Abstract
Detection of human cytomegalovirus (CMV) DNA in clinical specimens is considered a cornerstone in the diagnosis of CMV disease. The aim of this study was to evaluate a newly designed LightCycler-based quantitative CMV PCR. Specimens of human origin (n = 200) were tested using the LightCycler PCR, the quantitative COBAS AMPLICOR CMV MONITOR (CACM) assay, and a qualitative in-house PCR assay for the presence of CMV DNA. Samples that were reactive in at least two of the three assays were considered CMV DNA positive (n = 95 [47.5%]), while samples that were nonreactive in two of the three assays were considered CMV DNA negative (n = 105 [52.5%]). Using the LightCycler assay, CMV DNA was detected in 91 of the 95 CMV DNA-positive human specimens (sensitivity, 95.8%; 95% confidence interval [CI], 89.6 to 98.8) and in 1 of the CMV DNA-negative specimens (specificity, 99%; 95% CI, 94.8 to 99.8). Results of CMV load determination as assessed by both quantitative test systems were correlated (r = 0.73; P < 0.0001; 95% CI, 0.61 to 0.81). Results for undiluted samples containing a high CMV load were more accurate with the LightCycler test than were results obtained with the CACM test, which underestimated the viral load of samples containing high DNA copy numbers. The high level of sensitivity, specificity, accuracy, and rapidity provided by the LightCycler technology are favorable for the use of this system in the detection of CMV DNA in clinical specimens.
Diagnosis of primary and reactivated human cytomegalovirus (CMV) infections often requires the detection of virus by cell culture, antigenemia assay, or nucleic acid amplification techniques, because neither seroconversion nor determination of the presence of immunoglobulin M antibodies reliably reflects viral replication.
Previous studies have shown that qualitative PCR is not an appropriate tool for the diagnosis of CMV disease versus latent viral infection because of its low predictive value (3). However, the level of CMV DNAemia plays a critical role in the pathogenesis of CMV disease. It is considered a major risk factor for the development of CMV disease (8) and has been shown to predict CMV disease in AIDS patients (11) and renal-transplant patients (1).
Previous studies on the replication rate of CMV in vivo have substantially contributed to the understanding of the natural history of CMV disease (5). CMV replication is more dynamic than previously thought, and therefore short-term follow-up intervals for CMV DNAemia can improve the ability to predict the course of CMV disease. Thus, an accurate, rapid, and cost-effective assay is required for the frequent determination of CMV load in patients at increased risk of CMV disease.
A real-time quantitative PCR using the LightCycler instrument applies fluorescence resonance energy transfer technology with two fluorophore-labeled hybridization probes. The purpose of this study was to develop a quantitative CMV PCR assay using the LightCycler instrument and to compare the detection and quantification of CMV DNA in human specimens assessed with this assay to the results obtained with the commercially available COBAS AMPLICOR CMV MONITOR (CACM) test, a sensitive and specific test for the determination of CMV load (10).
MATERIALS AND METHODS
Specimens.
In our routine diagnostic laboratory, all specimens from patients with suspected CMV disease are tested by the CACM test. Based on the results of CACM tests, 100 consecutive positive and 100 consecutive negative plasma and urine samples were chosen for the study. The plasma (n = 128) and urine (n = 72) specimens were further evaluated by the LightCycler PCR and an in-house CMV PCR. Blood samples that were known to be positive for DNA from herpes simplex virus type 1 or 2 (n = 5), varicella-zoster virus (n = 5), Epstein-Barr virus (n = 5), human herpesvirus 8 (n = 1), and hepatitis B virus (n = 5) were analyzed as specificity controls by using the LightCycler PCR exclusively.
LightCycler CMV PCR.
Nucleic acid was extracted from 0.2 ml of each specimen by using the Qiagen (Hilden, Germany) blood kit according to the manufacturer's protocol. DNA was eluted from the column with 50 μl of PCR-grade H2O. This procedure is also suitable for the extraction of viral DNA from urine because of the high DNA yield and the removal of PCR inhibitors (4). An aliquot of 5 μl of the extracted nucleic acid was added to 15 μl of reaction mixture containing 4 mM MgCl2, a 0.66 μM concentration of each primer for the glycoprotein B gene, 0.4 μM fluorescein hybridization probe (TIB MOLBIOL, Berlin, Germany), 0.4 μM LC-Red 640 probe (TIB MOLBIOL), 5% formamide, and 2 μl of LightCycler-FastStart DNA (Master Hybridization Probes kit; Roche Molecular Biochemicals, Mannheim, Germany). Sequences of the primers for the glycoprotein B gene, which yield a 254-bp product, were published previously (GenBank accession no. A13758) (2). The hybridization probe sequences (5′ to 3′ direction) were as follows: donor fluorophore probe, CGTTTCGTCGTAGCTACGCRTACAT-fluorescein; acceptor fluorophore probe, LC-Red 640-ACACCACTTATCTYCTGGGCAGC-phosphate. Reaction capillaries were loaded, centrifuged, and placed into the carousel of a LightCycler instrument (Roche Molecular Biochemicals). The experimental PCR protocol was as follows: an initial 10 min at 95°C for FastStart Taq DNA polymerase activation, followed by 45 cycles of 10-s denaturation at 95°C, 15-s annealing at 58°C, and 12-s extension at 72°C. Data were obtained during the annealing period in the “single” mode, with the channel setting F2/F1. Fluorescence settings were as follows: F1 gain, 1; F2 gain, 14; and F3 gain, 10. The specificity of the obtained fluorescence signal was checked by a melting-curve analysis after each run. This analysis was initiated at a temperature of 45°C, which was gradually raised. During this process, the fluorescence signal was continuously monitored. The melting temperature of the specific probes was 59.2°C.
For data analysis, the baseline adjustment was carried out in the “proportional” mode, and the fluorescence curve analyses were carried out in the “fit points” mode with two points of the LightCycler software (version 3). The noise band for the crossing-point determination was adjusted at a mean F2/F1 fluorescence of three blanks plus 2 standard deviations at cycle 45. Samples with a fluorescence signal higher than the background signal were considered reactive.
Quantification of CMV DNA was performed with five 10-fold serial dilutions of a plasmid standard containing the primer-spanning region of the glycoprotein B gene. The amplicon was generated by PCR using CMV-infected diploid human fibroblasts and was subsequently cloned into the pCR2.1 vector (Invitrogen, Groningen, The Netherlands) in order to construct the plasmid pCR-gpB. The plasmid standard DNA concentration was calibrated by spectrophotometry at 260 nm.
As an inhibition control, 5-μl aliquots of nucleic acid preparations known to be negative for CMV DNA were spiked with 10,000 copies of the plasmid standard.
CACM test.
The CACM test (Roche Diagnostics, Mannheim, Germany) was chosen as an established and reliable method that is commercially available for the detection and quantification of CMV DNA. The primer set used in the CACM assay amplifies a 365-bp region of the CMV DNA polymerase gene. Preparation of undiluted samples, amplification, and detection were performed according to the manufacturer's advice, using only the provided components of the test kit.
Qualitative CMV PCR for resolution of discrepant results.
Nucleic acid was extracted from the specimens with the Qiagen blood kit, as described above. A 10-μl aliquot was added to 40 μl of reaction mixture. Components were adjusted to the following final concentrations: a 200 μM concentration of each deoxyribonucleoside triphosphate (Roche Molecular Biochemicals), a 0.4 μM concentration of each primer, 50 mU of Taq DNA polymerase (Roche Molecular Biochemicals) per μl, and 1.5 μM MgCl2. The primers CCGCAACCTGGTGCCCATGG (upstream primer) and CGTTTGGGTTGCGCAGCGGG (downstream primer) that had been validated previously for the detection of CMV late antigen gp64 were used (9). Cycling conditions included an initial 30-s step at 94°C followed by 40 cycles of 60-s denaturation at 94°C, 30-s annealing at 63°C, and 60-s extension at 72°C. The PCR was completed by a terminal extension step of 7 min at 72°C. The 139-bp amplicon was visualized by agarose gel electrophoresis and ethidium bromide staining.
Determination of detection limit.
The supernatant of CMV-infected diploid human fibroblast cultures was calibrated with the plasmid standard by LightCycler PCR in four independent PCR runs. The cell culture supernatant was serially diluted and tested by LightCycler PCR and the CACM test. The lowest concentration that still contained detectable CMV DNA was assigned as the detection limit.
Statistical analysis.
For all statistical tests and graphs, a statistical software package was used (MedCalc, Mariakerke, Belgium). In box-and-whisker plots, outside values were defined as values higher than the upper quartile plus 1.5 times the interquartile range. These values were plotted with a separate square marker.
RESULTS
Cycle conditions for a CMV-specific PCR on the LightCycler instrument were substantially improved as described above, using the primers and fluorophore probes described above. The correlation between the noise band crossing points and the log template concentration was determined with a series of 10-fold dilutions of the standard quantification plasmid, pCR-gpB. Statistical analysis revealed a negative linear correlation (r = −0.99; P < 0.0001; 95% confidence interval [CI], −0.999 to −0.990) between both parameters in a range between 102 and 108 plasmid copies per ml of standard. To evaluate the specificity of the assay, a panel of 21 samples positive for herpesvirus or hepatitis B virus DNA was tested concomitantly. All samples were nonreactive by the LightCycler PCR. No PCR inhibition was observed in samples with negative test results.
The endpoint detection limit of the different tests was determined using a cell culture supernatant that had been previously calibrated with the plasmid pCR-gpB. A serial dilution of this standard allows the comparison of all three tests evaluated in this study because it offers target sequences that can be amplified by the different primer-probe combinations. The endpoint detection limit of the LightCycler test was ≤100 copies per ml of standard solution, and for the CACM test, it was ≤500 copies per ml of standard solution.
Out of 72 urine specimens, 60 had positive results by the CACM assay and 54 had positive test results by the LightCycler PCR. Out of the six urine samples that were positive only by the CACM assay, five had negative test results by the in-house PCR. Out of 128 plasma specimens, 40 samples were positive by the CACM assay, and 36 of these 40 samples were also positive by the LightCycler PCR. Of the four plasma samples with discordant results in the two tests, three were positive by the in-house PCR. Positive test results by both the LightCycler PCR and the qualitative in-house PCR, but not by the CACM PCR, were found only for a single plasma sample. Additionally, a single plasma sample was positive by the LightCycler PCR only.
Human specimens that were positive in two of the three assays were classified as CMV DNA positive (n = 95 [47.5%]). Samples that were negative in two of the three assays were classified as CMV DNA negative (n = 105 [52.5%]).
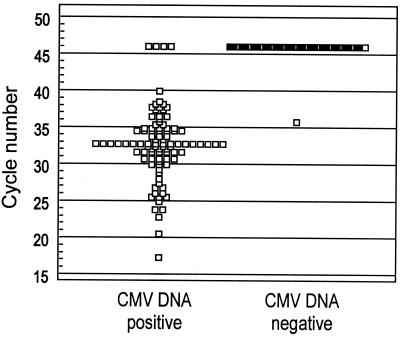
The noise band crossing points of the CMV DNA-positive and -negative samples by the LightCycler PCR are displayed in Fig. 1. All CMV DNA-positive samples that were reactive in the LightCycler PCR crossed the baseline prior to cycle 41. CMV DNA-negative samples with nonreactive results did not cross the noise band within the 45 cycles of the PCR run. CMV DNA was detected by the LightCycler test in 91 of the 95 CMV DNA-positive samples, yielding a sensitivity of 95.8% (95% CI, 89.6 to 98.8), and in 1 of the CMV DNA-negative samples, yielding a specificity of 99% (95% CI, 94.8 to 99.8). The positive-likelihood ratio was 100.58 and the negative-likelihood ratio was 0.04.
FIG. 1.
Detection of CMV DNA in human specimens by the LightCycler PCR. The dot diagram shows the cycle number of the crossing point with the noise band of each sample classified as CMV DNA positive or CMV DNA negative. Samples that did not cross the noise band during the 45 PCR cycles are displayed with a dot at cycle 46.
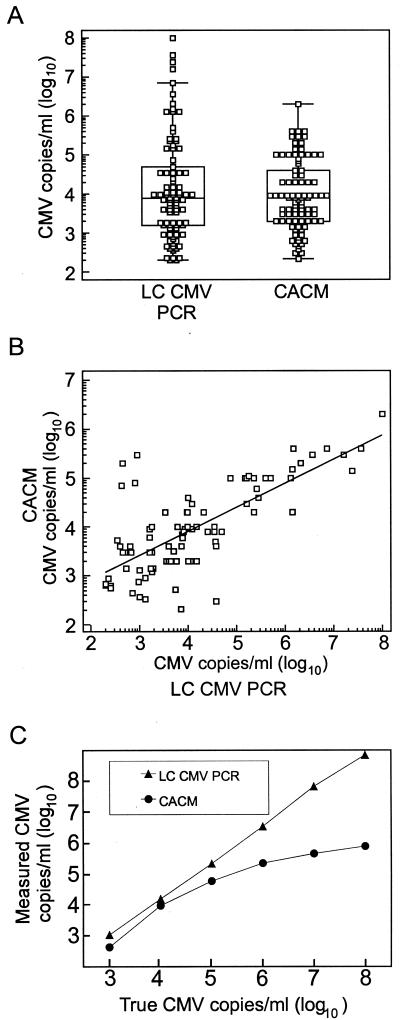
Results for CMV load determination in 90 congruent positive human specimens obtained by the LightCycler PCR and by the CACM test, respectively, are shown in Fig. 2A and B. In this study, the viral load detected by the LightCycler assay ranged from 100 to 1 × 108 copies per ml of sample and the load detected by the CACM assay ranged from 210 to 2 × 106 copies per ml of sample. The median CMV load for all samples tested was 7,800 copies per ml of sample for the LightCycler test and 8,000 copies per ml of sample for the CACM test. Results of CMV load determination from both test systems were correlated (r = 0.73; P < 0.001; 95% CI, 0.61 to 0.81). However, for undiluted samples with high CMV loads, the LightCycler test revealed higher loads than did the CACM test. CMV loads determined from a diluted series of the CMV DNA standard, which was derived from cell culture, are shown in Fig. 2C. Results of the LightCycler test correlated to the true copy number per milliliter (r = 1; P < 0.0001; 95% CI, 0.99 to 1.00), while the CACM test underestimated high target concentrations (r = 0.88; P = 0.02; 95% CI, 0.24 to 0.99).
FIG. 2.
Correlation between the quantitative CMV DNA values measured by the LightCycler (LC) CMV PCR and by the CACM test. (A) The combined box-and-whisker/dot plot diagram shows CMV DNA values for 90 samples that were reactive in the LightCycler CMV PCR and in the CACM test. (B) The scatter diagram and the regression line show the relation of quantitative CMV DNA values for each sample measured by both the LightCycler PCR and the CACM (r = 0.73; P < 0.0001; 95% CI, 0.61 to 0.81). (C) Quantitative measurement of CMV DNA in a serially diluted cell culture standard by the LightCycler PCR (r = 1; P < 0.0001; 95% CI, 0.99 to 1.00) and by the CACM test with undiluted samples (r = 0.88; P = 0.02; 95% CI, 0.24 to 0.99).
DISCUSSION
The LightCycler PCR used in this study allows the accurate detection and quantitative assessment of CMV DNA in human specimens, as demonstrated by the low detection limit and excellent linear correlation between target concentration and test results. Results for the CMV load generated by the LightCycler PCR correlate with the results generated by the commercially available CACM test. Additionally, the CACM test, which uses undiluted samples, does not properly estimate the viral load when concentrations greater than 105 copies per ml are present in the sample, as demonstrated by testing a calibrated CMV DNA standard. These findings are in agreement with the CACM manufacturer's recommendation to repeat every PCR run with diluted samples when viral loads greater than 105 copies per ml are detected. Even though results generated with diluted specimens tend to reach the expected viral load, this approach is rather time-consuming and expensive.
While CMV loads as high as 107 to 108 copies per ml of blood or urine have previously been found by other authors and were shown to be an important predictor of CMV disease (6), CMV loads below 103 to 104 copies per ml of blood or urine rarely have been associated with CMV disease (8, 11, 12). Thus, the proper quantification of CMV DNA above 105 copies per ml of sample should be a major criterion for the evaluation of a quantitative CMV assay.
Considering the dynamics of CMV replication in vivo (5), the frequent follow-up of CMV load can further improve the clinical management of patients at high risk of CMV disease. Thus, the selected assay should be time- and cost-effective by covering the entire range of relevant CMV loads in a one-step experiment. The entire test procedure of the LightCycler CMV PCR takes up to 150 min, while the CACM procedure requires 450 min for a single run and an additional 450 min for a second run with diluted samples when viral loads greater than 105 copies per ml of sample are detected. Currently, the costs for the commercially available CACM test exceed those for the supplies required for the LightCycler test by a factor of 20. Furthermore, the procedure for nucleic acid amplification and detection by the LightCycler assay is in a closed system that prevents cross-contamination of samples during postamplification steps (7). Based on the high level of sensitivity and specificity, accurate quantification, time- and cost-effectiveness, and low contamination risk, the LightCycler technology is a favorable test platform in the diagnostic virology laboratory.
REFERENCES
- 1.Aitken C, Barrett-Muir W, Millar C, Templeton K, Thomas J, Sheridan F, Jeffries D, Yaqoob M, Breuer J. Use of molecular assays in diagnosis and monitoring of cytomegalovirus disease following renal transplantation. J Clin Microbiol. 1999;37:2804–2807. doi: 10.1128/jcm.37.9.2804-2807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai X, Hosler G, Barton Rogers B, Dawson D B, Scheuermann R H. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem. 1997;43:1843–1849. [PubMed] [Google Scholar]
- 3.Delgado R, Lumbreras C, Alba C, Pedraza M A, Otero J R, Gómez R, Moreno E, Noriega A R, Payá C V. Low predictive value of polymerase chain reaction for diagnosis of cytomegalovirus disease in liver transplant recipients. J Clin Microbiol. 1992;30:1876–1878. doi: 10.1128/jcm.30.7.1876-1878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echavarria M, Forman M, Ticehurst J, Dumler J S, Charache P. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol. 1998;36:3323–3326. doi: 10.1128/jcm.36.11.3323-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emery V C, Cope A V, Bowen E F, Gor D, Griffiths P D. The dynamics of human cytomegalovirus replication in vivo. J Exp Med. 1999;190:177–182. doi: 10.1084/jem.190.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery V C. Relative importance of cytomegalovirus load as a risk factor for cytomegalovirus disease in the immunocompromised host. Monogr Virol. 1998;21:288–301. [Google Scholar]
- 7.Espy M J, Uhl J R, Mitchell P S, Thorvilson J N, Svien K A, Wold A D, Smith T F. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:795–799. doi: 10.1128/jcm.38.2.795-799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan-Walker A F, Kidd I M, Sabin C, Sweny P, Griffiths P D, Emery V C. Quantity of human cytomegalovirus (CMV) DNAemia as a risk factor for CMV disease in renal allograft recipients: relationship with donor/recipient CMV serostatus, receipt of augmented methylprednisolone and antithymocyte globulin (ATG) J Med Virol. 1999;58:182–187. [PubMed] [Google Scholar]
- 9.Shibata D, Martein W J, Appleman M D, Causey D M, Leedom J M, Arnheim N. Detection of cytomegalovirus DNA in peripheral blood of patients infected with human immunodeficiency virus. J Infect Dis. 1988;158:1185–1192. doi: 10.1093/infdis/158.6.1185. [DOI] [PubMed] [Google Scholar]
- 10.Sia I G, Wilson J A, Espy M J, Paya C V, Smith T F. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J Clin Microbiol. 2000;38:600–606. doi: 10.1128/jcm.38.2.600-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spector S A, Wong R, Hsia K, Pilcher M, Stempien M J. Plasma cytomegalovirus (CMV) load predicts CMV disease and survival in AIDS patients. J Clin Investig. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg A, Hodges T N, Li S, Cai G, Zamora M R. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol. 2000;38:768–772. doi: 10.1128/jcm.38.2.768-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]