Abstract
Feces are enriched with microRNAs (miRNAs) that shape the gut microbiota. These miRNAs are differentially expressed in the feces of healthy and diseased subjects. However, whether fecal miRNAs in subjects with inflammatory bowel diseases are involved in regulating microbiota composition and whether they have any beneficial effects remains unknown. Here, we studied the fecal microbiome composition and miRNA abundance in mice with dextran sulfate sodium (DSS)-induced colitis and mice at the recovery phase to explore different miRNAs expressed, their relations with microbial abundance, and their effects on colitis. We found that miR-142a-3p expression was significantly increased in the feces of mice recovered from colitis and that it could alleviate disease symptoms in mice treated with DSS in a microbiome-dependent manner. Specifically, miR-142a-3p promoted the growth of Lactobacillus reuteri, which had a high abundance in the feces of mice recovered from colitis, by regulating transcripts of polA and locus tag LREU_RS03575. Moreover, L. reuteri, as well as its metabolite reuterin, could alleviate DSS-induced disease symptoms. These results highlight the role of fecal miR-142a-3p in the prevention of colitis. We propose that the feces of subjects who have recovered from diseases might be enriched with miRNAs with preventive effects against those diseases.
Keywords: colitis, Lactobacillus reuteri, microbiome, miRNAs, reuterin
Graphical abstract
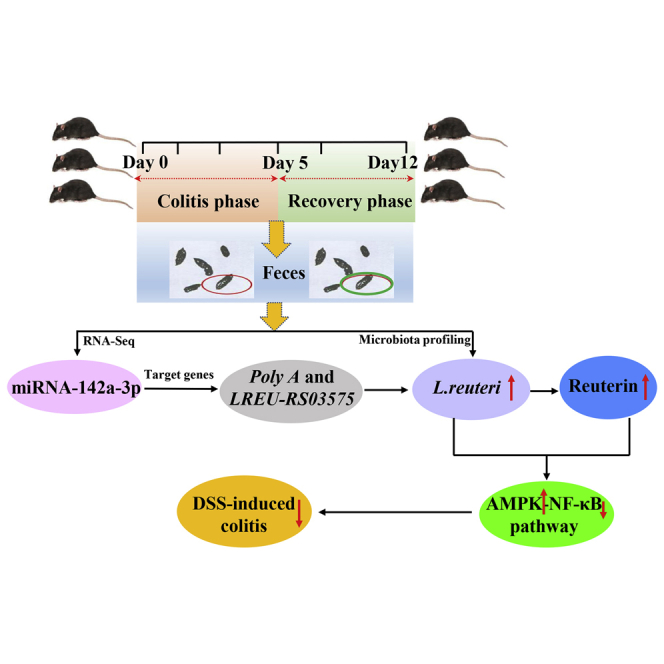
Feces are enriched with miRNAs that shape the gut microbiota. These miRNAs are differentially expressed in the feces of healthy and diseased subjects. This study proposed that the feces of subjects who have recovered from diseases might be enriched with miRNAs with preventive effects against those diseases via targeting microbiota.
Introduction
Inflammatory bowel disease (IBD) is a clinically important, chronic inflammatory disorder of the gastrointestinal tract. IBD is highly prevalent in Europe and North America, and its incidence has been rising in developing countries in recent times.1 Although the pathogenesis of IBD is still poorly understood, increasing evidence suggests that microbiota and their derived metabolites play a key role in the development of this disease, based on the results from several animal models.2, 3, 4 Genetically susceptible mice under germ-free conditions do not develop colitis.5 whereas healthy mice developed inflammation after transplantation of microbiota from mice with colitis.6,7 Moreover, colonization of mice with intestinal microbiota from diseased mice exacerbates colitis.8 However, although it is commonly considered that the bidirectional host-microbiome interactions play key roles in the pathogenesis of IBD, the related mechanisms remain to be uncovered. Consequently, it is important to explore the mechanisms by which the host modulates the microbiome.
MicroRNAs (miRNAs) are noncoding RNAs that are approximately 22 nucleotides long. Traditionally, miRNAs are produced as precursor miRNAs in the nucleus, which are then processed to generate mature miRNAs that become functional in the cytoplasm.9 However, miRNAs have also been found extracellularly in the body fluids in recent years.10 Importantly, a recent study found that gut miRNAs exists in feces and can regulate the gut microbiome.11 Surprisingly, the feces of multiple sclerosis patients are enriched with miRNAs that exert therapeutic effects via a miRNA-microbiome axis.12 However, whether miRNAs in the feces of subjects with IBD or of subjects recovered from the disease had preventive or therapeutic properties, and the mechanisms by which miRNAs regulate the development and pathogenesis of IBD, remains unknown.
Therefore, we developed a widely acceptable animal model of IBD via administration of dextran sulfate sodium (DSS) in mice. Then, we studied the gut microbiome and miRNAs in mice with DSS-induced colitis and in well-recovered mice to explore different miRNAs expressed and their relations with microbial abundance, as well as their effects on colitis. We found that miR-142a-3p expression was significantly increased in the feces of mice recovered from colitis and that it could alleviate disease symptoms in mice treated with DSS via regulating Lactobacillus reuteri. Specifically, miR-142a-3p could promote the growth of L. reuteri by targeting transcripts of polA and locus tag LREU_RS03575. Our results suggest that fecal miR-142a-3p plays key roles in the alleviation of colitis. We propose that the feces of subjects who have recovered from diseases might be enriched with miRNAs with preventive effects against those diseases.
Results
Changes in fecal microbiota composition during DSS-induced colitis and the recovery phase
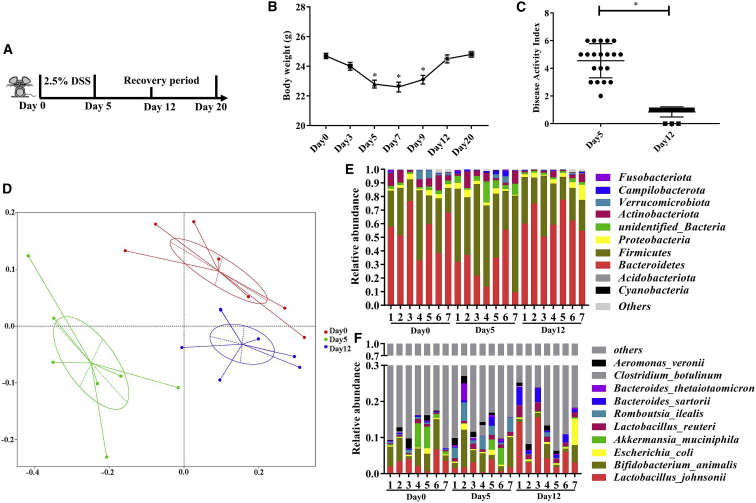
Initially, the mice were treated with 2.5% DSS in the drinking water for 5 days. Then, all the mice were provided DSS-free drinking water for the following days. On day 12, 38.5% of mice (20 of 52) whose body weight returned to the same level as on day 0 (the change of body weight was less than 1.0 g) and exhibited a disease activity index (DAI) of 0 or 1.00 were used as “well-recovered” mice. We found that their body weight was significantly decreased on day 5 (Figures 1A and 1B). Moreover, these mice showed an average DAI of 4.55 on day 5 (Figure 1C). These results suggested that the mice were experiencing acute colitis on day 5.
Figure 1.
Changes in fecal microbiota composition during DSS-induced colitis and the recovery phase
(A) Schematic design. (B) Body weight during the experiment (one-way ANOVA). (C) Disease activity index after DSS treatment and at recovery phase (one-tailed t test). (D–F) Microbiome analyses based on bacterial 16S rDNA sequence were performed. (D) Principal coordinates analysis (PCoA) based on weighted UniFrac distance. (E) Relative abundance of bacteria classified at a phylum-level taxonomy. (F) Relative abundance of bacteria classified at a species-level taxonomy. Values are mean ± SEM, n = 7. ∗p < 0.05. Abbreviations: Day 0, naive mice before treated with DSS; Day 5, mice treated with DSS for 5 consecutive days; Day 12, mice treated with DSS-free water for another 7 days after DSS treatment.
We then investigated the intestinal microbial alteration via 16S rDNA sequencing of feces collected from well-recovered mice on days 0, 5, and 12 of the treatment regimen (Figure 1A). Although no significant difference was observed in the alpha diversity (Figures S1A and S1B), the analysis of the microbial beta-diversity metric using the principal coordinates analysis (PCoA) based on weighted UniFrac distance revealed that the overall microbial structure in mice feces on day 5 was segregated away from the microbial structure in mice feces on day 0 and day 12 (Figure 1D). Additionally, no significant beta-diversity differences were observed using unweighted UniFrac distance among the feces collected from mice on days 0, 5, and 12 of the treatment regimen, since they were not separated into distinct clusters (Figure S1C).
To identify the specific intestinal microbes that were influenced by DSS treatment, we compared the relative abundance of the intestinal microbial species identified in the feces of well-recovered mice collected on days 0, 5, and 12. DSS decreased the abundance of Bacteroidetes, while increasing that of Firmicutes at the phylum level (Figure 1E); however, the abundance of Bacteroidetes and Firmicutes recovered on day 12. Additionally, our results show that the relative abundance of Romboutsia ilealis was increased, whereas that of Lactobacillus johnsonii was decreased in mice on day 5 compared with the abundance in mice on day 0 of the treatment regimen (Figure 1F). Notably, the relative abundance of L. reuteri and L. johnsonii in mice on day 12 was significantly increased compared to that in mice on day 0 and day 5. We further analyzed the changes in the abundance of L. reuteri via fecal microbe quantification using qPCR, and the results showed that its abundance was significantly higher on days 12 and 20 than that on days 0, 5, and 7 (Figure S1D).
Oral transfer of feces and fecal RNA from recovered mice prevents DSS-induced colitis
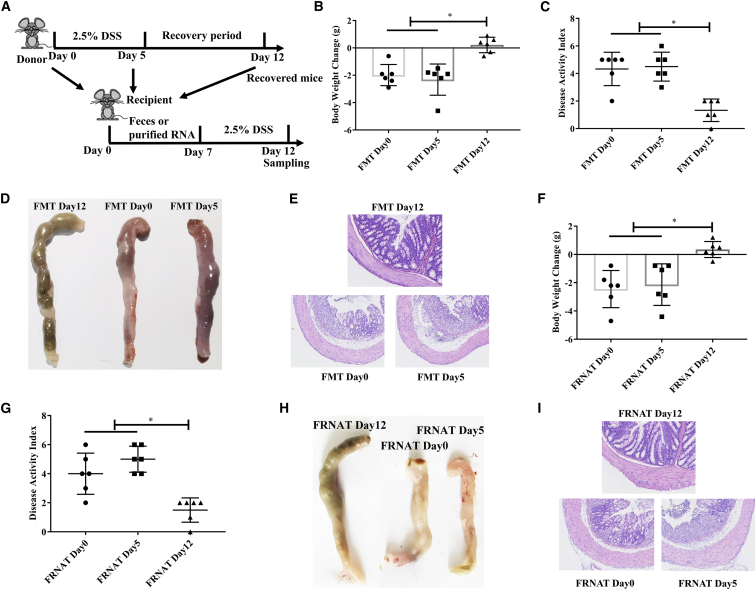
To explore whether the gut microbiome from mice recovered from DSS-induced colitis had any effect on DSS-induced colitis, we transplanted feces obtained from those well-recovered mice on days 0, 5, and 12 (Figure 2A). The results showed that the transfer of feces from mice obtained on day 12, but not on day 0 and day 5, alleviated body weight loss (Figure 2B), decreased the DAI (Figure 2C), and prevented the colon from swelling and bleeding (Figures 2D and 2E) in DSS-treated mice.
Figure 2.
Oral transfer of feces and fecal RNA from recovered mice prevents DSS-induced colitis
(A) Schematic design. (B–D) The effects of the transfer of feces from different stages (day 0, day 5, and day 12) of DSS-induced colitis mice on colitis in recipient mice. (B) Body weight change. (C) Disease activity index. (D) Representative colons. (E). Representative colonic morphology. Abbreviations: FMT Day 0, FMT Day 5, and FMT Day 12, mice were transplanted with feces collected from well-recovered mice on days 0, 5, and 12 of the treatment regimen. (F–I) The effects of oral administration of fecal RNA on colitis. (F) Body weight change. (G) Disease activity index. (H) Representative colons. (I) Representative colonic morphology. Abbreviations: FRNAT Day 0, FRNAT Day 5, and FRNAT Day 12, mice were transplanted with RNA extracted from feces collected from well-recovered mice on days 0, 5, and 12 of the treatment regimen. Data were analyzed with one-way ANOVA. Values are mean ± SEM, n = 6. ∗p < 0.05.
To investigate whether live bacteria in well-recovered mice were responsible for these effects, we heat-inactivated feces obtained from mice on days 0, 5, and 12 and then transplanted them. The results showed that only heat-inactivated feces obtained from mice on day 12 alleviated body weight loss and decreased the DAI (Figure S2). These results suggested that live bacteria are not indispensable for the alleviation of colitis.
It is suggested that miRNAs are heat resistant,13 and miRNAs could prevent DSS-induced colitis in mice.14 Consequently, to further explore whether fecal miRNAs were responsible for these effects, RNAs were purified from feces obtained from well-recovered mice on days 0, 5, and 12 of the treatment regimen. The results showed that RNA purified from the feces of mice obtained on day 12, but not on day 0 and day 5, alleviated body weight loss (Figure 2F), decreased the DAI (Figure 2G), and prevented the colon from swelling and bleeding (Figures 2H and 2I) in DSS-treated mice. These results suggested that RNA in feces may play a crucial role in alleviation of DSS-induced colitis.
Identification of differentially expressed fecal miRNA during DSS-induced colitis and the recovery phase
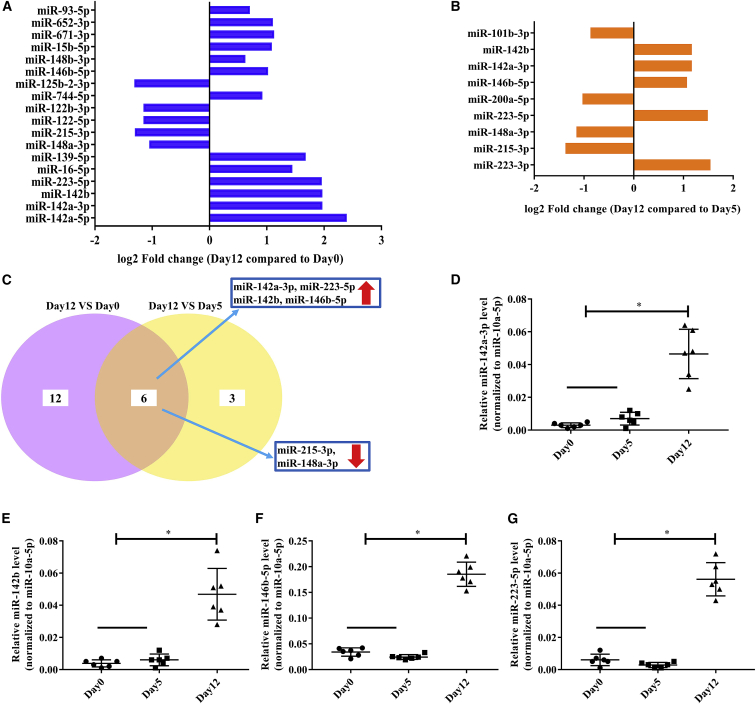
Previous studies have shown that DSS-induced colitis results in altered fecal miRNA profiles.11 To further identify which fecal miRNAs were affected during DSS-induced colitis and the recovery phase, we performed small RNA sequencing in feces obtained from well-recovered mice on days 0, 5, and 12 of the treatment regimen. The results showed that the levels of thirteen miRNAs were increased and those of five miRNAs were decreased in feces obtained from mice on day 12 relative to those in the feces of mice on day 0 of the treatment (Figure 3A), whereas the levels of five miRNAs were increased and those of four miRNAs were decreased in feces obtained from mice on day 12 relative to those in the feces of mice on day 5 of the treatment regimen (Figure 3B). Among these changes, the levels of four miRNAs (miR-142a-3p, miR-223-5p, miR-142b, and miR-146b-5p) were increased, and those of two miRNAs (miR-215-3p and miR-148a-3p) were decreased in feces obtained from mice on day 12 (compared with feces obtained from mice on days 0 or 5 of the treatment regimen; Figure 3C). Then, we confirmed the increase in the levels of those four miRNAs in the feces of recovered mice by qPCR (Figures 3D–3G). Additionally, we found that miR-142a-3p expression on day 20 was the same as that on day 12, but it was higher than that on day 7 (Figure 3D; Figure S3).
Figure 3.
Identification of differentially expressed fecal miRNA during DSS-induced colitis and the recovery phase
(A–G) RNA was isolated from feces collected from well-recovered mice on days 0, 5, and 12 of the treatment regimen and analyzed by small RNA sequencing. (A) Differentially expressed miRNAs in feces collected on day 12 compared to those of on day 0. (B) Differentially expressed miRNAs in feces collected on day 12 compared to those of on day 5. (C) Differentially expressed miRNAs in feces collected on day 12 when compared to those of either day 0 or day 5. Relative expression of miR-142a-3p (D), miR-142b (E), miR-146b-5p (F), and miR-223-5p (G) were verified by qPCR when normalized by miR-10a-5p. Data were analyzed with one-way ANOVA. Values are mean ± SEM, n = 6. ∗p < 0.05.
Previous study suggested that intestinal epithelial cells are the main source of fecal miRNA.11 We next treated Caco-2 cells with 2% DSS (w/v) to determine the changes in miRNA levels. The results showed that DSS enhanced the secretion of inflammatory cytokines from Caco-2 cells, as concentrations of TNF-α, IL-6, and IL-8 in the culture medium were significantly increased (Figure S4A). This suggested that DSS induced inflammatory responses in intestinal epithelial cells. Moreover, we found that miR-142a-3p was upregulated, while the expression of miR-223-5p, miR-142b, and miR-146b-5p was not significantly altered, in DSS-treated Caco-2 cells (Figure S4B). We selectively transfected miR-142a-3p agomir and antagomir in Caco-2 cells. The results showed that agomir upregulated miR-142a-3p, while antagomir downregulated miR-142a-3p (Figure S4C). Thus, epithelial cells might account for the change in miR-142a-3p level in feces.
Oral administration of miR-142a-3p alleviates DSS-induced colitis in a microbiome-dependent manner
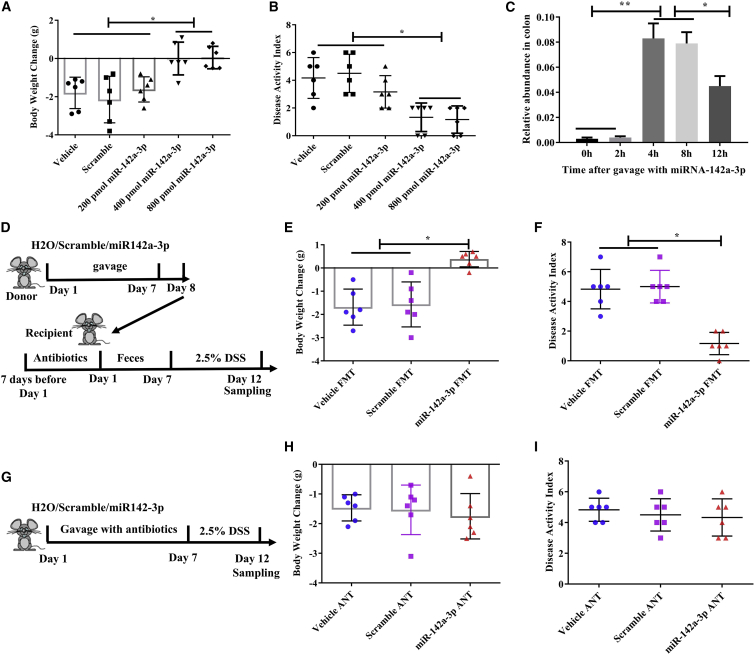
To investigate whether miR-142a-3p, miR-223-5p, miR-142b, and miR-146b-5p affect DSS-induced colitis, we synthesized these four miRNAs and individually gavaged mice with these miRNAs for 7 consecutive days. We first performed a dose-response experiment (0, 200, 400, and 800 pmol) with miR-142a-3p to determine the optimal dose. The results showed that 400 and 800 pmol miR-142a-3p exhibited the same beneficial effects on body weight change and DAI in mice treated with DSS, but the 200-pmol dose did not have any effect (Figures 4A and 4B). Then, we found that miR-223-5p, miR-142b, and miR-146b-5p (400 pmol and 1,000 pmol) and their associated scramble molecules had no effect on body weight change and DAI in mice treated with DSS (Figure S5).
Figure 4.
Oral administration of miR-142a-3p alleviates DSS-induced colitis in a microbiome-dependent way
(A–C) The effects of oral administration of miR-142a-3p on DSS-induced colitis. (A) Body weight change. (B) Disease activity index. (C) Relative abundance of miR-142a-3p in colon contents after oral administration. (D) Schematic design of fecal microbiota transplantation. (E and F) The effects of the transfer of feces from miR-142a-3p-treated mice on colitis. (E) Body weight change. (F) Disease activity index. (G) Schematic design. (H and I) The effects of miR-142a-3p on colitis in antibiotics pretreated mice. (H) Body weight change. (I) Disease activity index. Data were analyzed with one-way ANOVA. Values are mean ± SEM, n = 6. ∗p < 0.05.
We observed that the level of miR-142a-3p was significantly increased in the colon contents after 4 h of oral gavage (Figure 4C), suggesting that miR-142a-3p could reach the colon. Furthermore, we found that the level of fecal miR-142a-3p in mice on day 5 and day 7 during miRNA administration was higher than that in the control mice (Figure S6), confirming the effect of miR-142a-3p on the alleviation of colitis. Previous study has demonstrated that miRNA could shape intestinal microbiota.11 To explore whether miR-142a-3p engendered a protective phenotype, we gavaged the mice with miR-142a-3p for 7 consecutive days, and subsequently fresh feces were obtained and transferred to animals pretreated with antibiotics (Figure 4D). The results showed that microbiome transferred from miR-142a-3p-treated mice prevented the DSS-induced changes in body weight and DAI (Figures 4E and 4F). To further explore whether miR-142a-3p alleviated DSS-induced colitis in a microbiome-dependent manner, we administered antibiotics along with miR-142a-3p (Figure 4G). The results showed that the beneficial effects of miR-142a-3p on body weight and DAI were abrogated by antibiotics (Figures 4H and 4I).
Additionally, the mice were intravenously administered miR-142a-3p agomir via the tail vein injection before DSS treatment; we found that miR-142a-3p agomir increased miR-142a-3p expression in the feces and exhibited protective effects against DSS-induced colitis, while miR-142a-3p agomir did not exhibit any protective effects when the mice were pretreated with antibiotics (Figure S7). We also transfected Caco-2 cells with miR-142a-3p agomir and found that the expression of miR-142a-3p was significantly increased, while no inhibitory effects were observed on DSS-induced secretion of inflammatory cytokines (Figure S8). These results further indicated that miR-142a-3p by itself did not have any protective effects on DSS-induced colitis, and it may exert its effects in a microbiome-dependent manner.
miR-142a-3p alleviates DSS-induced colitis by promoting the growth of L. reuteri
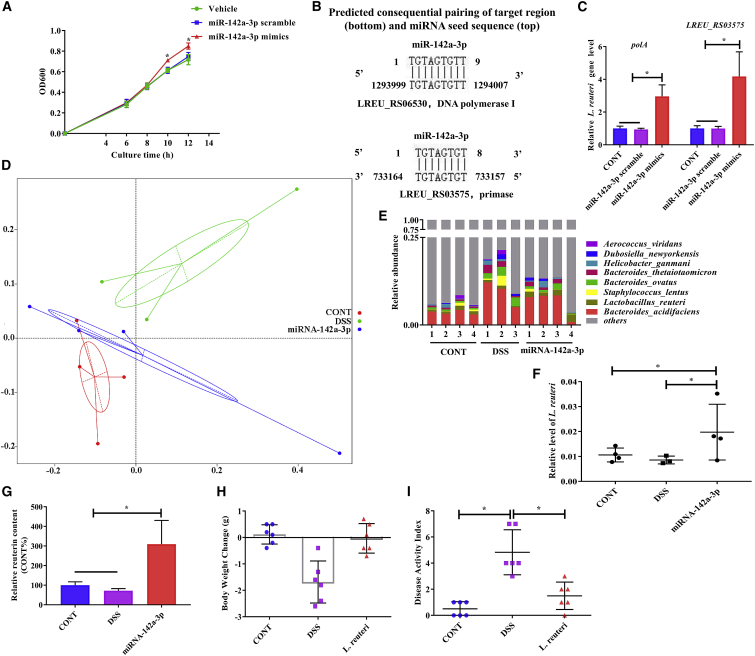
Previous studies have demonstrated that fecal miRNA could regulate gene transcription and growth of bacteria.11,12 Our above-mentioned results suggested that the relative abundance of L. reuteri and L. johnsonii in feces of mice recovered from DSS-induced colitis was significantly increased, and an increase in the expression of miR-142a-3p was observed. We first detected whether miR-142a-3p could promote the growth of these two microbes. We cultured L. reuteri or L. johnsonii with miR-142a-3p mimics and found that miR-142a-3p mimics promote the growth of L. reuteri (Figure 5A), while they did not have any effect on the growth of L. johnsonii (Figure S9). To explore how miR-142a-3p regulates L. reuteri, we BLAST-searched the seed sequence of miR-142a-3p against the whole genome sequence of L. reuteri. We found that two genes (locus tag LREU_RS06530 [polA] and locus tag LREU_RS03575) were potential targets of miR-142a-3p (Figure 5B). These two genes encode DNA polymerase I and primase, respectively. As expected, the relative expression of these two genes (polA and locus tag LREU_RS03575) was significantly higher when L. reuteri was incubated with miR-142a-3p mimics (Figure 5C).
Figure 5.
miR-142a-3p alleviates DSS-induced colitis by promoting the growth of L. reuteri
(A) L. reuteri was grown in culture media with 0.5 μM miR-142a-3p mimics and scramble. Growth was monitored as absorbance at 600 nm every 2 h (n = 3). (B) Schematic diagram of the putative binding sites of the seed sequence of miR-142a-3p in the two genes (LREU_RS06530 and LREU_RS03575) of L. reuteri. (C) Relative expression of the two genes of L. reuteri incubated with miR-142a-3p mimics (n = 3). (D–G) The effects of miR-142a-3p on microbiota composition in DSS-treated mice (n = 3–4). (D) Principal coordinates analysis (PCoA) based on weighted UniFrac distance. (E) Relative abundance of bacteria classified at a species-level taxonomy. (F) Relative abundance of L. reuteri determined by qRT-PCR. (G) Quantification of reuterin in feces by HPLC. (H and I) The effects of oral administration of L. reuteri (1 × 1011 colony-forming units [CFU]/mL) on DSS-induced colitis (n = 6). (H) Body weight change. (I) Disease activity index. Data were analyzed with one-way ANOVA. Values are mean ± SEM. ∗p < 0.05.
We investigated whether oral administration of miR-142a-3p affected the composition of the intestinal microbiota. PCoA based on weighted UniFrac distance suggested that the overall microbial structure in DSS-treated mice was clustered away from that in control mice and miR-142a-3p-treated mice (Figure 5D). Importantly, the relative abundance of L. reuteri was significantly higher in miR-142a-3p-treated mice than that in the other two groups (Figure 5E). We further confirmed the change in the abundance of L. reuteri via fecal microbe quantification using qPCR (Figure 5F). Notably, the concentration of reuterin, which is produced by L. reuteri, was also increased in the feces of miR-142a-3p-treated mice (Figure 5G). Previous studies have reported that L. reuteri could alleviate DSS-induced colitis.15,16 We further confirmed this effect of L. reuteri, as oral treatment of mice with L. reuteri for 7 consecutive days alleviated DSS-induced body weight loss and increase in the DAI (Figures 5H and 5I).
Reuterin alleviates DSS-induced colitis by activating the AMPK pathway
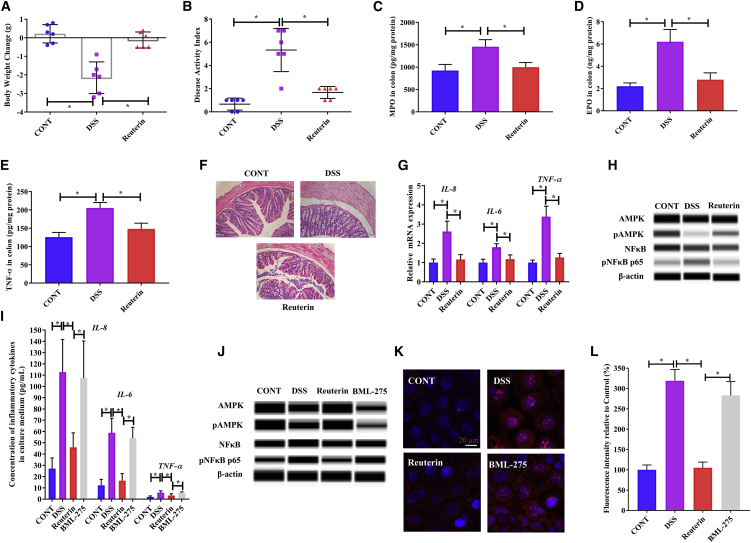
Reuterin is a potential antimicrobial and anti-inflammatory compound. To explore whether reuterin could alleviate DSS-induced colitis, we obtained reuterin produced by bacterial cultures and then orally administrated reuterin to the mice for 7 consecutive days. We found that reuterin-pretreated mice had unaltered body weight and a lower DAI upon treatment with DSS for 5 days (Figures 6A and 6B). We found that myeloperoxidase (MPO), eosinophil peroxidase (EPO), and TNF-α concentrations were significantly decreased upon pre-treatment of DSS-treated mice with reuterin (Figures 6C–6E), suggesting that reuterin alleviated DSS-induced colonic infiltration and inflammation. In addition, the mucous layer in the distal colon tissue did not exhibit marked edema in reuterin-pretreated mice (Figure 6F). Moreover, we found that reuterin pre-treatment inhibited the expression of inflammatory cytokines (Figure 6G), increased phosphorylated AMPK expression, and decreased phosphorylated NF-κB expression in the colonic tissue (Figure 6H), suggesting that reuterin may inhibit the inflammatory response through the AMPK-NF-κB pathway.
Figure 6.
Reuterin alleviates DSS-induced colitis via activating the AMPK pathway
(A–G) The effects of reuterin on DSS-induced colitis in mice (n = 3 for protein qualification and n = 6 for other data). (A) Body weight change. (B) Disease activity index. (C) Myeloperoxidase (MPO) concentration in colon. (D) Eosinophil peroxidase (EPO) concentration in colon. (E) TNF-α concentration in colon. (F) Representative colonic morphology. (G) Relative gene expression of inflammatory cytokines in colonic tissue. (H) Protein qualification by the Wes Simple Western System. (I–L) The effects of reuterin on DSS-treated Caco-2 cells (n = 3). (I) Concentration of inflammatory cytokines in culture medium. (J) Protein qualification using the Wes Simple Western System. (K) Reactive oxygen species (ROS) stained with MitoSOX reagent (red, reactive oxygen species; blue, nucleus). (L) Relative fluorescence intensity calculated according to the results of (J). Data were analyzed with one-way ANOVA. Values are mean ± SEM. ∗p < 0.05.
To further confirm this, we treated Caco-2 cells with BML-275, an inhibitor of AMPK. We found that upon pharmacological inhibition of AMPK, reuterin did not inhibit the DSS-induced section of inflammatory cytokines (IL-8, IL-6, and TNF-α) and expression of phosphorylated NF-κB in Caco-2 cells (Figures 6I and 6J). Further, our results showed that AMPK inhibition abrogated the inhibitory effects of reuterin on the over-production of reactive oxygen species (ROS) (Figures 6K and 6L).
Discussion
DSS-induced colitis is one of the most widely characterized experimental models of ulcerative colitis.17 It is commonly accepted that DSS significantly impacts the composition of the intestinal microbiota.18 Germ-free mice are more susceptible to DSS-induced colitis,19 suggesting that the microbiota plays a critical role during the self-preventive and self-healing processes in mice exposed to DSS. A previous study showed that the microbial composition in mice after recovering from DSS-induced colitis for 9 days began to resemble the microbial community in healthy mice.20 However, the recovering mice did not recover the original microbial community structure even after 20 days. These studies involved the collection of the luminal contents in the cecum and colon for the analysis of the microbial composition, a process that required the mice to be euthanized at each sampling point. Here, we collected the feces at the three different phases: before inflammation, the acute inflammatory phase, and the recovery phase. In this way, we could observe DSS-induced changes in the composition of microbiota during the recovery phase in the same mice. In line with a previous study,20 we found that the overall microbial structure of mice with acute inflammation was different from that of healthy mice, who share similarities with mice that had recovered from DSS-induced inflammation. However, differences in changes at different levels of taxonomy were observed between our results and those of a previous study,20 as DSS triggered different alterations in the composition of the microbiota in the feces and colon.18 Notably, we found that the abundance of L. reuteri and L. johnsonii was increased in the recovery phase, indicating that these two species may play an important role in the recovery of mice from DSS-induced colitis.
The microbiome is involved in the development of disease and during the recovery phase. Diet has been shown to modulate the function of the gut microbiome; however, there is an urgent need to explore methods that specifically modulate the microbiome. Fecal microbiota transplantation has been demonstrated to be effective at treating several diseases, while the targets during the intervention cannot be well defined.21 Recently, miRNAs were shown to exist in human and animal feces, and these molecules are well characterized.11 Importantly, miRNAs could enter the bacteria, although with differing efficacy based on the miRNA molecules.11 Previous studies have provided sufficient data to show that miRNAs can modulate bacterial gene expression through base pairing with specific bacterial genes and then modulate bacterial growth and metabolites.11,12,14 We found that miR-142a-3p promoted the growth of L. reuteri by targeting two genes involved in DNA replication. These results further suggested that miRNA could serve as a promising approach to specifically manipulate the microbiome. However, whether the two genes (polA and locus tag LREU_RS03575) are indispensable for the promoted growth of L. reuteri by miR-142a-3p needs further investigation.
A recent study found that miRNAs were differentially expressed in the feces of healthy mice and the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis.12 Importantly, the authors found that miR-30d, a highly expressed miRNA in the feces of diseased subjects, had therapeutic properties. In our study, we found that the levels of several miRNAs were significantly increased in the feces of mice recovered from DSS-induced colitis. Specifically, among these miRNAs, miR-142a-3p could alleviate DSS-induced colitis in a microbiome-dependent manner. These results increase our knowledge for modulating the microbiome and indicate that the feces of subjects recovered from diseases might be enriched with miRNAs exhibiting preventive effects against the disease.
L. reuteri is widely considered to be a probiotic microbe that exerts beneficial effects in the intestinal tract. Several studies have reported that L. reuteri could be used as a supplement for the prevention of ulcerative colitis.15,16,22 These studies suggest that L. reuteri alleviated DSS-induced colitis by decreasing the inflammatory response and leukocyte- and platelet-endothelial cell interactions and increasing P-selectin expression. However, whether L. reuteri exerts such effects through its metabolites was not well explored. We found that the increased abundance of L. reuteri, which was affected by miR-142a-3p, was associated with increased production of reuterin, which is a broad-spectrum antimicrobial and anti-inflammatory compound.23 Interestingly, reuterin itself could alleviate DSS-induced body weight loss and decrease the DAI. Moreover, reuterin decreased the expression of inflammatory cytokines through the AMPK-NF-κB signaling pathway, although the interactions between reuterin and AMPK need further investigation.
In conclusion, we unexpectedly found that the feces collected from animals well-recovered from colitis could alleviate the disease. We further identified that the expression of miRNAs and microbiome composition were differentially influenced in the feces of diseased and well-recovered subjects. Notably, we identified a highly expressed miRNA in recovered subjects that could alleviate colitis by promoting the growth of a specific bacterium and the production of its associated metabolite, further influencing the expression of inflammatory genes in intestinal epithelial cells. The results presented here provide insights into miRNAs enriched in the feces of animals recovered from disease; this might exhibit preventive effects against the disease.
Materials and methods
Animal experiments
Animal procedures were approved by the Protocol Management and Review Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences on Animals. Eight-week-old C57BL/6 mice were purchased from Slac Laboratory Animal Central (Changsha, China) and acclimated in plastic cages under standard conditions for 1 week before experiments. All mice used were of the same sex (male) and age (9 weeks), and they were randomly assigned into treatment groups.
DSS-induced colitis
Mice were exposed to 2.5% DSS (molecular weight [MW], 36,000–50,000; MP Biomedicals, Shanghai, China) prepared in autoclaved drinking water for 5 days, after which they were provided DSS-free drinking water. Colitis development was monitored daily by determining the change in body weight and DAI using a standard scoring system (Table S1).24
Fecal microbiota profiling
Fecal DNA was extracted and isolated using the QIAampDNA stoolMiniKit (QIAGEN, Shanghai, China). Bacterial 16S rDNA gene sequences (V3–V4 region) were amplified using specific primers 341F: 5′-CCTAYGGGRBGCASCAG-3′ and 806R: 5′-GGACTACNNGGGTATCTAAT-3′. PCR reactions were performed in 50 μL mixture consisting of 12.5 μL of Phusion High-Fidelity PCR Master Mix (New England BioLabs), 1 μL of each primer, 50 ng of template DNA, and PCR-grade water. Amplicons purified using QIAGEN Gel Extraction Kit (QIAGEN) were sequenced using the Illumina HiSeq2500 platform. Quality filtering and analysis were performed using USEARCH, while adhering to the QIIME quality-controlled process based on 97% sequence similarity (Novogene, Beijing, China).25
Fecal RNA isolation
Total RNA was extracted from fecal samples using a miRNA isolation kit with phenol (Invitrogen, Shanghai, China), as previously described.11 Briefly, feces were homogenized in PBS for extraction of RNA using acid-phenol:chloroform. Subsequently, the samples were mixed with 1.25 volumes of 100% ethanol for aqueous phase precipitation, followed by RNA purification.
Fecal transplantation and fecal RNA transplantation
First, freshly collected feces (5 mg) from donor mice were diluted and suspended in 200 μL nuclease-free water and orally gavaged into recipient mice once daily for 7 days prior to DSS treatment. Second, fecal samples were heated at 80°C for 60 min to kill bacteria while retaining miRNA before gavage. Finally, RNA isolated (10 μg) from feces was eluted in 200 μL nuclease-free water and orally gavaged into recipient mice once daily for 7 days prior to DSS treatment.
Small RNA sequencing
Three micrograms of total RNA per sample was used as input for the small RNA library. Sequencing libraries were generated using NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, Ipswich, MA, USA). Following PCR amplification using LongAmp Taq 2× Master Mix, DNA fragments corresponding to 140‒160 bp were purified, and library quality was assessed on an Agilent Bioanalyzer 2100 system. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2500 platform, and 50-bp-long single-end reads were generated. The small RNA tags were mapped to the reference sequence using Bowtie26 without mismatch to analyze their expression and distribution in the reference sequence.
miRNA measurement by qPCR
Fecal RNA (200 ng) was used for miRNA cDNA synthesis by using a miRNA cDNA Synthesis kit (Thermo Fisher). Real-time PCR was performed to quantify miRNA cDNAs using Fast Master Mix and Advanced miRNA Assays (Thermo Fisher, Shanghai, China) on Roche LightCycler 480 II (Indianapolis, IN, USA), according to the manufacturer’s protocol. Primers used for quantification of miRNA expression are listed in Table S2. As no reference gene has been established for quantifying fecal miRNA using qPCR, we used miR-10a-5p, which is present in the feces at a high level, and no difference in the expression of miR-10a-5p was observed between mice exposed to DSS for 5 days and those well-recovered mice on day 12, according to small RNA-seq data. Thus, miR-10a-5p was used as a reference to measure the relative abundance of miR-142a-3p, miR-142b, miR-146b-5p, and miR-223-5p.
miRNA treatment
miRNA mimics of miR-142a-3p, miR-142b, miR-146b-5p, miR-223-5p, and related scramble miRNA were purchased from Sangon Biotech (Shanghai, China); their sequences are described in Table S3. The mice were administered doses of 200, 400, 800, and 1,000 pmol miRNA mimics and related scramble miRNA by oral gavage for 7 consecutive days. Then, they were further administered 2.5% DSS in drinking water for 5 days.
Quantification of fecal microbes by qPCR
DNA was extracted from fecal pellets using a QIAamp Fast DNA Stool Mini Kit (QIAGEN), and the operational taxonomic units (OUTs) were quantified using qPCR; the reaction mixture contained primer pairs, genomic DNA, and TaqMan Universal PCR Master Mix (Applied Biosystems, Shanghai, China). Primer sequences are shown in Table S4.
Bacterial gene transcript quantification
Total bacterial RNA was extracted using TRIzol (Invitrogen). RNA was reverse-transcribed into cDNA using a cDNA Reverse Transcription Kit (Applied Biosystems). For the qPCR, the reaction contained primer pairs, cDNA, and Universal PCR Master Mix (Applied Biosystems). Primer sequences are shown in Table S4.
Antibiotic treatment
The mice were orally gavaged with a mixture of antibiotics (metronidazole 1 mg/mL, ampicillin 1 mg/mL, vancomycin 0.5 mg/mL, neomycin 1 mg/mL, and streptomycin 1 mg/mL) in 200 μL nuclease-free water for 7 consecutive days to deplete the gut bacteria.27
miRNA target prediction
The seed sequence of miR-142a-3p (gtagtgt) was BLAST-searched against the Lactobacillus reuteri genome for sequence pairing using the NCBI blast tool.
In vitro growth measurements of L. reuteri
L. reuteri (ATCC 23272) and L. johnsonii (ATCC 33200) were cultured in Man, Rogosa, and Sharpe medium (MRS) supplemented with miRNA mimics and scramble miRNAs at 37°C and then monitored by measuring absorbance at 600 nm (OD600).
Reuterin production
Reuterin was produced using bacterial cultures as previously described.28 Briefly, L. reuteri was grown overnight at 37°C in 10 mL MRS broth with 20 mM glycerol. Then, cells were harvested and re-suspended in sterile 600 mM glycerol. Conversion of glycerol to reuterin was conducted at 25°C for 3 h.
Quantification of reuterin by HPLC and absorbance spectrophotometry
Reuterin was chromatographically separated as previously described.29 In this method, the conversion of glycerol to reuterin proceeds at a ratio of 1:1, and reuterin level was assayed by subtracting the remaining amount of glycerol from its concentration in the starting solution. Reuterin samples were analyzed colorimetrically, and the accuracy of colorimetric results was confirmed by high-performance liquid chromatography (HPLC), as described previously.29
In vivo and in vitro experiments with reuterin
The mice were orally gavaged with 1.5 mg/d reuterin for 7 consecutive days. Then, they were further administered 2.5% DSS in drinking water for 5 days. For in vitro experiments, Caco-2 cells were incubated with 10 mM reuterin for 16 h and then treated with 2% DSS for 24 h. Additionally, Caco-2 cells were incubated with 10 μM BML-275 (Selleck, Shanghai, China) for 24 h for the inhibition of AMPK.
Intestinal morphology
Colon samples were fixed with 4% formaldehyde, embedded in paraffin, and then sections were stained with hematoxylin and eosin to observe morphological changes.30
Determination of gene expression by quantitative real-time PCR
Total RNA was isolated from the colonic tissue using TRIzol (Invitrogen). cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Dalian, China). Quantitative real-time PCR was performed using SYBR Green qPCR Master Mix (Takara), as previously described.31 The primer sequences are presented in Table S5.
Protein qualification using Wes Simple Western System
Protein was quantified by using the Wes Simple Western System (ProteinSimple) with automated capillary gel electrophoresis, as previously described.32 Primary antibodies used included antibodies against AMPK, pAMPK, NF-κB, and pNF-κB (Abcam, Cambridge, MA, USA). Results were obtained using the “gel view” function of the Protein Simple software (ProteinSimple).
Measuring the levels of EPO, MPO, and inflammatory cytokines
The levels of EPO, MPO, and inflammatory cytokines (TNF-α, IL-6, and IL-8) were determined using commercial assay kits obtained from Beyotime Biotechnology (Shanghai, China) according to the manufacturer’s instructions.
Determination of reactive oxygen species
Reactive oxygen species in mitochondria were determined as previously described.33 Briefly, Caco-2 cells were treated with 5 μM MitoSOX reagent (Invitrogen) for 10 min at 37°C in the dark. Then, cells were observed using a Zeiss LSM880 confocal microscope (Shanghai, China) following treatment with an anti-fluorescence quenching agent.
Statistical analysis
Significance between treatments was analyzed using a t test or one-way ANOVA followed by Student-Newman-Keuls post hoc test, using the data statistics software SPSS 18.0. All data are expressed as the mean ± SEM. p value < 0.05 was considered significant.
Availability of data
The 16S rDNA gene sequence data have been deposited in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA693887 and PRJNA693962. Small RNA sequencing data have been deposited in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE165648.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (U2002206), the Huxiang Young Talents Plan Project of Hunan Province (2019RS2046), the Natural Science Foundation for Outstanding Young Scholars of Hunan Province (2019JJ30017), the Hunan High-Level Talent Gathering Project (2018RS3111), and the Young Elite Scientists Sponsorship Program by CAST (20191QNRC001). The authors wish to thank Jinzhen Jiao, ISA Changsha, for her technical help.
Author contributions
X.Z. designed the research study; X.Z., L.H., Y.L., and L.Z. performed the research; X.Z., L.H., and F.L. analyzed and interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.08.025.
Contributor Information
Xihong Zhou, Email: xhzhou@isa.ac.cn.
Fengna Li, Email: lifengna@isa.ac.cn.
Supplemental information
References
- 1.Ng S.C., Shi H.Y., Hamidi N., Underwood F.E., Tang W., Benchimol E.I., Panaccione R., Ghosh S., Wu J.C.Y., Chan F.K.L., et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 3.Caruso R., Lo B.C., Núñez G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020;20:411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 4.Glassner K.L., Abraham B.P., Quigley E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020;145:16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Veltkamp C., Tonkonogy S.L., De Jong Y.P., Albright C., Grenther W.B., Balish E., Terhorst C., Sartor R.B. Continuous stimulation by normal luminal bacteria is essential for the development and perpetuation of colitis in Tg(epsilon26) mice. Gastroenterology. 2001;120:900–913. doi: 10.1053/gast.2001.22547. [DOI] [PubMed] [Google Scholar]
- 6.Ohkusa T., Okayasu I., Ogihara T., Morita K., Ogawa M., Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaubeck M., Clavel T., Calasan J., Lagkouvardos I., Haange S.B., Jehmlich N., Basic M., Dupont A., Hornef M., von Bergen M., et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65:225–237. doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton G.J., Contijoch E.J., Mogno I., Vennaro O.H., Llewellyn S.R., Ng R., Li Z., Mortha A., Merad M., Das A., et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice. Immunity. 2019;50:212–224.e4. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaich O., Parikh S., Bell R.E., Mekahel K., Donyo M., Leader Y., Shayevitch R., Sheinboim D., Yannai S., Hollander D., et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat. Commun. 2019;10:5657. doi: 10.1038/s41467-019-13527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S., Rezende R.M., Moreira T.G., Tankou S.K., Cox L.M., Wu M., Song A., Dhang F.H., Wei Z., Costamagna G., Weiner H.L. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe. 2019;26:779–794.e8. doi: 10.1016/j.chom.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung M., Schaefer A., Steiner I., Kempkensteffen C., Stephan C., Erbersdobler A., Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 14.Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C., et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe. 2018;24:637–652.e8. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber O., Petersson J., Phillipson M., Perry M., Roos S., Holm L. Lactobacillus reuteri prevents colitis by reducing P-selectin-associated leukocyte- and platelet-endothelial cell interactions. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G534–G542. doi: 10.1152/ajpgi.90470.2008. [DOI] [PubMed] [Google Scholar]
- 16.Sun M.C., Zhang F.C., Yin X., Cheng B.J., Zhao C.H., Wang Y.L., Zhang Z.Z., Hao H.W., Zhang T.H., Ye H.Q. Lactobacillus reuteri F-9-35 Prevents DSS-Induced Colitis by Inhibiting Proinflammatory Gene Expression and Restoring the Gut Microbiota in Mice. J. Food Sci. 2018;83:2645–2652. doi: 10.1111/1750-3841.14326. [DOI] [PubMed] [Google Scholar]
- 17.Jurjus A.R., Khoury N.N., Reimund J.M. Animal models of inflammatory bowel disease. J. Pharmacol. Toxicol. Methods. 2004;50:81–92. doi: 10.1016/j.vascn.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Munyaka P.M., Rabbi M.F., Khafipour E., Ghia J.E. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J. Basic Microbiol. 2016;56:986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- 19.Kitajima S., Morimoto M., Sagara E., Shimizu C., Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp. Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 20.Schwab C., Berry D., Rauch I., Rennisch I., Ramesmayer J., Hainzl E., Heider S., Decker T., Kenner L., Müller M., et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J. 2014;8:1101–1114. doi: 10.1038/ismej.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ianiro G., Bibbò S., Scaldaferri F., Gasbarrini A., Cammarota G. Fecal microbiota transplantation in inflammatory bowel disease: beyond the excitement. Medicine (Baltimore) 2014;93:e97. doi: 10.1097/MD.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dicksved J., Schreiber O., Willing B., Petersson J., Rang S., Phillipson M., Holm L., Roos S. Lactobacillus reuteri maintains a functional mucosal barrier during DSS treatment despite mucus layer dysfunction. PLoS ONE. 2012;7:e46399. doi: 10.1371/journal.pone.0046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prisciandaro L., Geier M., Butler R., Cummins A., Howarth G. Probiotics and their derivatives as treatments for inflammatory bowel disease. Inflamm. Bowel Dis. 2009;15:1906–1914. doi: 10.1002/ibd.20938. [DOI] [PubMed] [Google Scholar]
- 24.Wirtz S., Neufert C., Weigmann B., Neurath M.F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 25.Geng S., Cheng S., Li Y., Wen Z., Ma X., Jiang X., Wang Y., Han X. Faecal Microbiota Transplantation Reduces Susceptibility to Epithelial Injury and Modulates Tryptophan Metabolism of the Microbial Community in a Piglet Model. J. Crohn’s Colitis. 2018;12:1359–1374. doi: 10.1093/ecco-jcc/jjy103. [DOI] [PubMed] [Google Scholar]
- 26.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W., Zhai S., Xia Y., Wang H., Ruan D., Zhou T., Zhu Y., Zhang H., Zhang M., Ye H., et al. Ochratoxin A induces liver inflammation: involvement of intestinal microbiota. Microbiome. 2019;7:151. doi: 10.1186/s40168-019-0761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asare P.T., Greppi A., Stettler M., Schwab C., Stevens M.J.A., Lacroix C. Decontamination of Minimally-Processed Fresh Lettuce Using Reuterin Produced by Lactobacillus reuteri. Front. Microbiol. 2018;9:1421. doi: 10.3389/fmicb.2018.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spinler J.K., Taweechotipatr M., Rognerud C.L., Ou C.N., Tumwasorn S., Versalovic J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., He L., Wu C., Zhang Y., Wu X., Yin Y. Serine alleviates oxidative stress via supporting glutathione synthesis and methionine cycle in mice. Mol. Nutr. Food Res. 2017;61:1700262. doi: 10.1002/mnfr.201700262. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X., He L., Zuo S., Zhang Y., Wan D., Long C., Huang P., Wu X., Wu C., Liu G., Yin Y. Serine prevented high-fat diet-induced oxidative stress by activating AMPK and epigenetically modulating the expression of glutathione synthesis-related genes. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:488–498. doi: 10.1016/j.bbadis.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 32.He L., Long J., Zhou X., Liu Y., Li T., Wu X. Serine is required for the maintenance of redox balance and proliferation in the intestine under oxidative stress. FASEB J. 2020;34:4702–4717. doi: 10.1096/fj.201902690R. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X.H., Zhang Y., He L., Wan D., Liu G., Wu X., Yin Y. Serine prevents LPS-induced intestinal inflammation and barrier damage via p53-dependent glutathione synthesis and AMPK activation. J. Funct. Foods. 2017;39:225–232. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rDNA gene sequence data have been deposited in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA693887 and PRJNA693962. Small RNA sequencing data have been deposited in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE165648.