Abstract
Background
Red ginseng extract boosts immunity against inflammation and cancer in the human body. However, studies on the effects of red ginseng extract on the gut microbiome remain unexplored.
Methods
In 2019, the positive effects and changes in the gut microbiome after administering 1 pack (3 g) of red ginseng extract per day to 53 adults aged 40 to 75 for 24 weeks were investigated. The gut microbial environment changes were qualitatively and quantitatively analyzed using next-generation sequencing and real-time polymerase chain reaction technology.
Results
On comparing and analyzing alpha diversity and beta diversity, the microbial pattern showed significant differences (OTUs p = 0.003, chao1 p < 0.001, Bray-Curtis p = 0.001) before and after ingestion of red ginseng extract, indicating that gut microbial richness increased after ingestion. Moreover, after comparing and analyzing the gut microbiome's differences after red ginseng extract intake, significant differences were noted between three strains at the phylum level and among 57 strains at the genus level.
Conclusion
This study proposes the potential use of red ginseng extract as a prebiotic after confirming its positive effects, including increasing gut microbiome richness, reducing harm to the gut microbiome, and increasing the number of some strains in the gut microbiome.
Keywords: dietary supplements, gut microbiome, gut microbiome analysis, prebiotics, red ginseng extract
Graphical abstract
1. Introduction
At birth, the human intestinal tract is sterile, and at approximately 2.5 years of age, 1010 to 1012 intestinal bacteria per gram are found in the intestines [1,2]. The role of these intestinal bacteria is not fully understood. However, these bacteria form intestinal colonies that use all available nutrients, secrete inhibitory compounds to protect against pathogens, thicken the mucous membrane, and directly regulate antibody production [1,3,4]. In addition, some intestinal bacteria are also involved in the metabolism and fermentation of carbohydrates with short-chain fatty acids such as acetic acid and butyric acid to provide nutrients for human host cells [1]. Active research is being conducted on the roles of these intestinal bacteria, and in particular, several studies over the past 10 years have changed the paradigm by showing that intestinal bacteria affect brain function [5]. The brain-gut-microbiome axis is a comprehensive concept of biochemical signals and interactions between the gastrointestinal tract, enterobacteria, and brain, and bidirectional interactions in the axis have been shown in recent studies [5,6]. Thus, the brain regulates the autonomic nervous system, local intestinal motility, intestinal passage, intestinal secretion, and intestinal permeability, potentially affecting the microbial community's structure and function. Studies have also shown that intestinal bacteria's byproducts affect the onset of various diseases such as irritable bowel syndrome, autism spectrum disorder, Parkinson's disease, and multiple sclerosis [5,7,8]. The leaky gut syndrome refers to the inflammatory and allergic reactions caused by an impaired intestinal defense system, which leads to increased permeability and penetration of the endotoxins and antigens through mucosal cells [9]. Intestinal bacteria maintain intestinal defense ability by maintaining the balance between beneficial and harmful bacteria. Interruption of this balance may cause leaky gut syndrome, which may ultimately lead to autoimmune diseases, such as type 1 diabetes and lupus, as well as inflammatory and allergic diseases [4,9]. In vivo studies have also shown the role of intestinal bacteria; following transplantation of stool from healthy patients and those with major depressive disorder in the mice kept in aseptic conditions, those that received stool from patients with major depressive disorder showed higher anxiety levels and depression-related behaviors [10]. Recently, due to a plunge in the high cost of precision medicine and the ease of performing genetic analysis for the intestinal bacterial environment, many studies have analyzed the intestinal bacterial environments and the effects of changes in this environment on the human body [11,12].
Red ginseng extract has been traditionally consumed in several Eastern countries [13]. Recent ongoing research has shown that saponins, Rh2 and Rg3, components of red ginseng extract, increase the human body's anti-inflammatory and anticancer immunity [[14], [15], [16]]. Furthermore, in vivo and in vitro effects of red ginseng extract on intestinal bacteria have been assessed since 1990 [17]. In 2014, the effects of red ginseng extract and Semen coicis on ulcerative colitis were assessed [18]. Moreover, the effects of red ginseng extract and mushrooms on type 2 diabetic mice's intestinal bacteria were evaluated in 2015 [19]. As such, most studies have assessed the effects of red ginseng extract on intestinal bacteria in animals and patients. In a recent clinical study, ginseng extract was administered with lactic acid bacteria, and the effects of culturing said bacteria with red ginseng extract were assessed. Furthermore, red ginseng extract was administered with a supplement for liver improvement in patients with nonalcoholic fatty liver disease, and the effects were compared in the treatment, control, and placebo groups [20].
To study the effects of red ginseng extract on the gut microbiome and to investigate its potential as a prebiotic, red ginseng extract was ingested for 24 weeks by subjects who met the inclusion criteria, and changes in beneficial and harmful bacteria were qualitatively and quantitatively observed by studying their effects on the human intestinal bacterial environment. In particular, changes in diversity at the genus and phylum levels were assessed.
2. Materials and methods
2.1. Study subjects
In this study, 53 men and women aged 40 to 75 years who visited the Department of Family Medicine at a university hospital for health checkups from June 2018 to May 2019 were randomly selected. The exclusion criteria were: 1) those who were suspected of acute diseases (severe infection, severe trauma, severe diarrhea, etc.); 2) those who have received antibiotics for at least 1 month within the last 6 months; 3) those who have a gastrointestinal disease (inflammatory bowel disease, active peptic ulcer, etc.) or have undergone gastrointestinal surgery (except for simple appendix or hernia surgery); 4) those who have taken dietary supplements, such as lactic acid bacteria, or red ginseng extract within the past year; and 5) those who are currently participating in other clinical or human trials or have participated in clinical or human trials within the last month. This study was conducted in conformance with the ethical guideline from the 1975 Helsinki Declaration and reviewed by the Institutional Review Board of the Catholic Central Medical Center (approval number: UC18HESI0114; ClinicalTrial.gov ID: NCT03865745).
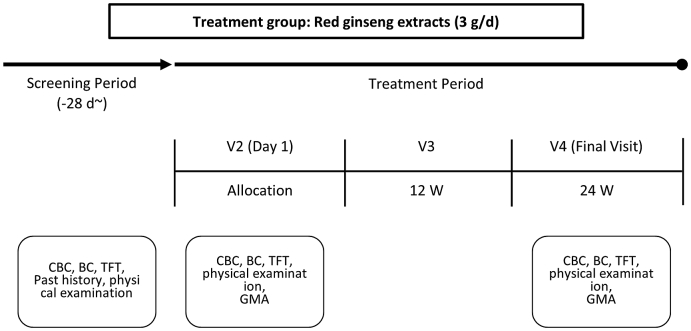
Written informed consent was obtained from each subject, and screening, such as blood tests, was conducted before administering red ginseng extract. Body mass index was obtained by weight (kg)/height (m2), and medical history, such as hypertension, diabetes, and hyperlipidemia, drug history, smoking history, and alcohol consumption history, was assessed using a structured questionnaire survey. The results were recorded after an interview with a researcher. The subject was considered a smoker if they smoked currently, regardless of smoking history. If the subject consumed alcohol more than once a week, the subject was considered an alcoholic. Blood pressure was measured using an automatic blood pressure monitor (Jawon FT-500, Korea) twice at 10-min intervals while seated, after at least 15 min of relaxation, and the median value was calculated. Venous blood was collected for blood tests after 12 h of fasting. Total and differential leukocyte count, red blood cell count, hematocrits, serum glucose levels, total cholesterol, triglycerides, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, and thyroid hormone were measured. Following the screening, intestinal bacteria were analyzed, and 1 pack (3g, Everytime®; Korea Ginseng Corp., Daejeon, Korea) of red ginseng extract, consisting of Rg1, 0.60 mg; Re, 0.63 mg; Rf, 0.99 mg; Rh, 1.17 mg; Rg2s, 1.11 mg; Rb1, 5.10 mg; Rc, 2.07 mg; Rb2, 1.77 mg; Rd, 0.96 mg; Rg3s, 2.67 mg; and Rg3r, 1.32 mg; was administered once per day for 24 weeks. Analysis of intestinal bacteria was performed again 24 weeks later (Fig. 1).
Fig. 1.
Flow chart and study design. CBC, complete blood cell count; BC, blood chemistry; TFT, thyroid function test; GMA, gut microbial analysis.
2.2. Analysis of intestinal bacteria
The QIAamp Fast DNA Stool Mint Kit (Qiagen, 2010, CA, USA) was used to extract microbial DNA from human stool samples. The extracted microbial DNA concentration was measured using a Quantus Fluorometer (Promega, NY, USA), and the 16S metagenomic primers were used to amplify the 16S ribosomal RNA v3-v4 region of microbial DNA using polymerase chain reaction (PCR). AMpure XP beads (Beckman, CA, USA) were used to remove primers and primer dimers from amplified products, and PCR was performed using the Nextera XT Index Kit v2 (Illumina, CA, USA) to attach dual indices and Illumina sequencing adapters to PCR products. The concentration of purified PCR products was accurately measured again using the Quantus Fluorometer. Each sample was subsequently loaded onto the MiSeq Reagent v2 or v3 cartridge and sequenced using MiSeq next-generation sequencing (NGS) equipment (Illumina, CA, USA). Data obtained from NGS equipment were first analyzed for the intestinal microbiome using MiSeq NGS equipment. Data analysis was performed using Mothur (ver. 1.44.2) or the QIIME program (ver.2). A unique sequence was selected and aligned on the reference sequence, forming clusters for each sequence. Last, intestinal microbial taxa were analyzed by classifying sequences that formed clusters. These classified sequences were analyzed by sequencing only those in the data to form operational taxonomic units (OTUs). Intestinal microbial taxa were subsequently analyzed.
2.3. Statistical analysis
The number of samples was not calculated by a statistical method, as there was no data on existing studies related to sample size calculation. However, in a similar randomized, double-blinded, placebo-controlled study on wrinkle improvements after administering mixed red ginseng root extract [21], type 1 procollagen mRNA increased 94% and 7% on the baseline and placebo groups, respectively, showing a statistically significant difference between the two groups.
Based on Cho et al.’s [21] findings, this current study assumed that the intestinal flora's change in composition using NGS would increase 94% from the baseline time point and allowed an error of 8%, which is within 16% of the 95% confidence interval, having an increase of 83.0%–98.8% in the intestinal flora. Therefore, 47 subjects were required; however, 53 subjects were calculated after considering a 10% dropout rate. Following the screening, red ginseng extract was administered for 24 weeks, and at the end, the same tests were performed and analyzed for comparison.
A test of homogeneity for general and health-related characteristics and screening items according to the gender of the subject was performed using a t-test or Mann-Whitney U test and χ2 test (Fisher's exact test). Frequency (%), mean value, and standard deviations are presented. The difference between the measured values related to the living body and the level of Lactobacillus in the intestine measured before and after red ginseng consumption was tested using parametric tests (two-sample t-test, paired t-test, ANOVA) and non-parametric tests (Mann-Whitney U test, Kruskal-Wallis H test) depending on the results of the normality test for each measurement (Kolmogorov-Smirnov test). IBM SPSS Statistics (version 25.0) was used for statistical analysis. Analysis of beta diversity was performed using PERMA-ANOVA, and analysis of phylum and genus levels was performed using SILVA 132v. The significance level of all statistics was set at p < 0.05.
3. Results
3.1. General characteristics of subjects
According to the gender of the study subjects, the mean age was 49.13 years for men and 48.05 years for women, with 37 subjects in their 40s (71.2%) and 15 subjects over the age of 50 (28.8%). The mean body mass index does not have much difference between genders, with 26.19 kg/m2 and 24.57 kg/m2 for men and women, respectively. However, there was a significant difference in those who drank alcohol, with 86.7% for men and 43.2% for women (p = 0.011). The results show that 53.3% of men and 0% of women smoked (p < 0.001). There were no gender differences in hypertension, dyslipidemia, diabetes, coronary artery disease, chronic heart failure, cerebrovascular disease, or gastrointestinal diseases (Table 1). Therefore, the correction of gender differences was not considered in this study. Moreover, blood tests during screening showed no specific abnormalities.
Table 1.
General characteristics (including homogeneity by gender) of study subjects
| Variable | Classification | n (%) | Sex |
p-value | |
|---|---|---|---|---|---|
| Male (n = 15) | Female (n = 37) | ||||
| Age (year)a | Mean ± SD | 49.13 ± 7.02 | 48.05 ± 7.03 | 0.591 | |
| ≤49 | 37 (71.2) | 10 (66.7) | 27 (73.0) | 0.740 | |
| ≥50 | 15 (28.8) | 5 (33.3) | 10 (27.0) | ||
| BMIaM(kg/m2) | Mean ± SD | 26.19 ± 3.77 | 24.57 ± 3.34 | 0.132 | |
| <23 | 10 (19.2) | 0 (0.0) | 10 (27.0) | 0.077 | |
| 23≤, <25 | 21 (40.4) | 8 (53.3) | 13 (35.2) | ||
| ≥25 | 21 (40.4) | 7 (46.7) | 14 (37.8) | ||
| Alcohol | No | 23 (44.2) | 2 (13.3) | 21 (56.8) | 0.011 |
| Yes | 29 (55.8) | 13 (86.7) | 16 (43.2) | ||
| Smoking | No | 44 (84.6) | 7 (46.7) | 37 (100.0) | <0.001 |
| Yes | 8 (15.4) | 8 (53.3) | 0 (0.0) | ||
| Medication | No | 29 (55.8) | 7 (46.7) | 22 (59.5) | 0.594 |
| Yes | 23 (44.2) | 8 (53.3) | 15 (40.5) | ||
| Hypertension | No | 43 (82.7) | 9 (60.0) | 34 (91.9) | 0.052 |
| Yes | 9 (17.3) | 6 (40.0) | 3 (8.1) | ||
| Dyslipidemia | No | 40 (76.9) | 10(66.7) | 30 (81.1) | 0.293 |
| Yes | 12 (23.1) | 5 (33.3) | 7 (18.9) | ||
| Diabetes Mellitus | No | 49 (94.2) | 14 (93.3) | 35 (94.6) | 1.000 |
| Yes | 3 (5.8) | 1 (6.7) | 2 (5.4) | ||
| CAD | No | 51 (98.1) | 15 (100.0) | 36 (97.3) | 1.000 |
| Yes | 1 (1.9) | 0 (0.0) | 1 (2.7) | ||
| CHF | No | 51 (98.1) | 15 (100.0) | 36 (97.3) | 1.000 |
| Yes | 1 (1.9) | 0 (0.0) | 1 (2.7) | ||
| CVA | No | 51 (98.1) | 15 (100.0) | 36(97.3) | 1.000 |
| Yes | 1 (1.9) | 0 (0.0) | 1 (2.7) | ||
| GI ds. | No | 49 (94.2) | 15 (100.0) | 34 (91.9) | 0.548 |
| Yes | 3 (5.8) | 0 (0.0) | 3 (8.1) | ||
| Other diseases | No | 7 (13.5) | 1 (6.7) | 6 (16.2) | 0.658 |
| Yes | 45 (86.5) | 14 (93.3) | 31 (83.8) | ||
Based on two-sample t-test, χ2-test.
BMI, body mass index; CAD, coronary artery disease; CHF, chronic heart failure; CVA, cerebrovascular accident; GI, gastrointestinal disease.
Mann-Whitney-sample t.
3.2. Differences in alpha diversity before and after red ginseng intake
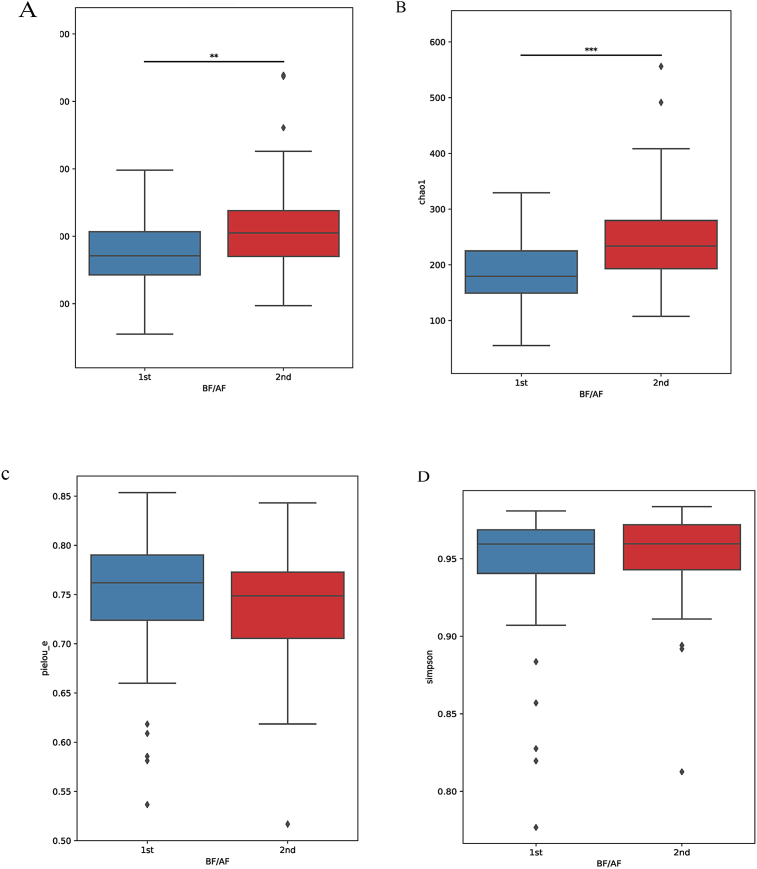
To analyze changes in the diversity of intestinal bacteria before and after consumption of red ginseng, alpha diversity, which indicates the diversity of microbial distribution in the sample, was measured. Observed OTUs (p = 0.003) and chao1 (p < 0.001) significantly increased (Table 2, Fig. 2).
Table 2.
Diversity and microbiome differences
| Variable | Before (M ± SD) | After (M ± SD) | Kruskal-Wallis H | p-value |
| Observed OTUsa | 171.85 ± 54.53 | 212.48 ± 75.26 | 8.4464 | 0.003 |
| Chao1a | 183.69 ± 60.16 | 244.04 ± 90.31 | 12.1428 | <0.001 |
| Pielou E. | 0.7446 ± 0.0673 | 0.7380 ± 0.0579 | 1.1229 | 0.289 |
| Simpsona | 0.9441 ± 0.0422 | 0.9523 ± 0.0292 | 0.5301 | 0.466 |
| Shannon |
5.4825 ± 0.7455 |
5.6524 ± 0.6523 |
0.9009 |
0.342 |
| Taxon |
Before (M ± SD) |
After (M ± SD) |
Kruskal-Wallis H |
p-value |
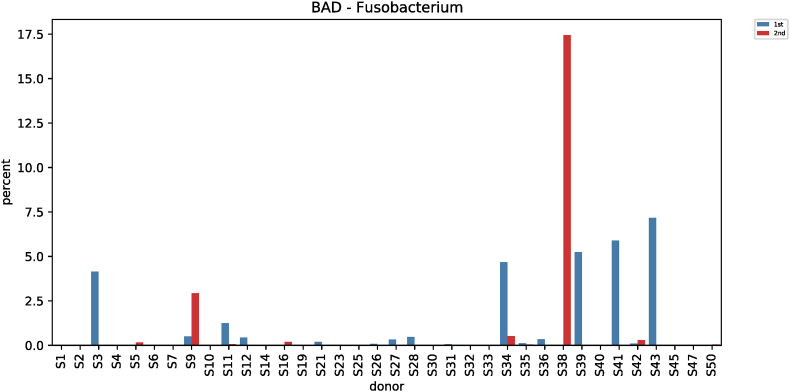
| Fusobacterium (BAD) | 0.6044 ± 1.6369 | 0.4219 ± 2.4430 | 5.3045 | 0.021 |
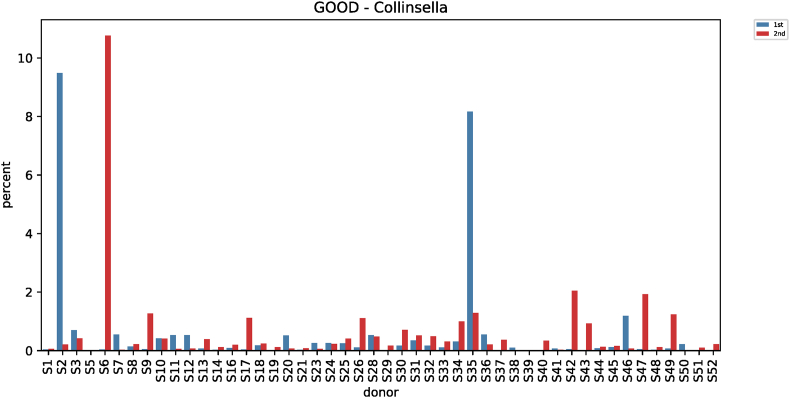
| Collinsella (GOOD) | 0.5173 ± 1.7006 | 0.5905 ± 1.5190 | 4.1222 | 0.042 |
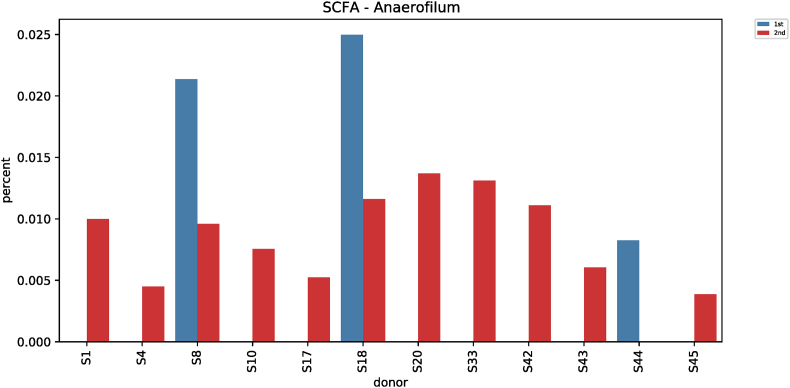
| Anaerofilum (SCFA) | 0.0010 ± 0.0046 | 0.0018 ± 0.0039 | 4.6843 | 0.030 |
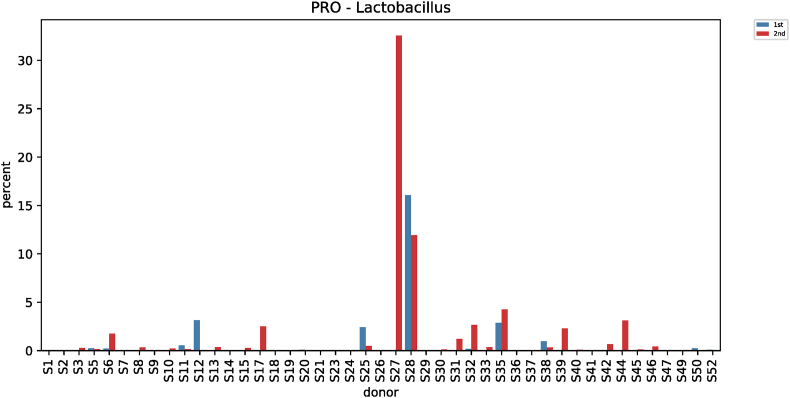
| Lactobacillus (PRO) | 0.5388 ± 2.2972 | 1.2943 ± 4.7879 | 3.9555 | 0.046 |
| Enterococcus (PRO) | 0.0411 ± 0.2326 | 0.0716 ± 0.2052 | 5.4974 | 0.019 |
Based on the ANOVA.
Observed OTUs: Number of distinct features, Chao1 index: Estimates diversity from abundant data, Pielou's evenness: Measure of relative evenness of species richness, Simpson's index: Measures the relative abundance of the different species making up the sample richness, Shannonelouovas: Accounts for both abundance and evenness of the taxa present SCFA, short-chain fatty acids.
Kruskal-Wallis H test.
Fig. 2.
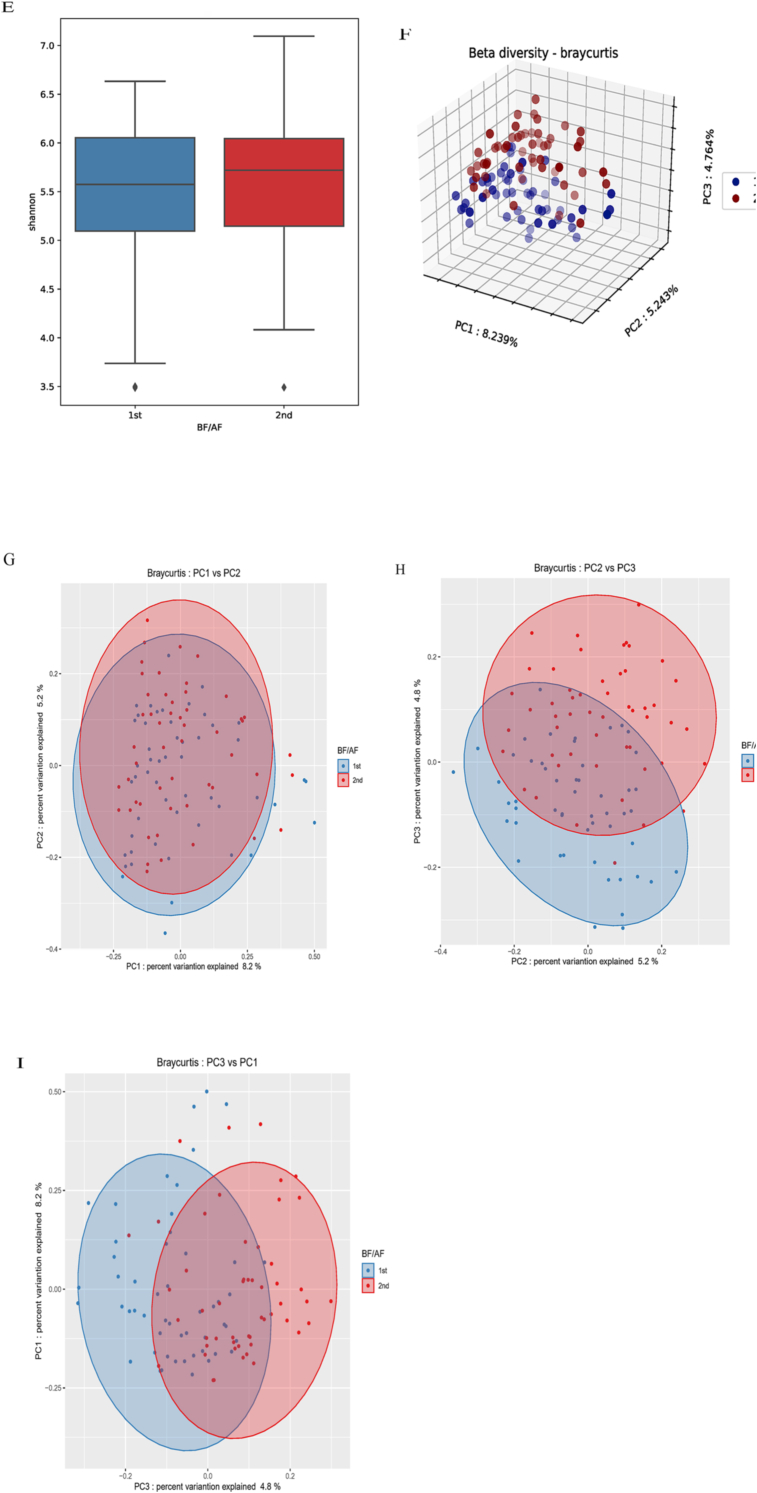
Differences in alpha diversity and beta diversity according to the pre-test. ∗ Alpha diversity: (A) Observed OTUs, (B) Chao 1, (C) Pielou evenness, (D) Simpson, (E) Shannon. ∗ Beta diversity: Bray-Curtis, Method name: PERMANOVA, Sample size: 104, Number of groups: 2, Test statistic: 3.423, p-value: 0.001, Number of permutations: 999; (F) Three-dimensional principal coordinates analysis plots using Bray-Curtis: Principal coordinates analysis depicting the beta diversity before and after ingestion of red ginseng BF: before (G), AF: after (H), PC: principal coordinate (I).
3.3. Differences in beta diversity before and after red ginseng intake
Beta diversity, which checks for similar sample diversity [22], was analyzed before and after red ginseng intake. Significant differences were noted in the microbial patterns in Bray-Curtis before and after red ginseng intake (p = 0.001). However, there were no significant differences in unweighted UniFrac distance metrics, weighted UniFrac distance, and Jaccard (Fig. 2).
3.4. Differences in phylum levels according to taxonomic classification before and after red ginseng intake
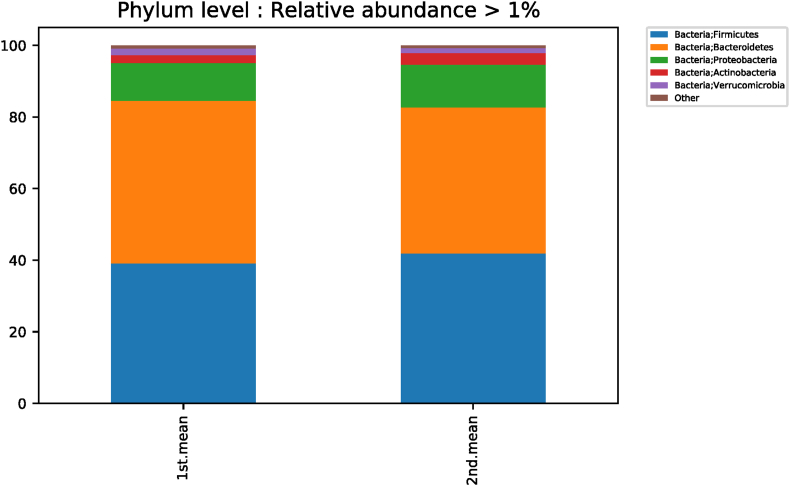
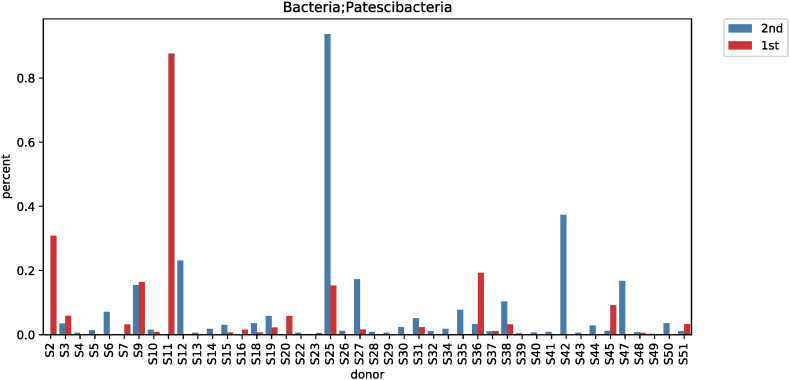
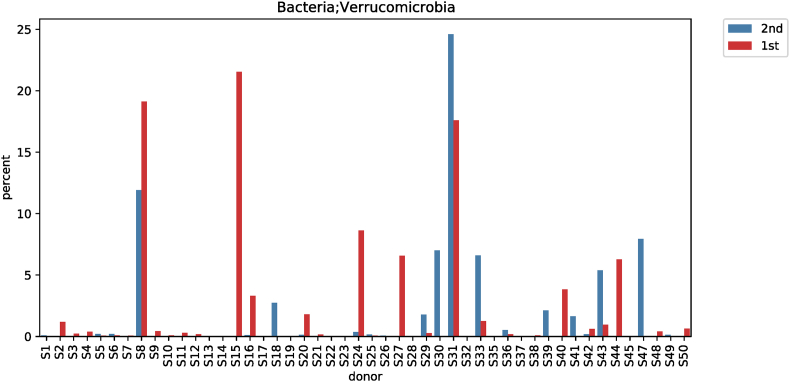
According to the taxonomic classification of subjects before and after red ginseng intake, a comparative analysis of different phylum levels showed significant differences among the three strains (Fig. S1). Fusobacteria decreased from 0.6158% ± 1.6365% to 0.4432% ± 2.4457% (p < 0.001; Fig. S2), while Patescibacteria increased from 0.0410% ± 0.1316% to 0.0543% ± 0.1426% (p = 0.008; Fig. S3). In comparison, Verrucomicrobia decreased from 1.8669% ± 4.7497% to 1.4285% ± 4.0707% (p = 0.030; Fig. S4) (Table 3).
Table 3.
Phylum and genus level difference according to taxonomic classification
| Taxon (Phylum) | Before (M ± SD) | After (M ± SD) | Kruskal-Wallis H | p-value | |
|---|---|---|---|---|---|
| Fusobacteria | 0.6158 ± 1.6365 | 0.4432 ± 2.4457 | 13.6573 | <0.001 | |
| Patescibacteria | 0.0410 ± 0.1316 | 0.0543 ± 0.1426 | 6.9591 | 0.008 | |
| Verrucomicrobia |
1.8669 ± 4.7497 |
1.4285 ± 4.0707 |
4.7015 |
0.030 |
|
| Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae |
Before (mean) |
After (mean) |
Kruskal -Wallis H |
p-value |
|
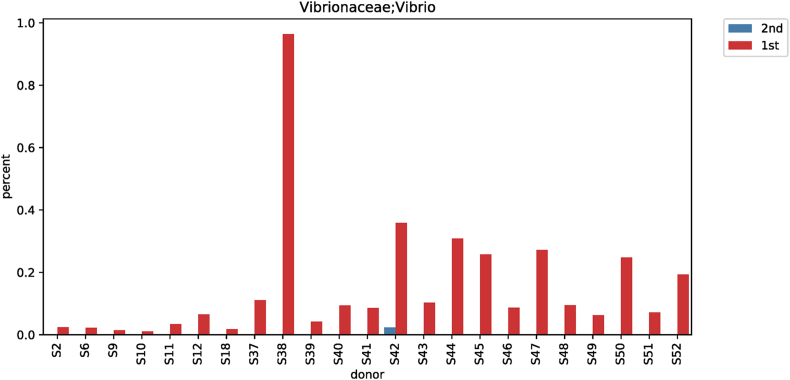
| Genus | Vibrio | 0.06825 | 0.00045 | 26.049 | <0.001 |
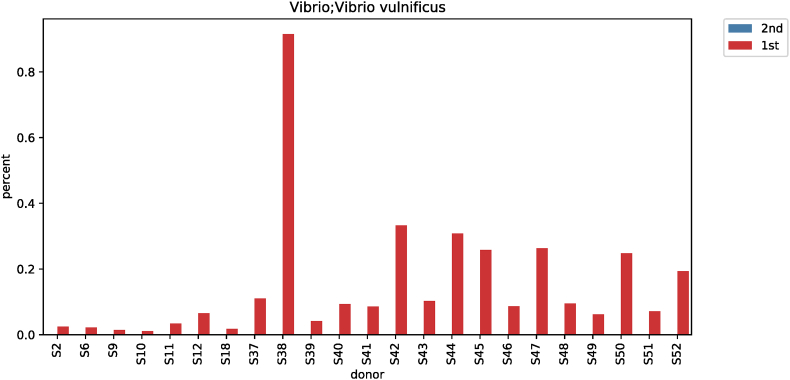
| Species | Vibrio vulnificus | 0.06665 | 0 | 28.648 | <0.001 |
Based on Kruskal-Wallis H test, Figure: Statistical package (SILVA 132v).
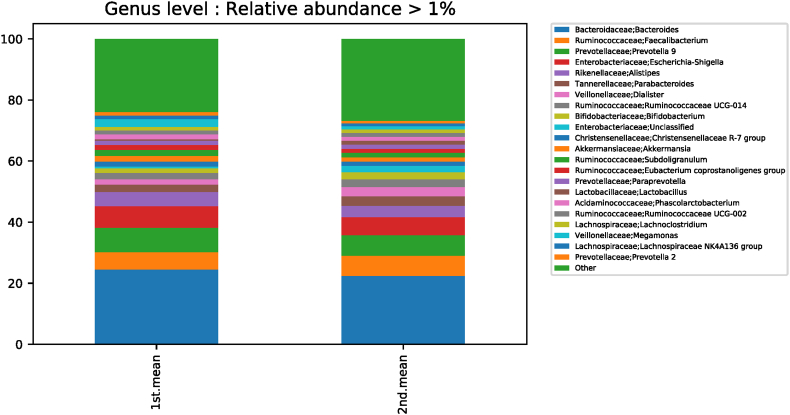
3.5. Differences in genus levels according to taxonomic classification before and after red ginseng intake
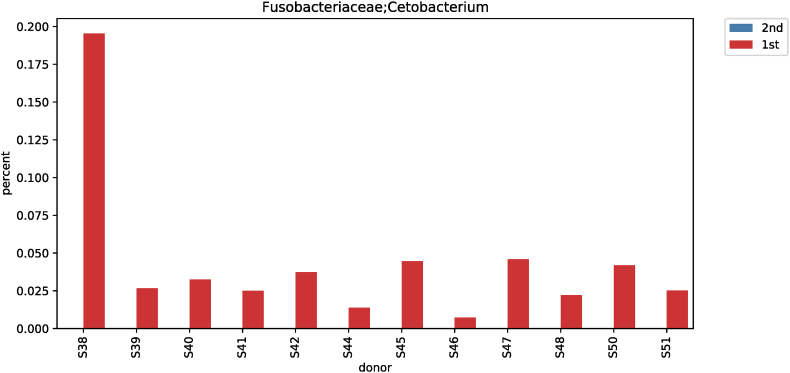
In terms of taxonomic classification, differences in genus levels before and after red ginseng intake were compared and analyzed. At the genus level, the Kruskal-Wallis test showed significant differences for 57 strains before and after red ginseng intake. In addition, an ANOVA showed a significant difference at the corresponding genus level, and statistical tests for strains with genus are shown in Table S1 and Fig. S5. Moreover, Cetobacterium was not detected after red ginseng intake in 12 subjects with Cetobacterium (0.0099% ± 0.0294%). Thus, a significant difference was noted before and after red ginseng intake for the Cetobacterium strain (p < 0.001) (Fig. S6).
3.6. Differences in vibrio (genus-, species-level) according to taxonomic classification before and after red ginseng intake
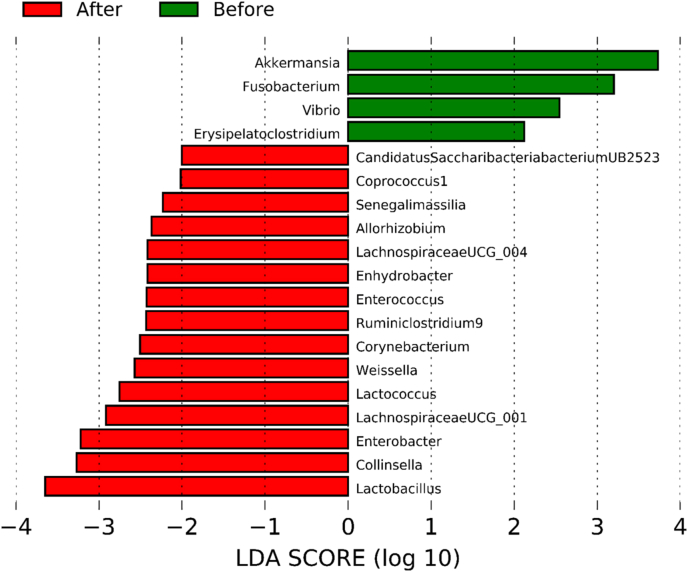
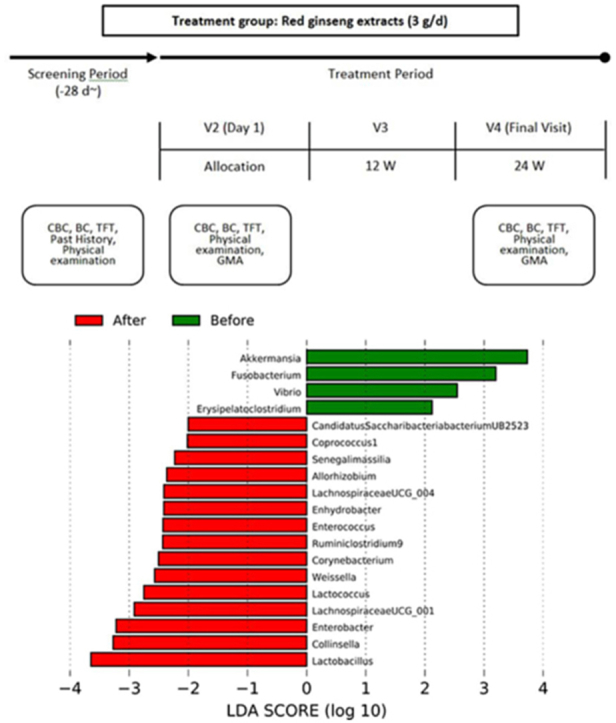
The results of analyzing species of strains were confirmed significant (p < 0.001) in Vibrio for subjects before and after their red ginseng intake (Table 3). In 23 of 52 subjects, Vibrio at the genus level decreased from 0.06825% to 0.00045% after red ginseng intake (p < 0.001; Fig. S7), and at the species level, Vibrio vulnificus was not detected after red ginseng intake in 23 patients (p < 0.001; Fig. S8). Moreover, the harmful bacteria's reduction and beneficial bacteria's increase were plotted based on each strain's differences based on the linear discriminant analysis score, which analyzed data distribution using log. The harmful bacteria that decreased were Akkermansia, Fusobacterium, Vibrio, Erysipelatoclostridium, and beneficial bacteria that increased were Candidatus Saccharibacteria bacterium UB2523, Coprococcus 1, Senegalimassilia, Allorhizobium, LachnospiraceaeUCG_004, Enhydrobacter, Enterococcus, Ruminiclostridium 9, Corynebacterium, Weissella, Lactococcus, LachnospiraceaeUCG_001, Enterobacter, Collinsella, and Lactobacillus (Fig. 3).
Fig. 3.
Differential taxonomy: linear discriminant analysis (LDA) score. The reduction of harmful bacteria and an increase in beneficial bacteria.
3.7. Comparative analysis of microbial strains before and after red ginseng consumption
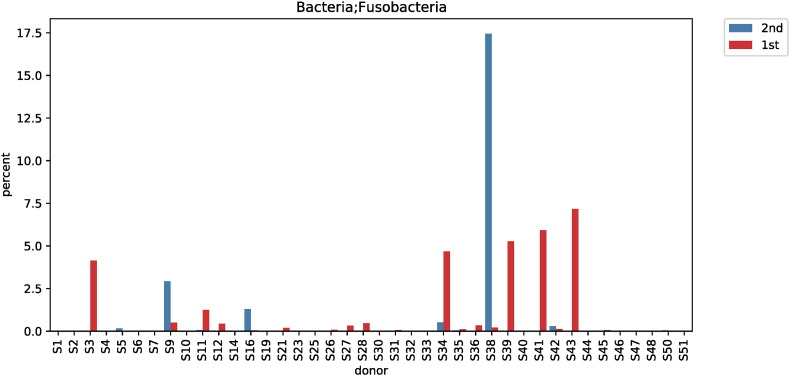
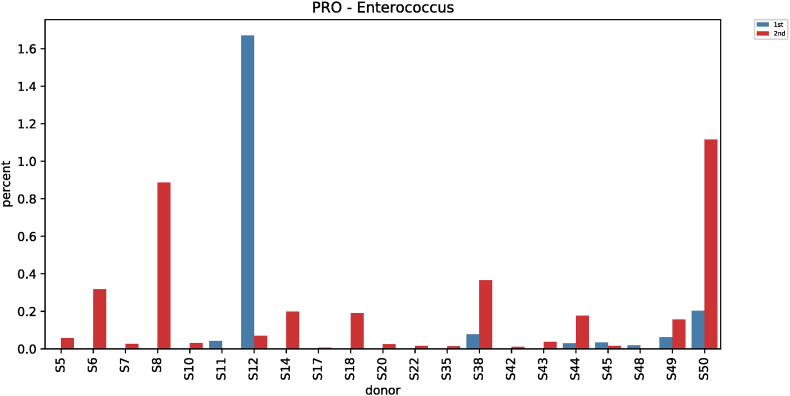
Comparative analysis of microbial strains before and after red ginseng intake showed changes in Fusobacterium in 36 subjects. In 25 of the 36 subjects, Fusobacterium decreased (0.006%–7.17%) after red ginseng intake, and increased (0.002%–17.43%) in 11 subjects. On average, Fusobacterium decreased from 0.6044% ± 1.6369% to 0.4219% ± 2.4430% (p = 0.021; Fig. S9). In addition, Collinsella was altered in 48 subjects, where it increased in 31 subjects (0.004%–10.72%) and decreased in 17 subjects (0.01%–9.27%). On average, Collinsella increased from 0.5173% ± 1.7006% to 0.5905% ± 1.5190% (p = 0.042; Fig. S10). Meanwhile, Anaerofilum, which makes short-chain fatty acids (SCFA), also increased from 0.0010 ± 0.0046 to 0.0018 ± 0.0039 on average (p = 0.030; Fig. S11). Moreover, Lactobacillus increased in 26 of the 45 test subjects (0.0002%–32.54%) and decreased in 19 subjects (0.004%–4.16%), with an average increase of 0.5388% ± 2.2972% to 1.2943 ± 4.7879% (p = 0.046; Fig. S12). Furthermore, Enterococcus was altered in 21 subjects. It increased in 17 subjects (0.007%–0.914%) and decreased in 4 subjects (0.019%–1.60%), increasing from 0.0411% ± 0.2326% to 0.0716% ± 0.2052% on average (p = 0.019; Fig. S13).
4. Discussion
Red ginseng extract was administered for 6 months, and changes in the intestinal microflora before and after intake were statistically analyzed. The observed OTUs (p = 0.003) and chao1 (p < 0.001) of alpha diversity showed significant differences, indicating a difference in richness before and after red ginseng intake. Comparative analysis of beta diversity showed significantly altered microbial patterns in Bray-Curtis before and after red ginseng intake (p = 0.001). Prebiotics are defined as a group of nutrients with beneficial effects on the human body. They improve digestion by stimulating intestinal bacteria's growth or activity in the large intestine [23]. This study showed that the intestinal bacteria's richness increased after red ginseng extract intake, which confirmed the possibility of red ginseng extract as a prebiotic.
Comparative analysis of differences before and after red ginseng intake showed significant differences among the three strains at the phylum level. Fusobacteria are harmful and are associated with colon cancer as they invade colon epithelial cells to enable their survival and maintenance [24,25]. Here, Fusobacteria significantly decreased (p < 0.001) and Patescibacteria, which prevents pathogenic Escherichia coli O157 infection, significantly increased (p = 0.008) [26]. In addition, Verrucomicrobia, which is found in increased amounts in patients with Parkinson's disease, affects neurodegeneration via neuritis, significantly decreased (p = 0.030) [27]. At the genus level, 57 strains showed significant differences. In particular, Vibrio, which was detected in 23 of 52 subjects before and after red ginseng intake, significantly decreased (p < 0.001). At the species level, V. vulnificus, which can cause wound infection, diarrhea, and sepsis, was not detected in 23 subjects after red ginseng intake. These findings follow previous reports by Na et al [28], who showed the antimicrobial activity of red ginseng extract against Vibrio. Moreover, a significant decrease in Fusobacterium and a significant increase in Collinsella, which is beneficial for producing short-chain fatty acids that correlate with the amount of insulin circulating in the body [29], were observed. In addition, the abundance of Enterococcus significantly increased (p = 0.019). Enterococcus is involved in the fermentation of dairy products, meat, and vegetables. Moreover, it is used as a probiotic, defined as living a microorganism that provides health benefits when administered in appropriate amounts by the Food and Agriculture Organization/World Health Organization. Enterococcus is effective in relieving irritable bowel syndrome symptoms and antibiotic-induced diarrhea and prevents various functional bowel diseases [30,31]. Lactobacillus, which is commonly used as a probiotic and has an antagonistic effect against pathogenic strains, also significantly increased (p = 0.046) [32,33].
In a recent study by Hong et al [20] on the association between red ginseng extract, nonalcoholic fatty liver disease, and intestinal bacteria, red ginseng extract was administered to hospitalized patients for 4 weeks. Decreases in aspartate transaminase, alanine transaminase, and gamma-glutamyl transferase (γ-GT or GGT) and changes in intestinal bacteria were observed, suggesting that red ginseng extract improved nonalcoholic fatty liver by changing the composition of intestinal bacteria. In addition, they observed that red ginseng extract increased Lactobacillus and decreased Verrucomicrobia, suggesting red ginseng extract's positive effects on intestinal bacteria. These findings follow the results of the current study. However, in this study, red ginseng extract was consumed for 24 weeks, which was 4 weeks longer than Hong et al.’s study [20]. Thus, relatively long-term effects were analyzed in the present study. Moreover, Hong et al.’s study had the strength of being conducted in an environment where lifestyle, including diet, can be easily controlled for subjects. In general, red ginseng extract is consumed in normal daily life; thus, the current study reflects the same results.
This study is noteworthy as it is the first to qualitatively and quantitatively confirm the positive effects of red ginseng extract on changes in the intestinal bacteria, using the latest technology in genetic analysis, besides the immunity and anti-inflammatory effects already known. Previous studies have assessed the effects of red ginseng extract on intestinal bacteria in animal experiments or specific diseases. In contrast, this study is meaningful as the effects of red ginseng extract on intestinal bacteria in humans were evaluated. In conclusion, the possibility of red ginseng extract as a probiotic was observed by assessing positive effects, such as an increase in the richness of certain strains, a reduction of harmful bacteria, and an increase of beneficial bacteria.
Other variables, such as lifestyle modification or diet, were not controlled and are considered limitations of this study. Moreover, a control group was not established due to the characteristics of the intestinal bacterial analysis. However, clinical significance can be found in that red ginseng extract can be consumed to give humans beneficial effects with no dietary restrictions.
Future studies that control for the number of subjects, the period of administration, control groups, lifestyle habits, and food intake are considered necessary for studying the more diverse effects of red ginseng extract on intestinal bacteria in the human body.
Data availability
Supplementary data related to this article can be found in the file attached to this manuscript: OVVPQ_8_4_Supplementary_data.
Authors' contribution
Young Kyun Kim: analysis and interpretation of data, collection and assembly of data, and drafting of the article. Keun Sang Yum: conception and design, critical revision of the article for important intellectual content, and final approval of the article.
Declaration of competing interest
The authors declare that there are no conflicts of interest concerning relevant financial interests, activities, relationships, affiliations, or any other as explicitly and implicitly expressed in the Editorial Policies for Authors.
Acknowledgment
This research was supported by the Korea Ginseng Corporation (KGC G18-Ef-12; 2018).
Footnotes
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
Fig. S6.
Fig. S7.
Fig. S8.
Fig. S9.
Fig. S10.
Fig. S11.
Fig. S12.
Fig. S13.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Quigley E.M.M. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y) 2013;9:560–569. [PMC free article] [PubMed] [Google Scholar]
- 2.Collins S., Reid G. Distant site effect of ingested prebiotics. Nutrients. 2016;8:523. doi: 10.3390/nu8090523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 4.Yoon M.Y., Lee K., Yoon S.S. Protective role of gut commensal microbes against intestinal infections [review] J Microbiol. 2014;52:983–989. doi: 10.1007/s12275-014-4655-2. [DOI] [PubMed] [Google Scholar]
- 5.Martin C.R., Osadchiy V., Kalani A., Mayer E.A. The brain-gut-microbiome axis. Cell Mol Gastroenterol Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carabotti M., Scirocco A., Maselli M.A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28:203–209. [PMC free article] [PubMed] [Google Scholar]
- 7.Vuong H.E., Hsiao E.Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol Psychiatry. 2017;81:411–423. doi: 10.1016/j.biopsych.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V., et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff S.C., Barbara G., Buurman W., Ockhuizen T., Schulzke J.D., Serino M., Tilg H., Watson A., Wells J.M. Intestinal permeability–a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., Zeng L., Chen J., Fan S., Du X., et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 11.Rennstam Rubbmark O., Sint D., Cupic S., Traugott M. When to use next generation sequencing or diagnostic PCR in diet analyses. Mol Ecol Resour. 2019;19:388–399. doi: 10.1111/1755-0998.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams A., Storton D., Buckles J., Llinas M., Wang W. Improvement of PCR-free NGS library preparation to obtain uniform read coverage of genome with extremely high AT content. J Biomol Tech. 2012;23(Suppl):S34. [Google Scholar]
- 13.Xiaoguang C., Hongyan L., Xiaohong L., Zhaodi F., Yan L., Lihua T., Rui H. Cancer chemopreventive and therapeutic activities of red ginseng. J Ethnopharmacol. 1998;60:71–78. doi: 10.1016/s0378-8741(97)00133-5. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen C.T., Luong T.T., Kim G.L., Pyo S., Rhee D.K. Korean Red Ginseng inhibits apoptosis in neuroblastoma cells via estrogen receptor β-mediated phosphatidylinositol-3 kinase/Akt signaling. J Ginseng Res. 2015;39:69–75. doi: 10.1016/j.jgr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H.Y., Qi L.W., Wang C.Z., Li P. Bioactivity enhancement of herbal supplements by intestinal microbiota focusing on ginsenosides. Am J Chin Med. 2011;39:1103–1115. doi: 10.1142/S0192415X11009433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S., Min H. Ginseng, the ‘immunity boost’: the effects of Panax ginseng on immune system. J Ginseng Res. 2012;36:354–368. doi: 10.5142/jgr.2012.36.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn Y.J., Kim M.J., Kawamura T., Yamamoto T., Fujisawa T., Mitsuoka T. Effect of Panax ginseng extract on growth responses of human intestinal bacteria and bacterial metabolism. J Ginseng Res. 1990;14:253–264. [Google Scholar]
- 18.Guo M.Z., Ding S., Zhao C., Gu X., He X., Huang K., Luo Y., Liang Z., Tian H., Xu W. Red ginseng and semen coicis can improve the structure of gut microbiota and relieve the symptoms of ulcerative colitis. J Ethnopharmacol. 2015;162:7–13. doi: 10.1016/j.jep.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Yang H.J., Kim M.J., Kwon D.Y., Kim D.S., Zhang T., Ha C., Park S. Combination of aronia, red ginseng, shiitake mushroom, and nattokinase potentiated insulin secretion and reduced insulin resistance with improving gut microbiome dysbiosis in insulin-deficient type 2 diabetic rats. Nutrients. 2018;10:948. doi: 10.3390/nu10070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong J.T., Lee M.J., Yoon S.J., Shin S.P., Bang C.S., Baik G.H., Kim D.J., Youn G.S., Shin M.J., Ham Y.L., et al. Effect of Korea red ginseng on nonalcoholic fatty liver disease: an association of gut microbiota with liver function. J Ginseng Res. 2020;7:4. doi: 10.1016/j.jgr.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S., Won C.H., Lee D.H., Lee M.J., Lee S., So S.H., Lee S.K., Koo B.S., Kim N.M., Chung J.H. Red ginseng root extract mixed with Torilus fructus and Corni fructus improves facial wrinkles and increases type I procollagen synthesis in human skin: a randomized, double-blind, placebo-controlled study. J Med Food. 2009;12:1252–1259. doi: 10.1089/jmf.2008.1390. [DOI] [PubMed] [Google Scholar]
- 22.Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography. 2010;33:2–22. doi: 10.1111/j.1600-0587.2009.05880.x. [DOI] [Google Scholar]
- 23.Davani-Davari D., Negahdaripour M., Karimzadeh I., Seifan M., Mohkam M., Masoumi S.J., Berenjian A., Ghasemi Y. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8:92. doi: 10.3390/foods8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z., Ji G. Fusobacterium nucleatum-positive colorectal cancer [review] Oncol Lett. 2019;18:975–982. doi: 10.3892/ol.2019.10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Zou Y., Wang J., Ma H., Zhang B., Wang S. The protective effects of 2’-fucosyllactose against E. coli O157 infection are mediated by the regulation of gut microbiota and the inhibition of pathogen adhesion. Nutrients. 2020;12:1284. doi: 10.3390/nu12051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C.H., Chen C.C., Chiang H.L., Liou J.M., Chang C.M., Lu T.P., Chuang E.Y., Tai Y.C., Cheng C., Lin H.Y., et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflammation. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Na H.S., Lim Y.J., YunYS, Choi Y.H., Oh J.S., Rhee J.H., Lee H.C. Protective effect of Ginsan against Vibrio vulnificus infection. J Bacteriol Virol. 2009;39:113–118. doi: 10.4167/jbv.2009.39.2.113. [DOI] [Google Scholar]
- 29.Qin P., Zou Y., Dai Y., Luo G., Zhang X., Xiao L. Characterization a novel butyric acid-producing bacterium Collinsella aerofaciens subsp. Shenzhenensis subsp. nov. Microorganisms. 2019;7:78. doi: 10.3390/microorganisms7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanchi H., Mottawea W., Sebei K., Hammami R. The genus Enterococcus: between probiotic potential and safety concerns—an update. Front Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braïek O.B., Smaoui S. Enterococci: between emerging pathogens and potential probiotics. Biomed Res Int. 2019:5938210. doi: 10.1155/2019/5938210. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walters J. Ecological role of Lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shokryazdan P., Sieo C.C., Kalavathy R., Liang J.B., Alitheen N.B., Faselah Jahromi M., Ho Y.W. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int. 2014;2014:927268. doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary data related to this article can be found in the file attached to this manuscript: OVVPQ_8_4_Supplementary_data.