Abstract
Genetic factors influence blood pressure (BP) response to the cold pressor test (CPT), which is a phenotype related to hypertension risk. We examined the association between variants of the α-adducin (ADD1) and guanine nucleotide binding protein (G protein) β-polypeptide 3 (GNB3) genes and BP response to the CPT. A total of 1998 Han Chinese participants from the Genetic Epidemiology Network of Salt Sensitivity completed the CPT. The area under the curve (AUC) above the baseline BP during the CPT was used to measure the BP response. Twelve single-nucleotide polymorphisms (SNPs) of the ADD1 and GNB3 genes were selected and genotyped. Both single-marker and haplotype association analyses were conducted using linear mixed models. The rs17833172 and rs3775067 SNPs of the ADD1 gene and the rs4963516 SNP of the GNB3 gene were significantly associated with the BP response to CPT, even after adjusting for multiple testing. For the ADD1 gene, the AA genotype of SNP rs17833172 was associated with lower systolic BP (SBP) reactivity (P<0.0001) and faster BP recovery (P=0.0003). The TT genotype of rs3775067 was associated with slower SBP recovery (P=0.004). For the GNB3 gene, the C allele of SNP rs4963516 was associated with faster diastolic BP recovery (P=0.002) and smaller overall AUC (P=0.003). Haplotype analysis indicated that the CCGC haplotype of ADD1 constructed by rs1263359, rs3775067, rs4961 and rs4963 was significantly associated with the BP response to CPT. These data suggest that genetic variants of the ADD1 and GNB3 genes may have important roles in BP response to the CPT. Future studies aimed at replicating these novel findings are warranted.
Keywords: α-adducin, blood pressure, cold pressor test, G protein, single-nucleotide polymorphism
INTRODUCTION
Hypertension is an important worldwide public health challenge, because of its high prevalence and the concomitant increase in the risk of vascular disease. Elevated blood pressure (BP) response to the cold pressor test (CPT) has been demonstrated by prospective studies to be a significant risk factor for hypertension in normotensive persons.1-3 A large body of evidence indicates that genetic factors have an important role in determining the individual BP response to CPT. For example, our previous study demonstrated that the heritabilities of systolic BP (SBP) and diastolic BP (DBP) responses to CPT are 29% and 28%, respectively.4 Similar findings were also reported in Caucasians.5,6
The CPT is known to cause global sympathetic activation and to result in significant arteriolar vasoconstriction with a subsequent increase in BP. Most previous association studies of the CPT have targeted candidate genes from the sympathetic nervous system because of its direct biological relevance for the BP response to cold stress.7 Although several genes from this system, such as those that code for β1-, β2-, and α2C-adrenergic receptors, have shown important roles in the BP response to the CPT;8-10 these genes explain only a fraction of the observed heritability, suggesting that further variants are yet to be discovered. To better understand the genetic mechanism of this significant predictor of hypertension, it is still highly necessary to discover novel genetic variants associated with BP response to the CPT.
In this study, we aimed to explore the role of two important cellular signal transduction genes, namely, the α-adducin (gene: ADD1) and guanine nucleotide binding protein (G protein) β-polypeptide 3 (gene: GNB3) genes. Both of these genes are involved in hypertension and salt sensitivity of BP, which has been demonstrated to share some genetic components with the phenotypes of BP response to the CPT.11 The current study was conducted in a large, homogeneous sample of Han Chinese participants who took part in the Genetic Epidemiology Network of Salt Sensitivity (GenSalt). The single-nucleotide polymorphisms (SNPs) of the ADD1 and GNB3 genes were tested to determine their associations with the BP response to the CPT.
MATERIALS AND METHODS
Study participants
The GenSalt study was conducted in a Han Chinese population from rural areas of northern China. A community-based BP screening was conducted among persons aged 18–60 years in the study villages to identify potential probands and their families. Those individuals with a mean SBP between 130 and 160mmHg and/or a DBP between 85 and 100mmHg, no use of antihypertensive medications, as well as their parents, siblings, offspring and spouses, were recruited. Detailed eligibility criteria for the participants have been presented elsewhere.12 In general, individuals who had stage 2 hypertension, secondary hypertension, use of antihypertensive medications, history of clinical cardiovascular disease, diabetes, chronic kidney disease, pregnancy or heavy alcohol use were excluded from the CPT. Overall, 1998 participants from 631 families completed the CPT and were included in the current analysis.
Institutional review boards at all of the participating institutes approved the GenSalt study. Written informed consent was obtained from each participant.
Data collection and CPT
A standard questionnaire was administered by a trained staff member to collect information on demographic characteristics, personal and family medical history and lifestyle risk factors. Body weight and height were measured twice in light indoor clothing without shoes. Body-mass index was calculated as weight (in kilograms) divided by the squared height (in meters). BP measurements were obtained by trained and certified observers using a standard mercury sphygmomanometer according to a common protocol adapted from procedures recommended by the American Heart Association.13 Participants were advised to avoid alcohol, cigarette smoking, coffee/tea and exercise for at least 30 min before their CPT. Three baseline BP measurements were obtained after the participant was seated for 20 min. Next, the participants immersed their left hands in the ice water bath (3–5 °C) to just above the wrist for 1 min. BP measurements at 0, 1, 2 and 4 min were obtained after the left hand had been removed from the ice water bath. The CPT was well tolerated in all subjects, and no side effects were reported.
SNP selection and genotyping
TagSNPs of the ADD1 and GNB3 genes were selected to cover their common variants fully. Tagger software (http://www.broad.mit.edu/mpg/tagger/) was used to select tagSNPs (r2⩾0.8) from each gene based on the empirical patterns of linkage disequilibrium (LD) structure in the Chinese population, as determined by Chinese HapMap data.14 In addition, potential functional SNPs were also included based on previously reported associations with BP. A total of 15 SNPs from these two genes were selected. Genotyping was performed using SNPlex assays based on the oligonucleotide ligation assay for capillary electrophoresis on an automated DNA sequencer (Applied Biosystems 3700 DNA Analyzer; Applied Biosystems, Carlsbad, CA, USA).
Of the 15 genotyped SNPs, two monomorphic SNPs and one SNP with a low genotyping call rate (53.1%) were excluded from the analysis. Table 1 lists detailed information for the 12 analyzed SNPs, including physical position, alleles and minor allele frequency.
Table 1.
Information of the analyzed SNPs of the ADD1 and GNB3 genes
| Gene | SNP | Chr |
Physical
location |
Region in gene |
Alleles (major/ minor) |
MAF |
HW P-value |
|---|---|---|---|---|---|---|---|
| ADD1 | rs4690002 | 4 | 281 1038 | 5′ Flanking | C/T | 0.44 | 0.60 |
| rs12503220 | 4 | 281 9940 | intron 1 | G/A | 0.14 | 0.70 | |
| rs1263359 | 4 | 283 1824 | intron 1 | T/C | 0.43 | 0.93 | |
| rs17833172 | 4 | 283 8312 | intron 1 | G/A | 0.04 | 0.21 | |
| rs3775067 | 4 | 285 8420 | intron 4 | C/T | 0.34 | 0.77 | |
| rs4961 | 4 | 287 6505 | exon 9(missense) | T/G | 0.48 | 0.20 | |
| rs4963 | 4 | 288 6560 | exon12(missense) | G/C | 0.48 | 0.54 | |
| GNB3 | rs4963516 | 12 | 681 8289 | 5′ flanking | A/C | 0.32 | 0.12 |
| rs1129649 | 12 | 681 8729 | 5′ flanking | T/C | 0.31 | 0.61 | |
| rs3213431 | 12 | 681 8789 | 5′ flanking | T/C | 0.11 | 0.82 | |
| rs2301339 | 12 | 682 4885 | intron 9 | G/A | 0.45 | 0.66 | |
| rs5446 | 12 | 682 6723 | 3′ UTR | C/T | 0.20 | 0.68 |
Abbreviations: ADD1, α-adducin; GNB3, guanine nucleotide binding protein (G protein) β-polypeptide 3; H–W, Hardy–Weinberg equilibrium test; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Statistical analysis
The Mendelian consistency of the SNP genotype data was assessed by PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/).15 The Hardy–Weinberg Equilibrium for each SNP and the extent of pairwise LD between SNPs were examined using Haploview software (version 4.2, http://www.broad.mit.edu/mpg/haploview).16 LD blocks were defined based on the confidence intervals method implemented in the Haploview software.17 The most likely haplotypes within each block for individuals were inferred using the MERLIN program (http://www.sph.umich.edu/csg/abecasis/Merlin).18
Both single-marker and haplotype association analyses were conducted using linear mixed models. In this model, a sandwich estimator was used to account for the non-independence of family members. All analyses were carried out separately for SBP and DBP. We used the average of three BP measurements taken before the CPT as the pre-CPT baseline BP. The area under the curve (AUC) above the baseline BP was calculated and taken as the measurement variable for BP responses to the CPT. In addition, the AUC of BP response to CPT was further divided into BP reactivity (from baseline to time 0) and recovery (from time 0 to time 4). The association between SNP genotypes and BP responses to CPT were tested under general (df=2), additive (df=1), dominant (df=1) and recessive (df=1) genetic models. Age, gender, body-mass index, room temperature of BP measurement, and the study site were adjusted in multivariable analyses. The P-values were adjusted for the total number of tested SNPs using the Bonferroni correction method (α=0.05/12=0.004). Association analyses were conducted using SAS statistical software (version 9.1; SAS Institute, Cary, NC, USA).
RESULTS
At the baseline examination, on average, study participants were 40.1 years of age, had a body-mass index of 23.4 kgm −2, and a mean pre-CPT baseline SBP and DBP of 120.1 and 74.7 mm Hg, respectively (Table 2). Approximately 52.5% of the study population was male. Table 2 also shows the overall AUC, as well as the AUC of BP reactivity and the AUC of BP recovery for SBP and DBP, respectively.
Table 2.
Characteristics and blood pressure responses to cold pressor test of 1998 study participants
| Variable | All subjects |
|---|---|
| No. of participants | 1998 |
| Age, years | 40.1±10.3 |
| Male, % | 52.5 |
| Body-mass index, kgm−2 | 23.4±3.2 |
| Baseline blood pressure, mmHg | |
| Systolic | 120.1±15.6 |
| Diastolic | 74.7±10.9 |
| Reactivity area under the curve | |
| Systolic | 6.8±5.3 |
| Diastolic | 3.5±3.4 |
| Recovery area under the curve | |
| Systolic | 10.4±19.0 |
| Diastolic | 7.3±14.3 |
| Overall area under the curve | |
| Systolic | 17.1±22.9 |
| Diastolic | 10.8±16.7 |
Data are presented as mean±s.d. or percentage.
The rs17833172 and rs3775067 SNPs of the ADD1 gene and the rs4963516 SNP of the GNB3 gene were significantly associated with SBP or DBP responses to the CPT under at least one of the four genetic models after adjusting for multiple testing. P-values for the associations between these SNPs and the BP responses to the CPT are listed in Table 3.
Table 3.
SNPs associated with blood pressure responses to cold pressor test
| Gene | SNP | P-values |
||||
|---|---|---|---|---|---|---|
|
General
model |
Additive
model |
Dominant
model |
Recessive
model |
|||
| Systolic blood pressure | ||||||
| Reactivity AUC | ADD1 | rs17833172 | <0.0001* | 0.68 | 0.50 | <0.0001* |
| rs3775067 | 0.03 | 0.61 | 0.49 | 0.03 | ||
| GNB3 | rs4963516 | 0.64 | 0.72 | 0.95 | 0.40 | |
| Recovery AUC | ADD1 | rs17833172 | 0.002 * | 0.66 | 0.90 | 0.0003 * |
| rs3775067 | 0.01 | 0.15 | 0.88 | 0.004 * | ||
| GNB3 | rs4963516 | 0.26 | 0.47 | 0.19 | 0.64 | |
| Overall AUC | ADD1 | rs17833172 | 0.0002 * | 0.77 | 0.96 | <0.0001* |
| rs3775067 | 0.01 | 0.21 | 0.94 | 0.005 | ||
| GNB3 | rs4963516 | 0.31 | 0.63 | 0.28 | 0.55 | |
| Diastolic blood pressure | ||||||
| Reactivity AUC | ADD1 | rs17833172 | 0.13 | 0.88 | 0.98 | 0.04 |
| rs3775067 | 0.02 | 0.23 | 0.93 | 0.008 | ||
| GNB3 | rs4963516 | 0.40 | 0.18 | 0.27 | 0.29 | |
| Recovery AUC | ADD1 | rs17833172 | 0.38 | 0.60 | 0.73 | 0.17 |
| rs3775067 | 0.05 | 0.08 | 0.49 | 0.02 | ||
| GNB3 | rs4963516 | 0.006 | 0.002 * | 0.002 * | 0.09 | |
| Overall AUC | ADD1 | rs17833172 | 0.36 | 0.62 | 0.75 | 0.16 |
| rs3775067 | 0.04 | 0.09 | 0.57 | 0.01 | ||
| GNB3 | rs4963516 | 0.01 | 0.003 * | 0.005 | 0.10 | |
Abbreviations: ADD1, α-adducin; AUC, area under the curve; GNB3, guanine nucleotide binding protein (G protein) β-polypeptide 3; SNP, single-nucleotide polymorphism.
P-value <0.05 after Bonferroni correction. Original P-values <0.05 are indicated in bold.
Compared with the individuals carrying the major G allele of rs17833172 for ADD1, participants who were homozygous for the minor A allele had much lower SBP reactivity (2.83±0.22 vs. 6.75±0.15, P<0.0001), recovery (−12.21±6.10 vs. 9.76±0.50, P=0.0003), and an overall AUC (−9.28±6.33 vs. 16.50±0.61, P<0.0001) (Figure 1). The rs3775067 SNP of the ADD1 gene was associated with the AUC for SBP recovery in a recessive model. The participants homozygous for the T allele of rs3775067 had a greater AUC for SBP recovery (13.06±1.26 vs. 9.31±0.53, P=0.004) than carriers of the C allele (Figure 2). For the rs4963516 SNP of GNB3, the AUC of DBP recovery decreased with the number of C alleles (P=0.002). The mean AUC of DBP recovery for the AA, AC and CC genotypes was 8.08±0.55, 6.13±0.53 and 5.23±0.99, respectively (Figure 3). A similar trend was observed for the mean AUC for DBP overall (P=0.003).
Figure 1.
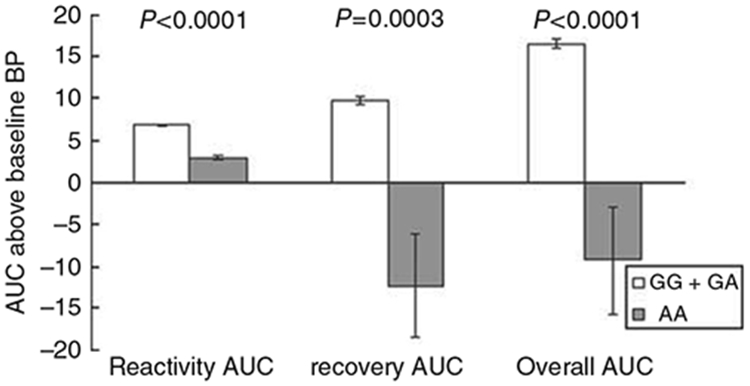
Systolic blood pressure responses to the cold pressure test according to rs17833172 genotypes of the ADD1 gene (recessive model). Tests adjusted for covariates, including age, gender, body-mass index, study site and room temperature of blood pressure measurements.
Figure 2.
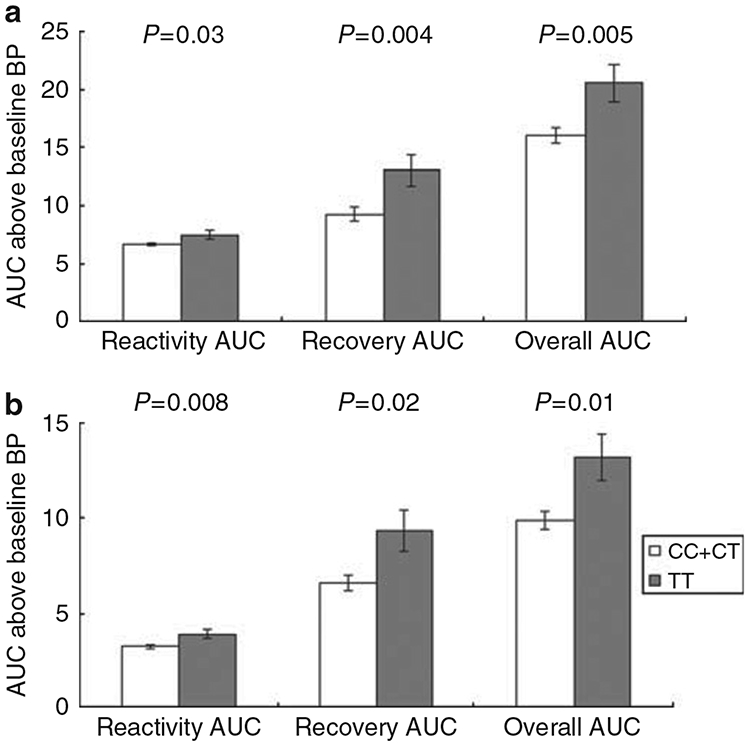
Systolic (a) and diastolic (b) blood pressure responses to the cold pressor test according to rs3775067 genotypes of the ADD1 gene (recessive model). Tests adjusted for covariates, including age, gender, body-mass index, study site and room temperature of blood pressure measurements.
Figure 3.
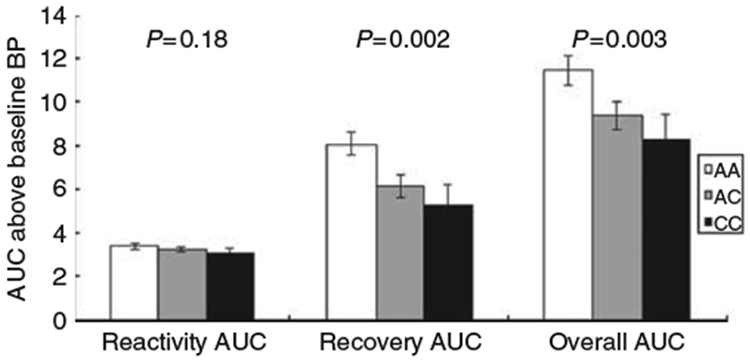
Diastolic blood pressure responses to the cold pressor test according to rs4963516 genotypes of the GNB3 gene (general model). Tests adjusted for covariates, including age, gender, body-mass index, study site and room temperature of blood pressure measurements.
We also performed haplotype analysis within LD blocks of the two genes (Figure 4). Only one LD block was identified for each gene. For the ADD1 gene, three common haplotypes within the LD block (including rs1263359, rs3775067, rs4961 and rs4963) were identified. Of these three haplotypes, CCGC was significantly associated with the AUC of DBP recovery and overall (P=0.0005 and 0.0007, respectively), even after adjusting for multiple testing (Table 4). For the GNB3 gene, no haplotypes were significantly associated with the BP response to CPT.
Figure 4.
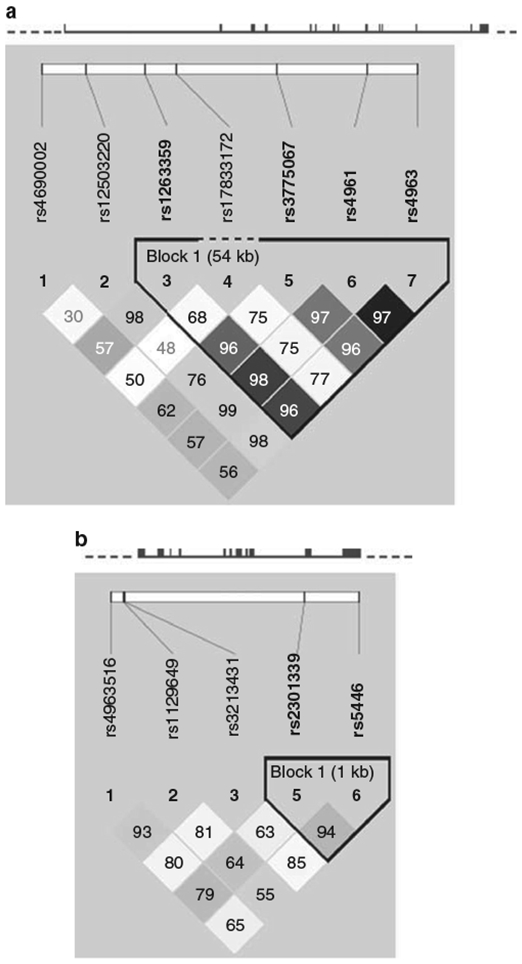
The structures of the ADD1 (a) and GNB3 (b) genes and the position of single-nucleotide polymorphisms. The black boxes in each gene are exons. Solid lines show introns, and dashed lines show flanking regions. Pairwise linkage disequilibrium (LD) coefficients D′ ×100 are shown in each cell (D′ values of 1.0 are not shown). The r2 color scheme of Haploview was applied (r2=0 shown in white, 0<r2<1 shown in shades of gray, and r2=1 shown in black). A full color version of this figure is available at the Hypertension Research journal online.
Table 4.
Association analysis of haplotypes with BP response to cold pressor test
| Genes |
Haplotype
sequence |
Haplotype
frequency |
P-values |
||
|---|---|---|---|---|---|
|
Reactivity
AUC |
Recovery
AUC |
Overall
AUC |
|||
| Systolic blood pressure | |||||
| ADD1 | TCTG | 0.52 | 0.83 | 0.85 | 0.85 |
| CTGC | 0.33 | 0.86 | 0.004 c | 0.008 c | |
| CCGC | 0.09 | 0.67 | 0.18 | 0.25 | |
| GNB3 | GC | 0.54 | 0.80 | 0.38 | 0.54 |
| AC | 0.26 | 0.23 | 0.06 | 0.07 | |
| AT | 0.20 | 0.16 | 0.53 | 0.37 | |
| Diastolic blood pressure | |||||
| ADD1 | TCTG | 0.52 | 0.19 | 0.46 | 0.37 |
| CTGC | 0.33 | 0.009 c | 0.02 a /0.01 c | 0.02 a /0.008 c | |
| CCGC | 0.09 | 0.04 b | 0.001 a * /0.0005 b * | 0.002 a * /0.0007 b * | |
| GNB3 | GC | 0.54 | 0.74 | 0.22 | 0.27 |
| AC | 0.26 | 0.03 b | 0.03 b | 0.04 a /0.03 b | |
| AT | 0.20 | 0.10 | 0.03 c | 0.04 c | |
Abbreviations: ADD1, α-adducin; AUC, area under the curve; BP, blood pressure; GNB3, guanine nucleotide binding protein (G protein) β-polypeptide 3.
For association not significant under any model, P-values for additive model are listed.
For ADD1 gene, haplotype sequence is in the order of rs1263359–rs3775067–rs4961–rs4963. For GNB3 gene, haplotype sequence is in the order of rs2301339-rs5446. Original P-value <0.05 is indicated in bold.
P-values are significant after Bonferroni correction (P<0.004).
Additive model.
Dominant model.
Recessive.
DISCUSSION
In this large family study, we identified novel genetic variants in the ADD1 and GNB3 genes that may contribute to the variation of the BP response to CPT among this population. These findings may provide valuable insight into the genetic regulation of the BP response to stress and the pathogenesis of hypertension.
In this study, we observed that the rs17833172 and rs3775067 SNPs of the ADD1 gene were associated with BP response to the CPT. Interestingly, rs17833172 has been found to be associated with salt sensitivity in our previous study.19 This finding may suggest that the ADD1 gene contributes to the genetic correlation between salt sensitivity and BP response to the CPT. Functional studies indicated that mutated forms of ADD1 could increase Na/K pump activity and the number of pump units in renal cell lines. It has also been suggested that the constitutive reduction of Na/K pump endocytic rate induced by mutated adducin variants might influence renal tubular sodium reabsorption and result in salt-sensitive hypertension.20 In addition, adducin has been involved in signal transduction, cell-to-cell contact formation and cell migration.21 However, the mechanism by which adducin regulates the BP response to stress is still not clear. Understanding how ADD1 is involved in the BP response to cold stress will require further experimental studies.
To speculate the potential functions of these two significant intronic SNPs of the ADD1 gene, we conducted an analysis using the SNP Function Prediction (FuncPred) (http://snpinfo.niehs.nih.gov/snpfunc.htm) database. Unfortunately, we could not find any functional prediction for these two SNPs. However, we found a functional SNP, rs16843169, which is in perfect LD with rs17833172 (D′ and r2=1) based on the HapMap data of Beijing Han Chinese individuals. SNP rs16843169 is located at a splice site of the ADD1 gene and may affect splicing of the gene. Future studies are warranted to identify the causal variants that lead to these association signals.
The GNB3 gene encodes a β-subunit of G proteins that mediates intracellular signaling transduction of adrenergic receptors in the sympathetic nervous system. This gene is an attractive candidate gene for the BP response to cardiovascular stress reactivity. A recent study indicated that the C825T variant (rs5443) might affect heart rate response to the CPT but not BP response to the CPT.10 Our study did not examine C825T directly, but we did investigate SNP rs2301339 of GNB3, which is in perfect LD with the C825T polymorphism (D′ and r2=1.0) among the HapMap Chinese Han and Caucasian populations. Consistently, we could not detect the association between this agent variant of C825T and BP response to the CPT. However, our study identified another variant, rs4963516, to be significantly associated with DBP response to CPT. Our previous report supports a role for this SNP in regulating resting BP.22 SNP rs4963516 is located at the 5′-flanking region of the GNB3 gene. Additionally, this SNP has been reported as a variation of intron 13 in the leprecan-like 2 (LEPREL2) gene. It should be noted that the GNB3 and LEPREL2 genes overlap on chromosome 12. FuncPred analysis shows that SNP rs4963516 lies in the transcription factor binding site and may alter the transcription level of either of the two genes or both. Certainly, further functional studies are warranted to determine whether this SNP regulates the transcription of the GNB3 or LEPREL2 genes.
In this study, rs4963516 in the GNB3 gene is significantly associated with DBP response to CPT; however, rs17833172 and rs3775067 in the ADD1 gene are significantly associated with the SBP response to the CPT. SBP may be more related to aortic elasticity and pulse wave velocity, whereas DBP may be more closely associated with peripheral vascular resistance.23 The magnitude of increase in SBP and DBP response to cold stimulus might be regulated by different genes. Such an influence may explain why certain SNPs were related to SBP, while others were associated with DBP.
Our study has several important strengths. To date, the GenSalt study is the largest study to investigate the genetic components of BP response to the CPT. This study is the most comprehensive one to examine the association between genetic variants of the ADD1 and GNB3 genes and BP response to the CPT. Additionally, the homogeneous sample of Han Chinese participants make our analysis robust for population stratification. Finally, stringent quality control procedures were employed during the performance of the CPT, genotyping and genetic data cleaning based on family relationships.
In summary, we identified a significant association between several genetic variants of the ADD1 and GNB3 genes and BP response to the CPT. These findings may contribute to a better understanding of the genetic mechanism of BP response to the CPT, which is a significant predictor for the risk of cardiovascular disease. Future studies are necessary to replicate these findings and to clarify their role in BP regulation.
ACKNOWLEDGEMENTS
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD.
APPENDIX
GenSalt Collaborative Research Group
Tulane University Health Sciences Center, New Orleans, LA, USA: Jiang He (PI), Lydia A Bazzano, Chung-Shiuan Chen, Jing Chen, Hao Mei, L Lee Hamm, Tanika N Kelly, Paul Muntner, Kristi Reynolds, Paul K Whelton, Wenjie Yang, and Qi Zhao.
Washington University School of Medicine, St Louis, MO, USA: Dabeeru C Rao (PI), Matthew Brown, Charles Gu, Hongyan Huang, Treva Rice, Karen Schwander, Shiping Wang, and Wei Yang.
University of Texas Health Sciences Center at Houston, Houston, TX, USA: James E Hixson (PI) and Lawrence C Shimmin.
National Heart, Lung and Blood Institute, Bethesda, MD, USA: Cashell E Jaquish.
Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (PI), Jie Cao, Jichun Chen, Jingping Chen, Zhenhan Du, Jianfeng Huang, Hongwen Jiang, Jianxin Li, Xiaohua Liang, De-Pei Liu, Xiangfeng Lu, Donghua Liu, Qunxia Mao, Dongling Sun, Hongwei Wang, Qianqian Wang, Xigui Wu, Ying Yang and Dahai Yu.
Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (PI), Zhendong Liu, Shikuan Jin, Yingxin Zhao, Shangwen Sun, Shujian Wang, Qengjie Meng, Baojin Liu, Zhaodong Yang and Chuanrui Wei.
Shandong Center for Diseases Control and Prevention, Shandong, China: Jixiang Ma (PI), Jiyu Zhang and Junli Tang.
Zhengzhou University, Henan, China: Dongsheng Hu (PI), Hongwei Wen, Chongjian Wang, Minghui Shen, Jingjing Pan and Liming Yang.
Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (PI), Rongyan Li, Haijun Zu and Junwei Song.
Ganyu Center for Disease Control and Prevention, Jiangsu, China: Delin Wu (PI), Xushan Wang and Xiaofeng Zhang.
Xi’an Jiaotong University, Shaanxi, China: Jianjun Mu (PI), Enrang Chen, Fuqiang Liu and Guanji Wu.
Chinese National Human Genome Center at Beijing, Beijing, China: Zhi-Jian Yao (PI), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang and Penghua Zhang.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Kasagi F, Akahoshi M, Shimaoka K. Relation between cold pressor test and development of hypertension based on 28-year follow-up. Hypertension 1995; 25: 71–76. [DOI] [PubMed] [Google Scholar]
- 2.Menkes MS, Matthews KA, Krantz DS, Lundberg U, Mead LA, Qaqish B, Liang KY, Thomas CB, Pearson TA. Cardiovascular reactivity to the cold pressor test as a predictor of hypertension. Hypertension 1989; 14: 524–530. [DOI] [PubMed] [Google Scholar]
- 3.Wood DL, Sheps SG, Elveback LR, Schirger A. Cold pressor test as a predictor of hypertension. Hypertension 1984; 6: 301–306. [DOI] [PubMed] [Google Scholar]
- 4.Mei H, Gu D, Rice TK, Hixson JE, Chen J, Jaquish CE, Zhao Q, Chen CS, Chen JC, Gu CC, Kelly TN, He J. Heritability of blood pressure responses to cold pressor test in a Chinese population. Am J Hypertens 2009; 22: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choh AC, Czerwinski SA, Lee M, Demerath EW, Wilson AF, Towne B, Siervogel RM. Quantitative genetic analysis of blood pressure response during the cold pressor test. Am J Hypertens 2005; 18: 1211–1217. [DOI] [PubMed] [Google Scholar]
- 6.Roy-Gagnon MH, Weir MR, Sorkin JD, Ryan KA, Sack PA, Hines S, Bielak LF, Peyser PA, Post W, Mitchell BD, Shuldiner AR, Douglas JA. Genetic influences on blood pressure response to the cold pressor test: results from the Heredity and Phenotype Intervention Heart Study. J Hypertens 2008; 26: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu T, Snieder H, de Geus E. Genetic influences on cardiovascular stress reactivity. Neurosci Biobehav Rev 2010; 35: 58–68. [DOI] [PubMed] [Google Scholar]
- 8.Li GH, Faulhaber HD, Rosenthal M, Schuster H, Jordan J, Timmermann B, Hoehe MR, Luft FC, Busjahn A. Beta-2 adrenergic receptor gene variations and blood pressure under stress in normal twins. Psychophysiology 2001; 38: 485–489. [PubMed] [Google Scholar]
- 9.Chotani MA, Flavahan S, Mitra S, Daunt D, Flavahan NA. Silent alpha(2C)-adrenergic receptors enable cold-induced vasoconstriction in cutaneous arteries. Am J Physiol Heart Circ Physiol 2000; 278: H1075–H1083. [DOI] [PubMed] [Google Scholar]
- 10.Kurnik D, Friedman EA, Muszkat M, Sofowora GG, Xie HG, Dupont WD, Wood AJ, Stein CM. Genetic variants in the alpha2C-adrenoceptor and G-protein contribute to ethnic differences in cardiovascular stress responses. Pharmacogenet Genomics 2008; 18: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei H, Rice TK, Gu D, Hixson JE, Jaquish CE, Zhao Q, Chen JC, Cao J, Li J, Kelly TN, Rao DC, He J. Genetic correlation of blood pressure responses to dietary sodium and potassium intervention and cold pressor test in Chinese population. J Hum Hypertens 2011; 25: 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GenSalt. Rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 2007; 21: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation 1993; 88: 2460–2470. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Consortium. The International HapMap Project. Nature 2003; 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 17.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 18.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101. [DOI] [PubMed] [Google Scholar]
- 19.Kelly TN, Rice TK, Gu D, Hixson JE, Chen J, Liu D, Jaquish CE, Bazzano LA, Hu D, Ma J, Gu CC, Huang J, Hamm LL, He J. Novel genetic variants in the alpha-adducin and guanine nucleotide binding protein beta-polypeptide 3 genes and salt sensitivity of blood pressure. Am J Hypertens 2009; 22: 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torielli L, Tivodar S, Montella RC, Iacone R, Padoani G, Tarsini P, Russo O, Sarnataro D, Strazzullo P, Ferrari P, Bianchi G, Zurzolo C. Alpha-Adducin mutations increase Na/K pump activity in renal cells by affecting constitutive endocytosis: implications for tubular Na reabsorption. Am J Physiol Renal Physiol 2008; 295: F478–F487. [DOI] [PubMed] [Google Scholar]
- 21.Chen CL, Lin YP, Lai YC, Chen HC. Alpha-Adducin translocates to the nucleus upon loss of cell-cell adhesions. Traffic 2011; 12: 1327–1340. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Wang H, Lu X, Liu DP, Chen J, Jaquish CE, Rao DC, Hixson JE, Kelly TN, Hou L, Wang L, Huang J, Chen CS, Rice TK, Whelton PK, He J, Gu D. Polymorphisms in the GNB3 and ADD1 genes and blood pressure in a Chinese population. Hum Genet 2010; 128: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safar ME, London GM, Asmar R, Frohlich ED. Recent advances on large arteries in hypertension. Hypertension 1998; 32: 156–161. [DOI] [PubMed] [Google Scholar]


