a significantly longer OS and CIR than AML with CEBPAsm out-of bZIP. The median OS of the group of patients aged ≤70 years and with intermediate-risk karyotype was as follows: AML with CEBPAsm in bZIP, not reached; AML with CEBPAsm out-of bZIP, 1410 days (P = .008). The median time to relapse in the group of patients aged ≤70 years and with intermediate-risk karyotype was as follows: AML with CEBPAsm in bZIP, not reached; AML with CEBPAsm out-of bZIP, 560 days (P = .026). These findings suggest that CEBPAmu in bZIP is a favorable prognostic marker in patients with AML with CEBPAsm.
Figure 3.
Kaplan-Meier survival curves for OS and CIR of patients with CEBPAsm comparing CEBPAsm in bZIP and CEBPAsm out-of bZIP. Analyses were performed for 54 of 59 patients with CEBPAsm AML who were followed up. Kaplan-Meier curves were stratified according to whether CEBPA mutation was “in” or “out-of” the bZIP domain: red, CEBPAmu in bZIP; blue, CEBPAmu out-of bZIP. (A) Kaplan-Meier curve of OS for all patients (A), Kaplan-Meier curve of CIR for all patients (B), Kaplan-Meier curve of OS for patients aged ≤70 years and with intermediate-risk karyotype (C), and Kaplan-Meier curve of CIR for patients aged ≤70 years and with intermediate-risk karyotype (D).
We compared 3 groups (CEBPAsm out-of bZIP/FLT3-ITD negative, CEBPAsm out-of bZIP/FLT3-ITD positive, and CEBPAsm in bZIP/FLT3-ITD negative) to investigate the effect of FLT3ITD, which is an adverse prognostic factor (there was only 1 case of CEBPAsm in the bZIP/FLT3-ITD positive group, and thus it was not included in the analysis) (supplemental Figure 3). FLT3-ITD tends to have a poor prognosis, but there was no significant difference in OS and CIR between the CEBPAsm out-of bZIP/FLT3-ITD–negative group and the CEBPAsm out-of bZIP/FLT3-ITD–positive group. When we compared the CEBPAsm in bZIP/FLT3-ITD–negative group and the CEBPAsm out-of bZIP/FLT3-ITD–negative group, AML with CEBPAsm in bZIP had significantly longer OS than AML with CEBPAsm out-of bZIP, even in the FLT3-ITD–negative group (all patients with CEBPAsm positive/FLT3-ITD negative, P = .018; patients aged ≤70 years and with CEBPAsm positive/FLT3-ITD negative, P = .022). Furthermore, we analyzed the OS in a subgroup who were FLT3-ITD negative, NPM1 mutation negative, and CEBPAsm positive, to eliminate the effect of FLT3-ITD and NPM1 mutation; CEBPAsm in bZIP remained a significantly favorable prognostic marker for predicting OS (all patients with CEBPAsm positive/FLT3-ITD negative/NPM1 mutation negative, P = .049; patients aged ≤70 years and with CEBPAsm positive/FLT3-ITD negative/NPM1 mutation negative: P = .018) (supplemental Figure 4).
Overlapping genetic mutations with CEBPAmu
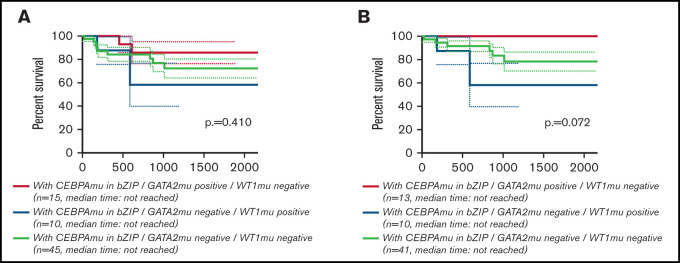
Global overlapping mutations were assessed by retrospective analysis of preserved DNA samples where possible, using next-generation sequencing analysis. The analysis was performed in 107 of 151 patients with AML with CEBPAmu, 66 patients with AML with CEBPAdm, and 41 patients with AML with CEBPAsm. The overview of individual somatic mutations detected in AML with CEBPAmu and a correlation matrix showing the Pearson correlation coefficient among CEBPAmu and common concurrent mutations are shown in Figure 4. The gene mutations were described in ELN 2017 and detected at a frequency ≥3% in CEBPAmu cases. In the Pearson correlation analysis, a higher frequency of GATA2, WT concurrent mutations and a lower frequency of NPM1, TET2, DNMT3A, FLT3-ITD, and IDH2 concurrent mutations were noted in AML with CEBPAmu in bZIP. The GATA2 mutation and WT1 mutation, which are frequently detected in AML with CEBPAmu in bZIP, were exclusively mutually detected. When we investigated the prognosis of GATA2/WT1, the 3-year OS in genotypes that were GATA2 negative/WT1 negative, GATA2 positive/WT1 negative, and GATA2 negative/WT1 positive was 72.27% (n = 45), 85.71% (n = 15), and 58.33% (n = 10), respectively, for all patients, and 78.53% (n = 41), 100% (n = 13), and 58.33% (n = 10) for patients aged ≤70 years and with intermediate-risk karyotype. Comparison of the 3 groups did not reveal any significant difference, but comparison of genotypes that were GATA2 positive/WT1 negative and GATA2 negative/WT1 positive in patients aged ≤70 years and with an intermediate-risk karyotype found a significant difference, with the GATA2 positive/WT1 negative genotype having a favorable prognosis (P = .016) (Figure 5).
Figure 4.
The spectrum of concurrent mutations among different genes. (A) The overview of individual somatic mutations detected in AML with CEBPAmu. Columns represent patients with CEBPA mutations (66 patients with CEBPAdm and 41 patients with CEBPAsm), and rows represent the genotypes. (B) Correlation matrix based on the Pearson correlation coefficient analyses. The Pearson product-moment correlation coefficients were calculated, and correlation matrices were constructed for 107 patients with AML with CEBPAmu. Different colors are used to represent different correlation strengths. The color scale is defined by the color bar legend. Here, the red color suggests a strong positive correlation, whereas the blue color indicates a strong negative correlation. The underline represents the P value <.05.
Figure 5.
Kaplan-Meier survival curves for OS of patients with CEBPAmu in bZIP comparing the 3 genotypes (GATA2 positive/WT1 negative, GATA2 negative/WT1 positive, GATA2 negative/WT1 negative). Kaplan-Meier curves were stratified according to the 3 genotypes: red, GATA2 positive/WT1 negative; blue, GATA2 negative/WT1 positive; and gray, GATA2 negative/WT1 negative. There was no genotype of GATA2 positive/WT1 positive. (A) Kaplan-Meier curve of OS for all patients (among the 3 groups P = .410; GATA2 positive/WT1 negative vs GATA2 negative/WT1 positive, P = .154; GATA2 negative/WT1 positive vs GATA2 negative/WT1 negative, P = .483; GATA2 positive/WT1 negative vs GATA2 negative/WT1 negative, P = .366). (B) Kaplan-Meier curve of OS for patients aged ≤70 years and with intermediate-risk karyotype (among the 3 groups, P = .072; GATA2 positive/WT1 negative vs GATA2 negative/WT1 positive, P = .016; GATA2 negative/WT1 positive vs GATA2 negative/WT1 negative, P = .208; GATA2 positive/WT1 negative vs GATA2 negative/WT1 negative, P = .113).
Multivariate analyses for OS and CIR
Multivariate analyses for OS and CIR were conducted on patients of the transplantation-adapted age of ≤70 years using the forward-backward stepwise method to evaluate the effect of year of diagnosis (2001-2009 or 2010-2019), age (>60 years or ≤60 years), sex, white blood cell count at first visit (≥20 000/μL or <20 000/μL), favorable-risk karyotype, adverse-risk karyotype, FLT3-ITD high allelic ratio, FLT3-ITD low allelic ratio, NPM1 mutation (positive or negative), CEBPAmu (positive or negative), CEBPAdm (positive or negative), and CEBPAmu in bZIP (positive or negative). The Akaike information criterion was adopted for model selection. Multivariate analysis for OS in 744 patients who were aged ≤70 years and had also undergone CEBPA mutation analysis, NPM1 mutation analysis, and FLT3-ITD fragment analysis revealed that CEBPAmu in bZIP was the strongest favorable prognostic factor of OS (hazard ratio, 0.3287; 95% confidence interval, 0.1852-0.5834; P < .001) (Table 1). In addition, multivariate analysis for CIR in 525 patients who achieved CR showed that CEBPAmu in bZIP was an independent favorable prognostic factor of CIR hazard ratio (0.6157; 95% confidence interval, 0.3931-0.9644; P = .034) (Table 2). On the other hand, CEBPAdm was not selected as an independent prognostic factor, indicating that it was a confounding factor for CEBPAmu in bZIP.
Table 1.
Multivariate analysis for OS in patients of the transplantation-adapted age of ≤70 years
| Variable | Hazard ratio | 95% confidence interval | P |
|---|---|---|---|
| CEBPAmu in bZIP domain | 0.3287 | 0.1852-0.5834 | <.001 |
| Favorable-risk karyotype | 0.5349 | 0.3539-0.8084 | <.001 |
| Year of diagnosis (in and after 2010) | 0.6804 | 0.4845-0.9554 | .026 |
| Female sex | 0.7218 | 0.5608-0.9291 | .011 |
| FLT3-ITD high AR | 1.917 | 1.34-2.743 | <.001 |
| Age ≥60 y | 1.959 | 1.525-2.517 | <.001 |
| Adverse-risk karyotype | 2.397 | 1.791-3.208 | <.001 |
n = 744, number of events = 265.
Table 2.
Multivariate analysis for CIR in patients of the transplantation-adapted age of ≤70 years
| Variable | Hazard ratio | 95% confidence interval | P |
|---|---|---|---|
| Year of diagnosis (in and after 2010) | 0.5284 | 0.3656-0.7639 | <.001 |
| CEBPAmu in the bZIP domain | 0.6157 | 0.3931-0.9644 | .034 |
| White blood cell count ≥20 000/μL | 1.325 | 0.9956-1.763 | .054 |
| Adverse-risk karyotype | 1.549 | 0.9442-2.54 | .083 |
| FLT3-ITD high AR | 1.762 | 1.164-2.667 | <.001 |
| Age ≥60 y | 1.93 | 1.456-2.559 | <.001 |
n = 525, number of events = 210 (219 cases were excluded from the analysis due to not achieving CR).
Discussion
In this study, we analyzed a large-scale cohort of 1028 cases of de novo AML, and the results showed that CEBPAdm is a favorable prognostic marker, as indicated in previous reports.4-9 CEBPAmu in bZIP, which is abundant in CEBPAdm, is a strong favorable prognostic factor. It was noted that if CEBPAsm was in the bZIP domain, it serves as a favorable prognostic factor. The results of this analysis reinforce the usefulness of the previous findings stating that CEBPAdm is a favorable prognostic factor, and it clarifies the importance of CEBPAmu in bZIP.
This study also conducted comprehensive retrospective genetic mutation analysis of preserved DNA samples where possible, using a next-generation sequencer on 107 patients with AML with CEBPAmu. We discovered that there were differences in the genetic background of AML with CEBPAmu in bZIP and AML without CEBPAmu in bZIP. GATA2 and WT1, which are frequently detected in AML with CEBPAmu in bZIP, allow for further stratification of the prognosis of AML with CEBPAmu in bZIP. Previous retrospective studies on small numbers of patients with AML with CEBPAdm have reported the importance of GATA2 mutations as a favorable prognostic factor and WT1 mutations as a poor prognostic factor. The results of this study can be regarded as data that support the previous findings.14,15 Interestingly, GATA2 and WT1 mutations were exclusively found in cases with CEBPAdm but not in cases with CEBPAsm. This finding suggests that CEBPAdm including bZIP mutations and CEBPAsm in bZIP were still separate biological entities.
The most important finding in this analysis is that CEBPAmu in bZIP was detected as a strong independent favorable prognostic factor even in multivariate analyses conducted on patients of the transplantation-adapted age of ≤70 years. CEBPAdm, which has been recommended in many previous guidelines, was found to be a confounding factor in the CEBPAmu in bZIP group and was not obtained as an independent prognostic factor. Given the prognostic importance, an accurate testing method is necessary to assess CEBPAmu for all patients with AML, but many laboratories struggle with implementing a reliable and sensitive CEBPA mutation assay for routine diagnostic purposes.16-18 The development of a reliable and sensitive CEBPA mutation assay is complicated by the GC-rich DNA sequences of the gene (75% in the coding region), presence of a trinucleotide repeat region, and frequent occurrence of mutations in mononucleotide repeats. Similarly, inclusion of CEBPA as part of next-generation sequencing panels has been interrupted by poor amplicon coverage and misidentification of variants. Consequently, an extensive whole exon screening by Sanger sequencing, which is labor intensive and expensive, is needed to identify CEBPAdm. A mutation assay for CEBPAmu in bZIP is very simple, as it only analyzes the 89 bp bZIP coding region. The simplification of CEBPA mutation analysis, as proposed in this study, is of great significance for AML treatment.
Previous studies suggest that CEBPAmu out-of bZIP is deficient in full-length p42 CEBPA and produces p30CEBPA without a N-terminal transactivation domain-1 region, but CEBPAmu in bZIP forms a bZIP domain mutant with an altered bZIP function.19-21 We are speculating that this function acquired by the CEBPA bZIP mutant determines the sensitivity of the disease to chemotherapy. The mechanisms underlying this biological paradox, which has previously been difficult to explain, states that a group with biallelic mutations is associated with a better prognosis than a group with monoallelic mutations, and this may be elucidated through further functional analysis of CEBPAmu in bZIP.
The limitations of this study include its retrospective nature and a selection bias for different treatment regimens. In addition, the presented outcomes are limited to the Japanese population and, thereby, a restricted sample size. In particular, the number of samples used for next-generation sequencing was too small to clarify the heterogeneity of CEBPA-mutated AML. To confirm the prognostic value of CEBPAmu for AML and the details of the genetic background of CEBPA-mutated AML, further studies are required on larger groups of patients with AML from diverse populations.
In summary, this study suggests that CEBPAmu in bZIP is strongly associated with a favorable prognosis, whereas CEBPAmu out-of bZIP is not. CEBPAmu in bZIP is a better prognostic marker than CEBPAdm. It is a strong indicator for achievement of complete remission, better survival outcome, and cumulative incidence of relapse. This study aids in better prognosis and development of treatment stratification for improved management of AML.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors are very grateful to all the physicians who managed the patients and collected clinical data in this study.
Authorship
Contribution: S.W. and M. Sakaguchi were the principal investigators and take primary responsibility for the paper; H.Y. designed the study and supervised the experiments; S.W. and M. Sakaguchi designed and performed the experiments, interpreted the data, and wrote the manuscript; I. Oh, S. Kako, T.T., Y. Najima, N.D., J. Kanda, J. Kuroda, S. Mori, A.S., K.U., T.U., N. Uoshima, Y. Kobayashi, E.K., K. Tajika, Y. Nagao, K.S., M. Shibusawa, J.T., K.K., M.H., H.U., N. Uchida, Y. Kubota, S. Kimura, H.N., T.I., S. Kurosawa, S. Motomura, A.H., H.M., E.S., M.O., K.M., J.A., N.K., T.F., K.O, Y. Kanda, and K.I. contributed to patient recruitment and data extraction; K.A., T.K, M.M., A.K., and A. Mizoguchi performed the laboratory work for the study; and A. Marumo, I. Omori, Y.F., K. Terada, and S.Y. interpreted the molecular data and performed the statistical analysis; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroki Yamaguchi, Department of Hematology, Nippon Medical School, 1-1-5 Sendagi, Bunkyo-Ku, Tokyo 113-8603, Japan; e-mail: y-hiroki@fd6.so-net.ne.jp.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136-1152. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurosawa S, Yakushijin K, Yamaguchi T, et al. Changes in incidence and causes of non-relapse mortality after allogeneic hematopoietic cell transplantation in patients with acute leukemia/myelodysplastic syndrome: an analysis of the Japan Transplant Outcome Registry. Bone Marrow Transplant. 2013;48(4):529-536. [DOI] [PubMed] [Google Scholar]
- 4.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100(8):1343-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol. 2010;28(16):2739-2747. [DOI] [PubMed] [Google Scholar]
- 7.Dufour A, Schneider F, Metzeler KH, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28(4):570-577. [DOI] [PubMed] [Google Scholar]
- 8.Taskesen E, Bullinger L, Corbacioglu A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469-2475. [DOI] [PubMed] [Google Scholar]
- 9.Schlenk RF, Taskesen E, van Norden Y, et al. The value of allogeneic and autologous hematopoietic stem cell transplantation in prognostically favorable acute myeloid leukemia with double mutant CEBPA. Blood. 2013;122(9):1576-1582. [DOI] [PubMed] [Google Scholar]
- 10.Avellino R, Delwel R. Expression and regulation of C/EBPα in normal myelopoiesis and in malignant transformation. Blood. 2017;129(15):2083-2091. [DOI] [PubMed] [Google Scholar]
- 11.Wakita S, Yamaguchi H, Omori I, et al. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27(5):1044-1052. [DOI] [PubMed] [Google Scholar]
- 12.Sakaguchi M, Yamaguchi H, Najima Y, et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2(20):2744-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okabe M, Yamaguchi H, Usuki K, et al. Clinical features of Japanese polycythemia vera and essential thrombocythemia patients harboring CALR, JAK2V617F, JAK2Ex12del, and MPLW515L/K mutations. Leuk Res. 2016;40:68-76. [DOI] [PubMed] [Google Scholar]
- 14.Fasan A, Eder C, Haferlach C, et al. GATA2 mutations are frequent in intermediate-risk karyotype AML with biallelic CEBPA mutations and are associated with favorable prognosis. Leukemia. 2013;27(2):482-485. [DOI] [PubMed] [Google Scholar]
- 15.Tien F-M, Hou H-A, Tang J-L, et al. Concomitant WT1 mutations predict poor prognosis in acute myeloid leukemia patients with double mutant CEBPA. Haematologica. 2018;103(11):e510-e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L-I, Lin T-C, Chou W-C, Tang J-L, Lin D-T, Tien H-F. A novel fluorescence-based multiplex PCR assay for rapid simultaneous detection of CEBPA mutations and NPM mutations in patients with acute myeloid leukemias [published correction appears in Leukemia. 2007;21(10):2236]. Leukemia. 2006;20(10):1899-1903. [DOI] [PubMed] [Google Scholar]
- 17.Benthaus T, Schneider F, Mellert G, et al. Rapid and sensitive screening for CEBPA mutations in acute myeloid leukaemia. Br J Haematol. 2008;143(2):230-239. [DOI] [PubMed] [Google Scholar]
- 18.Behdad A, Weigelin HC, Elenitoba-Johnson KS, Betz BL. A clinical grade sequencing-based assay for CEBPA mutation testing: report of a large series of myeloid neoplasms [published correction appears in J Mol Diagn. 2015;17(2):206]. J Mol Diagn. 2015;17(1):76-84. [DOI] [PubMed] [Google Scholar]
- 19.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27(3):263-270. [DOI] [PubMed] [Google Scholar]
- 20.Cleaves R, Wang QF, Friedman AD. C/EBPalphap30, a myeloid leukemia oncoprotein, limits G-CSF receptor expression but not terminal granulopoiesis via site-selective inhibition of C/EBP DNA binding. Oncogene. 2004;23(3):716-725. [DOI] [PubMed] [Google Scholar]
- 21.Kato N, Kitaura J, Doki N, et al. Two types of C/EBPα mutations play distinct but collaborative roles in leukemogenesis: lessons from clinical data and BMT models. Blood. 2011;117(1):221-233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.