Abstract
CRISPR-Cas9 is rapidly entering molecular biology and biomedicine as a promising gene-editing tool. A unique feature of CRISPR-Cas9 is a single-guide RNA directing a Cas9 nuclease toward its genomic target. Herein, we highlight new approaches for improving cellular uptake and endosomal escape of CRISPR-Cas9. As opposed to other recently published works, this review is focused on non-viral carriers as a means to facilitate the cellular uptake of CRISPR-Cas9 through endocytosis. The majority of non-viral carriers, such as gold nanoparticles, polymer nanoparticles, lipid nanoparticles, and nanoscale zeolitic imidazole frameworks, is developed with a focus toward optimizing the endosomal escape of CRISPR-Cas9 by taking advantage of the acidic environment in the late endosomes. Among the most broadly used methods for in vitro and ex vivo ribonucleotide protein transfection are electroporation and microinjection. Thus, other delivery formats are warranted for in vivo delivery of CRISPR-Cas9. Herein, we specifically revise the use of peptide and nanoparticle-based systems as platforms for CRISPR-Cas9 delivery in vivo. Finally, we highlight future perspectives of the CRISPR-Cas9 gene-editing tool and the prospects of using non-viral vectors to improve its bioavailability and therapeutic potential.
Keywords: CRISPR-Cas9, gene editing, guide RNA, cellular uptake, endocytosis, ribonucleotide protein transfection, endosomal escape, non-viral carriers
Graphical abstract
In this review, we describe recent advances in CRISPR-Cas9 technology for gene therapy and research applications. In particular, we describe novel peptides and inorganic materials for the specific delivery of CRISPR-Cas9 that allow for efficient cellular uptake and endosomal escape.
Introduction
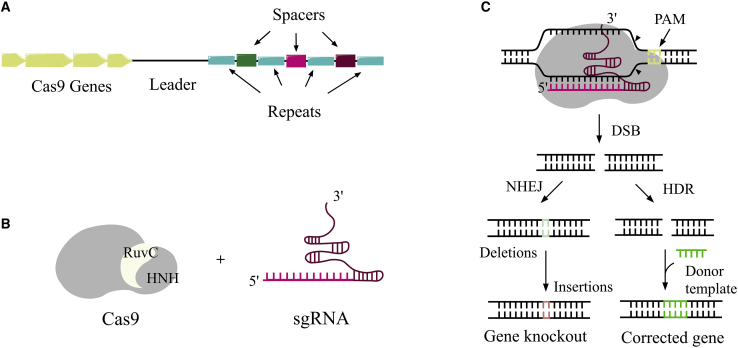
CRISPR and its associated protein (Cas) complex were originally discovered in bacteria and archaea.1,2 A groundbreaking gene-editing application of CRISPR-Cas in mammalian cells was reported 30 years later.3, 4, 5, 6, 7 The CRISPR-Cas locus consists of clustered Cas genes followed further downstream by palindromic DNA repeat sequences interspaced by variable sequences (spacers) (Figure 1A). The direct repeats ensure that the CRISPR system can distinguish between self and non-self-recognition.8,9 Spacers derived from foreign DNA dictate the targeting specificity of the nuclease domain in the Cas protein responsible for catalyzing double-strand breaks (DSBs) in the targeted genomic sequence.8
Figure 1.
CRISPR-Cas9 structure and function
(A) A generalized CRISPR locus in bacteria. Cas genes encode for Cas proteins in which their functions are to add new spacer repeats, process the CRISPR transcript, and cleave the DNA upon recognition. The spacers are variable sequences derived from encounters with DNA phages.4 CRISPR arrays are first transcribed as single RNA before they are further processed into CRISPR RNAs (crRNAs), which direct the activity of the Cas enzyme.10 (B) CRISPR-Cas9 consists of a Cas9 protein with two cutting domains (RuvC and HNH) and a single guide RNA (sgRNA) that recognizes the complementary DNA sequence. (C) Repair mechanisms result in double-stranded breaks on DNA catalyzed by CRISPR-Cas9: the non-homologous end joining (NHEJ) pathway creates an insertion thereby knocking out a functional gene; homology-directed repair (HDR) corrects the gene in the presence of a donor template.
The majority of CRISPR-based gene-editing tools is derived from the type II adaptive immune defense system evolved in bacteria to combat invading viruses.11, 12, 13 In type II CRISPR, the trans-activating CRISPR RNA (crRNA; tracrRNA) forms an RNA duplex with crRNA by hybridizing with the direct repeats within the CRISPR array.8 This RNA duplex, called guide RNA (gRNA), forms a complex with the Cas9 protein. When the gRNA recognizes a viral DNA, it directs the Cas9 complex to the target strand, which matches a spacer in the CRISPR array. The nuclease domains of Cas9 subsequently catalyze the DNA DSB causing a silencing effect on the target gene.8
In 2013, this CRISPR method was successfully applied to multiple locations of DNA in eukaryotic cells, by changing the single gRNA (sgRNA) to match the desired target genetic sequence.14 The sgRNA is produced by fusing a crRNA, containing a targeting guide sequence, to a tracrRNA, which facilitates DNA cleavage by Cas9.8 The Cas protein efficiently scans DNA for the presence of short protospacer adjacent motif (PAM) sequences upstream of the target recognition site. Upon recognition of PAM and binding, Cas unwinds the adjacent DNA, which becomes available for hybridization with the target domain of the crRNA (or sgRNA) producing a triple-stranded R-loop structure. If the match is complete, then the Cas nuclease domains (HNH and RuvC) will catalyze the splitting of the bonds in the two DNA strands,15 thereby introducing DSB.16
The repair mechanism in the cell nucleus aims to recover the damaged DNA through one of two cellular repair mechanisms: the non-homologous end-joining (NHEJ) pathway or the homology-directed repair (HDR) pathway (Figure 1C).8,17 The NHEJ pathway is prone to errors and often leads to either insertion or deletion of bases during repair. The repair mechanism of HDR has previously been used to introduce defined alterations, such as insertions, into the genetic sequence.10,18,19 In doing so, a target DNA must be added to the CRISPR-Cas9 complex containing the desired genetic sequence. By pairing up with the cut ends, the target DNA is inserted into the original DNA strand. Application wise, much effort has been put into better understanding a way to control the two repair pathways depending on which type of mutation is desired. The NHEJ pathway is primed for investigating the function of a gene by disrupting it, e.g., by creating knockout genes in mice.20, 21, 22, 23 On the other hand, the inherent function of the HDR pathway has potential to replace the dysfunctional and mutated gene with a wild-type genetic sequence.24, 25, 26, 27 For example, the HDR-directed CRISPR-Cas9 technology can in this way be used to correct mutations in disease-causing genes or to engineer T cells for cancer immunotherapy.28, 29, 30, 31, 32, 33, 34 One way to promote the HDR pathway is to inhibit the NHEJ pathway either by gene silencing or by the action of small molecule inhibitors.30,31 However, so far, this approach has only been partially efficient.17
CRISPR-Cas9 still needs to meet several requirements to become an effective therapeutic tool. As in the case for other gene therapies,35, 36, 37 the targeted delivery of CRISPR-Cas9 remains a major challenge. Selected attempts to deliver CRISPR-Cas9 into eukaryotic cells are discussed below.
Current challenges of CRISPR-Cas9
There are several considerations and challenges regarding future therapeutic use of the CRISPR-Cas9 system. First, the clinical field so far has focused on identifying ex vivo delivery options. In vivo delivery remains more challenging from a technical point of view and is associated with multiple potential side effects.23,26
In relation to improving the bioavailability of CRISPR-Cas9, major obstacles include (1) efficient uptake in the eukaryotic cell;38 (2) efficient and reliable endosomal escape;39 (3) chemical and enzymatic stability;40 and (4) specificity of the CRISPR-Cas9 complex for target DNA.41, 42, 43, 44 This review focuses on addressing (1) and (2), while also providing future prospects on how to solve these issues.
Intracellular delivery of CRISPR-Cas9
Owing to its large size and polyionic nature, the CRISPR-Cas9 protein complex does not readily cross the cellular membrane and is in need of assistance in order to do so.45,46 In the following section, different methods for cellular uptake of the Cas9/sgRNA complex are reviewed.
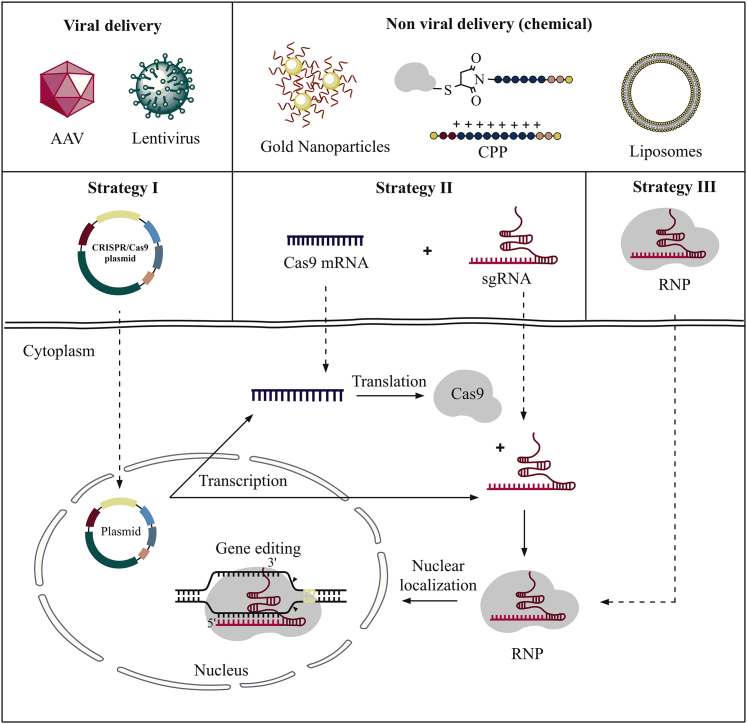
Various delivery methods have previously been investigated as a means to facilitate cellular uptake of Cas9/sgRNA. Essentially, two main parameters are known to affect uptake: cargo design and a delivery system.47 The three main strategies typically adopted for CRISPR-Cas9 cargo design include plasmid-based CRISPR-Cas9, a mixture of Cas9 mRNA and sgRNA, and Cas9/sgRNA (i.e., complex) (Figure 2). The delivery systems may further be divided into two main categories: namely, viral vectors and non-viral vectors.29,47
Figure 2.
Cargo-based delivery methods of the CRISPR-Cas9 complex
Strategy I: plasmid-based CRISPR-Cas9 system encoding both Cas9 and sgRNA. Strategy II: cellular delivery of Cas9 encoding mRNA in combination with sgRNA. Inside the cell, the mRNA is translated to Cas9 protein, and the Cas9/sgRNA complex is formed. Strategy III: direct delivery of the fully assembled Cas9/sgRNA complex into the cell.29
The first strategy involves a DNA plasmid-based CRISPR-Cas9 system,48, 49, 50 where the Cas9 protein and the sgRNA are encoded into the same plasmid vector. The second strategy aims at delivering mRNA for Cas9 (to be translated inside the cell)51,52 in conjunction with a separate sgRNA. The common denominator for these two approaches is the post-uptake biosynthesis of the desired CRISPR-Cas9 complex. Finally, a third strategy seeks to deliver fully assembled Cas9/sgRNA complexes by use of non-viral vectors.29
In strategy I, a well-defined plasmid encoding both the Cas9 protein and the sgRNA is delivered to the cell.29 One main drawback of this approach is the need for efficient intranuclear delivery—a task generally considered difficult.53, 54, 55 Furthermore, the plasmid must be translated into Cas9 mRNA inside the cell, which requires a long target-editing time.29 This can lead to overexpression of the Cas protein, which may further result in unwanted immune responses caused by the bacterial sequences as well as off-target effects and suboptimal safety.29,56,57
In strategy II, genome editing takes place after the Cas9 mRNA expression, and the Cas9/sgRNA complex is formed inside the cell. Advantages of this strategy include reduced off-target effects compared to the plasmid-based strategy due to the use of mRNA and its reportedly low cytotoxicity in primary cells and cell lines.58,59 In addition, the mRNA only needs to enter the cytoplasm rather than the nucleus, an inherently easier task to achieve.60 A disadvantage with the mRNA-based strategy is the poor circulatory and intracellular stability of mRNA.29 Nevertheless, this is the strategy that is being actively pursued in on-going research and clinical trials.61,62
Strategy III concerns the delivery of exogenous Cas9/sgRNA. The overall positive charge of the Cas9 protein enables it to form strong complexation to negatively charged sgRNA.63,64 Delivery of the Cas9/sgRNA complex (also known as RNA-guided endonuclease [RGEN]52 or ribonucleoprotein [RNP] complex29) has many advantages such as reduced off-target effects, lowered toxicity and immune responses, rapid action, and high gene-editing efficiency.65 Further, there is no need for codon optimization and promoter selection.29 However, many obstacles are associated with transfecting large RNPs.29 Next, we will revise representative methods previously employed as tools to accommodate transfection of CRISPR-Cas9.
Delivery systems for CRISPR-Cas9
As described above, the CRISPR-Cas9 complex and/or its components need a delivery system in order to cross the cell membrane.29,57 Current strategies for CRISPR-Cas9 delivery are based mainly on viral vectors. In the following, viral and non-viral delivery systems (including physical and chemical approaches) for CRISPR-Cas9 are reviewed.
Viral vectors
Viral vectors have been developed to effectively deliver nucleic acid-based therapeutics in vitro, ex vivo, and in vivo, and they can be applied to deliver plasmid-based CRISPR-Cas9 to mammalian cells.66 Important examples of viral vector delivery systems are adeno-associated virus (AAV) and lentivirus, reviewed below.67
Delivery based on AAV
AAV is a small, non-enveloped parvovirus that contains a single-stranded (ss)DNA. It is serotype specific and can infect both dividing and non-dividing cells. Owing to the high efficiency of transduction of a broad range of target tissues and good safety profile, AAV has been actively explored for gene editing in vivo.29 AAVs exhibit excellent genotoxicity profiles, which is also supported by multiple studies in vivo,68 and by clinical trials, e.g., with no tumor formation documented >7 years post-gene transfer in hemophilia B subjects.69 Nevertheless, immune responses to both AAV capsid and the encapsulated transgene have been reported.68,70 Therefore, immunogenicity remains a key point in assessing the safety profile of AAV gene therapy.68
Immune responses to the AAV capsid can be divided into two groups: pre-existing neutralizing antibodies to AAV and CD8+ T cell-mediated cytotoxic immune response toward transduced cells presenting AAV capsid antigens. Among other strategies, use of immunosuppression has been suggested to manage immune response to the AAV gene.71,72 Furthermore, Xiang et al.73 showed that by reducing the number of CpG motifs from the AVV genome, the expansion of naive T cells directed against an epitope within the capsid was lowered.
Directing AAV to specific tissues and cells is highly desired for in vivo applications. For example, Bengtsson et al.74 developed an approach for editing the mutation in dystrophic mdx4cv mice using systemic administration of single and dual AAV vectors carrying a muscle-specific Cas9 cassette together with sgRNA cassettes. Muscle-restricted Cas9 expression enabled direct editing of the mutation causing Duchenne muscular dystrophy, multi-exon deletion, or complete gene correction via homologous recombination in myogenic cells.74
Directing AAV to the CNS is another exciting approach with multiple potential applications. Nonnenmacher et al.75 recently reported an efficient screening model for peptide components that allow for blood brain barrier (BBB) penetration by AAVs. Ten individual variants were characterized in vivo and showed up to 400-fold higher brain transduction over AAV9 following systemic administration.
It is well known that the majority of plasmid vectors exists in an extrachromosomal state. However, it has been shown previously that a fraction of AAV vectors integrates into pre-existing DSB.76,77 Furthermore, NHEJ-mediated integration of AAV vectors was also observed after DSBs induced by zinc-finger nuclease (ZFN) or CRISPR in liver, muscle, and eye.78, 79, 80
Hanlon et al.81 observed high levels of AAV integration (up to 47%) into Cas9-induced DSBs in therapeutically relevant genes in cultured murine neurons, mouse brain, muscle, and cochlea. The impact of AAV transfection on the genome in mouse brain showed no overall increase of AAV integration except at the CRISPR-Cas9 target site. The integration profile of the developed vector AAV465λ in cultured cells was analyzed by sequencing. It revealed both full-length and fragmented AAV genomes at Cas9 on-target sites. The authors underline that based on their results, AAV integration is common, and it should be considered for AAV applications in translational studies.
In the context of Cas delivery, another critical limitation of viral vectors is the need for a self-inactivating system to limit overexpression and potential immunogenicity against bacterial Cas protein and potential elimination of edited cells by cell immunity. Li et al.82 described a self-deleting AAV-CRISPR system that introduces insertion and deletion mutations into AAV episomes. This system dramatically reduces the level of Cas (>79%) at high rates of on-target editing in the liver. Off-target mutagenesis was not observed for the self-deleting Cas9 gRNA at any of the predicted potential off-target sites examined.
Another limit of AAVs for CRISPR-Cas9 delivery is the packing limit, which is approximately 4.5 kb. The sequence encoding SpCas9 protein and sgRNA is approximately 4.2 kb.29
Lentivirus-based delivery
Lentivirus is another viral delivery system often applied in CRISPR. Lentivirus belongs to retroviruses, where the ssRNA genome is being reverse transcribed into cDNA upon cellular entry. Lentiviruses can integrate a significant amount of viral cDNA into the DNA of the host cell and can efficiently infect non-dividing cells. By integrating their genome into the host germline genome, lentiviruses can also become endogenous, causing the virus to be inherited by the host’s offspring cells.83
Generally, most of the lentiviral vectors tested thus far induced undetectable activation of innate immune responses.84, 85, 86 Stimulation of adaptive immune responses against lentiviral vectors was effective in causing a decrease in transgene expression only if the immune response was directed against the transgene.87 In the case of tissue-expressed, vector-derived epitopes, this could lead to immunogenicity. The lentivirus is characterized by long-term expression of transduced genes. Long-term toxicity is further expected for gene products with an immunogenic profile.87
Tissue and cell specificity has been explored for lentiviruses. Recently, Park et al.88 designed a lentivirus-RNA system, although not CRISPR, to target overexpressed vascular endothelial growth factor (VEGF) A in human gastric cancer cells and proved its tumor specificity in vivo. In another recent work, Raikwar et al.89 directed CRISPR to reactive glial cells surrounding the amyloid plaques in the mouse model of Alzheimer’s disease. Microglia cells are the major source of proinflammatory cytokines and chemokines including glia maturation factor (GMF). The developed lentiviral vectors expressed either Streptococcus pyogenes Cas9 (SpCas9) or GMF-sgRNAs. With the use of this system, targeting of GMF was achieved selectively in glial cells.
The amount and persistence of Cas expression in target cells have also been correlated to off-target mutation rate. Thus, high concentrations of the Cas nuclease are reported to increase off-site cleavage, whereas lower amounts improve the specificity.90,91 Petris et al.92 developed a self-limiting “hit and go” system, Self-Limiting Cas9 Circuit for Enhanced Safety and Specificity (SLiCES), which consists of an expression unit for SpCas9, a self-targeting sgRNA, and a second sgRNA targeting a chosen genomic locus. SLiCES was tested in lentiviral delivery system. The self-limiting circuit resulted in increased genome-editing specificity by controlling Cas9 levels, with off-target gene editing reduced up to 5-fold by the self-limiting system.
A promising hybrid approach to sgRNA/Cas9 delivery ex vivo has been demonstrated by Shifrut et al.33 In this work, sgRNA-loaded lentivirus was first delivered to T cells, and then Cas9 protein was delivered to these cells by electroporation.33 The rationale behind this approach is that by directly delivering Cas9, the mutation is induced more quickly than when mRNA encoding for Cas9 is applied. This strategy allowed for the identification of gene modifications that promote T cell proliferation in response to stimulation.33
Immune-cell engineering with CRISPR opens new capabilities for fundamental immunology research and immunotherapy. Among others, systems for efficient generation of chimeric antigen receptor (CAR)-engineered T cells (CAR-T cells) are in development. Herein, viral delivery of CRISPR, mainly done ex vivo, has been dominating the field. The success of CAR-T cells for the treatment of B cell leukemias and lymphomas has been remarkable, and this approach has relied heavily on lentivirus-mediated gene transfer.93, 94, 95
Lentiviruses and other retroviruses have also been explored for editing natural killer (NK) cells in the context of immuno-oncology applications.96 Contrary to T cells, NKs eliminate their targets in a non-antigen-specific manner and do not carry the risk of inducing graft-versus-host disease, allowing application of donor-derived cells in an allogenic setting.96 Hence, unlike autologous CAR-T cells, therapeutic CD19-CAR-NK cells can be generated as an off-the-shelf product from healthy donors.
Overall, viral vectors are generally considered efficient. However, several drawbacks to this technique include undesired genetic alterations and host immune responses, which lead to potential issues with applications in vivo, as well as limitations of insertion size.17 These issues are being addressed in part by non-viral delivery systems reviewed below.
Non-viral vectors
The main gain of using non-viral vectors over viral vectors is the higher safety.97,98 Furthermore, there is no size limit on the transgenic DNA to be transported by non-viral vectors, and non-viral delivery systems are readily available and cost effective.29 On the contrary, non-viral vectors have a poor delivery efficacy, which limits in vivo applications.97,98 However, different ligands, such as antibodies and aptamers, can be implemented into the non-viral vector design to induce cellular uptake through receptor-mediated endocytosis.17 Physical delivery of CRISPR-Cas9 is limited to cellular and ex vivo applications.99,100 RNPs are more complicated than other programmable nucleases such as ZFNs and cell-penetrating peptide (CPP)-conjugated transcription activator-like effector nucleases (TALENs).56 This is because RNPs contain both a protein and a sgRNA. Physical delivery of RNPs, along with a hybrid delivery by physical methods with viral carries, has been used to generate modified cells, which can be applied for studying genetic diseases and specific gene functions.33
Overall, non-viral delivery systems include physical delivery methods such as electroporation, microinjection, and hydrodynamic injection, whereas chemical delivery methods cover ligand-mediated delivery systems, CPPs, and nanoparticles, the latter of which includes exosomes, lipid nanoparticles (LNPs), and organic and inorganic polymers. The physical delivery methods cannot be translated to in vivo applications. In the remaining sections, we focus on the non-viral delivery methods, more specifically, CPP conjugates and nanoparticles, as tools for CRISPR-Cas9 delivery, which potentially could be applied in vivo.
Ligand-mediated targeted delivery of CRISPR-Cas9
Receptor-facilitated and cell type-selective gene editing by CRISPR-Cas has been explored by Rouet et al.101 With the focus on hepatocyte selective delivery, CRISPR-Cas9 RNP harboring a ligand for the asialoglycoprotein receptor (ASPGR) was engineered and tested in cells. ASPGR is expressed almost exclusively by liver-derived cells (e.g., HEPG2). Ligand variants for ASPGR were designed and synthesized to be attached to Cas9 via a cleavable disulfide linker. The product conjugates were tested in HEPG2 cells with sgRNA targeting fluorescent protein reporters. It was shown that ASPGR ligand-Cas RNPs were internalized via receptor-mediated endocytosis. Subsequent endosomal escape, assisted by an endosomolytic agent, followed by nuclear transport, ultimately resulted in selective gene editing.101
CPPs for CRISPR-Cas9 delivery
CPP is a short peptide sequence consisting of highly abundant, positively charged amino acids or alternating polar and nonpolar amino acid residues. These peptides have the ability to cross the cell membrane by different mechanisms including direct penetration, translocation mediated by endocytosis, or formation of pores.36
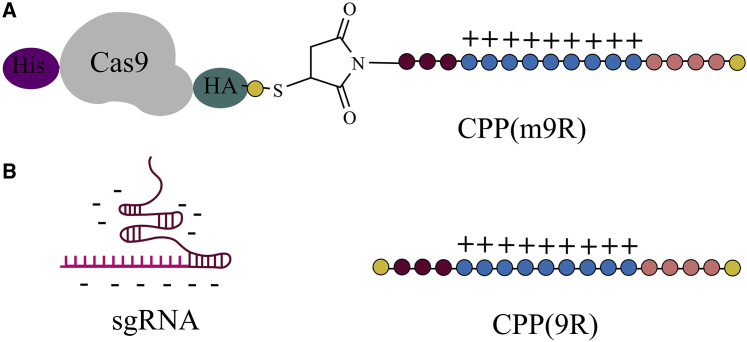
The delivery of Cas by non-viral vectors reduces mutation rates by circumventing potential overexpression of Cas reported in strategy I (plasmid delivery).90,91 The covalent attachment of CPPs onto the Cas protein allows plasmid transfection to proceed without the need for exogenously added transfection reagents. Ramakrishna et al.64 successfully produced a CPP-Cas9 construct by conjugating CPP to the C terminus end of Cas9 (Figure 3A). Specifically, introducing a C-terminal cysteine residue enabled its conjugation with a maleimide-functionalized CPP through a thioether bond. They also introduced a His tag on the N terminus and a hemagglutinin (HA) tag on the C terminus on the Cas9 protein for purification and detection purposes (Figure 3A).64 Additionally, an sgRNA was co-formulated by complexation to a naked CPP (Figure 3B). The overall positive charge of the CPP surface can facilitate cellular uptake through interaction with the negatively charged plasma membrane.64
Figure 3.
CPP-mediated delivery of Cas9/sgRNA system
(A) Cas9 protein conjugated to a CPP (m9R) via a thioether bond. (B) Subsequently, sgRNA forms a complex with the CPP domain of CPP(m9R) due to electrostatic attractions.64 Amino acids indicated by yellow (Cys), brown (Gly), blue (Arg), and orange (Lys).
The electrostatics favor 9R and sgRNA to condense into positively charged nanoparticles, which were found to efficiently transfect sgRNA into cells.64 Experiments showed that efficiency (on-target effects) and specificity (off-target effects) are strongly affected by Cas9 protein and sgRNA concentrations. Furthermore, the mutation frequency was found to depend on various factors such as number of treatments, exposure time, incubation temperatures, and the sgRNA-to-m9R ratios.64
Production of recombinant Cas9 with CPP has the disadvantage of being difficult to handle, mainly due to the large size of Cas (>150 kDa), and to be labor intensive and expensive.102
A supramolecular strategy for the direct delivery of Cas9 by an amphiphilic-penetrating peptide has been explored.103 The peptide/Cas9 non-covalent nanoparticles performed with similar efficiency and less toxicity than one of the best methods described to date. Notably, the complex also efficiently escaped from the endosomes. The results reported in this work confirmed that peptide amphiphilic vectors could deliver Cas9 in a single incubation step, via a macropinocytic uptake mechanism, with good efficiency and low toxicity, so far confirmed only in cell lines (HeLa, A549, and DF1).102,103
One disadvantage of using CPP-mediated delivery is that for most CPP sequences, the delivery is non-specific. Furthermore, extended structure-property studies and optimizations of CPPs are needed to achieve adequate protection against protease degradation in vivo.104,105
Another major limitation of CPPs to be overcome is potential toxicity.106 Moreover, Venit et al.107 recently reported on gene-expression alteration as a response to CPPs. For this study, four CPPs were selected, i.e., penetratin, PepFect14, mtCPP1, and TP10. After treatment with these CPPs, HeLa cells were transcriptionally profiled by RNA sequencing. Results from these analyses showed a time-dependent response to CPPs, with genes related to ribosome biogenesis, microtubule dynamics, and long-noncoding RNAs being differentially expressed compared to untreated controls. The authors speculate that the gene-expression alterations are a part of a natural response to such compounds. However, it potentially raises issues of gene toxicity toward CPPs that needs to be further assessed.107 It is also noteworthy that the toxicity of CPPs is dose dependent.
Campeiro et al.104 in the aforementioned study reduced the dose of the CPP down to 1 μg/animal/day for recurrent dosing or did a high single dose (30 μg/animal) of acute intraperitoneal (i.p.) administration of crotamine. Klein et al.108 reported on an arginine-rich Pip6a CPP and showed that Pip6a-conjugated morpholino phosphorodiamidate oligomer (PMO) dramatically enhanced antisense oligonucleotide (ASO) delivery into striated muscles of DM1 mice following systemic administration in comparison with unconjugated PMO and other ASO strategies. Low-dose treatment with Pip6a-PMO-CAG targeting pathologic expansions (single dose, 12.5 mg/kg) was sufficient to reverse both splicing defects and myotonia in DM1 mice and normalized the overall disease transcriptome. Pip6 CPP series for in vivo gene therapy has been patented in 2016 by Gait et al.109 Studies are currently underway focusing on the in vivo toxicological profile of these compounds and understanding the minimal requirements of CPP composition to maintain robust activity while minimizing organ toxicity, especially nephrotoxicity.110
Nanoparticles for CRISPR-Cas9 delivery
Complexation of the CRISPR-Cas9 with non-viral vectors has previously shown to promote cellular uptake of CRISPR-Cas9.17 Based on the nature of the applied materials, nanoparticle systems for CRISPR delivery can be divided into four main categories: (1) exosomes, (2) artificial LNPs, (3) organic, and (4) inorganic artificial nanoparticles. Notably, there are examples of combining these platforms; i.e., organic-inorganic and exosome-LNP fusions.
Exosomes are nanoscale membrane vesicles secreted by most cells and function to transmit different signaling molecules such as mRNA, proteins, and lipids. Due to the small size of exosomes (30−100 nm), they can escape via phagocytosis by mononuclear phagocytes and pass through the vascular endothelium to reach the target cells. They are considered rather effective drug-delivery systems, as they exhibit a high degree of cellular targeting due to their surface proteins. However, exosomes are limited by size and their low efficiency to encapsulate large nucleic acids. Furthermore, exosomes are rather difficult to purify, which is considered a major barrier for their clinical use.111 Lack of standardized isolation and purification methods and insufficient clinical grade production are other challenges to be overcome in order to make exosome-based delivery systems more accessible.
As with CPPs, nanoparticles for CRISPR-Cas9 delivery need to exhibit good tissue and cell specificity. Gulei and Berindan-Neagoe112 proposed cancer-engineered exosomes as delivery vesicles for necroptosis activation via CRISPR-Cas9. Targeted delivery could be achieved by developing exosomes bearing a specific ligand on their surface. Once these exosomes were produced and selected, they were successfully applied as a delivery vesicle for CRISPR-Cas9 vectors targeting necroptotic pathway IAP1/2 and caspase-8.
LNPs are artificial lipid-based delivery vehicles with the ability to encapsulate large oligonucleotides, such as the CRISPR-Cas9 expression vector. It is generally acknowledged that LNPs possess limited cellular uptake and have a tropism to liver.113,114 This property has previously been exploited for editing the mouse transthyretin (Ttr) gene in the liver, with a >97% reduction in serum protein levels that persisted for at least 12 months. The developed CRISPR-LNP delivery system was biodegradable and well tolerated.59 Nevertheless, LNPs need optimization and potentially surface decoration to target other tissues and cells.
Rosenblum et al.115 reported on successful delivery of Cas9 mRNA and sgRNAs by using a novel ionizable lipid. In vivo, the system proved to be safe and efficient; a single intracerebral injection of CRISPR-LNPs against PLK1 (sgPLK1-cLNPs) into aggressive orthotopic glioblastoma enabled up to ∼70% gene editing, which caused tumor cell apoptosis, inhibited tumor growth by 50%, and improved survival by 30%. To reach disseminated tumors, CRISPR-LNPs were also engineered for antibody-targeted delivery. Intraperitoneal injections of EGFR-targeted sgPLK1-cLNPs caused their selective uptake into disseminated ovarian tumors and enabled up to ∼80% gene editing in vivo, inhibited tumor growth, and increased survival by 80%.115
In another recent work, Qui et al.116 explored LNPs for repair of loss-of-function mutations in angiopoietin-like 3 (Angptl3) that is associated with lowered blood lipid levels. In this study, Cas9 mRNA and gRNA for CRISPR-Cas9-based genome editing of Angptl3 were delivered by LNPs in vivo. This system mediated specific and efficient Angptl3 gene knockdown in the liver of wild-type C57BL/6 mice, resulting in profound reductions in serum ANGPTL3 protein, low-density lipoprotein cholesterol, and triglyceride levels. The delivery platform was significantly more efficient than the US Food and Drug Administration (FDA)-approved MC-3 LNP, the current gold standard.117 Neither off-target effects nor evidence of liver toxicity were detected. Remarkably, remission upon CRISPR-Cas gene editing was stable for >100 days after a single dose treatment.
An interesting approach has been taken by Lin et al.118 who reported on a fused hybrid exosome-liposome nanoparticle system for CRISPR-Cas9 delivery. By fusing the exosome and liposome together via incubation, a hybrid nanoparticle is created primed for encapsulating plasmids encoding the desired CRISPR-Cas9 during incubation.118 Lin et al.118 found that the resulting hybrid nanoparticles underwent cellular uptake to mesenchymal stem cells due to specific vesicle proteins in the surface of the exosome. It was further noticed that the hybrid nanoparticles had similar toxic effects as the liposomes, and it was concluded that the liposomes need to be further optimized in order to resolve this issue.118
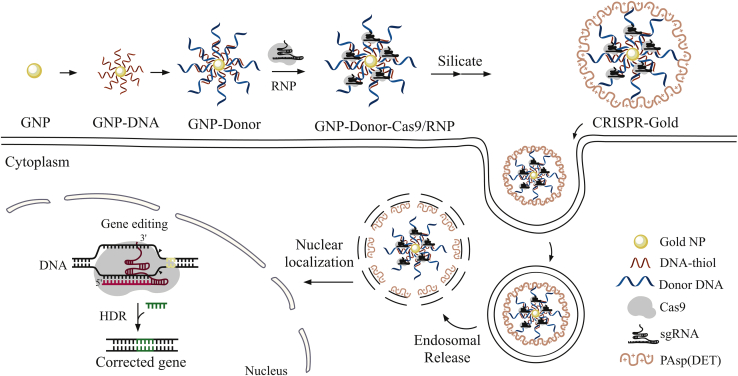
Lee et al.119 developed a gold nanoparticle system with a HDR-directing plasmid containing Cas9 mRNA, sgRNA, and a DNA template. Specifically, this system was composed of gold nanoparticles conjugated to DNA and complexed with the cationic endosomal disruptive polymer PAsp(DET).119 They found that this method readily delivers Cas9 protein and donor DNA into different cell types and correct DNA mutation of a given disease through the HDR pathway. This strategy, coined CRISPR-Gold, exhibits minimal off-target DNA damage and immunogenicity. The polymer PAsp(DET), which complexes the other components of the system, is the main mediator causing the endocytosis of CRISPR-Cas9 components while also triggering endosomal disruption and release of cargo. Here, glutathione cleaves DNA from the gold core of the complex leading to release of the Cas9 RNP and donor DNA119 (Figure 4). In this system, gold nanoparticles were coated with a densely packed layer of donor DNA with affinity for the Cas9-gRNA complex.
Figure 4.
Synthesis of the CRISPR-Gold system
Illustration of the cellular uptake and endosomal escape mechanism by the CRISPR-Gold nanoparticle delivery system. The CRISPR-Gold complex is uptaken by the cell and triggers endosomal escape by the PAsp(DET) polymer. The Cas9-sgRNA and donor DNA migrate to the nucleus to perform gene editing through the HDR pathway.119
Thus, a number of non-viral vectors for CRISPR-Cas9 delivery exist. However, these systems need further development before they can replace the currently used viral vectors such as AAVs.120
Endosomal escape of CRISPR-Cas9
Degradation in the acidic environment of the endo-lysosomes (endosomes fused with lysosomes) located in the cytoplasm must be avoided in order for the CRISPR-Cas9 system to reach the nucleus and the target DNA sequence.17,29
Different methods to induce endosomal escape are outlined above, for example, the gold nanoparticle system using PAsp(DET) polymers that disrupt the vesicle after endocytosis.119 In the following section, methods for endosomal escape are explored in more detail, focusing on three main strategies: proton sponge, pH responsive nanoparticles, and peptide-mediated systems, including dynamic polyconjugates.
A common strategy to measure oligonucleotide uptake and intracellular distribution involves fluorescence read-out of fluorophore-labeled sequences that have been incubated with cells.121 Confocal fluorescence microscopy studies can be used to examine co-localization of a fluorophore-labeled oligonucleotide and dye-labeled antibodies of endocytic markers. In these co-localization experiments, chloroquine is used as a control additive to halt endosomal maturation. With the use of this approach, scientists in the Crooke group122 at Ionis Pharmaceuticals identified coat protein complex II (COPII)-containing vesicles and associated tethering protein STX5 as facilitators of endosomal escape of ss phosphorothioate (PS) oligonucleotides.
However, it is rather challenging to quantify weak and diffuse signals indicative of cytosolic uptake in the presence of a bright signal in endo- and lysosomes.123 Furthermore, any punctate signal that does not co-localize with common endosomal markers such as Rab5 or EEA1 (early endosome), Rab7 (late endosome), and LAMP1 (lysosome) is often mischaracterized as endosomal escape. Co-incubation of samples with calcein prior to transfection is often another method used to overcome the requirement for fluorescent labeling. Calcein is a small membrane-impermeable dye that appears punctate when binding to endosomal and lysosomal compartments. However, this approach does not provide a direct measurement of cargo escape. Moreover, photobleaching of many common dyes used in fluorescence microscopy complicates the measurements.124 Quantitative measurements of endosomal escape are therefore very difficult.
In spite of technical issues with direct measurement of efficacy, endosomal escape has been widely approached and characterized qualitatively, using fluorescence-based methods such as those mentioned above. A general approach explored for a vast number of oligonucleotide-based delivery systems is to utilize the change in pH in the acidic environment within the endo-lysosomes. The “proton-sponge effect” is a phenomenon that takes advantage of the pH gradient change inside the endo-lysosome as it transgresses into the late-stage lysosome.125,126 This effect is accessed by use of polymers with a high proton buffering capacity. Most notably, polyamines of secondary and/or tertiary nature are protonated inside the acidic endo-lysosomes, thus inducing a flow of chloride ions into the vesicles as counterbalance. This event creates an osmotic gradient across the vesicle125,126 and consequently, the swelling of the endo-lysosome that eventually leads to a burst to release its contents into the cytoplasm.17 Different non-viral vectors exhibit this proton-sponge effect. For example, compounds such as chloroquine, imidazole, and zinc chloride can be combined with cationic vectors in order to enhance the endosomal rupture.127,128
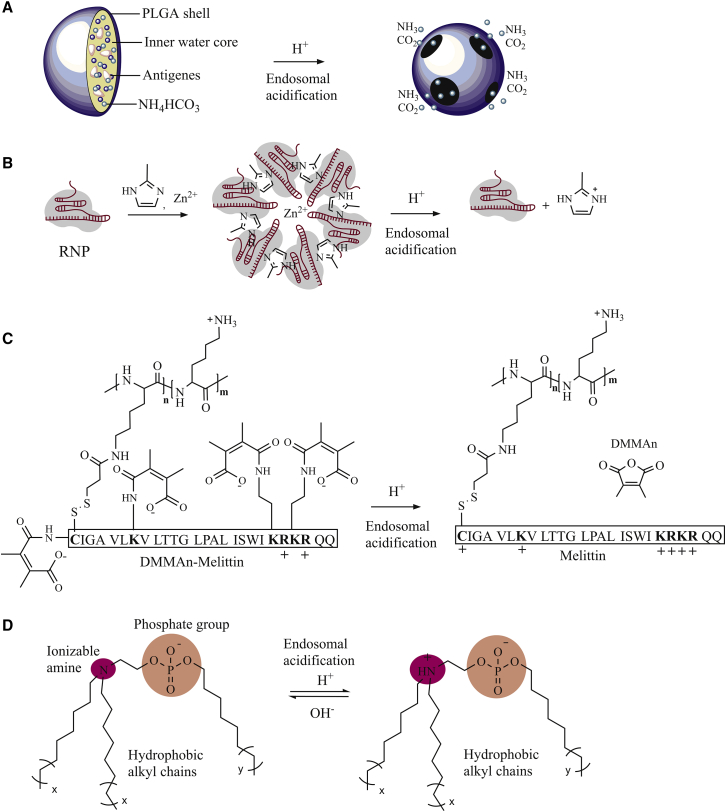
Liu et al.129 designed a pH-responsive, gas-generating polynanoparticle composed of poly(D,L-lactic-co-glycolic acid) (PLGA) and ammonium bicarbonate as a stabilizer. Once the particles enter the acidic environment of an endo-lysosome, ammonium bicarbonate undergoes a chemical reaction to produce carbon dioxide and ammonia, causing the particle to rupture (Figure 5A). Liu et al.129 used this platform to introduce antigens into dendritic cells, but the system might be interesting for a non-viral CRISPR-Cas9 delivery carrier.
Figure 5.
Endosomal escape methods applied to CRISPR-Cas9
(A) Proton sponge: when ammonium bicarbonate (NH4HCO3) reacts with protons in the acidic environment in endo-lysosomes, the products formed (CO2 and NH3) disrupt the vesicle resulting in endosomal escape.129 (B) Synthesis of CC-ZIFs and endosomal escape induced by CC-ZIFs through protonation of the imidazole ring.130 (C) Principles for function of dynamic conjugates with pH labile dimethylmaleic anhydride (DMMAn)-protected melittin,131 and (D) ionizable phospholipids (iPhos) adopting a cone shape once entering the acidic environments of the endosome making the endosomal release due to interacting with cell membrane.132
Another option is to design membrane fusion peptides that undergo a structural change in acidic environments and to implement these in non-viral vectors.17 Such peptides may include GALA peptides and acid-activated melittin. They can undergo a conformational change from a random coil to a structured α-helix when they enter acidic pH, and this promotes endosomal fusion.18 However, toxicity is a potential risk with peptide-based systems as mentioned above and reviewed before.133 Therefore, a careful assessment of toxicity, ideally in vivo, is needed prior to applications.104
Dynamic polyconjugates for endosomal escape have been explored by several research groups and industry. The technology was initially demonstrated by Meyer et al.131 in 2007. It is based on using a functionalized poly-lysine backbone typically modified covalently with an acid labile poly(ethylene glycol) (PEG) functionality along with masked peptide melittin (Figure 5C). The obtained conjugate was activated in acidic compartments to facilitate endosomal escape for RNA therapeutics. In addition, positively charged amino groups in the poly-lysine framework were able to electrostatically bind negatively charged RNA. Later on, it was reported that these constructs caused substantial liver necrosis with abdominal bleeding in mice.134 The likely reason for high toxicity of these dynamic conjugates was the risk for electrostatic aggregation into micrometer-sized particles with accumulation in liver.135
Rozema et al.136 used a similar approach for targeted liver delivery of gene therapeutics. The technology was based on poly butyl and amino vinyl ethers generating a masked endosomolytic polymer derived from melittin. The system included a PEG functionality and targeted the liver via the apolipoprotein B receptor. This approach was initially developed with support from Mirus Bio, later absorbed by Roche, and then transferred to Arrowhead.137 With the use of the optimized melittin variant, Arrowhead’s technology demonstrated >90% knockdown in hepatocytes at low dose (3 mg/kg) in non-human primates. Unfortunately, clinical trials conducted in 2015−2018 showed toxicity of the dynamic conjugates, likely occurring from melittin.138 This led to Arrowhead dropping all three clinical programs that rely on melittin. In 2019, Colletti et al.139 at Merck filed a patent on a similar technology. To date, no information on initiated clinical trials has been disclosed.
With the aim for a safe solution to the problem of endosomal escape, Liu et al.132 took inspiration from natural phospholipids that comprise biological membranes. They prepared and tested in vivo multi-tailed ionizable phospholipids (iPhos) capable of delivering mRNA or mRNA/sgRNA (Figure 5D).132 Optimized iPhos were composed of one pH-switchable zwitterion and three hydrophobic tails adopting a cone shape once entering the acidic environments of the endosome. The lowering of pH facilitates membrane hexagonal transformation and subsequent cargo release from the endosomes (Figure 5D). A structure-activity relationship study revealed that iPhos modulates in vivo efficacy and organ selectivity. In this system, zwitterionic and ionizable cationic helper lipids aided delivery of mRNA and CRISPR-Cas9 selectively to spleen, liver, and lungs, where gene editing took place.
Another rapidly growing field uses inorganic chemistry approaches for creating nanoscale frameworks with tunable biological properties including propensity for endosomal escape. Alsaiari et al.130 designed a nanoscale zeolitic imidazole framework (ZIF) able to encapsulate the CRISPR-Cas9 complex. It is a subclass of metal-organic frameworks that are formed by the coordination between Zn2+ ions and 2-methylimidazole.130 ZIFs exhibit good biocompatibility and have adjustable pore openings, which can accommodate compounds of different shapes and sizes. Imidazole linkers have a decent pH-buffering capacity and the ability to promote endosomal escape. For example, a CRISPR-Cas9 ZIF (CC-ZIF) (Figure 5B) has previously been prepared by mixing Cas9 protein and sgRNA in PBS. Next, a 2-methylimidazole solution (pH 7) was added, followed by an aqueous solution of zinc nitrate. The CC-ZIF compound was found to be stable at neutral pH; however, gradual degradation of the CC-ZIF complex accelerated at lower pH, likely due to protonation of the imidazole ring causing the destabilization of CC-ZIF. They found that at the point of rupture, the Cas9-sgRNA complex was released into the cytoplasm from where it migrated into the nucleus. One major advantage of the CC-ZIF complex is that it is biodegradable and non-toxic at lower concentrations (below 100 μg/mL).130
Although being a hotspot in nucleic acid research, endosomal escape is still rather poorly characterized quantitatively. This is mainly due to technical issues with existing fluorescence-based methods, as mentioned above. To address this, Teo et al.123 recently reported on a quantitative strategy, called Split Luciferase Endosomal Escape Quantification (SLEEQ), for measuring endosomal escape. In brief, the split luciferase assay is comprised of two subunits: large binary technology protein (LgBiT; 17.8 kDa) and a high-affinity complementary peptide (HiBiT; 1.3 kDa). LgBiT was expressed as a fusion protein with actin, causing the transport of LgBiT into the cytosol, and the attachment to green fluorescent protein (GFP) further enabled its quantification in the cytosol. Teo et al.123 demonstrated that SLEEQ could be used to detect picomolar concentrations of proteins delivered to the cytosol and to quantify the efficiency of endosomal escape.
SLEEQ was used to evaluate the endosomal escape of a range of widely studied putative endosomal escape peptides (EEPs). Putative EEPs were fused to GFP as a model delivery cargo. Eight widely used EEPs were tested, including polycationic TAT, polyarginine (R9), 5.3, ZF5.3, and amphiphilic peptides pHlip, melittin-derived pHD118, and HA2.
Overall, endosomal escape was shown to be a highly inefficient process, with only ∼2% of GFP reaching the cytosol in HEK293 cells, and ∼7% of GFP reaching the cytosol in HeLa cells. Positively charged EEPs increased the total amount of protein delivered to the cytosol. However, according to the SLEEQ data, the efficiency of endosomal escape was the same or lower than the efficiency of GFP escape without EEPs. This suggests that the positively charged EEPs increased cytosolic accumulation mostly through non-specific association with the cells, rather than inducing an active mechanism of endosomal escape. The authors argue that since the selected EEPs did not increase endosomal escape, a more appropriate name for this group of peptides would be membrane adsorptive peptides (MAPs).
Discussion
The concept of solving two of the major challenges associated with effective CRISPR-Cas9 gene modification, namely cellular uptake and endosomal escape, by using a single delivery system, seems promising. Along with toxicity and specificity, addressing these two major issues potentially would allow for systemic administration of CRISPR-Cas9 in patients.
In this work, we review several existing methods to shorten the gap between preclinical and clinical trials. Thus far, viral vectors have been dominating in the clinical exploration of CRISPR, with CAR-T and applications in cancer immunotherapy being ultimate successes achieved to date.93, 94, 95 High doses of viral vectors are typically needed for successful transfection.33,93, 94, 95,140 Moreover, lentiviruses that are currently used ex vivo also have a substantial risk of unspecific gene editing due to prolonged transduction times.90,91 In vivo applications of viral carriers have also raised concerns in terms of toxicity.141
A promising new direction would be to switch to non-viral delivery methods, both for ex vivo and in vivo delivery routes. Inducing cellular uptake through endocytosis by non-viral vectors can reduce immunogenicity, unspecific gene editing, and toxicity compared to the use of viral vectors. Systems with the ability to encapsulate the entire Cas9 protein and sgRNA now exist.101 Contrary to the plasmid vector strategy, these components are readily primed to perform gene-editing functions once entering the cell.33,101
Non-viral delivery systems are represented by a variety of approaches, with LNPs59,115,116 and exosomes112 being among the most advanced ones. Issues relating to production and scale-up have been partly solved for LNP-based technologies; however, reliable scale-up procedures for exosomes still need further development.
Peptide-based systems64 and inorganic frameworks119 compatible with CRISPR delivery are under rapid development, with several successful examples already demonstrated in vivo.60,142 These systems possess many advantages including robustness, large loading capacity, and the possibility of scale-up for translational applications. Tissue and cell specificity, potential toxicity, and biological stability are critical parameters for these systems to be taken into consideration.101,104 To assess these metrics, most studies have been limited to in vitro examinations and cellular assays. However, in vivo data are still needed to fully evaluate the delivery potential of these systems.104
Suboptimal endosomal escape is still hampering the advancement of nucleic acid therapeutics in a clinical setting. Taking inspiration from natural vesicles such as exosomes and phospholipids that comprise biological membranes, successful systems delivering CRISPR-Cas components have been proposed and tested also in vivo.132
Furthermore, the pH in the endo-lysosomes is acidic, and this can be used as a trigger for endosomal escape for the Cas9-sgRNA complex by use of components that are responsive to change in pH from neutral to acidic.139 It is important to ensure that the components used for non-viral transport of the CRISPR-Cas9 complex are non-toxic and biodegradable and that such excipients do not cause immune response activation.131,133, 134, 135
Structure-activity relationship studies are critical to advance further the field of non-viral delivery systems for CRISPR gene editing. A few such studies conducted on LNPs reveal that rather minute details in chemical structure (of the lipid components) control in vivo efficacy and organ selectivity.132 As an example, zwitterionic, ionizable cationic, and permanently cationic helper lipids enable tissue-selective mRNA delivery and CRISPR-Cas9 gene editing in spleen, liver, and lungs (respectively) following intravenous administration. This rational design of functional phospholipids demonstrates a substantial value for gene-editing research and therapeutic applications.
Conclusions
Despite the promising outlook for the CRISPR-Cas9 gene-editing tool, different challenges still need to be overcome. This review has focused on two of these problems, namely, cellular uptake and endosomal escape. To date, viral vectors account for the majority of reported delivery systems employed for CRISPR-Cas9.93, 94, 95 However, these systems’ propensity to activate immune defenses in vivo141 has also partly justified research into other delivery technologies, such as non-viral vectors. Regardless of the delivery system used, key properties of any CRISPR vector include targeted and efficient delivery, low toxicity, and a high gene-editing rate. In several works, the cell’s own defense mechanism against macromolecules—endocytosis—is being used as a means to promote cellular uptake of the Cas9-sgRNA complex.101,132
Endosomal escape is critical for non-viral vectors. Once the Cas9-sgRNA resides inside the cell’s endosome, the transgression from endosome into the acidic lysosome promotes pH sensitive compounds to react, either by chemical reactions or conformational changes, such as peptide triggered cytosolic release of Cas9-sgRNA.17,129 Moreover, various non-viral delivery vectors can be tailored to fit a certain tissue or cell-specific release, such as surface-modified nanoparticles112 or conjugates of Cas and/or sgRNA with a specific ligand.64,101
Herein, we have reviewed some of the most recent methods employed to address these challenges. Given the attractiveness of CRISPR for medical applications, multiple approaches are expected to be explored in the future. Ex vivo delivery of CRISPR-Cas is currently one of the most studied approaches, as it solves issues relating to cell specificity and the inherent immunogenicity of CRISPR-Cas.93, 94, 95 A shift toward systemic administration of CRISPR-Cas would be a major breakthrough in the field and requires development of more sophisticated delivery routes with high efficacy and safety profiles,101,103 self-limiting systems,92 etc. Herein, non-viral systems are promising candidates with the potential of tunable cell and tissue specificity101 and high loading capacity59 and the possibility to include endosomolytic agents101 and lower immunogenicity.101,132 In the future, to secure translation into the clinic, it is important to consider toxicity, immune response, and an efficient metabolism of new non-viral vectors for CRISPR-Cas9 gene editing.
Acknowledgments
S.S. and K.A. acknowledge support from the Technical University of Denmark, Elite PhD Scholarship Programme.
Author contributions
M.v.H. wrote the initial draft of the paper. M.v.H. and S.S. did the graphical work. All authors contributed to revision of the paper and approved the final version.
Declaration of interests
The authors declare no competing interests.
References
- 1.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mojica F.J., Juez G., Rodríguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993;9:613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Bell J., Lau B.T., Whittaker T., Stapleton D., Ji H.P. A functional CRISPR/Cas9 screen identifies kinases that modulate FGFR inhibitor response in gastric cancer. Oncogenesis. 2019;8:33. doi: 10.1038/s41389-019-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang F., Doudna J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 8.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huai C., Li G., Yao R., Zhang Y., Cao M., Kong L., Jia C., Yuan H., Chen H., Lu D., Huang Q. Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nat. Commun. 2017;8:1375. doi: 10.1038/s41467-017-01496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan X., Luo Z., Sun W. A peptide delivery system sneaks CRISPR into cells. J. Biol. Chem. 2018;293:17306–17307. doi: 10.1074/jbc.H118.006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Hu S., Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials. 2018;171:207–218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y., Zhang L., Huang X. Genome modification by CRISPR/Cas9. FEBS J. 2014;281:5186–5193. doi: 10.1111/febs.13110. [DOI] [PubMed] [Google Scholar]
- 16.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell. 2018;70:327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J.H., Adikaram P., Pandey M., Genis A., Simonds W.F. Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered. 2016;7:166–174. doi: 10.1080/21655979.2016.1189039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Modzelewski A.J., Chen S., Willis B.J., Lloyd K.C.K., Wood J.A., He L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc. 2018;13:1253–1274. doi: 10.1038/nprot.2018.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G., et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wefers B., Bashir S., Rossius J., Wurst W., Kühn R. Gene editing in mouse zygotes using the CRISPR/Cas9 system. Methods. 2017;121-122:55–67. doi: 10.1016/j.ymeth.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Meng X., Li J., Gao C. Gene Replacement by Intron Targeting with CRISPR-Cas9. Methods Mol. Biol. 2019;1917:285–296. doi: 10.1007/978-1-4939-8991-1_21. [DOI] [PubMed] [Google Scholar]
- 25.Miki D., Zhang W., Zeng W., Feng Z., Zhu J.K. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 2018;9:1967. doi: 10.1038/s41467-018-04416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCullough K.T., Boye S.L., Fajardo D., Calabro K., Peterson J.J., Strang C.E., Chakraborty D., Gloskowski S., Haskett S., Samuelsson S., et al. Somatic Gene Editing of GUCY2D by AAV-CRISPR/Cas9 Alters Retinal Structure and Function in Mouse and Macaque. Hum. Gene Ther. 2019;30:571–589. doi: 10.1089/hum.2018.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 29.Liu C., Zhang L., Liu H., Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karakashev S., Fukumoto T., Zhao B., Lin J., Wu S., Fatkhutdinov N., Park P.H., Semenova G., Jean S., Cadungog M.G., et al. EZH2 Inhibition Sensitizes CARM1-High, Homologous Recombination Proficient Ovarian Cancers to PARP Inhibition. Cancer Cell. 2020;37:157–167.e6. doi: 10.1016/j.ccell.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayavaradhan R., Pillis D.M., Goodman M., Zhang F., Zhang Y., Andreassen P.R., Malik P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019;10:2866. doi: 10.1038/s41467-019-10735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowther M.D., Dolton G., Legut M., Caillaud M.E., Lloyd A., Attaf M., Galloway S.A.E., Rius C., Farrell C.P., Szomolay B., et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 2020;21:178–185. doi: 10.1038/s41590-019-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shifrut E., Carnevale J., Tobin V., Roth T.L., Woo J.M., Bui C.T., Li P.J., Diolaiti M.E., Ashworth A., Marson A. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell. 2018;175:1958–1971.e15. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H., et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taskova M., Mantsiou A., Astakhova K. Synthetic Nucleic Acid Analogues in Gene Therapy: An Update for Peptide-Oligonucleotide Conjugates. ChemBioChem. 2017;18:1671–1682. doi: 10.1002/cbic.201700229. [DOI] [PubMed] [Google Scholar]
- 36.Guan X., Chang Y., Sun J., Song J., Xie Y. Engineered Hsp Protein Nanocages for siRNA Delivery. Macromol. Biosci. 2018;18:e1800013. doi: 10.1002/mabi.201800013. [DOI] [PubMed] [Google Scholar]
- 37.Moss K.H., Popova P., Hadrup S.R., Astakhova K., Taskova M. Lipid Nanoparticles for Delivery of Therapeutic RNA Oligonucleotides. Mol. Pharm. 2019;16:2265–2277. doi: 10.1021/acs.molpharmaceut.8b01290. [DOI] [PubMed] [Google Scholar]
- 38.Zhen S., Li X. Liposomal delivery of CRISPR/Cas9. Cancer Gene Ther. 2020;27:515–527. doi: 10.1038/s41417-019-0141-7. [DOI] [PubMed] [Google Scholar]
- 39.Wang H.X., Song Z., Lao Y.H., Xu X., Gong J., Cheng D., Chakraborty S., Park J.S., Li M., Huang D., et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. USA. 2018;115:4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domljanovic I., Hansen A.H., Hansen L.H., Klitgaard J.K., Taskova M., Astakhova K. Studies of Impending Oligonucleotide Therapeutics in Simulated Biofluids. Nucleic Acid Ther. 2018;28:348–356. doi: 10.1089/nat.2017.0704. [DOI] [PubMed] [Google Scholar]
- 41.Kim D., Kim D.E., Lee G., Cho S.I., Kim J.S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 2019;37:430–435. doi: 10.1038/s41587-019-0050-1. [DOI] [PubMed] [Google Scholar]
- 42.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamal M., Ullah A., Ahsan M., Tyagi R., Habib Z., Rehman K. Improving CRISPR-Cas9 On-Target Specificity. Curr. Issues Mol. Biol. 2018;26:65–80. doi: 10.21775/cimb.026.065. [DOI] [PubMed] [Google Scholar]
- 45.Flynt A.S., Rao M., Patton J.G. Blocking Zebrafish MicroRNAs with Morpholinos. Methods Mol. Biol. 2017;1565:59–78. doi: 10.1007/978-1-4939-6817-6_6. [DOI] [PubMed] [Google Scholar]
- 46.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lino C.A., Harper J.C., Carney J.P., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Quan R., Wang J. Development and application of CRISPR/Cas9 technologies in genomic editing. Hum. Mol. Genet. 2018;27(R2):R79–R88. doi: 10.1093/hmg/ddy120. [DOI] [PubMed] [Google Scholar]
- 49.Lombardi L., Oliveira-Pacheco J., Butler G. Plasmid-Based CRISPR-Cas9 Gene Editing in Multiple Candida Species. MSphere. 2019;4:e00125-19. doi: 10.1128/mSphere.00125-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura M., Okamura Y., Iwai H. Plasmid-based and -free methods using CRISPR/Cas9 system for replacement of targeted genes in Colletotrichum sansevieriae. Sci. Rep. 2019;9:18947. doi: 10.1038/s41598-019-55302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Li L., Zheng G., Jiang W., Deng Z., Wang Z., Lu Y. CRISPR/dCas9-Mediated Multiplex Gene Repression in Streptomyces. Biotechnol. J. 2018;13:e1800121. doi: 10.1002/biot.201800121. [DOI] [PubMed] [Google Scholar]
- 53.Hao X., Li Q., Guo J., Ren X., Feng Y., Shi C., Zhang W. Multifunctional Gene Carriers with Enhanced Specific Penetration and Nucleus Accumulation to Promote Neovascularization of HUVECs in Vivo. ACS Appl. Mater. Interfaces. 2017;9:35613–35627. doi: 10.1021/acsami.7b11615. [DOI] [PubMed] [Google Scholar]
- 54.Feng G., Chen H., Li J., Huang Q., Gupte M.J., Liu H., Song Y., Ge Z. Gene therapy for nucleus pulposus regeneration by heme oxygenase-1 plasmid DNA carried by mixed polyplex micelles with thermo-responsive heterogeneous coronas. Biomaterials. 2015;52:1–13. doi: 10.1016/j.biomaterials.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 55.Zhang C., Gao S., Jiang W., Lin S., Du F., Li Z., Huang W. Targeted minicircle DNA delivery using folate-poly(ethylene glycol)-polyethylenimine as non-viral carrier. Biomaterials. 2010;31:6075–6086. doi: 10.1016/j.biomaterials.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 56.Fong S., Liu Y., Heath T., Fong P., Liggitt D., Debs R.J. Membrane-permeant, DNA-binding agents alter intracellular trafficking and increase the transfection efficiency of complexed plasmid DNA. Mol. Ther. 2004;10:706–718. doi: 10.1016/j.ymthe.2004.06.1016. [DOI] [PubMed] [Google Scholar]
- 57.Song H., Yu M., Lu Y., Gu Z., Yang Y., Zhang M., Fu J., Yu C. Plasmid DNA Delivery: Nanotopography Matters. J. Am. Chem. Soc. 2017;139:18247–18254. doi: 10.1021/jacs.7b08974. [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M. Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 2019;31:e1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 60.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Intellia Therapeutics Study to Evaluate Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of NTLA-2001 in Patients With Hereditary Transthyretin Amyloidosis With Polyneuropathy (ATTRv-PN). ClinicalTrials.gov: NCT04601051. First posted October 23, 2020, last update posted December 4, 2020. https://clinicaltrials.gov/ct2/show/NCT04601051
- 62.Yu H., Rimbert A., Palmer A.E., Toyohara T., Xia Y., Xia F., Ferreira L.M.R., Chen Z., Chen T., Loaiza N., et al. BIOS Consortium GPR146 Deficiency Protects against Hypercholesterolemia and Atherosclerosis. Cell. 2019;179:1276–1288.e14. doi: 10.1016/j.cell.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mout R., Ray M., Yesilbag Tonga G., Lee Y.W., Tay T., Sasaki K., Rotello V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao J., Bergmann T., Zhang W., Schiwon M., Ehrke-Schulz E., Ehrhardt A. Viral Vector-Based Delivery of CRISPR/Cas9 and Donor DNA for Homology-Directed Repair in an In Vitro Model for Canine Hemophilia B. Mol. Ther. Nucleic Acids. 2019;14:364–376. doi: 10.1016/j.omtn.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehrke-Schulz E., Schiwon M., Leitner T., Dávid S., Bergmann T., Liu J., Ehrhardt A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 2017;7:17113. doi: 10.1038/s41598-017-17180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D., et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verdera H.C., Kuranda K., Mingozzi F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 72.Mingozzi F., Chen Y., Murphy S.L., Edmonson S.C., Tai A., Price S.D., Metzger M.E., Zhou S., Wright J.F., Donahue R.E., et al. Pharmacological modulation of humoral immunity in a nonhuman primate model of AAV gene transfer for hemophilia B. Mol. Ther. 2012;20:1410–1416. doi: 10.1038/mt.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiang Z., Kurupati R.K., Li Y., Kuranda K., Zhou X., Mingozzi F., High K.A., Ertl H.C.J. The Effect of CpG Sequences on Capsid-Specific CD8+ T Cell Responses to AAV Vector Gene Transfer. Mol. Ther. 2020;28:771–783. doi: 10.1016/j.ymthe.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nonnenmacher M., Wang W., Child M.A., Ren X.Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2020;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller D.G., Petek L.M., Russell D.W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 2004;36:767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- 77.Miller D.G., Petek L.M., Russell D.W. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A., et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 79.Anguela X.M., Sharma R., Doyon Y., Miller J.C., Li H., Haurigot V., Rohde M.E., Wong S.Y., Davidson R.J., Zhou S., et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122:3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Castellanos Rivera R.M., Collier J.H., et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanlon K.S., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Volak A., Spirig S.E., Muller A., Sousa A.A., Tsai S.Q., Bengtsson N.E., et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019;10:4439. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li A., Lee C.M., Hurley A.E., Jarrett K.E., De Giorgi M., Lu W., Balderrama K.S., Doerfler A.M., Deshmukh H., Ray A., et al. A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol. Ther. Methods Clin. Dev. 2018;12:111–122. doi: 10.1016/j.omtm.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cockrell A.S., Kafri T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 84.Hetzel M., Suzuki T., Hashtchin A.R., Arumugam P., Carey B., Schwabbauer M., Kuhn A., Meyer J., Schambach A., Van Der Loo J., et al. Function and Safety of Lentivirus-Mediated Gene Transfer for CSF2RA-Deficiency. Hum. Gene Ther. Methods. 2017;28:318–329. doi: 10.1089/hgtb.2017.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lwin S.M., Syed F., Di W.L., Kadiyirire T., Liu L., Guy A., Petrova A., Abdul-Wahab A., Reid F., Phillips R., et al. Safety and early efficacy outcomes for lentiviral fibroblast gene therapy in recessive dystrophic epidermolysis bullosa. JCI Insight. 2019;4:e126243. doi: 10.1172/jci.insight.126243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kohn D.B., Booth C., Kang E.M., Pai S.Y., Shaw K.L., Santilli G., Armant M., Buckland K.F., Choi U., De Ravin S.S., et al. Net4CGD consortium Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 2020;26:200–206. doi: 10.1038/s41591-019-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abordo-Adesida E., Follenzi A., Barcia C., Sciascia S., Castro M.G., Naldini L., Lowenstein P.R. Stability of lentiviral vector-mediated transgene expression in the brain in the presence of systemic antivector immune responses. Hum. Gene Ther. 2005;16:741–751. doi: 10.1089/hum.2005.16.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park J.H., Seo J.H., Jeon H.Y., Seo S.M., Lee H.K., Park J.I., Kim J.Y., Choi Y.K. Lentivirus-Mediated VEGF Knockdown Suppresses Gastric Cancer Cell Proliferation and Tumor Growth in vitro and in vivo. OncoTargets Ther. 2020;13:1331–1341. doi: 10.2147/OTT.S234344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raikwar S.P., Thangavel R., Dubova I., Selvakumar G.P., Ahmed M.E., Kempuraj D., Zaheer S.A., Iyer S.S., Zaheer A. Targeted Gene Editing of Glia Maturation Factor in Microglia: a Novel Alzheimer’s Disease Therapeutic Target. Mol. Neurobiol. 2019;56:378–393. doi: 10.1007/s12035-018-1068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petris G., Casini A., Montagna C., Lorenzin F., Prandi D., Romanel A., Zasso J., Conti L., Demichelis F., Cereseto A. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat. Commun. 2017;8:15334. doi: 10.1038/ncomms15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kochenderfer J.N., Dudley M.E., Carpenter R.O., Kassim S.H., Rose J.J., Telford W.G., Hakim F.T., Halverson D.C., Fowler D.H., Hardy N.M., et al. Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood. 2013;122:4129–4139. doi: 10.1182/blood-2013-08-519413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee D.W., Kochenderfer J.N., Stetler-Stevenson M., Cui Y.K., Delbrook C., Feldman S.A., Fry T.J., Orentas R., Sabatino M., Shah N.N., et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F., et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boroughs A.C., Larson R.C., Choi B.D., Bouffard A.A., Riley L.S., Schiferle E., Kulkarni A.S., Cetrulo C.L., Ting D., Blazar B.R., et al. Chimeric antigen receptor costimulation domains modulate human regulatory T cell function. JCI Insight. 2019;5:e126194. doi: 10.1172/jci.insight.126194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jain P.K., Lo J.H., Rananaware S., Downing M., Panda A., Tai M., Raghavan S., Fleming H.E., Bhatia S.N. Non-viral delivery of CRISPR/Cas9 complex using CRISPR-GPS nanocomplexes. Nanoscale. 2019;11:21317–21323. doi: 10.1039/c9nr01786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D.K., Théberge V., Hallée S., Duperré A.M., Del’Guidice T., Lepetit-Stoffaes J.P., et al. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 2019;10:4906. doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shin G., Grimes S.M., Lee H., Lau B.T., Xia L.C., Ji H.P. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat. Commun. 2017;8:14291. doi: 10.1038/ncomms14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou B., Ho S.S., Greer S.U., Zhu X., Bell J.M., Arthur J.G., Spies N., Zhang X., Byeon S., Pattni R., et al. Comprehensive, integrated, and phased whole-genome analysis of the primary ENCODE cell line K562. Genome Res. 2019;29:472–484. doi: 10.1101/gr.234948.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rouet R., Thuma B.A., Roy M.D., Lintner N.G., Rubitski D.M., Finley J.E., Wisniewska H.M., Mendonsa R., Hirsh A., de Oñate L., et al. Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing. J. Am. Chem. Soc. 2018;140:6596–6603. doi: 10.1021/jacs.8b01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suresh B., Ramakrishna S., Kim H. Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA for Genome Editing. Methods Mol. Biol. 2017;1507:81–94. doi: 10.1007/978-1-4939-6518-2_7. [DOI] [PubMed] [Google Scholar]
- 103.Lostalé-Seijo I., Louzao I., Juanes M., Montenegro J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem. Sci. (Camb.) 2017;8:7923–7931. doi: 10.1039/c7sc03918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Campeiro J.D., Dam W., Monte G.G., Porta L.C., Oliveira L.C.G., Nering M.B., Viana G.M., Carapeto F.C., Oliveira E.B., van den Born J., Hayashi M.A.F. Long term safety of targeted internalization of cell penetrating peptide crotamine into renal proximal tubular epithelial cells in vivo. Sci. Rep. 2019;9:3312. doi: 10.1038/s41598-019-39842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McErlean E.M., Ziminska M., McCrudden C.M., McBride J.W., Loughran S.P., Cole G., Mulholland E.J., Kett V., Buckley N.E., Robson T., et al. Rational design and characterisation of a linear cell penetrating peptide for non-viral gene delivery. J. Control. Release. 2021;330:1288–1299. doi: 10.1016/j.jconrel.2020.11.037. [DOI] [PubMed] [Google Scholar]
- 106.Tarvirdipour S., Huang X., Mihali V., Schoenenberger C.A., Palivan C.G. Peptide-Based Nanoassemblies in Gene Therapy and Diagnosis: Paving the Way for Clinical Application. Molecules. 2020;25:3482. doi: 10.3390/molecules25153482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Venit T., Dowaidar M., Gestin M., Mahmood S.R., Langel Ü., Percipalle P. Transcriptional Profiling Reveals Ribosome Biogenesis, Microtubule Dynamics and Expression of Specific lncRNAs to be Part of a Common Response to Cell-Penetrating Peptides. Biomolecules. 2020;10:1567. doi: 10.3390/biom10111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klein A.F., Varela M.A., Arandel L., Holland A., Naouar N., Arzumanov A., Seoane D., Revillod L., Bassez G., Ferry A., et al. Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. J. Clin. Invest. 2019;129:4739–4744. doi: 10.1172/JCI128205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gait M.J., Arzumanov A.A., Saleh A.F., Wood M.J., Betts C., Koo T. 2016. Cell-penetrating peptides having a central hydrophobic domain. US patent US9302014B2, filed August 29, 2012, and issued April 5, 2016. [Google Scholar]
- 110.Gait M.J., Arzumanov A.A., McClorey G., Godfrey C., Betts C., Hammond S., Wood M.J.A. Cell-Penetrating Peptide Conjugates of Steric Blocking Oligonucleotides as Therapeutics for Neuromuscular Diseases from a Historical Perspective to Current Prospects of Treatment. Nucleic Acid Ther. 2019;29:1–12. doi: 10.1089/nat.2018.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meng W., He C., Hao Y., Wang L., Li L., Zhu G. Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv. 2020;27:585–598. doi: 10.1080/10717544.2020.1748758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gulei D., Berindan-Neagoe I. Activation of Necroptosis by Engineered Self Tumor-Derived Exosomes Loaded with CRISPR/Cas9. Mol. Ther. Nucleic Acids. 2019;17:448–451. doi: 10.1016/j.omtn.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hajj K.A., Melamed J.R., Chaudhary N., Lamson N.G., Ball R.L., Yerneni S.S., Whitehead K.A. A Potent Branched-Tail Lipid Nanoparticle Enables Multiplexed mRNA Delivery and Gene Editing In Vivo. Nano Lett. 2020;20:5167–5175. doi: 10.1021/acs.nanolett.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramishetti S., Hazan-Halevy I., Palakuri R., Chatterjee S., Naidu Gonna S., Dammes N., Freilich I., Kolik Shmuel L., Danino D., Peer D. A Combinatorial Library of Lipid Nanoparticles for RNA Delivery to Leukocytes. Adv. Mater. 2020;32:e1906128. doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 115.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M., Schubert M.S., Friedmann-Morvinski D., Cohen Z.R., Behlke M.A., et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020;6:eabc9450. doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qiu M., Glass Z., Chen J., Haas M., Jin X., Zhao X., Rui X., Ye Z., Li Y., Zhang F., Xu Q. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020401118. e2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lindfors L., Kjellman T. 2017. Lipid nanoparticles comprising lipophilic anti-inflammatory agents and methods of use thereof. US patent US20170367988A1 (International publication WO2017194454A1), filed May 8, 2017, and publication December 8, 2017. [Google Scholar]
- 118.Lin Y., Wu J., Gu W., Huang Y., Tong Z., Huang L., Tan J. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. (Weinh.) 2018;5:1700611. doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A., Mackley V.A., Chang K., Rao A., Skinner C., et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.LaFleur M.W., Nguyen T.H., Coxe M.A., Yates K.B., Trombley J.D., Weiss S.A., Brown F.D., Gillis J.E., Coxe D.J., Doench J.G., et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 2019;10:1668. doi: 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deprey K., Batistatou N., Kritzer J.A. A critical analysis of methods used to investigate the cellular uptake and subcellular localization of RNA therapeutics. Nucleic Acids Res. 2020;48:7623–7639. doi: 10.1093/nar/gkaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liang X.H., Sun H., Nichols J.G., Allen N., Wang S., Vickers T.A., Shen W., Hsu C.W., Crooke S.T. COPII vesicles can affect the activity of antisense oligonucleotides by facilitating the release of oligonucleotides from endocytic pathways. Nucleic Acids Res. 2018;46:10225–10245. doi: 10.1093/nar/gky841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Teo S.L.Y., Rennick J.J., Yuen D., Al-Wassiti H., Johnston A.P.R., Pouton C.W. Unravelling cytosolic delivery of endosomal escape peptides with a quantitative endosomal escape assay (SLEEQ) bioRxiv. 2020 doi: 10.1038/s41467-021-23997-x. 2020.08.20.258350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Astakhova K. Toward non-enzymatic ultrasensitive identification of single nucleotide polymorphisms by optical methods. Chemosensors. 2014;2:193–206. [Google Scholar]
- 125.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q., et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hosseini E.S., Nikkhah M., Hosseinkhani S. Cholesterol-rich lipid-mediated nanoparticles boost of transfection efficiency, utilized for gene editing by CRISPR-Cas9. Int. J. Nanomedicine. 2019;14:4353–4366. doi: 10.2147/IJN.S199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wojnilowicz M., Glab A., Bertucci A., Caruso F., Cavalieri F. Super-resolution Imaging of Proton Sponge-Triggered Rupture of Endosomes and Cytosolic Release of Small Interfering RNA. ACS Nano. 2019;13:187–202. doi: 10.1021/acsnano.8b05151. [DOI] [PubMed] [Google Scholar]
- 128.Creusat G., Rinaldi A.S., Weiss E., Elbaghdadi R., Remy J.S., Mulherkar R., Zuber G. Proton sponge trick for pH-sensitive disassembly of polyethylenimine-based siRNA delivery systems. Bioconjug. Chem. 2010;21:994–1002. doi: 10.1021/bc100010k. [DOI] [PubMed] [Google Scholar]
- 129.Liu Q., Chen X., Jia J., Zhang W., Yang T., Wang L., Ma G. pH-Responsive Poly(D,L-lactic-co-glycolic acid) Nanoparticles with Rapid Antigen Release Behavior Promote Immune Response. ACS Nano. 2015;9:4925–4938. doi: 10.1021/nn5066793. [DOI] [PubMed] [Google Scholar]
- 130.Alsaiari S.K., Patil S., Alyami M., Alamoudi K.O., Aleisa F.A., Merzaban J.S., Li M., Khashab N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018;140:143–146. doi: 10.1021/jacs.7b11754. [DOI] [PubMed] [Google Scholar]
- 131.Meyer M., Zintchenko A., Ogris M., Wagner E. A dimethylmaleic acid-melittin-polylysine conjugate with reduced toxicity, pH-triggered endosomolytic activity and enhanced gene transfer potential. J. Gene Med. 2007;9:797–805. doi: 10.1002/jgm.1075. [DOI] [PubMed] [Google Scholar]
- 132.Liu S., Cheng Q., Wei T., Yu X., Johnson L.T., Farbiak L., Siegwart D.J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hou K.K., Pan H., Schlesinger P.H., Wickline S.A. A role for peptides in overcoming endosomal entrapment in siRNA delivery - A focus on melittin. Biotechnol. Adv. 2015;33:931–940. doi: 10.1016/j.biotechadv.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meyer M., Dohmen C., Philipp A., Kiener D., Maiwald G., Scheu C., Ogris M., Wagner E. Synthesis and biological evaluation of a bioresponsive and endosomolytic siRNA-polymer conjugate. Mol. Pharm. 2009;6:752–762. doi: 10.1021/mp9000124. [DOI] [PubMed] [Google Scholar]
- 135.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 136.Rozema D.B., Lewis D.L., Wakefield D.H., Wong S.C., Klein J.J., Roesch P.L., Bertin S.L., Reppen T.W., Chu Q., Blokhin A.V., et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rozema D.B., Wakefield D.H. 2016. Peptide-based in vivo siRNA delivery system. US patent US9526796B2, filed July 1, 2015, and granted December 27, 2016. [Google Scholar]
- 138.Arrowhead Pharmaceuticals A Study of ARC-AAT in Healthy Volunteer Subjects and Patients With Alpha-1 Antitrypsin Deficiency (AATD). ClinicalTrials.gov: NCT02363946. First posted February 16, 2015, last update posted August 3, 2018. https://clinicaltrials.gov/ct2/show/NCT02363946
- 139.Colletti S.L., Tucker T.J., Tellers D.M., Boyoung K., Burke R., Calati K.B., Stanton M.G., Parmar R.G., Aaronson J.G., Wang W. 2019. Peptide containing conjugates for dual molecular delivery of oligonucleotides. US patent US10239957B2, filed November 3, 2014, and granted March 26, 2019. [Google Scholar]
- 140.Müller S., Bexte T., Gebel V., Kalensee F., Stolzenberg E., Hartmann J., Koehl U., Schambach A., Wels W.S., Modlich U., Ullrich E. High Cytotoxic Efficiency of Lentivirally and Alpharetrovirally Engineered CD19-Specific Chimeric Antigen Receptor Natural Killer Cells Against Acute Lymphoblastic Leukemia. Front. Immunol. 2020;10:3123. doi: 10.3389/fimmu.2019.03123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Majowicz A., Nijmeijer B., Lampen M.H., Spronck L., de Haan M., Petry H., van Deventer S.J., Meyer C., Tangelder M., Ferreira V. Therapeutic hFIX Activity Achieved after Single AAV5-hFIX Treatment in Hemophilia B Patients and NHPs with Pre-existing Anti-AAV5 NABs. Mol. Ther. Methods Clin. Dev. 2019;14:27–36. doi: 10.1016/j.omtm.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alyami M.Z., Alsaiari S.K., Li Y., Qutub S.S., Aleisa F.A., Sougrat R., Merzaban J.S., Khashab N.M. Cell-Type-Specific CRISPR/Cas9 Delivery by Biomimetic Metal Organic Frameworks. J. Am. Chem. Soc. 2020;142:1715–1720. doi: 10.1021/jacs.9b11638. [DOI] [PubMed] [Google Scholar]