Abstract
Genome editing in the lung has the potential to provide long-term expression of therapeutic protein to treat lung genetic diseases. Yet efficient delivery of CRISPR to the lung remains a challenge. The NIH Somatic Cell Genome Editing (SCGE) Consortium is developing safe and effective methods for genome editing in disease tissues. Methods developed by consortium members are independently validated by the SCGE small animal testing center to establish rigor and reproducibility. We have developed and validated a dual adeno-associated virus (AAV) CRISPR platform that supports effective editing of a lox-stop-lox-Tomato reporter in mouse lung airway. After intratracheal injection of the AAV serotype 5 (AAV5)-packaged S. pyogenes Cas9 (SpCas9) and single guide RNAs (sgRNAs), we observed ∼19%–26% Tomato-positive cells in both large and small airways, including club and ciliated epithelial cell types. This highly effective AAV delivery platform will facilitate the study of therapeutic genome editing in the lung and other tissue types.
Keywords: lung editing, AAV5, CRISPR, Cas9, club cells, ciliated cells
Graphical abstract
Efficient delivery of CRISPR to the lung remains a challenge. We have developed and validated a dual adeno-associated virus (AAV) CRISPR platform that supports effective editing of a lox-stop-lox-Tomato reporter in mouse lung airway. We observed ∼19%–26% Tomato-positive cells in both large and small airways, including club and ciliated epithelial cell types. This highly effective AAV delivery platform will facilitate the study of therapeutic genome editing in the lung and other tissue types.
Introduction
Genome editing1 in the lung has the potential to provide long-term therapeutic protein expression after a single administration. However, efficient delivery of genome editing machinery to disease-relevant cell types in vivo remains a major challenge for the field.2,3 adeno-associated virus (AAV) is a commonly used viral vehicle for gene therapy.4,5 The lung was the target of the first human clinical treatment using recombinant adeno-associated virus (rAAV) to treat a monogenic disorder.6 Despite this, achieving effective and sustained correction of cystic fibrosis and other lung diseases remains elusive. Currently, the most promising AAV vectors for lung delivery include AAV1, 5, 6, 6.2, and 9.7 AAV1 and 5 have both been shown to target lung airways in mice.7 However, it is well known that mouse models may not predict the transduction efficiency in primates and humans.8,9 For example, AAV1 has been used to successfully transduce human airway epithelial cells in vitro, and chimpanzee airways in vivo following bronchoscopic aerosol delivery.8,9 In contrast, AAV5 appears to be more effective in mice and lower primate models.7
Due to the relatively high turnover rate of airway epithelial cells in the lung, sustained correction of genetic diseases will require either repeated administration and/or permanent genetic changes that are heritable to daughter cells. The physiology of the lung also strongly suggests that an apical route of administration through aerosols or infusions will be more effective at mediating delivery to the airways. Although AAV has been explored extensively for gene replacement therapy, efficient AAV-based delivery of CRISPR (clustered regularly interspaced short palindromic repeats) to lung airways has not been reported. Optimizing rAAV vector design and identifying the best AAV serotypes is needed to achieve efficient genome editing in the lung. The commonly used S. pyogenes Cas9 (SpCas9)1,2 is more than 4 kb in length, making it difficult to package in a single AAV vector together with the necessary guide RNA. To address this challenge, we have developed a highly efficient dual AAV platform to co-express SpCas9 and two sgRNAs.
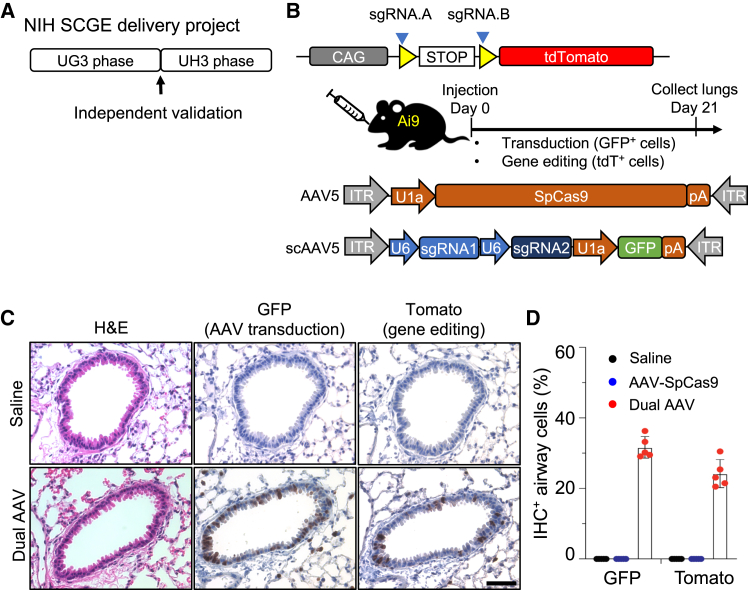
This work was initiated and completed as part of our involvement in the NIH Somatic Cell Genome Editing (SCGE) Consortium,10 which was launched in 2018 to accelerate the development of safe and effective genome editing methods. One of the SCGE initiatives is to develop innovative technologies to deliver genome editing machinery to disease-relevant cells and tissues (Figure 1A).10 The NIH UG3 funding phase (3 years) is to support proof-of-concept studies in mouse models in vivo. To ensure the rigor of the in vivo gene editing toolkit, SCGE requires validation of the delivery reagents through an independent small animal testing center (SATC) before the funding can be transitioned to the next NIH UH3 grant phase. The 2-year UH3 phase will support the scale up and testing of the genome editing delivery in a large animal model (such as non-human primate and pig models), in collaboration with the SCGE large animal testing centers.
Figure 1.
Dual AAV5 delivery of spCas9 and sgRNAs supports genome editing in mouse lung
(A) The SCGE UG3/UH3 delivery program. The UG3 funding phase is to support proof-of-concept studies in mouse models in vivo. To ensure the rigor of the in vivo gene editing toolkit, SCGE requires validation of the delivery reagents at an independent small animal testing center before the funding can be transitioned to the UH3 phase. (B) Measuring CRISPR-mediated NHEJ using lox-STOP-lox Tomato (Ai9) reporter mice. Two sgRNAs (A and B) will delete the STOP cassettes and activate the Tomato reporter. (C) Dual AAVs were intratracheally injected in Ai9 mice. Lung sections were stained for GFP and Tomato. Saline serves as a negative control. Scale bar, 100 μm. (D) Quantification of IHC in (C). Each dot is the average percentage of a mouse (n = 5 mice). Error bars are SD.
Here, we demonstrate that direct administration of AAV5 encoding CRISPR-Cas9 can mediate highly efficient genome editing in several cell types of mouse lung airways. Our results were independently validated through an SATC within the SCGE, further establishing their rigor and reproducibility.
Results
Development of AAV5 delivery platform for spCas9
To monitor genome editing in vivo, we used a fluorescent reporter mouse to detect CRISPR cutting events. The Ai9 reporter mice harbors a lox-STOP-lox (LSL)-tdTomato reporter knockin at the Rosa26 locus11 (Figure 1). Using two sgRNAs flanking LSL, Cas9 will delete the STOP cassette by NHEJ, resulting in tdTomato expression.12 This system allows direct detection of cells edited by CRISPR in vivo.
We cloned a self-complementary (sc) AAV vector to express two sgRNAs under the control of U6 promoters (Figure 1). A U1A.GFP reporter facilitates detection of AAV-infected cells (Figure 1). SpCas9 is expressed from our published single-stranded AAV vector with U1A promoter.13 We packaged both vectors as AAV5, a serotype capable of infecting lung airway in mice.7
Eight-week-old LSL-tdTomato mice were injected intratracheally with 6e10 vg of each AAV, and lungs sections were analyzed by immunohistochemistry (IHC) 21 days later. As shown in Figures 1C and 1D, GFP+ cells were detected in the airway, indicating successful AAV5 transduction. Moreover, 24.2% ± 3.6% tdTomato+ cells (n = 5 mice) were detected in the airway, indicating genome editing. These data showed that our AAV-based spCas9 delivery vehicles can induce efficient genome editing in lung airways.
Independent validation of AAV reagents at the Baylor-Rice SATC
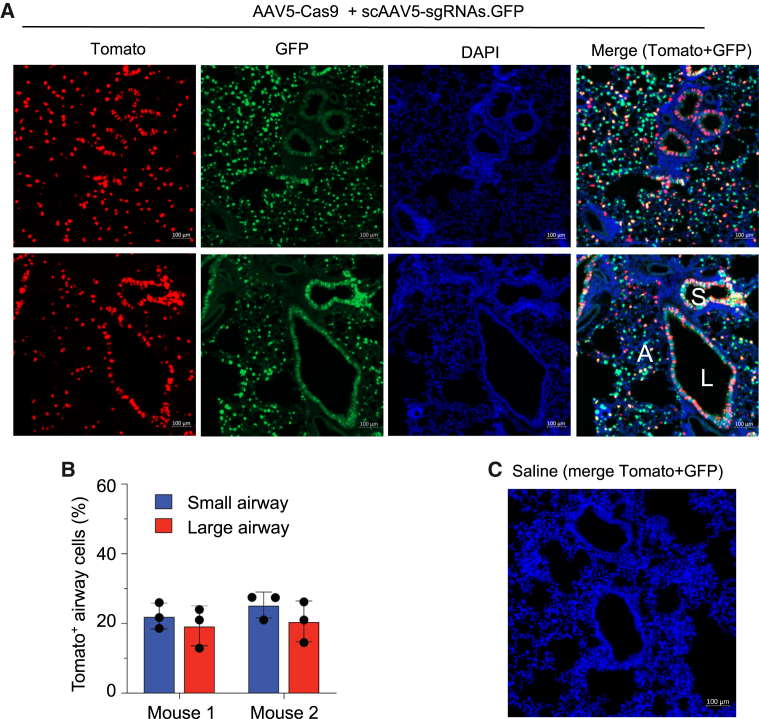
AAV5 prepared at UMass was shipped to Baylor College of Medicine for independent validation by an SCGE-supported SATC. Two Ai9 mice were dosed with 1.7 × 1011 vg each AAV per mouse by intratracheal injection. Four weeks after administration, lung and various non-target tissues were imaged for allele activation (tdTomato fluorescence), viral transduction (GFP fluorescence), and nuclei (DAPI). The percentage of genome edited cells were then counted in the large and small airways (Figure S1). In both Ai9 mice, administration of dual AAVs resulted in GFP+ and tdTomato+ cells in the airway epithelia, indicating AAV5 infection and genome editing, respectively (Figure 2A). Critically, the dual AAVs achieved >19% tdTomato+ cells in large airways (19.3% ± 5.7% mouse 1, 20.6% ± 5.8% mouse 2) and >22% in small airways (22.1% ± 3.7% mouse 1, 25.3% ± 3.7% mouse 2) (Figure 2B). These data are consistent with the results observed at UMass (Figure 1). As a negative control, saline-treated mice (n = 6) showed neither GFP nor tdTomato signals (Figure 2C), confirming the specificity of fluorescence signal in dual AAV-treated mice.
Figure 2.
Independent validation of AAV5 reagents at SCGE small animal testing center
(A) Ai9 mice were injected with dual AAVs and harvested 4 weeks after administration. Representative native fluorescence images of lung sections are shown. Tomato indicates genome editing. GFP indicates AAV.sgRNA infection. L, large airway; S, small airway; A, alveolar region. (B) Quantification of Tomato+ cells in n = 3 airways per mouse. Error bars are SD. (C) Saline control mice showed neither GFP nor Tomato signal. Merged image of Tomato, GFP, and DAPI from a representative mouse is shown.
Fluorescent imaging of sections from non-target (i.e., non-lung) organs revealed extremely rare tdTomato+ cells. In the first mouse, a cluster of a few (<10) tdTomato+ cells was observed in the liver (Figures S2A and S2B). No tdTomato+ were observed in other organs. In the second mouse, single tdTomato+ cells were observed in sections from the brain, heart, and trachea (Figure S2B). Notably, neither mouse produced tdTomato+ cells in reproductive organs, confirming that genome editing was restricted to somatic tissues. These results suggest that AAV5 delivered by intratracheal injection does not efficiently enter the bloodstream and transduce organs other than lung. To assess the safety of AAV5 delivery and CRISPR-mediated gene editing, we measured body weights, liver weights, and spleen weights and performed hematoxylin and eosin staining in these tissues from dual AAV-treated mice. No anomalies were observed. Microscopic examination of the lung, liver, and spleen revealed normal histology, with no evidence of inflammation or toxicity (data not shown). Together, these data independently confirm that AAV5 can deliver spCas9 to mouse lung airways and mediate highly efficient genome editing.
Characterization of genome editing in lung club and ciliated cells
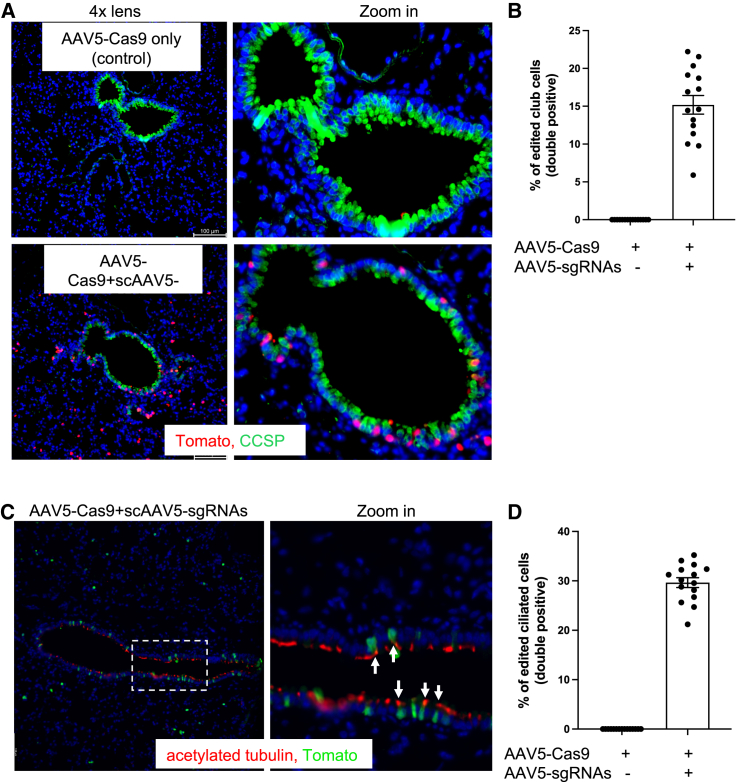
Club and ciliated cells are major subtypes of airway epithelial cells in the lung.14,15 Clara cell secretory protein (CCSP) is a marker of club cell (formerly known as clara cells). We stained lung sections with CCSP antibody to characterize club cells that are positive for tdTomato (Figure 3A). We detected double-positive cells (Figure 3B), suggesting that AAV5 can mediate genome editing in a subset of club cells. We also stained for acetylated tubulin, a marker of ciliated cells. Double-positive cells were observed (Figures 3C and 3D).
Figure 3.
AAV5 mediates genome editing in club and ciliated cells
(A) Paraffin-embedded lung sections were stained with antibodies for CCSP (green), a marker of club/clara cells, and Tomato. Native GFP from AAV.sgRNA is quenched in paraffin slides. (B) Quantification of edited (Tomato+) CCSP+ cells. Error bars are SEM. (C) Lung sections were stained with antibodies for acetylated tubulin (red, a marker of ciliated cell) and Tomato (green). Arrows denote double-positive cells. (D) Quantification of edited (Tomato+) ciliated cells. (B and D) n = 15 sections from three mice. Error bars are SEM.
We observed GFP or tdTomato+ cells in the alveolar region (Figure 2A). Future studies will characterize whether alveolar cells, immune cells, or endothelial cells are infected or edited by AAV5.
Discussion
In this study, we showed that AAV5 can efficiently deliver CRISPR-Cas9 to mouse lung airway. Because the dual AAV5 can mediate genome editing in both large and small airways, this platform is suitable to study genome editing for a variety of lung diseases affecting airway cells. Adenovirus and AAV (e.g., AAV9 and AAV-DJ) have been used to infect mouse lung to generate tumor models,16, 17, 18, 19 but they have not been used to systematically measure genome editing in disease-relevant cell types in the lung airway. To our knowledge, this study is the first to achieve ∼20% editing efficiency (based on reporter expression) in the mouse lung airway. Importantly, our results were confirmed through independent experiments at two different institutes, reinforcing their validity.
McCray and coworkers have recently shown that intratracheal delivery of Cas9 or Cas12a ribonucleoproteins (RNPs) with a shuttle peptide supports editing in the mouse lung airway20; 12%–13% reporter positive cells were reported.20 Our AAV platform provides a viral delivery toolkit to complement the RNP technique. This dual AAV design is also amenable to target other organ types using different AAV serotypes.
We showed that AAV5 can mediate genome editing in club and ciliated cells in the lung airway. Recent studies have used AAV serotypes, including AAV1,7 5,7 6,21 and 9,22 for lung delivery. Our data demonstrated that AAV5 supports high-level genome editing in both large and small airways. Minimal infection was observed in other organs (Figure S2). In addition, the Engelhardt lab showed that both AAV5 and AAV1 can infect a subset of long-lived club/clara cells and alveolar type II cells in mice.7 Future studies are needed to test whether AAV5 can transduce mouse lung basal cells23 in the trachea or in primate models. Persistent expression from singly AAV transduced cells may cause an immune response against Cas9 and GFP. Assaying additional safety profiles, such as off-target editing, is also needed in future experiments.
In summary, our dual AAV vectors provide a blueprint for designing future AAV-based CRISPR delivery systems in the lung. This highly efficient dual AAV platform will facilitate the study of genome editing in the lung and other tissue types.
Methods
Generation of plasmid
pAAV-sgA-sgB was generated through Gibson assembly, by combining the following three DNA fragments: (1) gBlock sgAi9L driven by U6, (2) gBlock sgAi9R driven by U6, (3) a MluI/EagI-digested AAV backbone. SpCas9 is expressed from our published single-stranded AAV vector with U1A promoter.13 Target sequences of the gRNAs are:
sgRNA.A: aaagaattgatttgataccg
sgRNA.B: gtatgctatacgaagttatt
AAV vector production
AAV vectors (AAV5 or scAAV5 capsids) were packaged at the Viral Vector Core of the Horae Gene Therapy Center at the University of Massachusetts Medical School. In brief, rAAV vector plasmid carrying an expression cassette for the gene of interest flanked by AAV2 ITRs is co-transfected into HEK293 cells with a packaging plasmid and adenovirus helper plasmid. The packaging plasmid expresses regulatory proteins of AAV2 and capsid proteins of AAV5 serotype, which will excise the recombinant genome from the rAAV vector plasmid, replicate the genome, and package the genome into AAV virions. Adenovirus serotype 5 E1, E2a, and E4 proteins, and VA I and II RNAs expressed from the adenovirus helper plasmid provides helper functions essential for rAAV rescue, replication, and packaging.24 The recombinant viruses are purified by standard CsCl gradient sedimentation method and desalted by dialysis.25 The vectors are quality control tested by ddPCR titration for DNase-resistant vector genome (vg) concentration using probe and primers targeting the poly(A) region of the vg26 and silver-stained SDS-polyacrylamide gel analysis to establish the purity of each lot.27
Animal studies
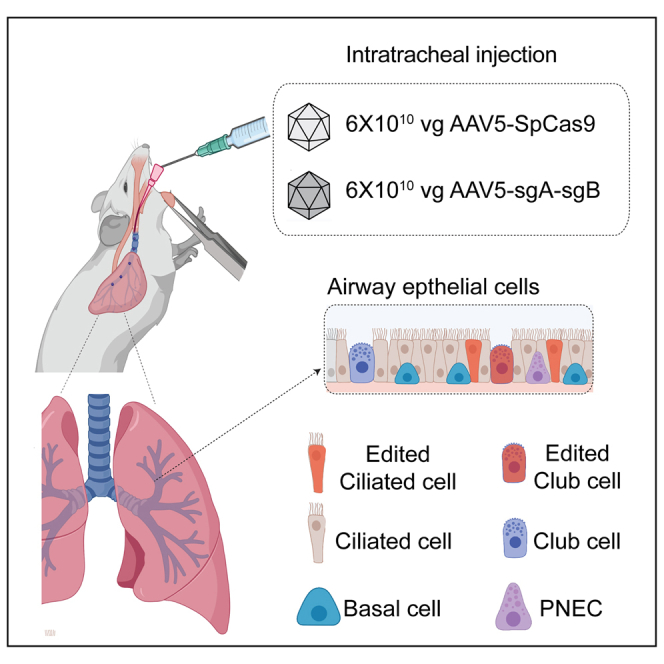
All animal experiments were authorized by the Institutional Animal Care and Use Committee (IACUC) at UMass medical school. For in vivo gene editing, Ai9 mice (strain no. 007909) were purchased from Jackson Laboratories. Eight-week-old mice were randomly allocated into two groups. The AAVs were then delivered to mouse lung through intratracheal intubation. Each mouse was injected with 60 μL PBS containing 6 × 1010 vg of scAAV5-sgA-sgB virus and 6 × 1010 vg of AAV5-SpCas9 virus or control at UMass. Animals were sacrificed at the end of each experiment (21 days after injection). Lungs were fixed with formalin or stored at −80°C with OTC (TissueTek) freezing compound until further analyses. No sample size calculation was performed and each group consisted of at least three mice for statistical analysis. All animal experiments at the Baylor College of Medicine were performed under an IACUC-approved protocol (AN-8084).
IHC and immunofluorescence
For IHC studies, formalin-fixed, paraffin-embedded (FFPE) mouse lung samples were sectioned at 4 μm, deparaffinized, and the antigens were retrieved with 10 mM citrate buffer for 9 min at 95°C. Then the slides were incubated overnight at 4°C with anti-GFP (CST, cat. no. 2956, 1:200) or anti-RFP (Rockland, cat. no. 600-401-379, 1:300). Visualization was performed using the DAB Quanto kit (Fisher Scientific, cat. no. TA-125-QHDX) as instructed by the manufacturer. In Figures 1D, 20 airways from 5 IHC images for each mouse were used to generate the average percentage.
For CCSP double staining, FFPE mouse lung samples were sectioned at 4 μm, deparaffinized and the antigen were retrieved with 10 mM citrate buffer for 9 min at 95°C. Then the slides were incubated overnight at 4°C with anti-CCSP antibody (Santa Cruz, cat. no. SC-25555, 1:2,000) and anti-RFP antibody (Thermo Fisher Scientific, cat. no. MA5-15257, 1:300), then the sections were incubated 1 h at room temperature with Alexa Fluor 488 donkey anti-rabbit IgG (CCSP) and Alexa Fluor 647 donkey anti-mouse IgG (tdTomato). Nuclei were counterstained with DAPI. For acetylated tubulin double staining, Alexa Fluor 647 anti-acetylated tubulin antibody (Santa Cruz, cat. no. SC-23950, 1:200) and anti-RFP antibody (Rockland, cat. no. 600-401-379) were used. Nuclei were counterstained with DAPI. Images were acquired on a Leica DMi8 imaging microscope.
For direct fluorescence imaging at the Baylor-Rice SATC, treated mice were weighed, then euthanized, and a panel of organs dissected. Liver, lungs and spleens were weighed immediately following dissection. Each organ was fixed in 4% buffered paraformaldehyde overnight at 4°C, then equilibrated in 30% sucrose overnight at 4°C before freezing in OCT. Three non-consecutive sections from each organ sample were mounted with DAPI to visualize nuclei, and imaged for DAPI, tdTomato, and GFP. An AxioScan.Z1 (Zeiss) scanner at 20× magnification was used in conjunction with ZEN Blue software to obtain and process the images. For quantification of editing, airways were identified in lung sections by morphology (thick ring of DAPI-stained nuclei), and their individual area measured for classification into small (<50,000 μm2) and large (>50,000 μm2). In each airway, the total number of tdTomato+ cells and DAPI-stained nuclei were counted. Editing efficiency was calculated as tdTomato+ cells/DAPI-stained nuclei and expressed as a percentage. Three large and three small airways were quantified per mouse.
For histological analysis, small portions of lung liver and spleen were fixed in neutral-buffered formalin overnight, then stored in 70% ethanol prior to paraffin embedding, sectioning, and hematoxylin and eosin staining. Sections were imaged with a scanned as above with a bright-field light source.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.4. Sample size was not pre-determined by statistical methods, but rather, based on preliminary data. Group allocation was performed randomly. In all studies, data represent biological replicates (n) and are depicted as mean ± SD as indicated in the figure legends.
Data availability
The authors declare that all other data supporting the findings of this study are available within the paper and its supplemental information files or upon request.
Acknowledgments
We thank S. Wolfe, T. Flotte, D. Anderson, and E. Sontheimer for discussions. We thank Marrah Lachowicz-Scroggins and P.J. Brooks at NIH for advice and discussions. We thank Y. Liu in the UMass Morphology for support. We also thank Cecilia Ljungberg in the BCM RNA In Situ Hybridization Core for support. This core receives funding from the Baylor College of Medicine Intellectual and Developmental Disabilities Center (IDDRC) (P50HD103555) and the Texas Medical Center Digestive Diseases Center (DDC) (P30DK056338). S.L., D.W., G.G., and W.X. were supported by NIH UG3HL147367 and UH3HL147367. W.X. was supported by grants from the National Institutes of Health (DP2HL137167 and P01HL131471), the American Cancer Society (129056-RSG-16-093), and the Cystic Fibrosis Foundation. C.J.W., A.E.M., D.G.L., J.R.S., M.E.D., and W.R.L. were supported by a grant from the National Institutes of Health (U42OD026645).
Author contributions
S.Q.L. and C.J.W. performed the experiments, analyzed data, and wrote the manuscript. A.E.M., Q.S., M.E.D., and D.W. performed the experiments. W.R.L., J.D.H., G.P.G., and W.X. supervised the study and wrote the manuscript with all co-authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.10.023.
Contributor Information
Guangping Gao, Email: Guangping.Gao@umassmed.edu.
Wen Xue, Email: wen.xue@umassmed.edu.
Supplemental information
References
- 1.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Zhang F., Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeler A.M., Flotte T.R. Recombinant adeno-associated virus gene therapy in light of luxturna (and zolgensma and glybera): where are we, and how did we get here? Annu. Rev. Virol. 2019;6:601–621. doi: 10.1146/annurev-virology-092818-015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flotte T., Carter B., Conrad C., Guggino W., Reynolds T., Rosenstein B., Taylor G., Walden S., Wetzel R. A phase I study of an adeno-associated virus-CFTR gene vector in adult CF patients with mild lung disease. Hum. Gene Ther. 1996;7:1145–1159. doi: 10.1089/hum.1996.7.9-1145. [DOI] [PubMed] [Google Scholar]
- 7.Gruntman A.M., Mueller C., Flotte T.R., Gao G. Gene transfer in the lung using recombinant adeno-associated virus. Curr. Protoc. Microbiol. 2012 doi: 10.1002/9780471729259.mc14d02s26. Chapter 14, Unit14D.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Luo M., Trygg C., Yan Z., Lei-Butters D.C., Smith C.I., Fischer A.C., Munson K., Guggino W.B., Bunnell B.A., Engelhardt J.F. Biological differences in rAAV transduction of airway epithelia in humans and in old world non-human primates. Mol. Ther. 2007;15:2114–2123. doi: 10.1038/sj.mt.6300277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flotte T.R., Fischer A.C., Goetzmann J., Mueller C., Cebotaru L., Yan Z., Wang L., Wilson J.M., Guggino W.B., Engelhardt J.F. Dual reporter comparative indexing of rAAV pseudotyped vectors in chimpanzee airway. Mol. Ther. 2010;18:594–600. doi: 10.1038/mt.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha K., Sontheimer E.J., Brooks P.J., Dwinell M.R., Gersbach C.A., Liu D.R., Murray S.A., Tsai S.Q., Wilson R.C., Anderson D.G., et al. The NIH somatic cell genome editing program. Nature. 2021;592:195–204. doi: 10.1038/s41586-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A., et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D., Li J., Song C.Q., Tran K., Mou H., Wu P.H., Tai P.W.L., Mendonca C.A., Ren L., Wang B.Y., et al. Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat. Biotechnol. 2018;36:839–842. doi: 10.1038/nbt.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rock J.R., Randell S.H., Hogan B.L. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Models Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C., Niklason L., Calle E., Le A., Randell S.H., et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maddalo D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A., Han Y.-C., Ogrodowski P., Crippa A., Rekhtman N., de Stanchina E., et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau C.-H., Suh Y. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res. 2017;6:2153. doi: 10.12688/f1000research.11243.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthelsen M.F., Leknes S.L., Riedel M., Pedersen M.A., Joseph J.V., Hager H., Vendelbo M.H., Thomsen M.K. Comparative analysis of stk11/lkb1 versus pten deficiency in lung adenocarcinoma induced by CRISPR/Cas9. Cancers. 2021;13:974. doi: 10.3390/cancers13050974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann O., Reissland M., Maier C.R., Fischer T., Prieto-Garcia C., Baluapuri A., Schwarz J., Schmitz W., Garrido-Rodriguez M., Pahor N., et al. Implementation of CRISPR/Cas9 genome editing to generate murine lung cancer models that depict the mutational landscape of human disease. Front. Cel. Dev. Biol. 2021;9:641618. doi: 10.3389/fcell.2021.641618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D.K., Théberge V., Hallée S., Duperré A.M., Del'Guidice T., Lepetit-Stoffaes J.P., et al. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 2019;10:4906. doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbert C.L., Allen J.M., Miller A.D. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell C.L., Vandenberghe L.H., Bell P., Limberis M.P., Gao G.P., Van Vliet K., Agbandje-McKenna M., Wilson J.M. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J. Clin. Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Q., Sena-Esteves M., Gao G. Analysis of recombinant adeno-associated virus (rAAV) purity using silver-stained SDS-PAGE. Cold Spring Harb. Protoc. 2020;2020:095679. doi: 10.1101/pdb.prot095679. [DOI] [PubMed] [Google Scholar]
- 25.Su Q., Sena-Esteves M., Gao G. Purification of recombinant adeno-associated viruses (rAAVs) by cesium chloride gradient sedimentation. Cold Spring Harb. Protoc. 2020;2020:095604. doi: 10.1101/pdb.prot095604. [DOI] [PubMed] [Google Scholar]
- 26.Su Q., Sena-Esteves M., Gao G. Titration of recombinant adeno-associated virus (rAAV) genome copy number using real-time quantitative polymerase chain reaction (qPCR) Cold Spring Harb. Protoc. 2020;2020:095646. doi: 10.1101/pdb.prot095646. [DOI] [PubMed] [Google Scholar]
- 27.Su Q., Sena-Esteves M., Gao G. Production of recombinant adeno-associated viruses (rAAVs) by transient transfection. Cold Spring Harb. Protoc. 2020;2020:095596. doi: 10.1101/pdb.prot095596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all other data supporting the findings of this study are available within the paper and its supplemental information files or upon request.