Summary
Two type II‐C Cas9 orthologs (Nm1Cas9 and Nm2Cas9) were recently identified from Neisseria meningitidis and have been extensively used in mammalian cells, but whether these NmCas9 orthologs or other type II‐C Cas9 proteins can mediate genome editing in plants remains unclear. In this study, we developed and optimized targeted mutagenesis systems from NmCas9s for plants. Efficient genome editing at the target with N4GATT and N4CC protospacer adjacent motifs (PAMs) was achieved with Nm1Cas9 and Nm2Cas9 respectively. These results indicated that a highly active editing system could be developed from type II‐C Cas9s with distinct PAM preferences, thus providing a reliable strategy to extend the scope of genome editing in plants. Base editors (BEs) were further developed from the NmCas9s. The editing efficiency of adenine BEs (ABEs) of TadA*‐7.10 and cytosine BEs (CBEs) of rat APOBEC1 (rAPO1) or human APOBEC3a (hA3A) were extremely limited, whereas ABEs of TadA‐8e and CBEs of Petromyzon marinus cytidine deaminase 1 (PmCDA1) exhibited markedly improved performance on the same targets. In addition, we found that fusion of a single‐stranded DNA‐binding domain from the human Rad51 protein enhanced the base editing capability of rAPO1‐CBEs of NmCas9s. Together, our results suggest that the engineering of NmCas9s or other type II‐C Cas9s can provide useful alternatives for crop genome editing.
Keywords: CRISPR, NmCas9, type II‐C, genome editing, rice
Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR)‐associated protein 9 (Cas9) from the type II CRISPR‐Cas bacterial adaptive immune system has been engineered as a versatile genome‐editing platform for a wide range of organisms, including plants. The Cas9 endonuclease is an RNA‐programmable site‐specific nuclease, and the specificity of Cas9 is dictated by a customizable guide RNA (gRNA) with a sequence complementary to the genome target. A protospacer adjacent motif (PAM) immediately downstream of the target sequence is strictly required for target recognition of the gRNA‐Cas9 complex, which greatly restricts the available editing scope of the CRISPR‐Cas9 system. Notably, the PAM sequence shows variations among Cas9 orthologs identified from different bacterial strains (Chylinski et al., 2014). Therefore, the diverse PAM preferences of different Cas9s would facilitate extension of the range of CRISPR‐editing tools in the genome.
Type II CRISPR systems are divided into three subtypes (type II‐A, II‐B, and II‐C). All of these subtypes contain the Cas1, Cas2, and Cas9 proteins, whereas Csn2 and Cas4 are present in the type II‐A and II‐B systems, respectively, and no additional Cas protein is found in the type II‐C system (Chylinski et al., 2014; Wei et al., 2015). Abundant and divergent Cas9 orthologs of the three subtypes have been identified in a large variety of bacteria (Chylinski et al., 2014; Karvelis et al., 2015; Siksnys and Gasiunas, 2016). However, only a few of these orthologs have been functionally characterized and engineered for eukaryotic genome‐editing tools. Type II‐A Streptococcus pyogenes Cas9 (SpCas9) was the first reported Cas9 and is responsible for the majority of the current CRISPR applications (Jinek et al., 2012). Several type II‐A Cas9 orthologs with altered PAM specificity, for example, SaCas9 from Staphylococcus aureus, SauriCas9 from Staphylococcus auricularis, and St1Cas9 and St3Cas9 from Streptococcus thermophilus, have also been adopted for genome editing in human cells (Hu et al., 2020; Kleinstiver et al., 2015; Ran et al., 2015; Xu et al., 2015). A SpCas9 ortholog with high sequence similarity, Streptococcus canis Cas9 (ScCas9), accepts the much more relaxed NNG PAM than the NGG PAM of SpCas9, which enables a broad range of targeting capabilities in vivo (Chatterjee et al., 2018). An A‐rich PAM‐preferred iSpyMacCas9 was developed by engineering the PAM‐interacting domain of Streptococcus macacae Cas9 (SmacCas9) to SpCas9 (Chatterjee et al., 2020). In the type II‐B system, a Cas9 from Francisella novicida (FnCas9) with sequence‐specific DNA and RNA dual‐nuclease activities has also been harnessed for the genetic modification of human cells and mouse zygotes (Harrington et al., 2017; Hirano et al., 2016; Sampson et al., 2013). In addition, a growing number of CRISPR genome‐editing tools have been developed using type II‐C Cas9 proteins, including Nm1Cas9 and Nm2Cas9 from Neisseria meningitidis, GeCas9 from Geobacillus stearothermophilus, CcCas9 from Clostridium cellulolyticum, CdCas9 from Corynebacterium diphtheriae, and CjCas9 from Campylobacter jejuni (Amrani et al., 2018; Edraki et al., 2019; Fedorova et al., 2020; Harrington et al., 2017; Hirano et al., 2019; Hou et al., 2013; Kim et al., 2017). The compact protein size, diverse PAM compatibility, and accurate target specificity of type II‐C Cas9 proteins provide alternative solutions for genome editing in mammals (Mir et al., 2018).
In plants, CRISPR‐Cas9 systems have been confirmed to be powerful tools for basic research as well as breeding applications. Similar in mammalian cells, SpCas9 is the most commonly used CRISPR tool in different plant species (Chen et al., 2019). The rest of the identified type II‐A Cas9 proteins, for example, SaCas9, iSpyMacCas9, St1Cas9, and ScCas9, have also been applied and significantly expand the editing scope of plant genomes (Hua et al., 2019; Kaya et al., 2016; Qin et al., 2019a; Sretenovic et al., 2020; Steinert et al., 2015; Wang et al., 2020; Xu et al., 2020). Type II‐B FnCas9 has been reprogrammed to obtain an RNA‐targeting system to confer virus resistance in Arabidopsis and tobacco (Zhang et al., 2018). However, very few studies have developed plant genome‐editing systems adopted from type II‐C Cas9, although the type II‐C subtype comprises nearly half of the Cas9 proteins discovered in the sequence database (Mir et al., 2018). Here, we established and optimized two type II‐C Cas9 systems (Nm1Cas9 and Nm2Cas9) for efficient targeted mutagenesis in rice. Several adenine base editors (adenine BEs, ABEs) and cytosine BEs (CBEs) of NmCas9s were constructed to introduce precise base conversions. Our data reveal the functional pattern of NmCas9s in rice, suggesting the great potential of developing plant genome‐editing tools from type II‐C subtype CRISPR‐Cas9 systems.
Results
Engineering Nm1Cas9 systems for targeted mutagenesis in rice
Genome‐editing systems engineered from type II‐C Nm1Cas9 have been widely applied in mammals (Amrani et al., 2018; Esvelt et al., 2013; Hoffmann et al., 2019; Hou et al., 2013; Ibraheim et al., 2018; Lee et al., 2016, 2019). To develop tools for plant genome editing, the coding sequence of Nm1Cas9 was codon optimized for monocot expression and was placed under the control of the maize ubiquitin (ZmUBI) promoter in a pHUC400 backbone (Figure 1a). The rice U3 (OsU3) promoter was used to express the single guide RNA (sgRNA) in the plant Nm1Cas9 vector, namely, pHUC7a11. The 5′‐NNNNGATT‐3′ (N4GATT) sequence is the canonical PAM for Nm1Cas9 in nature (Zhang et al., 2013). To validate the editing capacity of the Nm1Cas9 system, eight targets with the N4GATT PAM were selected in seven rice genes. The 24‐nt gRNA sequence was individually added to the pHUC7a11 vector with a 101‐nt sgRNA scaffold (Figure 1b) (Nowak et al., 2016). The vectors were introduced into the japonica rice cultivar Nipponbare via agrobacterium‐mediated transformation. After genotyping the target sites in regenerated plants, mutants were found at only two loci with extremely limited frequency (3/48 at the BADH2‐1 site and 1/48 at the WX‐1 site, Figure 1c), and no mutants were detected at the remaining genomic loci. To improve the efficiency of the plant Nm1Cas9 system, a 121‐nt extended sgRNA (esgRNA) scaffold was further tested (Hou et al., 2013). The targeted mutants were induced at frequencies greater than 60.4% in all of the tested targets (Figure 1c and Figure S1), and frequencies as high as 89.6% (43/48) could be achieved at the BADH2‐1 site. Moreover, biallelic/homozygous mutants accounted for 52.1%, 64.6%, and 66.7% of the transgenic plants induced by the BEL‐1, IPA1‐1, and BADH2‐1 esgRNAs (Figure 1c), confirming the efficient mutagenesis capability of the Nm1Cas9 system at plant genomic sites. The off‐target effect was further examined in the transgenic plants. The three most likely off‐target sites of IPA1‐1 and WX‐1 were predicted by the Cas‐OFFinder tool and examined by site‐specific sequencing (Bae et al., 2014), and no mutants were detected (Table S1). Consistent with previous studies in mammalian cells, these results suggested that off‐target effects may not be a serious concern in Nm1Cas9‐induced genome editing.
Figure 1.
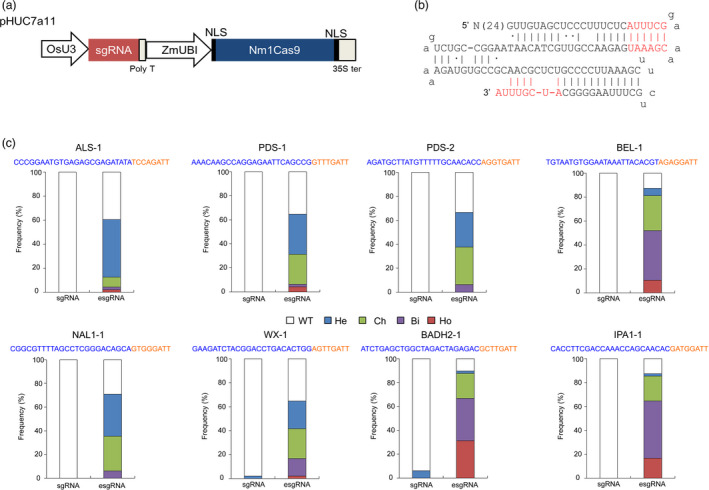
Targeted mutagenesis induced by the CRISPR‐Nm1Cas9 system in rice. (a) Schematic illustration of the expression cassettes of the CRISPR‐Nm1Cas9 system in the pHUC7a11 binary vector. OsU3, rice U3 promoter; ZmUBI, maize ubiquitin‐1 promoter; NLS, nuclear location sequence; poly T, poly T terminator; 35S ter, CaMV 35S terminator. (b) Schematic representation of the 121‐nt sgRNA (esgRNA) scaffold of Nm1Cas9. The red letters indicate the discrepant nucleotides existing in the 121‐nt scaffold but not in the 101‐nt sgRNA scaffold. (c) Identification of targeted mutants in CRISPR‐Nm1Cas9 transgenic plants. The target sequence is presented above the column. Blue letters indicate the protospacer, and the PAM is labelled in orange. For each vector, 48 regenerated T0 plants were examined, and the ratio of the number of plants with certain zygotypes to the total plant number was considered the frequency. WT, wild type at the target; He, heterozygous mutant (a mutated allele and a WT allele); Ch, chimeric mutant (more than three different alleles at the target in a single plant); Bi, biallelic mutant (two different mutated alleles); Ho, homozygous mutant (single mutated allele); sgRNA, 101‐nt sgRNA scaffold used in the CRISPR‐Nm1Cas9 system; esgRNA, 121‐nt sgRNA scaffold.
At all eight targets, the predominant mutation induced by Nm1Cas9 was a single base insertion 3 bp upstream of the PAM (Figure S1). To further elucidate the editing pattern of Nm1Cas9 in plants, a NAL1‐2 target with a CGGAGATT PAM was independently targeted by Nm1Cas9 and SpCas9 (Figure S2). In collections of stably transformed calli, the targeting was examined by amplicon next‐generation sequencing (NGS). The efficiency of mutagenesis induced by SpCas9 was slightly higher than that induced by Nm1Cas9 (P < 0.05). However, the frequency of Nm1Cas9‐introduced insertions was 4.5‐fold higher than that of insertions introduced by SpCas9 (P < 0.01), whereas the deletion frequency of NmCas9 was 17.6‐fold lower than that of SpCas9 (P < 0.01). We also found that the single base insertions 3 bp upstream of the PAM were the most frequent mutations induced by Nm1Cas9, accounting for 92.8% of the total edits at the NAL1‐2 target. In contrast, 1‐bp insertions accounted for only 17.8% of the mutations induced by SpCas9, and the major types of frequently occurring mutations were short deletions. Together, our results indicate an intriguing bias towards insertions among the mutagenesis products of Nm1Cas9 in rice.
In bacterial and mammalian cell systems, nucleotide A at the first position of the PAM (AN3GATT) is not preferred for the activity of Nm1Cas9 (Esvelt et al., 2013; Lee et al., 2016). However, no apparent difference in the frequency of mutant occurrence was observed between the AN3GATT PAM targets (PDS‐2, BEL‐1 and WX‐1) and the BN3GATT (B = G/T/C) PAM targets. In addition to the N4GATT PAM, Nm1Cas9 was able to tolerate certain single or double mutations in the conserved PAM sequence. According to previous results from mammalian cells, N4GCTT is a well‐identified noncanonical PAM in which Nm1Cas9 could still exhibit high levels of activity (Esvelt et al., 2013; Hou et al., 2013). To validate the PAM compatibility of Nm1Cas9 in plants, six additional sgRNAs were designed to target rice genome sites with the N4GCTT PAM. The targeted mutants failed to be induced by two sgRNAs (ALS‐2 and BEL‐2), and 2, 3, 9, and 14 mutants were identified from 48 plants with the NAL1‐3, PDS‐3, WX‐2, and BADH2‐2 sgRNAs, resulting in 4.2%, 6.3%, 18.8%, and 29.2% frequencies respectively (Figure S3). These results showed that Nm1Cas9 can exploit the N4GCTT PAM in plants, whereas its activity would be highly likely to be influenced by other factors.
Development and optimization of Nm2Cas9 systems for rice gene editing
Nm2Cas9 is a type II‐C Cas9 ortholog that exhibits high cleavage activity in mice and human cells (Edraki et al., 2019; Sun et al., 2019). Compared with the optimal N4GATT PAM of Nm1Cas9, the Nm2Cas9 system recognizes a short N4CC PAM, which increases the density of targetable genomic sites. Similar to the plant Nm1Cas9 system, the coding sequence of Nm2Cas9 was codon optimized for monocot expression and inserted into the pHUC400 backbone to construct a pHUC7b11 binary vector (Figure 2a). Then, 24‐nt gRNAs were designed for six targets with the N4CC PAM in the rice genome. It has been revealed that both Nm1Cas9 and Nm2Cas9 can be guided by the same sgRNA scaffold (Edraki et al., 2019). Therefore, the 121‐nt esgRNA scaffold was fused with the gRNAs for evaluation of the plant Nm2Cas9 system. In Nm2Cas9 plants with the PDS‐4 and ALS‐3 sgRNAs, mutations were not observed in the target genome region (Figure 2b). At the remaining four sites, targeted mutations were induced in transgenic plants at variable frequencies (from 6.25 to 77.1%). Detailed genotyping results indicated that biallelic or homozygous mutants were lacking in the transgenic populations (Figure 2b).
Figure 2.
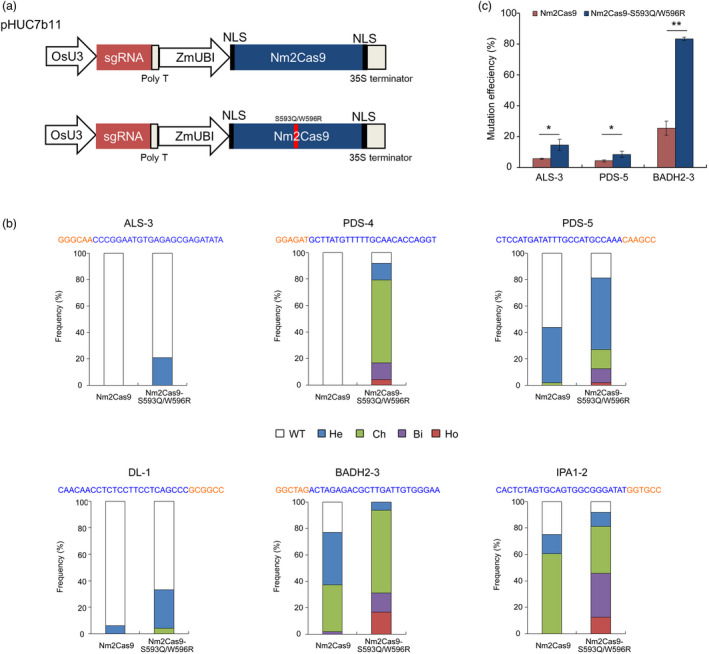
Targeted mutagenesis induced by CRISPR‐Nm2Cas9 systems in rice. (a) Schematic illustration of the expression cassettes of the CRISPR‐Nm2Cas9 system in the pHUC7b11 binary vectors. The S593Q/W596R double mutations are labelled by a red rectangle inside Nm2Cas9. (b) Mutagenesis efficiencies induced by Nm2Cas9 and Nm2Cas9‐S593Q/W596R in rice. In individual transformants, more than 200 newly emerged calli were collected after 2 weeks of selection under 50 mg/L hygromycin and mixed as a sample for the extraction of genomic DNA. The target region was then analysed by amplicon NGS. The mutagenesis efficiency of Nm2Cas9s was calculated by counting the mutated reads compared with the total clean reads. ±SD values were generated from three biological replicates. *, P < 0.05; **, P < 0.01; two‐tailed t‐test. (c) Identification of targeted mutants in transgenic plants of Nm2Cas9 or Nm2Cas9‐S593Q/W596R.
Although the activity of Nm2Cas9 and Nm1Cas9 is similar in human cells (Edraki et al., 2019), our results suggest that the targeted mutagenesis efficiency of the Nm2Cas9 system may be restricted in plants. The efficiency of Nm2Cas9 is affected by the length of the protospacer (Edraki et al., 2019). To optimize the gRNA length for the plant Nm2Cas9 system, a series of 5’ truncated gRNAs ranging from 24 to 21 nt in length were further tested for two target sites. At the BADH2‐3 target, comparable activities could be obtained with the 24‐nt full‐length gRNA and the truncated gRNAs with a length of up to 21 nt, whereas the mutant occurrence frequency at the PDS‐5 target generated with the 21‐nt gRNA was markedly lower than that obtained with the longer gRNAs (Figure S4). Therefore, reliably enhancing the activity of the plant Nm2Cas9 system by optimizing the gRNA length may be difficult.
Structural and experimental analyses have indicated that mutations at Ser593 and Trp596 of the HNH domain can significantly improve the double‐stranded DNA (dsDNA) cleavage activity of Nm1Cas9 in vivo (Sun et al., 2019). Nm2Cas9 shares high sequence identity with Nm1Cas9 except in the PAM interaction domain, and it is logical to presume that the identical amino acids have conserved roles in the function of both NmCas9s. To enhance the targeted mutagenesis efficiency of the plant Nm2Cas9 system, a S593Q/W596R double mutant of Nm2Cas9 was generated (Figure 2a). The editing efficiency of Nm2Cas9‐S593Q/W596R was evaluated in newly transformed rice calli by amplicon NGS (Figure 2c). Compared with the results obtained with wild‐type Nm2Cas9, the ALS‐3, PDS‐5, and BADH2‐3 esgRNAs significantly increased the mutagenesis efficiency of the S593Q/W596R variant by 2.0–3.3‐fold (P < 0.05, two‐tailed t‐test). The mutagenesis mediated by Nm2Cas9‐S593Q/W596R was further examined in the regenerated plants (Figure 2b and Figure S5). The frequencies of the mutants were 20.8%–100% higher in the regenerated plants with Nm2Cas9‐S593Q/W596R at the six targets. At the IPA1‐2 site, up to 50% (22 out of 44) of mutants carried biallelic/homozygous mutations. These results indicate that editing at the N4CC PAM target can be efficiently achieved by optimizing the Nm2Cas9 system in plants. In the transgenic population of PDS‐4 and BADH2‐3 with high on‐target mutation frequencies, the off‐target effects were further examined. At the three most likely potential off‐target sites of each gRNA, we did not find any possible off‐target events (Table S2).
Nm1Cas9‐ and Nm2Cas9‐mediated cytosine and adenine base editing in the rice genome
Base editors have been derived from type II‐A CRISPR‐Cas9 or type V CRISPR‐Cas12a systems to mediate targeted precise nucleotide substitutions without double‐strand breaks (DSBs) (Rees and Liu, 2018). To test whether ABEs can be developed from type II‐C Cas9 proteins, the nickase mutant of Nm1Cas9 (nNm1Cas9 D16A) or Nm2Cas9‐S593Q/W596R (nNm2Cas9 D16A/S593Q/W596R) was fused with a simplified version of the evolved adenine deaminase TadA*7.10 at the N terminus (Hua et al., 2020), and triplet repeats of a nuclear location sequence (NLS) were added at the C terminus to increase the editing efficiency (Figure 3a) (Li et al., 2018). The editing ability of TadA*7.10‐nNm1Cas9 (Nm1‐ABE) was tested at three canonical N4GATT PAM sites via stable rice transformation. A‐to‐G conversion was only detected at the WX‐3 target at a frequency of 1.0% (Figure 3b and Table S3). With TadA*7.10‐nNm2Cas9 (Nm2‐ABE), editing was also detected at one out of three targets (the Hd6‐1 target) with a frequency of 2.1%.
Figure 3.
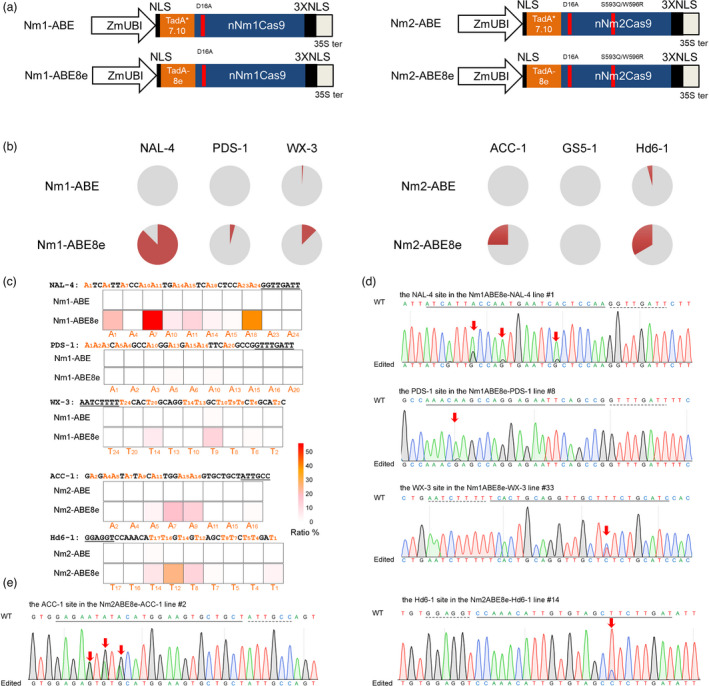
Nm‐ABEs introduce A‐to‐G conversions in rice. (a) Schematic representation of the ABEs derived from nNmCas9s. TadA*7.10 and TadA‐8e, evolved DNA adenine deaminases from E. coli TadA. (b) Frequency of mutants induced by Nm‐ABEs in the rice genome. For each sgRNA, 48–96 independent T0 lines were randomly selected and genotyped. Grey, wild type, no mutation was found at the target; red, plants harbouring base editing. (c) Heatmap of adenine base conversions induced by Nm‐ABEs. The target regions are indicated, and the positions of targetable nucleotides distal to the PAM are labelled. The substitution frequency of each base was calculated as the percentage of edited plants within the total plant population. (d) Sequencing chromatogram of edited plants. The A∙T‐to‐G∙C base conversion is labelled by a red arrow.
To improve the editing efficiency of the ABEs, TadA*7.10 in Nm1‐ABE and Nm2‐ABE was separately replaced by the highly active deaminase TadA‐8e to generate Nm1‐ABE8e and Nm2‐ABE8e respectively (Figure 3a). In the stable transgenic lines of Nm1‐ABE8e, 87.5%, 4.2%, and 12.5% of the regenerated plants carried A•T‐to‐G•C conversions in the target region of NAL1‐4, PDS‐1, and WX‐3 respectively (Figure 3b,c,d and Table S3). The base editing of Nm2‐ABE8e was also examined. At the targets of ACC‐1 and Hd6‐1, substitutions were detected in 25.0% and 33.3% of the transformed plants (Figure 3b,c,e and Table S3). No mutants were detected at the GS5‐1 target, possibly because the OsU3 promoter‐driven expression of the GS5‐1 sgRNA was inhibited by a 6‐nt poly (T) sequence in the protospacer. Targeted genotyping showed that base editing induced by Nm1‐ABE8e and Nm2‐ABE8e occurred in the region between positions 1–18 and positions 1–16, respectively (Figure 3c), suggesting a markedly wider editing window than that of SpCas9‐based ABE8e in plants (Li et al., 2021; Yan et al., 2021).
To develop CBEs from NmCas9s (Nm‐CBEs), the nickase of Nm1Cas9 or Nm2Cas9‐S593Q/W596R was applied to fuse rat APOBEC1 (rAPO1) cytosine deaminase and triplet copies of an uracil glycosylase inhibitor (UGI) at the N and C termini, constructing enhanced base editor 3 of Nm1Cas9 and Nm2Cas9 (Nm1‐enBE3 and Nm2‐enBE3, Figure 4a). For each vector, three esgRNAs were designed to examine the editing activity in stable transgenic plants. However, no desired base conversions were observed in the corresponding targeted regions (Figure 4b and Table S4). To optimize the BE3s, hyper BE3 of NmCas9s (Nm1‐hyBE3 and Nm2‐hyBE3) was constructed by inserting a non‐sequence‐specific single‐stranded DNA‐binding domain from the human Rad51 protein (RAD51‐ssDBD) between rAPO1 and nNmCas9s (Zhang et al., 2020). For Nm1‐hyBE3, C‐to‐T edits were detected in 12.5% and 10.4% of plants at the NAL1‐5 and PDS‐6 targets respectively (Figure 4b‐d and Table S4). Similarly, Nm2‐hyBE3 successfully mediated cytosine editing at all three targets in rice and achieved up to 14.6% base conversion frequency at the GRF6‐1 target.
Figure 4.
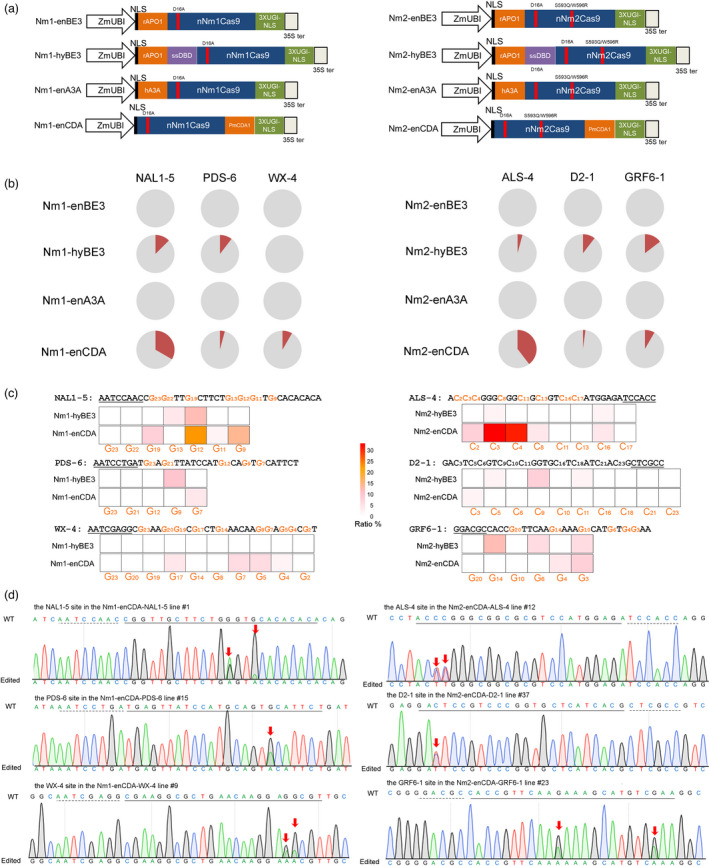
Nm‐CBEs introduce C‐to‐T conversions in rice. (a) Schematic representation of the CBEs derived from nNmCas9s. rAPO1, rat APOBEC1; hA3A, human APOBEC3a; PmCDA1, Petromyzon marinus cytidine deaminase 1; 3XUGI‐NLS, three copies of UGI‐NLS separated by a T2A self‐cleaving peptide; ssDBD, single‐stranded DNA‐binding domain from human Rad51 protein. (b) Frequency of mutants induced by Nm‐CBEs in the rice genome. For each sgRNA, 48–96 independent T0 lines were randomly selected and genotyped. Grey, wild‐type plants; red, plants harbouring base editing. (c) Heatmap of cytosine base conversions induced by Nm‐CBEs. The target regions are indicated, and the positions of targetable nucleotides distal to the PAM are labelled. The substitution frequency of each base was calculated as the percentage of edited plants among the total plant population. (d) Sequencing chromatogram of the edited plants. The C∙G‐to‐T∙A base conversions are labelled by a red arrow.
In addition to rAPO1, a variety of cytidine deaminases, such as human APOBEC3A (hA3A) and Petromyzon marinus cytidine deaminase 1 (PmCDA1), have been harnessed to achieve flexible editing in plants. Subsequently, Nm‐enA3As were developed by replacing rAPO1 with hA3A. Nm‐enCDAs were produced by fusing PmCDA1 to the C terminus of the nickase of NmCas9s with additional copies of UGI (Figure 4a). All of the vectors were examined at the three targets in the transgenic plants. Consistent with the editing mediated by Nm‐enBE3s, no edits were achieved with Nm‐enA3As. However, C‐to‐T conversions were introduced by Nm‐enCDAs at all tested targets. Targeted sequencing showed that 4.2%–33.3% and 2.1%–39.6% of the plants harboured edits in the protospacer region (Figure 4b‐d and Table S4). No indels or non‐C‐to‐T conversions were detected at the targets. The base substitutions induced by Nm1‐enCDA and Nm2‐enCDA were found in regions ranging from position 4 to 19 and from position 2 to 16, respectively, which indicates that Nm‐CBEs have a wide editing window (Figure 4c).
Discussion
A number of Cas9 proteins have been programmed to target the mammalian genome, and the majority of these Cas9s belong to the type II‐C subtype. Previously, a preliminary study using particle bombardment‐mediated transient expression suggested that a type II‐C Cas9 from Brevibacillus laterosporus (BlCas9) could induce genome mutations in immature maize embryos (Karvelis et al., 2015), but the mutagenesis mediated by type II‐C Cas9s in transgenic plants have not yet been reported. Here, we showed that rice mutants can be induced by type II‐C Nm1Cas9 or Nm2Cas9 in a targeted manner via stable transformation. Both Nm1Cas9 and Nm2Cas9 were 1082 aa in length, equivalent to ~78.5% the length of SpCas9. It has been reported that Nm1Cas9 exhibits PAM‐ and tracrRNA‐independent ‘DNase H‐like’ activity in vitro (Zhang et al., 2015), but no significant growth abnormality was observed in the transformed rice calli or regenerated lines, which suggested that Nm1Cas9 is not genotoxic to plant cells. Intriguingly, due to DNase H activity, an enhancement of systematic resistance to single‐stranded DNA (ssDNA) viruses (e.g. plant geminiviruses) can be expected in transgenic plants generated by Nm1Cas9. Our results indicated that the frequency of mutants induced by Nm1Cas9‐esgRNA was as high as 60.4%–89.6% in the regenerated population, which is comparable to the efficiency of the SpCas9 system in rice. However, the targetable sites of Nm1Cas9 would be much lower than those of SpCas9 because the N4GATT PAM occurred once in a 128‐bp random sequence (Lee et al., 2016). In mammals, Nm1Cas9 tolerates mutated PAMs, such as N4GCTT, N4GTTT, N4GACT, N4GATA, N4GACA, and N4GTCT, and could achieve efficiencies approaching those generated at the N4GATT PAM (Amrani et al., 2018). Unfortunately, Nm1Cas9 exhibited reduced editing activity at the N4GCTT PAM in rice, which suggested that the application may be limited to noncanonical PAMs in plants. On the other hand, the dinucleotide PAM of Nm2Cas9 represents a much higher target density than Nm1Cas9 in the genome (Edraki et al., 2019). Although Nm2Cas9 may be less active in plants, its targeting efficiency can be greatly enhanced by the S593Q/W596R double mutation in the HNH domain (Figure 2c). Together, our results demonstrate that the engineered and optimized Nm1Cas9 and Nm2Cas9 could complement the convenient SpCas9 system by providing efficient and reliable editing at their unique PAMs in plants. Moreover, these data strongly suggest that type II‐C orthologs, in addition to type II‐A Cas9s, could be used for the engineering of plant genome‐editing tools. Considering the large numbers of type II‐C cas9 orthologs with divergent PAM preferences and varied biochemical identities (Mir et al., 2018), we can anticipate the development of a wide variety of orthogonal tools for implementation in different scenarios of plant genome editing.
The high frequency of base conversions by BEs was generally restricted to a narrow ‘window,’ for example, ~5 bp for BE3 or ABE, in the protospacer distal to PAM (Rees and Liu, 2018). Therefore, a BE toolbox harbouring flexible PAM compatibility is desired to access a high density of genomic targets. To increase the scope of base editing, SpCas9 in the convenient BE system was replaced by various SpCas9 variants, type II‐A Cas9 orthologs, and Cas12a members with relaxed or alternative PAM recognition (Rees and Liu, 2018; Richter et al., 2020). However, few studies have shown the development of BEs from type II‐C Cas9 systems in plants or mammals. Due to weak DNA helicase activity (Ma et al., 2015), type II‐C Cas9s may form an R‐loop structure at lower frequencies than type II‐A Cas9s, which is essential to expose the ssDNA in the editing window to the deaminase of the BE complex. In this study, our data showed extremely limited activity of Nm‐ABEs, Nm‐enBE3s, and Nm‐enA3As, confirming the low compatibility of type II‐Cas9s in BE engineering. Recently, the deamination reaction catalysed by ABE8e was found to be more than 1000‐fold faster than that catalysed by conventional ABEs (Lapinaite et al., 2020), and PmCDA1‐based CBEs also robustly exhibited enhanced editing capability compared with those of rAPO1 and hA3A in plants (Ren et al., 2021). Consistent with the results, we also found that the Nm‐ABE8es and Nm‐enCDAs could introduce targeted nucleotide substitutions at frequencies as high as 87.5% and 39.6% in the regenerated plants respectively. Although the base editing frequencies may be locus dependent, we believe that more efficient and robust BEs could be established from type II‐C Cas9s with more active deaminases. In addition, the fusing of a RAD51‐ssDBD in mammalian cells increased the editing frequencies of BE4max and eA3A‐BE4max by up to 18‐fold and 257‐fold by (Zhang et al., 2020). Our data confirmed that the insertion of RAD51‐ssDBD between rAPO1 and the nickase also improved the editing activity of Nm‐enBE3s. Based on this strategy, it is logical to presume that optimized Nm‐BEs with RAD51‐ssDBD may further improve the base editing activity. During the preparation of this manuscript, a patent was claimed by Keith Joung for constructing CBEs from type II‐C Cas9 orthologs by further fusing an adjacently targeted transcription activator‐like effector or zinc finger DNA‐binding domain (Joung et al., 2020), providing another development strategy for Nm‐BEs. Taken together, the results show that NmCas9s and other type II‐C Cas9 orthologs can potentially be used to develop BEs for efficient base substitutions in plants and other organisms.
Methods
Vector construction
Monocot codon‐optimized Nm1Cas9 and Nm2Cas9 sequences were synthesized (Appendix S1; Genscript, Nanjing, China). The fragment of Nm1Cas9 or Nm2Cas9 (Appendix S1) was inserted into the pHUC400 backbone to replace the SpCas9 fragment via NotI/SacI digestion and thus construct pHUC7a00 or pHUC7b00 respectively (Xing et al., 2014). For the Nm2Cas9‐S593Q/W596R mutant, site‐specific mutations were induced with the Fast Mutagenesis System (Transgen, Beijing, China). The sgRNA scaffolds of NmCas9s were also synthesized (Figure 1b). The expression cassette of OsU3‐sgRNA was then inserted into the HindIII‐predigested pHUC7a00 or pHUC7b00 vector by Gibson cloning. In the resulting pHUC7a11 (Nm1Cas9) and pHUC7b11 (Nm2Cas9) constructs, a spectinomycin resistance (SpR) gene was placed between the OsU3 promoter and sgRNA scaffold by using two BsaI digestion sites, similar to the structure of the pHUN series plasmids (Xing et al., 2014). For the construction of gRNAs, the sense and antisense strands of the 24‐nt protospacer were synthesized with the appropriate adapter. The oligos were then annealed and linked to BsaI‐predigested pHUC711 vectors. Kanamycin‐positive and spectinomycin‐negative clones were selected and confirmed by Sanger sequencing.
To construct the BEs, a D16A mutation was introduced into Nm1Cas9 or Nm2Cas9‐S593Q/W596R by site‐specific mutation. The fragment of the TadA*7.10 mutant but not the wild‐type TadA was amplified from the previously reported rice codon‐optimized SpCas9‐ABE (Li et al., 2019). TadA*7.10 was added to the 5’ termini of the nNm‐Cas9s by Gibson cloning. NLSs were then further added by direct PCR to develop Nm‐ABEs (Li et al., 2018). For the construction of Nm‐ABE8es, the TadA‐8e sequence was amplified from SpG‐ABE8e (Li et al., 2021) and assembled with nNmCas9s‐3XNLS.
The sequences of rAPO1‐XTEN and 3XUGI (UGI‐NLS‐T2A‐UGI‐NLS‐T2A‐UGI‐NLS) were amplified from a formerly constructed SpCas9‐eBE3 plasmid (Qin et al., 2019b) and assembled into nNm‐Cas9s to generate Nm‐enBE3s. Similarly, the hA3A and PmCDA1 fragments were amplified from SpG‐eA3A and SpCas9‐eCDA (Li et al., 2021; Qin et al., 2019b) respectively. Then, they were separately assembled with nNmCas9s and 3XUGI to form Nm‐enA3As and Nm‐enCDAs. To generate Nm‐hyBE3, a japonica rice codon‐optimized RAD51‐ssDBD sequence was synthesized and inserted between rAPO1 and nNmCas9 with 32‐aa linkers on both sides (Appendix S1). All assembled sequences were confirmed by Sanger sequencing and inserted into the pHUC400 backbone by NotI/SacI or PstI/SacI digestion.
Rice transformation
The binary vectors were transformed into Agrobacterium strain EHA105 via the freeze–thaw method. The positive clones were confirmed by Sanger sequencing of the sgRNA region. Rice transformation was performed following a previously described protocol with some modifications (Hu et al., 2016). Mature seeds of Oryza sativa L. sub. japonica Nipponbare were dehulled and sterilized for callus induction. The embryotic calli were infected by agrobacteria and selected by 50 mg/L hygromycin for 4 weeks. For each resistance event, two to three well‐grown calli were selected and transformed for regeneration. After another 4–5 weeks, the regenerated shoots were transformed to rooting medium. In each event, only one line was selected for further growth and genotyping. All rice material was incubated in a growth chamber with a 16‐h light:8‐h dark photoperiod at 30 °C–32 °C.
Sampling and genotyping
For each transgenic event, more than three leaves from different tillers were collected as a single sample. Genomic DNA was extracted following the cetyltrimethylammonium bromide (CTAB) method. The targeted mutations were identified by amplicon Sanger sequencing and/or NGS‐based high‐throughput tracking of mutation (Hi‐TOM) assays with a 5% threshold (Liu et al., 2019). To determine the efficiency of Cas9 in rice cells, >200 newly emerged resistant calli were collected from each transformant. The activities of SpCas9 versus Nm1Cas9 or Nm2Cas9 versus Nm2Cas9‐S593Q/W596R were then compared by amplicon NGS using an Illumina HiSeq X Ten platform, and the data can be retrieved from the SRA database (No. PRJNA742144).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
P.W. designed the experiments and wrote the manuscript with input from all the authors. P.W., P.L., and Y.Y. supervised the project. R.X., R.Q., H.X., and J.L. performed the vector construction and genotyping. X.L., M.Z., and Y.S. transformed the plants. R.X., R.Q., and P.L. analysed the data.
Supporting information
Appendix S1 Supplemental sequences.
Figure S1 Alignment of mutations in the transgenic plants of the Nm1Cas9‐esgRNA complex.
Figure S2 Targeted mutagenesis on the NAL1‐2 site by SpCas9 and Nm1Cas9.
Figure S3 Identification of targeted mutants at N4GCTT PAM targets in CRISPR‐Nm1Cas9 transgenic plants.
Figure S4 Identification of targeted mutants induced by Nm2Cas9 with different lengths of gRNAs
Figure S5 Alignment of mutations in the Nm2Cas9‐S593Q/W596R transgenic plants.
Table S1 Off‐target analysis of Nm1Cas9 in transgenic plants.
Table S2 Off‐target analysis of Nm2Cas9 in transgenic plants.
Table S3 Targeted editing induced by Nm‐ABEs in transgenic rice plants.
Table S4 Targeted editing induced by Nm‐CBEs in transgenic rice plants.
Table S5 Oligos used in the study.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. U19A2022 and No. 32000284), the Natural Science Foundation of Anhui Province (No. 2108085Y07, No. 2008085QC101 and No. 2008085MC71), the Open Project Funding of the State Key Laboratory of Crop Stress Adaptation and Improvement (No. 2020KF04), the Key Laboratory of Indica Rice Genetics and Breeding in the Middle and Lower Reaches of the Yangtze River Valley Foundation (No. 2018KLMA01), the Science and Technology Major Projects of Anhui Province (No. 202003a06020009), the University Synergy Innovation Program of Anhui Province (No. GXXT‐2021‐056), and the Open Project Program of the State Key Laboratory of Rice Biology (No. 20210103).
Xu, R. , Qin, R. , Xie, H. , Li, J. , Liu, X. , Zhu, M. , Sun, Y. , Yu, Y. , Lu, P. and Wei, P. (2022) Genome editing with type II‐C CRISPR‐Cas9 systems from Neisseria meningitidis in rice. Plant Biotechnol. J., 10.1111/pbi.13716
[Correction added on 15 January 2022, after first online publication: The affiliations of Yang Sun and Pingli Lu have been corrected in this version.]
Contributor Information
Pingli Lu, Email: pinglilu@henu.edu.cn.
Pengcheng Wei, Email: weipengcheng@gmail.com.
References
- Amrani, N. , Gao, X.D. , Liu, P. , Edraki, A. , Mir, A. , Ibraheim, R. , Gupta, A. et al. (2018) NmeCas9 is an intrinsically high‐fidelity genome‐editing platform. Genome Biol. 19, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S. , Park, J. and Kim, J.‐S. (2014) Cas‐OFFinder: a fast and versatile algorithm that searches for potential off‐target sites of Cas9 RNA‐guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, P. , Jakimo, N. and Jacobson, J.M. (2018) Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 4, eaau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, P. , Lee, J. , Nip, L. , Koseki, S.R.T. , Tysinger, E. , Sontheimer, E.J. , Jacobson, J.M. et al. (2020) A Cas9 with PAM recognition for adenine dinucleotides. Nat. Commun. 11, 2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Chylinski, K. , Makarova, K.S. , Charpentier, E. and Koonin, E.V. (2014) Classification and evolution of type II CRISPR‐Cas systems. Nucleic Acids Res. 42, 6091–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edraki, A. , Mir, A. , Ibraheim, R. , Gainetdinov, I. , Yoon, Y. , Song, C.‐Q. , Cao, Y. et al. (2019) A compact, high‐accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell, 73, 714–726.e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, K.M. , Mali, P. , Braff, J.L. , Moosburner, M. , Yaung, S.J. and Church, G.M. (2013) Orthogonal Cas9 proteins for RNA‐guided gene regulation and editing. Nat. Methods, 10, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova, I. , Arseniev, A. , Selkova, P. , Pobegalov, G. , Goryanin, I. , Vasileva, A. , Musharova, O. et al. (2020) DNA targeting by Clostridium cellulolyticum CRISPR–Cas9 Type II‐C system. Nucleic Acids Res. 48, 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.B. , Paez‐Espino, D. , Staahl, B.T. , Chen, J.S. , Ma, E. , Kyrpides, N.C. and Doudna, J.A. (2017) A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 8, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, H. , Gootenberg, J.S. , Horii, T. , Abudayyeh, O.O. , Kimura, M. , Hsu, P.D. , Nakane, T. et al. (2016) Structure and engineering of Francisella novicida Cas9. Cell, 164, 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S. , Abudayyeh, O.O. , Gootenberg, J.S. , Horii, T. , Ishitani, R. , Hatada, I. , Zhang, F. et al. (2019) Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9. Nat. Commun. 10, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M.D. , Aschenbrenner, S. , Grosse, S. , Rapti, K. , Domenger, C. , Fakhiri, J. , Mastel, M. et al. (2019) Cell‐specific CRISPR–Cas9 activation by microRNA‐dependent expression of anti‐CRISPR proteins. Nucleic Acids Res. 47, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z. , Zhang, Y. , Propson, N.E. , Howden, S.E. , Chu, L.‐F. , Sontheimer, E.J. and Thomson, J.A. (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl Acad. Sci. 110, 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Li, H. , Qin, R. , Xu, R. , Li, J. , Li, L. , Wei, P. and et al. (2016) Plant phosphomannose isomerase as a selectable marker for rice transformation. Sci. Rep. 6, 25921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Wang, S. , Zhang, C. , Gao, N. , Li, M. , Wang, D. , Wang, D. et al. (2020) A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 18, e3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. and Zhu, J.‐K. (2019) Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 17, 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. , Liang, W. , Zhang, Z. , Gou, R. and Zhu, J.‐K. (2020) Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J. 18, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheim, R. , Song, C.‐Q. , Mir, A. , Amrani, N. , Xue, W. and Sontheimer, E.J. (2018) All‐in‐one adeno‐associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 19, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, K.J. , Angstman, J. , Gehrke, J.M. and Grunewald, J. (2020) Bipartite Base Editor (BBE) Architectures and TYPE‐II‐C‐CAS9 Zinc Finger Editing. US Patent App. 16/616,014. [Google Scholar]
- Karvelis, T. , Gasiunas, G. , Young, J. , Bigelyte, G. , Silanskas, A. , Cigan, M. and Siksnys, V. (2015) Rapid characterization of CRISPR‐Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, H. , Mikami, M. , Endo, A. , Endo, M. and Toki, S. (2016) Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6, 26871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. , Koo, T. , Park, S.W. , Kim, D. , Kim, K. , Cho, H.‐Y. , Song, D.W. et al. (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Topkar, V.V. , Nguyen, N.T. , Zheng, Z. , Gonzales, A.P.W. et al.(2015) Engineered CRISPR‐Cas9 nucleases with altered PAM specificities. Nature, 523, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinaite, A. , Knott, G.J. , Palumbo, C.M. , Lin‐Shiao, E. , Richter, M.F. , Zhao, K.T. , Beal, P.A. et al. (2020) DNA capture by a CRISPR‐Cas9–guided adenine base editor. Science, 369, 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.M. , Cradick, T.J. and Bao, G. (2016) The Neisseria meningitidis CRISPR‐Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 24, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Mou, H. , Ibraheim, R. , Liang, S.‐Q. , Liu, P. , Xue, W. and Sontheimer, E.J. (2019) Tissue‐restricted genome editing in vivo specified by microRNA‐repressible anti‐CRISPR proteins. RNA, 25, 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zong, Y. , Wang, Y. , Jin, S. , Zhang, D. , Song, Q. , Zhang, R. et al. (2018) Expanded base editing in rice and wheat using a Cas9‐adenosine deaminase fusion. Genome Biol. 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Qin, R. , Liu, X. , Liao, S. , Xu, R. , Yang, J. and Wei, P. (2019) CRISPR/Cas9‐mediated adenine base editing in rice genome. Rice Sci. 26, 125–128. [Google Scholar]
- Li, J. , Xu, R. , Qin, R. , Liu, X. , Kong, F. and Wei, P. (2021) Genome editing mediated by SpCas9 variants with broad non‐canonical PAM compatibility in plants. Molecular Plant, 14, 352–360. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Ma, E. , Harrington, L.B. , O’Connell, M.R. , Zhou, K. and Doudna, J.A. (2015) Single‐stranded DNA cleavage by divergent CRISPR‐Cas9 enzymes. Mol. Cell, 60, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir, A. , Edraki, A. , Lee, J. and Sontheimer, E.J. (2018) Type II‐C CRISPR‐Cas9 biology, mechanism, and application. ACS Chem. Biol. 13, 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, C.M. , Lawson, S. , Zerez, M. and Bleris, L. (2016) Guide RNA engineering for versatile Cas9 functionality. Nucleic Acids Res. 44, 9555–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, R. , Li, J. , Li, H. , Zhang, Y. , Liu, X. , Miao, Y. , Zhang, X. et al. (2019a) Developing a highly efficient and wildly adaptive CRISPR‐SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 17, 706–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, R. , Liao, S. , Li, J. , Li, H. , Liu, X. , Yang, J. and Wei, P. (2019b) Increasing fidelity and efficiency by modifying cytidine base‐editing systems in rice. Crop J. 8, 396–402. [Google Scholar]
- Ran, F.A. , Cong, L. , Yan, W.X. , Scott, D.A. , Gootenberg, J.S. , Kriz, A.J. , Zetsche, B. et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, H.A. and Liu, D.R. (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Q. , Sretenovic, S. , Liu, G. , Zhong, Z. , Wang, J. , Huang, L. , Tang, X. et al. (2021) Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol. J, 19, 2052–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M.F. , Zhao, K.T. , Eton, E. , Lapinaite, A. , Newby, G.A. , Thuronyi, B.W. , Wilson, C. et al. (2020) Phage‐assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, T.R. , Saroj, S.D. , Llewellyn, A.C. , Tzeng, Y.‐L. and Weiss, D.S. (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature, 497, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksnys, V. and Gasiunas, G. (2016) Rewiring Cas9 to target new PAM sequences. Mol. Cell, 61, 793–794. [DOI] [PubMed] [Google Scholar]
- Sretenovic, S. , Yin, D. , Levav, A. , Selengut, J.D. , Mount, S.M. and Qi, Y. (2020) Expanding plant genome‐editing scope by an engineered iSpyMacCas9 system that targets A‐rich PAM sequences. Plant Commun. 2, 100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert, J. , Schiml, S. , Fauser, F. and Puchta, H. (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Sun, W. , Yang, J. , Cheng, Z. , Amrani, N. , Liu, C. , Wang, K. , Ibraheim, R. et al. (2019) Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti‐CRISPR‐inhibited states. Mol. Cell, 76, 938–952.e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Xu, Z. , Gosavi, G. , Ren, B. , Cao, Y. , Kuang, Y. , Zhou, C. et al. (2020) Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J, 18(8), 1645–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. , Terns, R.M. and Terns, M.P. (2015) Cas9 function and host genome sampling in Type II‐A CRISPR–Cas adaptation. Genes Dev. 29, 356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.‐L. , Dong, L. , Wang, Z.‐P. , Zhang, H.‐Y. , Han, C.‐Y. , Liu, B. , Wang, X.‐C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Ren, C. , Liu, Z. , Zhang, T. , Zhang, T. , Li, D. , Wang, L. et al. (2015) Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell. Mol. Life Sci. 72, 383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Meng, X. , Wang, J. , Qin, B. , Wang, K. , Li, J. , Wang, C. et al. (2020) ScCas9 recognizes NNG protospacer adjacent motif in genome editing of rice. Sci. China Life Sci. 63, 450–452. [DOI] [PubMed] [Google Scholar]
- Yan, D. , Ren, B. , Liu, L. , Yan, F. , Li, S. , Wang, G. , Sun, W. et al. (2021) High‐efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant, 14, 722–731. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Zheng, Q. , Yi, X. , An, H. , Zhao, Y. , Ma, S. and Zhou, G. (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Chen, L. , Zhu, B. , Wang, L. , Chen, C. , Hong, M. , Huang, Y. et al. (2020) Increasing the efficiency and targeting range of cytidine base editors through fusion of a single‐stranded DNA‐binding protein domain. Nat. Cell Biol. 22, 740–750. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Heidrich, N. , Ampattu, B.J. , Gunderson, C.W. , Seifert, H.S. , Schoen, C. , Vogel, J. et al. (2013) Processing‐independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell, 50, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Rajan, R. , Seifert, H.S. , Mondragón, A. and Sontheimer, E.J. (2015) DNase H activity of Neisseria meningitidis Cas9. Mol. Cell, 60, 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplemental sequences.
Figure S1 Alignment of mutations in the transgenic plants of the Nm1Cas9‐esgRNA complex.
Figure S2 Targeted mutagenesis on the NAL1‐2 site by SpCas9 and Nm1Cas9.
Figure S3 Identification of targeted mutants at N4GCTT PAM targets in CRISPR‐Nm1Cas9 transgenic plants.
Figure S4 Identification of targeted mutants induced by Nm2Cas9 with different lengths of gRNAs
Figure S5 Alignment of mutations in the Nm2Cas9‐S593Q/W596R transgenic plants.
Table S1 Off‐target analysis of Nm1Cas9 in transgenic plants.
Table S2 Off‐target analysis of Nm2Cas9 in transgenic plants.
Table S3 Targeted editing induced by Nm‐ABEs in transgenic rice plants.
Table S4 Targeted editing induced by Nm‐CBEs in transgenic rice plants.
Table S5 Oligos used in the study.


