Abstract
Background
Circulating RNAs (Circ-RNAs) are tightly related to the processes of neuroblastoma. The circ-ACAP2 has been reported as dysregulated in various cancers; however, its biological roles and mechanisms in neuroblastoma remain largely unclear.
Methods
We collected 40 neuroblastoma tissues and adjacent noncancerous tissues. Quantitative reverse transcription polymerase chain reaction (qRT-RCR) or western blot were used to examine ACAP2, miR-143-3p, and HK2 abundances. Cell migration, invasion, glycolysis, and apoptosis were assessed via wound healing, transwell, glucose uptake and lactate, 3-(4,5-diamethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay, and flow cytometry. The association between circRNA, microRNA (miRNA), and messenger RNA (mRNA) was examined by dual-luciferase reporter analysis and RNA immunoprecipitation.
Results
The abundances of ACAP2 and HK2 were remarkedly increased in neuroblastoma tissues and cell lines. Silencing ACAP2 significantly constrained neuroblastoma cell migration, invasion, and glycolysis, and promoted apoptosis. Bioinformatics prediction, luciferase assay, and RNA pull-down assay consistently demonstrated that ACAP2 sponged miR-143-3p to downregulate its expression in neuroblastoma cells. Furthermore, we identified that hexokinase 2, a glycolysis key enzyme, was a direct target of miR-143-3p in neuroblastoma cells. Rescue of miR-143-3p in ACAP2-overexpressing cells effectively mitigated the influence of ACAP2 on neuroblastoma cell processes.
Conclusions
Our study revealed biological roles and molecular mechanisms for circ-ACAP2 in the oncogenic characteristics of neuroblastoma, facilitating the development of circRNA-based treatment approaches for anti-brain tumor therapy.
Keywords: Neuroblastoma, circulating RNA (Circ-RNAs), ACAP2, miR-143-3p, glucose metabolism
Introduction
Neuroblastoma is an embryonal malignancy, which leads to cancer-related death in early childhood (1). Moreover, the clinical symptoms of neuroblastoma are highly variable and dependent on diverse factors (2). Currently, the genetic basis of neuroblastoma is still under investigation. Surgical resection, chemo- or/and radiotherapy are frequently applied to improve the survival rate of neuroblastoma patients (3); however, the development of recurrence and metastasis has impaired treatment efficiency (4). Thus, the identification of new molecular targets and therapeutic strategies for the treatment of neuroblastoma is of pressing importance.
Circular RNAs (circRNAs), which are abundantly expressed in the cytoplasm of eukaryotic cells, are created by back-splicing of pre-messenger RNA (mRNA) transcripts, resulting in formation of a covalently closed-loop (5). We know that circRNAs play essential roles in regulating the oncogenesis and progressions of various cancers (6), suggesting that targeting circRNA-mediated signaling pathways is a potential therapeutic strategy against cancer. Accumulating studies have demonstrated that circRNAs function as post-transcriptional regulators through sponging target microRNAs (miRNAs) to form competing endogenous RNAs (ceRNAs) complexes (7). Moreover, roles of the ceRNA network have been highlighted in a range of cancers, including brain tumors (8-10). For instance, Chen et al. established that circPTN worked as a ceRNA for miR-145-5p/miR-330-5p to contribute to the proliferation and stemness of glioma (11). Specifically, Circ-ACAP2 has been reported to promote the development and radioresistance of colorectal cancer cells partly through targeting miR-143-3p/FZD4 axis (12), suggesting an oncogenic role of ACAP2 in colon cancers. However, the precise biological roles and molecular mechanisms of circRNA-APAC2 in neuroblastoma have not been fully elucidated.
Cancer cells display significantly elevated glucose metabolism, characterized by increased glucose uptake and lactic acid production, but diminished oxidative phosphorylation (13). This phenomenon is called the “Warburg effect”. Furthermore, inhibiting glucose metabolism has been shown to effectively suppress the progression of cancer cells (14). MicroRNAs were known to associate with 3'-UTR of their target mRNAs to block gene expressions (15). Multiple studies unveiled that miR-143-3p specifically targets 3'-UTR of HK2 in cancers (16,17), leading to inhibition of cellular glucose metabolism. Yet, the upstream regulator of the miR-143-3p-HK2 in neuroblastoma has not been elucidated. In the present study, we investigated the roles and mechanisms of the circRNA-ACAP2 in neuroblastoma. We aimed to identify the molecular targets of ACAP2, contribute to understanding the critical roles of circRNA-ACAP2 in metastasis and apoptosis of neuroblastoma cells, and provide new therapeutic strategies against neuroblastoma. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-527).
Methods
Clinical tissues collection
The study was approved by the Ethics Committee of Children’s Hospital of Soochow University (No. 2020CS053). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Neuroblastoma tissues and their matched non-tumorous neuronal tissues were obtained from 40 patients during surgical treatment. Tissue samples were immediately placed in liquid nitrogen then stored at −80 °C after resection. Written informed consent was taken from parents/guardians of the patients.
Cell culture and transfection
Human astrocytes, NHA, which is a normal human cell line derived from human brain tissue, and two neuroblast cell lines, IMR-32 and SK-N-SH were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37 °C under 5% CO2 supply. The short hairpin RNA (shRNA) against ACAP2 (sh ACAP2), miR-143-3p mimics, and overexpression plasmids of ACAP2 (pcDNA3,1) and HK2 (pcDNA3.1) as well as their negative controls were obtained from GenePharma (Shanghai, China). The shRNA sequences were: shcirc_ACAP2: Forward: 5'-GGCAGCATACAGGAAGATGAA-3', Reverse: 5'-UUCAUCUUCCUGUAUGCUGCC-3'; Negative control: Forward: 5'-UUCUCCGAACGUGUCACGUUUC-3', Reverse: 5'-GAAACGUGACACGUUCGGAGAA-3'. Cells were seeded onto 6-well plates and cultured to 60–70% confluence, then transfected with shRNA, overexpression plasmid, or miRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfections were conducted for 48 h, followed by cell collections for downstream experiments. Rabbit anti-β-actin (#4970) and anti-HK2 (#2867S) were purchased from Cell Signaling Technology (CST; Danvers, MA, USA).
Wound healing assay
The migration capacities of IMR-32 and SK-N-SH cells were evaluated by wound healing assay. Cells (1×105/well) were seeded into 12-well plates for 24 h to reach >95% confluence. Then, a straight scratch was conducted using a 200 µL pipette tip. After 16 h, migration of cells was monitored under bright field microscope (magnification ×100). Experiments were repeated 3 times.
Transwell analysis
For detection of invasion capacity, the transwell chamber (Becton, Dickinson, and Co., Franklin Lakes, NJ, USA) was pre-coated with Matrigel (Invitrogen). Lower chambers were filled with DMEM with 10% serum. We added IMR-32 and SK-N-SH cells (8×104) in DMEM medium (200 µL) without serum into the upper chambers. After 16 h culture at 37 °C in a humidified atmosphere with 5% CO2, cells were fixed with methanol and stained with 0.2% crystal violet. Non-migrated cells from the upper surfaces of the filters were removed with cotton swabs. The invasive cells were stained with 0.25% crystal violet in 2% ethanol for 5 min and monitored under a microscope (magnification ×100, Olympus IX71). The relative invasive capacity was normalized to the control group. Experiments were conducted 3 times.
RNA isolation and quantitative reverse transcription polymerase chain reaction
Total RNAs from neuroblastoma cells were isolated using the TRIzol™ Plus RNA Purification Kit (Invitrogen, USA) according to the manufacturer’s protocols. Concentrations and qualities of RNA samples were examined using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Circular DNA (cDNA) was synthesized using the SuperScript™ IV First-Strand Synthesis System (Invitrogen). SuperScript™ III Platinum™ One-Step quantitative reverse transcription polymerase chain reaction (qRT-PCR) kit (Invitrogen) was applied to detect the miRNA and circRNA expressions. Real-time PCR reactions were performed using the SYBR Green Real-Time SYBR Green master mix (TaKaRa, Kusatsu, Shiga, Japan). The primers used in this study were as follows: miR-143-3p: forward: 5'-GTGAGATGAAGCACTGTAGC-3' and reverse: 5'-GTGCAGGGTCCGAGGT-3'; ACAP2: forward: 5'-GAATGGGATTCGAGACCTG-3' and reverse: 5'-TTCTTCCAAAGCTGCCTGT-3'; HK2: forward: 5'-ATTGTCCAGTGCATCGCGGA-3' and reverse: 5'-AGGTCAAACTCCTCTCGCCG-3'; U6: forward: 5'-GGCAACATTCAACGCTGTC-3' and reverse: 5'-GTGCAGGGTCCGAGGT-3'; β-actin: forward: 5'-CTGTCTGGCGGCACCACCAT-3' and reverse: 5'-GCAACTAAGTCATAGTCCGC-3'. We used U6 small nuclear RNA and β-actin as internal controls. The cycling conditions were set as follows: 1 cycle at 95 °C for 15 min, then 40 cycles at 95 °C for 10 s and at 60 °C for 60 s. The relative expressions were calculated using the 2−ΔΔct method. Experiments were performed in triplicate.
Luciferase assay
The binding sites on ACAP2 or HK2 3'-UTR by miR-143-3p were predicted by starBase (https://bio.tools/starbase). Wild-type (WT) or binding sites mutant (Mut) DANCR or 3'-UTR sequences of lactate dehydrogenase A (LDHA) were cloned into the pmirGLO vectors (Promega, Madison, WI, USA). Neuroblastoma cells were co-transfected with control miRNA or miR-143-3p with WT- or Mut- 3'-UTR of HK2 or ACAP2 using Lipofectamine 2000 for 48 h. Luciferase activities were then determined by a luciferase assay kit (Promega, USA) according to the manufacturer’s protocols. Experiments were performed in triplicate.
RNA pull-down assay
The circRNA-miRNA association was detected by RNA pull-down assay. Control, antisense- and sense-APAC2 probes were biotin-labeled by RiboBio Co. Ltd (Guangzhou, China). Cells were collected and whole cell lysates were extracted, followed by incubation with each probe at 4 °C for 8 h. Streptavidin-coupled agarose beads were added into cell lysates for another 2 h. After washing, the abundance of miR-143-3p in the RNA-RNA complex was detected by qRT-PCR.
Evaluation of glucose metabolism
The glucose metabolism rate was assessed by glucose uptake (#ab136955) and lactate products (#ab65331) from Abcam (Cambridge, MA, USA) according to the manufacturer’s instructions. Relative glucose metabolism rate was normalized to protein amount. This process was replicated 3 times.
Cell viability assay
Cells were collected after transfection and seeded onto 96-well culture plates. After culture for 0, 24, 48, and 72 h, Cell viability was evaluated by 3-(4,5-diamethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Sigma, Shanghai, China) according to the manufacturer’s instructions. The optical density (OD) was measured at 450 nm of each group. Relative cell proliferation rate was calculated by the following formula: cell proliferation rate = (OD450 nm value of 24, 48, or 72 h days/OD450 nm value of 0) × 100%.
Flow cytometry
Cell apoptosis was determined using the Annexin V-FITC apoptosis kit (Sigma, China) following the manufacturer’s instructions. In brief, 2×105 cells (IMR-32 and SK-N-SH) were seeded in 6-well plates and then incubated for 24 h. Subsequently, cells were harvested at 0, 24, 48, and 72 h and incubated with Annexin V binding buffer from the kit. Cells were incubated with Annexin V FITC (3 mL) and 5 mL of propidium iodide (PI) staining solution for 30 min. The apoptosis rate was detected by a flow cytometer (Agilent, Hangzhou, China). Experiments were repeated 3 times.
Western blot
Total protein from neuroblastoma cells were isolated by radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma, China). Protein concentrations were determined by Bradford assay. Equal amounts of protein of each group was loaded into 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) then transferred onto polyvinylidene fluoride (PVDF) membranes followed by blocking with 5% bovine serum albumin (BSA) for 1 h at room temperature. Membranes were incubated with primary antibodies (1:1,000) at 4 °C overnight. After complete washing by phosphate-buffered saline with Tween 20 (PBST), membranes were incubated with secondary antibody (horseradish peroxidase conjugated) for 1 h at room temperature. Protein bands were detected using the enhanced chemiluminescence chromogenic substrate kit (Pierce, Waltham, MA, USA). Experiments were repeated 3 times.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA). Data of were expressed as the mean ± standard deviation (SD) Statistical analysis between 2 groups was conducted by Student’s t-test and the comparison among multiple groups were analyzed using one-way analysis of variance (ANOVA). A P value <0.05 was considered statistically significant.
Results
CircRNA-ACAP2 was upregulated in neuroblastoma patients and cells
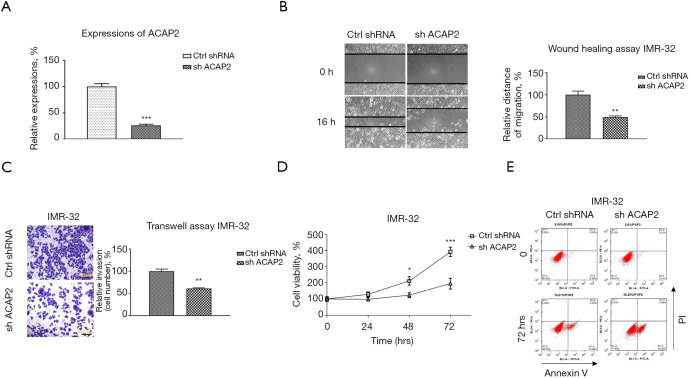
To explore the clinical roles of circRNA-ACAP2 in neuroblastoma development, expressions of ACAP2 were examined in neuroblastoma tissues. Compared with normal neuron tissues, abundance of ACAP2 was evidently upregulated in tumor tissues (n=40) (Figure 1A). Moreover, higher level of ACAP2 was detected in neuroblastoma cell lines, IMR-32 and SK-N-SH cells which showed markedly increased ACAP2 expressions compared to that from primary neuronal cells (Figure 1B). These results suggested that ACAP2 might play oncogenic roles in neuroblastoma.
Figure 1.
Circ-ACAP2 is upregulated in neuroblastoma tissues and cells. (A) Abundance of ACAP2 in neuroblastoma and adjacent noncancerous tissues (n=40) was examined by qRT-PCR; (B) expressions of ACAP2 in human primary neuron cell and neuroblastoma cells detected by qRT-PCR. ***, P<0.001. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
ACAP2 promoted the invasion, migration, and anti-apoptosis of neuroblastoma cells
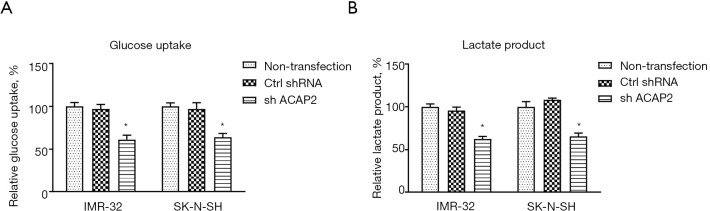
To analyze the biological functions of circRNA-ACAP2 on neuroblastoma development in vitro, IMR-32 cells were transfected with control (shNC) or sh-ACAP2. Results from wound healing assay and transwell assay showed silencing of ACAP2 (Figure 2A) effectively reduced the in vitro migration and invasion capacities of neuroblastoma cells (Figure 2B,2C). Besides, cell viability assay and flow cytometry analysis displayed that silence of ACAP2 effectively promoted apoptosis of IMR-32 cells (Figure 2D,2E). Consistently, the oncogenic roles of ACAP2 were validated in another neuroblastoma cell line, SK-N-SH (data not shown). Studies have shown that tumor cells display dysregulated cellular metabolism (13). Furthermore, the influence of ACAP2 on glycolysis in neuroblastoma cells was evaluated. As shown in Figure 3A,3B, blocking ACAP2 effectively lowered the glucose uptake and lactate product, which are 2 glycolysis speed-limiting reactions in glucose metabolism (13). Taken together, the above results demonstrated that ACAP2 promotes various neuroblastoma processes in vitro, suggesting targeting ACAP2 might be a potentially therapeutic approach against neuroblastoma.
Figure 2.
Oncogenic functions of ACAP2 in neuroblastoma. (A) IMR-32 cells were transfected with control (shNC) or sh-ACAP2, expressions of ACAP2 were examined by qRT-PCR. (B) IMR-32 cells without or with ACAP2 knockdown were subjected to wound healing assay which was monitored under microscope (C) transwell assay (cells were stained with 0.25% crystal violent), (D) cell viability assay and (E) apoptosis assay. *, P<0.05; **, P<0.01; ***, P<0.001. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Figure 3.
ACAP2 promotes glucose metabolism of neuroblastoma cells. (A) SK-N-SH and IMR-32 cells were transfected with control (shNC) or sh-ACAP2, the glucose uptake and (B) lactate product were measured. *, P<0.05.
ACAP2 associated with miR-143-3p to downregulate its expression
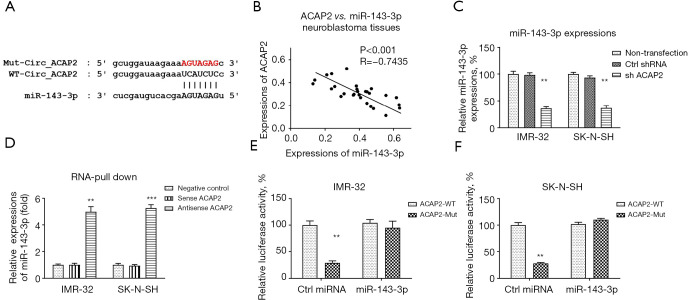
To investigate the underlying mechanisms of the ACAP2-promoted oncogenic phenotypes in neuroblastoma cells, we searched the potential target of ACAP2 since accumulating studies have described that circRNAs sponge target miRNAs to form a ceRNA network, leading to downregulation of target miRNAs (7). Bioinformatics analysis showed that miRNA-143-3p, which has been reported to act as a tumor suppressor in cancers (18), contains putative ACAP2 binding sites (Figure 4A). As expected, a markedly negative correlation between ACAP2 and miR-143-3p was detected in neuroblastoma tissues via Pearson’s correlation coefficient analysis (Figure 4B). To evaluate whether ACAP2 could inhibit miR-143-3p expression, ACAP2 was blocked in IMR-32 and SK-N-SH cells. Predictably, the results showed that neuroblastoma cells with lower ACAP2 expressions were accompanied by higher miR-143-3p expressions (Figure 4C). To test whether ACAP2 binds to the predicted regions of miR-143-3p, we performed RNA-pull down assay which showed that miR-143-3p bond with ACAP2 antisense probe but not control or ACAP2 sense probe (Figure 4D). Furthermore, luciferase assay results demonstrated that IMR-32 and SK-N-SH cells with co-transfection of miR-143-3p plus luciferase vector containing WT-ACAP2 had significantly suppressed luciferase activities (Figure 4E,4F). The luciferase activity of cells transfected with miR-143-5p plus mutant ACAP2 was not affected (Figure 4E,4F). In summary, these results validated that ACAP2 inhibits miR-143-3p expression through sponging it in neuroblastoma cells.
Figure 4.
ACAP2 associates with miR-143-3p to downregulate it expression. (A) Prediction of the ACAP2-miR-143-3p interaction from starBase 2.0. (B) The correlation between ACAP2 and miR-143-3p from neuroblastoma tissues was analyzed by Pearson’s correlation test. (C) IMR-32 and ACAP2 cells were transfected with control shRNA or sh-ACAP2 for 48 h, expressions of miR-143-3p were examined by qRT-PCR. (D) RNA pull-down assay was performed in IMR-32 and SK-N-SH cells by incubation with control, sense ACAP2, or antisense ACAP2 probes. The abundance of miR-143-3p in the RNA-RNA complex was determined by qRT-PCR. (E) Luciferase assay was performed in IMR-32 and SK-N-SH cells which were co-transfected with luciferase vector containing WT- or Mut- ACAP2 plus control miRNA or miR-143-3p. *, P<0.05; **, P<0.01; ***, P<0.001. qRT-PCR, quantitative reverse transcription polymerase chain reaction; WT, wild type; miRNA, microRNA.
MiR-143-3p targeted HK2 to inhibit the oncogenic characteristics of neuroblastoma cells
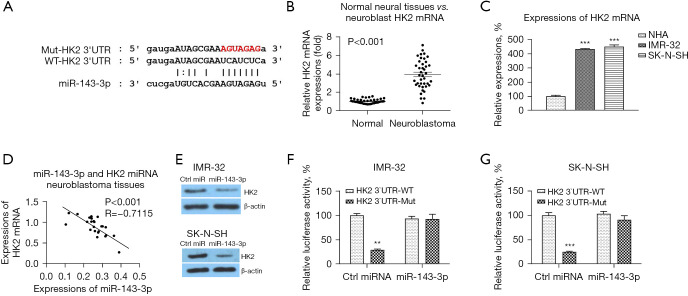
We then assessed the mRNA targets of miR-143-3p. Interestingly, the 3’UTR of hexokinase 2 (HK2), which catalyzes the convert of glucose to glucose-6-phosphate in glycolysis pathway (13), was predicted to contain miR-143-3p binding sites (Figure 5A). Expectedly, HK2 was significantly upregulated in neuroblastoma tissues compared with normal neuron tissues (Figure 5B). In addition, HK2 was apparently upregulated in neuroblastoma cell lines compared with normal neuron cells (Figure 5C). To evaluate the clinical relevance of the miR-143-3p-HK2 interaction, the correlation between miR-143-3p and HK2 expressions was analyzed by Pearson’s correlation coefficient analysis. The results displayed in Figure 5D illustrated that higher miR-143-3p expression was associated with lower HK2 expression in neuroblastoma tissues. We then examined whether miR-143-3p could target HK2 in neuroblastoma cells. Overexpression of miR-143-3p effectively suppressed HK2 protein expressions (Figure 5E). Subsequently, to validate whether miR-143-5p could directly target the 3'-UTR of HK2, we performed luciferase reporter assay. The results demonstrated that co-transfection of miR-143-3p with luciferase vector containing WT-HK2 3'-UTR significantly inhibited the luciferase activity (Figure 5F,5G), while co-transfection of miR-143-3p with Mut-HK2 3'-UTR did not change the luciferase activity (Figure 5F,5G). Taken together, these results validated that miR-143-3p direct targeted 3’UTR of HK2 mRNA to block its expression in neuroblastoma cells.
Figure 5.
miR-143-3p targets 3’UTR of HK2 mRNA in neuroblastoma cells. (A) Prediction of the miR-143-3p-HK2 3'-UTR interaction from starBase 2.0. (B) Expressions of miR-143-3p in neuroblastoma and adjacent noncancerous tissues (n=40) was examined by qRT-PCR. (C) Expressions of miR-143-3p in human primary neuron cell and neuroblastoma cells detected by qRT-PCR. (D) The correlation between miR-143-3p and HK2 mRNAs from neuroblastoma tissues was analyzed by Pearson correlation test. (E) IMR-32 and SK-N-SH cells were transfected with control miRNA or miR-143-3p for 48 h, protein expressions of HK2 were examined by western blot. (F) Luciferase assay was performed in IMR-32 and SK-N-SH cells which were co-transfected with luciferase vector containing WT- or Mut- HK2 3'-UTR plus control miRNA or miR-143-3p. **, P<0.01; ***, P<0.001. qRT-PCR, quantitative reverse transcription polymerase chain reaction; mRNA, messenger RNA; miRNA, microRNA; WT, wild type.
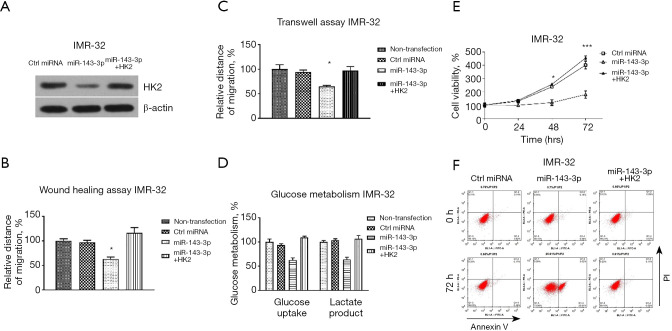
To test whether the miR-143-3p-supressed migration, invasion, glucose metabolism, and anti-apoptosis were through targeting HK2, rescue experiments were subsequently performed. We transfected IMR-32 cells with control miRNAs, miR-143-3p alone or with an HK2 overexpression vector. Western blot results showed that co-transfection of miR-143-3p with HK2 successfully recovered HK2 protein expression (Figure 6A). Overexpression of miR-143-3p alone significantly inhibited cell migration (Figure 6B), invasion (Figure 6C), and glucose metabolism (Figure 6D) and promoted cell apoptosis (Figure 6E,6F). As predicted, the above phenotypes in HK2-rescued IMR-32 cells were markedly recovered to the normal levels (Figure 6B-6F). In summary, these rescue experiments successfully verified that miR-143-3p suppresses oncogenic characteristics of neuroblastoma cells through targeting HK2.
Figure 6.
Restoration of HK2 rescues the miR-143-3p-suppressed processes in neuroblastoma cells. (A) IMR-32 cells were transfected with control miRNA, miR-143-3p or miR-143-3p plus HK2 overexpression vector. Protein expressions of HK2 were examined by western blot. (B) The above transfected cells were subjected to wound healing assay, (C) transwell assay, (D) glycolysis metabolism, (E) cell viability assay and (F) apoptosis assay. *, P<0.05; ***, P<0.001.
The ACAP2-miR-143-3p-HK2 axis is responsible for the migration, invasion glucose, metabolism and anti-apoptosis of neuroblastoma cells
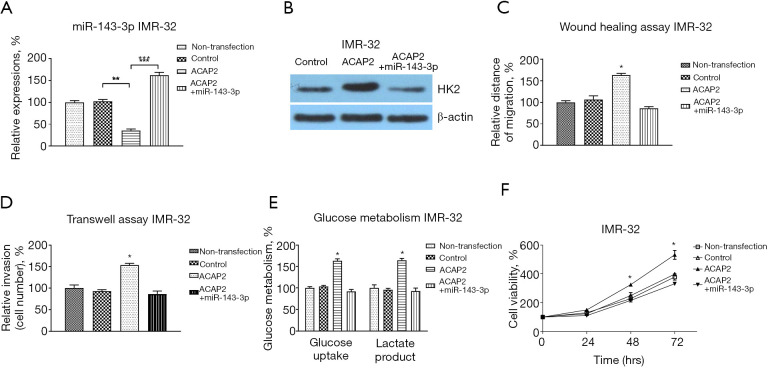
We then evaluated whether the ACAP2-promoted migration, invasion glucose metabolism, and anti-apoptosis was through the miR-143-3p-HK2 axis. The IMR-32 cells were co-transfected with control vector, ACAP2 overexpression vector alone or with miR-143-3p. Western blot and qRT-PCR results consistently showed that overexpression of ACAP2 effectively blocked the miR-143-3p expression and upregulated HK2 expression in IMR-32 cells (Figure 7A,7B). These regulations were further reversed by restoration of miR-143-3p (Figure 7A,7B). As expected, the ACAP2-promoted oncogenic phenotypes were rescued by miR-143-3p restoration (Figure 7C-7F). In summary, the above results unveiled an ACAP2-miR-143-3p-HK2 axis in regulating neuroblastoma processes.
Figure 7.
The influence of miR-143-3p-HK2 axis on ACAP2-mediated processes in neuroblastoma cells. (A) IMR-32 cells were transfected with control vector, ACAP2 (alone or plus miR-143-3p) for 48 h. miR-143-3p and (B) protein expressions of HK2 were examined by qRT-PCR and western blot, respectively. (C) The above transfected cells were subjected to wound healing assay, (D) transwell assay, (E) glycolysis metabolism and (F) cell viability assay. *, P<0.05; **, P<0.01; ***, P<0.001. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Discussion
Neuroblastoma, which is associated with poor prognosis in pediatrics, currently remains a challenge for clinical therapy due to recurrence of tumor and metastasis (1-3). Our study aimed to investigate the non-coding RNA-mediated molecular mechanisms for the migration, invasion, glycolysis, and anti-apoptosis processes of neuroblastoma cells. The results showed that the circulating RNA, ACAP2, was significantly upregulated in neuroblastoma tumors as well as cells compared with normal neuronal cells. Silencing ACAP2 effectively suppressed neuroblastoma cell migration, invasion, and glycolysis but significantly promoted cell apoptosis. We therefore proposed a circRNA/miRNA-based molecular axis which is responsible to characteristics and progressions of neuroblastoma.
Accumulating studies have revealed that circRNAs primarily regulate downstream gene expressions through formation of ceRNA networks with miRNAs to regulate the expressions of their target genes (7). Clusters of differentially expressed circRNAs in neuroblastoma have been reported (19), suggesting circRNAs play an essential role in regulating the tumorigenesis and progressions of neuroblastoma. However, the biological roles and molecular targets of ACAP2 have not been fully elucidated. In this study, we observed that the expressions of ACAP2 and miR-143-3p had a remarkedly negative pattern in neuroblastoma tumors. Silencing ACAP2 effectively upregulated miR-143-3p expressions. Importantly, RNA pull-down assay and luciferase assay consistently demonstrated that ACAP2 interacted with miR-143-3p, indicating that ACAP2 sponges miR-143-3p in neuroblastoma cells. These results suggest that the ACAP2-miR-143-3p axis is a potential target for neuroblastoma therapy.
Cancer cells display apparently elevated anaerobic glycolysis and impaired mitochondrial oxidative respiration although under sufficient oxygen supply (13). The phenomenon of cancer cells is called the “Warburg effect”. In addition, reprogramming of glucose metabolism provides an advantage for proliferation, migration, invasion, and anti-apoptosis of cancer cells (14). The HK2, which catalyzes conversion of glucose to glucose-6-phosphate, is known to be upregulated in diverse cancer and positively associated with cancer progressions. This study illustrated that HK2 was directly targeted by miR-143-3p in neuroblastoma cells by luciferase assay and western blot. Rescue experiments showed that the miR-143-3p-suppressed neuroblastoma progressions were through targeting HK2. Importantly, results demonstrated that circRNA ACAP2-promoted migration, invasion, glycolysis, and anti-apoptosis of neuroblastoma cells through the miR-143-3p-HK2 axis. Although the miR-143-3p-HK2 axis has been reported in other cancers (16,17), our data firstly uncovered that miR-143-3p directly targeted HK2 in neuroblastoma and integrated the ACAP2-miR-143-3p-HK2-glycolysis pathway to the progressions of neuroblastoma cells. However, this study still had some limitations in that all the conclusions were from an in vitro model and an in vivo xenograft mice model could further consolidate the discoveries.
In conclusion, this study demonstrated that circRNA ACAP2 promoted the migration, invasion, and glycolysis but inhibited apoptosis of human neuroblastoma through modulation of the miR-143-3p-HK2 axis. We propose that APAP2 is an effective therapeutic target for neuroblastoma treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Children’s Hospital of Soochow University (No. 2020CS053). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was taken from Written informed consent was taken from parents/guardians of the patients.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-527
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tp-21-527
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-527). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Mallepalli S, Gupta MK, Vadde R. Neuroblastoma: An Updated Review on Biology and Treatment. Curr Drug Metab 2019;20:1014-22. 10.2174/1389200221666191226102231 [DOI] [PubMed] [Google Scholar]
- 2.Tsubota S, Kadomatsu K. Origin and initiation mechanisms of neuroblastoma. Cell Tissue Res 2018;372:211-21. 10.1007/s00441-018-2796-z [DOI] [PubMed] [Google Scholar]
- 3.Newman EA, Abdessalam S, Aldrink JH, et al. Update on neuroblastoma. J Pediatr Surg 2019;54:383-9. 10.1016/j.jpedsurg.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 4.Swift CC, Eklund MJ, Kraveka JM, et al. Updates in Diagnosis, Management, and Treatment of Neuroblastoma. Radiographics 2018;38:566-80. 10.1148/rg.2018170132 [DOI] [PubMed] [Google Scholar]
- 5.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 6.Pellecchia S, Quintavalle C, Pallante P. The control of tumor progression by circular RNAs: novel prognostic and therapeutic insights resulting from the analysis of the circAGO2/human antigen R complex. Transl Cancer Res 2019;8:S211-5. 10.21037/tcr.2019.03.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol 2018;1087:67-79. 10.1007/978-981-13-1426-1_6 [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110:304-15. Erratum in: J Natl Cancer Inst 2018;110:1147. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin W, Wang Z, Wang J, et al. circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3: Selected Key circRNAs in Neuroblastoma and Their Associations with Clinical Features. Cancer Manag Res 2021;13:4271-81. 10.2147/CMAR.S297316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Yang F, Hu A, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med 2019;11:e10835. 10.15252/emmm.201910835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Chen T, Zhu Y, et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res 2019;38:398. 10.1186/s13046-019-1376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, Liu Z, Zhong J, Lin L. Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. Eur J Clin Invest 2021;51:e13607. 10.1111/eci.13607 [DOI] [PubMed] [Google Scholar]
- 13.Abbaszadeh Z, Çeşmeli S, Biray Avcı Ç. Crucial players in glycolysis: Cancer progress. Gene 2020;726:144158. 10.1016/j.gene.2019.144158 [DOI] [PubMed] [Google Scholar]
- 14.Akins NS, Nielson TC, Le HV. Inhibition of Glycolysis and Glutaminolysis: An Emerging Drug Discovery Approach to Combat Cancer. Curr Top Med Chem 2018;18:494-504. 10.2174/1568026618666180523111351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali Syeda Z, Langden SSS, Munkhzul C, et al. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci. 2020;21:1723. 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Liang F, Zhao F, et al. Resibufogenin suppresses tumor growth and Warburg effect through regulating miR-143-3p/HK2 axis in breast cancer. Mol Cell Biochem 2020;466:103-115. 10.1007/s11010-020-03692-z [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer 2019;18:33. 10.1186/s12943-019-0947-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu T, Qiu T, Han B, et al. Circular RNA circCSNK1G3 induces HOXA10 signaling and promotes the growth and metastasis of lung adenocarcinoma cells through hsa-miR-143-3p sponging. Cell Oncol (Dordr) 2021;44:297-310. 10.1007/s13402-020-00565-x [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhou H, Li J, et al. Comprehensive Characterization of Circular RNAs in Neuroblastoma Cell Lines. Technol Cancer Res Treat 2020;19:1533033820957622. 10.1177/1533033820957622 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as