Abstract
Colorectal cancer (CRC) is one of the most common malignancies and has been a leading cause of cancer-related death worldwide in recent years. N6-methyladenosine (m6A) methylation is the most abundant epigenetic modification of various types of RNAs, and it plays a vital role in promoting cancer development. Here, we obtained SNV and transcriptome data of CRC from The Cancer Genome Atlas (TCGA). We demonstrated that most m6A methylation regulators were aberrantly expressed in individuals with CRC. The abnormal expression of m6A regulators was caused by their different copy number variation (CNV) patterns, and alteration of m6A regulators was significantly correlated with prognosis and tumor stage. By using weighted coexpression network analysis (WGCNA), we identified m6A-related long noncoding RNAs (lncRNAs) and mRNAs; then we used least absolute shrinkage and selection operator (LASSO) Cox regression analysis to construct m6A-related lncRNA and mRNA prognostic signatures in the TCGA dataset. Furthermore, a nomogram with clinicopathological features, lncRNA risk scores, and mRNA risk scores was established, which showed a strong ability to forecast the overall survival of the individuals with CRC in training and testing sets. In conclusion, m6A methylation regulators played a vital role in affecting the prognosis of subjects with CRC, and m6A-related lncRNAs and mRNAs revealed underlying mechanisms in CRC tumorigenesis and progression.
Keywords: colorectal cancer, TCGA, N6-methylandenosine, m6A-related lncRNA, m6A-related mRNA, WGCNA, prognosis signature, nomogram
Graphical Abstract

N6-methyladenosine (m6A) modification plays a key role in the post-transcriptional modification of RNAs. In this work, we illustrate the expression landscape of m6A regulators and how m6A-related RNAs forecast prognosis among individuals with CRC. The results expand existing knowledge about m6A modification and might aid in the identification of novel prognostic biomarkers of CRC.
Introduction
In a 2018 global burden of disease study, colorectal cancer (CRC) was identified as one of the most common types of cancer worldwide and the third leading cause of cancer-related death.1 CRC comprises of rectal adenocarcinoma (READ) and colon adenocarcinoma (COAD). It has been identified as the third most prevalent cancer in men and the second in woman globally.2 Although tremendous progress in diagnostic methods and therapeutic strategies has been made, the clinical prognosis of individuals with CRC is still far from satisfactory because of advanced stage, rapid progression, and early metastasis.3,4 Many risk factors, such as environmental and genetic factors, have been found to be significantly associated with CRC. Approximately 10% of individuals with CRC can be classified as having hereditary CRC, while because of the accumulation of a multistep carcinogenic transformation affected by diet and lifestyle, the cancerization and progression of CRC occur gradually.5 Instability of genomic activation of oncogenes and tumor suppressor genes mutational inactivation are all related to cancer development.6 Therefore, extensive and in-depth research on the molecular biology of CRC is indispensable for identification of diagnostic and therapeutic biomarkers and for improvements in prognosis.
Different from the reversible epigenetic modification of DNA and histone protein to regulate gene expression, RNA methylation modification represents another layer of gene expression regulation.7 N6-methyladenosine (m6A) modification, which is the most prevalent epigenetic methylated modification of mRNAs and noncoding RNAs (ncRNAs), has a profound impact on RNA translation, splicing, transportation, and stability.8,9 It was found that m6A modification could exert essential functions in diverse biological processes, especially carcinogenesis.10,11 In addition, there is overwhelming evidence that abnormal methylation of m6A is clearly associated with variety of cancers.12 m6A regulators, which consist of methyltransferases (writers), RNA-binding proteins (readers), and demethylases (erasers), regulate invertible and dynamic RNA epigenetic modification.13 Writers are composed of METTL3, METTL14, KIAA1429, RBM15, WTAP, and ZC3H13. Readers are composed of YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPC, and HNRNPA2B1. ALKBH3, ALKBH5, and FTO make up erasers to carry out demethylation activity.14
A large number of studies have revealed that m6A RNA methylation had a significant relationship with the development and progression of multiple malignant tumors, including breast cancer (BC), hepatocellular carcinoma (HCC), glioma, and CRC.15, 16, 17, 18 Hou et al.19 highlighted the profound implication of m6A modification in hepatocellular carcinoma. They indicated that HIF-2α-mediated inhibition of YTHDF2 in HCC plays an important role in hypoxia, adapting the epitranscriptome and cancer progression. With respect to CRC, Zhu et al.20 demonstrated that METTL3 promotes CRC proliferation by stabilizing the CCNE1 mRNA by means of methylating the m6A site in the 3′ untranslated region (UTR). Recently, some investigations have shown that long noncoding RNAs (lncRNAs) regulate the expression and stabilization of m6A regulators and the impact of m6A modification on lncRNA transcription in cancer progression. By masking site K139, the lncRNA LINRIS suppressed the ubiquitination of IGF2BP2 and prevented it from degrading through the ALP, sustaining MYC-mediated glycolysis and significantly influencing the prognosis of CRC.21 Combined with lncRNA GAS5, the expression of YAP is degraded via its phosphorylation and ubiquitination and thereby downregulated YAP-mediated YTHDF3 transcription, which reversibly binds m6A-methylated GAS5, resulting in its attenuation and causing a negative functional feedback loop.22 Thus, a deeper understanding of how m6A modifications interact with lncRNAs and mRNAs in CRC progression may contribute to identifying biomarkers that can be used as effective therapeutic targets.
In this study, we performed a comprehensive evaluation to show the landscape of m6A RNA methylation in CRC and to explore the underlying mechanisms. We obtained RNA expression levels, copy number variation (CNV), and clinical phenotypes from The Cancer Genome Atlas (TCGA) database (n = 582). The association between m6A regulator expression and clinicopathological parameters was analyzed in individuals with CRC. Using weighted coexpression network analysis (WGCNA) and least absolute shrinkage and selection operator (LASSO) Cox regression analysis, we constructed m6A-related lncRNA and mRNA prognostic signatures to predict overall survival (OS). Furthermore, we internally validated the accuracy and efficiency of the m6A-related lncRNA and mRNA signatures to demonstrate their prognostic significance. Finally, a clinicopathologic-genomic nomogram was developed to predict OS in individuals with CRC.
Results
Expression landscape of m6A RNA methylation regulators in CRC
We first compared the expression of 19 m6A RNA methylation regulators in CRC in the TCGA dataset. The results revealed that all 19 genes except for YTHDC2 were differentially expressed in CRC tissues compared with normal tissues (Figure 1A). METTL3, WTAP, KIAA1429, RBM15, ZC3H13, YTHDC1, YTHDF1, YTHDF2, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, ALKBH3, and FTO in CRC tissues were significantly upregulated compared with in normal tissues, whereas METTL14, YTHDF3, and ALKBH5 were significantly downregulated in CRC tissues (Figure 1B). We further evaluated relationships between CNV patterns and the expression of m6A RNA methylation regulators. The results indicated that m6A regulator expression levels were significantly associated with different CNV patterns. Our findings revealed that, except for IGF2BP1 and IGF2BP3, CNV gains correlated remarkably well with higher gene expression, whereas CNV deletions caused a decrease in gene expression (Figure S1). Then we identified the correlations among 19 m6A RNA methylation regulators. The correlations among m6A RNA methylation regulators were mostly positive, and KIAA1429 and YTHDF3 were the most positively correlated (Cor = 0.81) (Figure 1C). In addition, we comprehensively displayed the relationships between m6A RNA methylation regulators and the clinical features of CRC, including gender, age, tumor stage, T stage, M stage, N stage, and vital status (Figure 1D).
Figure 1.
The landscape of m6A RNA methylation regulators in CRC
(A) Expression of 19 m6A RNA methylation regulators in CRC. (B) Violin plots of the 19 m6A RNA methylation regulators’ differential expression in CRC. (C) Spearman’s correlation analysis of the 19 m6A RNA modification regulators in CRC. (D) Expression of m6A RNA modification regulators in CRC with different clinicopathological features.
Associations between m6A RNA methylation regulator expression and clinicopathological features and OS
We then evaluated associations between m6A RNA methylation regulator expression levels and clinicopathological features in CRC. Expression of KIAA1429, ZC3H13, YTHDC1, YTHDF1, YTHDF3, and FTO was associated with age, whereby individuals older than 65 years exhibited significantly decreased expression of these genes (Figure S2A). Expression of WTAP and HNRNPC were significantly upregulated in men (Figure S2B). For tumor stage, expression of KIAA1429, ZC3H13, YTHDF1, IGF2BP1, and FTO were significantly upregulated in individuals with stage III and IV CRC. Conversely, expression of METTL14, YTHDC2, and YTHDF2 exhibited significant downregulation in individuals with advanced CRC (Figure 2A). We further analyzed relationships between m6A regulators and T, N, and M stages. We found that m6A regulators might have potential associations with lymphatic and distant metastasis in CRC (Figures 2C and 2D). Because different mutations, such as KRAS, BRAF, and PIK3CA mutations, have a significant impact on treatment strategy and prognosis in individuals with CRC, we further explored the relationship between these three gene mutations and the expression of m6A regulators. The results showed that the expression levels of some m6A regulators were significantly correlated with the mutation status of KRAS, BRAF, and PIK3CA (Figures S2C–S2E). Next, we investigated the relationship between CNV of 19 m6A regulators and clinicopathological features of individuals with CRC. The results showed that CNV alterations to m6A regulators were significantly correlated with tumor stage, involving METTL14, KIAA1429, ZC3H13, YTHDF1, HNRNPA2B1, IGF2BP3, ALKBH3, and FTO (Table 1); that is, individuals with advanced-stage tumors had more CNV events of m6A regulators. High expression of IGF2BP1, IGF2BP2, and ALKBH5 predicted poor OS, and individuals with higher expression of METTL14, WTAP, RBM15, YTHDC1, YTHDC2, YTHDF1, YTHDF2, ALKBH3, and FTO had better clinical outcomes (Figure 3). Furthermore, we explored relationships between CNV events of m6A regulators and OS and displayed the prognostic value of CNV events in CRC. We found that CNV events of m6A regulators, such as METTL14, WTAP, KIAA1429, RBM15, YTHDC1, YTHDF2, IGF2BP2, and ALKBH3, were significantly associated with OS (Figure S3).
Figure 2.
The 19 m6A RNA modification regulators correlated with clinicopathological features in the TCGA CRC cohort
(A) Correlation between m6A regulators and tumor stage. (B) Correlation between m6A regulators and T stage. (C) Correlation between m6A regulators and N stage. (D) Correlation between m6A regulators and M stage.
Table 1.
Relationship between tumor stage and m6A regulator CNV alterations in CRC subjects
| Gene | Stages Ⅰ + Ⅱ |
Stages Ⅲ + Ⅳ |
Chi-Square Value | p Value | ||
|---|---|---|---|---|---|---|
| CNV | Wild | CNV | Wild | |||
| METTL3 | 45 | 275 | 46 | 216 | 1.08 | 0.3 |
| METTL14 | 15 | 305 | 34 | 228 | 11.79 | 0.0006 |
| WTAP | 17 | 303 | 23 | 239 | 2.19 | 0.14 |
| KIAA1429 | 107 | 213 | 114 | 148 | 5.79 | 0.02 |
| RBM15 | 18 | 302 | 26 | 236 | 3.22 | 0.07 |
| ZC3H13 | 133 | 187 | 150 | 112 | 13.57 | 0.0002 |
| YTHDC1 | 14 | 306 | 20 | 242 | 2.22 | 0.14 |
| YTHDC2 | 32 | 288 | 37 | 225 | 1.96 | 0.16 |
| YTHDF1 | 197 | 123 | 188 | 74 | 6.24 | 0.01 |
| YTHDF2 | 34 | 286 | 43 | 219 | 3.71 | 0.05 |
| YTHDF3 | 98 | 222 | 101 | 161 | 3.68 | 0.06 |
| HNRNPA2B1 | 106 | 214 | 110 | 152 | 4.47 | 0.03 |
| HNRNPC | 45 | 275 | 49 | 213 | 1.96 | 0.16 |
| IGF2BP1 | 28 | 292 | 35 | 227 | 2.71 | 0.1 |
| IGF2BP2 | 19 | 301 | 20 | 242 | 0.42 | 0.52 |
| IGF2BP3 | 104 | 216 | 108 | 154 | 4.36 | 0.04 |
| ALKBH3 | 5 | 315 | 18 | 244 | 9.34 | 0.002 |
| ALKBH5 | 107 | 213 | 101 | 161 | 1.42 | 0.23 |
| FTO | 17 | 303 | 32 | 230 | 8.03 | 0.005 |
Figure 3.
Correlation between the expression levels of individual m6A RNA modification regulators and overall survival (OS) of CRC subjects in the TCGA database
Kaplan-Meier survival plots show OS of CRC subjects with high (red lines) and low (blue lines) expression of the 12 m6A regulators. Kaplan-Meier survival curves for (A) METTL14, (B) WTAP, (C) RBM15, (D) YTHDC1, (E) YTHDC2, (F) YTHDF1, (G) YTHDF2, (H) IGF2BP1, (I) IGF2BP2, (J) ALKBH3, (K) ALKBH5, and (L) FTO. p < 0.05 by log rank test.
Detection of m6A-related lncRNAs and mRNAs by WGCNA
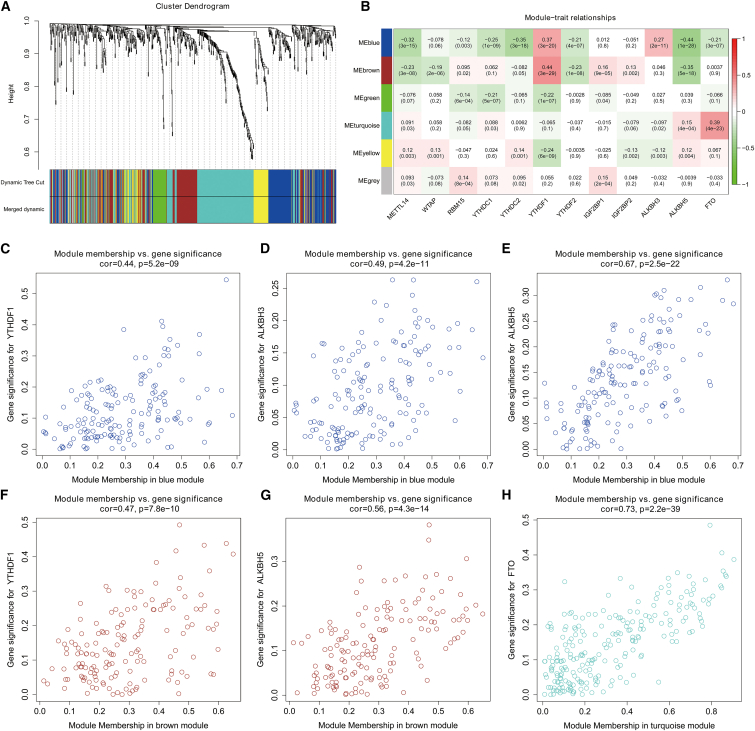
Using the R package edgeR, we identified 700 lncRNAs and 3,637 mRNAs in the TCGA dataset for further analysis (Figure S4). By WGCNA, we confirmed six lncRNA co-expression modules and evaluated their associations with 12 OS-related m6A RNA methylation regulators (Figures 4A and 4B). We found that blue, brown, and turquoise modules were significantly correlated with m6A regulators, including YTHDF1, ALKBH3, ALKBH5, and FTO (Figures 4C–4H). A total of 544 lncRNAs were involved in the three modules; we defined those lncRNAs relevant to m6A regulators as m6A-related lncRNAs. As for mRNAs, 10 modules were identified for further analysis (Figures 5A and 5B). Moreover, compared with other modules, the red and turquoise modules were significantly associated with m6A RNA regulators (Figures 5C–5H). A total of 1,292 mRNAs in these two modules were regarded as m6A-related mRNAs.
Figure 4.
Identification of m6A-related lncRNAs of CRC in the TCGA dataset through WGCNA
(A) Dendrogram of all DElncRNAs clustered on the basis of a dissimilarity measure. (B) Heatmap of the correlation between module eigengenes and 12 OS-related m6A regulators of CRC. Each cell contains the correlation coefficient and p value. (C–H) Scatterplots of module eigengenes in the selected modules.
Figure 5.
Identification of m6A-related mRNAs of CRC in the TCGA dataset through WGCNA
(A) Dendrogram of all DEmRNAs clustered on the basis of a dissimilarity measure. (B) Heatmap of the correlation between module eigengenes and 12 OS-related m6A regulators of CRC. Each cell contains the correlation coefficient and p value. (C–H) Scatterplots of module eigengenes in the selected modules.
Construction and validation of the prognostic m6A-related lncRNA signature
Taking advantage of the prognostic information, we applied univariate Cox regression to identify prognostic m6A-related lncRNAs from the 544 m6A-related lncRNAs in the training TCGA dataset. We found that 37 m6A-related lncRNAs were significantly related to the OS of CRC individuals in the training dataset. Then we constructed LASSO Cox analysis on the basis of the 37 prognostic m6A-related lncRNAs, and it generated the m6A-related lncRNA signature, which contains 24 m6A-related lncRNAs (Table S1; Figures S5A and S5B). According to the coefficient of each lncRNA, a risk score was computed for each affected individual. Subjects with CRC were classified into low-risk and high-risk groups on the basis of the median of risk scores. Kaplan-Meier (K-M) survival curves showed that CRC subjects in the low-risk group had better clinical outcomes than those in the high-risk group: training set (p = 9.5 × 10−7) (Figure 6A), testing set (p = 2.44 × 10−4) (Figure S6A), and total TCGA cohort (p = 1.79 × 10−9) (Figure S6E). Also, the receiver operating characteristic (ROC) curves revealed that the m6A-related lncRNA signature exhibits strong accuracy to predict OS in the training set (Figure 6B), testing set (Figure S6B), and total TCGA cohort (Figure S6F). Risk scores and vital status scattergrams are also shown in Figures 6 and S6.
Figure 6.
Construction and validation of the m6A-related lncRNA and mRNA signatures in the TCGA training dataset
(A) Kaplan-Meier survival curves show OS of high- and low-risk CRC subjects in the training dataset on the basis of the m6A-related lncRNA signature. (B) Time-dependent ROC curves show the accuracy of OS prediction for the training dataset on the basis of the m6A-related lncRNA signature. (C) Distribution of risk scores for the CRC subjects in the training dataset on the basis of the m6A-related lncRNA signature. (D) The survival status of 393 CRC subjects in the training dataset belonging to the high- and low-risk groups on the basis of the m6A-related lncRNA signature. (E) Kaplan-Meier survival curves show OS of high- and low-risk CRC subjects in the training dataset on the basis of the m6A-related mRNA signature. (F) Time-dependent ROC curves show the accuracy of OS prediction for the training dataset on the basis of the m6A-related mRNA signature. (G) Distribution of risk scores for the CRC subjects in the training dataset on the basis of the m6A-related mRNA signature. (H) Survival status of 393 CRC subjects in the training dataset belonging to the high- and low-risk groups on the basis of the m6A-related mRNA signature.
Construction and validation of the prognostic m6A-related mRNA signature
In the training dataset, 75 mRNAs were identified as prognostic m6A-related mRNAs by using univariate Cox regression. Furthermore, LASSO Cox analysis was used to construct a m6A-related mRNA signature, involving 33 prognostic m6A-related mRNAs (Table S2; Figures S5C and S5D). On the basis of the median risk scores, CRC subjects were divided into low-risk and high-risk groups. Kaplan-Meier survival curves indicated that CRC subjects with higher risk scores had worse clinical outcomes (shorter OS time and lower OS rates) (Figure 6E). The ROC analysis showed that the m6A-related mRNA signature harbored a promising ability to predict OS in the training dataset (1-year area under the curve [AUC] = 0.853, 3-year AUC = 0.891, 5-year OS = 0.892) (Figure 6F). Risk score and survival status distributions are plotted in Figures 6G and 6H. We validated the prognostic ability of m6A-related mRNA signature in the testing set (Figures S6I–S6L) and total TCGA cohort (Figures S6M–S6P). These results showed a robust OS-predictive ability of the signature.
The m6A-related lncRNA and mRNA signatures are independent prognostic factors for individuals with CRC
Using the TCGA data after adjusting for clinicopathological parameters such as age, gender, tumor stage, T stage, N stage, and M stage, we conducted univariate and multivariate Cox regression analyses to further identify whether the risk scores calculated by the m6A-related lncRNA and mRNA signatures accurately predicted prognosis of individuals with CRC. The results showed that the m6A-related lncRNA and mRNA signatures correlated remarkably well with OS, and multivariate Cox analysis showed that the signatures were independent prognosis factors of OS (Figures 7A and 7B) in the training dataset. These conclusions were validated in the testing set (Figures S7A and S7B) and total TCGA cohort (Figures S7C and S7D). Moreover, we attempted to explore whether the risk scores were associated with clinicopathological features. The results revealed that compared with the low-risk group, the high-risk group was significantly correlated with a higher death rate and higher tumor stage but was not associated with age and gender (Figures 7C and 7D). In addition, we also analyzed the relationship between m6A-related lncRNA and mRNA signatures and stem cell phenotype and drug sensitivity. We found that m6A-related lncRNA and mRNA signatures were significantly associated with CRC stem cell phenotype (Figure S8) and that the high-risk subjects in both signatures may be more sensitive to drug therapy (Figure S9). These results indicated that our m6A-related lncRNA and mRNA signatures, as independent prognostic indicators, might have great clinical application and significance.
Figure 7.
Association of the m6A-related lncRNA and mRNA signatures with OS of CRC subjects
(A) Univariate Cox regression analysis shows the clinicopathological parameters associated with OS among CRC subjects in the training dataset. (B) Multivariate Cox regression analysis shows clinicopathological parameters associated with OS among CRC subjects in the training dataset. (C) Correlation analysis results show the relationship between the m6A-related lncRNA signature and the clinicopathological parameters in the training dataset. (D) Correlation analysis results show the relationship between the m6A-related mRNA signature and the clinicopathological parameters in the training dataset.
Nomogram construction and validation based on the m6A-related lncRNA and mRNA signatures
To create an applicable clinical evaluation instrument to enhance the accuracy of predicting OS among individuals with CRC, we constructed a nomogram, including age, gender, T stage, tumor stage, lncRNA risk scores, and mRNA risk scores, to predict 1-, 3-, and 5-year OS probability in the training dataset (Figure 8A). As demonstrated by the calibration plots, the performance of the nomogram was best in forecasting 3-year OS (Figure 8B). As Figure 8C illustrates, the nomogram exhibited an improved net benefit with a broader scope of threshold probability in the decision curve analysis (DCA) for forecasting the corresponding 1-, 3-, and 5-year OS. As the decision curve analysis verified, Figure 8D depicts the nomogram's greater net clinical benefit compared with m6A-related lncRNA and mRNA signature-based risk scores. The nomogram's C-index in the training set was 0.848 (95% confidence interval [CI], 0.792–0.904). Then time-dependent ROC curves were applied to estimate the prognostic capacity of the nomogram, and the results showed that the nomogram had excellent prognostic accuracy regarding 1-, 3-, and 5-year OS (AUC = 0.844, 0.855, and 0.824, respectively) (Figure 8F). On the basis of the nomogram's risk score, subjects were stratified into two groups by the median value. The results suggest that subjects in the high-risk group had significantly worse outcomes compared with those in the low-risk group (p < 0.001) (Figure 8E). Using the testing set and total TCGA cohort for validation, we verified the same results, as demonstrated in Figure S10.
Figure 8.
Establishment and validation of the nomogram for predicting OS of CRC subjects in the TCGA training dataset
(A) Nomogram with age, gender, T stage, tumor stage, lncRNA risk scores, and mRNA risk scores for predicting 1-year, 3-year, and 5-year OS among CRC patients. (B) Calibration plot shows the comparison between nomogram-predicted and actual 3-year OS. (C) Decision curve analysis shows predicted 1-year, 3-year, and 5-year OS among CRC subjects on the basis of the nomogram. (D) Decision curve analysis shows predicted 3-year OS among CRC subjects on the basis of the nomogram, lncRNA risk scores, and mRNA risk scores. (E) Kaplan-Meier survival curves show OS of CRC subjects on the basis of the nomogram. (F) Time-dependent ROC curves show the accuracy of OS prediction on the basis of the nomogram.
Discussion
Aberrant expression of m6A RNA methylation regulators is related to carcinogenesis of various cancers. However, different m6A regulators may lead to distinct functions in diverse tumors.23 In our research, we aim to estimate the feasibility of m6A regulators to predict CRC prognosis. We found that, except for YTHDC2, expression levels of all 19 m6A regulator were significantly different between tumor and normal samples. With respect to CNV, the results showed that CNV gains were associated with higher expression of m6A regulators and vice versa, indicating that mRNA expression was significantly correlated with various CNV mutation patterns. In addition, alteration of m6A regulator expression and CNV events were significantly correlated with clinicopathological features and OS. To some extent, our findings suggest that m6A regulation disorder, initiated by CNV events, might have a significant impact on the development and progression of CRC. Consider the “writer” gene METTL14, for example. CNV deletion was the main mutation pattern of METTL14; hence the expression level of METTL14 was decreased in tumor samples, and more CNV events were detected in subjects with advanced-stage tumors. Moreover, individuals with CRC with METTL14 CNV events and lower gene expression exhibited poor clinical outcomes. Recently, Chen et al. demonstrated that the progression of CRC can be inhibited by METTL14 through the SOX4-mediated EMT process and PI3K/Akt signals.24 It has also been demonstrated that METTL14 inhibits the CRC malignant process via the miR-375/YAP1 pathway and suppresses CRC cell migration and invasion through the miR-375/SP1 pathway.25 With respect to the “reader” gene IGF2BP2, CNV gain was the major mutation pattern, so the expression level of IGF2BP2 was significantly upregulated in CRC, and it was positively correlated with tumor stage and OS. Xu et al. revealed that IGF2BP2 overexpression is partially caused by genomic amplification, and the oncogenic role of IGF2BP2 in pancreatic cancer contributed to the miR-141-mediated PI3K/Akt signaling pathway.26 In CRC, HK2 and SLC2A1, stabilized by the m6A reader IGF2BP2, might activate the glycolysis pathway and induce tumorigenesis.27 Thus, these m6A regulators have an essential function in the emergence and development of CRC.
Moreover, m6A RNA methylation regulators' mRNA expression levels and their post-transcriptional regulations were also affected by a variety of factors. Yang et al.28 found that YTHDF2 was significantly associated with hepatocellular carcinoma, that miR-145 could target the 3′ UTR of YTHDF2 mRNA, and that miR-145 was negatively correlated with the expression level of YTHDF2 mRNA in HCC tissues. In non-small cell lung cancer (NSCLC), miR-33a downregulates the expression of METTL3, then decreases expression of the downstream genes EGFR, DNMT3A, and TAZ and inhibits NSCLC cell proliferation.29 Du et al.30 discovered that SUMOylation of METTL3 significantly reduces its m6A methyltransferase activity, leading to decreases of m6A levels in mRNAs, which directly enhanced tumorigenesis in lung cancer. Other investigations have also shown that m6A mRNA expressions and m6A enzyme activity are regulated by a variety of molecular mechanisms, and these specific mechanisms need further study.
Numerous studies have shown that m6A modification regulators might play a role in cancer pathogenesis, but how they interact with lncRNAs during CRC progression is still unclear. lncRNAs, which were extensively modified by m6A regulators, controlled gene expression and cellular biology at both transcriptional and post-transcriptional levels.31 IGF2BP2 serves as a reader for stabilizing DANCR RNA and regulating the expression of lncRNA DANCR; then IGF2BP2 and DANCR work together to stimulate stem cell properties and carcinogenesis in pancreatic cancer.32 METTL3-mediated modification led to LINC00958 upregulation by stabilizing its RNA transcript, and LINC00958 sponged miR-3619-5p to upregulate hepatoma-derived growth factor (HDGF) expression, thereby facilitating HCC lipogenesis and progression.33 Yang et al. identified that low expression of METTL14 substantially abolished the m6A level of XIST, and increased XIST expression caused proliferation and metastasis of CRC. Furthermore, they found that m6A reader YTHDF2 could recognize m6A-methylated XIST to mediate the degradation of that lncRNA.34 lncRNA FOXM1-AS could enhance the coaction of ALKBH5 and FOXM1 and lead to the sustainability of glioblastoma cell stemness.35 We demonstrated that m6A modification of lncRNAs could affect the occurrence and progression of cancers, and lncRNAs might act as competing endogenous RNA (ceRNAs), targeting m6A regulators to influence tumor invasiveness. Therefore, we believe that lncRNA is one of the important targets of m6A modification regulators, and close attention should be paid to the interactions of m6A modifications and lncRNAs so that potential prognostic markers or therapeutic targets of cancers may be identified.
We explored m6A-related lncRNAs by WGCNA and LASSO Cox analysis; 37 prognostic m6A-related lncRNAs were identified from CRC subjects in the training dataset, and 24 of them were selected to generate the m6A-related lncRNA signature.
The expression of LINC00702 was decreased in CRC and promoted cell proliferation, migration, and invasion by enhancing the PI3K/AKT pathway via inhibition of PTEN expression.36 It has been reported that lncRNA SNHG3 increases the malignancy of CRC by regulating the miR-539/RUNX2 axis37 and promotes stem cell-like properties of gastric cancer by regulating the miR-3619-5p/ARL2 axis.38 Vishnubalaji et al. demonstrated that LINC01614 was an unfavorable prognostic marker in breast cancer, which was associated with the HR+/HER2+ BC molecular subtype and regulation by TGFβ and FAK signaling.39 Except for the above lncRNAs, reports on how the 21 residual m6A-related lncRNAs are associated with cancer progression and how the lncRNAs interact with m6A regulators are rare. Thus, we expect that the prognostic lncRNAs we identified in this study might have potential associations with m6A regulators, thereby providing insights into their underlying mechanisms in CRC tumorigenesis and progression.
Similar to lncRNAs, m6A modification was also significant for fate decision of regulation of mRNAs.40 However, the m6A modification of mRNA under various circumstances was largely unknown. We report here that 33 mRNAs correlated with m6A regulators, and we constructed a powerful prediction model to assess OS among individuals with CRC. SOSTDC1, a potential therapeutic target in metastatic colorectal cancer, promotes invasion and liver metastasis by inhibiting BMP4-specific antimetastatic signals and inducing ALCAM-mediated Src and PI3K/AKT activation.41 Wang et al. demonstrated that POLR1D-induced promotion of G1-S cell-cycle transition was mediated by activation of wnt-β-catenin signaling and inactivation of p53 signaling.42 In addition, POLR1D, as a subunit of RNA polymerases I and III, plays an important role in the oncogenesis of CRC by affecting VEGFA and EREG expression.43 In cooperation with Wip1, KPNA2 could modulate CRC cell proliferation and migration through the downstream AKT/GSK-3β pathway, in a p53-dependent manner.44 SEZ6L2 was significantly upregulated in tumor tissues and correlated with poor prognosis in individuals with CRC, and downregulation of SEZ6L2 impairs the growth of the CRC cells by inducing caspase-dependent apoptosis, which was mediated by mitochondria-related proteins.45 Several of the 33 mRNAs were found to be associated with oncogenesis and development, but there have been few studies involving CRC, and investigations of how the mRNAs interact with m6A regulators rarely have been seen. Therefore, our results might help identify the m6A regulators that target prognostic mRNAs, consequently providing a deep understanding of their underlying mechanisms in CRC tumorigenesis and progression.
Furthermore, we constructed and validated prognostic m6A-related lncRNA and mRNA signatures in CRC. In K-M survival plots, ROC curve analyses, and univariate and multivariate Cox regression analyses, our signatures exhibited great capability in predicting OS among individuals with CRC. We observed that high-risk groups were correlated with more deaths, higher tumor stage, and lymph node and distant metastasis of CRC. These clinicopathological features are considered determinants of OS and especially contribute to the deterioration of individuals with CRC. We also established a nomogram on the basis of the m6A-related signatures and clinicopathological features, which are integrated into a single numerical algorithm to predict the prognosis of every individual with CRC. These results were validated in testing and total TCGA cohorts to ensure the accuracy. The prognostic m6A-related RNA signatures should be validated independently in more CRC cohorts. However, because of the different types and numbers of RNAs annotated by different microarray chip platforms and because inclusion criteria vary among different datasets, it is difficult to validate the same RNA signature across different datasets. In the process of establishing a signature, one should fully consider the heterogeneity among different datasets, optimize the process and algorithm of data processing, and improve the predictive efficiency and application value of the signature. In addition, the mechanisms of the RNAs and their interactions with m6A-related regulators should be verified by experiments both in vitro and in vivo. Our results may open a path for further investigation, concentrating on the mechanism underlying m6A modification of RNAs.
In this study, we conducted a comprehensive analysis of the landscape of 19 m6A RNA methylation regulators by evaluating the RNA expression, CNVs, and clinicopathological features in CRC. The expression and CNV patterns of m6A RNA methylation regulators were closely related to the malignant clinicopathological features of CRC. The prognostic m6A-related lncRNA and mRNA signatures might contribute to the personalized prediction of CRC prognosis, and these RNAs are capable of serving as potential biomarkers of CRC that specifically target m6A. Finally, further research is necessary to provide solid confirmation of how m6A methylation regulators affect RNAs and subsequently are associated with CRC prognosis.
Materials and methods
Datasets acquisition
The CRC FPKM (fragments per kilobase of transcript per million mapped reads) normalized mRNA expression files, somatic mutation data, and corresponding clinicopathological data were downloaded from the TCGA dataset (https://portal.gdc.cancer.gov/) in 2020.46 For CNV data, we obtained "Masked Copy Number Segment" subtype of copy number data from TCGA. Stem-like phenotype data, including DNAs and RNAs, were downloaded from the UCSC (University of California, Santa Cruz) database (http://xena.ucsc.edu/) for each CRC subject. Our study was in accordance with the TCGA publication guidelines (https://cancergenome.nih.Gov/publications/publicationguidelines). All of those data in the study were retrieved from TCGA; therefore, informed consent and ethics approval were not needed.
Data processing
A total of 618 samples were gathered in this study, including 582 primary tumor tissues and 36 normal tissues. Using the annotation database of the Ensembl Genome Browser 99 (GRCh38.p13),47 14,176 lncRNAs and 19,645 mRNAs were identified on the basis of the gene symbols in the TCGA dataset. We used the R package edgeR to analyze differentially expressed lncRNAs (DElncRNAs) and differentially expressed mRNAs (DEmRNAs) between tumor and normal tissues for further analysis. The thresholds were set as |log2 fold change (FC)| > 1 and adjusted p < 0.05. In addition, 19 m6A regulators’ expression values were extracted from the TCGA mRNA matrix according to previous publications. For CNV analysis of the 19 m6A regulators, we applied segmentation analysis and GISTIC2.0 algorithm to identify the gain and loss of copy number of each sample.
Bioinformatics analysis
To compare the expression of 19 m6A RNA methylation regulators between CRC and normal tissues, we used the R package limma. Next, vioplot and pheatmap were used to visualize the expression of the 19 regulators in 582 tumor tissues and 36 normal tissues. Then the expression data profile of DElncRNAs and DEmRNAs was extracted from differential expression analysis to conduct WGCNA using the R package WGCNA.48 At first, sample clustering of all DElncRNAs and DEmRNAs was applied to test whether they were good samples and good RNAs. Next, we selected soft threshold power β values of 4 (lncRNAs) and 3 (mRNAs) (scale-free R2 = 0.95) to determine the scale-free topology model. The adjacency matrix was transformed into topological overlap matrix (TOM). On the basis of the TOM-based dissimilarity measure, lncRNAs and mRNAs were classified into different modules. Here, we define minimal module size as 30 and cut height as 0.25 to confirm key modules. The module eigengene (ME) was considered as the major principal component of a given RNA module. It could be known as a specific gene expression pattern of a module; the ME can summarize the gene expression profiles, and the correlation between ME and m6A regulators’ expression levels was calculated to identify the m6A-related lncRNA and mRNA modules.
Afterward, we divided the subjects with CRC from TCGA randomly into a training set (n = 393) and a testing set (n = 189). Then univariate Cox regression analysis was performed to select the prognostic m6A-related lncRNAs and mRNAs in the training dataset (with p < 0.05), among all lncRNAs and mRNAs in the m6A-related modules. Using the least absolute shrinkage and selection operator Cox regression analysis, we constructed prognostic m6A-related lncRNA and mRNA signatures for the CRC subjects. Risk scores were calculated by sum of the coefficients and products of each RNA expression level for each patient involved in our study. On the basis of the median risk scores, subjects were divided into high-risk and low-risk groups. To evaluate the distinguishing and prognostic capacities of the risk score classifier, we performed the Kaplan-Meier survival analysis and time-dependent ROC analysis. The R package pRRophetic was used to evaluate the sensitivity of chemotherapy and targeted drugs in high- and low-risk groups of m6A-related lncRNA and mRNA signatures. Furthermore, we used the R package rms to generate a nomogram by including factors of multivariate Cox regression results to evaluate the OS probability of each CRC subject. Decision curve analysis and calibration plots were applied to measure the predictive performance of the nomogram.
Statistical analysis
All statistical analyses were conducted using R (version 4.0.2). We used Wilcoxon's test to compare the expression levels of m6A RNA methylation regulators between CRC and normal tissues. The correlation between m6A RNA methylation regulators and the clinical factors and CNV of CRC subjects was analyzed using the Kruskal-Wallis test. Kaplan-Meier survival analysis and the log rank test were used to analyze the prognosis of m6A regulators. The optimal cutoff value of each gene was determined using X-tile.49 Univariate and multivariate Cox analyses were performed to identify the prognostic determinants from clinicopathological features and m6A-related lncRNA and mRNA signatures. The association between m6A regulators and m6A-related lncRNAs and mRNAs was evaluated using Spearman’s correlation coefficients.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572758); the Foundation for Distinguished Young Talents in Higher Education of Hebei (BJ2018042); the National Natural Science Foundation of Hebei (H2020206374 and H2021206306); 2018 Hebei Provincial Government Funded Specialist Leader Training Program (361034); and other Hebei Province Projects (LS2020001, 2019YX006A, YZ201802, 20180222, 20180213, LNB201911, zh2018002, and G2018019).
Author contributions
W.L., Y.G., and X. Jiang contributed to the planning of the study. W.L., Y.G., and X. Jin conducted data analysis. Y.G., T.L., Z.L., and M.W. drafted the manuscript and revised the manuscript. W.L., W.Y., and G.W. prepared all the figures and tables. Z.Z., Z.L., and X. Jiang contributed to interpretation of data and review of the manuscript. All authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.007.
Contributor Information
Zengren Zhao, Email: zhaozengren@hebmu.edu.cn.
Xia Jiang, Email: jiangxia0925@hebmu.edu.cn.
Supplemental information
References
- 1.Fitzmaurice C., Akinyemiju T.F., Al Lami F.H., Alam T., Alizadeh-Navaei R., Allen C., Alsharif U., Alvis-Guzman N., Amini E., Anderson B.O., et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4:1553–1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hissong E., Pittman M.E. Colorectal carcinoma screening: Established methods and emerging technology. Crit. Rev. Clin. Lab. Sci. 2020;57:22–36. doi: 10.1080/10408363.2019.1670614. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y., Yang B., Lin M., Yu H., Chen H., Zhang Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24:299–305. doi: 10.3233/CBM-182129. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka T. Colorectal carcinogenesis: Review of human and experimental animal studies. J. Carcinog. 2009;8:5. doi: 10.4103/1477-3163.49014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okugawa Y., Grady W.M., Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N., Pan T. RNA epigenetics. Transl. Res. 2015;165:28–35. doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai D., Wang H., Zhu L., Jin H., Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F., Cheng W., Zhao F., Tang M., Diao Y., Xu R. Association of N6-methyladenosine with viruses and related diseases. Virol. J. 2019;16:133. doi: 10.1186/s12985-019-1236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z.X., Li L.M., Sun H.L., Liu S.M. Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 2018;6:89. doi: 10.3389/fbioe.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Sun C., Li J., Zhang E., Ma Z., Xu W., Li H., Qiu M., Xu Y., Xia W., et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 14.Meyer K.D., Jaffrey S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., Wang Y., Li X., Xiong X.F., Wei B., et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Li F., Peng Y., Fang J., Zhou J. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 2020;9:1877–1889. doi: 10.1002/cam4.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G., et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng W., Li J., Chen R., Gu Q., Yang P., Qian W., Ji D., Wang Q., Zhang Z., Tang J., et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., Hu B., Zhou J., Zhao Z., Feng M., et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer. 2019;18:163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W., Si Y., Xu J., Lin Y., Wang J.Z., Cao M., Sun S., Ding Q., Zhu L., Wei J.F. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J. Cell Mol. Med. 2020;24:3521–3533. doi: 10.1111/jcmm.15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Lu J.H., Wu Q.N., Jin Y., Wang D.S., Chen Y.X., Liu J., Luo X.J., Meng Q., Pu H.Y., et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer. 2019;18:174. doi: 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni W., Yao S., Zhou Y., Liu Y., Huang P., Zhou A., Liu J., Che L., Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng X., Su R., Feng X., Wei M., Chen J. Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Xu M., Xu X., Zeng K., Liu X., Pan B., Li C., Sun L., Qin J., Xu T., et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer. 2020;19:106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Xu M., Xu X., Zeng K., Liu X., Sun L., Pan B., He B., Pan Y., Sun H., et al. METTL14 suppresses crc progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol. Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Xu X., Yu Y., Zong K., Lv P., Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:497. doi: 10.1186/s13046-019-1470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., Zhang X., Cao Y., Ma D., Zhu X., et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z., Li J., Feng G., Gao S., Wang Y., Zhang S., Liu Y., Ye L., Li Y., Zhang X. MicroRNA-145 modulates N(6)-methyladenosine levels by targeting the 3'-untranslated mrna region of the N(6)-methyladenosine binding yth domain family 2 protein. J. Biol. Chem. 2017;292:3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du M., Zhang Y., Mao Y., Mou J., Zhao J., Xue Q., Wang D., Huang J., Gao S., Gao Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 2017;482:582–589. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 30.Du Y., Hou G., Zhang H., Dou J., He J., Guo Y., Li L., Chen R., Wang Y., Deng R., et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46:5195–5208. doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X., Peng W.X., Zhou H., Jiang J., Zhou X., Huang D., Mo Y.Y., Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., Cao M., Cai J., Wu J., Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X., Zhang S., He C., Xue P., Zhang L., He Z., Zang L., Feng B., Sun J., Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bogler O., et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D., Wang X.Y., Jin Z.L. Linc00702 inhibits cell growth and metastasis through regulating PTEN in colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3624–3632. doi: 10.26355/eurrev_202004_20824. [DOI] [PubMed] [Google Scholar]
- 37.Dacheng W., Songhe L., Weidong J., Shutao Z., Jingjing L., Jiaming Z. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed. Pharmacother. 2020;125:110039. doi: 10.1016/j.biopha.2020.110039. [DOI] [PubMed] [Google Scholar]
- 38.Sun B., Han Y., Cai H., Huang H., Xuan Y. Cellular oncology; Dordrecht): 2020. Long non-coding RNA SNHG3, induced by IL-6/STAT3 transactivation, promotes stem cell-like properties of gastric cancer cells by regulating the miR-3619-5p/ARL2 axis. [DOI] [PubMed] [Google Scholar]
- 39.Vishnubalaji R., Shaath H., Elkord E., Alajez N.M. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov. 2019;5:109. doi: 10.1038/s41420-019-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roignant J.Y., Soller M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Bartolomé R.A., Pintado-Berninches L., Jaén M., de Los Ríos V., Imbaud J.I., Casal J.I. SOSTDC1 promotes invasion and liver metastasis in colorectal cancer via interaction with ALCAM/CD166. Oncogene. 2020;39:6085–6098. doi: 10.1038/s41388-020-01419-4. [DOI] [PubMed] [Google Scholar]
- 42.Wang M., Niu W., Hu R., Wang Y., Liu Y., Liu L., Zhong J., Zhang C., You H., Zhang J., et al. POLR1D promotes colorectal cancer progression and predicts poor prognosis of patients. Mol. Carcinog. 2019;58:735–748. doi: 10.1002/mc.22966. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Q., Perakis S.O., Ulz P., Mohan S., Riedl J.M., Talakic E., Lax S., Tötsch M., Hoefler G., Bauernhofer T., et al. Cell-free DNA analysis reveals POLR1D-mediated resistance to bevacizumab in colorectal cancer. Genome Med. 2020;12:20. doi: 10.1186/s13073-020-0719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P., Zhao Y., Liu K., Liu X., Liang J., Zhou H., Wang Z., Zhou Z., Xu N. Wip1 cooperates with KPNA2 to modulate the cell proliferation and migration of colorectal cancer via a p53-dependent manner. J. Cell. Biochem. 2019;120:15709–15718. doi: 10.1002/jcb.28840. [DOI] [PubMed] [Google Scholar]
- 45.An N., Zhao Y., Lan H., Zhang M., Yin Y., Yi C. SEZ6L2 knockdown impairs tumour growth by promoting caspase-dependent apoptosis in colorectal cancer. J. Cell Mol. Med. 2020;24:4223–4232. doi: 10.1111/jcmm.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Research N., Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S., et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.