Abstract
Most gene editing technologies introduce breaks or nicks into DNA, leading to the generation of mutagenic insertions and deletions by non-homologous end-joining repair. Here, we report a new, cleavage-free gene editing approach based on replication interrupted template-driven DNA modification (RITDM). The RITDM system makes use of sequence-specific DLR fusion molecules that are specifically designed to enable localized, temporary blockage of DNA replication fork progression, thereby exposing single-stranded DNA that can be bound by DNA sequence modification templates for precise editing. We evaluate the use of zinc-finger arrays for sequence recognition. We demonstrate that RITDM can be used for gene editing at endogenous genomic loci in human cells and highlight its safety profile of low indel frequencies and undetectable off-target side effects in RITDM-edited clones and pools of cells.
Keywords: genome editing, human cells, cleavage-free and off-targets
Graphical abstract

Genome editing by RITDM does not require DNA cleavage and achieves good editing frequencies while avoiding the generation of significant amounts of indels and other forms of off-target effects.
Introduction
Many current gene correction systems make use of nuclease or nickase activity to introduce DNA breaks. The generation of DNA breaks has the implicit disadvantage that DNA repair processes, such as non-homologous end-joining (NHEJ), are being triggered, which often results in the generation of mutagenic indels.1,2 Minimizing mutational damage is an essential feature that will enable a wider range of genome editing-based therapies to progress to the clinic. In addition to the current gene editing technologies, it would be pragmatically useful to have an alternative gene editing system that does not depend on double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA) breakage. We report the development of a new nuclease-free gene editing system—RITDM (replication interrupted template-driven DNA modification)—based on sequence-specific chimeric molecules that interact with replication forks, which, in combination with DNA modification polynucleotides, enables editing without DNA cleavage.
During cell division, the replication fork complex transiently exposes short stretches of ssDNA.3 Genome-wide slowing and stalling of replication forks leads to a lingering ssDNA presence within cells and can generate signals equivalent to those of damaged DNA to evoke DNA repair.4 Introduction of ssODNs can result in their annealing at DNA replication forks, which can be used for genome editing in lower eukaryotes.5 We hypothesized that we could convert these genome-wide effects to a locus-specific approach by blocking replication forks specifically in the vicinity of a target locus, and that this approach could be developed for usage in the larger genomes of mammalian cells. Such a system could enable locus-specific gene editing in human cells.
We first developed sequence-specific blocking agents (named DLRs) by designing fusion proteins in which one module (D: DNA binding) binds sequence specifically to one strand of DNA, and, through a linker (L: linker), connects with a second DNA binding module (R: replication fork block) that binds to the opposite strand of DNA. The hypothesis underpinning this design was that such a molecule could (reversibly) bind to both DNA strands and thereby form a temporary “clamp” at the target site. For the sequence-specific D-module, we used zinc-finger arrays, which have been used extensively in the creation of various types of fusion proteins (Figure 1A).6 In the R-module, we used a domain that can bind to the phosphate backbone of the opposing DNA strand. When introduced into a cell, DLR fusion proteins can bind to a specific genomic locus and obstruct the progression of a replication fork, thereby exposing single-stranded sequences to which complementary oligonucleotides with (a) desired sequence modification(s) can bind (Figure 1B). After the binding of a DNA modification template, the DNA contains (a) sequence mismatch(es). The cellular DNA repair mechanisms can resolve these mismatches. Depending on which strand is used for correction, part of the cells will be converted to have the desired sequence modification.
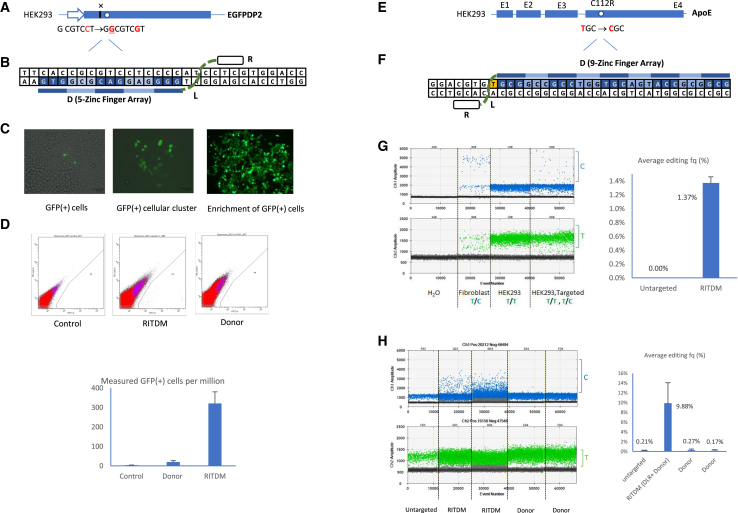
Figure 1.
RITDM gene editing strategy
(A) A RITDM system consists of a programmable DLR molecule containing a sequence-specific recognition domain (D), such as a zinc-finger array, fused to a linker domain (L), and a non-sequence-specific DNA binding domain (R). (B) Schematic of RITDM editing. A locus-specific DLR molecule binds DNA at the target site to block replication fork progression temporarily, and a DNA correction template anneals and incorporates at the DNA replication fork. The resulting intermediary DNA heteroduplex can be repaired to enable a permanent genetic modification.
We demonstrated the direct interaction between DLR fusion molecules and the replication fork, enabling a novel mechanism for gene editing in mammalian cells. Next, we applied RITDM at various disease-relevant loci in the human genome and showed that RITDM can be used to introduce heritable and precise genetic modifications. We achieved precise editing involving (1) single base pair changes, (2) restoration of reading frames by inserting nucleotides, and (3) gene knockouts by the introduction of stop codons and/or restriction enzyme sites. Since RITDM does not require DNA cleavage at the target site, the mutational indel incidence remains at a low level on a par with that of the background. Moreover, both DLR and the donor template must work in concert, implying that off-target effects are of second-order kinetics. This results in a favorable safety profile of RITDM. Taken together, RITDM is a novel DNA cleavage-free editing system that can be used to achieve gene editing while avoiding the generation of indels and/or other off-target effects. This approach thus provides an additional option in the development of gene correction therapies.
Results
Development of sequence-specific blocking DLR fusion molecules
When designing a DLR molecule for a specific locus, the D domain must be programmable to recognize the target sequence on one strand of DNA. In principle, any DNA sequence recognition platform can be used, including zinc-finger arrays, TALE effectors, and dCAS9 along with a gRNA. In consideration of keeping the size of DLR constructs as small as possible to also allow the use of in vivo delivery vectors, we chose to use zinc-finger arrays.7, 8, 9, 10 Zinc fingers measure only about 30 amino acids long and can target 3 nucleotides, making it the most compact DNA binding element. The L domain is an orientation enabler for the R domain reaching the opposite DNA strand. For the R unit, we ideally wanted to design a structure that is (1) as compact as possible, (2) able to bind to DNA in a non-sequence-specific manner (e.g., via phosphate backbone and/or major or minor groove interaction), (3) have (a) crystal structure(s) available for modeling, and that is (4) present in a broad range of proteins for non-specific DNA binding, assuming such structures could also serve a similar function in our DLR fusion protein. A structure fulfilling these requirements is the PD-(D/E)xK fold that is present in a superfamily of proteins.11 Members of this protein family are involved in numerous activities that involve DNA interaction.12 A feature of this fold is that it can bind to the phosphate backbone, thereby providing non-sequence-specific binding. Thus, we set out to design the first DLR fusion protein using a zinc-finger array linked to a PD-(D/E)xK domain.
Of the number of zinc-finger crystal structures that have been elucidated, we used the 1MEY structure as a reference.13 This crystal structure of a designed zinc-finger protein provided useful information on amino acid side-chain interactions with nucleotides as well as details on how its C terminus was positioned in the major DNA groove. For the design of the R unit, we made use of the crystal structure of FokI, as it contains a PD-(D/E)xK fold in its catalytic domain.12 In addition to looking at the potential suitability of FokI-derived R unit structures, we also included another naturally existing PD-(D/E)xK fold, appearing in the small unit of BtsI, in our analysis.14 Not only does its size and stability make it a good candidate for use in protein engineering, but amino acid sequence alignments also showed similarities between BtsI and FokI (Figure S1A). From BtsI, we identified the PD-(D/E)xK fold located within the β2 and β3 sheets of a three-antiparallel β sheet structure and selected this three-antiparallel β sheet structure for further development of the R unit. Moreover, the crystal structure of FokI indicated that this structure can orient perpendicularly on the DNA phosphate backbone.12 In FokI, the loop between β1 and β2 in the PD-(D/E)xK fold participates in DNA major groove binding,12 which would support the desired orientation of the R unit versus that of the zinc-finger DNA binding D domain. To enable this desired orientation, we replaced the loop of BtsI with that of FokI to create an R unit that combines the major groove-binding loop as found in FokI with the compact β sheet structure (β1, β2, and β3) of BtsI (Figures 1A and S1A).
When considering how we could link the N-terminal zinc finger to the C-terminal R domain, we again looked at the crystal structures. The C terminus (in MEY1) of the zinc finger bound to DNA is positioned in the major groove of the DNA it is bound to. The crystal structure of the antiparallel β sheet structure (as in Fok1), when it is oriented on top of the phosphate backbone of the opposing strand, has its N terminus in close proximity to the major groove. The N terminus of the R unit could thus be fused to the C terminus of the zinc-finger array using a relatively short linker. We then used a four-amino-acid LRGS linker, which is a prototypical linker used in synthetic zinc-finger biology.15
To avoid the risk of residual nuclease activity, amino acid residues with a negatively charged side chain, an aspartic acid (D) residue from the β2 sheet, or glutamic acid (E) from the β3 at the catalytically active sites in the PD-(D/E)xK fold were replaced by amino acids either with an uncharged side chain, such as serine (S), glutamine (Q), asparagine (N), or threonine (T), a hydrophobic chain, such as alanine (A), valine (V), leucine (L), isoleucine (I), or methionine (M), or with a positively charged chain, such as histamine (H), arginine (R), or lysine (K), which, in native FokI and in BtsI, eliminates nuclease activity (Figure S1B).11
Mutant EGFP reporter gene editing by RITDM
We developed a gene correction reporter system to test if our approach using DLR fusion proteins in combination with a modification template could be used to perform gene editing. For this proof-of-principle validation we developed a cell line with a GFP reporter (Figure S2A). The integrated mutant EGFP (EGFPDP2) gene bears a frameshift causing a deletion (G) in combination with a G-to-C point mutation, rendering the expressed protein nonfunctional (Figures 2A and S2B). We obtained a clone with two inserted copies of the EGFPDP2 construct with the idea that, if one copy was corrected, the other copy could serve as an indicator for potential off-target effects. As a first attempt, a DLR fusion protein with a 5-zinc-finger array, recognizing a unique 15-nucleotide sequence in EGFPDP2, was developed (Figure 2B). A 142-nucleotide ssDNA correction template was designed that incorporated the corrections for a “G insertion” and a “C-to-G conversion” (Figure S2B).
Figure 2.
Genome editing by RITDM
(A) Diagram of a mutated EGFP (EGFPDP2) reporter gene in the HEK293 reporter cell line. The EGFDP2 reporter has a deletion of single nucleotide (G) and a G-to-C point mutation. Restoration of EGFP reading frame after RITDM editing. (B) Schematic depicting that a mutated EGFP (EGFPDP2) reporter gene was targeted and repaired using a specifically designed RITDM system. The DLR molecule recognizes 15-nucleotide sequences, specifically. (C) EGFP was restored to express functionally in the reporter cells, as shown in forms of positive cells (left), cellular cluster (middle), and enrichment of positive cells (right). (D) Representative flow plots from flow cytometric analysis of alive EGFP cells 7 days post-genomic editing with the indicated conditions of control, donor alone, RITDM genome editing. Quantification of GFP (+) cells (per million) from the indicated conditions. Bar graphs represent two independent experiments with standard error shown. (E) The endogenous ApoE gene was genetically modified at codon 112 by RITDM in HEK293 cells. (F) The DLR molecule 27 nucleotide sequences at the target. (G) Single-nucleotide T-to-C conversion was detected by ddPCR. The bar graph represents the editing frequencies at codon 112 of the ApoE gene achieved by RITDM in HEK293 cells measured and calculated by ddPCR. (H) Enhanced genome editing frequencies were detected by ddPCR, while ssODN alone did not induce T-to-C conversion. The bar graph represented the editing frequencies at codon 112 of the ApoE gene achieved by RITDM in HEK293 cells measured and calculated by ddPCR.
Five days after the transfection of thymidine synchronized cells, fluorescent green cells and cellular clusters could be observed using fluorescent microscopy (Figure 2C, left and middle). Following reseedings, we could obtain pure green cell populations (Figure 2C, right). Cells were also analyzed by flow cytometry 7 days after transfection. This showed that, under these initial conditions, RITDM editing resulted in 300 ± 60 cells per million with bright green fluorescence in a normal cellular morphology, whereas the “ssODN alone” control showed a frequency as low as 19 ± 7 per million cells (Figure 2D), which is in line with published observations.16,17 These observations confirmed that RITDM could be used for gene editing.
To further evaluate the nature of the genetic conversion at the GFP reporter locus, we extracted genomic DNA from a pure green cellular population that was obtained after repeated reseeding. We used Sanger sequencing to confirm the genetic modifications (Figure S2C) followed by next-generation sequencing. Approximately 59.4% of the reads had the anticipated G insertion and C-to-G conversion (Figures S2D–S2F), indicating that one allele of EGFPDP2 was corrected into EGFP by RITDM gene editing.
We tested a series of donors with lengths between 60 and 200 nucleotides that also varied the position of the editing sequences. All donors with a length between 90 and 200 nucleotides yielded green cells or green clusters. We did not observe a meaningful pattern relevant to the positions of genetic modification within the donor either symmetrically or asymmetrically. These observations were in line with results obtained when we started using RITDM to edit codon 112 of the ApoE gene. When using a correction oligonucleotide of 60 nucleotides, we only observed a weak ddPCR signal, while a 71-nucleotide oligonucleotide allowed an increase in signal (Figure S15A). Therefore, we subsequently designed donor templates with an approximate length of 150 nucleotides with the modification sequence in its middle.
Conversion of codon 112 of human ApoE by RITDM gene editing
To confirm that RITDM could also modify an endogenous human gene, we selected the ApoE gene and aimed to convert a “T” to “C” at codon 112 in ApoE in HEK293 cells. ApoE has three major alleles (E2, E3, and E4) that differ at codons 112 and 158. E4 is a risk allele for late-onset Alzheimer disease (LOAD), E3 is the dominant allele in the human population, and E2 reduces the likelihood and age of onset of LOAD.18,19 A DLR fusion protein was developed with an array of 9 zinc fingers recognizing a 27-nucleotide sequence on the leading strand of human ApoE gene (Figures 2E, 2F, and S3A). We evaluated a number of potential binding sites close to the target site and found this 27-nucleotide sequence to be unique in the human genome, even when we allowed for multiple mismatches in BLAST searches. For the donor template, a 129-nucleotide ssDNA with a T-to-C substitution located in its middle was used.
Detection of T-to-C conversion was performed by droplet digital PCR (ddPCR). One common primer was located inside this ssDNA template sequence while the other was located outside. Allele-specific probes conjugated with fluorophores FAM and HEX were designed to distinguish between C and T, respectively (Figure S3A). Following transfection of HEK293 cells with the DLR and correction template, cells were allowed to recover and grow on complete culture medium for 7 days, after which the genomic DNA was isolated and used in ddPCR analysis. Raw droplet data are shown in Figure 2G, where C droplets are displayed in the top panel and T droplets are in the lower panel. “No DNA” was used as negative control, showing neither C nor T droplets. Wild-type fibroblast was used as a positive control because of its heterozygous T/C genotype for codon 112 of human ApoE, showing both C and T droplets. The untreated HEK293 only had T droplets, demonstrating a homozygous T/T genotype. After RITDM editing, C droplets appeared, demonstrating successful T-to-C genetic conversion at codon 112 of human ApoE with an average DNA editing frequency of 1.37% ± 0.09% (Figure 2G; Table S3), obtained from three independent experiments. We also confirmed the successful T-to-C conversion by next-generation sequencing. Cells edited by RITDM showed a T-to-C conversion at the expected nucleotide position with a frequency of 1.6%. Since HEK293 has five to six copies of chromosome 19, a DNA conversion rate of 1.5%–1.6% implies that the frequency of cells containing at least one corrected copy will be higher and may be in the 7.5%–9.6% range. Compared with untransfected cells, no obvious unwanted single-nucleotide polymorphisms (SNPs) were detected (Figure S3B). We further optimized the transfection protocol by increasing the amount of DNA plasmid and ssODN, which resulted in an increase of the correction frequency to 9.88% ± 4.19%, as measured by ddPCR (Figure 2H; Table S3).
Following these experiments, we used the human U937 cell line, which is homozygous ApoE4/E4.20 We aimed to convert ApoE4 to E3 by a C-to-T conversion at codon 112 by co-transfection of the same DLR used in the experiments above. For a DNA modification template we used a 150-nucleotide ssODN with a desired C-to-T substitution (Figures 3A and S5A). As detected by ddPCR, RITDM targeting resulted in C-to-T gene conversion, which was verified by next-generation sequencing (Figures 3A, S5B, and S5C). We repeated these experiments multiple times and obtained an average 16.67% ± 7.92% of DNA correction frequencies (Figure S5B).
Figure 3.
Illustrations of the scope of genome editing by RITDM
A number of disease-related genomic loci were modified by RITDM. For each case, the genomic target sequence is shown in blue and the sequence to be changed in red. The donor template with the desired sequence modification (in green) is shown. For each case the type of editing and the intended consequences are indicated. A representative ddPCR plot illustrates typical experimental data obtained, while the bar charts show the percentage of cells without indels, the percentage of cells edited as intended, respectively, the percentage of cells with indels, as determined by deep sequencing. Experimental results are shown for ApoE (A), Bcl11a (B), and PDCD1 (C). Bar graphs represent at least two independent experiments with standard errors shown.
We performed a number of controls, including one that contained the zinc-finger array without the R unit (Figure S6C). This was done to evaluate if the zinc-finger array by itself could block replication fork progression and elicit gene editing. Blocking of replication fork progression by deadCAS9 (dCAS9) has been described.21,22 Performing this control, we did not observe gene conversion, indicating that the R unit is needed to achieve the intended effect. In addition, the donor alone did not induce genome conversion at significant levels. Also, in subsequent experiments, we never observed gene conversion using zinc-finger array controls (plus modification oligonucleotide) without an R unit (Figures S7C, S8C, and S9C). This is in line with gene editing experiments in which dCAS9 is used as a control where, typically, very low or no gene editing frequencies are observed.23,24
In addition, we further ruled out the possibility of false positivity stemming from residual donor template surviving in cells and being co-isolated with chromosomal DNA. Shown in Figure S6A, we performed ddPCR assay using only one end primer. If there was donor template left in the genomic DNA extraction, the donor template then could pair with one end primer to amplify the target region, leading to false-positive droplets that could be detected by the highly sensitive ddPCR assay. Clearly, we did not find any positive droplets in any condition tested that involved donor template alone, chromosomal DNA from mixed positive pooled cells, and positive single cellular clones (Figure S6B).
Generation and analysis of ApoE-converted clones
Having established that RITDM can be used to convert the ApoE gene at codon 112, we wanted to generate clones to enable more detailed genetic and functional characterization of the gene editing effects. In subsequent transfections, single clones were obtained by seeding 0.5 transfected cell/well and continued culturing for 2–3 weeks. Positive clones were identified by ddPCR (Figures S4A and S4B). Positive clone frequencies obtained averaged 11.6% ± 5.3% in three independent experiments (Figure S4C). Sanger sequencing results of representative clones confirmed the desired T-to-C conversions (Figure S4D). HEK293 has five to six copies of the section of chromosome 19 that contains the ApoE gene.25 Next-generation sequencing results indicated that 14.7% of reads had the desired T-to-C conversion (Figure S4E), indicating that one of the six copies was modified in the clones analyzed. In addition, we continued culturing positive clones for more than 50 passages, and found that this conversion is stable over time. Collectively, it demonstrates that RITDM can achieve gene editing in a human genome.
Having established that a DLR design can be used for endogenous genomic editing, we tested if specific active site amino acid substitutions influenced the observed gene correction frequency (Figure S1B) by analyzing the generation of ApoE-converted clones. Substituting aspartic acid (D) residue from β2 sheet or the glutamic acid (E) from β3 in the PD-(D/E)xK fold with serine (S), glutamine (Q), asparagine(N), valine (V), leucine (L), histidine (H), or alanine (A) allowed 31%–163% relative correction frequency, confirming that the gene editing activity is not dependent on active site residues or nickase activity (Figures S1C, S1D, and S1E; Table S1). We also tested if changing the loop between the β2 and β3 sheets would influence the observed frequency and if we could replace the β2 and β3 sheets from FokI with that of BtsI. These experiments were done to test if the performance was a function of the overall 3D architecture, rather than it being dependent on a specific amino acid sequence (Figure S1B). The conversion frequencies observed were not significantly different. Therefore, we settled on using a single construct in which a 33-amino-acid R-module contains valine (V) in the β3 sheet (Figure 1A) in all further experiments of this study.
Gene editing of human disease loci
We explored the generalizability of RITDM by editing additional genes that are of therapeutic interest. In addition to showing that other loci can be converted, we also wanted to explore if we could perform edits other than single-nucleotide conversions.
As the first additional example, we chose to target the enhancer within intron 2 of the BCL11A gene, a potential therapeutic target of sickle cell anemia and β-thalassemia.26 The aim was to disrupt the critical GATAA box in the enhancer and to convert it into an EcoRI restriction enzyme recognition sequence, generating the potential for RFLP analysis (Figures 3B and S7A). We designed a DLR fusion molecule with a DNA recognition domain comprised of an array of 7 zinc fingers specifically designed to recognize a 21-nucleotide sequence close to the target site. The correction template was a 140-nucleotide ssDNA containing the TTATC-to-GAATTC substitution roughly located in its middle.
As a second example, we targeted exon 51 of the human Dystrophin gene. A common type of mutation in the Dystrophin gene are deletions of exons which disrupt the reading frame and can cause Duchenne muscular dystrophy (DMD).27 We wanted to demonstrate that RITDM can be used in such cases to restore the reading frame via the insertion of one or two nucleotides. To do so, we designed a DLR molecule with a 10-zinc-finger array recognizing a 30-nucleotide sequence at the beginning of the exon 51 of Dystrophin. The recognition site was selected to be unique in the human genome, even when we allowed for multiple mismatches. It was used it in combination with a 137-nucleotide ssDNA template with a 2-nucleotide “GA” insertion at the desired position, enabling a TTACTCT-to-TTAGACTCT substitution (Figures 4A and S8A).
Figure 4.
RITDM editing of genomic DNA in human B lymphocytes
RITDM editing efficiencies and indel frequencies achieved at two genomic sites in human B lymphocytes. We used RITDM to convert T to C at codon 112 of ApoE (A) in human B lymphocytes respectively to insert two nucleotides, “GA,” at the beginning of exon 51 of the Dystrophin gene (B) both in the U937 cell line and in human B lymphocytes. The bar charts show the percentages of deep-sequencing reads without indels (the intended 2-nucleotide insertion is scored as indel), with genomic edits as intended (2-nucleotide insertion), respectively, with unintended indels from pooled RITDM-edited cells. Editing efficiencies and indel frequencies from each independent experiment are shown. Results obtained from the various RITDM editing experiments in human B lymphocytes were calculated from all biological replicates and displayed as mean ± SD (n = 3 independent experiments for ApoE and n = 4 independent experiments for Dystrophin genomic editing by RITDM. ∗p < 0.05).
The third example involved the human PDCD1 gene. For immune-oncology therapies, the PDCD1 gene is frequently inactivated using nucleases that generate a wide spectrum of indels.28 We wanted to show that RITDM could be used for “knockout” purposes by introducing a stop codon. This will allow for a more homogeneous population (versus semi-random indel generation by nucleases) and can be combined with introducing a restriction enzyme site to allow for RFLP analysis. A DLR fusion protein was designed to recognize a 21-nucleotide sequence in exon 1, using a 7-zinc-finger array. For the DNA donor template, a 149-nucleotide sequence modification polynucleotide was used, which had a substitution sequence of “AATTCAT” that was intended to replace “CA” at its targeting locus, leading to a stop codon (TGA) in frame (Figures 3C and S9A).
We co-transfected DLR-encoding plasmids and ssDNAs into HEK293 or U937 cells and measured gene editing effects by ddPCR (Figures 3B, 3C, and 4A; Table S3). We observed the desired modifications at all three targets. Validation was done by deep sequencing. The gene editing efficiencies achieved averaged 10.3% ± 5.28% in HEK293 cells for BCL11A (Figures 3B, S7B, and S7C), 30.3% in the case of Dystrophin (Figure 4B and S8B), and 11.07% ± 7.88% for PDCD1 in U937 (Figure 3C, S9B and S9C; Table S3), consistent with those frequencies as determined by ddPCR from multiple independent experiments (Figure S7B, S8B, and S9B; Table S3). Part of the variation observed in the editing frequencies stems from the fact that we used results obtained from low-dose and high-dose electroporation protocols, and that the high-dose protocols yielded higher frequencies. These results collectively establish that RITDM can be used to edit genes in the human genome, not only for single nucleotide conversions, but other purposes such as to restore reading frames, introduce stop codons, and to create RFLPs.
RITDM gene editing in human B lymphocytes
We wanted to explore the potential of RITDM gene editing in human primary cells. For this purpose, we selected B lymphocytes (B cells). They can be therapeutically relevant as they can be isolated from peripheral blood mononuclear cells, they can secrete large amounts of protein, and they are metabolically active and can migrate throughout the body, including the CNS.29, 30, 31 To avoid any potential issues resulting from cell-cycle synchronization when considering therapeutic developments, a B cell transfection protocol was developed that omitted this step.
B cells were transfected with the same DLR construct and ssODN used for ApoE codon 112 conversion. Transfected cells were analyzed by ddPCR followed by deep-sequencing validation (Figures 4, S10A–S10C, and S11A–S11C). A conversion rate of 18.5% ± 6.9% was obtained in three independent experiments (Figure 4A). As further illustration of the potential, we transfected B cells obtained from a DMD patient (with exons 46–50 deleted) and wild-type B cells with the same components used in U937 cells previously. Overall Dystrophin gene conversion frequencies of 38.8% ± 29.2% were achieved in B cells from two independent donors (Figure 4B).
Indels and SNP profiles generated by RITDM genomic editing
Following the successful gene conversions, we wanted to study, in more detail, the molecular nature of these conversions and explore potential side effects. In the case of the EGFP reporter we had two copies of the reporter gene and in the case of ApoE we could use the additional five copies to analyze if any of them displayed signs of indels. Indel levels at the EGFPDP2 locus were minimal and showed no significant differences between HEK293-negative and -positive cellular populations. Next-generation sequencing analysis revealed that the overall indel percentages among three cell populations were 1.07%, 1.06%, and 0.53%, respectively, when analyzing a 171-bp window (Figures 5A and S2D). The indel profiles were almost indistinguishable, except for the positive population, which had an insertion at the expected position (Figure S2D). Detailed SNP analysis showed no SNPs above background levels (Figure S2E). The indel length histogram (Figure 5B) showed mostly single-nucleotide deletions, followed by two-nucleotide deletions and very low numbers of other types of indels. Since indel sizes formed by NHEJ following the action of nucleases typically have a wider size distribution, this suggests that even the very low indel frequencies measured might be more indicative of background noise resulting from the analysis methodology rather than a reflection of NHEJ activity.
Figure 5.
RITDM results in low indels at the target sites
(A) Overall indel frequencies at the EGFPDP2 target site comparing control, positive, and negative cell populations. (B) Overall indel distribution over a 171-nucleotide window in positive and negative cell populations after RITDM gene editing. (C) Overall indel frequencies (mean ± SEM) at ApoE codon 112, comparing positive (n = 3) and negative clones (n = 2). (D) Indel frequencies in a 108-nucleotide window in representative positive and negative clones of ApoE codon 112 conversion by RITDM. The y axis scale maximum is 0.25%.
Next, we followed similar analysis protocols for ApoE codon 112 conversions in clones and in pools of cells. We observed a very low frequency of indels using a number of randomly selected cellular clones. The overall indel frequencies measured in a 108-nucleotide window were an average of 1.69% for two negative clones and 0.6% for three positive clones (Figure 5C). Patterns and measured frequencies of indels at each position in this 108-nucleotide window obtained from positive and negative clones were not significantly different (Figure 5D). Similarly, low frequencies of indels were observed when analyzing pooled HEK293 cells after editing. The highest level of change measured at any position was a one-nucleotide insertion (position 52) with a frequency of 0.15%, which most likely reflected background signal and/or a technical artifact (Figure S3D). The overall indel frequency was 0.59% in pooled U937 cells (Figures 3A and S5C). Next-generation sequencing was also used to determine the indel frequencies for the experiments involving BCL11A, Dystrophin, and PDCD1. The frequencies of unintended insertions and deletions remained below 2.09% (Figures 3B and S7C), 0.97% (Figures 4B and S8C), and 1.72% (Figures 3C and S9D), respectively. In B cells, the observed average indel frequencies were 1.3% and 1.7% in ApoE and Dystrophin respectively as determined by deep sequencing. Collectively, this indicates that indel frequencies can remain low after RITDM editing.
Accessing off-target effects of RITDM
Having established that RITDM generates (very) low levels of indels and SNPs, we also wanted to analyze potential off-target events. To investigate putative off-targets of the 27-nuclotide recognition sequence “GCGGCCGCCTGGTGCAGTACCGCGGCG” from ApoE 112 site on the human genome, we enumerated all the possible sites with up to 10 mismatches. FASTA files for the hg38 assembly were obtained from https://hgdownload.soe.ucsc.edu/goldenPath/hg38/chromosomes/. Using the query sequence “GCGGCCGCCTGGTGCAGTACCGCGGCG,” matches were enumerated using CRISPRitz version 1.1.1.32 Given that this tool was originally designed for CRISPR applications, we removed any PAM constraint and allowed sequences longer than 20 bp. Using this strategy, no matches were recovered with up to 4 mismatches, and a total of 79,987 matches were recovered with up to 10 mismatches, as shown in Table S2. We set to explore the off-target events after RITDM editing in pooled B cells by studying indels and SNPs at each site with 5 and 6 mismatches. All sites of up to 6 mismatches are shown in Table 1, in which lowercase nucleotides in the “Match” column correspond to mismatches.
Table 1.
Off-target analysis after RITDM genetic editing at codon 112 of human ApoE gene in primary B cells
| OT# | Chromosome | Position | Strand | Match | Mismatches | Gene | wt |
RITDM |
RITDM donor insertion | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Depth | Indels | Depth | Indels | ||||||||||||
| L1 | chr19 | 1975511 | – | GtGcCCGCCTGGTGCtGTgCCGCGGCt | 5 | CSNK1G2/XM_005259498.1/XP_005259555.1 | 304,901 | 0.59% | 264,232 | 0.52% | N/D | ||||
| L2 | chr9 | 14322823 | + | GCGGCCGCaTGGgGCcGgAgaGCGGCG | 6 | NFIB/NM_001369468.1/NP_001356397.1 | 37,659 | 0.31% | 98,144 | 0.26% | N/D | ||||
| L3 | chr11 | 74006321 | – | cgGcCCGgCTcGTGCAGTACCGtGGCG | 6 | UCP3/NM_003356.4/NP_003347.1 | 199,825 | 0.06% | 291,333 | 0.05% | N/D | ||||
| L4 | chr1 | 155071301 | + | GCGGgCGgCTGaTtCAGTcCCGgGGCG | 6 | EFNA4/NM_005227.3/NP_005218.1 | 75,705 | 0.34% | 36,817 | 0.20% | N/D | ||||
| L5 | chr7 | 1503036 | – | GCGGCCGCgTtGTcCAGTgCCtCGGCc | 6 | INTS1/NM_001080453.3/NP_001073922.2 | 318,258 | 0.45% | 218,258 | 0.44% | N/D | ||||
| L6 | chr12 | 125331217 | – | GCGGaCtCCTGGgGCAGcAaCGCGGaG | 6 | TMEM132B/NM_001366854.1/NP_001353783.1 | 187,872 | 0.71% | 192,460 | 0.18% | N/D | ||||
| L7 | chr7 | 47560011 | – | GCGGCCGgCTctTGCAGTcCtGCGGgG | 6 | TNS3/XM_011515477.2/XP_011513779.1 | 143,380 | 0.18% | 140,906 | 0.16% | N/D | ||||
| L8 | chr3 | 183447638 | – | cCGGCCGCtTGGTGaAGTgCgGCtGCG | 6 | LINC00888/NR_038302.1 | 154,259 | 0.26% | 156,735 | 0.26% | N/D | ||||
| L9 | chr15 | 89371748 | + | GaGGCCGCgcGGTGCAGaACCGCcGgG | 6 | MIR9-3HG/NR_133001.1 | 33,500 | 0.15% | 59,190 | 0.15% | N/D | ||||
| L10 | chr10 | 97882322 | – | aCtGCaGCCTGGTGCAGTACtGgGGCa | 6 | CRTAC1/XM_011539917.1/XP_011538219.1 | 106,612 | 0.23% | 93,726 | 0.16% | N/D | ||||
| L11 | chr3 | 49724183 | – | GgGGCCGgCgGcTGCtGgACCGCGGCG | 6 | RNF123/XM_017007018.1/XP_016862507.1 | 59,636 | 0.51% | 53,979 | 0.61% | N/D | ||||
| L12 | chr19 | 45153753 | + | GCGGCCtCCTGcTGCAtcAgCGCtGCG | 6 | NKPD1/NM_198478.4/NP_940880.3 | 271,874 | 0.18% | 235,883 | 0.30% | N/D | ||||
| L13 | chr6 | 35688953 | – | GCGGCCGgCTGGgGCgGgACgGCGcCG | 6 | FKBP5/NM_001145775.3 | 127,998 | 0.42% | 131,188 | 0.47% | N/D | ||||
| L14 | chr14 | 67600139 | – | GCGGCCGCCTGGcGCtGcAgCGCcGCt | 6 | PIGH/XM_017021371.2/XP_016876860.1 | 99,453 | 0.07% | 86,034 | 0.05% | N/D | ||||
| L15 | chr8 | 54021997 | + | GCGGCCcCCTcGgGCcGgACCGCGGCc | 6 | TCEA1/NM_201437.3/NP_958845.1 | 190,996 | 0.56% | 111,968 | 0.57% | N/D | ||||
| L16 | chr6 | 41921800 | – | cCaGCCGCgTGGTGCgtTcCCGCGGCG | 6 | MED20/NM_001305455.2 | 84,296 | 0.81% | 88,668 | 0.51% | N/D | ||||
| L17 | chr6 | 41895056 | – | GCGGCgGCagGGgGCgGgACCGCGGCG | 6 | USP49/NM_001286554.2 | PCR failure: no PCR products | ||||||||
| L18 | chr6 | 136792483 | – | GCGGCgcCCTtGaGCt GcACCGCGGCG |
6 | MAP3K5/XM_017010875.1/XP_016866364.1 | 39,159 | 0.75% | 87,160 | 0.65% | N/D | ||||
| L19 | chr7 | 159024680 | – | GCGcCCtgCTGcTGCA GgACCGCtGCG |
6 | LINC00689/NR_024394.1 | Deep-sequencing failure: no mapping | ||||||||
| L20 | chr16 | 22827283 | + | GgGGCCGCCTGGTGCAGgACCctGtaG | 6 | HS3ST2/NM_006043.2/NP_006034.1 | 57,200 | 0.16% | 48,433 | 0.32% | N/D | ||||
| L21 | chr19 | 1249701 | + | GCtGCCaCgTGGgGCgGTgCCGCGGCG | 6 | LOC102723811/XR_002958449.1 | PCR failure: no PCR products | ||||||||
| L22 | chr9 | 86945861 | + | GCGGCCGCCcGaTGCAGcAgCGCcGCt | 6 | GAS1/NM_002048.3/NP_002039.2 | 48,731 | 1.77% | 61,970 | 1.47% | N/D | ||||
| L23 | chr15 | 73052768 | + | GCGGCCGCCaGGaGCgGctCCGCGcCG | 6 | NEO1/XM_005254408.1/XP_005254465.1 | 183,646 | 0.34% | 174,440 | 0.41% | N/D | ||||
| L24 | chr4 | 2354008 | + | GCGGCCaCCTGGTtCAGctCCtCGtCG | 6 | ZFYVE28/XM_006713902.3/XP_006713965.1 | 68,034 | 0.36% | 103,538 | 0.26% | N/D | ||||
| L25 | chr3 | 47497960 | + | GacGaCGCCTGcTGCAGTACtGCGGaG | 6 | ELP6/XM_005265241.4/XP_005265298.1 | 12,736 | 15.21% | 85,908 | 13.72% | N/D | ||||
| L26 | chr10 | 97185738 | + | GCcGCCtCCaGGTGCAGTcCCGgGGCa | 6 | SLIT1/NM_003061.3/NP_003052.2 | 267,330 | 0.45% | 340,331 | 0.39% | N/D | ||||
| L27 | chr22 | 46064562 | + | tCGGCgcCCTGGTGCcGcACCGCGcCG | 6 | Uncharactorized gene LOC642648 | 73,324 | 0.15% | 78,854 | 0.12% | N/D | ||||
| L28 | chr7 | 27200182 | – | GCGGCCGCCTGGcGCgaccCCGCGGgG | 6 | HOXA13/NM_000522.5/NP_000513.2 | 273,228 | 1.71% | 262,149 | 1.59% | N/D | ||||
| L29 | chr2 | 53787274 | + | GCGGCCtCCTGGaGgcGTcCgGCGGCG | 6 | ASB3/NM_145863.3 | 15,695 | 3.14% | 19,711 | 2.57% | N/D | ||||
| L30 | chr19 | 48729172 | – | GCGGCCGCggccTGCAGgAgCGCGGCG | 6 | Ras interacting protein | 127,710 | 0.26% | 165,265 | 0.17% | N/D | ||||
| L31 | chr5 | 179821117 | + | GCGGCttCCaGGcGCAcTACCGCGGtG | 6 | SQSTM1/NM_001142298.2/NP_001135770.1 | 31,757 | 0.35% | 123,314 | 0.33% | N/D | ||||
Genomic deep-sequencing analysis of PCR amplicons generated for 31 (OT#) predicated off-target sites in the human genome. Nucleotide mismatches are indicated and represented in lowercase. The chromosomal location and gene involved for each predicated off-targeted site are displayed. The (+) and (−) represent whether the off-target site involves the leading or lagging strand, respectively. Depth refers to the total reads obtained from deep sequencing. The genomic DNA inputs were extracted from untargeted and combined pools (n = 3) of RITDM-edited human B lymphocytes. N/D, no data.
Targeted amplification followed by deep sequencing of the 31 potential off-target sites with 5 or 6 mismatches was performed. For each site, a set of primers was designed and amplification conditions were optimized. Two out of the 31 sites did not yield amplification products, neither did the wild-type, untargeted B cells, nor the edited cells. One site did not yield mapped sequence reads from either untargeted or edited B cells. Deep-sequence analysis results for the 28 sets of sequence analysis are shown in Table 1. The sequence depths at all 28 sites were beyond 10,000 reads per site per sample. There were no significant differences in indel frequencies between any of the evaluated 28 potential off-target sites. In addition, no significant signs of DNA conversion were observed that could indicate the modification oligo being involved in any way (e.g., a T-to-C conversion corresponding to codon 112). We did not observe DNA conversion at the potential off-target binding sites. While further detailed analysis may be warranted, this analysis did not reveal signs of off-target effects occurring at significant levels at the sites analyzed.
In addition, a single-stranded “Circular-Seq” method was deployed (Figures S12A–S12E) to test for random integration or recombination involving the oligonucleotide. Out of a total of 22,043 reads, no sequences were obtained that differed from ApoE sequences, which would have been an indication of potentially off-target effects of correction templates. Of 124 reads analyzed containing the T-to-C conversion, 65 were long enough to extend beyond the sequence of the oligonucleotide used. If integration of a correction template had occurred at a site other than an ApoE site, flanking DNA sequences would have been different from ApoE sequences. All sequences obtained from these 65 reads corresponded to expected ApoE sequences, indicating that neither off-target integration had been found nor that any genome rearrangements had happened at a significant frequency (Figures S12 and S13).
DLR fusion molecules directly interact with replication forks
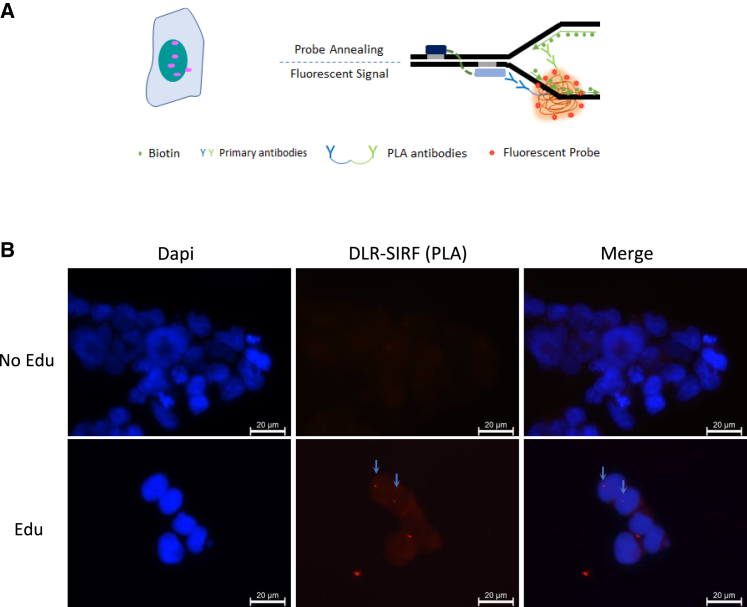
To demonstrate a direct interaction between the DLR fusion protein and the replication fork, analyses were done that made use of an in situ interaction at replication fork (“SIRF”) methodology (Figure 6A).33 SIRF assays confirmed direct association of a Flag-tagged DLR fusion protein with EdU-labeled nascent DNA at replication forks (Figures S14A and S14B), as the red fluorescent puncta could clearly be detected in transfected cells (Figure 6B). Since the mutant EGFP reporter cell line had two copies of the EGFPDP2 gene, one on each chromosome, SIRF would be expected to detect up to two red puncta per cell. We observed multiple cells with one red puncta in their nucleus (Figure 6B), supporting the mechanism that RITDM is rooted in locus-specific replication fork interaction via programmable DLR molecules.
Figure 6.
SIRF analysis showing the interaction between the DLR fusion protein and nascent replication forks
(A) Schematic of detection of interaction of DLR with a replication fork by SIRF assay (in situ analysis of protein interactions at the DNA replication fork). (B). Arrows point to red puncta indicating DLR interactions with the replication fork.
Discussion
The results presented here describe a novel gene editing method. Differing from nuclease-driven gene editing tools, RIDTM is not reliant on either dsDNA cleavage or ssDNA nicking to induce genetic modifications in targeted genes, allowing for gene editing with low indel formation and off-target effects. The editing frequencies achieved in the B cell examples are high enough to be of practical use, with the real benefit coming from (much) lower levels of indel generation and other off-target effects. These features are of great importance, especially when developing therapies. The safety profile is the result of a number of factors. First, the conversion mechanism does not make use of nucleases or nickases and does not trigger NHEJ, thus avoiding a major pathway contributing to indel formation. Second, the technology requires both the DLR molecule and the modification template to operate in concert, thereby minimizing off-target effects of single components.
We have demonstrated that RITDM can be used not only for single nucleotide changes but also for more complex changes, allowing, for example, the introduction of stop codons, the creation of restriction enzyme sites for RFLP analysis, and the introduction of additional nucleotides to restore reading frames. Given the range of possibilities that will each require optimization for its specific purpose, we illustrated the possibilities using model cell lines and B cells, realizing that application in other primary cells and optimization of conditions will be required for each given target.
RITDM uses a single zinc-finger array recognizing one strand of DNA. Compared with zinc-finger nucleases, which require two zinc-finger arrays that recognize sequences on opposite DNA strands and are separated by a constraint spacing of five to six nucleotides, this architecture significantly reduces the complexity of designing and evaluating usable zinc-finger arrays. Given the size of the human genome, zinc-finger arrays should comprise seven fingers or more. The donor template plays an equally important role for a successful genomic editing. We have successfully used ssDNA templates to modify genomes for various SNPs, small deletions, insertions, and combinations thereof. A length of 90–200 nucleotides has proven to be effective. Other sizes and configurations of donor templates should also be usable, and this will be a topic for further research.
Like most other gene editing tools, RITDM depends on cellular DNA repair processes to permanently modify target genes. As observed for other gene editing technologies, editing efficiencies can be locus- and cell-type-dependent and can be influenced by variations in cell status, cell cycle, and the types of desired genome modification. For each specific case, RITDM may require additional optimization and calibration. There could be several approaches to enhancing editing frequencies, also depending on ex vivo or in vivo delivery requirements. Identification and standardization of cell synchronization regimes for ex vivo editing, which is suitable for different cell types, could be useful in increase the numbers of cells at a similar stage in the cell cycle that favors RITDM targeting. We have used thymidine synchronization in cell lines HEK293 and U937. For therapeutic purposes, other ex vivo synchronization protocols may be preferential. In B cells, we achieved editing frequencies in the range of 15%–40% without deploying cell synchronization. For in vivo purposes, cell-cycle-specific promoters could be used. Mobilization and activation of cells may be deployed to make cells more amenable to editing, making the chromosomal DNA more accessible to RITDM components. Therapeutic protocols may also evaluate differences in effectiveness if the DLR construct is added as mRNA or protein. Donor template design could also enhance editing efficiencies.
A limitation of RITDM is that it operates within the context of replicating cells. This constrains its use to gene and cell therapy applications that can make use of a replicating cell type, ex vivo or potentially in vivo. In pediatric and adult settings, there are still a number of genetic diseases in which gene editing of stem cells, progenitor cells, and dividing cells can provide therapeutic relief. In vivo delivery and effectiveness still need to be further explored. In addition, it will be incredibly interesting to explore the feasibilities of using mRNA or protein instead of DNA plasmid for RITDM genomic editing.
The small footprint of the DLR design makes RITDM compact enough to fit into various delivery vectors. Even our largest DLR molecule, containing a 12-zinc-finger array, still requires less than 1.6 kB for its entire coding sequence. RITDM is a simple system of two components, which can also be delivered using liquid nanoparticles and other types of delivery vehicles.
Additional research is needed to elucidate more details of the mechanisms and DNA repair pathways involved. The mechanism(s) of oligonucleotide integration also warrant(s) further investigation. Although RITDM can edit target genes with single-nucleotide substitutions, small insertions or deletions, or combinations thereof, it will be worth further investigation using dsDNA as donor template and to explore the feasibility of knocking in larger DNA constructs at target sites. Further research is also needed on the effectiveness of RITDM in various types of primary cells and its applicability in in vivo applications.
Materials and methods
General methods and agents
All chemicals, reagents, and buffers were purchased from MilliporeSigma (Burlington, MA), Thermo Fisher Scientific (Waltham, MA), and Boston BioProducts (Ashland, MA). DNA amplification was carried out by PCR using Phusion Hi-Fidelity Polymerase or Taq DNA polymerase (New England Biolabs [Ipswich, MA], Thermo Fisher Scientific), unless otherwise noted. DNA oligonucleotides, including single-stranded donor templates, were obtained from Integrated DNA Technologies. GenScript (Piscataway, NJ) made all DNA plasmids. Mammalian expression vector pVAX1 was obtained from Thermo Fisher Scientific. The EGFPDP2 reporter cell line in HEK293 was generated using an Flp-In cell line development kit (Life Technologies, Carlsbad, CA). Puresyn (Malvern, PA) manufactured all plasmids for mammalian cell expression experiments. Genomic DNA extractions were conducted using a Wizard Genomic DNA Extraction Kits from Promega (Madison, WI). The PCR amplicons were purified and recovered using GeneJet DNA purification columns (Thermo Fisher Scientific). Restriction enzymes and DNA modification enzymes were obtained from New England Biolabs. Endonucleases I and III and ssDNA ligase were purchased from Lucigen (Middleton, WI). Sanger sequencing was performed by the Tufts University Genomic Core (Boston, MA) or by GENEWIZ (South Plainfield, NJ). Next-generation sequencing and bioinformatic analysis were provided by GENEWIZ. No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Zinc-finger array designs
Zinc-finger arrays were designed to recognize sequences of genes of interest in the genome using guidance described in the literature. Potential recognition sequences close to the target site were first evaluated by BLAST searches to confirm sufficient sequence uniqueness within the genome. The seven amino acids of α helices in each finger were selected from sequences available in the literature. The final zinc-finger array was then fitted into a ZF-RITDM framework for complete DLR plasmid design. All constructs containing zinc-finger arrays were de novo synthesized.
The 5-zinc-finger array that recognizes the 15-nucleotide sequence of 5′-GGGGAGGACGCGGTG-3′ in EGFPDP2 was designed to contain recognition α helices from finger 1 to finger 5: RSSALTR, RSDTLTR, DRSNLTR, RSDNLTR, and RSDHLTR.
The 9-zinc-finger array that recognizes the 27-nucleotide sequence 5′-GCGGCCGCCTGGTGCAGTACCGCGGCG-3′ from ApoE was designed to contain recognition α helices from finger 1 to finger 9: RSSDLTR, RSDTLTR, QSGDLSE, TSGHLTT, DSSHLTT, RSSHLTT, DRSDLTR, DRSDLTR, and RSDTLTR.
The 7-zinc-finger array that recognizes the 21-nucleotide sequence 5′-GAGGCCAAACCCTTCCTGGAG-3′ from BCL11A was designed to contain recognition α helices from finger 1 to finger 7: RSSNLTR, RSDALSE, DSSALTT, DSSDLSE, QSGNLSQ, DRSDLTR, and RSDNLTR.
The 10-zinc-finger array that recognizes the 30-nucleotide sequence 5′-CTGGTGACACAACCTGTGGTTACTAAGGAA-3′ from the exon51 of Dystrophin gene was designed to contain recognition α helices from finger 1 to finger10: QSGNLTR, RSDNLSQ, TSGDLSQ, TSGSLTR, RSDALTR, TSGDLSE, QSGNLSE, QSGDLSQ, RSSALTR, and RSDALSE.
The 7-zinc-finger array that recognizes the 21-nucleotide sequence 5′-CTGGTGGGGCTGCTCCAGGCA-3′ from PDCD-1 was designed to contain recognition α helices from finger 1 to finger 7: QSGDLTR, RSDNLSE, DRSALSE, RSSALSE, RSSHLTR, RSDALTR, and RSDALSE.
RITDM targeting vector designs
All RITDM targeting vectors were designed using modules with a common D-L-R architecture. For the DLR fusion proteins containing one zinc-finger array, the DLR molecules were engineered, from N-terminal to C-terminal, as D (zinc-finger arrays)-L (shorter linker)-R (non-sequence-specific DNA binding domain). The cDNAs encoding the DLRs were de novo synthesized and cloned into a mammalian expression vector under the control of a CMV promoter. In a number of experiments, a FLAG tag and NLS sequence were inserted at the N-terminal in front of the coding sequence for the DLR constructs. DLR single-amino acid variations in R-elements were engineered by mutagenesis. The DLR containing dual zinc-finger arrays was engineered as D (zinc-finger array)-L (longer linker)-R (zinc-finger array). The cDNA was de novo synthesized and cloned into the mammalian expression. The full amino acid sequences of the DLR molecules are given in the supplemental information.
EGFPDP2 reporter cells and cell cultures
The development of the EGFPDP2 reporter cell line was carried out using a HEK293 FlpIN system (Life Technologies). The HEK293 FlpIN host cell line contained a fusion gene of lacZ-zeocin that was stably inserted into its genome by a transfection of plasmid of pFRT/lacZeo resistant to zeocin-containing medium. Plasmid pcDNA5/FRT/EGFP-DP2 was constructed by cloning a defective EGFP reporter, referred as to EGFPDP2, and coding sequencing into plasmid vector pcDNA5/FRT under the control of a CMV promoter. Plasmids pcDNA5/FRT/EGFP-DP2 were then co-transfected together with plasmid pOG44 into this HEK293 host cell line. Since pOG44 expresses a recombinase, it induced recombination occurring at two FRT sites present in this system: one in the genome and one on plasmid pcDNA5/FRT/EGFPDP2. Successful recombination resulted in lacZ-zeocin moving out of frame and simultaneously enabling functional expression of a hygromycin resistance gene upstream, thus allowing cells become resistant to hygromycin-containing medium. Cells expressing the EGFPDP2 gene could survive in hygromycin. The reporter cell line was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (v/v), GlutaMAX, penicillin-streptomycin, and hygromycin. Wild-type HEK293T (ATCC CRL3216) was maintained in DMEM supplemented with 10% FBS (v/v) at 37°C CO2. U937 (ATCC CRL1593.2) cells were maintained in RPMI 1640 supplemented with 10% FBS. All B lymphocytes were obtained from Coriell Institute (Camden, NJ). B/DMD, GM032929 had a deletion from exon 46 to exon 50 in the DYSTROPHIN gene. Normal B cells used in the study were ND23350, from a healthy male donor. All B lymphocytes were cultured in RPMI supplemented with 15% FBS, 2 mM L-glutamine, and penicillin-streptomycin. All culture medium and supplements were obtained from Gibco Thermo Fisher Scientific.
RITDM gene targeting and editing
The flow of RITDM targeting in mammalian cell lines is schematically as follows: seeding cells > thymidine synchronization > nucleofection/electroporation > recovery > cell growth > analysis, unless otherwise noted. 48h before experiments, approximately two million cells were seeded on 100-mm culture plates and cultured in the maintenance medium supplemented with 10% FBS. 24h before nucleofection, cells were exposed to thymidine at a concentration of 2–5 mM for 6–18 h, then released into normal maintenance medium. Nucleofection was performed by Nucleofector 4D (Lonza, Walkersville, MD) using a nucleofection kit (SF Cell Line 4D-NucleofectorX Kit). Each nucleofection reaction was set up as follows: one million detached cells, a nucleofection agent, 5 μg DLR plasmid, and 0.3 μmol ssDNA donor template. After nucleofection, transfected cells were placed onto a plate pre-coated with 0.1% gelatin (to enhance survival and adherence). Culturing continued at 5% CO2 in a 37°C incubator for at least 5–7 days for downstream genetic analysis. The high-dose editing protocol for cell lines and primary B cells is as follows: five million detached cells were electroporated with a combination of 30 μg DLR plasmid and 1.5 μmol ssDNA donor template.
For B lymphocyte editing, a high-dose editing protocol was applied. No cell-cycle synchronization reagent, such as thymidine, was administrated for B cell editing. The “1.5 kv” electroporation protocol was modified from the manufacturer's instructions for the Neon system (Thermo Fisher Scientific) using settings of 1.5 kv, 30 ms, and 1 pulse.
Enriching GFP-positive cells after RITDM gene editing
Under fluorescent microscopy, the green cellular clusters were circled and marked in the bottom of culture vessels, such as 6-well plates or 100-mm Petri dishes. In the sterile environment, the clone cylinders were placed onto the marked areas. The cells in the cylinders were then trypsinized, combined, and transferred to a fresh culture well filled with complete growth medium and continued culturing to semi-confluency. Such procedures were repeated three to four times to obtain highly enriched green cellular populations.
Generation of single-cellular clones
For the generation of cellular clones from single cells, nucleofected cells were grown for 5 days in a complete growth medium supplemented with 15% FBS. Cells were dissociated with 0.25% trypsin/EDTA solution and plated in 96-well plates at a density of 0.5–1.0 cells per well. Cells were allowed to grow into clones, which usually took about 3–4 weeks. Wells containing clones were marked, and individual clones were then dissociated and transferred into 24-well plates for expansion. Prior to genomic analysis of individual clones, the cells were split into two parts: one for continuation of culture and the other for genomic DNA extraction.
Detection of genomic modifications by ddPCR
Allele-specific TaqMan probes and end primers were synthesized by Integrated DNA Technologies (Coralville, IA). The probes were conjugated with fluorophores FAM or HEX to distinguish between wild-type or edited loci. Various end primer sets were designed to amplify regions of genomic area containing the genetic modifications, yielding a length of around 100–200 bp. For each genomic detection, the probes and end primers were mixed at a ratio of 3.6:1, along with genomic DNA and 2×ddPCR Supermix for Probes (Bio-Rad, Hercules, CA). Droplet generation with a QX200DG ddPCR system was performed according to the manufacturer's instructions (Bio-Rad), and the reaction was then transferred into a 96-well PCR plate for standard PCR protocol on a T100 Thermal Cycler (Bio-Rad). The thermal cycling program for optimization was: 95°C for 10 min; followed by 39 cycles of 94°C for 30 s and 50°C–60°C for 1 min; and a final step of 98°C for 10 min. After the PCR reaction was completed, the samples were cooled to 4°C overnight to stabilize the droplets. Droplets were analyzed using a QX200 Droplet Reader (Bio-Rad) using the “Rare Mutation Detection” setup, and data were analyzed using QuantaSoft (Bio-Rad). Genomic DNA from human fibroblasts (ATCC, Manassas, VA) with appropriate genotypes were used as a positive control. The optimal annealing temperatures for droplet PCR amplification for specific applications were selected based on the best separation of the positive and negative droplets.
Flow cytometry
HEK293 reporter cells were harvested 7 days after RITDM genome editing by trypsinization and resuspended in PBS without fixation. GFP-positive cells were analyzed using a 488-nm laser for excitation and a 530/30 filter. Data were analyzed using FloJo software. For all flow cytometry analysis, experiments were performed on BD FACS ARIA at Tufts University Flow cytometry Core facility.
Library preparation for single-stranded circular-seq
Genomic DNA was extracted from a specific single clone, sheared into fragments approximately 500 bp in length, as verified by gel electrophoresis, and purified. DNA fragments were phosphorylated by T4 PNK (New England Biolabs), denatured into ssDNA fragments, and purified again. Three hundred nanograms of DNA were circularized in a 20-μL reaction volume using CircLigase II ssDNA ligase (Lucigen) twice, and uncircularized DNA was removed with exonuclease I and III (Lucigen). This was followed by DNA purification using a GeneJet spin column (Thermo Fisher Scientific). The circular ssDNAs were then used as PCR templates, using various sets of PCR primers. Amplification primers containing Illumina forward and reverse adapters were used for the first round of PCR to amplify the genomic regions that contained sequences corresponding to the donor template. The amplification primers were designed to orient facing away from each other. As a result, the amplicons covered the donor template region and continued into the flanking regions, potentially stopping only at the annealing site of the other primer binding site. PCR reactions were performed with 0.5 μM of both forward and reverse primer, 2 μL circular ssDNA template, and Phusion PCR components. PCR reactions were carried out as follows: 98°C for 5 min, then 45 cycles of 98°C for 20 s and 72°C for 45 s, followed by a 72°C final extension for 2 min. The PCR products were purified using a GeneJet PCR purification kit (Thermo Fisher Scientific). The DNA concentration was determined by NanoDrop (Thermo Fisher Scientific) and samples were sequenced using an Amplicon-EZ platform by GENEWIZ. Unique sequence reads were provided by GENEWIZ. After the removal of reads shorter than the length of the donor template, sequences that contained a signature of the donor template modification were selected. From this pool we analyzed each read using the Blastn program to enable the alignment of each segment from individual reads. If the flanking sequences were ApoE sequences precisely aligned with the target site, they were scored as precision editing on the target. If a flanking sequence was different from the expected, aligned ApoE sequence, then it was scored as an off-target effect.
High-throughput DNA sequencing of genomic DNA samples
Genomic sites of interest were amplified from genomic DNA samples and sequenced on an Illumina platform. In brief, amplification primers were used for the first round of PCR to amplify the genomic region of interest. The PCR reactions were performed using a Phusion High Fidelity amplification kit according to the manufacturer's instructions. The PCR reactions were carried out as follows: 98°C for 2 min, then 30–35 cycles of 98°C for 10 s and 72°C for 20 s, followed by a final 72°C extension for 2 min. The PCR samples were then purified using a GeneJet DNA purification kit. The SNPs and indels were analyzed using an Amplicon-EZ NGS from GENEWIZ. Indel frequencies were calculated by the percentage derived from the (number of reads with indels)/(number of total reads).
SIRF
To detect a direct interaction between DLR fusion proteins and the replication fork, in situ analysis of protein interactions at DNA replication forks (SIRF) was used. In short, HEK293 cells were transfected with Flag-tagged DLRs, grown in microchamber slides, and pulsed with 100 μM EdU for 8 min, followed by a step in which EdU is biotinylated using click chemistry. Cells were incubated with primary antibodies overnight at 4°C with 1:250 rabbit anti-biotin antibody with 1:1,000 mouse anti-Flag antibody (M2, Sigma). Cells were washed twice with PBS and incubated with pre-mixed Duolink PLA plus and minus probes for 1 h at 37°C. The subsequent steps in proximal ligation assay were carried out using the Duolink PLA Fluorescence Kit (MilliporeSigma) according to the manufacturer's instructions. Slides were stained with 4′,6-diamidino-2-phenylindole and imaged using an upright fluorescent microscope.
Acknowledgments
We thank Stuart Orkin and Farren Isaacs for their insightful comments.
Author contributions
C.K. and Y.X. designed the research, performed experiments, and analyzed the data. C.K. and D.H. designed the research, analyzed the data, wrote the manuscript, and supervised the research. D.H. oversaw and managed the research program. All of the authors contributed to editing the manuscript.
Declaration of interests
The authors have declared conflicting financial interests. A patent application has been filed relating to this work through Peter Biotherapeutics, Inc. The authors are employees and equity holders of Peter Biotherapeutics, Inc.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.12.001.
Supplemental information
and Tables S1–S3
References
- 1.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pannunzio N.R., Watanabe G., Lieber M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018;293:10512–10523. doi: 10.1074/jbc.TM117.000374TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickman K., Smogorzewska A. Advances in understanding DNA processing and protection at stalled replication forks. J. Cell Biol. 2019;218:1096–1107. doi: 10.1083/jcb.201809012jcb.201809012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri E.M., Muir P., Akhuetie-Oni B.O., Yellman C.M., Isaacs F.J. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell. 2017;171:1453–1467.e13. doi: 10.1016/j.cell.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 7.Sera T., Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41:7074–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 8.Choo Y., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl. Acad. Sci. U S A. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu C., Gupta A., Hall V.L., Rayla A.L., Christensen R.G., Dake B., Lakshmanan A., Kuperwasser C., Stormo G.D., Wolfe S.A. Using defined finger-finger interfaces as units of assembly for constructing zinc-finger nucleases. Nucleic Acids Res. 2013;41:2455–2465. doi: 10.1093/nar/gks1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo Y., Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc. Natl. Acad. Sci. U S A. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wah D.A., Bitinaite J., Schildkraut I., Aggarwal A.K. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. U S A. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C.A., Berg J.M. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 14.Xu S.Y., Zhu Z., Zhang P., Chan S.H., Samuelson J.C., Xiao J., Ingalls D., Wilson G.G. Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res. 2007;35:4608–4618. doi: 10.1093/nar/gkm481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright D.A., Thibodeau-Beganny S., Sander J.D., Winfrey R.J., Hirsh A.S., Eichtinger M., Fu F., Porteus M.H., Dobbs D., Voytas D.F., Joung J.K. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 16.Igoucheva O., Alexeev V., Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 17.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 18.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 19.Chartier-Harlin M.C., Parfitt M., Legrain S., Pérez-Tur J., Brousseau T., Evans A., Berr C., Vidal O., Roques P., Gourlet V., et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum. Mol. Genet. 1994;3:569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- 20.Schaffer S., Lam V.Y., Ernst I.M., Huebbe P., Rimbach G., Halliwell B. Variability in APOE genotype status in human-derived cell lines: a cause for concern in cell culture studies? Genes Nutr. 2014;9:364. doi: 10.1007/s12263-013-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whinn K.S., Kaur G., Lewis J.S., Schauer G.D., Mueller S.H., Jergic S., Maynard H., Gan Z.Y., Naganbabu M., Bruchez M.P., et al. Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci. Rep. 2019;9:13292. doi: 10.1038/s41598-019-49837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi G., Okada S., Yasukawa T., Sugiyama Y., Bala S., Miyazaki S., Kang D., Ito T. Catalytically inactive Cas9 impairs DNA replication fork progression to induce focal genomic instability. Nucleic Acids Res. 2021;49:954–968. doi: 10.1093/nar/gkaa12416062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y.C., Boone M., Meuris L., Lemmens I., Van Roy N., Soete A., Reumers J., Moisse M., Plaisance S., Drmanac R., et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014;5:4767. doi: 10.1038/ncomms5767ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P., et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amoasii L., Long C., Li H., Mireault A.A., Shelton J.M., Sanchez-Ortiz E., McAnally J.R., Bhattacharyya S., Schmidt F., Grimm D., et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl Med. 2017;9:eaan8081. doi: 10.1126/scitranslmed.aan80819/418/eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beane J.D., Lee G., Zheng Z., Mendel M., Abate-Daga D., Bharathan M., Black M., Gandhi N., Yu Z., Chandran S., et al. Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor Infiltrating lymphocytes for the treatment of metastatic melanoma. Mol. Ther. 2015;23:1380–1390. doi: 10.1038/mt.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anthony I.C., Crawford D.H., Bell J.E. B lymphocytes in the normal brain: contrasts with HIV-associated lymphoid infiltrates and lymphomas. Brain. 2003;126:1058–1067. doi: 10.1093/brain/awg118. [DOI] [PubMed] [Google Scholar]
- 30.Ortega S.B., Torres V.O., Latchney S.E., Whoolery C.W., Noorbhai I.Z., Poinsatte K., Selvaraj U.M., Benson M.A., Meeuwissen A.J.M., Plautz E.J., et al. B cells migrate into remote brain areas and support neurogenesis and functional recovery after focal stroke in mice. Proc. Natl. Acad. Sci. U S A. 2020;117:4983–4993. doi: 10.1073/pnas.19132921171913292117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blauth K., Owens G.P., Bennett J.L. The ins and outs of B cells in multiple sclerosis. Front Immunol. 2015;6:565. doi: 10.3389/fimmu.2015.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancellieri S., Canver M.C., Bombieri N., Giugno R., Pinello L. CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing. Bioinformatics. 2020;36:2001–2008. doi: 10.1093/bioinformatics/btz8675640496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy S., Luzwick J.W., Schlacher K. SIRF: quantitative in situ analysis of protein interactions at DNA replication forks. J. Cell Biol. 2018;217:1521–1536. doi: 10.1083/jcb.201709121jcb.201709121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and Tables S1–S3