Abstract
Background
Soft-tissue sarcomas (STSs) are heterogeneous and aggressive tumors, with high metastatic risk. The immunologic constant of rejection (ICR) 20-gene signature is a signature of cytotoxic immune response. We hypothesized that ICR might improve the prognostic assessment of early-stage STS.
Methods
We retrospectively applied ICR to 1455 non-metastatic STS and searched for correlations between ICR classes and clinicopathological and biological variables, including metastasis-free survival (MFS).
Results
Thirty-four per cent of tumors were classified as ICR1, 27% ICR2, 24% ICR3, and 15% ICR4. These classes were associated with patients’ age, pathological type, and tumor depth, and an enrichment from ICR1 to ICR4 of quantitative/qualitative scores of immune response. ICR1 class was associated with a 59% increased risk of metastatic relapse when compared with ICR2-4 class. In multivariate analysis, ICR classification remained associated with MFS, as well as pathological type and Complexity Index in Sarcomas (CINSARC) classification, suggesting independent prognostic value. A prognostic clinicogenomic model, including the three variables, was built in a learning set (n=339) and validated in an independent set (n=339), showing greater prognostic precision than each variable alone or in doublet. Finally, connectivity mapping analysis identified drug classes potentially able to reverse the expression profile of poor-prognosis tumors, such as chemotherapy and targeted therapies.
Conclusion
ICR signature is independently associated with postoperative MFS in early-stage STS, independently from other prognostic features, including CINSARC. We built a robust prognostic clinicogenomic model integrating ICR, CINSARC, and pathological type, and suggested differential vulnerability of each prognostic group to different systemic therapies.
Keywords: gene expression profiling, sarcoma
Background
Soft-tissue sarcomas (STSs) are rare and aggressive tumors with high metastatic risk. They constitute a heterogeneous group with at least 100 different pathological subtypes.1 Despite complete surgical resection of the tumor, ~50% of patients with early-stage STS develop metastatic relapse within 5 years,2 from which they will die. The results of adjuvant chemotherapy remain conflicting, with negative results from the largest randomized study, but positive results in term of relapses in meta-analyses. Today, adjuvant chemotherapy is not a standard treatment in adult-type STS: it can be proposed in high-risk situations (high-grade, deep-seated tumor, and tumor size >5 cm) after a shared decision-making with the patient. Improving the prognostic factors is crucial in order to better define the role, if any, of adjuvant chemotherapy by better selecting the ‘high-risk’ patients who will benefit from this potentially toxic strategy.
Currently, the prognostic assessment in early-stage STS is mainly driven by three tumor factors: pathological grade (most commonly based on the Federation Française des Centers de Lutte Contre le Cancer (FNCLCC) grading system3), pathological size, and depth.1 The grade is the most influential for the decision of adjuvant chemotherapy, but displays several limitations making imperfect the predictions. Since the 2000s, advances in high-throughput expression profiling technologies such as DNA microarrays4 allowed to tackle the molecular heterogeneity of cancers. In STS, they led to the refinement of molecular classification of certain sarcoma types5 and the development of prognostic signatures.6 Initial studies suggested the potential prognostic value of multigene signatures in STS, but were based on relatively small sample numbers and lacked validation set.7 8 Today, the most developed one is the 67-gene CINSARC signature.9 This signature, mainly based on genes involved in mitosis and maintenance of chromosomes integrity, classifies the tumors into high or low risk of relapse and outperforms the performances of pathological grade. The Genomic Grade Index (GGI), initially identified in breast cancer, is another proliferation-based prognostic signature that refines the prediction of metastasis-free survival in operated STS.10 11 Such signatures might reconcile the interest of adjuvant chemotherapy in STS, and two prospective clinical trials recently launched by the French Sarcoma Group are testing this hypothesis around CINSARC (NCT03805022, NCT04307277). Other reported prognostic signatures were mainly related to tumor hypoxia.12–14
In parallel, recent data suggested that the immune system might impact the outcome of patients with STS,15 16 and clinical trials testing immunotherapy are ongoing.17 Based on the notion that the composition of tumor-infiltrating immune cells and/or their functional orientation might impact the clinical outcome, immune multigene prognostic/predictive signatures have been developed,18 notably in breast,19 20 lung,21 and colon22 cancers. One of them is the immunologic constant of rejection (ICR) signature,23 which reflects the strength of the cytotoxic response. This 20-gene signature includes genes involved in Th-1 signaling (IFNG, TBX21, CD8A/B, IL12B, STAT1, and IRF1), Th-1 chemoattraction (such as the CXCR3 and CCR5 ligands, respectively, CXCL9 and CXCL10, and CCL5), cytotoxic functions (GNLY, PRF, GZMA, GZMB, and GZMH), immune checkpoints (IDO1, CTLA4, CD274/PDL1, PDCD1/PD1), and inhibition of T-cell function (FOXP3). In breast cancer, we showed that ICR added prognostic information to the current proliferation-based signatures, to which its integration improved the prognostication.24 Regarding STS, recent studies reported immune gene signatures associated with survival. The Cancer Genome Atlas (TCGA) study was the first one to suggest an association between infiltration score of immune cell types and survival in STS.25 Several other prognostic immune signatures were then reported.26–30 To our knowledge, none of them is currently used in clinical practice.
Here, we retrospectively applied the ICR signature to a dataset of 1455 non-metastatic STS, and searched for correlations between ICR-based classification and clinicopathological and biological variables, including metastasis-free survival (MFS).
Methods
STS samples and gene expression profiling
We retrospectively gathered clinicopathological and mRNA expression data of clinical STS samples from 16 public datasets.9 25 31–44 Data were collected from the National Center for Biotechnology Information (NCBI)/GenBank GEO, Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/), and ArrayExpress databases and authors’ websites (online supplemental file 1). The selection of datasets was based on the availability of clinical and expression data, including the expression level of 20 genes included in the ICR signature. Samples had been profiled using DNA microarrays or RNA-sequencing. The pooled dataset contained 1455 clinical samples of primary STS.
jitc-2021-003687supp001.pdf (866.2KB, pdf)
Gene expression data analysis
The pre-analytic processing first included normalization of each dataset separately, by using robust multichip average with the non-parametric quantile algorithm for the raw Affymetrix data and quantile normalization for the available processed non-Affymetrix microarray data. Normalization was done in R using Bioconductor and associated packages. Then, we mapped hybridization probes across the different technological platforms as reported.45 When multiple probes mapped to the same GeneID, we retained the one with the highest variance in each dataset. We log2-transformed the already normalized TCGA RNAseq data. Next, the batch effects were corrected across the 16 studies using standardization. Briefly, for each expression value in each study separately, all values were transformed by subtracting the mean of the gene in that dataset divided by its SD, mean and SD being measured on leiomyosarcoma samples.
We then applied several multigene signatures to each dataset separately. First, the ICR classifier based on consensus clustering (CC) analysis of the expression levels of 20 immune genes (namely, CCL5, CD274, CD8A, CD8B, CTLA4, CXCL9, CXCL10, FOXP3, GNLY, GZMA, GZMB, GZMH, IDO1, IFNG, IL12B, IRF1, PDCD1, PRF1, STAT1, and TBX21) as previously described.23 We applied several other expression signatures: CINSARC,9 TP53 activation pathway signature,46 MAPK-mut score, reflection of the degree of MAPK deregulation,23 and many immunity-related signatures, including the metagenes associated with the T-cell-inflamed signature (TIS),47 the tertiary lymphoid structures (TLS) signature,48 the signatures of 24 different innate and adaptative immune cell subpopulations defined by Bindea et al49 the cytolytic activity score,50 the pathway activation score of interferon-α (IFNα), IFNγ, and tumor necrosis factor-α (TNFα),46 and the antigen processing machinery (APM) score.51 Briefly, the CINSARC classifier was based on Spearman correlation to the nearest centroid using genes, data, and parameters described by authors.9 The MAPK-mut score, the signatures of TIS, TLS, and 24 immune cell subpopulations, the cytolytic activity and APM scores23 47–51 were based on a Z-score metagene using the gene lists described in each respective study. The pathway activation scores were measured with data and methodology using binary regression model described by authors.46 Finally, to explore more in depth the biological features related to the two prognostic groups defined by our clinicogenomic model and to broaden the therapeutic perspectives, we applied a supervised analysis to the largest dataset, the French Sarcoma Group series,9 divided in a learning set (n=148) and a validation set (n=141). In the learning set, we compared the tumor expression profiles between the ‘poor-prognosis’ (n=76) and ‘good-prognosis’ (n=72) groups using a moderated t-test and the following significance thresholds, p<1%, q<1%, and IFCI >2. The resulting 252-gene signature was analyzed using Gene Set Enrichment Analysis (GSEA)52 applied to the 50 hallmark gene sets from the Molecular Signatures Database (MSigDB). We also submitted it to Connectivity Map (CMap) algorithm against L1000 profiles/signatures present in the CMap dataset (https://clue.io/cmap), which catalog the transcriptional responses of human cells to a variety of chemical or genetic perturbations. The resulting connectivity scores (CSs) reflect the level of agreement between the tested signature and the L1000 profiles/signatures. Analysis was limited to the chemical perturbations by 2919 drugs/compounds with known mechanisms of action and targets through 54 cultured human cell lines (38,516 signatures). Typically, a drug is considered potentially useful for treating a B group tumor if the drug-induced differential gene expression profile is negatively correlated with the differentially expressed genes between the B versus A tumor groups. Because each drug might have been used through multiple (from 2 to 381) different culture conditions, we focused our analysis on the drug classes used in at least 50 conditions, and computed for each class the average of normalized connectivity scores (average ncs) and assessed its statistical significance by one sample Student’s t-test.
Statistical analysis
Correlations between tumor classes and clinicopathological variables were analyzed using the one-way analysis of variance or the Fisher’s exact test when appropriate. MFS was calculated from the date of diagnosis until the date of distant relapse or death from any cause, whichever occurred first. Follow-up was measured from the date of diagnosis to the date of last news for event-free patients. Survivals were calculated using the Kaplan-Meier method and curves were compared with the log-rank test. Univariate and multivariate prognostic analyses were done using Cox regression analysis (Wald test). The variables tested in univariate analysis included patients’ age and gender, pathological tumor type, grade, and size, tumor depth and site, the CINSARC-based risk (high vs low) and the ICR-based classification. Multivariate analysis incorporated all variables with a p value inferior to 5% in univariate analysis. We then built a clinicogenomic model based on the variables retained in multivariate analysis as follows. The patients’ population was divided into randomly selected learning and validation sets. In the learning set, and by starting from the three variables (pathological type, CINSARC, and ICR) found as significant in multivariate analysis, we searched for the best variable combination associated with MFS by using Akaike information criterion (AIC) stepwise regression analysis: those variables were then combined to build the clinicogenomic model. This classifier defined two groups of patients defined as ‘good-prognosis group’ and ‘poor-prognosis group’. Its robustness was then tested in the remaining validation set. The likelihood ratio (LR) tests were used to assess the prognostic information provided beyond that of each variable included in the model, assuming a X2 distribution. Changes in the LR values (LR-ΔX2) quantified the relative amount of information of one model compared with another. A resampling scheme was used to generate 100,000 random learning and validation sets allowing to test the clinicogenomic model in each validation set and to measure the proportion of random sets with significant p value for MFS. A diagram of analytic workflow (online supplemental file 2) summarizes all analyses.
Results
STS population and ICR classification
We analyzed 1455 clinical samples of STS primary tumors. Patients’ and tumor characteristics are summarized in table 1. The median patients’ age was 63 years (range 2–93) and 49% were females. The most frequent tumor sites were extremities, then internal trunk; 84% of tumors were deeply seated, below, or through the superficial fascia. As expected, the most frequent pathological types were liposarcomas (LPS), leiomyosarcomas (LMS) and undifferentiated pleomorphic sarcomas (UPS). The median pathological tumor size on the operative specimen was 9 cm. The FNCLCC pathological grade 3 was the most represented (59%), and 48% of samples were classified as high risk according to CINSARC. ICR classification defined 491 tumors as ICR1 (34%), 390 as ICR2 (27%), 353 as ICR3 (24%), and 221 as ICR4 (15%), with progressive increase of the enrichment of the immune signature from ICR1 to ICR4.
Table 1.
Clinicopathological characteristics
| Characteristics | N (%) |
| Median age (range), years | 63 (2–93) |
| Sex | |
| Female | 333 (49%) |
| Male | 348 (51%) |
| Tumor site | |
| Extremity | 208 (42%) |
| Head and neck | 9 (2%) |
| Internal trunk | 195 (39%) |
| Superficial trunk | 84 (17%) |
| Pathological type | |
| Leiomyosarcoma | 341 (24%) |
| Liposarcoma | 476 (33%) |
| Undifferentiated sarcoma | 330 (23%) |
| Myxofibrosarcoma | 105 (7%) |
| Other | 177 (12%) |
| Depth | |
| Deep | 195 (84%) |
| Superficial | 37 (16%) |
| Pathological tumor size, median (range), cm | 9 (1.2–39.5) |
| Pathological FNCLCC grade | |
| 1–2 | 165 (41%) |
| 3 | 240 (59%) |
| CINSARC risk | |
| Low | 763 (52%) |
| High | 692 (48%) |
FNCLCC, Fédération Nationale des Centres de Lutte Contre le Cancer.
ICR classification and clinicopathological and biological correlations
Differences were observed between the four ICR classes regarding the patients’ age (p=1.4E–04), the tumor depth (p=4.42E–02), and the pathological type (p=2.55E–10; online supplemental file 3): ICR1 class was associated with younger age, tumors deeply located, and less frequent UPS and myxofibrosarcoma types. No difference was found regarding the gender, the tumor site, the pathological tumor size and grade, and the CINSARC risk.
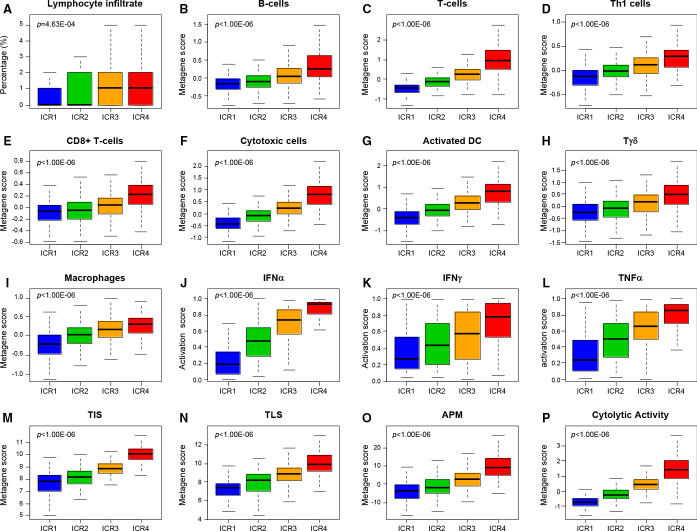
We also searched for correlations between these classes and immune-related factors and other expression signatures such as CINSARC (figure 1; online supplemental files 4 and 5). The lymphocyte infiltration percentage positively increased with increasing ICR classes (p=4.63E–04). We also found strong positive correlations (p<1.00E–06) with several immune gene expression signatures: the cytolytic activity score50 which increased from ICR1 to ICR4, as did the activation score of IFNα, IFNγ, and TNFα pathways,46 the TLS signature,48 the T-cell-inflamed signature,47 and the APM score.51 This immune pattern was further confirmed and refined using the Bindea’s signatures for 24 immune cell subsets,49 showing a strong enrichment from ICR1 to ICR4 for T-cells, cytotoxic T-cells, CD8+ T cells, T-helper cells, Tγδ cells, B-cells, activated dendritic cells, macrophages, neutrophils, and eosinophils. Of note, for all tested signatures, a continuum was present between the four classes. Regarding the other non-immune signatures tested, no correlation was found with the CINSARC classification (high vs low risk), whereas the activation score of TP53 pathway decreased from ICR1 to ICR4, as well as the MAPK-mut score, as previously reported.23
Figure 1.
Correlations of ICR classes with immunity-related parameters. For each ICR class, they indicated the percentage of samples with lymphocyte infiltrate (A), metagene expression scores of B-cells (B), T-cells (C), Th1 cells (D), CD8 +T cells (E), cytotoxic cells (F), activated dendritic cells (DC) (G), Tγδ cells (H), macrophages (I), activation score of IFNα (J), IFNγ (K), and TNFα (L) pathways, TIS (M) and TLS (N) signatures, antigen processing machinery score (O), and the cytolytic activity score (P). The p values are indicated (Fisher’s exact test or analysis of variance test when appropriate). APMS, Antigen Processing Machinery Score; DC, dendritic cell; IFN, interferon; Th1, T helper 1; TIS, T-cell-inflamed signature; TLS, tertiary lymphoid structure; TNF, tumor necrosis factor.
ICR classification and MFS
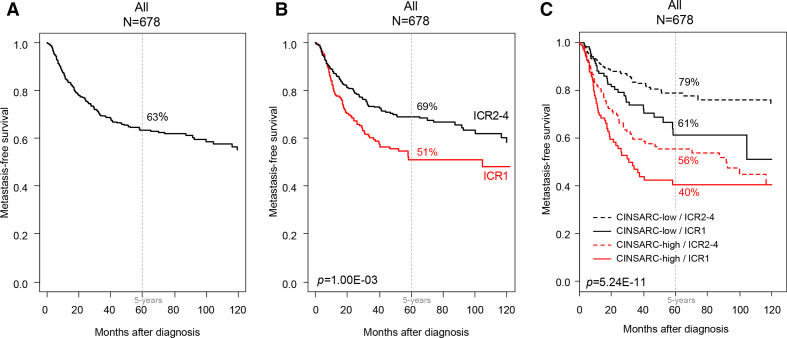
MFS data were available for 678 operated patients. With a median follow-up of 32 months (range 1–222), 209 patients displayed a metastatic relapse, and the 5-year MFS was 63% (95% CI 59% to 68%; figure 2A). No difference in MFS was observed between the ICR2, 3, and 4 classes (p=0.259, log-rank test; online supplemental file 6). These classes were thus pooled in an ICR2-4 class that we compared with ICR1 class in the subsequent analyses. These two classes were associated with the patients’ age (p=4.65E−06), the tumor depth (p=1.92E−02), and the pathological type (p=2.59E−11; table 2).
Figure 2.
Metastasis-free survival (MFS) in patients with soft-tissue sarcoma (STS) according to the immunologic constant of rejection (ICR) signature. Kaplan-Meier MFS curves in all patients (A), according to the ICR classification (B), and according to the four groups based on both ICR classes and CINSARC (C). The p values for the log-rank test are indicated.
Table 2.
ICR classification and correlations with clinicopathological characteristics
| Characteristics | N | ICR1 | ICR2-4 | P value* |
| n=491 (34%) | n=964 (66%) | |||
| Median age (range), years | 604 | 57.76 (2–91) | 63.86 (11–93) | 4.65E−06 |
| Sex | ||||
| Female | 333 | 106 (54%) | 227 (47%) | 0.0853 |
| Male | 348 | 89 (46%) | 259 (53%) | |
| Tumor site | ||||
| Extremity | 208 | 58 (40%) | 150 (43%) | 0.782 |
| Head and neck | 9 | 2 (1%) | 7 (2%) | |
| Internal trunk | 195 | 57 (39%) | 138 (39%) | |
| Superficial trunk | 84 | 28 (19%) | 56 (16%) | |
| Depth | ||||
| Deep | 195 | 61 (94%) | 134 (80%) | 1.92E−02 |
| Superficial | 37 | 4 (6%) | 33 (20%) | |
| Pathological type | ||||
| Leiomyosarcoma | 341 | 119 (25%) | 222 (23%) | 2.59E−11 |
| Liposarcoma | 476 | 151 (31%) | 325 (34%) | |
| Undifferentiated sarcoma | 330 | 86 (18%) | 244 (26%) | |
| Myxofibrosarcoma | 105 | 25 (5%) | 80 (8%) | |
| Other | 177 | 100 (21%) | 77 (8%) | |
| Pathological tumor size, median (range), cm | 241 | 10 (1.6–39.5) | 9 (1.2–36) | 0.113 |
| Pathological FNCLCC grade | ||||
| 1–2 | 165 | 52 (40%) | 113 (41%) | 0.92 |
| 3 | 240 | 78 (60%) | 162 (59%) | |
| CINSARC risk | ||||
| Low | 763 | 242 (49%) | 521 (54%) | 0.0963 |
| High | 692 | 249 (51%) | 443 (46%) |
*Student’s t-test for continuous variables and Fisher’s exact test for discrete variables.
FNCLCC, Fédération Nationale des Centers de Lutte Contre le Cancer.
In univariate analysis (table 3), they were associated with MFS: patients in the ICR1 class showed shorter 5-year MFS (51%, 95% CI 44% to 60%) than patients in the ICR2-4 class (69%, 95% CI 64% to 74%; p=1.00E−03, log-rank test; figure 2B), representing a 59% increased risk of event (HR=1.59, 95% CI 1.20 to 2.08; p=1.11E−03, Wald test; table 3). The other variables associated with shorter MFS included the pathological type (p=1.35E−06) and CINSARC classification (p=2.03E−10). In multivariate analysis (table 3), ICR classification remained associated with MFS (p=3.54E−03, Wald test), as well as pathological type and CINSARC, suggesting independent prognostic value. As shown in figure 2C, there was a relationship between ICR classification and MFS within each CINSARC class: the 5-year MFS was 79% (95% CI 73% to 85%) in the ‘CINSARC-low/ICR2-4’ group vs 61% (95% CI 50% to 75%) in the ‘CINSARC-low/ICR1’ group, and 56% (95% CI 48% to 64%) in the ‘CINSARC-high/ICR2-4’ group vs 40% (95% CI 31% to 53%) in the ‘CINSARC-high/ICR1’ group (p=5.24E−11; log-rank test). Similarly, ICR classification was associated with the clinical outcome of patients in each of the three major pathological types (online supplemental file 7): LMS (p=0.075), LPS (p=2.18E−02), and UPS (p=0.079). Of note, in the 181 TCGA samples informative for both lymphocyte infiltration and ICR, the lymphocyte infiltration, a relatively simple measure of immune response, was not associated with MFS (p=0.920) in univariate analysis, whereas ICR classification was associated (p=7.14E−03).
Table 3.
Univariate and multivariate prognostic analyses for MFS
| Characteristics | Univariate | Multivariate | ||||
| N | HR (95% CI) | P value | N | HR (95% CI) | P value* | |
| Age (years) | 371 | 1.00 (0.99 to 1.01) | 0.902 | |||
| Gender | ||||||
| Male vs female | 371 | 1.02 (0.70 to 1.49) | 0.909 | |||
| Tumor site | ||||||
| Head and neck vs extremity | 382 | 0.00 (0.00 to Inf) | 0.66 | |||
| Internal trunk vs extremity | 0.77 (0.50 to 1.18) | |||||
| Superficial trunk vs extremity | 0.81 (0.47 to 1.40) | |||||
| Depth | ||||||
| Superficial vs deep | 196 | 0.78 (0.38 to 1.61) | 0.495 | |||
| Pathological type | ||||||
| Liposarcoma vs leiomyosarcoma | 678 | 0.48 (0.35 to 0.67) | 1.35E−06 | 678 | 0.58 (0.41 to 0.81) | 1.51E−03 |
| Myxofibrosarcoma vs leiomyosarcoma | 0.45 (0.24 to 0.86) | 678 | 0.52 (0.27 to 0.98) | 4.44E−02 | ||
| Other vs leiomyosarcoma | 0.21 (0.08 to 0.57) | 678 | 0.25 (0.09 to 0.69) | 7.65E−03 | ||
| Undifferentiated sarcoma vs leiomyosarcoma | 0.43 (0.30 to 0.61) | 678 | 0.47 (0.33 to 0.68) | 4.40E−05 | ||
| Pathological tumor size, cm | 210 | 1.00 (0.96 to 1.04) | 0.898 | |||
| Pathological FNCLCC grade | ||||||
| 3 vs 1–2 | 307 | 1.43 (0.95 to 2.17) | 0.088 | |||
| CINSARC risk | ||||||
| High vs low | 678 | 2.48 (1.87 to 3.28) | 2.03E−10 | 678 | 2.19 [1.65 to 2.92) | 8.14E−08 |
| ICR classification | ||||||
| 1 vs 2–3 | 678 | 1.59 (1.20 to 2.08) | 1.11E−03 | 678 | 1.52 (1.15 to 2.00) | 3.54E−03 |
*Wald test.
FNCLCC, Fédération Nationale des Centres de Lutte Contre le Cancer; ICR, immunologic constant of rejection; MFS, metastasis-free survival.
Construction of a prognostic clinicogenomic model
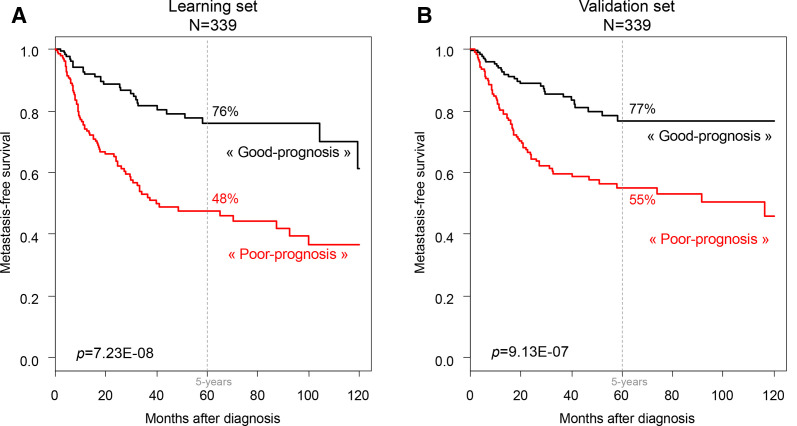
Starting from the three variables (pathological type, CINSARC, and ICR) found as significant in multivariate analysis, we then built a prognostic clinicogenomic model in a randomly defined learning set of 339 samples and tested its robustness in the validation set of 339 remaining samples. In the learning set, the three variables were retained after AIC stepwise regression analysis and thus included in the clinicogenomic model. Of note, the same variables were retained by using shrinkage methods such as Lasso (Least Absolute Shrinkage and Selection) and Ridge regression (data not shown). As expected, this model displayed prognostic value in the learning set (figure 3A), with 48% 5-year MFS (95% CI 40% to 57%) in the ‘poor-prognosis’ group (n=178) and 76% (95% CI 68% to 85%) in the ‘good-prognosis’ group (n=161; p=7.23E−08, log-rank test). The ROC AUC, measured using the R package timeROC (V.0.4), was 0.708 (p<1.00E−06, estimated using bootstrap resampling). Importantly, this prognostic value was maintained in the independent validation set, suggesting its robustness (figure 3B): the 5-year MFS was 55% (95% CI 47% to 64%) in the ‘poor-prognosis’ group (n=169) and 77% (95% CI 69% to 85%) in the ‘good-prognosis’ group (n=170; p=9.13E−07, log-rank test). Here, the ROC AUC was 0.659 (p=1.49E−04, estimated using bootstrap resampling). In the validation set, the clinicogenomic model provided more prognostic information than the models with pathological type alone (LR-ΔX2=20.22, p=4.08E−05), with CINSARC alone (LR-ΔX2=18.96, p=2.04E−03), with ICR alone (LR-ΔX2=31.18, p=8.64E−06), and with combined pathological type and CINSARC (LR-ΔX2=5.54, p=1.86E−02; online supplemental file 8). Analysis of the 100,000 random validation sets showed statistical significance of the model in 97% of iterations (p=9.88E−324, binomial test), further reinforcing its robustness.
Figure 3.
Metastasis-free survival (MFS) in patients with soft-tissue sarcoma according to the clinicogenomic model integrating the pathological type, CINSARC, and immunologic constant of rejection (ICR). Kaplan-Meier MFS curves in the learning set (A) and the validation set (B). The p values for the log-rank test are indicated.
Biological and therapeutic correlates of the two prognostic groups
We compared the gene expression profiles of the two model-based prognostic groups in the French Sarcoma Group samples9: 252 genes (118 overexpressed and 134 underexpressed in the poor-prognosis group) were identified as differentially expressed in the learning set and were validated in the validation set (online supplemental file 9). This 252-gene signature was submitted to GSEA applied to the 50 hallmark pathway signatures. Twenty-six pathways were significant (p≤0.01 and q≤0.01; online supplemental files 10 and 11): those associated with the poor-prognosis group included, for example, E2F targets, G2M checkpoint, MYC targets, DNA repair, mitotic spindle, MTORC signaling, and glycolysis, whereas those associated with the good-prognosis group included many signatures related to immune response such as IFN gamma response, allograft rejection, and TNF alpha signaling via NFKB, and metabolism and differentiation.
From the CMap database, we extracted 38,516 ranked gene lists corresponding to signatures induced by 2919 drugs and tested their correlation with our ranked 252-gene signature. A significant negative or positive correlation (q≤0.05) was obtained for 7211 signatures. Because each drug might have been used through multiple (from 2 to 381) different culture conditions, we focused our analysis on the drug classes used in at least 50 conditions; we computed for each class the average of normalized connectivity scores (mean ncs) and assessed its statistical significance. Thirty drug classes were significant (p≤0.05; table 4): two classes (adrenergic receptor agonist and acetylcholine receptor antagonist) showed positive correlation with our 252-gene signature, suggesting potential therapeutic value in the good-prognosis group, whereas 28 showed negative correlation suggesting potential therapeutic value in the poor-prognosis group. The top 10 classes, in terms of decreasing absolute value of average ncs, included inhibitors of HDAC, CDK, AKT, topoisomerase, MTOR, FGFR, EGFR, VEGFR, ATPase, tyrosine kinase. Other classes comprised PI3K, PDGFR, KIT, MET, Src, or JAK inhibitors.
Table 4.
Predicted candidate drugs for each prognostic group according to CMap
| Drug class | Cultured cell lines tested (n) | Mean normalized connectivity scores (95% CI) | P value* | Prognostic group candidate for this drug class |
| Adrenergic receptor agonist | 82 | 0.40 (0.16 to 0.63) | 1.11E−03 | Good-prognosis group |
| Acetylcholine receptor antagonist | 75 | 0.31 (0.06 to 0.56) | 1.70E−02 | Good-prognosis group |
| Dopamine receptor antagonist | 203 | −0.24 (−0.40 to 0.08) | 3.61E−03 | Poor-prognosis group |
| DNA inhibitor | 103 | −0.31 (−0.55 to 0.07) | 1.12E−02 | Poor-prognosis group |
| Retinoid receptor agonist | 76 | −0.35 (−0.62 to 0.08) | 1.26E−02 | Poor-prognosis group |
| Bcr-Abl inhibitor | 92 | −0.37 (−0.62 to 0.11) | 5.53E−03 | Poor-prognosis group |
| Aurora kinase inhibitor | 90 | −0.42 (−0.69 to 0.15) | 2.65E−03 | Poor-prognosis group |
| Calcium channel blocker | 104 | −0.42 (−0.64 to 0.21) | 1.99E−04 | Poor-prognosis group |
| Abl inhibitor | 108 | −0.49 (−0.72 to 0.26) | 5.11E−05 | Poor-prognosis group |
| NFKB inhibitor | 72 | −0.52 (−0.76 to 0.27) | 7.51E−05 | Poor-prognosis group |
| JAK inhibitor | 79 | −0.52 (−0.79 to 0.24) | 3.35E−04 | Poor-prognosis group |
| Glucocorticoid receptor agonist | 102 | −0.53 (−0.73 to 0.33) | 1.11E−06 | Poor-prognosis group |
| FLT3 inhibitor | 103 | −0.56 (−0.80 to 0.32) | 8.98E−06 | Poor-prognosis group |
| PDGFR inhibitor | 134 | −0.58 (−0.78 to 0.39) | 1.26E−08 | Poor-prognosis group |
| KIT inhibitor | 96 | −0.63 (−0.87 to 0.39) | 7.22E−07 | Poor-prognosis group |
| Src inhibitor | 76 | −0.65 (−0.91 to 0.39) | 4.28E−06 | Poor-prognosis group |
| RAF inhibitor | 58 | −0.65 (−0.95 to 0.35) | 6.01E−05 | Poor-prognosis group |
| MEK inhibitor | 88 | −0.66 (−0.91 to 0.41) | 1.14E−06 | Poor-prognosis group |
| PI3K inhibitor | 182 | −0.67 (−0.84 to 0.50) | 7.61E−13 | Poor-prognosis group |
| IKK inhibitor | 55 | −0.67 (−0.97 to 0.36) | 4.96E−05 | Poor-prognosis group |
| Tyrosine kinase inhibitor | 63 | −0.70 (−0.97 to 0.42) | 3.59E−06 | Poor-prognosis group |
| ATPase inhibitor | 81 | −0.71 (−0.93 to 0.50) | 2.41E−09 | Poor-prognosis group |
| VEGFR inhibitor | 196 | −0.73 (−0.88 to 0.58) | 3.05E−18 | Poor-prognosis group |
| EGFR inhibitor | 381 | −0.77 (−0.87 to 0.66) | 1.15E−37 | Poor-prognosis group |
| FGFR inhibitor | 58 | −0.77 (−1.04 to 0.50) | 4.86E−07 | Poor-prognosis group |
| MTOR inhibitor | 131 | −0.81 (−1.00 to 0.63) | 1.37E−14 | Poor-prognosis group |
| Topoisomerase inhibitor | 136 | −0.89 (−1.06 to 0.72) | 7.29E−19 | Poor-prognosis group |
| AKT inhibitor | 60 | −0.99 (−1.20 to 0.78) | 1.84E−13 | Poor-prognosis group |
| CDK inhibitor | 129 | −1.09 (−1.24 to 0.94) | 5.76E−28 | Poor-prognosis group |
| HDAC inhibitor | 148 | −1.19 (−1.27 to 1.11) | 5.63E−62 | Poor-prognosis group |
*One sample Student’s t-test.
AKT, protein kinase B; CDK, cyclin-dependant kinase; EGFR, epidermal growth factor receptor; HDAC, histone deacetylase; MTOR, mechanistic target of rapamycin kinase; VEGFR, vascular endothelial growth factor receptor.
Discussion
The absence of accurate prognostic/predictive features in patients with STS and the scarcity and heterogeneity of disease explain in part the difficulty to demonstrate the benefit, if any, of adjuvant chemotherapy. Given the potential prognostic value of immune parameters in STS and that of ICR signature in breast cancer, we tested its prognostic value in a series of operated STS samples. We demonstrated that ICR, reflect of an antitumor cytotoxic immune response, defines clinically and biologically relevant classes of STS. The signature is associated with clinicopathological and immunity-related tumor characteristics, and more importantly with MFS, refining the prognostic value of CINSARC. We built a robust clinicogenomic model combining pathological type, ICR, and CINSARC, and suggested potential targeted therapies in the poor-prognosis group. To our knowledge, this is the largest series reporting the prognostic value of an immune signature and of a clinicogenomic predictor in patients with early-stage STS.
The ICR signature was previously defined in breast cancer. The scarcity of STS explains the relatively small number of samples profiled in previous prognostic studies of molecular profiling, 310 in the largest one.9 To overcome this problem, we pooled 16 public sets including the multicentric prospective TCGA series representing a total of 1455 operated primary cancers. The whole series displayed the expected clinicopathological characteristics, including poor prognosis with 63% 5-year MFS. Our approach allowed avoiding any problem of overfitting since none of the STS samples had been used to generate the signature. More than 760 patients were informative for MFS, allowing the test of our prognostic hypothesis in multivariate analysis and to build and validate a robust prognostic model in independent datasets. Moreover, the whole-genome expression data provided opportunity to apply several gene signatures and modules potentially relevant to STS, including the CINSARC signature nowadays recognized as more prognostic than the pathological grade, as well as to search for potential therapeutic vulnerabilities in the two prognostic groups.
ICR classification defined 34% of STS samples as ICR1, 27% as ICR2, 24% as ICR3, and 15% as ICR4. There was an immunological continuum with an enrichment from ICR1 to ICR4 of scores reflecting the presence of an efficient antitumor immune response. These latter included not only the lymphocyte infiltrate and signatures of immune cell types, such as T-cells, cytotoxic T-cells, CD8+ T cells, T-helper cells, Tγδ cells, and antigen-presenting cells, but also more functional scores, such as those of IFNγ pathway activation, cytolytic activity, antigen presentation, and scores predictive for response to ICI (TIS and TLS). The decrease of TP53 pathway activation score46 observed from ICR1 to ICR4 agreed with the higher rate of inactivating TP53 mutations reported in ICR4 in breast cancer23 and in ‘immune-high’ classes (D and E) of STS.26 Such immune continuum, previously reported in breast cancer,24 suggested the biological relevance of ICR in STS.
The ICR classes did not correlate with the CINSARC classes, nor with two major prognostic features of STS (pathological grade and size). ICR1 class was associated with younger age, deep location, and less frequent UPS and myxofibrosarcoma types. Similar data have been reported with lower immune scores in younger patients, female patients, and UPS.30 Of note, proportions of the four ICR classes were not different between the myxofibrosarcoma and UPS types, whereas they were different between the other pathological types and myxofibrosarcoma or UPS type (data not shown), in agreement with the molecular similarities between myxofibrosarcoma and UPS.25 This low degree of correlation with the major prognostic variables of STS suggested a possible prognostic complementarity. The four ICR classes displayed different MFS, but no significant difference existed between ICR2, 3, and 4 that were thus merged (ICR2-4). The ICR1 class displayed shorter 5-year MFS than the ICR2-4 class (51% vs 69%, respectively: HR for relapse equal to ~1.6). Importantly, such prognostic value was independent of CINSARC, suggesting that the immune response (reflected by ICR) and the tumor cell proliferation (reflected by CINSARC) provide complementary prognostic information in STS. Interestingly, the 5-year MFS of patients classified as CINSARC-low/ICR1 was similar to that of patients classified as CINSARC-high/ICR2-4 (61% vs 56%, respectively). This is a major finding since CINSARC remains to date the most promising prognostic signature of STS. Of note, the lymphocyte infiltration, relatively simple measure of immune response, was not associated with MFS, whereas ICR classification, a more complex measure of immune response, was associated with MFS, as already observed in breast cancer.24
Our results refine the literature data. The immune tumor microenvironment of STS has been little studied to date,16 but the presence of tumor-infiltrating lymphocytes and expression of immune checkpoints has been associated with clinical outcome.53–56 For example, the high density of CD20+ lymphocytes was an independent favorable prognostic indicator for survival,54 as was the high density of CD8+ T cells in a series of 110 patients with UPS independently from the pathological grade.27 By contrast, macrophages have been associated with an immune-hostile tumor microenvironment promoting the progression of STS.55 57 We recently showed the independent unfavorable prognostic value of PDL1 mRNA expression in localized STS.58 The gene expression of 93 immune checkpoints and membrane markers of immune cells, tested in 253 STS (synovial sarcoma, myxoid LPS, sarcoma with complex genomic, and GIST),59 was heterogeneous between the pathological types and is associated with MFS for a few genes in certain types in univariate analysis. Further evidence of the prognostic value of immune variables in STS was provided by multigene signatures. The first one reported by TCGA showed the association of infiltration score of immune cell types, estimated using Bindea’s signatures,49 with survival in certain pathological types,25 but no multivariate analysis was done. A similar approach, based on the composition of tumor microenvironment estimated using the MCP-Counter deconvolution tool, defined five biologically and clinically relevant classes: two classes were ‘immune-low’ (A and B), two were ‘immune-high’ (D and E), and one was highly vascularized (class C). Class E, characterized by the presence of TLS and rich in B-cells, was associated with better survival in multivariate analysis including the clinicopathological factors and with response to PD1 blockade.26 However, CINSARC was not included in the multivariate analysis. Similarly, the CIBERSORT signatures of 22 immune cell types, assessed in 253 STS,59 showed heterogeneous profiles between the four pathological types represented and prognostic value for a few of them, but no multivariate analysis was done. In a reanalysis of the TCGA dataset, an association between shorter overall survival and lower immune scores, CD4+ T cells and CD8+ T cells estimated using ESTIMATE and TIMER deconvolution algorithms was reported.30 In a study dedicated to UPS, two groups (‘immune-high’ and ‘immune-low’) were identified from gene expression profiles and confirmed at the proteomic level.27 When compared with the ‘immune-low’ group, the ‘immune-high’ group showed longer overall survival, higher tumor mutational burden, and lower copy number alterations rate, and lower sensitivity to FGFR inhibition. No multivariate prognostic analysis was done. Thus, to our knowledge, our series is the largest one reported to date showing the independent prognostic value of an immune signature in adult-type STS. Our ICR signature is different from the deconvolution algorithms such as MCP-Counter.26 A comparison of their prognostic value in the same series of samples is warranted. MCP-Counter provides a quantification of the absolute abundance of eight immune cell (B-cells, T-cells, CD8+ T cells, cytotoxic lymphocytes, NK cells, monocytic lineage, myeloid dendritic cells, and neutrophils) and two stromal cell populations (endothelial cells and fibroblasts) in heterogeneous tissues, whereas ICR in addition provides a functional orientation (Th1, chemokine, cytotoxicity, adhesion) of the immune contexture.18 Furthermore, ICR (20 genes) includes a smaller number of genes than MCP-Counter (101 genes), which should facilitate its clinical application. Both methods, based on the bulk transcriptomics, have the drawback of losing the spatial organization. We also applied our ICR signature to publicly available RNA-seq data of primary and transplant STS developed in mice (50 samples, including 13 baseline samples untreated with radiotherapy and anti-PD1 drug) from a high-mutation mouse model of sarcoma60: in this study, the authors showed that the baseline transplant tumors from mice exhibited enrichment of immune-related pathways and resembled the ‘immune-high’ E class described by Petitprez et al26), while baseline primary tumors resembled the less inflamed sarcoma immune classes. In agreement with this study, we found that ICR score/class was associated with the type of baseline samples (online supplemental file 12): ICR score was higher in transplant tumors (n=4) than in primary tumors (n=9; p=0.036, Wilcoxon’s test) and only 25% of transplant tumors were classified as ICR1 vs 89% of primary tumors (p=0.052, Fisher’s exact test).
Given the independent prognostic value of ICR, we built a clinicogenomic model combining ICR and the two other variables significant in multivariate analysis (pathological type and CINSARC). Each of them provided a biological and prognostic information complementary from the others, leading to greater prognostic precision for the clinicogenomic model than for each variable alone or in doublet. The potential of clinicogenomic models over clinical or genomic models alone has been reported in breast cancer with models such as ROR-P,61 or recently RSClin.62 In STS too, recent clinicogenomic models have been published as nomograms through analysis of immune genes in the TCGA series, but without effort of validation in an independent set.28–30 Our model is the first one to show robustness in a validation set (339 samples), in which we confirmed better prediction accuracy than that achieved by using either clinical data or genomic signature alone.
CMap analysis identified potential therapeutic avenues in each prognostic group based on our model. Systemic therapies potentially more efficient in the poor-prognosis patients included chemotherapy and targeted therapies. Chemotherapy corresponded to topoisomerase inhibitors such as anthracyclines that represent the blackbone of chemotherapy for STS. The most significant targeted therapy was the class of HDAC (histone deacetylase) inhibitors (HDIs). These latter are epigenetic-modifying agents that inhibit sarcoma growth and progression in vitro and in vivo by inducing tumor cell apoptosis, causing cell cycle arrest, impairing tumor invasion, and preventing metastasis.63 Preclinical studies have also revealed that HDIs can sensitize sarcomas to chemotherapy, targeted therapies and immunotherapy, and clinical trials are ongoing either as monotherapy or in combination in sarcomas. Other targeted therapies included notably vascular endothelial growth factor receptor (VEGFR) inhibitors such as pazopanib already marketed in non-adipocyte sarcomas or regorafenib under development in STS,64 and many drugs under clinical trials in patients with STS such as CDK inhibitors,65 FGFR inhibitors,27 66 AKT inhibitor,67 or mTOR inhibitor.68 CMap analysis, based on analysis of cultured cancer cell lines cannot directly predict potential sensitivity to immune therapy. However, it may be anticipated that the good-prognosis tumors, which were associated with many signatures related to cytotoxic immune response and expression of genes involved in T-cell exhaustion (CD160, CTLA4, EOMES and IKZF1, HAVCR2, TIGIT, etc) should more benefit from immune checkpoint inhibitors than the poor-prognosis tumors.
Conclusion
In conclusion, we showed that the 20-gene ICR signature is associated with postoperative MFS of patients with early-stage STS independently from other prognostic features such as CINSARC, the most promising prognostic signature reported to date. We built a clinicogenomic model integrating ICR, CINSARC, and pathological type: it showed a robust prognostic value, and in silico analysis suggested differential vulnerability of each prognostic group to different systemic therapies. The strength of our results lies in (1) the number of 678 samples that, to our knowledge, makes our series the largest prognostic gene expression study reported so far in early-stage STS; (2) its originality, being the first one describing ICR signature in STS; (3) the biological and clinical relevance of ICR classification and its independent prognostic value; (4) the small number of genes in ICR (20 genes), which should facilitate its clinical application by using other tests applicable to formaldehyde-fixed paraffin-embedded samples as done with CINSARC69; and (5) the construction of a robust clinicogenomic model that provides more individualized prognostic information than either clinicopathological or genomic data alone. Limitations include the retrospective nature of our series and associated statistical biases, and heterogeneity with several different pathological types, and regarding CMap analysis, the possibility of false positives among the potentially useful drugs identified. Of course, analysis of larger series, retrospective, then prospective, is warranted to confirm our observation and to assess each pathological type independently, as well as analysis of STS preclinical models. The perspectives are therapeutic. If validated, ICR or our clinicogenomic model should help to better select the patients candidate to adjuvant chemotherapy and might thus help reconcile the disparate results of adjuvant/neoadjuvant chemotherapy in STS. Furthermore, since ICR has been associated with the response to ICIs in other cancers,70 good-prognosis patients with high ICR score (ICR four notably) might be suitable for testing immunotherapy. Poor-prognosis patients with low ICR score are less suitable for immunotherapy, but more candidates to chemotherapy and/or targeted therapies, such as those suggested by CMap analysis.
Acknowledgments
The authors wish to thank the patients and families who consented to donate tumor samples for research. We also thank the computing facilities DISC (Datacenter IT and Scientific Computing, CRCM) and DSIO (Institut Paoli Calmettes) for their technical support.
Footnotes
VN, AdN and PF contributed equally.
Contributors: FB and PF designed the study and performed data analyses; FB, PF, VN, AdN, and EM interpreted the results and wrote the manuscript; LM, OM, AI, ALC, J-YB, MC, D Bedognetti, and D Birnbaum contributed materials and analysis tools. All authors reviewed the manuscript and approved the final manuscript. FB is guarantor of the work.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository.
Data availability statement
All expression and clinicopathological data analyzed in the present study are available in the GEO, ArrayExpress, EGA, and TCGA databases, as indicated in supplemental table 1.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This in silico meta-analysis was based on data from published studies for which the informed patients’ consent to participate and the ethics approval from the institutional review board were already obtained by the authors. Participants gave informed consent to participate in the study before taking part.
References
- 1.Casali PG, Abecassis N, Bauer S, et al. Soft tissue and visceral sarcomas: ESMO–EURACAN clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. Elsevier 2018;29:iv51–67. [DOI] [PubMed] [Google Scholar]
- 2.ESMO/European Sarcoma Network Working Group . Soft tissue and visceral sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:102–12. 10.1093/annonc/mdu254 [DOI] [PubMed] [Google Scholar]
- 3.Guillou L, Coindre JM, Bonichon F, et al. Comparative study of the National Cancer Institute and French Federation of cancer centers sarcoma group grading systems in a population of 410 adult patients with soft tissue sarcoma. J Clin Oncol 1997;15:350–62. 10.1200/JCO.1997.15.1.350 [DOI] [PubMed] [Google Scholar]
- 4.Bertucci F, Houlgatte R, Nguyen C, et al. Gene expression profiling of cancer by use of DNA arrays: how far from the clinic? Lancet Oncol 2001;2:674–82. 10.1016/S1470-2045(01)00557-5 [DOI] [PubMed] [Google Scholar]
- 5.Guo X, Jo VY, Mills AM, et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin Cancer Res 2015;21:3501–11. 10.1158/1078-0432.CCR-14-3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merry E, Thway K, Jones RL, et al. Predictive and prognostic transcriptomic biomarkers in soft tissue sarcomas. NPJ Precis Oncol 2021;5:17. 10.1038/s41698-021-00157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y-F, John M, Falconer A, et al. A gene expression signature associated with metastatic outcome in human leiomyosarcomas. Cancer Res 2004;64:7201–4. 10.1158/0008-5472.CAN-04-1673 [DOI] [PubMed] [Google Scholar]
- 8.Francis P, Namløs HM, Müller C, et al. Diagnostic and prognostic gene expression signatures in 177 soft tissue sarcomas: hypoxia-induced transcription profile signifies metastatic potential. BMC Genomics 2007;8:73. 10.1186/1471-2164-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chibon F, Lagarde P, Salas S, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med 2010;16:781–7. 10.1038/nm.2174 [DOI] [PubMed] [Google Scholar]
- 10.Bertucci F, De Nonneville A, Finetti P, et al. The genomic grade index predicts postoperative clinical outcome in patients with soft-tissue sarcoma. Ann Oncol 2018;29:459–65. 10.1093/annonc/mdx699 [DOI] [PubMed] [Google Scholar]
- 11.Bertucci F, Finetti P, Monneur A, et al. Pathological grade-independent prediction of chemosensitivity by CINSARC should rehabilitate adjuvant chemotherapy in soft tissue sarcomas of any grade. Ann Oncol 2019;30:342–3. 10.1093/annonc/mdy528 [DOI] [PubMed] [Google Scholar]
- 12.Harris BHL, Barberis A, West CML, et al. Gene expression signatures as biomarkers of tumour hypoxia. Clin Oncol 2015;27:547–60. 10.1016/j.clon.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 13.Aggerholm-Pedersen N, Sørensen BS, Overgaard J, et al. A prognostic profile of hypoxia-induced genes for localised high-grade soft tissue sarcoma. Br J Cancer 2016;115:1096–104. 10.1038/bjc.2016.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Forker L, Irlam JJ, et al. Validation of a hypoxia related gene signature in multiple soft tissue sarcoma cohorts. Oncotarget 2018;9:3946–55. 10.18632/oncotarget.23280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C-H, Espinosa I, Vrijaldenhoven S, et al. Prognostic significance of macrophage infiltration in leiomyosarcomas. Clin Cancer Res 2008;14:1423–30. [DOI] [PubMed] [Google Scholar]
- 16.Koumarianou A, Duran-Moreno J. The sarcoma immune landscape: emerging challenges, prognostic significance and prospective impact for immunotherapy approaches. Cancers 2021;13 10.3390/cancers13030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siozopoulou V, Domen A, Zwaenepoel K, et al. Immune checkpoint inhibitory therapy in sarcomas: is there light at the end of the tunnel? Cancers 2021;13:360. 10.3390/cancers13020360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galon J, Angell HK, Bedognetti D, et al. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 2013;39:11–26. 10.1016/j.immuni.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 19.Sabatier R, Finetti P, Mamessier E, et al. Kinome expression profiling and prognosis of basal breast cancers. Mol Cancer 2011;10:86. 10.1186/1476-4598-10-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teschendorff AE, Miremadi A, Pinder SE, et al. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 2007;8:R157. 10.1186/gb-2007-8-8-r157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Cui Y, Diehn M, et al. Development and validation of an individualized immune prognostic signature in early-stage Nonsquamous non-small cell lung cancer. JAMA Oncol 2017;3:1529–37. 10.1001/jamaoncol.2017.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mlecnik B, Bifulco C, Bindea G, et al. Multicenter International Society for immunotherapy of cancer study of the consensus immunoscore for the prediction of survival and response to chemotherapy in stage III colon cancer. J Clin Oncol 2020;38:3638–51. 10.1200/JCO.19.03205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrickx W, Simeone I, Anjum S, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology 2017;6:e1253654. 10.1080/2162402X.2016.1253654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertucci F, Finetti P, Simeone I, et al. The immunologic constant of rejection classification refines the prognostic value of conventional prognostic signatures in breast cancer. Br J Cancer 2018;119:1383–91. 10.1038/s41416-018-0309-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network . Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017;171:950–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petitprez F, de Reyniès A, Keung EZ, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. 10.1038/s41586-019-1906-8 [DOI] [PubMed] [Google Scholar]
- 27.Toulmonde M, Lucchesi C, Verbeke S, et al. High throughput profiling of undifferentiated pleomorphic sarcomas identifies two main subgroups with distinct immune profile, clinical outcome and sensitivity to targeted therapies. EBioMedicine 2020;62:103131. 10.1016/j.ebiom.2020.103131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu C, Chen B, Huang Z, et al. Comprehensive profiling of immune-related genes in soft tissue sarcoma patients. J Transl Med 2020;18:337. 10.1186/s12967-020-02512-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen R, Liu B, Li X, et al. Development and validation of an immune gene-set based prognostic signature for soft tissue sarcoma. BMC Cancer 2021;21:144. 10.1186/s12885-021-07852-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai D, Xie L, Shui Y, et al. Identification of tumor microenvironment-related prognostic genes in sarcoma. Front Genet 2021;12:620705. 10.3389/fgene.2021.620705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baird K, Davis S, Antonescu CR, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 2005;65:9226–35. 10.1158/0008-5472.CAN-05-1699 [DOI] [PubMed] [Google Scholar]
- 32.Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 2010;42:715–21. 10.1038/ng.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck AH, Lee C-H, Witten DM, et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene 2010;29:845–54. 10.1038/onc.2009.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Detwiller KY, Fernando NT, Segal NH, et al. Analysis of hypoxia-related gene expression in sarcomas and effect of hypoxia on RNA interference of vascular endothelial cell growth factor A. Cancer Res 2005;65:5881–9. 10.1158/0008-5472.CAN-04-4078 [DOI] [PubMed] [Google Scholar]
- 35.Gibault L, Pérot G, Chibon F, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol 2011;223:64–71. 10.1002/path.2787 [DOI] [PubMed] [Google Scholar]
- 36.Gobble RM, Qin L-X, Brill ER, et al. Expression profiling of liposarcoma yields a multigene predictor of patient outcome and identifies genes that contribute to liposarcomagenesis. Cancer Res 2011;71:2697–705. 10.1158/0008-5472.CAN-10-3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hajdu M, Singer S, Maki RG, et al. IGF2 over-expression in solitary fibrous tumours is independent of anatomical location and is related to loss of imprinting. J Pathol 2010;221:300–7. 10.1002/path.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson SR, Guiliano D, Presneau N, et al. A molecular map of mesenchymal tumors. Genome Biol 2005;6:R76. 10.1186/gb-2005-6-9-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama R, Nemoto T, Takahashi H, et al. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Mod Pathol 2007;20:749–59. 10.1038/modpathol.3800794 [DOI] [PubMed] [Google Scholar]
- 40.Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet 2002;359:1301–7. 10.1016/S0140-6736(02)08270-3 [DOI] [PubMed] [Google Scholar]
- 41.Renner M, Wolf T, Meyer H, et al. Integrative DNA methylation and gene expression analysis in high-grade soft tissue sarcomas. Genome Biol 2013;14:r137. 10.1186/gb-2013-14-12-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skubitz KM, Francis P, Skubitz APN, et al. Gene expression identifies heterogeneity of metastatic propensity in high-grade soft tissue sarcomas. Cancer 2012;118:4235–43. 10.1002/cncr.26733 [DOI] [PubMed] [Google Scholar]
- 43.West RB, Nuyten DSA, Subramanian S, et al. Determination of stromal signatures in breast carcinoma. PLoS Biol 2005;3:e187. 10.1371/journal.pbio.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ylipää A, Hunt KK, Yang J, et al. Integrative genomic characterization and a genomic staging system for gastrointestinal stromal tumors. Cancer 2011;117:380–9. 10.1002/cncr.25594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertucci F, Finetti P, Viens P, et al. EndoPredict predicts for the response to neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer. Cancer Lett 2014;355:70–5. 10.1016/j.canlet.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 46.Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A 2010;107:6994–9. 10.1073/pnas.0912708107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coppola D, Nebozhyn M, Khalil F, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol 2011;179:37–45. 10.1016/j.ajpath.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 50.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JC, Davis C, Deshpande C, et al. Gene signature of antigen processing and presentation machinery predicts response to checkpoint blockade in non-small cell lung cancer (NSCLC) and melanoma. J Immunother Cancer 2020;8. 10.1136/jitc-2020-000974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Y, Shen J, Gao Y, et al. Expression of programmed cell death ligand 1 (PD-L1) and prevalence of tumor-infiltrating lymphocytes (TILs) in chordoma. Oncotarget 2015;6:11139–49. 10.18632/oncotarget.3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorbye SW, Kilvaer T, Valkov A, et al. Prognostic impact of lymphocytes in soft tissue sarcomas. PLoS One 2011;6:e14611. 10.1371/journal.pone.0014611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee C-H, Espinosa I, Vrijaldenhoven S, et al. Prognostic significance of macrophage infiltration in leiomyosarcomas. Clin Cancer Res 2008;14:1423–30. 10.1158/1078-0432.CCR-07-1712 [DOI] [PubMed] [Google Scholar]
- 56.Nabeshima A, Matsumoto Y, Fukushi J, et al. Tumour-associated macrophages correlate with poor prognosis in myxoid liposarcoma and promote cell motility and invasion via the HB-EGF-EGFR-PI3K/Akt pathways. Br J Cancer 2015;112:547–55. 10.1038/bjc.2014.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganjoo KN, Witten D, Patel M, et al. The prognostic value of tumor-associated macrophages in leiomyosarcoma: a single institution study. Am J Clin Oncol 2011;34:82–6. 10.1097/coc.0b013e3181d26d5e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertucci F, Finetti P, Perrot D, et al. PDL1 expression is a poor-prognosis factor in soft-tissue sarcomas. Oncoimmunology 2017;6:e1278100. 10.1080/2162402X.2016.1278100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dufresne A, Lesluyes T, Ménétrier-Caux C, et al. Specific immune landscapes and immune checkpoint expressions in histotypes and molecular subtypes of sarcoma. Oncoimmunology 2020;9:1792036. 10.1080/2162402X.2020.1792036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wisdom AJ, Mowery YM, Hong CS, et al. Single cell analysis reveals distinct immune landscapes in transplant and primary sarcomas that determine response or resistance to immunotherapy. Nat Commun 2020;17:6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. 10.1200/JCO.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sparano JA, Crager MR, Tang G, et al. Development and validation of a tool integrating the 21-gene recurrence score and clinical-pathological features to Individualize prognosis and prediction of chemotherapy benefit in early breast cancer. J Clin Oncol 2021;39:557–64. 10.1200/JCO.20.03007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang F, Choy E, Tu C, et al. Therapeutic applications of histone deacetylase inhibitors in sarcoma. Cancer Treat Rev 2017;59:33–45. 10.1016/j.ctrv.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mir O, Brodowicz T, Italiano A, et al. Safety and efficacy of regorafenib in patients with advanced soft tissue sarcoma (REGOSARC): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2016;17:1732–42. 10.1016/S1470-2045(16)30507-1 [DOI] [PubMed] [Google Scholar]
- 65.Kohlmeyer JL, Gordon DJ, Tanas MR, et al. CDKs in sarcoma: mediators of disease and emerging therapeutic targets. Int J Mol Sci 2020;21 10.3390/ijms21083018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dadone-Montaudié B, Laroche-Clary A, Mongis A, et al. Novel therapeutic insights in dedifferentiated liposarcoma: a role for FGFR and MDM2 dual targeting. Cancers 2020;12 10.3390/cancers12103058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayasaka N, Takada K, Nakamura H, et al. Combination of eribulin plus AKT inhibitor evokes synergistic cytotoxicity in soft tissue sarcoma cells. Sci Rep 2019;9:5759. 10.1038/s41598-019-42300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slotkin EK, Patwardhan PP, Vasudeva SD, et al. MLN0128, an ATP-competitive mTOR kinase inhibitor with potent in vitro and in vivo antitumor activity, as potential therapy for bone and soft-tissue sarcoma. Mol Cancer Ther 2015;14:395–406. 10.1158/1535-7163.MCT-14-0711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Guellec S, Lesluyes T, Sarot E, et al. Validation of the complexity index in sarcomas prognostic signature on formalin-fixed, paraffin-embedded, soft-tissue sarcomas. Ann Oncol 2018;29:1828–35. 10.1093/annonc/mdy194 [DOI] [PubMed] [Google Scholar]
- 70.Roelands J, Hendrickx W, Zoppoli G, et al. Oncogenic states dictate the prognostic and predictive connotations of intratumoral immune response. J Immunother Cancer 2020;8. 10.1136/jitc-2020-000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003687supp001.pdf (866.2KB, pdf)
Data Availability Statement
Data are available in a public, open access repository.
All expression and clinicopathological data analyzed in the present study are available in the GEO, ArrayExpress, EGA, and TCGA databases, as indicated in supplemental table 1.