Abstract
Purpose
Glioblastoma (GBM) patients suffer from a dismal prognosis, with standard of care therapy inevitably leading to therapy-resistant recurrent tumors. The presence of cancer stem cells (CSCs) drives the extensive heterogeneity seen in GBM, prompting the need for novel therapies specifically targeting this subset of tumor-driving cells. Here, we identify CD70 as a potential therapeutic target for recurrent GBM CSCs.
Experimental design
In the current study, we identified the relevance and functional influence of CD70 on primary and recurrent GBM cells, and further define its function using established stem cell assays. We use CD70 knockdown studies, subsequent RNAseq pathway analysis, and in vivo xenotransplantation to validate CD70’s role in GBM. Next, we developed and tested an anti-CD70 chimeric antigen receptor (CAR)-T therapy, which we validated in vitro and in vivo using our established preclinical model of human GBM. Lastly, we explored the importance of CD70 in the tumor immune microenvironment (TIME) by assessing the presence of its receptor, CD27, in immune infiltrates derived from freshly resected GBM tumor samples.
Results
CD70 expression is elevated in recurrent GBM and CD70 knockdown reduces tumorigenicity in vitro and in vivo. CD70 CAR-T therapy significantly improves prognosis in vivo. We also found CD27 to be present on the cell surface of multiple relevant GBM TIME cell populations, notably putative M1 macrophages and CD4 T cells.
Conclusion
CD70 plays a key role in recurrent GBM cell aggressiveness and maintenance. Immunotherapeutic targeting of CD70 significantly improves survival in animal models and the CD70/CD27 axis may be a viable polytherapeutic avenue to co-target both GBM and its TIME.
Keywords: antigens, neoplasm, cell engineering, brain neoplasms, immunotherapy, receptors, chimeric antigen
CD70 as an actionable immunotherapeutic target.
Introduction
Glioblastoma (GBM) is the most common malignant brain tumor in adults accounting for approximately 14.6% of all brain tumors.1 Despite an aggressive standard of care (SoC) including maximal surgical resection and chemoradiotherapy, GBM patients have a median survival time of less than 15 months, and a 5-year survival rate of less than 6.8%.2–4 GBM often recurs 7–9 months after resection of the primary tumor, at which point the tumor is often non-resectable, and poorly responsive to chemoherapy and/or radiotherapy, leaving patients with therapeutic options limited to clinical trial enrollment.5
In the past three decades, survival rates across several cancers have improved significantly, due in part to major advances in technology allowing for early detection, as well as significant leaps in targeted and novel therapeutic strategies.6 However, despite these advances, little to no improvement has been made in prognosis for GBM patients, who continue to suffer from dismal outcomes.
Therapeutic failure, in part, is due to extensive intratumoral heterogeneity at the cellular, genetic, and functional levels.7–9 This heterogeneity may be explained by a distinct subset of cells coined cancer stem cells (CSCs),8 10 11 which possess stem cell-like traits such as self-renewal, therapy evasion and multilineage differentiation.12–14 It is believed that this subpopulation of CSCs, after undergoing selective pressures from primary GBM (pGBM) SoC therapy, become chemotherapy and radiotherapy resistant, and seed formation of the therapy-resistant recurrent tumors.15 16 Expression of GBM CSC markers such as CD133, CD15, and CD44 are generally associated with worse clinical outcome.17 Thus, novel therapeutic interventions to target not only the tumor bulk, but the treatment resistant CSC population that seeds recurrence is necessary.
Immunotherapy holds great promise in cancer treatment, and recent studies in gliomas provide encouraging results.18–20 Among various immunotherapeutic approaches are adoptive T cell therapies, including chimeric antigen receptor (CAR)-T therapy. CAR-Ts are T-cells expressing a recombinant cell-surface receptor that directs these cells to specific tumor associated antigens (TAAs). On binding to the TAA, T-cells undergo major histocompatibility-independent activation and induce apoptosis of the target cell.21 22 However, to develop safe and effective CAR-T cells, novel tumor-specific antigens with a sufficient therapeutic window are required.
Genomic and proteomic data from a multiomic target development pipeline revealed CD70 as a suitable therapeutic target in recurrent GBM (rGBM). Our data show that CD70 is more highly expressed in rGBM samples compared with pGBM samples. Moreover, CD70 is absent on normal human astrocytes and neural stem cells, as is supported by the literature,23 thereby presenting a novel opportunity to target rGBM. CD70 is a transmembrane glycoprotein and a member of the tumor necrosis factor (TNF) superfamily, and is the only known ligand for CD27. While CD70 is transiently expressed on activated T- B-cells, as well as mature dendritic cells, it is minimally expressed in most normal tissues.24 25 Similarly, CD27 is primarily only expressed on specific subsets of T-cells B-cells, and NK cells.26–28 The CD70/CD27 signaling axis leads to differentiation, proliferation, and T- and B-cell survival and proliferation.29–31 Prolonged expression of CD70 has been shown to elicit lethal immunosuppression in mice,32 and result in exhaustion of effector memory T-cells in B-cell non-Hodgkin’s lymphoma.33
CD70 displays aberrant constitutive expression in a variety of cancers, include renal cell carcinoma, leukemia, non-small cell lung cancer, melanoma, GBM, and others.24 34–39 In 2005, researchers showed that in B-cell lymphoma, CD70 and CD27 are mutually overexpressed, resulting in increased proliferation and survival of tumor cells via amplified signaling through the CD70/CD27 axis.40 In the context of GBM, CD70 has been shown to promote tumor progression and invasion.41 While in healthy individuals CD70 plays a role in eliciting an immune response, its role in the tumor microenvironment is far more multi-faceted. Within the GBM microenvironment, CD70 mediates immune escape,42 and its overexpression leads to recruitment and activation of immunosuppressive T regulatory cells (Tregs)43 and tumor associated macrophages (TAMs).41 Together, these studies suggest that CD70 plays a major role in the recruitment and maintenance of the GBM immunosuppressive microenvironment, while promoting protumorigenic processes.
The soluble form of CD27 (sCD27) is detected at high levels in the blood of cancer patients.44 45 Currently, there are multiple therapeutic strategies targeting CD70-expressing malignancies,46–48 however, the prevalence of the CD70/CD27 interaction provides a rationale for synergistic therapeutic opportunities targeting both tumor cells and the immune microenvironment.
To our knowledge, this is the first time CD70 has been identified as an immunotherapeutic target on CSCs from patient-derived GBM samples. In the presented work, we conduct a systematic study evaluating the efficacy of CD70 CAR-T cells in using our established patient-derived GBM mouse model, illustrating the potential of a CD70-directed CAR-T therapy to offer hope to GBM patients suffering from a dismal prognosis.
Material and methods
Dissociation and culture of pGBM tissue
Human GBM samples (online supplemental table S1) were obtained from consenting patients, as approved by the Hamilton Health Sciences/McMaster Health Sciences Research Ethics Board. Brain tumor samples were dissociated in phosphate buffered saline (PBS) (ThermoFisher, Cat#10010049) containing 0.2 Wunsch unit/mL Liberase Blendzyme 3 (Millipore Sigma, Cat#5401119001), and incubated in a shaker at 37°C for 15 min. The dissociated tissue was filtered through a 70 µm cell strainer (Falcon, Cat#08-771-2) and collected by centrifugation (475 g, 3 min). Red blood cells were lysed using ammonium chloride solution (STEMCELL Technologies, Cat#07850). GBM cells were resuspended in Neurocult complete (NCC) media, a chemically defined serum-free neural stem cell medium (STEMCELL Technologies, Cat#05751), supplemented with human recombinant epidermal growth factor (20 ng/mL: STEMCELL Technologies, Cat#78006), basic fibroblast growth factor (20 ng/mL; STEMCELL Technologies Cat#78006), heparin (2 mg/mL 0.2% Heparin Sodium Salt in PBS; STEMCELL technologies, Cat#07980), antibiotic-antimycotic (1X; Wisent, Cat# 450-115-EL), and plated on ultra-low attachment plates (Corning, Cat#431110) and cultured as neurospheres. GBM8 and GBM4 was a kind gift from Dr. Hiroaki Wakimoto (Massachusetts General Hospital, Boston, MA, USA), RN1, S2b2 and WK1 were gifts from Dr Andrew Boyd (QIMR Berghofer Medical Research Institute, Australia).
jitc-2021-003289supp002.pdf (61.3KB, pdf)
Propagation of brain tumor stem cells
Neurospheres derived from minimally cultured (<20 passages) human GBM samples were plated on polyornithine-laminin coated plates for adherent growth. Adherent cells were replated in low-binding plates and cultured as tumorspheres, which were maintained as spheres on serial passaging in vitro. As shown before, compared with commercially available GBM cell lines, patient derived 3D cultures represent the variety of heterogeneous clones present within patient samples.49 These models recapitulate the key GBM morphological, architectural and expression features that are present in pGBM. These cells retained their self-renewal potential and were capable of in vivo tumor formation.
Glycocapture proteomics
Briefly, cells were lysed in PBS:TFE (trifluoroethanol) (50:50) using pulse sonication and by incubating the lysates at 60°C for 2 hours (lysates were vortexed every 30 min). Protein concentration was determined using the BCA assay (Pierce). Cysteines were reduced with DTT (5 mM final concentration) at 60°C for 30 min and alkylation was performed by adding iodoacetamide (25 mM final concentration) to the cooled lysates and subsequent incubation at room temperature for 30 min. Trypsin was added at a 1:500 dilution and protein digestion was performed overnight at 37°C. Tryptic peptides were desalted on C18 Macrospin columns (Nest Group), lyophilized and resuspended in coupling buffer (0.1M Sodium Acetate, 0.15M Sodium Chloride, pH 5.5). Glycan chains were oxidized using 10 mM NaIO4 for 30 min in the dark and peptides were again desalted. Lyophilized peptides were resolubilized in coupling buffer and oxidized glycopeptides were captured on hydrazide magnetic beads (Chemicel, SiMAG Hydrazide) for 12 hours at room temperature. The coupling reaction was catalyzed by adding aniline (50 mM) and the reaction was allowed to continue for 3 hours at room temperature.
Hydrazide beads containing the covalently coupled oxidized glycopeptides were thoroughly washed (2 × coupling buffer; 5×1.5M NaCl; 5 × HPLC H2O; 5 × methanol; 5×80% acetonitrile; 3 × water; 3×100 mM NH4OH, pH 8.0) to remove non-specific binders. N-glycopeptides were eluted off the hydrazide beads using 5U PNGase F in 100 mM ammonium bicarbonate at 37°C overnight. The deglycosylation reaction converts the asparagine residue, covalently linked to a glycan chain, to aspartic acid, the process carrying a signature mass shift of 0.98 Da.
Eluted (ie, deamidated) glycopeptides were recovered and the hydrazide beads were additionally washed 2× with 80% acetonitrile solution. Glycopeptides were desalted using C18 stage tips, eluted using 80% acetonitrile, 0.1% F.A. and lyophilised. The purified glycopeptides were dissolved in 21 µL 3% acetonitrile, 0.1% F.A. Peptide concentration was determined using a NanoDrop 2000 (Thermo) spectrophotometer.
RNA sequencing and Gene Set Enrichment Analysis/cytoscape analysis
Total RNA was extracted using the Norgen Total RNA isolation kit (Cat #48400) and quantified using a NanoDrop Spectrophotometer ND-1000. The RNA was sequenced using single-end 50 bp reads on the Illumina HiSeq platform (Illumina, San Diego CA, USA). FASTQ files were filtered to remove reads with length less than 36 bp using a bespoke Perl script. Filtered reads were then mapped to the human reference genome (GRCh38/hg38) and Gencode transcript models (V.25) using the STAR short-read aligner (V.2.4.2a).50 Gene-level read counts were exported by STAR, and merged with the Ensembl gene annotations into a count matrix in R. The count data matrix was then filtered to remove genes whose expression did not exceed a ‘counts per million’ (cpm) threshold of 0.5 in at least two samples. Filtered gene count data was depth-normalized using the calcNormFactors() function from edgeR (V.3.30.3),51 prepared for linear modeling using the voom() function from limma (V.3.44.3),52 and the main differential expression effect (shCD70) between cell lines was determined using the limma functions lmFit() and eBayes(). Individual cell line comparisons were subsequently performed using the exactTest() function from edgeR.
Differential gene expression profiles were generated by DESeq2 using the Galaxy online suite () and as imput of the Gene Set Enrichment Analysis (GSEA). Gene sets were randomized at 2000 permutations per analysis against Oncogenic (C6), Curated (C2) and Hallmark MSigDB collections of gene sets (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Secondary sphere formation assay
Tumorspheres were dissociated using 5–10 µL Liberase Blendzyme3 (0.2 Wunsch unit/mL) in 1 mL PBS for 5 min at 37°C. Based on each cell line’s growth kinetics, cells were plated at 200–1000 cells per well in 200 µL of NCC media in a 96-well plate. Cultures were left undisturbed at 37°C, 5% CO2. After 4 days, the number of secondary spheres formed were counted.
Limiting dilution analysis
GBM cells (CD70+/CD70- or Control/CD70 Knockout cells) were plated at varying densities (1, 3, 5, 10, 15, 25, 50, 75, 100, 125, 150, 200 cells/well) were flow-sorted onto a low-binding 96-well plate. Cells were cultured in 200 µL of media and incubated for 4 days at 37°C and 5% CO2. Following incubation, spheres formed were counted by microscopy. For each sample, the frequency of sphere-forming cells in each cell line was determined using the elda() function from statmod (V.1.4.36). A single-hit model with a log-log binomial regression was applied to determine CIs for sphere-forming cell frequency. A χ2 likelihood ratio test statistic was applied to signify difference between samples.
Cell proliferation assay
Single cells were plated in a 96-well plate at a density of 200–1000 cells/200 µL (based on each cell line’s growth kinetics) per well in quadruplicate and incubated for 5 days. 20 µL of Presto Blue (ThermoFisher, Cat#A13262), a fluorescent cell metabolism indicator, was added to each well approximately 4 hours prior to the readout time point. Fluorescence was measured using a FLUOstar Omega Fluorescence 556 Microplate reader (BMG LABTECH) at excitation and emission wavelengths of 535 nm and 600 nm, respectively. Readings were analyzed using Omega analysis software.
Receptor internalization and antibody drug conjugate assay
For detection of internalization, 200,000 cells were being used for each condition, where both were incubated with antibody 30 min on ice, rinsed twice and left toincubate for 2 hours either at 37°C or at 4°C, before being analyzed under flow cytometry.
GBM CSCs expressing CD70 on their cell surface were seeded and incubated for 30 min with different concentrations of he-Lm-Fab’2 anti-CD70, followed by addition of 13 nM of 2ºADC α-HFab-NC-MMAF (conjugated with Monomethyl auristatin F) (Moradec, Cat# AH-121-AF) and proliferation was measured after 5 days (n=3 for BT241s and n=2 for HEK293s). The manufacturer’s protocol was followed directly, with the exception of using twice the initial amount of recommended antibody (40 nM).
In vivo intracranial injections and H&E/immunostaining of xenograft tumors
Animal studies were performed according to guidelines under Animal Use Protocols of McMaster University Central Animal Facility. Intracranial injections in 6–8 week NOD/SCID gamma (NSG) mice were performed as previously described53 using either BT241, GBM8 or GBM4 cells (1 00 000 cells/mouse). Briefly, a burr hole is drilled at the point located 2 mm behind the coronal suture, and 3 mm to the right of the sagittal suture and GBM cells suspended in 10 µL PBS are intracranially injected with a Hamilton syringe (Hamilton, Cat#7635-01) into the right frontal lobes of 6–8 weeks NSG mice. For CAR-T treatment, ConCAR-T or CD70CAR-T cells were injected intratumorally once a week for 2 weeks (for BT241, 1M first week then 0.5M; for GBM8 0.75M first week then 1M for GBM8). For tumor volume evaluation, animals were sacrificed when control mice reached endpoint. When mice reached endpoint, they were perfused with 10% formalin and collected brains were sliced at 2 mm thickness using a brain-slicing matrix for paraffin embedding and H&E staining. Images were captured using an Aperio Slide Scanner (Leica Biosystems) and analyzed using ImageScope v11.1.2.760 software (Aperio). For survival studies, all the mice were kept until they reached endpoint and number of days of survival were noted for Kaplan-Meier analysis. CD3 stained slides were scanned and captured using an Aperio Slide Scanner and analyzed using ImageScope V.11.1.2.760 software (Aperio). Tumor areas were generated using Aperio Membrane Algorithm.
Generation of knockdown/knockout Lentivirus
Guide RNAs (gRNAs) targeting AAVS1 (5’-GGGGCCACTAGGGACAGGAT-3’) and CD70 (A: 5’- GCTGAGCCTGTGCGAAGCGC-3’; B: 5’-ATGGGACCAAAGCAGCCCGC-3’ were obtained from TKOv354 and cloned into a single-gRNA lentiCRISPRv2 construct (Addgene 52961). Sequences were verified using Sanger sequencing. Lentiviral vectors shCD70-1 and shCD70-2, expressing short hairpin RNA (shRNAs) targeting human CD70 (5’ CCATCGTGATGGCATCTACAT3’ and 5’TGGCATCTACATGGTACACAT3’, respectively), and the control vector, shGFP (5’ACAACAGCCACAACGTCTATA 3’), were gifts from Dr. Jason Moffat. Each construct was packaged independently into lentivirus using second-generation packaging constructs. Briefly, HEK293T cells were seeded into T75 cm2 flasks at a density of 10 million cells per flask and cultured in high-glucose DMEM with 2 mM L-glutamine and 1 mM sodium pyruvate supplemented with 1% non-essential amino acid solution and 10% fetal bovine serum. The following day, the HEK293T media was replaced with viral harvesting media (HEK culture media supplemented with 10 mM HEPES and 1 mM sodium butyrate). Next, 15 µg of transfer plasmid (lentiCRISPRv2, AAVS1, crCD70-A or crCD70-B), 7.2 µg of psPAX2 (Addgene), and 4.8 µg of pMD2.G (Addgene) were mixed with polyethylenimin at a 1:3 ratio (m:v) in 1.3 mL of Opti-MEM. After complexing for 15 min at room temperature, the PEI/DNA mixture was transferred dropwise into the HEK293T-containing flasks. Viral supernatants were collected 24 and 48 hours after transfection and then concentrated using ultracentrifugation (41 832 g for 2 hours at 4°C) before being aliquoted and stored at −80°C.
Generation of CAR lentivirus
Human anti-CD70 (he_L and he_Lm) scFv sequences were synthesized with a 5’ leader sequence and 3’ Myc tag by Genescript. The scFv was cloned into the lentiviral vector pCCL ΔNGFR (kindly provided by Dr. Bramson, McMaster University, Hamilton, ON, Canada) downstream of the human EF1α promoter leaving ΔNGFR intact downstream of the minimal cytomegalovirus promoter. Empty pCCL ΔNGFR was used as a control vector. Replication-incompetent lentiviruses were produced by cotransfection of the CAR vectors and packaging vectors pMD2G and psPAX2 in HEK293FT cells using Lipofectamine 3000 (ThermoFisher, Cat#L3000075) as recommended by the manufacturer. Viral supernatants were harvested 24 and 48 hours after transfection and concentrated by ultracentrifugation at 41 832 g for 2 hours at 4°C. The viral pellet was resuspended in 1.0 mL of T cell media, aliquoted and stored at −80°C.
Generation of CAR-T cells
Peripheral blood mononuclear cells (PBMCs) from consenting healthy blood donors were obtained using SepMate (STEMCELL technologies, Cat#85450). This research was approved by the McMaster Health Sciences Research Ethics Board. 1×105 cells in XSFM media (Irvine Scientific, Cat#91141) were activated with anti-CD3/CD28 beads at a 1:1 ratio (Dynabeads, GIBCO, Cat#113.31D) in a 96-well round bottom plate with 100 U/mL rhIL-2 (Peprotech, Cat#200-02). Twenty-four hours after activation, T cells were transduced with lentivirus at an MOI~1. CAR-T cell cultures were expanded into fresh media (XSFM media supplemented with 100 U/mL rhIL-2) as required for a period of 6–8 days prior to experimentation.
Evaluation of cytokine release
NGFR+ sorted CAR-T cells (CD70CAR or ConCAR) were co-incubated with GBM cells at a 1:1 ratio for 24 hours. Supernatants were collected in duplicate for each condition and stored at 80°C for analysis of cytokines. Human TNF-α DuoSet ELISA kit (R & D Systems, Cat #: DY210-05) and IFN-γ DuoSet ELISA kit (R & D Systems, Cat #: DY285B-05) were used for quantification of the two cytokines by ELISA, according to manufacturer’s recommendation.
Flow cytometric analysis and sorting
GBM cells and T cells in single cell suspensions were resuspended in PBS+2 mM EDTA. GBM cells were stained with he_L or he_Lm IgG (0.064–1000 nM) or by IgG control AffiniPure Goat Anti-Human IgG, F(ab’)2 fragment specific, Jackson ImmunoResearch, Cat#109-005-006) or APC-conjugated anti-CD70 antibody (Miltenyi Biotech, REA 292) and incubated for 30 min on ice. CAR-T cells were stained with fluorescent tagged anti-CD3 (BD Biosciences, Cat#557851), anti-NGFR (Miltenyi Biotech, Cat#130-112-790) and anti-c-Myc (Miltenyi Biotech, Cat#130-116-653). Samples were run on a MoFlo XDP Cell Sorter (Beckman Coulter). Dead cells were excluded using the viability dye 7AAD (1:10; Beckman Coulter, A07704). Compensation was performed using mouse IgG CompBeads (BD Biosciences, Cat#552843).
Cytotoxicity assays
Luciferase-expressing GBM cells at a concentration of 30 000 cells/well were plated in 96-well plates in triplicates. In order to establish the BLI baseline reading and to ensure equal distribution of target cells, D-firefly luciferin potassium salt (15 mg/mL) was added to the wells and measured with a luminometer (Omega). Subsequently, effector cells were added at 4:1, 3:1, 2:1, 1:1, and 0:1 effector-to-target (E:T) ratios and incubated at 37°C for 4–8 hours. BLI was then measured for 10 seconds with a luminometer as relative luminescence units (RLU). Cells were treated with 1% Nonidet P-40 (NP40, Thermofisher, Cat#98379) to measure maximal lysis. Target cells incubated without effector cells were used to measure spontaneous death RLU. The readings from triplicates were averaged and percent lysis was calculated with the following equation:
Isolation and evaluation of immune cells from brain tumor samples
EasySep human CD45 Depletion kit II (Stem Cell Technology, Cat#: 17898) was used to extract immune cells from freshly dissected patient tumors, according to the manufacturer’s protocol but with slight modifications. A single-cell suspension from the tumor is first prepared and resuspended at 108 cells/mL in EasySep Buffer (Cat#: 20144). A 12.5 µL of EasySep Human CD45 Depletion Cocktail II (Cat#: 17 898C) is then added to the suspension and incubated at room temperature for 5 min. 20 µL of EasySep Dextran RapidSpheres (Cat #: 50101) is then added to the suspension and incubated at room temperature for 3 min. The final volume of the mixture is then brought up to 2.5 mL using EasySep Buffer, and placed in an EasySep Magnet (Cat #: 18000) for 5 min to allow for separation of bead-bound CD45+ cells. The solution containing CD45- cells is then poured off, and the CD45+ cells remaining in the tube are harvested for future experimentation. For optimal CD45+ cell recovery, the separation should be performed twice.
To identify individual tumor immune microenvironment (TIME) immune cell populations, CD45+ cells were thawed and used immediately to run a panel of antibodies in order to identify individual immune cell populations. Antibodies used are as follows and were used according to manufacturer’s protocol: PE-Cy7 Mouse Anti-Human CD3 (Cat 563423; BD Pharmingen), PE Mouse Anti-Human CD4 (Cat555347; BD Pharmingen), APC Mouse Anti-Human CD8 (Cat555369; BD Pharmingen), PE-CF594 Mouse Anti-Human CD68 (Cat564944; BD Horizon), APC-H7 Mouse Anti-Human HLA-DR (Cat561358; BD Pharmingen), BV421 Mouse Anti-Human CD27 (Cat562514; BD Horizon).
CAR-T fratricide
Jurkat human T lymphocytes (Cedarlane Cat#: TIB-152) were expanded and grown in RPMI 1640 (Gibco Cat#:11 875-093) with 10% FBS (Multicell Cat#:08105), 1% Penicillin-Streptomycin (Gibco Cat#:15 140-122) and 10 mM HEPES (Gibco Cat#: 15 630-080). Jurkat cells were then transduced with either an shGFP or shCD70 lentiviral construct and selected for by puromycin selection. shGFP Jurkat cells were sorted by flow cytometry to isolate a CD70hi shGFP population, and untransduced Jurkat cells were sorted to isolate a CD70hi population. shGFP and shCD70 Jurkat cells were then transduced with either Control CAR or CD70 CAR virus and allowed to expand. At 4 and 8 days after transduction, each population (shGFP ConCAR; shGFP CD70CAR; shCD70 ConCAR; shCD70 CD70CAR) was assessed by flow cytometry for the following markers: NGFR, CD70, CD69, viability.
SciRNA sequencing analyses
Data source
Single-cell RNAseq (scRNAseq) data from Neftel et al was obtained from Gene Expression Omnibus (GEO; accession number GSE131928)55; Richards et al from the Broad Institute Single-Cell Portal56; Ochocka et al (2021) from GEO (accession number GSE136001)57, Cao et al from GEO (accession number GSE156793)58; and Zeisel et al from http://mousebrain.org/downloads.html.59
Data preprocessing
ScRNAseq data sets were normalized, scaled, dimensionally-reduced and visualized on a UMAP using the Seurat (V.4.0.4) workflow. In brief, count matrices were loaded into a Seurat object and normalized using NormalizeData(…, normalization.method = ‘LogNormalize’, scale.factor=10 000). Variable features were identified using FindVariableFeatures (…, selection.method = ‘mvp’, mean.cutoff=c(0.1,8), dispersion.cutoff=c(1, Inf)) and then data were scaled using ScaleData(). Principal component analysis and UMAP embedding was performed using RunPCA() and RunUMAP(…, dims=1:30), respectively. Metadata from original publications were used to annotate cell types.
GBM subtype classification
To assign GBM subtypes to scRNAseq data from Neftel et aland Richards et al,55 56 the AddModuleScore() function in Seurat was used to compute gene signature scores for each gene panel, and for each cell, the signature with the highest score was taken as the subtype. The Neftel subtypes included MES1 (mesenchymal type 1), MES2 (mesenchymal type 2), NPC1 (neural projenitor type 1), NPC2 (neural progenitor type 2), OPC (oligodendrocyte progenitor cells), and AC (astrocyte-like), whereas the Richards subtypes included Developmental and Injury Response.
Expression analysis
To visualize the expression of CD70 and CD27 in the public scRNAseq datasets, cell-level expression was projected onto a UMAP using the FeaturePlot() function in Seurat. To evaluate CD70 and CD27 expression stratified by cell type, Neftel subtype or Richards subtype, the mean normalized expression and expression fraction for each subgroup was computed and visualized using barplots and dotplots overlaid on a common axis. Subgroups were arranged based on hierarchical clustering performed on normalized expression and expression fraction values.
Statistical analysis
Biological replicates from at least three patient samples were compiled for each experiment, unless otherwise specified in figure legends. Respective data represent mean±SD, n values are listed in figure legends. Student’s t test analyses were performed using GraphPad Prism V.5, p>0.05=n.s., p<0.05=*, p<0.01=**, p<0.001=***, p<0.0001=****.
Results
CD70 expression is a unique marker of rGBM
Between the underrepresentation of rGBM samples in biobanks, due to the relatively low reoperation rate at GBM recurrence, and the variable presence of CSCs within bulk tumor samples, rGBM targets are often overlooked.60 In this study, we leveraged an RNA sequencing platform using four in-house, low-passage CSC-enriched cell lines, derived from pGBM or rGBM patient samples in a way previously described by our lab.61
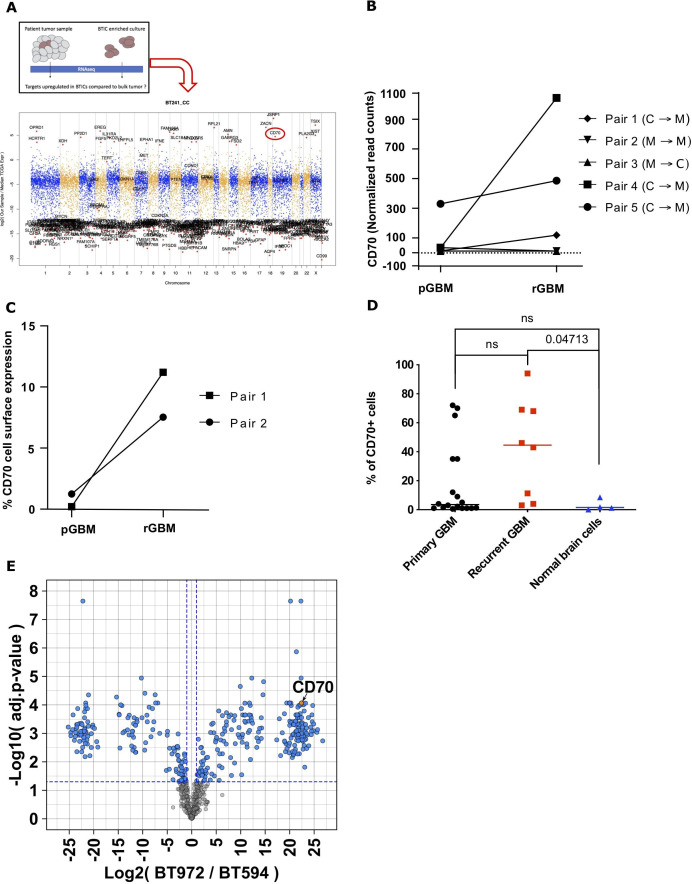
Using sequencing data from unmatched bulk tumor samples from the Cancer Genome Atlas (TCGA) GBM repository62 we identified genes over-represented in CSC-enriched populations. We uncovered upregulation of TNF superfamily member CD70, which was also found to be highly expressed in patient-derived pGBM and rGBM CSCs that were propagated in stem-cell enriching conditions (figure 1A, online supplemental figure 1A). In light of the growing body of evidence around sexual dimorphism in GBM,63 64 analysis of our patient-derived GBM samples yielded no significant differences in CD70 expression between male and female derived samples (data not shown). To further investigate the relevance of CD70 as a rGBM marker, we used six primary/recurrent pairs from patient-matched GBM samples present in the TCGA database to evaluate CD70 expression. In silico analysis of CD70 mRNA expression revealed increased levels in rGBM samples compared with their matched primaries for the majority of the pairs available, however this trend did not reach significance (figure 1B). Additionally, these same matched pairs exhibited a Classical (TCGA-CL) to Mesenchymal (TCGA-MES) subtype transition from primary to recurrence, indicating a shift towards a more aggressive and therapy-resistant subtype with poorer prognosis.65 66 Further in silico analysis using the Chinese Glioma Genome Atlas,67 TCGA and Ivy Glioma Atlas Project68 was performed to assess expression, correlation with prognosis, regional distribution within the tumor, and any age/sex differences in expression that may be present (online supplemental figure 2). Given that mRNA expression does not necessarily translate directly to cell-surface protein expression, we interrogated cell-surface CD70 protein levels on two in-house matched primary/recurrent patient derived CSC lines. We observed an increase in CD70 surface expression in both pairs by flow analysis (figure 1C) and a switch from the CL to MES subtype as seen in our bulk RNA sequencing samples (BT594/BT972, data not shown). We next screened a variety of unmatched primary and rGBMs, as well as normal human cells lines (neural stem cells and astrocytes) for CD70 expression. We demonstrate a clear trend towards increased CD70 expression in rGBM compared with pGBM in our CSC-enriched cell populations; though not statistically significant, bulk tumor data also demonstrates a trend toward increased CD70 expression in recurrent vs primary samples69 (figure 1D, online supplemental figure 1B). RGBMs displayed significantly higher CD70 expression than normal human cell lines (figure 1D). We identified a therapeutic window with normal human brain cells, which minimally express CD70 on their cell surface, in accordance with the existing literature.20 27 Single-cell RNA sequencing of publicly available GBM and non-malignant datasets yielded no significant expression of CD70 in non-malignant cells, with some residual expression on lymphoid-lineage cells, likely to be T cells (online supplemental figure 8). Lastly, using our BT594/BT972 matched-pair, we performed N-Glycocapture Proteomics which ranked CD70 among the top upregulated cell surface markers in rGBM compared with pGBM (figure 1E). This data led us to further inquiries about the functional role that CD70 plays in GBM progression and maintenance.
Figure 1.
CD70 expression is a relevant marker of rGBM. (A) Manhattan plot of the top upregulated RNAseq genes in the recurrent patient-derived GBM cancer stem cells (rGBM CSCs) BT241, compared with the publicly available the Cancer Genome Atlas database, identified CD70 (circled) as a target candidate upregulated in the CSC subpopulation, compared with tumor bulk. (B) Analysis of CD70 mRNA levels in the five TGCA primary/recurrent patient matched GBM pairs depicts an CD70 increase in three pairs on recurrence, alongside a switch towards the Mesenchymal subtype (GBM subtype classification: C, classical; P, proneural; M, mesenchymal). (C) Among GBM CSCs from figure 1C, the two in-house matched p/rGBM CSCs pairs display an increase of CD70 cell surface expression on tumor recurrence. (D) Dot plot representation of CD70 cell surface expression in rGBM CSCs compared with primary (p-) GBM CSCs and normal brain cells (astrocytes, neural stem cells), assessed by flow cytometry (Singh lab brain tumor database). (E) Volcano plot of the top up- and down-regulated cell surface proteins of pair two from figure 1D, as assessed by glycocapture proteomics. CSC, cancer stem cell; GBM, glioblastoma; rGBM, recurrent GBM.
jitc-2021-003289supp001.pdf (6.1MB, pdf)
CD70 is a key player in GBM maintenance and tumor formation
Given the upregulation of CD70 in GBM, specifically in rGBM, we sought to explore the role that CD70 expression plays in GBM maintenance and progression. We sorted pGBM and rGBM cells as CD70 positive or CD70 negative using FACS analysis and carried out a PrestoBlue cell viability assay. CD70 positive cells showed an increased signal compared with CD70 negative cells in the cell viability assay (PrestoBlue) that uses the reducing power of living cells to quantitatively measure the proliferation of cells (figure 2A, online supplemental figure 3A). Moreover, by limiting dilution analysis (LDA), CD70+ cells showed an increase in in vitro self-renewal capacity when compared with CD70negative cells in both GBM4 (frequency of 1/14 cells compared with 1/70 cells) and BT241 (frequency of 1/37 cells compared with 1/71 cells) (figure 2B). We aimed to determine the function of CD70 in tumorigenesis by shRNA mediated silencing. In order to exclude the possibility of off-target effects, we used two independent shRNA vectors and found both of them to be effective in reducing proliferation of GBM cells (online supplemental figure 3B). We moved forward using the shCD70-1 construct, which gave us better knockdown efficiency, for our further in vitro and in vivo studies.
Figure 2.
CD70 is a dedicated player in GBM maintenance and tumor formation. (A) GBM CSCs were sorted into positive and negative populations and proliferation was assessed by PrestoBlue assay (B) Limiting dilution analyses of CD70 positive and negative cells in pGBM (GBM8) and rGBM (BT241). (C) Cell surface CD70 expression after shRNA knockdown in three CD70HIGH GBM lines, as assessed by flow cytometry. (D) Silencing of CD70 expression by shRNA (shCD70) knockdown and sphere formation ability was assessed compared with shGFP (control shRNA). (E–I) Immunocompromised mice (NSG, a minimum of six mice per condition) were intracranially injected with shGFP or shCD70 CSCs. (E, F) Tumor area of CD70-silenced CSCs compared with control knockdown CSCs was measured using formalin-fixed, H&E-stained mouse brain slices (right, representative image). (G, H) Kaplan-Meier survival curves comparing mice engrafted with shCD70 CSCs compared with shGFP CSCs. The two remaining BT241 shCD70 mice at the end of experiment showed an absence of tumor by H&E staining at experimental endpoint (data not shown). (I) MRI images representative of xenografts from shGFP and shCD70 transduced GBM CSC line BT241. Images on the right are control images of normal mouse brain. (*P<0.05; **p<0.01; ***p<0.001; ****p<0.0001). CSCs, cancer stem cells; GBM, glioblastoma; pGBM, primary GBM; NSG, NOD/SCID gamma; shRNA, short hairpin RNA.
We next aimed to assess the role of CD70 in sphere formation, a stem-like trait that is typical of CSCs and correlates with self-renewal capacity in vitro and tumorigenesis in vivo.53 70 In both CD70HIGH CSC cell lines, silencing of CD70 using shRNA knockdown vector led to a significant decrease in sphere formation capacity compared with controls (figure 2C, D). We further validated the functional role of CD70 in contributing to stemness by performing LDA on rGBM cells that were either knocked out for CD70 and control cells that were transduced with a construct targeting the safe harbor locus, AAVS1. We found that AAVS1-transduced control cells had significantly higher self-renewal capacity (frequency of 1/5 cells) compared with CD70 knockout cells using either construct A or construct B (frequency of 1/25 cells and 1/26 cells, respectively). These data demonstrate the importance of CD70 expression in rGBM cells’ capacity for de novo tumor formation (online supplemental figure 3C).
Given the correlation of sphere formation with tumorigenesis in vivo, we investigated whether CD70 silencing limits GBM tumor formation in our patient-derived orthotopic xenograft animal model. We generated CD70 knockdown (shCD70) and control lines (shGFP) of three GBM CSC lines that naturally express high levels of CD70 (figure 2C), and intracranially injected these into immunodeficient mice, as previously described.53 We observed a significant decrease in the size of tumors formed by shCD70 cells compared with shGFP controls, as determined by H&E staining (figure 2E, F, online supplemental figure 3D) and MRI imaging (figure 2I). This was further reflected in a significant survival advantage for mice engrafted with shCD70 cells compared with controls (figure 2G, H, online supplemental figure 3E). These findings demonstrate that CD70 plays a key role in rGBM proliferation, tumor formation and survival both in vitro and in vivo.
CD70 plays a crucial role in cellular programs implicated in tumorigenesis
Previous studies have emphasized the function of CD70 in GBM as it contributes to T-cell apoptosis, and mediates tumor cell migration and invasion, a feature characteristic of mesenchymal-like cells.41 71 72 To further investigate the role of the CD70 signaling network in GBM, we investigated transcriptional changes and their predicted networks after CD70 silencing using RNA sequencing and subsequent GSEA. Using three GBM lines transduced with shCD70 or shGFP, wex observed strong downregulation of FOSL1, a gene recently discovered to play a pivotal role in stemness, migration, and epithelial-to-mesenchymal transition (EMT) (online supplemental figure 4A).73 74 Other slightly downregulated genes included CDH2 (N-Cadherin), PLAUR, and CXCR4; genes known to be associated with the Mesenchymal subtype in GBM and a worse overall prognosis.75–77 OLIG2, a transcription factor commonly associated with the proneural subtype and tumor recurrence,78 79 showed upregulation following CD70 knockdown, while expression of the proangiogenic factor vascular endothelial growth factor alpha (VEGFA) was depressed, indicating that CD70 may play a role in GBM angiogenesis, a characteristic previously documented in other pathologies, but not in cancer.80 81
GSEA was performed using Gene Ontology82 and MSigDB C2 and C6 gene sets,83 84 to gain a deeper understanding of the cellular programs associated with CD70 expression (online supplemental figure 4A). Top modulated pathways showed that silencing CD70 results in downregulation of EMT and hypoxia signatures, and upregulation of Interferon type I/interleukin-1 proinflammatory signatures85 (online supplemental figure 4A, B). Hypoxia and EMT pose major hurdles in GBM, as they promote migration of tumor cells further into the brain tissue, while proinflammatory signals are often depressed in GBM.86–88 While these data are limited, they do further implicate the role of CD70 in various processes linked to invasiveness, immunosuppression, and poor prognosis in GBM, as well as angiogenesis and stem-like characteristics of GBM CSCs.
Generation and characterization of CD70-directed CAR-T cells
Adoptive cell therapies have shown great promise in overcoming therapy resistance and providing a more specific targeted therapy in multiple cancers, including in GBM.89 However, despite significant global efforts to develop these therapies, they have only been approved for B cell malignancies thus far, and have yet to show efficacy in solid tumors such as GBM.90 It is believed that this lack of progress is in part due to the immunosuppressive microenvironment of solid tumors, particularly GBM, as well as antigen escape.91
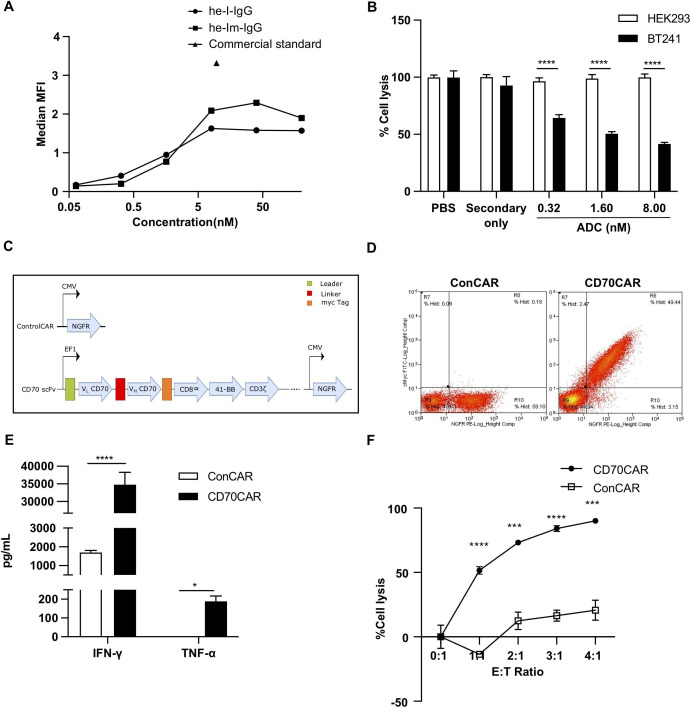
Given our data implicating CD70 as a key factor in GBM functionality, we tested two distinct in-house fragments antigen-binding (Fabs) for their ability to bind cell-surface CD70, and compared these to commercially available CD70 antibody (figure 3A). The Fab he-Lm was used to develop a non-covalently conjugated therapeutic antibody-drug conjugate (ADC) (figure 3B), which we postulated would be advantageous due to the rapid internalization of CD70 on ligand binding25 92 (online supplemental figure 5A). We incubated CD70HIGH GBM cells for 72 hours with our ADC, and observed a dramatic cytotoxic effect; an effect not seen in HEK293 control cells, indicating that our ADC is both specific and cytotoxic, and is suitable for developing adoptive cell therapies. Thus, we cloned the scFv region of he-Lm into a second-generation CAR linked to a truncated c-Myc tag (figure 3C), and achieved moderate CAR cell-surface expression 9 days post-transduction in human T-cells (figure 3D). To determine the efficacy of these anti-CD70 CAR-T (CD70CAR-T) cells, we cocultured them with CD70HIGH GBM cells. CD70CAR-T cells cocultured with CD70HIGH GBM cells released significantly more IFN-γ and TNF-α into culture supernatants compared with a control CAR-T (ConCAR-T) (figure 3E, online supplemental figure 5B, C). Additionally, CD70CAR-Ts demonstrated significant cytotoxicity against numerous CD70HIGH GBM cells at E:T ratios as low as 1:1 (figure 4F, online supplemental figure 5D). Together, these data indicate that CD70CAR-T cells are capable of mounting a robust and specific immune response against CD70-expressing GBMs.
Figure 3.
Generation and in vitro characterization of CD70-Specific CAR-T Cells. (A) Binding curve comparing CD70-specific Fabs to commercial standard antibody. (B) Anti-CD70 Fab’2 is specific against CD70, assessed by cytotoxicity assay under combination treatment with 2ºADC, against GBM cells expressing high (GBMCSC BT241) or no (HEK293) CD70. (C) Schematic representation of CAR structure. (D) Successful transduction of CAR-T vectors as observed by NGFR+ cells in ConCAR-T cells and NGFR+Myc+ cells in CD70 CAR-T cells, displayed as a representative flow plot. (E) Testing of CAR-T cell activation; IFN-γ and TNF-α cytokine release during coculture of GBM CSC BT241 with CD70 CAR-T, compared with ConCAR-T cells, as analyzed by ELISA (n=3). (F) Cytotoxicity assay to assess CD70CAR killing capacity compared with ConCAR after coculturing for 24 hours, tested at various effector to target (E:T) ratios (n=3). (***p<0.001; ****p<0.0001). CAR, chimeric antigen receptor; CSCs, cancer stem cells; GBM, glioblastoma; MFI, mean fluorescence intensity.
Figure 4.
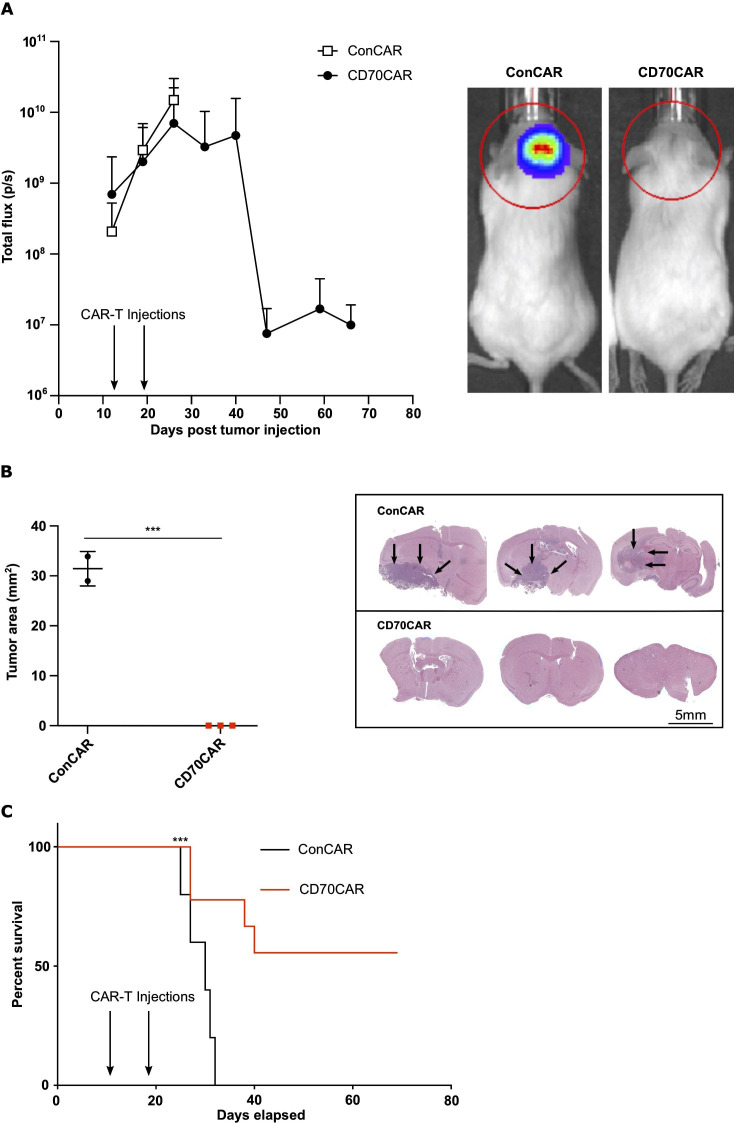
CD70 CAR-Ts are efficacious against recurrent GBM tumors in vivo. NSG mice (at least n=6 per group) were intracranially implanted with 100,000 human BT241 ffLuc GBM cells. Upon successful engraftment, mice were treated with 1×106 CD70CAR-T or ConCAR-T cells, delivered intracranially once a week for 2 weeks. (A) CD70 CAR-T treated mice showed decreased tumor signal, as assessed by bioluminescence measurement (right, representative image of radiance measurement in the region of interest). A lower tumor burden was observed in the CD70 CAR-T group compared with the control group, as measured on (B) formalin-fixed, H&E-stained mouse brain slices (representative image on the right), and (C) an extended survival (Kaplan-Meier curve) (***p<0.001). CAR, chimeric antigen receptor; GBM, glioblastoma; NSG, NOD/SCID Gamma.
Lastly, we assessed the antitumor potential of our CD70CAR-T cells in orthotopically xenografted NODSCID mice, using CD70HIGH BT241 rGBM CSCs. After confirming tumor engraftment using the in vivo imaging system, we intracranially injected 1M CD70CAR-T or ConCAR-T cells weekly over 2 weeks. Mice treated with CD70CAR-T cells displayed significantly lower tumor burden, as observed by bioluminescence signal, and a significant decrease in tumor volume as shown by H&E staining, confirming that CD70CAR-T cell-treated mice experience far less tumor growth compared with controls (figure 4A, B). Unsurprisingly based on our previous data, CD70CAR-T cell-treated animals had a significant survival advantage compared with control mice (figure 4C). Of note is that fact that the majority of animals (five out of nine) did not display any tumor-related symptoms post-treatment, nor did H&E staining display any presence of tumor at the end of study (figure 4B, representative image on the right). To further validate our CD70CAR-T cell therapy, we reproduced this with another GBM cell line, and observed similar results, indicating that this approach is efficacious in multiple CD70HIGH cell lines (online supplemental figure 6A-C).
CD70 and its role in the GBM TIME
CD70 is the only known ligand for the receptor CD27, a TNF receptor superfamily member, and is known to trigger T cell apoptosis and induce exhaustion,93 as well as recruit TAMs to the GBM microenvironment, contributing to the immunosuppressive nature of GBM.41 However, as far as its role in immune system functionality, no evidence has been found to date indicating it is essential.94 Thus, we elected to investigate the interaction between CD70-expressing GBM cells and CD27-expressing T cells, to see whether there would be any observed effect on T cell viability.95 Additionally, CD70 cleaved from the cell surface and present in the supernatant may act similarly to cell-surface CD70.96 We co-cultured CD70HIGH BT241 cells with CD27+ T cells, and observed a decrease in CD27+ T cell populations, an effect that was not seen when coculturing with CD70 knockdown BT241 cells, indicating that CD70/CD27 interaction between T cells and GBM cells may initiate T cell signaling programs, adding to the immunosuppressive capacity of GBM, as previously observed97 (online supplemental figure 7A). We then cultured CD27+ T cells with supernatant from CD70HIGH GBM cells to observe whether soluble CD70 cleaved into the supernatant could bind CD27+ T cells and initiate downstream apoptotic effects, however, we observed no decrease in CD27+ T cell populations (online supplemental figure 7A).
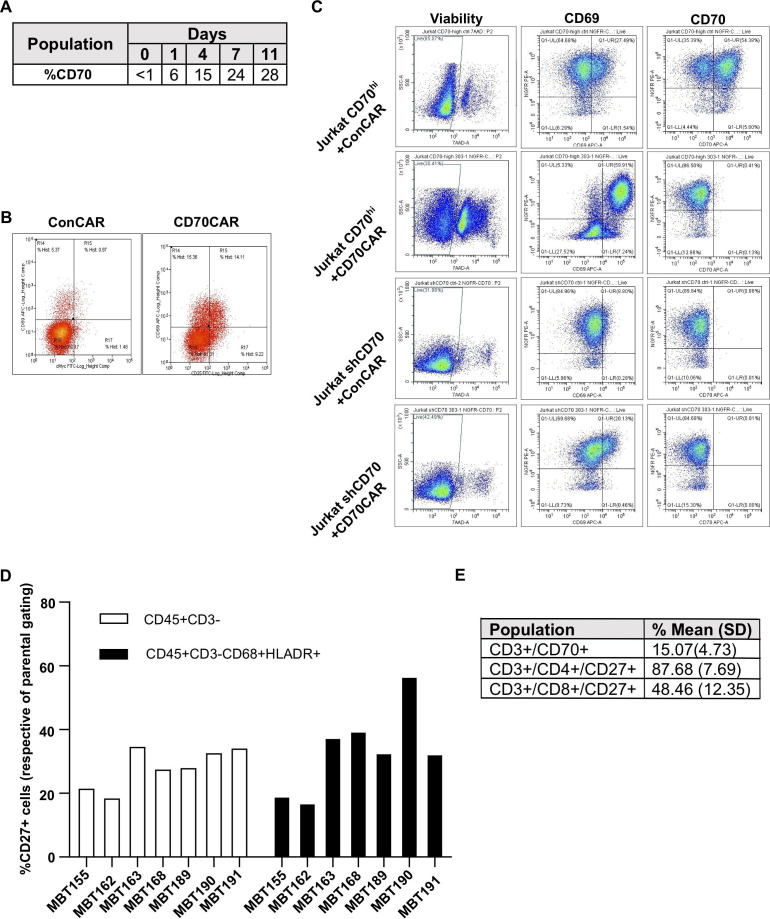
As seen in the literature,98 we observed increased expression of CD70 on activated T cells (figure 5A, B) and a subsequent decrease in CD70CAR-T cells count and viability, compared with ConCAR-T cells (data not shown). This insinuated that we could be observing fratricide between CAR-T cells, as seen previously with other targets.99 Thus, we elected to create a CD70 knockdown CD70CAR-T cell to overcome this problem. For this preliminary model we utilized Jurkat T cells due to their robustness compared with donor-derived T cells, and sorted them by flow cytometry to obtain a CD70HIGH Jurkat cell population. We also created an shCD70 knockdown Jurkat cell population. Both CD70HIGH and shCD70 Jurkats were transduced with CD70CAR construct or ConCAR construct, after which we examined any change in viability and presence of the activation marker CD69 (figure 5C). We were able to demonstrate that CD70HIGH CD70CAR Jurkat cells had decreased viability and increased expression of the activation marker CD69, compared with both ConCAR and shCD70 CD70CAR Jurkats. This indicates that silencing of CD70 may be a viable option for overcoming CD70CAR-T fratricide, improving viability of CD70CAR-T cells. However, as recent investigation noted the impact of CD70 deficiency on the function of CD8 T cells,100 the possibility of selectively expanding CD4+ CAR-T cells, already shown to be more effective against GBM,101 would be even more relevant.
Figure 5.
Modeling of CD70s influence on GBM TIME. (A) CD70 expression kinetics on in-house, activated T cells and (B) levels of CD69 and cMyc displayed by CD70CAR or ConCAR-T cells 9 days post-transduction, evaluated by flow cytometry. (C) CD70-enriched or -silenced Jurkat cells were transduced with either ConCAR or CD70CAR. After 8 days, CD69 and CD70 levels were assessed by flow cytometry. (D) TIME cells extracted from patient tumor samples were analyzed by flow cytometry, evaluating the pattern of expression of CD27 in non-lymphoid (CD45+CD3-) and M1 populations (CD45+CD3-CD68+HLADR+). (E) Average expression of CD27 on CD4/CD8 lymphoid population, and CD70 expression on the lymphoid population (CD3+). CAR, chimeric antigen receptor; GBM, glioblastoma; TIME, tumor immune microenvironment.
According to the literature, cancer cells may use the CD70/CD27 interaction to their advantage to modulate both T cells and macrophages to create more immunosuppressive or immune-evasive environments.41 72 We performed single-cell RNA sequencing analysis on pre-existing publicly available datasets55–59 and highlight CD70 expression on human GBM CSCs compared with bulk GBM cells. Additionally, CD70 expression was particularly increased for CSCs of the Mesenchymal subtype, as well as on cells of the lymphoid-lineage, most likely T cells. We also observed that CD27 expression primarily appears to be immune-cell specific, particularly within lymphoid and microglial cells (figure 6, online supplemental figure 8). These data support the potential for a CD70/CD27 signaling axis within the tumor microenvironment.
Figure 6.
CD70 and CD27 expression in GBM and immune cells. (A, B) ScRNAseq profile of human GBM and immune infiltrate (Neftel 2019). Cell-type annotated UMAP (A) and expression of CD70 (top) and CD27 (bottom) visualized on UMAP and stratified by cell type (all cells included), Neftel GBM subtype (GBM only), and Richards GBM subtype (GBM only) (B). (C, D) ScRNAseq profile of human glioblastoma stem cells (Richards 2021). Cell-type annotated UMAP (C) and expression of CD70 (top) and CD27 (bottom) visualized on UMAP and stratified by Neftel GBM subtype and Richards GBM subtype (D). (E, F) ScRNAseq profile of immune cell infiltrates in murine GL261 tumors (Ochocka 2021). Cell-type annotated UMAP (E) and expression of CD70 (top) and CD27 (bottom) visualized on UMAP and stratified by cell type (F). Astrocyte-like, BAM, border-associated macrophage; DC, dendritic cell; Develop., Developmental; Expr, expression; GBM, glioblastoma; Injury Resp, injury response; MES1, mesenchymal type 1; MES2, mesenchymal type 2; NK, natural killer; NPC1, neural progenitor cell type 1; NPC2, neural progenitor cell type 2; OPC, oligodendrocyte progenitor cell.
Due to the absence of an immune system in our immunodeficient mouse models, we elected to interrogate the GBM TIME using flow cytometry on CD45+ cells extracted from fresh patient-derived tumor samples.). From these data, we saw that CD27 had a similar expression pattern among non-T-cell populations (CD3- cells) from different tumor samples (figure 5D). Further, CD27 expression was found on putative ‘M1 proinflammatory’ (CD45+CD3-CD68+HLADR+) macrophages to a similar extent (figure 5D).102 It is interesting to note that CD27 expression was increased in this particular ‘M1’ population when their corresponding GBM cells were CD70HIGH (ie, MBT190); a phenomenon not seen when correspond GBM cells were CD70LOW (ie, MBT162). Interestingly, MBT190 had far fewer CD8+ cytotoxic tumor infiltrating lymphocytes than most samples, including MBT162 (online supplemental figure 7B). In the CD3+ lymphoid population, we found that while CD27 was expressed on the majority of these cells, very few of them expressed CD70 (figure 5E, online supplemental figure 7B), in agreement with existing literature.99 Lastly, we noted that CD27 was highly expressed on CD4 helper T cells, potentially making them more sensitive to CD70HIGH GBMs. Together, these data indicate that multiple CD27/CD70-axis interactions are occurring within the GBM TIME, likely contributing to the low immunogenicity of rGBM.
Discussion
Since the creation of the Stupp protocol in 2005, few advances have been made in bringing new therapeutics to market for GBM. In addition, resistance to SoC treatment has begun to direct therapeutic investigation towards a small reservoir of cells termed CSCs.103 CSCs display enhanced self-renewal properties and are believed to be capable of de novo tumor formation, and driving tumor recurrence.103 It is believed that SoC therapy helps drive recurrent tumors by creating a bottleneck, with cells that escape initial chemoradiotherapy driving formation of a therapy-resistant recurrent tumor. Thus, recent endeavors have sought to identify actionable targets on this subpopulation of cells. Numerous clinical trials against these targets have assessed the viability of CAR-T cells against different antigens, dendritic cell vaccines and immune checkpoint inhibitors, among others. However, despite many efforts little progress has been made, and most rGBM patients are destined for clinical trials or palliative care.
Here, we used a multiomics approach using our in-house collection of low-passage, patient-derived CSCs, and existing public datasets to identify a GBM cell surface marker of treatment-resistant CSCs. Using this approach, we identified and validated CD70 as a promising therapeutic target in rGBM.
From the literature, CD70 seems to have a confounding role in cancer, both stimulating and suppressing immune response in various cancers.96 104 With this in mind, we sought not only to validate CD70 as a therapeutic target in GBM, but also to better understand its role in GBM cell maintenance and progression, as well as the TIME. We demonstrate that CD70 is vital in sphere formation and proliferation of GBM cells, two essential CSC characteristics which correlate with ability to recapitulate GBM tumors in vivo. To follow, we show that CD70 silencing significantly reduce tumor burden and volume, and significantly increases survival time, with some animals showing no signs of disease up until the end of the experiment. These data represent similar findings to that seen in the literature, indicating that CD70 may play a crucial role in tumor-initiating cells in multiple cancers.105 106 To better understand the cellular pathways and programs involving CD70, we carried out GSEA using RNAseq of our CD70-silenced cell lines. We were able to contribute to the functional understanding of CD70 by showing that, as previously noted in RCC and other cancers, CD70 plays a role in controlling hypoxia.107 108 However, this is the first time to our knowledge that the regulatory role of CD70 in hypoxia has been demonstrated in GBM specifically, or brain cancer in general. Hypoxia is the consequence of poor vascularization, and thus poor blood delivery, within the tumor. This often promotes cancer cell spreading via invasion so that cells may escape the low-oxygen environment, thereby rendering the tumor diffuse and far more aggressive, a characteristic often seen in rGBM. Based on our GSEA, it is possible that the vascularization factor VEGF is dependent on CD70 expression and the CD70/CD27 signaling axis, and tumor neoangiogenesis, a role previously established in non-cancer pathologies.80 Our analysis also revealed that CD70 silencing appeared to depress pathways related to EMT signaling, a program that is linked to a more aggressive cancer capable of therapy evasion in multiple cancers, including GBM.109–112
By defining the tumorigenic significance of CD70 in GBM, we sought to develop potential therapeutic modalities directed against CD70. We developed a CAR against CD70 and demonstrated its efficacy both in vitro and in vivo, where it displayed high specificity for CD70, and conferred significantly extended survival in our orthotopically xenografted animal models, with some animals experiencing complete remission. CD70CAR-T cell efficacy varied slightly in vivo, which may be due to the extensive heterogeneity of GBM, particularly in early passage GBM CSCs which recapitulate intratumoral heterogeneity quite well. Changes in associated clonal dynamics which arise with therapeutic pressure may generate therapy-resistant CD70LOW subpopulations with antigen escape, as has been seen in the clinic.113 While antigen escape is a major problem at the moment, particularly as not all CSCs uniformly express CD70 (online supplemental figure 1B) or any other single targetable protein, it is may be possible to overcome this issue using a poly-therapeutic strategy. Examples of such a combinatorial strategy include a bispecific anti-CD70/SIRPα antibody which outperforms individually delivered antibodies in models of human Burkitt’s lymphoma, allowing for cotargeting of both tumor cells and TAMs in the TIME.114 We also detected a population of CD27+ proinflammatory putative ‘M1’ macrophages in our profiling of patient-derived rGBM TIME samples, indicating that while the TIME is immunosuppressive, it does contain traditionally antitumor components. CD27 is a marker of highly immunosuppressive Tregs,115 and it is possible that TAMs and other members of the TIME may exert some of their immunosuppressive effects through CD27/CD70 interactions, inducing a more immunosuppressive phenotype in tumor infiltrating lymphocytes.24 Among the TCGA matched pairs, we noticed a trend of subtype switching to the mesenchymal subtype on recurrence, as has been observed in the literature,106 116 and increased CD70 expression correlated with the mesenchymal subtype as well. The mesenchymal subtype is associated with a more aggressive tumor, as well as invasion, therapy evasion and a poorer prognosis,36 66 and has been shown to harbor a stronger immunosuppressive microenvironment. While initially this may seem to be negative as far as the impact cell therapies might have, some studies suggest the opposite, and that immunotherapies may have a more drastic impact on these tumors.117 118 In accordance with the literature, after silencing of CD70 expression in various GBM cell lines, we found that CD70 repression results in downregulation of IFN-α and IL-1, both of which play an important role in the Th1 lymphocyte response, and are known for their proinflammatory role in tumors.119 120 In conjunction with other work highlighting CAR-T cells’ ability to induce inflammation,121 we see the potential for CD70CAR-T cells to convert the GBM microenvironment from immunologically ‘cold’ to ‘hot’, eliciting an antitumor immune response of endogenous effector cells. It has previously been shown that certain aspects of T cell functionality are dependent on IFN-α and CD70,122 although to our knowledge this is the first time that this dependence has been shown in cancer cells.
Our work, as well as recent work from others,41 maintain that CD70 is a promising target for rGBM, and a better understanding of the role of CD70 in the GBM TIME is needed. In particular, we advocate for determining the role that CD70 plays in initiating GBM recurrence and potential mechanisms of therapy evasion. While the literature shows conflicting results regarding whether CD70 plays a protumorigenic or antitumorigenic effect in cancer, we postulate a novel mechanism, through which continued stimulation of the CD70/CD27 axis leads to continuous T cell activation by GBM cells, and subsequent exhaustion within the T-cell compartment, as previously observed in models of HIV.32
As has been investigated in the literature, CD70 is present on the cell surface of T-cells, however, it does not play a functional role, and as a result, other groups have noted fratricide in their CD70 CAR-T cell populations.123 Based on these reports, as well as our own observations collected from our CD70 CAR-T cell populations, we developed a model of CD70CAR-T cell fratricide using Jurkat cells, which may be used as a platform for future studies in adoptive cell therapy. Reasons for the observed decrease in viability of Jurkat cells, though not assessed experimentally here, have previously been speculated on by colleagues using CAR-transduced Jurkat cells,124 reporting that cell death occurs due to prolonged activation-induced paracrine and autocrine interactions. Compared with the flow cytometry-sorted CD70-enriched Jurkat cells used in our experiments, CD70 expression is relatively scarce on activated T cells, indicating that fratricide would occur to a far lesser extent on natural PBMC-derived CD70 CAR-T cells. In our hands, only small variations in cell viability were observed in our CD70 CAR-T cell experiments. Nonetheless, others recently reported an additional fitness benefit in a CD70 knockout CD70 CAR-T cell, which conferred various functional benefits including increased proliferation and cytotoxicity, and was far more advantageous than other obvious knockout targets such as PD-1 and LAG3.125 Thus, we highly encourage future studies exploring any potential benefit a CD70KO CD70 CAR-T cell may display in GBM, where the TIME is notoriously difficult to overcome. Targeted delivery of the CAR construct directly into the CD70 locus would disrupt CD70 expression, preventing undesirable stimulation and enhancing proliferation and cytotoxicity of CD70 CAR-T cells.126 Strategies involving targeted gene delivery have already been applied to an array of other targets to optimize adoptive cell therapies.127
Here, we investigated the functional benefit that CD70 expression confers in GBM cells, and the implied influence that CD70 expression may have on interactions with the immunosuppressive landscape. We employed a reverse translational approach128 to determine CD27’s expression pattern—CD70s only known receptor—in different compartments of the GBM TIME. Previous literature has identified CD27 expression on subsets of NK cells, B-cells and hematopoietic stem cells.129–132 This highlights the influence that CD70-expressing GBMs may have on the different subsets within its microenvironment by leveraging these potential interactions and associated blockade. Utilizing a dual-targeting therapeutic strategy—targeting both GBM cells and immunosuppressive microenvironment cells—may result in highly potent antitumor activity. Thus, future priorities would be to study the impact our CD70CAR-T therapy has on the GBM immune microenvironment in a murine model engrafted with human CSCs, harboring an HLA-compatible human immune system such as huMice, with a particular focus on models recapitulating a GBM-representative myeloid population.133
Considering our data and that of recent clinical trials targeting CD70 by systemic administration,134 we believe intracranially delivered CD70 CAR-T therapy holds great promise, and should be explored alone and in conjunction with TIME-targeting therapeutics.
Translational relevance
Glioblastoma is the most common adult malignant brain tumor, and is characterized by a dismal prognosis and poor response to therapy at recurrence. Little-to-no change in standard of care therapy in the last 15 years, despite numerous potential therapies entering clinical trials. Therapeutic failure is largely due to tumor heterogeneity, and a lack of unique tumor associated antigens. Here, we propose CD70 as an immunotherapeutic target in recurrent glioblastoma. CD70 plays a role in key pro-tumorigenic processes and is minimally expressed in normal tissues. We develop a CD70-directed CAR-T cell, which we show to be highly efficacious in extending survival in our intracranial mouse models of recurrent GBM. In addition, we identify CD27—the receptor for CD70—on the surface of multiple populations within the tumor immune microenvironment, implying that CD70/CD27 interactions may play a role in the tumor immune microenvironment.
Acknowledgments
MS was supported by MITACS fellowship in partnership with Longbow Therapeutics. SS is supported by the Terry Fox Program Project grant and McMaster University Department of Surgery. We thank Dr Mary Haak-Frendscho and Dr Alan Wahl for providing us with CD70 antibodies.
Footnotes
Twitter: @sheilasinghlab
MS and WTM contributed equally.
Correction notice: This article has been corrected since it was first published to correct author name Muhammad Vaseem Shaikh.
Contributors: MS, DU, PV, CV and SS contributed to the conception and design of the research; MS, WTM, NT, DU, CC, DP, NS, DM, LK and BA-B performed the experiments, acquired data; MS, WTM, VMS, MS, LK, AK, WG, DB, SS, NM, TK, JM, CV and SS contributed to analysis and interpretation of the data. KB provided support for bioinformatics analysis. MS and WTM wrote the manuscript. All authors critically revised the manuscript. SS is the guarantor for the manuscript and is accountable for ensuring the integrity and accuracy of the work, and read and approved the final manuscript.
Funding: Funding for this project was in part provided by Longbow Therapeutics and the Terry Fox Research Initiative.
Competing interests: DU, DB and PV are employees of Century Therapeutics Canada. The other authors declare no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by Hamilton Health Sciences Research ethics board at McMaster University.
References
- 1.Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol 2019;21:v1–100. 10.1093/neuonc/noz150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Taillibert S, Kanner A, et al. Effect of Tumor-Treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 2017;318:2306–16. 10.1001/jama.2017.18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rønning PA, Helseth E, Meling TR, et al. A population-based study on the effect of temozolomide in the treatment of glioblastoma multiforme. Neuro Oncol 2012;14:1178–84. 10.1093/neuonc/nos153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Linde ME, Brahm CG, de Witt Hamer PC, et al. Treatment outcome of patients with recurrent glioblastoma multiforme: a retrospective multicenter analysis. J Neurooncol 2017;135:183–92. 10.1007/s11060-017-2564-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 7.Bergmann N, Delbridge C, Gempt J, et al. The intratumoral heterogeneity reflects the intertumoral subtypes of glioblastoma multiforme: a regional immunohistochemistry analysis. Front Oncol 2020;10:494. 10.3389/fonc.2020.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soeda A, Hara A, Kunisada T, et al. The evidence of glioblastoma heterogeneity. Sci Rep 2015;5:7979. 10.1038/srep07979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Z, Yang Q, Li X. Effect of intra- and inter-tumoral heterogeneity on molecular characteristics of primary IDH-wild type glioblastoma revealed by single-cell analysis. CNS Neurosci Ther 2020;26:981–9. 10.1111/cns.13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017;23:1124–34. 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 11.Saygin C, Matei D, Majeti R, et al. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 2019;24:25–40. 10.1016/j.stem.2018.11.017 [DOI] [PubMed] [Google Scholar]
- 12.Cusulin C, Chesnelong C, Bose P, et al. Precursor states of brain tumor initiating cell lines are predictive of survival in xenografts and associated with glioblastoma subtypes. Stem Cell Reports 2015;5:1–9. 10.1016/j.stemcr.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol 2017;134:505–12. 10.1007/s11060-017-2379-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas TL, Sciuto MR, Brunetto L, et al. Integrin α7 is a functional marker and potential therapeutic target in glioblastoma. Cell Stem Cell 2017;21:35–50. 10.1016/j.stem.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006;5:67. 10.1186/1476-4598-5-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 17.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res 2008;14:123–9. 10.1158/1078-0432.CCR-07-0932 [DOI] [PubMed] [Google Scholar]
- 18.Vora P, Venugopal C, Salim SK, et al. The rational development of CD133-Targeting immunotherapies for glioblastoma. Cell Stem Cell 2020;26:832–44. 10.1016/j.stem.2020.04.008 [DOI] [PubMed] [Google Scholar]
- 19.Morgan RA, Johnson LA, Davis JL, et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum Gene Ther 2012;23:1043–53. 10.1089/hum.2012.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin L, Ge H, Long Y, et al. CD70, a novel target of CAR T-cell therapy for gliomas. Neuro Oncol 2018;20)::55–65. 10;. 10.1093/neuonc/nox116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priceman SJ, Forman SJ, Brown CE. Smart cars engineered for cancer immunotherapy. Curr Opin Oncol 2015;27:466–74. 10.1097/CCO.0000000000000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Liu J, Zhong JF, et al. Engineering CAR-T cells. Biomark Res 2017;5:22. 10.1186/s40364-017-0102-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grewal IS. CD70 as a therapeutic target in human malignancies. Expert Opin Ther Targets 2008;12:341–51. 10.1517/14728222.12.3.341 [DOI] [PubMed] [Google Scholar]
- 24.Jacobs J, Zwaenepoel K, Rolfo C, et al. Unlocking the potential of CD70 as a novel immunotherapeutic target for non-small cell lung cancer. Oncotarget 2015;6:13462–75. 10.18632/oncotarget.3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam PJ, Terrett JA, Steers G, et al. CD70 (TNFSF7) is expressed at high prevalence in renal cell carcinomas and is rapidly internalised on antibody binding. Br J Cancer 2006;95:298–306. 10.1038/sj.bjc.6603222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowman MR, Crimmins MA, Yetz-Aldape J, et al. The cloning of CD70 and its identification as the ligand for CD27. J Immunol 1994;152:1756–61. [PubMed] [Google Scholar]
- 27.Hintzen RQ, Lens SM, Koopman G, et al. CD70 represents the human ligand for CD27. Int Immunol 1994;6:477–80. 10.1093/intimm/6.3.477 [DOI] [PubMed] [Google Scholar]
- 28.Tesselaar K, Xiao Y, Arens R, et al. Expression of the murine CD27 ligand CD70 in vitro and in vivo. J Immunol 2003;170:33–40. 10.4049/jimmunol.170.1.33 [DOI] [PubMed] [Google Scholar]
- 29.Denoeud J, Moser M. Role of CD27/CD70 pathway of activation in immunity and tolerance. J Leukoc Biol 2011;89:195–203. 10.1189/jlb.0610351 [DOI] [PubMed] [Google Scholar]
- 30.Nolte MA, van Olffen RW, van Gisbergen KPJM, et al. Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev 2009;229:216–31. 10.1111/j.1600-065X.2009.00774.x [DOI] [PubMed] [Google Scholar]
- 31.Borst J, Hendriks J, Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol 2005;17:275–81. 10.1016/j.coi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 32.Tesselaar K, Arens R, van Schijndel GMW, et al. Lethal T cell immunodeficiency induced by chronic costimulation via CD27-CD70 interactions. Nat Immunol 2003;4:49–54. 10.1038/ni869 [DOI] [PubMed] [Google Scholar]
- 33.Yang Z-Z, Grote DM, Xiu B, et al. TGF-β upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia 2014;28:1872–84. 10.1038/leu.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diegmann J, Junker K, Gerstmayer B, et al. Identification of CD70 as a diagnostic biomarker for clear cell renal cell carcinoma by gene expression profiling, real-time RT-PCR and immunohistochemistry. Eur J Cancer 2005;41:1794–801. 10.1016/j.ejca.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 35.Lens SM, Drillenburg P, den Drijver BF, et al. Aberrant expression and reverse signalling of CD70 on malignant B cells. Br J Haematol 1999;106:491–503. 10.1046/j.1365-2141.1999.01573.x [DOI] [PubMed] [Google Scholar]
- 36.Pich C, Sarrabayrouse G, Teiti I, et al. Melanoma-expressed CD70 is involved in invasion and metastasis. Br J Cancer 2016;114:63–70. 10.1038/bjc.2015.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Held-Feindt J, Mentlein R. CD70/CD27 ligand, a member of the TNF family, is expressed in human brain tumors. Int J Cancer 2002;98:352–6. 10.1002/ijc.10207 [DOI] [PubMed] [Google Scholar]
- 38.Hishima T, Fukayama M, Hayashi Y, et al. CD70 expression in thymic carcinoma. Am J Surg Pathol 2000;24:742–6. 10.1097/00000478-200005000-00014 [DOI] [PubMed] [Google Scholar]
- 39.Pahl JH, Santos SJ, Kuijjer ML, et al. Expression of the immune regulation antigen CD70 in osteosarcoma. Cancer Cell Int 2015;15:31. 10.1186/s12935-015-0181-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsson A, de Milito A, Mowafi F, et al. Expression of CD27-CD70 on early B cell progenitors in the bone marrow: implication for diagnosis and therapy of childhood all. Exp Hematol 2005;33:1500–7. 10.1016/j.exphem.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 41.Ge H, Mu L, Jin L, et al. Tumor associated CD70 expression is involved in promoting tumor migration and macrophage infiltration in GBM. Int J Cancer 2017;141:1434–44. 10.1002/ijc.30830 [DOI] [PubMed] [Google Scholar]
- 42.Wischhusen J, Jung G, Radovanovic I, et al. Identification of CD70-mediated apoptosis of immune effector cells as a novel immune escape pathway of human glioblastoma. Cancer Res 2002;62:2592–9. [PubMed] [Google Scholar]
- 43.Claus C, Riether C, Schürch C, et al. CD27 signaling increases the frequency of regulatory T cells and promotes tumor growth. Cancer Res 2012;72:3664–76. 10.1158/0008-5472.CAN-11-2791 [DOI] [PubMed] [Google Scholar]
- 44.Purdue MP, Lan Q, Hoffman-Bolton J, et al. Circulating sCD27 and sCD30 in pre-diagnostic samples collected fifteen years apart and future non-Hodgkin lymphoma risk. Int J Cancer 2019;144:1780–5. 10.1002/ijc.31879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashima J, Okuma Y, Hosomi Y, et al. High serum soluble CD27 level correlates with poor performance status and reduced survival in patients with advanced lung cancer. Oncology 2019;97:365–72. 10.1159/000502441 [DOI] [PubMed] [Google Scholar]
- 46.Aftimos P, Rolfo C, Rottey S, et al. Phase I dose-escalation study of the anti-CD70 antibody ARGX-110 in advanced malignancies. Clin Cancer Res 2017;23:6411–20. 10.1158/1078-0432.CCR-17-0613 [DOI] [PubMed] [Google Scholar]
- 47.Squibb B-M, Phase A, Multicenter O-L. Dose-escalation, multidose study of MDX-1203 in subjects with advanced/recurrent clear cell renal cell carcinoma or relapsed/refractory B-cell non Hodgkin’s Lymphoma [online] Report No.: NCT00944905, 2013. Available: https://clinicaltrials.gov/ct2/show/NCT00944905 [Accessed 2 Dec 2020].
- 48.Seagen Inc . A phase 1 trial of SGN-CD70A in patients with CD70-positive malignancies [online] Report No.: NCT02216890, 2018. Available: https://clinicaltrials.gov/ct2/show/NCT02216890 [Accessed 2 Dec 2020].
- 49.Patrizii M, Bartucci M, Pine SR, et al. Utility of glioblastoma patient-derived orthotopic xenografts in drug discovery and personalized therapy. Front Oncol 2018;8:23. 10.3389/fonc.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- 54.Hart T, Tong AHY, Chan K, et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 2017;7:2719–27. 10.1534/g3.117.041277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richards LM, Whitley OKN, MacLeod G, et al. Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat Cancer 2021;2:157–73. [DOI] [PubMed] [Google Scholar]
- 57.Ochocka N, Segit P, Walentynowicz KA, et al. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat Commun 2021;12:1151. 10.1038/s41467-021-21407-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao J, O'Day DR, Pliner HA, et al. A human cell atlas of fetal gene expression. Science 2020;370:eaba7721. 10.1126/science.aba7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeisel A, Hochgerner H, Lönnerberg P, et al. Molecular architecture of the mouse nervous system. Cell 2018;174:999–1014. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robin AM, Lee I, Kalkanis SN. Reoperation for recurrent glioblastoma multiforme. Neurosurg Clin N Am 2017;28:407–28. 10.1016/j.nec.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 61.Venugopal C, McFarlane NM, Nolte S, et al. Processing of primary brain tumor tissue for stem cell assays and flow sorting. J Vis Exp 2012;67 10.3791/4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bowman RL, Wang Q, Carro A, et al. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 2017;19:139–41. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bayik D, Zhou Y, Park C, et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov 2020;10:1210–25. 10.1158/2159-8290.CD-19-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W, Warrington NM, Taylor SJ, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med 2019;11:eaao5253. 10.1126/scitranslmed.aao5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sa JK, Chang N, Lee HW, et al. Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol 2020;21:216. 10.1186/s13059-020-02140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017;32:42–56. 10.1016/j.ccell.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Z, Zhang K-N, Wang Q, et al. Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics 2021;19:00045 10.1016/j.gpb.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puchalski RB, Shah N, Miller J, et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018;360:660–3. 10.1126/science.aaf2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahman M, Kresak J, Yang C, et al. Analysis of immunobiologic markers in primary and recurrent glioblastoma. J Neurooncol 2018;137:249–57. 10.1007/s11060-017-2732-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirschhaeuser F, Menne H, Dittfeld C, et al. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol 2010;148:3–15. 10.1016/j.jbiotec.2010.01.012 [DOI] [PubMed] [Google Scholar]
- 71.Diegmann J, Junker K, Loncarevic IF, et al. Immune escape for renal cell carcinoma: CD70 mediates apoptosis in lymphocytes. Neoplasia 2006;8:933–8. 10.1593/neo.06451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inaguma S, Lasota J, Czapiewski P, et al. CD70 expression correlates with a worse prognosis in malignant pleural mesothelioma patients via immune evasion and enhanced invasiveness. J Pathol 2020;250:205–16. 10.1002/path.5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fiscon G, Conte F, Paci P. SWIM tool application to expression data of glioblastoma stem-like cell lines, corresponding primary tumors and conventional glioma cell lines. BMC Bioinformatics 2018;19:436. 10.1186/s12859-018-2421-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feldker N, Ferrazzi F, Schuhwerk H, et al. Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. Embo J 2020;39:e103209. 10.15252/embj.2019103209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilder AS, Natali L, Van Dyk DM, et al. The urokinase receptor induces a mesenchymal gene expression signature in glioblastoma cells and promotes tumor cell survival in neurospheres. Sci Rep 2018;8:2982. 10.1038/s41598-018-21358-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tao C, Huang K, Shi J, et al. Genomics and prognosis analysis of epithelial-mesenchymal transition in glioma. Front Oncol 2020;10:183. 10.3389/fonc.2020.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yi L, Tong L, Li T, et al. Bioinformatic analyses reveal the key pathways and genes in the CXCR4 mediated mesenchymal subtype of glioblastoma. Mol Med Rep 2018;18:741–8. 10.3892/mmr.2018.9011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouchart C, Trépant A-L, Hein M, et al. Prognostic impact of glioblastoma stem cell markers OLIG2 and CCND2. Cancer Med 2020;9:1069–78. 10.1002/cam4.2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu F, Chen Y, Zhao C, et al. Olig2-dependent reciprocal shift in PDGF and EGF receptor signaling regulates tumor phenotype and mitotic growth in malignant glioma. Cancer Cell 2016;29:669–83. 10.1016/j.ccell.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simons KH, Aref Z, Peters HAB, et al. The role of CD27-CD70-mediated T cell co-stimulation in vasculogenesis, arteriogenesis and angiogenesis. Int J Cardiol 2018;260:184–90. 10.1016/j.ijcard.2018.02.015 [DOI] [PubMed] [Google Scholar]
- 81.Winkels H, Meiler S, Smeets E, et al. CD70 limits atherosclerosis and promotes macrophage function. Thromb Haemost 2017;117:164–75. 10.1160/TH16-04-0318 [DOI] [PubMed] [Google Scholar]
- 82.Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guarda G, Braun M, Staehli F, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 2011;34:213–23. 10.1016/j.immuni.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 86.Monteiro AR, Hill R, Pilkington GJ, et al. The role of hypoxia in glioblastoma invasion. Cells 2017;6:45. 10.3390/cells6040045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010;463:318–25. 10.1038/nature08712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh A, Settleman J. Emt, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29:4741–51. 10.1038/onc.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bielamowicz K, Khawja S, Ahmed N. Adoptive cell therapies for glioblastoma. Front Oncol 2013;3:275. 10.3389/fonc.2013.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Rourke DM, Nasrallah MP, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 2017;9:eaaa0984. 10.1126/scitranslmed.aaa0984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sterner RC, Sterner RM. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J 2021;11:69. 10.1038/s41408-021-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McDonagh CF, Kim KM, Turcott E, et al. Engineered anti-CD70 antibody-drug conjugate with increased therapeutic index. Mol Cancer Ther 2008;7:2913–23. 10.1158/1535-7163.MCT-08-0295 [DOI] [PubMed] [Google Scholar]
- 93.Chahlavi A, Rayman P, Richmond AL, et al. Glioblastomas induce T-lymphocyte death by two distinct pathways involving gangliosides and CD70. Cancer Res 2005;65:5428–38. 10.1158/0008-5472.CAN-04-4395 [DOI] [PubMed] [Google Scholar]
- 94.Shaffer DR, Savoldo B, Yi Z, et al. T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood 2011;117:4304–14. 10.1182/blood-2010-04-278218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wajant H. Therapeutic targeting of CD70 and CD27. Expert Opin Ther Targets 2016;20:959–73. 10.1517/14728222.2016.1158812 [DOI] [PubMed] [Google Scholar]
- 96.Rowley TF, Al-Shamkhani A. Stimulation by soluble CD70 promotes strong primary and secondary CD8+ cytotoxic T cell responses in vivo. J Immunol 2004;172:6039–46. 10.4049/jimmunol.172.10.6039 [DOI] [PubMed] [Google Scholar]
- 97.Wang QJ, Hanada K-I, Robbins PF, et al. Distinctive features of the differentiated phenotype and infiltration of tumor-reactive lymphocytes in clear cell renal cell carcinoma. Cancer Res 2012;72:6119–29. 10.1158/0008-5472.CAN-12-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lens SM, Baars PA, Hooibrink B, et al. Antigen-presenting cell-derived signals determine expression levels of CD70 on primed T cells. Immunology 1997;90:38–45. 10.1046/j.1365-2567.1997.00134.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sánchez-Martínez D, Baroni ML, Gutierrez-Agüera F, et al. Fratricide-resistant CD1a-specific CAR T cells for the treatment of cortical T-cell acute lymphoblastic leukemia. Blood 2019;133:2291–304. 10.1182/blood-2018-10-882944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abolhassani H. Specific immune response and cytokine production in CD70 deficiency. Front Pediatr 2021;9:615724. 10.3389/fped.2021.615724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang D, Aguilar B, Starr R, et al. Glioblastoma-targeted CD4+ CAR T cells mediate superior antitumor activity. JCI Insight 2018;3:e99048. 10.1172/jci.insight.99048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma J, Liu L, Che G, et al. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer 2010;10:112. 10.1186/1471-2407-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest 2017;127:415–26. 10.1172/JCI89587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aulwurm S, Wischhusen J, Friese M, et al. Immune stimulatory effects of CD70 override CD70-mediated immune cell apoptosis in rodent glioma models and confer long-lasting antiglioma immunity in vivo. Int J Cancer 2006;118:1728–35. 10.1002/ijc.21544 [DOI] [PubMed] [Google Scholar]
- 105.Nakamura K, Sho M, Akahori T, et al. Clinical relevance of CD70 expression in resected pancreatic cancer: prognostic value and therapeutic potential. Pancreatology 2021;21:573–80. 10.1016/j.pan.2021.01.013 [DOI] [PubMed] [Google Scholar]
- 106.Liu L, Yin B, Yi Z, et al. Breast cancer stem cells characterized by CD70 expression preferentially metastasize to the lungs. Breast Cancer 2018;25:706–16. 10.1007/s12282-018-0880-6 [DOI] [PubMed] [Google Scholar]
- 107.Kitajima S, Lee KL, Fujioka M, et al. Hypoxia-inducible factor-2 alpha up-regulates CD70 under hypoxia and enhances anchorage-independent growth and aggressiveness in cancer cells. Oncotarget 2018;9:19123–35. 10.18632/oncotarget.24919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruf M, Moch H, Schraml P. Interaction of tumor cells with infiltrating lymphocytes via CD70 and CD27 in clear cell renal cell carcinoma. Oncoimmunology 2015;4:e1049805. 10.1080/2162402X.2015.1049805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tulchinsky E, Demidov O, Kriajevska M, et al. Emt: a mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev Cancer 2019;1871:29–39. Jan 1. 10.1016/j.bbcan.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 110.Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015;527:525–30. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perotti V, Baldassari P, Molla A, et al. An actionable axis linking NFATc2 to EZH2 controls the EMT-like program of melanoma cells. Oncogene 2019;38:4384–96. 10.1038/s41388-019-0729-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang H, Zhao J, Zhang L, et al. SRPX2 enhances the epithelial-mesenchymal transition and temozolomide resistance in glioblastoma cells. Cell Mol Neurobiol 2016;36:1067–76. 10.1007/s10571-015-0300-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim J, Lee I-H, Cho HJ, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 2015;28:318–28. 10.1016/j.ccell.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 114.Ring NG, Herndler-Brandstetter D, Weiskopf K, et al. Anti-SIRPα antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc Natl Acad Sci U S A 2017;114:E10578–85. 10.1073/pnas.1710877114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Starzer AM, Berghoff AS. New emerging targets in cancer immunotherapy: CD27 (TNFRSF7). ESMO Open 2020;4:e000629. 10.1136/esmoopen-2019-000629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Segerman A, Niklasson M, Haglund C, et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to Proneural-Mesenchymal transition. Cell Rep 2016;17:2994–3009. 10.1016/j.celrep.2016.11.056 [DOI] [PubMed] [Google Scholar]
- 117.Haddad AF, Chen J-S, Oh T, et al. Higher cytolytic score correlates with an immunosuppressive tumor microenvironment and reduced survival in glioblastoma. Sci Rep 2020;10:17580. 10.1038/s41598-020-73793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Z, Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front Immunol 2018;9:1004. 10.3389/fimmu.2018.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Biron CA. Role of early cytokines, including alpha and beta interferons (IFN-alpha/beta), in innate and adaptive immune responses to viral infections. Semin Immunol 1998;10:383–90. 10.1006/smim.1998.0138 [DOI] [PubMed] [Google Scholar]
- 120.Kim BS, Jin Y-H, Meng L, et al. IL-1 signal affects both protection and pathogenesis of virus-induced chronic CNS demyelinating disease. J Neuroinflammation 2012;9:217. 10.1186/1742-2094-9-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gajewski TF, Corrales L, Williams J, et al. Cancer immunotherapy targets based on understanding the T Cell-Inflamed versus non-T Cell-Inflamed tumor microenvironment. Adv Exp Med Biol 2017;1036:19–31. 10.1007/978-3-319-67577-0_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allam A, Swiecki M, Vermi W, et al. Dual function of CD70 in viral infection: modulator of early cytokine responses and activator of adaptive responses. J Immunol 2014;193:871–8. 10.4049/jimmunol.1302429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang QJ, Yu Z, Hanada K-I, et al. Preclinical evaluation of chimeric antigen receptors targeting CD70-expressing cancers. Clin Cancer Res 2017;23:2267–76. 10.1158/1078-0432.CCR-16-1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Raikar SS, Fleischer LC, Moot R, et al. Development of chimeric antigen receptors targeting T-cell malignancies using two structurally different anti-CD5 antigen binding domains in NK and CRISPR-edited T cell lines. Oncoimmunology 2017;7:e1407898. 10.1080/2162402X.2017.1407898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dequeant M-L, Sagert J, Kalaitzidis D, et al. CD70 knockout: a novel approach to augment CAR-T cell function. Am Assoc Cancer Res 2021. [Google Scholar]
- 126.Kumar S, Singh SK, Viswakarma N, et al. Rationalized inhibition of mixed lineage kinase 3 and CD70 enhances life span and antitumor efficacy of CD8+ T cells. J Immunother Cancer 2020;8:e000494. 10.1136/jitc-2019-000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cooper ML, Choi J, Staser K, et al. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia 2018;32:1970–83. 10.1038/s41375-018-0065-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goswami S, Walle T, Cornish AE, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med 2020;26:39–46. 10.1038/s41591-019-0694-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol 2006;176:1517–24. 10.4049/jimmunol.176.3.1517 [DOI] [PubMed] [Google Scholar]
- 130.Vossen MTM, Matmati M, Hertoghs KML, et al. CD27 defines phenotypically and functionally different human NK cell subsets. J Immunol 2008;180:3739–45. 10.4049/jimmunol.180.6.3739 [DOI] [PubMed] [Google Scholar]
- 131.Morimoto S, Kanno Y, Tanaka Y, et al. CD134L engagement enhances human B cell Ig production: CD154/CD40, CD70/CD27, and CD134/CD134L interactions coordinately regulate T cell-dependent B cell responses. J Immunol 2000;164:4097–104. 10.4049/jimmunol.164.8.4097 [DOI] [PubMed] [Google Scholar]
- 132.Nolte MA, Arens R, van Os R, et al. Immune activation modulates hematopoiesis through interactions between CD27 and CD70. Nat Immunol 2005;6:412–8. 10.1038/ni1174 [DOI] [PubMed] [Google Scholar]
- 133.Hanazawa A, Ito R, Katano I, et al. Generation of human immunosuppressive myeloid cell populations in human interleukin-6 transgenic NOG mice. Front Immunol 2018;9:152. 10.3389/fimmu.2018.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riether C, Pabst T, Höpner S, et al. Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat Med 2020;26:1459–67. 10.1038/s41591-020-0910-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003289supp002.pdf (61.3KB, pdf)
jitc-2021-003289supp001.pdf (6.1MB, pdf)
Data Availability Statement
Data are available on reasonable request.