Abstract
Biomedical research has been revolutionized by the introduction of many CRISPR-Cas systems that induce programmable edits to nearly any gene in the human genome. Nuclease-based CRISPR-Cas editors can produce on-target genomic changes but can also generate unwanted genotoxicity and adverse events, in part by cleaving non-targeted sites in the genome. Additional translational challenges for in vivo somatic cell editing include limited packaging capacity of viral vectors and host immune responses. Altogether, these challenges motivate recent efforts to control the expression and activity of different Cas systems in vivo. Current strategies utilize small molecules, light, magnetism, and temperature to conditionally control Cas systems through various activation, inhibition, or degradation mechanisms. This review focuses on small molecules that can be incorporated as regulatory switches to control Cas genome editors. Additional development of CRISPR-Cas-based therapeutic approaches with small molecule regulation have high potential to increase editing efficiency with less adverse effects for somatic cell genome editing strategies in vivo.
Keywords: Enter keywords here
Graphical abstract
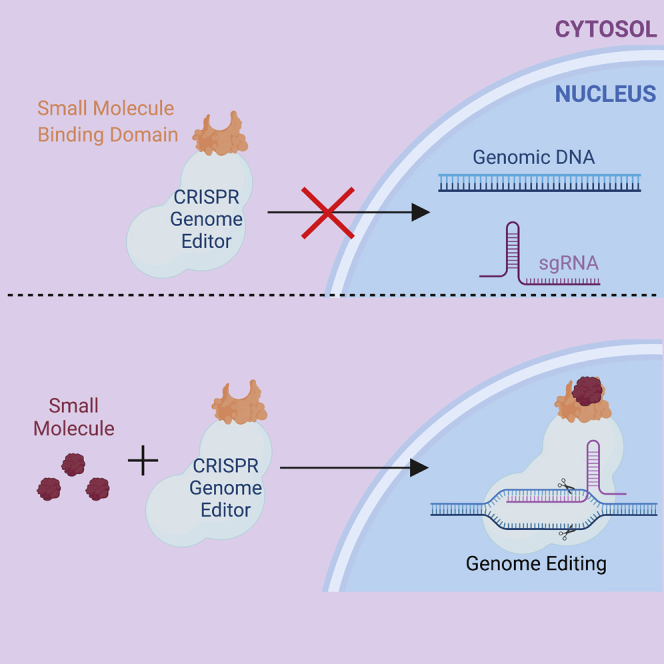
Khajanchi and Saha review recent advances in establishing small molecule control over CRISPR genome editors for gene therapy applications in vivo.
Introduction
CRISPR-Cas systems are now being translated into several gene therapeutic candidates, with several promising results coming out of clinical trials.1, 2, 3, 4, 5 Many therapeutic candidates to date have utilized Streptococcus pyogenes (Spy)Cas9 (referred to as Cas9 hereafter). Since 2012, when Cas9 was first implemented for genome editing,6 CRISPR-Cas9 has been applied in ex vivo clinical trials related to cancers,7, 8, 9 human immunodeficiency virus-1 (HIV) infection,3 sickle cell disease (ClinicalTrials.gov: NCT03745287),10 and β-thalassemia (ClinicalTrials.gov: NCT03655678) (with follow-up ClinicalTrials.gov: NCT04208529).10 There are now 37 Cas9-related clinical trials with 11 phase 2 studies in the US (as of May 2021; no phase 3 or 4 studies). In vivo editing of somatic cells within patients has been lagging behind many of the ex vivo editing strategies, which in part can be attributed to challenges posed by persistent Cas9 activity after delivery. However, in vivo strategies are emerging: in March 2020, a phase 1/phase 2 clinical trial (ClinicalTrials.gov: NCT03872479) targeted the CEP290 gene in vivo to treat a childhood blindness disease (Leber congenital amaurosis type 10), and another in vivo trial targeted the transthyretin (TTR) gene to treat hereditary transthyretin amyloidosis (ClincialTrials.gov: NCT04601051) using Cas9 delivered by lipid nanoparticles.
While traditional genome-editing systems such as zinc finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs) rely on protein-DNA interactions to promote sequence-specific DNA binding, Cas9 relies on short sequences of guide RNA to target the gene loci of interest.11 The targeted DNA sequence is adjacent to the protospacer adjacent motif (PAM) site, a 5-NGG-3 (where N is any nucleotide) sequence that binds and activates the Cas9 enzyme to induce a DNA double-stranded break (i.e., “cut” site). Following the cut, endogenous cellular DNA repair pathways can fix the cut primarily either through (1) non-homologous end joining (NHEJ) that leads to insertions or deletions, or (2) homology-directed repair (HDR)—particularly when an exogenous donor repair template is proximal to the cut DNA. While the CRISPR-Cas9 nuclease can modify genes, concern arises with cleavage at non-targeted sites, which can generate lethal, undesirable,12,13 unpredictable mutations, and toxicity in particular cells, such as neurons, from Cas9 activity.14 Given that constitutive expression of Cas systems may result in more unintended on-target and off-target site modification, there is a need to control CRISPR activity to address the in vivo translational challenges.15
This review focuses on efforts that attempt to control CRISPR activity, most particularly strategies using small molecules, to turn Cas9 “on” or “off.” Small molecules are of interest, as many can pass through the cell membrane, be administered orally, and pass the blood-brain barrier (BBB). Furthermore, established screening technologies can easily identify or repurpose small molecules that can target various cell machinery. Such small molecule regulation can be applicable to other CRISPR systems such as Cas12 (which generates staggered DNA breaks distal to the PAM site),16 dCas9 (catalytically inactive Cas9 that can be used for gene regulation, epigenetic editing, chromatin engineering, and even imaging),17,18 and base editors derived from Cas proteins.19
Key challenges for in vivo editing
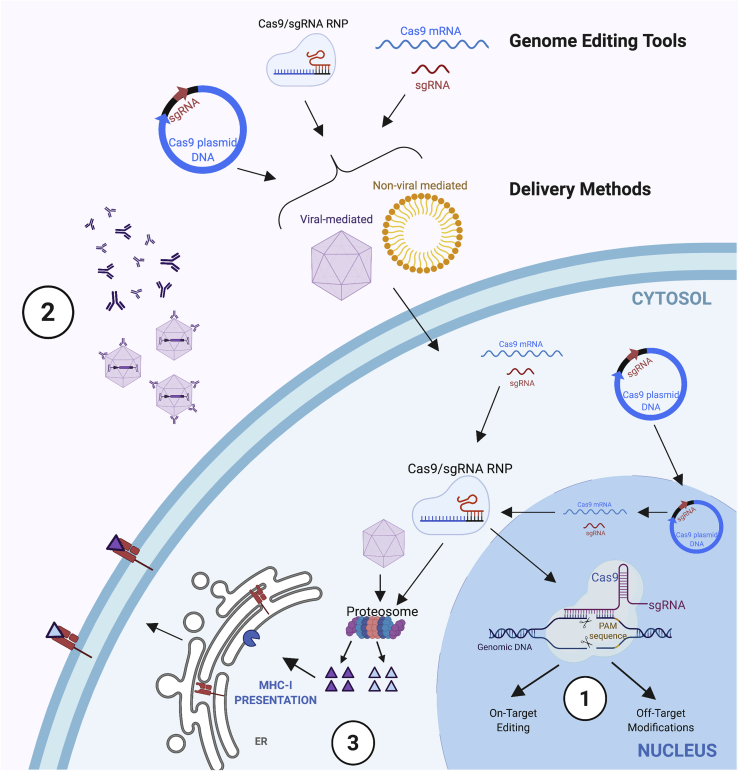
Even though the CRISPR-Cas system can modify DNA at high efficiencies up to 80%,20 there are several key challenges that need to be addressed to make CRISPR-Cas systems more versatile for in vivo somatic cell therapeutic applications. One prominent safety concern involves the consequences of off-target DNA cleavage (Figure 1).21, 22, 23, 24, 25 The single guide RNA (sgRNA) that targets a specific gene can tolerate up to five mismatches within the target site,24,26 and hence there are typically hundreds of off-target sites in the human genome for a particular sgRNA. Decreasing the Cas9-sgRNA complex concentration has shown to improve the on-target/off-target ratio (cleavage specificity).26 Tools to monitor off-target modifications have evolved quickly, and we refer the reader to several recent reviews on this topic.27, 28, 29 The risk of off-target cutting can be exacerbated by prolonged expression of Cas9 and can be mitigated with methods to turn the CRISPR-Cas9 system off. Second, since Cas9 and other CRISPR-based systems are derived from bacteria, host immune responses may be an obstacle for CRISPR technology to go into the clinic (Figure 1). Major histocompatibility complex class I (MHC class I) immune responses by the host cells to Cas9 have been observed,30 which can result in the elimination of Cas9-expressing cells. Hence, there is a need to develop methods to dispose of Cas9 immediately after editing has occurred; in this manner, the chance of Cas9 peptide presentation by MHC class I on cells would be lowered after delivery. Lower MHC class I presentation would reduce the chance that cytotoxic T cells would target these cells for elimination, thereby increasing the durability of any therapeutic effect from edited cells. In the extracellular space, intact Cas9 proteins can be recognized by pre-existing neutralizing antibodies and innate immune responses (Figure 1).31 Non-viral vector methods, such as lipid-based vectors or nanoparticles with polyethylene glycol (PEG),32,33 can be used to cloak Cas9 to potentially avoid these responses. For viral vectors, they can induce adaptive immune responses with repeated systemic administration.34, 35, 36, 37 Up to 50% of individuals are ineligible for AAV (adeno-associated virus) gene therapy through treatment or clinical trials because they have neutralizing antibodies to at least one of the AAV serotypes.38, 39, 40, 41 As a workaround in some cases, clinicians can immunosuppress a patient to avoid the initial immune response to AAV or other viral vectors.
Figure 1.
Three translational challenges for in vivo somatic cell genome editing with CRISPR-Cas9
(1) Off-target DNA double-strand break formation. The gene of interest is targeted by the single guide RNA (sgRNA) and cut by Cas9, and it results in double-stranded breaks that can subsequently be repaired to generate on-target edits. However, with prolonged exposure of Cas9 to the genome, there can be an increase in unwanted modifications at off-target sites. (2) Antibody neutralization to delivery vectors. Antigen-presenting cells may bind capsid proteins or other components from the delivery vector (e.g., polyethylene glycol [PEG]) to trigger an antibody-mediated immune response. (3) Immune response to Cas and vector proteins. MHC class I molecules may bind peptides from degradation of Cas9 and/or the viral vector and present them on the cell surface. These peptides could be presented to T cells and trigger an adaptive immune response.
Controlling Cas9 via delivery strategies
Inefficient delivery of therapeutic genome-editing payloads to defined cell types and tissues is still an outstanding challenge in the field,42 but delivery strategies themselves can be exploited to establish control of Cas9 activity. The most common delivery strategy for limiting Cas9 activity and off-target editing involves transient delivery of either Cas9-encoding plasmid or messenger RNA (mRNA) or direct transient delivery of Cas9-sgRNA ribonucleoproteins (RNPs).43 Transient delivery limits the levels of Cas9 proteins or active RNPs within cells to the lifetime of the protein or complex within the nucleus. For instance, mRNA encoding CRISPR base editors delivered to the livers of cynomolgus monkeys were cleared within 2 weeks but resulted in durable therapeutic responses for several months.20 However, transient non-viral delivery for many applications is generally less efficient than viral delivery.42 Adenoviruses (AdVs) and AAV vectors are the most prominent viral mode of Cas9 delivery. The packaging of CRISPR components into a single AAV vector is limited because the carrying capacity of AAV vectors is ∼4.7 kb,44 and the size of the Cas9 gene alone is ∼4.3 kb;30 additionally, in combination with the guide RNA and necessary elements (i.e., promoter, fluorescent proteins, and polyadenylation sequences), the size of the Cas9 system reaches more than 5 kb. Hence, the Cas9 system frequently needs to be split into two or more AAV vectors to be delivered.45,46 The vector size problem can also be resolved by utilizing a smaller Cas protein that can fit AAV vectors for in vivo delivery such as Cas12a that uses a single nuclease domain to cleave complementary and non-complementary strands of DNA47 or a “SauCas9” from Staphylococcus aureus.21,48 Alternatively, a larger vector such as AdV or lentivirus may be used to deliver large transgenes.49 The intein-mediated split-Cas9 is another way to reconstitute parts of Cas9.50,51 Inteins are proteins that can splice translated polypeptide into a functional protein. Cas9 halves are fused to split inteins that are co-expressed in dual recombinant AAV vectors, and the full Cas9 is expressed once intein-mediated trans-splicing occurs.
In addition to controlling Cas9 via delivery, inducible systems can be implemented to stimulate Cas9 to edit the genome at specific times and sites, turning the system on and off. Switches for inducible CRISPR-Cas9 systems include small molecules,52 light (including near infrared [IR]/UV),46,53, 54, 55, 56 magnetic fields,57 and temperature.58 These switches can easily be layered with other methods to control dose, timing, and localization of Cas9. Below, we focus on small molecule switches with CRISPR systems that have the potential to be implemented in vivo for therapeutic applications.
Inducers to regulate Cas9 activity and abundance
Switching on recombinases, promoters, and proteins fused to Cas9 are various ways to activate genome editing within cells. The Cre-recombinase system can give localized control over Cas9 expression via use of small molecules or tissue-specific promoters. In these strategies, the loxP-stop-loxP cassette is placed between the promoter of interest and the Cas9 coding sequence. Following administration of the small molecules, genomic recombination of the cassette within cells can result in the activation of Cas9. The bulk of research with Cre-dependent control of CRISPR-Cas9 has been to inactivate or knockout genes in various animal models.59, 60, 61, 62 The Cre-controlled CRISPR (3C) mutagenesis system uses a ligand-dependent chimeric Cre-recombinase, known as CreER recombinase.59 The CreER recombinase is inactive until the synthetic estrogen receptor ligand 4-hydroxytamoxifen (4-OHT) is added to induce recombination and activate Cas9 activity. This method provides both spatial and temporal control: floxed chromosomal DNA is excised at specific promoters once 4-OHT is added. Some challenges in using recombinases and CRISPR-Cas9 together for somatic cell editing include the need to engineer recombination sites into the genome, toxicity of inducers, leakiness of gene expression in the absence of the inducer, and promoters not being available for every tissue.63
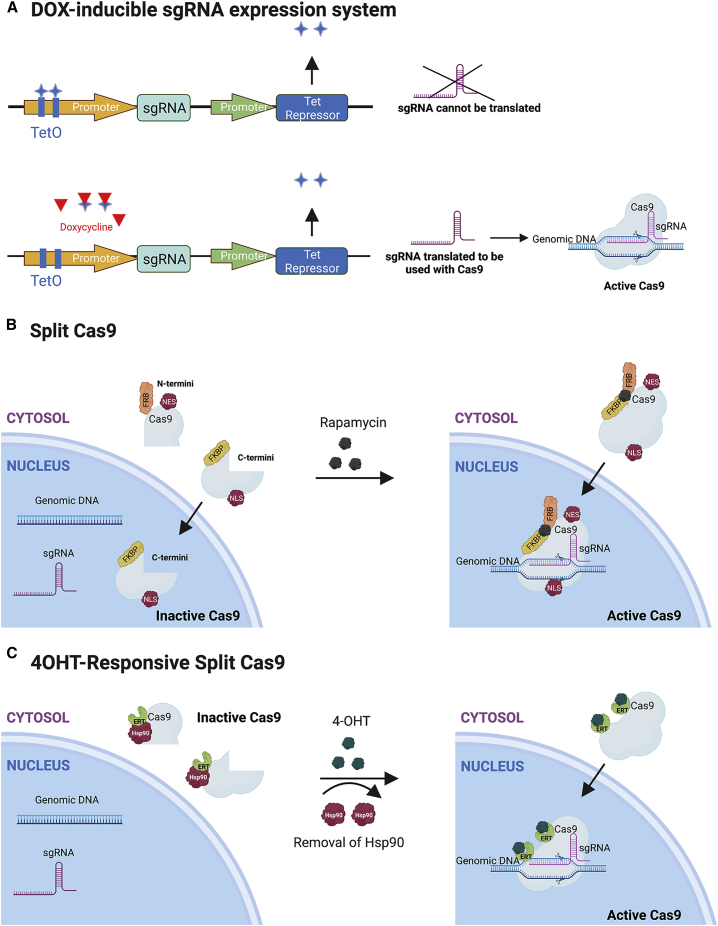
Cas9 activity can also be controlled by using small molecules to control sgRNA expression64 through aptamers,64, 65, 66, 67, 68 aptazymes,69 or other sgRNA-controlling switches.70 A doxycycline-inducible sgRNA expression system modulates the activity of the sgRNA rather than the Cas9 (Figure 2).71 The sgRNA is driven by a U6 promoter that contains either one or two TetO operator sites. The tightly controlled double TetO system showed high cleavage efficiency after induction with negligible background activity in 11 human and mouse cell lines in vitro and a hematopoietic reconstitution mouse model.
Figure 2.
Small molecule inducible activation and split systems to control sgRNA or Cas9 expression
(A) The doxycycline-inducible sgRNA expression system71 has a Tet repressor that keeps the sgRNA from being translated, so that Cas9 is not functional. However, once doxycycline is added, the repressors will no longer bind to the Tet-responsive sequence (TetO) and sgRNA will be expressed to use with Cas9. (B) The rapamycin-inducible Cas972 is split into two lobes, an N terminus lobe that has the FRB component and a nuclear export signal (NES), and the C terminus lobe that has FKBP and a nuclear localization signal (NLS). Without rapamycin, only the C terminus Cas9 goes into the nucleus and is not functional. With the addition of rapamycin, the lobes are fused via FRB and FKBP binding and the functional Cas9 is shuttled to the nucleus for activity. (C) The 4-hydroxytamoxifen (4-OHT)-responsive split Cas973 is sequestered in the cytoplasm and has multiple ligand domains of the estrogen receptor (ERT). Addition of 4-OHT releases the Cas9 fragments from a heat shock protein (Hsp90) and translocates the reconstituted Cas9 to the nucleus for genome editing.
Timing and exposure of small molecules such as doxycycline have been shown to reduce toxicity and off-target effects that occur with constitutive expression of Cas9 complexes.74 With a timed doxycycline system, in mouse embryonic stem cells (mESCs), there was only 1 out of 18 off-target sites that showed indels.74 Doxycycline has also been utilized in the “self-inactivating CRISPR (SiC)” system,75 which is composed of a sgRNA that can deactivate Cas9 in a timely manner through Tet-repressor based feedback.76 This system reduced off-target effects (with the on-target/off-target editing raising from 0.8 to 1.3 after the addition of doxycycline).75 Another recent study developed a Tet-On system called ObLiGaRe doxycycline inducible Cas9 (ODInCas9) that uses a doxycycline-inducible Cas9 to temporally regulate Cas9 in human induced pluripotent stem cells (iPSCs) and mESCs.77 This ODInCas9 cassette can induce Cas9 expression in vivo with no background Cas9 activity and no leaky or sustained expression of Cas9.77 Even though doxycycline-inducible promoters have been used to control CRISPR-Cas9 transcriptional activity,71 these systems take several days to achieve maximum Cas9 activity and thus do not provide the high temporal resolution needed in some studies.73 Regardless, tight control of Cas9 expression could be valuable for evading immune responses in vivo.
In an attempt to control the kinetics of genome editing, small molecules have also been used to drive the fusion between parts of Cas9.72,78 The Cas9 system was split into N and C termini with the FKBP rapamycin binding (FRB) domain and the FK506 binding protein 12 (FKBP) fused to each terminus, respectively (Figure 2).72 In the presence of the small molecule rapamycin, FKBP and FRB heterodimerize and reconstitute, making Cas9 functional. Without the addition of rapamycin, there was 10% indel frequency because of Cas9 auto-assembly.72 The leaky Cas9 issue led the authors to localize the Cas9(C)-FKBP fragment to the nucleus with a nuclear localization sequence (NLS) and sequester the Cas9(N)-FRB portion in the cytoplasm with a nuclear export sequence (NES).71,74,73,79,80 When rapamycin was present, there was inducible activation of Cas9. As a result, there were less off-target indels (5%–10%) as compared to wild-type (WT) Cas9 (27%) after half of Cas9 was nuclearized.72 Nguyen et al.73 added another layer of control by linking each half of Cas9 with the ligand-binding domain ERT from the estrogen receptor to sequester Cas9 in the cytoplasm; the synthetic ligand 4-OHT was needed to express the split Cas9 and translocate it to the nucleus (Figure 2). This method resulted in quick CRISPR activation with low background activity without rapamycin and high tunability. In this study, when rapamycin was administered, human embryonic kidney 293T (HEK293T) cells exhibited up to ∼25% WT Cas9 activity.73 By activating the system using 4-OHT and changing activation domains, background activity was reduced. The split Cas9s can be delivered in two viral or non-viral vectors, and therefore overcome the limitations imposed by the packaging capacity of a single vector. Even though there was minimal background activity for Cas9 genome editing in the study by Nguyen et al., the leakiness of the inducer could pose a challenge for translational somatic cell-editing applications.
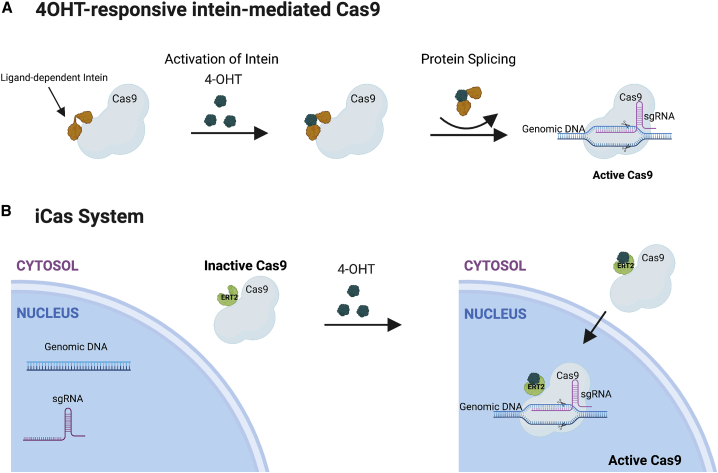
As mentioned previously, intein-mediated split-Cas9 can also be useful in addressing vector size issues. In a study with Neuro-2a (N2a) cells, split Cas9 indels were similar to WT Cas9 (23.1% and 22.7%, respectively), showing that split inteins did not affect endonuclease activity.50 Another study used a 4-OHT-responsive intein for Cas9 in HEK293 cells and found up to a 25-fold higher specificity (on-target/off-target indel frequency ratio) with low background activity in the absence of 4-OHT (Figure 3).81 This intein-inducible system also bypasses the packaging limits using one vector, as the split components can be delivered separately. In contrast to other split systems, once splicing occurs, the system is irreversible. The iCas system (Figure 3) also tightly controls Cas9 through a mutated ligand-binding 4-OHT, but, unlike the 4-OHT-responsive intein and the split-Cas9 method, it is reversible.82 Moreover, compared to the intein-Cas9 and split-Cas9, the iCas system has a higher editing efficiency (i.e., higher cleavage specificity),82 but it had only 60% of WT Cas9 activity at most. Oakes et al.83 added a ligand-binding domain of human estrogen receptor-α to Cas9, creating a 4-OHT-responsive Cas9 called allosterically regulated Cas9 (arC9). There was no background when 4-OHT was absent. arC9 exhibited only 30% of editing as compared to WT Cas9 once 4-OHT was added, and arC9 showed slow reversibility after removal of 4-OHT.83
Figure 3.
Small molecule inducible systems to control Cas9 expression
(A) The intein-mediated Cas981 has Cas9 fused to intein sequences. Once 4-OHT is added, the inteins are spliced out, leaving a functional Cas9. (B) The iCas system82 consists of Cas9 fused to the hormone-binding domain of the estrogen receptor (ERT2). Upon addition of 4-OHT, which binds to ERT2, Cas9 is translocated to the nucleus to partake in gene-editing activity.
Inducible degraders to remove Cas9
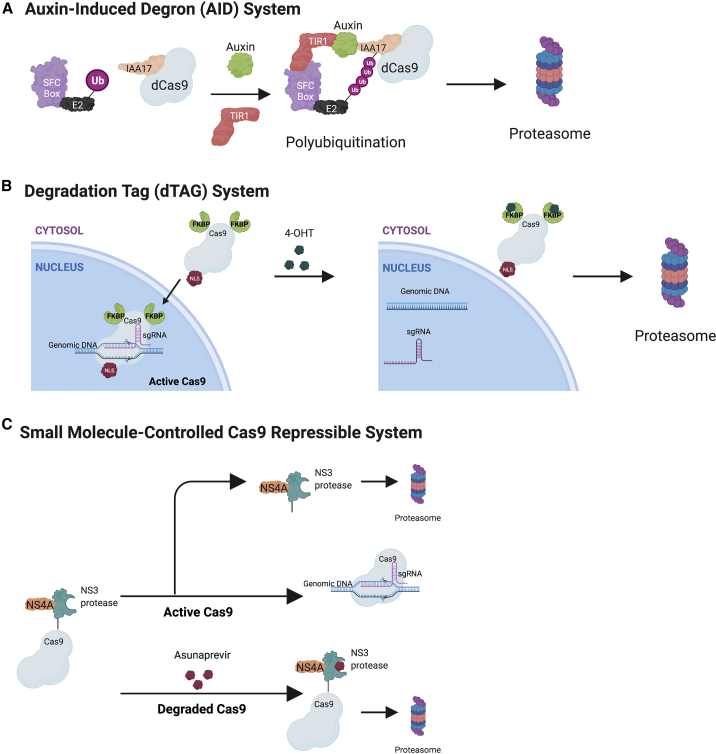
Inducible degradation strategies can be used to regulate Cas9. Degrons are typically short amino acid sequences located in the flexible regions of the protein with easy access to other proteins and transcriptional factors,84 which can assist with degradation. Fusion of degrons to Cas9 would be able to eliminate the protein from the cell. Examples of such systems that include degrons are the auxin-induced degradation (AID) systems,68,85 degradation tag (dTAG),86 and the small molecule-assisted shut-off (SMASh) system (Figure 4).87
Figure 4.
Inducible degradation systems to control CRISPR-Cas9 genome editing
(A) Auxin-induced degron (AID) system.68 dCas9 can be fused with the degron IAA17. T1R1 is the auxin-binding receptor from the rice plant, and auxin is a plant hormone. In non-plant cells, T1R1 complexes with the conserved ubiquitin E3 ligase and Skp1-Cul1-F (SFC) box. dCas9 has been shown to be degraded with the AID system but has not been demonstrated with the Cas9 nuclease. (B) Degradation tag (dTAG) system.88 Cas9 can be fused to multiple mutant FKBP12 (i.e. FKBP12F36V). Once FKBP12F36V is polyubiquitinated, Cas9 will be degraded along with the FKBP12F36V. (C) Small molecule-controlled Cas9 repressible system.87 Cas9 is fused to a SMASh tag. Upon the addition of the inhibitor asunaprevir (ASV) that attaches to the NS3 protease and NS4A degron, Cas9 is degraded. When asunaprevir is removed, the NS3 protease self-cleaves and is degraded with the NS4A, while Cas9 is fully functional.
In the AID system, the auxin-binding receptor (osTIR1) from the rice plant and the plant hormone auxin are key components. In non-plant cells, TIR1 complexes with the conserved ubiquitin E3 ligase Skp1-Cul1-F box (SFC),89, 90, 91, 92 which is only activated in the presence of a natural or synthetic auxin termed indole-3-acetic acid (IAA) or 1-naphthaleneacetic acid (NAA) (Figure 4).91 Kleinjan et al.68 showed that a IAA17-dCas9 can efficiently be degraded with co-transfection of the auxiliary protein (osTIR1) and auxin. Expressing the osTIR1 gene under the control of a tetracycline-inducible promoter gives dCas9 tissue specific control.68 However, this system requires a high dose of auxin and expression of osT1R1, which are not endogenous to humans, and currently, there is not sufficient research on the effects of these components on humans.
The dTAG system developed by Nabet et al.86 avoids the usage of exogenous co-expression of degradation factors that are needed for the AID system. The dTag system has recently been shown to induce Cas9 degradation in HEK293T, U2OS, and Drosophila’s S2 cell lines.88 The FKBP12F36V tag is fused to multiple parts of Cas9 (Figure 4).88 When the heterobifunctional small molecule degrader dTAG is added, it binds to a E3 ubiquitin ligase on one side and targets FKBP12F36V with the other. The E3 ligase hijacks the cell machinery and sends Cas9 for degradation. The FKBP-Cas9 fusion has shown a higher on-target/off- target ratio (cleavage specificity) than does WT Cas9. With the addition of the small molecule dTag-47, Cas9 shows 80%–90% degradation at doses as low as 12 nM.88 Similar to that of the AID system, the immunogenicity of the dTAG components in humans has not been well characterized.
An alternative destabilizing degron is called the SMASh, with a degron domain from the hepatitis C virus (HCV) with a protease domain. A small molecule-controlled Cas9 repressible system was created with the fusion of the SMASh tag with Cas9.87 Upon addition of a clinically approved HCV protease inhibitor called asunaprevir (ASV), Cas9 is degraded (Figure 4).87 However, in the absence of this inhibitor, the SMASh tag self cleaves and removes itself to stabilize Cas9. Adding the SMASh tag to both the N and C termini of Cas9 yielded more than 50% degradation and 50% less indel formation with the addition of ASV as compared to having the SMASh tag on only the C termini. Moreover, because ASV only degrades newly synthesized Cas9, removal of ASV leads to restoration of gene editing activity. As with the dTag system, limiting Cas9 duration also enhances specificity of gene editing. Cas9 has also been fused to the FKBP12-derived destabilizing domain system that is stabilized with a synthetic ligand, Shield-1.93,94 There are still many degrons and degrader systems that have not been tested in Cas9, and we refer the reader to a review on more degraders that can be used to control Cas9.95,96
Suitability for somatic cell genome editing
Applying control strategies for in vivo somatic editing may be able to address the key challenges described in the first section concerning off-target adverse events, delivery, and immunogenicity. In Table 1, we compare and contrast how the inducible CRISPR-Cas9 systems can be evaluated across three key categories: (1) the degree of uninduced editing, (2) the degree of editing upon inducer addition, and (3) the animal model/cell type used to test the system.71 Several activator strategies may be appropriate for translational studies. The doxycycline-inducible, Cre-recombinase system has been tested in mouse models on a variety of genes, but only now are doxycycline-inducible Cas9 systems being introduced into the field for therapeutic purposes. While there is high control over the timing of Cas9-mediated editing with doxycycline in Cre-recombinase systems without a significant reduction in editing activity, this strategy is usually irreversible upon activation. Additionally, diffusion of doxycycline into and out of cells can lead to genotoxicity related to off-target effects/recombinase.77,97 Moreover, doxycycline at higher concentrations has been shown to cause a cytotoxic effect or a decrease in mitotic activity in human cell lines.98,99 In general, diffusion and transport into various tissues is a concern for establishing broad control over genome editing inside the body, especially for the brain and spinal cord (Table 2). Both the SiC system and ODInCas9 have the ability to turn Cas9 on and off directly instead of constitutively expressing Cas9 such as the 3C system.75,77 The disadvantage to methods such as these is that the system takes days to achieve maximum Cas9 activity because the Tet-inducible system uses repressor-based feedback regulation of Cas9. Moreover, the SiC method only inactivates Cas9 and keeps the Cas9 protein still expressed in the system, which may lead to host immune responses.
Table 1.
Potential inducible CRISPR-Cas9 systems for somatic cell genome editing
| Inducer/small molecule | “Leakiness” (degree of uninduced editing) | Degree of editing upon inducer addition | Animal model(s)/cell type(s) used to test system | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| Cre-controlled CRISPR (3C) mutagenesis system | not reported | not reported | zebrafish embryos (RPE cells or in neural crest-derived melanocytes) | • heat treatment is also needed to display gene editing | • has not been tested in human cells | Hans et al.59 |
| • zebrafish line can be used for conditionally controlled tissue-specific biallelic gene inactivation | • cannot be used for somatic cell editing in humans due to their inability to produce cre-recombinase | |||||
| 2xTetO DOX-inducible sgRNA system | low (0%–14%) | 20%–80% (high cleavage activity) (activity score >0.5 for most cell types) | L-363, MC-38, A-498, LL/2, LP-1, 786-0, NCI-H1299, CT26, 4 TI, HEK293T, mouse (spleen cells, bone marrow cells, blood cells) | • can be used for high-throughput genetic screening | • variable editing efficiency in different cells | Sun et al.71 |
| • mouse model available for this method | ||||||
| • DOX can be paired with IPTG-inducible systems to enable modulation of two genes independently | ||||||
| Cre-recombinase doxycycline | not reported | not reported | mouse/mESCs | • can be light activated | • non-reversible | Dow et al.74 |
| • temporal advantage since system is regulated by doxycycline | • diffusion of small molecules in and out of cells causes off-target recombination | |||||
| • DOX at higher concentrations has been shown to cause cytotoxic effect in human cell lines | ||||||
| • cannot be used for somatic cell editing in humans due to their inability to produce cre-recombinase | ||||||
| Self-inactivating CRISPR (SiC) with doxycycline | not reported | on-target/off-target editing 0.8 to 1.3 after addition of doxycycline | myeloid and lymphoid cells in vivo, both in mouse peripheral blood and bone marrow as well as in vitro human lymphocytes (HL-60s) | • Cas9 on and off method exists | • system takes days to achieve max Cas9 activity, so there is a longer time for not only on-target editing but off-target cleavages as well | Kelkar et al.75 |
| ObLiGaRe doxycycline-inducible SpCas9 (OdInCas9) | no Cas9 background activity | not reported | hiPSC, HCT116, HEK293, HepG2, A549, OVCAR8, N2a cells | • inducible Cas9 expression | • off-target recombination | Lundin et al.77 |
| • reversibility with DOX withdrawal | • not able to use for somatic cell editing in humans due to Tet systems not being able to be reproduced in humans | |||||
| • low immune response with use of AAV to deliver sgRNA | ||||||
| Split FK506 binding protein 12/FKBP rapamycin binding (FKBP/FRB) system | 10% | fewer off-target indels (5%–10%) as compared to WTCas9 (27%) | HEK293FT | • inducible Cas9 expression | • auto-assembly problematic when half of Cas9 not nuclearized | Zetsche et al.72 |
| • rapamycin is crucial to provide temporary immunity against Cas9 | • non-covalent protein dimerization | |||||
| 4-Hydroxytamoxifen (4-OHT) with split FKBP/FRB system | none to low background activity | 25% Cas9 activity | HEK293T | • high tunability | • no reversibility | Nguyen et al.73 |
| • split systems can easily be delivered in multiple plasmids for efficient delivery | • Cas9 remains constitutively active | |||||
| • rapamycin could reduce the immune response against Cas9 | ||||||
| 4-OHT-responsive intein for Cas9 | low | 25-fold higher specificity (on-target/off-target indel frequency) | HEK293 cells | • improved specificity | • irreversible | Davis et al.81 |
| • split proteins can form aggregates due to exposed hydrophobic core | ||||||
| iCas system | low (but higher than split-Cas9 and comparable to intein-Cas9) | 38%–60% editing | HEK293 | • higher editing efficiency (as compared to split Cas9 and 4-OHT-responsive intein-mediated Cas9 at multiple loci) | • high background activity | Liu et al.82 |
| • reversible | ||||||
| Allosterically regulated Cas9 (arC9) | no background | 30% editing | E. coli, HEK293T, murine BNL CL.2 cells | • reversible | • slow reversibility (took 2 days) | Oakes et al.83 |
| Degradation tag (dTAG) system | low | on-target/off-target editing is enhanced (depending on the site, 1.5-fold to 4-fold as much compared to WT Cas9) | HEK293T, U2OS.eGFP.PEST, Drosophila S2 cells, mESC cell line | • small molecule activation provides temporal control | • while said to be reversible, there is no data on reversibility with Cas9 | Sreekanth et al.88 |
| • Cas9 target specificity is enhanced by addition of dTag | • tested in only a few organisms | |||||
| • degradation of Cas9 allows for control of editing outcome | ||||||
| • reversible | ||||||
| Small molecule-Controlled Cas9 repressible system | not reported | on-target/off-target editing is enhanced | HEK293T cells | • reversible | • existing Cas9 cannot be degraded | Wu et al.87 |
| • no structural modification from tagging | • exact mechanism is unknown | |||||
| • easy to measure half-life kinetics | ||||||
| Auxin-induced degron (AID) system | low background with auxin absent | not reported | HEK293FT, CHO-K1 | • makes tissue-specific activity controllable | • receptor (osTIR1) might be immunogenic | Kleinjan et al.68 |
| • reversibility with auxin removal when using miniAID system | • high dosages of auxin osTIR1 needed to attain degradation | Li et al.85 | ||||
| • minimal basal degradation | • IAA has been shown to cause toxicity at high amounts100 |
Selected genome editing studies that have been conducted using small molecule regulation are compared here. Leakiness, degree of editing upon inducer addition, and animal model/cell type used to test the system are summarized, with the reference indicated on the right. Leakiness is the degree of editing that occurs when the small molecule is not added to activate the system. Animal/cell models: RPE, retinal pigment epithelium; L-363; human plasma cell leukemia cell line; MC-38, C57BL/6 murine colon adenocarcinoma cell line; A-498, Homo sapiens kidney carcinoma cell line; LL/2, murine Lewis lung carcinoma cell line; LP-1, human myeloma cell line; 786-0, kidney adenocarcinoma cell line; NCI-H1299, human non-small cell lung carcinoma cell line; CT26, undifferentiated colon carcinoma cell line; 4T1, murine breast cancer cell line; HEK293T/FT, human embryonic kidney cell line; HL-60, human lymphocyte line; hiPSC, human-induced pluripotent stem cell; HCT116, human colon cancer cell line; HepG2, human liver cancer cell line; A549, adenocarcinoma human alveolar basal epithelial cell line; OVCAR8, human ovarian carcinoma cell line; N2a, mouse neuroblastoma cell line; U2OS, Homo sapiens bone osteosarcoma; CHO-K1, Chinese hamster ovary cell line.
Table 2.
Small molecules with potential to cross blood-brain barrier (BBB)
| Small Molecule | System(s) | Crosses BBB? | References |
|---|---|---|---|
| 4-Hydroxytamoxifen (4-OHT) | Cre-controlled CRISPR (3C), 4-OHT with split FKBP/FRB system, 4-OHT-responsive intein for Cas9, ERT2-based iCas system, allosterically regulated Cas9 (arC9) | yes | Rotheneichner et al.101 |
| Doxycycline (DOX) | 2xTetO DOX-inducible sgRNA system, self-inactivating CRISPR (SiC), ObLiGaRe doxycycline inducible Cas9 (ODInCas9) | yes | Norrby et al.102 |
| Isopropyl β-d-1-thiogalactopyranoside (IPTG) | 2xTetO DOX-inducible sgRNA system | yes | Morton et al.103 and Ryan and Scrable104 |
| Rapamycin | split FK506 binding protein 12/FKBP rapamycin binding (FKBP/FRB) system | yes | Majumder et al.105 |
| Auxin | auxin-induced degron (AID) system | possibly, shown to work in Drosophila melanogaster | McClure et al.106 |
| Indole-3-acetic acid (IAA) | AID system | possibly (but highly unlikely to use because of toxicity) | Lin et al.,107 Chen et al.,108 Hąc-Wydro and Flasiński,100 and Puurunen et al.109 |
| 1-Naphthaleneacetic acid (NAA) | AID system | possibly, shown to work in Drosophila melanogaster | Lin et al.107 and Chen et al.108 |
| dTag-47 | dTag System | unknown | N/A |
| Asunaprevir | SMASh | no | Koduri et al.110 |
| Shield-1 | mutated FKBP12-derived destabilization domain | yes | Froschauer et al.111 |
The ability for small molecules to penetrate the BBB could be helpful for controlling genome editing in the central nervous system.
The split systems are useful in terms of bypassing packaging limits, since multiple vectors deliver Cas9 components. The rapamycin-inducible split Cas9 could be a win-win for control and immunogenicity.72 Because rapamycin is an immunosuppressive drug,112 it can alter the immune response to the CRISPR-Cas9 editor during editing. However, with the initial FRB/FKBP and intein-mediated split Cas9,50,72 the systems were irreversible: once activated, Cas9 would remain constitutively active. Within a year, researchers overcame this issue by fusing estrogen receptors to split and intein-mediated Cas9,73,81 and they developed the iCas and arC9 to harness system reversibility by using 4-OHT.82,83
Advantages of a degrader, such as the dTAG system, are that it can be regulated via small molecules and/or light, does not need any exogenous elements like the AID system does, and has a cell-permeable small heterofunctional degrader. The AID system includes a receptor that cannot be found in the human body, but it can associate with the ubiquitin machinery within human cells. By placing the osTIR1 gene under tissue-specific promoters, Cas9 activity can be spatially controlled. However, even when condensing the IAA17 to miniIAA7,91,113 the tag is still 7.4 kDa and fusion to Cas9 may hinder Cas9’s editing ability and stability. The small molecules in both the AID and dTag systems have not been very well characterized in humans as compared to the SMASh system that uses a clinically approved drug. ASV cannot cross the BBB, so it would not be useful for control in the central nervous system, but the small molecule Shield-1 can cross the BBB (Table 2).110,111 Using the FKBP destabilizing domains fusion with Cas9 enables conditional and temporal control of Cas9 via Shield-1 that is necessary for Cas9 activity.93
Layered and combinatorial control
All of the above studies that use small molecules to control Cas9 or its sgRNA could be combined or layered with additional control strategies for CRISPR that do not rely on small molecule application. First, inhibitor proteins can be used to inactivate the functional Cas9, which would reduce off-targets related to prolonged Cas9 activity. Bacterial phages express anti-CRISPR (Acr) proteins to inhibit immune functionality.114, 115, 116, 117, 118 These natural Acrs can be useful in regulating Cas9 activity and acting as “off switches” for the CRISPR-Cas9 system.115,117 Most Acr proteins have been tested with dCas9. AcrIIA2 and AcrIIA4 inhibit dCas9, with AcrIIA2 blocking ∼25% of dCas9 function and AcrIIA4 blocking 85% of dCas9.115 In HEK293T cells, co-expression of either Acr protein reduced Cas9-based gene editing.115 In human K562 cells, AcrIIA4 shows nearly complete inhibition of Cas9 at three different target loci.119 Methods have also been developed to control the activity of Acrs spatiotemporally using light. AcrIIC3 has been engineered to be light-dependent to control Neisseria meningitidis Cas9,46 and AcrIIC4 has been engineered to be light-dependent to inhibit SpyCas9.53 Strategies where constitutively active Cas9 is functionally deactivated using inhibitors have high translational potential. However, photoactive Cas9 methods require laboratories to have specialized illumination devices, and penetration of light into tissue is limited.120 To avoid the use of illumination devices, inhibitors may be able to work in combination with one of the other small molecules mentioned previously or delivered in AAV vectors after efficient editing has occurred via Cas9. Furthermore, there could be immune responses to the Acr inhibitory proteins as well as to Cas9 because Acr proteins only inactivate Cas9 and do not repress the expression.
Second, translational control could be combined to avoid editing in nontarget cells/tissues.121,122 MicroRNAs (miRNAs) are short, single-stranded non-coding RNA molecules that can regulate gene expression post-transcriptionally by either inhibiting the translational pathway and/or targeting particular mRNAs for degradation.123,124 Since the activity of miRNAs differ among cell types, it makes miRNAs effective markers to track in targeted cells.121 The miR-Cas9 switch represses Cas9 when the target miRNA is expressed (OFF-state) and activates Cas9 when the target miRNA is absent (ON-state).125 The initial miRNA-Cas9 switch study by Hirosawa et al.125 was in HeLa cells, human iPSCs, and iPSC-derived differentiated neuronal cells. The authors initially found an ON-state that had leaky Cas9. To fix this issue, the researchers regulated Cas9 by using Acr protein that responds to miRNA. AcrIIA4 activity was regulated by miRNA to turn the system ON to achieve cell type-specific editing and activation in HeLa cells.121 Similarly, a miRNA-responsive AcrIIA4 system for cell-specific genome editing was developed by Hoffman et al.122 and tested in hepatocytes, cardiomyocytes, human hepatocellular carcinoma cells (Huh-7), human cervix carcinoma cells (HeLa), and human embryonic kidney cells (HEK293T).122 Unlike Hirosawa et al., the more recent Cas-ON-switch design had a post-translational negative feedback loop based on Acr proteins. This strategy was tried on variants of Cas9, including dCas9-effector fusions and NmeCas9. Developing systems that combine synthetic RNA switches that respond to internal endogenous signals or miRNAs with the CRISPR-Cas system are ideal as they circumscribe editing to target cells and, in theory, do not affect other cell types.
Lastly, all of the above studies use small molecules to control Cas9 or its sgRNA could also be layered upon additional control strategies that utilize endogenous or self-deleting/restrictive mechanisms. First, Oakes et al.126 use endogenous proteases to trigger activation of Cas9. Circular permutation was used to reengineer Cas9 for a diverse range of protease-sensing Cas9s (ProCas9s). Data showed that there was no background activity for the ProCas9s prior to proteolytic cleavage for two cell lines and up to 35% genome editing. Second, the self-deleting system was tested in vitro (HEK293T cells) as well as in vivo (male C57BL/6J mice) by Li et al.127 An AAV vector expressing the self-deleting guide RNA (gRNA) was co-injected with the AAV-Cas9 or delayed by 5 days. With the delayed injection, the vector was blocked from entering the murine liver because of host immune responses. When tested at other endogenous targets, there was similar editing efficiency between the WT AAV-Cas9 and self-deleting system.127 The self-deleting system decreased Cas9 levels by 70%–84%. However, the decrease occurred during a period of several weeks; a high amount of Cas9 was still present 4–6 weeks after AAV administration.127 Li et al.127 were unsuccessful in developing a single vector system for the self-deleting system so that the same gRNA sequence could destroy the target gene and prevent expression of Cas9. Third, Wang et al.128 were able to encode the self-restricting system within a single plasmid, avoiding the issue of an extra gRNA and observed no additional off-target modifications. Using the self-restricting system, Cas9 is reduced to 10% of peak levels 60 hours after transfection in HEK293T cells while editing efficiency is at 50%, with off-target formation down by 76.7%.128 Wang et al. did not systematically sequence for additional off-targets that may have been introduced by the extra self-deleting gRNA, but Li et al. found no modification of several additional off-target sites. Moreover, the self-restricting system may be easily loaded into viral vectors or nanoparticles for in vivo delivery since it is a single vector system.
Outlook
Controlling CRISPR-Cas9 activity is critical for these genome editing tools to have an impact in the clinic, and small molecule strategies may be able to address several in vivo translational challenges for somatic cell editing.129 Delivery of not only the inducer, but also the editor, is a challenge, because AAV, the most popular gene therapy delivery vector, is only 0.4 kb larger than Cas930,130 and many other editors, including base editors. Moreover, Cas9 shows immunogenicity in several studies,131,132 and any system that involves expression of a non-endogenous human protein is a major concern for gene therapy; thus, obtaining regulatory approval to test split or degradable Cas9 systems is a crucial challenge to overcome. Humanizing Cas9 has been proposed,133 but not extensively tested, to address this immune response to Cas9 editors. Furthermore, small molecules such as rapamycin can be used simultaneously as inducers and immunosuppressants.
Prolonged editor activity in vivo can be genotoxic or cytotoxic in addition to immunogenic. Hence, research has focused on methods to limit the life of Cas9 once editing is completed. Delivery itself can be engineered to control various parts of the CRISPR-Cas9 system, either through transient delivery or sequential delivery. Future studies that tackle these challenges could use combinations of strategies that include inducible activators, inhibitors, and degrons. Split variations of Cas9 can have little to no background editing but present a major challenge for clinical translation, as researchers have to ensure that all split vector systems are produced with the same purity, infectivity, and potency. While Acr protein inhibitors for Cas9 exist, none have been tested without light being the inducer. Using small molecules such as rapamycin or doxycycline to induce Acr protein activity is likely to be effective. Degrons have been relatively understudied in conjunction with CRISPR-Cas systems, and the different degraders suggested in this review may be further combined with other genome editing systems.
As the CRISPR-Cas editors continue to increase in precision, accuracy, and diversity, new editors may have different kinetics of editing and molecular targets for off-targets (e.g., base editors not utilizing DNA repair and off-targeting of RNA). These strategies would likely build upon control strategies established with Cas9. Tight control over both established and next-generation CRISPR genome editors will likely remain an important goal in the field in order to broaden therapeutic applications in vivo.
Acknowledgments
We thank Dr. Subhojit Roy for helpful discussions regarding viral delivery and degron strategies. This work was supported by the National Science Foundation (CBET-1350178 and EEC-1648035), the National Institutes of Health (1R35GM119644-01), the Wisconsin Alumni Research Foundation, and the Wisconsin Institute for Discovery. N.K. was supported by an NIH NHGRI training grant to the Genomic Sciences Training Program (5T32HG002760). All figures were prepared using BioRender.
Author contributions
N.K. and K.S. were involved in drafting and revising the manuscript. N.K. conducted the literature review, drafted the manuscript, and designed the tables and figures.
Declaration of interests
K.S. receives sponsored research support from Spotlight Therapeutics and Synthego. N.K. declares no competing interests.
References
- 1.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ledford H. Quest to use CRISPR against disease gains ground. Nature. 2020;577:156. doi: 10.1038/d41586-019-03919-0. [DOI] [PubMed] [Google Scholar]
- 3.Xu L., Wang J., Liu Y., Xie L., Su B., Mou D., Wang L., Liu T., Wang X., Zhang B., et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 4.Foss D.V., Hochstrasser M.L., Wilson R.C. Clinical applications of CRISPR-based genome editing and diagnostics. Transfusion. 2019;59:1389–1399. doi: 10.1111/trf.15126. [DOI] [PubMed] [Google Scholar]
- 5.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018;20:e3015. doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirakawa M.P., Krishnakumar R., Timlin J.A., Carney J.P., Butler K.S. Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 2020;40 doi: 10.1042/BSR20200127. BSR20200127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan T., Rindtorff N., Betge J., Ebert M.P., Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 2019;55:106–119. doi: 10.1016/j.semcancer.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 10.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 11.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Wang M., Zheng T., Hou Y., Zhang P., Tang T., Wei J., Du Q. Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing. Sci. Rep. 2020;10:2269. doi: 10.1038/s41598-020-58627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S., Li S., Li X.-J. Shortening the half-life of Cas9 maintains its gene editing ability and reduces neuronal toxicity. Cell Rep. 2018;25:2653–2659.e3. doi: 10.1016/j.celrep.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenblum D., Gutkin A., Dammes N., Peer D. Progress and challenges towards CRISPR/Cas clinical translation. Adv. Drug Deliv. Rev. 2020;154-155:176–186. doi: 10.1016/j.addr.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whinn K.S., Kaur G., Lewis J.S., Schauer G.D., Mueller S.H., Jergic S., Maynard H., Gan Z.Y., Naganbabu M., Bruchez M.P., et al. Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci. Rep. 2019;9:13292. doi: 10.1038/s41598-019-49837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 20.Musunuru K., Chadwick A.C., Mizoguchi T., Garcia S.P., DeNizio J.E., Reiss C.W., Wang K., Iyer S., Dutta C., Clendaniel V., et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 21.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243, 1 p. following 243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 23.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mout R., Ray M., Lee Y.W., Scaletti F., Rotello V.M. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjug. Chem. 2017;28:880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin F., Sánchez-Hernández S., Gutiérrez-Guerrero A., Pinedo-Gomez J., Benabdellah K. Biased and unbiased methods for the detection of off-target cleavage by CRISPR/Cas9: An overview. Int. J. Mol. Sci. 2016;17:1507. doi: 10.3390/ijms17091507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tadić V., Josipović G., Zoldoš V., Vojta A. CRISPR/Cas9-based epigenome editing: An overview of dCas9-based tools with special emphasis on off-target activity. Methods. 2019;164-165:109–119. doi: 10.1016/j.ymeth.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mout R., Ray M., Lee Y.-W., Scaletti F., Rotello V.M. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjug. Chem. 2017;28:880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wignakumar T., Fairchild P.J. Evasion of pre-existing immunity to Cas9: A prerequisite for successful genome editing in vivo? Curr. Transplant. Rep. 2019;6:127–133. [Google Scholar]
- 32.Wan T., et al. Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Mater. Today. 2019;26:40–66. [Google Scholar]
- 33.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundstrom K. Viral vectors in gene therapy. Diseases. 2018;6:42. doi: 10.3390/diseases6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauschhuber C., Noske N., Ehrhardt A. New insights into stability of recombinant adenovirus vector genomes in mammalian cells. Eur. J. Cell Biol. 2012;91:2–9. doi: 10.1016/j.ejcb.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Deyle D.R., Russell D.W. Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- 37.Park K., Kim W.J., Cho Y.H., Lee Y.I., Lee H., Jeong S., Cho E.S., Chang S.I., Moon S.K., Kang B.S., et al. Cancer gene therapy using adeno-associated virus vectors. Front. Biosci. 2008;13:2653–2659. doi: 10.2741/2872. [DOI] [PubMed] [Google Scholar]
- 38.Elmore Z.C., Oh D.K., Simon K.E., Fanous M.M., Asokan A. Rescuing AAV gene transfer from neutralizing antibodies with an IgG-degrading enzyme. JCI Insight. 2020;5:e139881. doi: 10.1172/jci.insight.139881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halbert C.L., Miller A.D., McNamara S., Emerson J., Gibson R.L., Ramsey B., Aitken M.L. Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2006;17:440–447. doi: 10.1089/hum.2006.17.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 42.Saha K., Sontheimer E.J., Brooks P.J., Dwinell M.R., Gersbach C.A., Liu D.R., Murray S.A., Tsai S.Q., Wilson R.C., Anderson D.G., et al. SCGE Consortium The NIH Somatic Cell Genome Editing program. Nature. 2021;592:195–204. doi: 10.1038/s41586-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S., et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maddalena A., Tornabene P., Tiberi P., Minopoli R., Manfredi A., Mutarelli M., Rossi S., Simonelli F., Naggert J.K., Cacchiarelli D., Auricchio A. Triple vectors expand AAV transfer capacity in the retina. Mol. Ther. 2018;26:524–541. doi: 10.1016/j.ymthe.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann M.D., Mathony J., Upmeier Zu Belzen J., Harteveld Z., Aschenbrenner S., Stengl C., Grimm D., Correia B.E., Eils R., Niopek D. Optogenetic control of Neisseria meningitidis Cas9 genome editing using an engineered, light-switchable anti-CRISPR protein. bioRxiv. 2019 doi: 10.1101/858589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao R., Liu D., Jia X., Zheng Y., Liu W., Xiao Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 2018;3:135–149. doi: 10.1016/j.synbio.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M., et al. Characterization of Staphylococcus aureus Cas9: A smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N., Cowan C.A., Rader D.J., Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truong D.-J.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose J.C., Stephany J.J., Valente W.J., Trevillian B.M., Dang H.V., Bielas J.H., Maly D.J., Fowler D.M. Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat. Methods. 2017;14:891–896. doi: 10.1038/nmeth.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bubeck F., Hoffmann M.D., Harteveld Z., Aschenbrenner S., Bietz A., Waldhauer M.C., Börner K., Fakhiri J., Schmelas C., Dietz L., et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat. Methods. 2018;15:924–927. doi: 10.1038/s41592-018-0178-9. [DOI] [PubMed] [Google Scholar]
- 54.Nihongaki Y., Kawano F., Nakajima T., Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 55.Lyu Y., He S., Li J., Jiang Y., Sun H., Miao Y., Pu K. A photolabile semiconducting polymer nanotransducer for near-infrared regulation of CRISPR/Cas9 gene editing. Angew. Chem. Int. Ed. Engl. 2019;58:18197–18201. doi: 10.1002/anie.201909264. [DOI] [PubMed] [Google Scholar]
- 56.Pan Y., Yang J., Luan X., Liu X., Li X., Yang J., Huang T., Sun L., Wang Y., Lin Y., Song Y. Near-infrared upconversion-activated CRISPR-Cas9 system: A remote-controlled gene editing platform. Sci. Adv. 2019;5:eaav7199. doi: 10.1126/sciadv.aav7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu H., Zhang L., Tong S., Lee C.M., Deshmukh H., Bao G. Spatial control of in vivo CRISPR-Cas9 genome editing via nanomagnets. Nat. Biomed. Eng. 2019;3:126–136. doi: 10.1038/s41551-018-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spera R., Apollonio F., Liberti M., Paffi A., Merla C., Pinto R., Petralito S. Controllable release from high-transition temperature magnetoliposomes by low-level magnetic stimulation. Colloids Surf. B Biointerfaces. 2015;131:136–140. doi: 10.1016/j.colsurfb.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 59.Hans S., Zöller D., Hammer J., Stucke J., Spieß S., Kesavan G., Kroehne V., Eguiguren J.S., Ezhkova D., Petzold A., et al. Cre-controlled CRISPR mutagenesis provides fast and easy conditional gene inactivation in zebrafish. Nat. Commun. 2021;12:1125. doi: 10.1038/s41467-021-21427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K., Jin Q., Ruan D., Yang Y., Liu Q., Wu H., Zhou Z., Ouyang Z., Liu Z., Zhao Y., et al. Cre-dependent Cas9-expressing pigs enable efficient in vivo genome editing. Genome Res. 2017;27:2061–2071. doi: 10.1101/gr.222521.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Du Y., He X., Huang X., Shi Y.S. A convenient Cas9-based conditional knockout strategy for simultaneously targeting multiple genes in mouse. Sci. Rep. 2017;7:517. doi: 10.1038/s41598-017-00654-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edwards W.F., Young D.D., Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem. Biol. 2009;4:441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- 64.Ferry Q.R.V., Lyutova R., Fulga T.A. Rational design of inducible CRISPR guide RNAs for de novo assembly of transcriptional programs. Nat. Commun. 2017;8:14633. doi: 10.1038/ncomms14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kundert K., Lucas J.E., Watters K.E., Fellmann C., Ng A.H., Heineike B.M., Fitzsimmons C.M., Oakes B.L., Qu J., Prasad N., et al. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 2019;10:2127. doi: 10.1038/s41467-019-09985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin B., An Y., Meng L., Zhang H., Song J., Zhu Z., Liu W., Song Y., Yang C. Control of CRISPR-Cas9 with small molecule-activated allosteric aptamer regulating sgRNAs. Chem. Commun. (Camb.) 2019;55:12223–12226. doi: 10.1039/c9cc05531b. [DOI] [PubMed] [Google Scholar]
- 67.Iwasaki R.S., Ozdilek B.A., Garst A.D., Choudhury A., Batey R.T. Small molecule regulated sgRNAs enable control of genome editing in E. coli by Cas9. Nat. Commun. 2020;11:1394. doi: 10.1038/s41467-020-15226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleinjan D.A., Wardrope C., Nga Sou S., Rosser S.J. Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nat. Commun. 2017;8:1191. doi: 10.1038/s41467-017-01222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang W., Hu J.H., Liu D.R. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chylinski K., Hubmann M., Hanna R.E., Yanchus C., Michlits G., Uijttewaal E.C.H., Doench J., Schramek D., Elling U. CRISPR-switch regulates sgRNA activity by Cre recombination for sequential editing of two loci. Nat. Commun. 2019;10:5454. doi: 10.1038/s41467-019-13403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun N., Petiwala S., Wang R., Lu C., Hu M., Ghosh S., Hao Y., Miller C.P., Chung N. Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening. BMC Genomics. 2019;20:225. doi: 10.1186/s12864-019-5601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zetsche B., Volz S.E., Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen D.P., Miyaoka Y., Gilbert L.A., Mayerl S.J., Lee B.H., Weissman J.S., Conklin B.R., Wells J.A. Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat. Commun. 2016;7:12009. doi: 10.1038/ncomms12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dow L.E., Fisher J., O’Rourke K.P., Muley A., Kastenhuber E.R., Livshits G., Tschaharganeh D.F., Socci N.D., Lowe S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelkar A., Zhu Y., Groth T., Stolfa G., Stablewski A.B., Singhi N., Nemeth M., Neelamegham S. Doxycycline-dependent self-inactivation of CRISPR-Cas9 to temporally regulate on- and off-target editing. Mol. Ther. 2020;28:29–41. doi: 10.1016/j.ymthe.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 77.Lundin A., Porritt M.J., Jaiswal H., Seeliger F., Johansson C., Bidar A.W., Badertscher L., Wimberger S., Davies E.J., Hardaker E., et al. Development of an ObLiGaRe doxycycline inducible Cas9 system for pre-clinical cancer drug discovery. Nat. Commun. 2020;11:4903. doi: 10.1038/s41467-020-18548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright A.V., Sternberg S.H., Taylor D.W., Staahl B.T., Bardales J.A., Kornfeld J.E., Doudna J.A. Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. USA. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu J., Zhao C., Zhao Y., Zhang J., Zhang Y., Chen L., Han Q., Ying Y., Peng S., Ai R., Wang Y. Multimode drug inducible CRISPR/Cas9 devices for transcriptional activation and genome editing. Nucleic Acids Res. 2018;46:e25. doi: 10.1093/nar/gkx1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bao Z., Jain S., Jaroenpuntaruk V., Zhao H. Orthogonal genetic regulation in human cells using chemically induced CRISPR/Cas9 activators. ACS Synth. Biol. 2017;6:686–693. doi: 10.1021/acssynbio.6b00313. [DOI] [PubMed] [Google Scholar]
- 81.Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu K.I., Ramli M.N., Woo C.W., Wang Y., Zhao T., Zhang X., Yim G.R., Chong B.Y., Gowher A., Chua M.Z., et al. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016;12:980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 83.Oakes B.L., Nadler D.C., Flamholz A., Fellmann C., Staahl B.T., Doudna J.A., Savage D.F. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mészáros B., Kumar M., Gibson T.J., Uyar B., Dosztányi Z. Degrons in cancer. Sci. Signal. 2017;10:eaak9982. doi: 10.1126/scisignal.aak9982. [DOI] [PubMed] [Google Scholar]
- 85.Li S., Prasanna X., Salo V.T., Vattulainen I., Ikonen E. An efficient auxin-inducible degron system with low basal degradation in human cells. Nat. Methods. 2019;16:866–869. doi: 10.1038/s41592-019-0512-x. [DOI] [PubMed] [Google Scholar]
- 86.Nabet B., Roberts J.M., Buckley D.L., Paulk J., Dastjerdi S., Yang A., Leggett A.L., Erb M.A., Lawlor M.A., Souza A., et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Y., Yang L., Chang T., Kandeel F., Yee J.-K. A small molecule-controlled Cas9 repressible system. Mol. Ther. Nucleic Acids. 2020;19:922–932. doi: 10.1016/j.omtn.2019.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sreekanth V., Zhou Q., Kokkonda P., Bermudez-Cabrera H.C., Lim D., Law B.K., Holmes B.R., Chaudhary S.K., Pergu R., Leger B.S., et al. Chemogenetic system demonstrates that Cas9 longevity impacts genome editing outcomes. ACS Cent. Sci. 2020;6:2228–2237. doi: 10.1021/acscentsci.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakano R., Ihara N., Morikawa S., Nakashima A., Kanemaki M.T., Ikegaya Y., Takeuchi H. Auxin-mediated rapid degradation of target proteins in hippocampal neurons. Neuroreport. 2019;30:908–913. doi: 10.1097/WNR.0000000000001299. [DOI] [PubMed] [Google Scholar]
- 90.Mendoza-Ochoa G.I., Barrass J.D., Terlouw B.R., Maudlin I.E., de Lucas S., Sani E., Aslanzadeh V., Reid J.A.E., Beggs J.D. A fast and tuneable auxin-inducible degron for depletion of target proteins in budding yeast. Yeast. 2019;36:75–81. doi: 10.1002/yea.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yesbolatova A., Natsume T., Hayashi K.I., Kanemaki M.T. Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods. 2019;164–165:73–80. doi: 10.1016/j.ymeth.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 92.Brosh R., Hrynyk I., Shen J., Waghray A., Zheng N., Lemischka I.R. A dual molecular analogue tuner for dissecting protein function in mammalian cells. Nat. Commun. 2016;7:11742. doi: 10.1038/ncomms11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., Sordella R. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell. 2018;70:801–813.e6. doi: 10.1016/j.molcel.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Röth S., Fulcher L.J., Sapkota G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019;76:2761–2777. doi: 10.1007/s00018-019-03112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu T., Yoon H., Xiong Y., Dixon-Clarke S.E., Nowak R.P., Fischer E.S. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020;27:605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Loonstra A., Vooijs M., Beverloo H.B., Allak B.A., van Drunen E., Kanaar R., Berns A., Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mhase P.P. Cytotoxicity assessment of nanoparticulate doxycycline (DH LNP) and rifampicin against human macrophage (U 937) Int. J. Pure Appl. Biosci. 2018;6:921–929. [Google Scholar]
- 99.Şekeroğlu Z.A., Afan F., Şekeroğlu V. Genotoxic and cytotoxic effects of doxycycline in cultured human peripheral blood lymphocytes. Drug Chem. Toxicol. 2012;35:334–340. doi: 10.3109/01480545.2011.621954. [DOI] [PubMed] [Google Scholar]
- 100.Hąc-Wydro K., Flasiński M. The studies on the toxicity mechanism of environmentally hazardous natural (IAA) and synthetic (NAA) auxin—The experiments on model Arabidopsis thaliana and rat liver plasma membranes. Colloids Surf. B Biointerfaces. 2015;130:53–60. doi: 10.1016/j.colsurfb.2015.03.064. [DOI] [PubMed] [Google Scholar]
- 101.Rotheneichner P., Romanelli P., Bieler L., Pagitsch S., Zaunmair P., Kreutzer C., König R., Marschallinger J., Aigner L., Couillard-Després S. Tamoxifen activation of Cre-recombinase has no persisting effects on adult neurogenesis or learning and anxiety. Front. Neurosci. 2017;11:27. doi: 10.3389/fnins.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Norrby R. Symposium on doxycycline: Held in Brussels, April 2–3, 1976. Scand. J. Infect. Dis. 1976;8:1–115. [Google Scholar]
- 103.Morton S.K., Chaston D.J., Baillie B.K., Hill C.E., Matthaei K.I. Regulation of endothelial-specific transgene expression by the LacI repressor protein in vivo. PLoS ONE. 2014;9:e95980. doi: 10.1371/journal.pone.0095980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ryan A., Scrable H. Visualization of the dynamics of gene expression in the living mouse. Mol. Imaging. 2004;3:33–42. doi: 10.1162/15353500200403193. [DOI] [PubMed] [Google Scholar]
- 105.Majumder S., Richardson A., Strong R., Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS ONE. 2011;6:e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McClure C.D., Hassan A., Duggal A., Sia C.Y., Southall T.D. AGES: An auxin-inducible, GAL4-compatible, gene expression system for Drosophila. bioRxiv. 2021 doi: 10.1101/2021.01.26.428209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin Y.-T., Wu P.H., Lee H.H., Mubanga M., Chen C.S., Kuo M.C., Chiu Y.W., Kuo P.L., Hwang S.J. Indole-3 acetic acid increased risk of impaired cognitive function in patients receiving hemodialysis. Neurotoxicology. 2019;73:85–91. doi: 10.1016/j.neuro.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 108.Chen W., Werdann M., Zhang Y. The auxin-inducible degradation system enables conditional PERIOD protein depletion in the nervous system of Drosophila melanogaster. FEBS J. 2018;285:4378–4393. doi: 10.1111/febs.14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Puurunen J., Sulkama S., Tiira K., Araujo C., Lehtonen M., Hanhineva K., Lohi H. A non-targeted metabolite profiling pilot study suggests that tryptophan and lipid metabolisms are linked with ADHD-like behaviours in dogs. Behav. Brain Funct. 2016;12:27. doi: 10.1186/s12993-016-0112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koduri V., McBrayer S.K., Liberzon E., Wang A.C., Briggs K.J., Cho H., Kaelin W.G., Jr. Peptidic degron for IMiD-induced degradation of heterologous proteins. Proc. Natl. Acad. Sci. USA. 2019;116:2539–2544. doi: 10.1073/pnas.1818109116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Froschauer A., Kube L., Kegler A., Rieger C., Gutzeit H.O. Tunable protein stabilization in vivo mediated by Shield-1 in transgenic medaka. PLoS ONE. 2015;10:e0131252. doi: 10.1371/journal.pone.0131252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harrar Y., Bellini C., Faure J.-D. FKBPs: At the crossroads of folding and transduction. Trends Plant Sci. 2001;6:426–431. doi: 10.1016/s1360-1385(01)02044-1. [DOI] [PubMed] [Google Scholar]
- 113.Yesbolatova A., Saito Y., Kanemaki M.T. Constructing auxin-inducible degron mutants using an all-in-one vector. Pharmaceuticals (Basel) 2020;13:103. doi: 10.3390/ph13050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stanley S.Y., Borges A.L., Chen K.H., Swaney D.L., Krogan N.J., Bondy-Denomy J., Davidson A.R. Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription. Cell. 2019;178:1452–1464.e13. doi: 10.1016/j.cell.2019.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rauch B.J., Silvis M.R., Hultquist J.F., Waters C.S., McGregor M.J., Krogan N.J., Bondy-Denomy J. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell. 2017;168:150–158.e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harrington L.B., Doxzen K.W., Ma E., Liu J.J., Knott G.J., Edraki A., Garcia B., Amrani N., Chen J.S., Cofsky J.C., et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell. 2017;170:1224–1233.e15. doi: 10.1016/j.cell.2017.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L., Davidson A.R. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016;167:1829–1838.e9. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borges A.L., Zhang J.Y., Rollins M.F., Osuna B.A., Wiedenheft B., Bondy-Denomy J. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell. 2018;174:917–925.e10. doi: 10.1016/j.cell.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hynes A.P., Rousseau G.M., Agudelo D., Goulet A., Amigues B., Loehr J., Romero D.A., Fremaux C., Horvath P., Doyon Y., et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 2018;9:2919. doi: 10.1038/s41467-018-05092-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsu M.-N., Hu Y.-C. Local magnetic activation of CRISPR. Nat. Biomed. Eng. 2019;3:83–84. doi: 10.1038/s41551-019-0354-y. [DOI] [PubMed] [Google Scholar]
- 121.Hirosawa M., Fujita Y., Saito H. Cell-type-specific CRISPR activation with microRNA-responsive AcrllA4 switch. ACS Synth. Biol. 2019;8:1575–1582. doi: 10.1021/acssynbio.9b00073. [DOI] [PubMed] [Google Scholar]
- 122.Hoffmann M.D., Aschenbrenner S., Grosse S., Rapti K., Domenger C., Fakhiri J., Mastel M., Börner K., Eils R., Grimm D., Niopek D. Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 2019;47:e75. doi: 10.1093/nar/gkz271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Visone R., Croce C.M. miRNAs and cancer. Am. J. Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 125.Hirosawa M., Fujita Y., Parr C.J.C., Hayashi K., Kashida S., Hotta A., Woltjen K., Saito H. Cell-type-specific genome editing with a microRNA-responsive CRISPR-Cas9 switch. Nucleic Acids Res. 2017;45:e118. doi: 10.1093/nar/gkx309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oakes B.L., Fellmann C., Rishi H., Taylor K.L., Ren S.M., Nadler D.C., Yokoo R., Arkin A.P., Doudna J.A., Savage D.F. CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell. 2019;176:254–267.e16. doi: 10.1016/j.cell.2018.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li A., Lee C.M., Hurley A.E., Jarrett K.E., De Giorgi M., Lu W., Balderrama K.S., Doerfler A.M., Deshmukh H., Ray A., et al. A self-deleting AAV-CRISPR system for in vivo genome editing. Mol. Ther. Methods Clin. Dev. 2018;12:111–122. doi: 10.1016/j.omtm.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang H., Lu H., Lei Y.S., Gong C.Y., Chen Z., Luan Y.Q., Li Q., Jian Y.Z., Wang H.Z., Wu F.L., et al. Development of a self-restricting CRISPR-Cas9 system to reduce off-target effects. Mol. Ther. Methods Clin. Dev. 2020;18:390–401. doi: 10.1016/j.omtm.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cox D.B.T., Platt R.J., Zhang F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A., et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chew W.L. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018;10:e1408. doi: 10.1002/wsbm.1408. [DOI] [PubMed] [Google Scholar]
- 132.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dai W.-J., Zhu L.Y., Yan Z.Y., Xu Y., Wang Q.L., Lu X.J. CRISPR-Cas9 for in vivo gene therapy: Promise and hurdles. Mol. Ther. Nucleic Acids. 2016;5:e349. doi: 10.1038/mtna.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]