Abstract
The nitroxide, Tempol, prevents obesity related changes in mice fed a high fat diet (HFD). The purpose of this study was to gain insight into the mechanisms that result in such changes by Tempol in female C3H mice. Microarray methodology, Western blotting, bile acid analyses, and gut microbiome sequencing were used to identify multiple genes, proteins, bile acids, and bacteria that are regulated by Tempol in female C3H mice on HFD. The effects of antibiotics in combination with Tempol on the gut microflora were also studied. Adipose tissue, from Tempol treated mice, was analyzed using targeted gene microarrays revealing up-regulation of fatty acid metabolism genes (Acadm and Acadl > 4-fold, and Acsm3 and Acsm5 > 10-fold). Gene microarray studies of liver tissue from mice switched from HFD to Tempol HFD showed down-regulation of fatty acid synthesis genes and up-regulation of fatty acid oxidation genes. Analyses of proteins involved in obesity revealed that the expression of aldehyde dehydrogenase 1A1 (ALDH1A1) and fasting induced adipose factor/angiopoietin-like protein 4 (FIAF/ANGPTL4) was altered by Tempol HFD. Bile acid studies revealed increases in cholic acid (CA) and deoxycholic acid (DCA) in both the liver and serum of Tempol treated mice. Tempol HFD effect on the gut microbiome composition showed an increase in the population of Akkermansia muciniphila, a bacterial species known to be associated with a lean, anti-inflammatory phenotype. Antibiotic treatment significantly reduced the total level of bacterial numbers, however, Tempol was still effective in reducing the HFD weight gain. Even after antibiotic treatment Tempol still positively influenced several bacterial species such as as Akkermansia muciniphila and Bilophila wadsworthia. The positive effects of Tempol moderating weight gain in female mice fed a HFD involves changes to the gut microbiome, bile acids composition, and finally to changes in genes and proteins involved in fatty acid metabolism and storage.
Keywords: Nitroxides, Tempol, Obesity, Antioxidant, Gut microbiome
1. Introduction
Obesity is a major health issue and a matter of global concern. The wide array of diseases obesity is associated with is well documented [1–3]. Consequently, the discovery of anti-obesity compounds is an ongoing effort by the research community world-wide [4–6].
Our group has been involved for a considerable time in studying the antioxidant properties of the free radical nitroxide, Tempol [7–15]. We have demonstrated that Tempol exerts a number of properties relevant to biological systems such as superoxide dismutase mimic activity [12, 13] a potent recycling antioxidant [14], and a radiation protector [16], both in vitro and in vivo. An unexpected observation was prevention of weight gain after supplementing the drinking water with sucrose and Tempol in C3H mice [16]. Further studies revealed a similar effect on weight gain in mice fed a HFD with Tempol [17]. For both of these studies, no untoward toxicity was observed in mice administered with Tempol for 16–20 weeks. A study by Ley et al. [18] laid the foundation that the gut microbiome is altered in obesity. Given the manner of administration of Tempol (food, water, gavage) the role of the gut microbiome in regulating systemic metabolism and controlling fat accumulation has been explored vigorously by various researchers [19, 20]. Li et al. [19] found that Tempol treatment alters the gut microbiome by decreasing Lactobacillus which led to a decrease in bile salt hydrolase, improving the metabolic phenotype of mice on HFD. This in turn led to an increase in the bile acid, tauro-β-muricholic acid, which is a substrate of bile salt hydrolase and a farnesoid X receptor antagonist. Thus, Tempol was postulated to exert its anti-obesity effects by inhibiting intestinal FXR via increased levels of tauro-β-muricholic acid. Further studies have demonstrated that Tempol treatment impacts liver genes involved in glucose and lipid metabolism [20]. One important point to note is that for Li et al. study [19] and others [20–22] only male mice were studied. Our group had previously shown that Tempol-mediated moderation of weight increase in male C3H mice was not as pronounced as that seen for female mice of the same strain [16]. Furthermore, since much of our previous work was carried out in C3H female mice to study the effects of Tempol on radiation-induced carcinogenesis [23] we wanted to study the gut microflora of C3H female mice in response to Tempol treatment. Lastly, since Tempol was able to modulate weight over the lifespan of C3H mice [16,23] we were interested if Tempol altered gene expression in liver and adipose tissue.
In the current study, a significant remodeling of the gut microflora was observed in female mice fed a Tempol/HFD. Bile acid analysis revealed that Tempol changed the composition of bile acids in both the liver and serum of HFD treated mice. In addition, gene expression studies in liver and white adipose tissue showed changes in fatty acid synthesis and oxidation genes. Finally, proteins involved in fatty acid storage and metabolism were altered by Tempol administration.
2. Methods
2.1. Animal studies
Female C3H/Hen−TacMTV− mice between 5 and 6 weeks of age were acquired from Taconic Farms, Hudson, NY. Mice were housed in a specific pathogen-free facility on a 12-h day/night cycle with chow and water ad libitum. When mice were 8–9 weeks old, they were divided into groups and fed either control chow diet (CD), a control high fat diet (CHFD), or a high fat diet containing Tempol (10 gm/kg) (THFD), all with added bacon flavor and purchased from Bio-Serv (Flemington, NJ). The components of the high fat diet from Bio-Serv are given in Table S4. Animal weights were recorded 2–3 times a week during the entire duration of each experiment. In one study, groups of mice consuming CHFD or THFD were switched after 70 days to THFD and CHFD, respectively. The animals were then followed for times up to 119 days. Liver tissue was collected at days 70, 85, and 119 days. In another experiment, plasma was collected and the weights of the liver and adipose tissue were recorded from animals sacrificed at day 70, 77, and 94 days. The liver and fat were divided into 2 parts and either fixed in formalin or flash frozen in liquid nitrogen. All animal experiments were carried out in accordance with protocols approved by the National Cancer Institute Animal Care and Use Committee (ACUC) and followed the Guide for the Care and Use of Laboratory Animal Resource (2011, National Research Council).
2.2. Gene arrays
Frozen liver and fat samples were homogenized in Trizol (Thermo Fisher Scientific, Waltham, MA). The subsequent steps were done using the RNeasy kit from Qiagen (Germantown, MD). The aqueous phase was transferred to RNeasy column, followed by the manufacturer’s instructions for the isolation of RNA.
For liver tissues cDNA was synthesized for use with microarrays developed by the Radiation Oncology Sciences Program, National Cancer Institute (NCI) [23]. Gene expressions were assayed using mouse NIA15K cDNA clone set microarrays containing 16,192 spotted features. Details of arrays, feature intensity determination and normalization may be found elsewhere [24]. Significantly differentially expressed genes were identified by one-way ANOVA of the expression data of 24 mice at time points I, II and III (on day 70, day 85 and last day respectively), where each time point had 4 mice on CHFD and 4 mice on THFD. There were 1353 features altered by 2-fold (maximum/minimum ratio >2) at p < 0.01 and their false discovery rates estimated by Benjamini & Hochberg method [25] were <0.0375. The original feature annotations from ROSP-NIA15K-v2px-32Bx23Cx22R.gal file provided by NCI were embedded in the intensity data files. These feature annotations were updated using S.O.U.R.C.E. database (https://source-search.princeton.edu/) and manual queries of UCSC Genome Browser (https://genome.ucsc.edu, Mouse Dec. 2011, GRCm38/mm10 Assembly). The annotations of 1265 of the above 1353 features were identified in which 598 altered by 2.5-fold. Hierarchical clustering of mean centered logarithmic data was performed by average linkage algorithm and 1-correlation coefficient as distance metric [26].
For adipose tissue gene expression studies RNA was extracted and shipped to Qiagen (Germantown, MD) for pathway-focused gene expression analysis with the RT2 Profiler PCR fatty acid metabolism array (PAMM-007Z). The data obtained was normalized to the housekeeping genes.
2.3. Oil Red O staining for lipid droplets
An earlier published protocol was followed for Oil Red O staining [27]. Frozen liver sections (5–6 μm) were air dried and fixed in 10% neutral buffered formalin for 10 min and then dipped in 60% isopropanol followed by staining in a working solution of Oil Red O for 15 min. The sections were then rinsed in 60% isopropanol very briefly followed by rinsing in distilled water. The sections were then counter-stained with Mayer’s Hematoxylin and then rinsed several times in distilled water. Cover slips were mounted on slides with an aqueous mounting medium.
2.4. Western blots
Frozen samples of adipose tissues were homogenized in ice cold RIPA buffer supplemented with protease and phosphatase inhibitors. The samples were incubated on ice for 30 min and then centrifuged at 10,000×g for 10 min. The supernatant was collected and centrifuged again at 10,000×g for 30 min. The supernatant was then pipetted into fresh tubes and stored at −70 °C. Protein samples of equal amounts were subjected to SDS PAGE on 4%–20% Tris-glycine acrylamide gels (Novex-Invitrogen). Following transfer to nitrocellulose, samples were probed with primary antibodies ALDH1A1 (ab52492) from Abcam (Waltham, MA), ANGPTL4/FIAF (PA1-1053) from Thermo Fisher Scientific (Waltham, MA) followed by rabbit secondary antibody from Santa Cruz Biotechnology (Dallas, TX), and were visualized by chemiluminescence (PerkinElmer; Billerica, MA). To confirm equal protein loading and transfer, membranes were stripped by ReBlot Plus from MilliporeSigma (Burlington,MA) and re-probed using anti-HPRT antibody (sc-376938) from Santa Cruz Biotechnology (Dallas, TX). Densitometric analysis was accomplished with image analyzer software coupled with the Fluorchem FC800 system (Alpha Innotech).
2.5. Bile acid analysis
Quantitative bile acid analysis was done on serum and liver samples using the Biocrates Bile Acids Kit. The Duke Proteomics and Metabolomics Shared Resource (Durham, NC) services were utilized for this purpose. The kit measures 20 bile acids out of which 3 are murine specific (Supplemental Table 5). Selective analyte detection was accomplished by use of a triple quadrupole tandem mass spectrometer operated in Multiple Reaction Monitoring (MRM) mode, in which specific precursor to product ion transitions are measured for every analyte and stable isotope labeled internal standard.
2.6. Gut microbiome analysis
A total of 120 mice equally divided into 8 groups (n=15 per group) were included to study the gut microbiome. Half of the groups were treated with antibiotic cocktail containing vancomycin (500 mg/L), neomycin (1 g/L) and primaxin (500 mg/L) by mixing in the animals’ drinking water [28] and replacing with fresh antibiotics on alternate days. All the mice were fed with control chow diet (CD) for 3 weeks. At the end of 3 weeks, one group without antibiotics treatment (3wkCD) and one with antibiotics treatment (3wkAbCD) were euthanized, their whole cecum contents were extracted and flash frozen. Beginning from 4th week, two groups were fed with high fat diet (CHFD) and two other groups were fed tempol high fat diet (THFD) instead of CD, and a group from each diet received antibiotics. At the end of 6 weeks all the six groups of mice were euthanized, and their cecal contents were extracted. The samples were labeled with their diet codes CD, CHFD and THFD and by prefixing with Ab to distinguish the antibiotics treated groups, (see Supplemental Fig. S2).
Microbial genomic DNA was extracted from the cecum contents using the DNeasy PowerSoil Kit (Qiagen, Germantown, MD). The extracted DNA was subjected to microbiome sequencing at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on Illumina MiSeq according to the manufacturer’s guidelines and the sequence data were processed by MR DNA analysis pipeline. Briefly, the microbiome was assayed by sequencing 16S rRNA gene V4 variable region using PCR primer pair 515/806, the sequences were joined, depleted of barcodes, sequences <150 bp and with ambiguous base calls were removed, denoised, and chimeras were removed.
About 13,000 Operational taxonomic units (OTUs) were obtained by clustering at 97% similarity, which were taxonomically classified using BLASTn against a curated database derived from RDPII and NCBI (http://rdp.cme.msu.edu and www.ncbi.nlm.nih.gov). The counts of species, genus, family, order, class, or phylum were calculated as aggregate sum of respective OTU counts. There were 656 bacterial species in 14 phyla present at least in one of the 120 samples. The total counts of individual samples widely varied between 700 and 300,000 due to antibiotics treatment and diet variations. The percent (%) abundances of flora in a sample were calculated relative to its total counts.
Statistical analysis of the gut microbiome data was performed in R software environment (http://www.cran.org). The Shannon diversity index (H), evenness and richness values were estimated using vegan package [29]. Principal component analysis (PCA) was performed using mixOmics package [30]. The count data were centered and scaled to have equal weights to all species. The projection on the 3 principal components (PCs) accounting for highest variance is shown as a 3-dimensional graph where the points represent samples. The samples within each group are enclosed by ellipsoids covering 95% probability. Hierarchical clustering of 50 species having significant differential abundances between any two of CHFD, THFD, AbCHFD and AbTHFD groups (ANOVA or Wilcoxon rank sum test p < 0.001, mean counts ≥100 and present in ≥7 samples in a group) was performed using the data transformed to logarithmic scale. The data were mean centered and clustered by average linkage algorithm using 1-correlation as distance metric [26].
2.7. Additional statistical analysis
Error bars shown for qPCR data were determined from 95% confidence intervals. Ratio values and error bars were determined by the propagation of error formula for the bile acids. All other statistics were calculated using the mean and SEM (standard error of mean). A Student’s t-test was used to the determine statistical significance.
3. Results
3.1. Tempol restricts weight gain and lipid accumulation in HFD fed mice
Fig. 1A shows the effect of using a CHFD (88% saturated fats/12% unsaturated fats) ± 10 mg/kg of Tempol bacon flavored chow in female C3H mice over a 120-day period. Mice on a CHFD gained significant weight for the first 70 days (phase I) (from 20 to 45 gm), while the mice on the THFD only gained 5 gm during this period.
Fig. 1. Tempol mitigates weight gain in HFD fed mice.
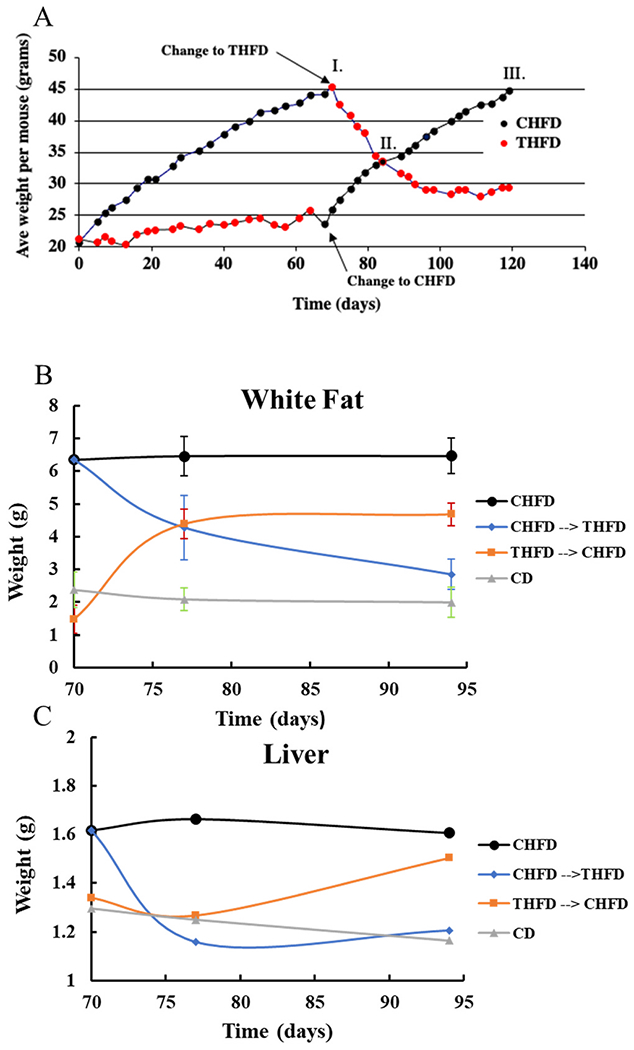
A) Mice fed CHFD chow gain more weight than those fed THFD. When the diets are switched the weights are also reversed. B) The weight of the inguinal white fat and C) weight of liver follows the same trend as the total body weight.
One clear difference in the livers between the THFD and CHFD animals on day 70 was the size difference of the lipid droplets measured. Figs. S1A and S1B shows Oil Red O staining of liver sections from the CHFD (A) and THFD mice (B). Animals on Tempol had significantly smaller lipid vesicles area (25 μ2) versus the lipid droplet area measured in the CHFD animals (>150 μ2) (Fig. S1E). In addition to the reduced i-WF measured in the THFD animals, the adipose cell size was also altered by Tempol (Figs. S1C and D). The adipose cell area measured for the Tempol animals was approximately 35% reduced compared to the CHFD animals (Fig. S1F). Overall, the adipose cell structure observed in the THFD animals was very similar to that found in the CHFD animals except that the size was reduced.
Starting at day 70 (phase I) RNA was isolated from the livers of both groups of mice (CHFD and THFD) and the diets were switched so that mice on the CHFD diet were switched to the THFD diet and the reverse for the mice on the THFD diet. On day 85 of the switched diets the animals in both groups had similar weights (phase II) and thus RNA was isolated from livers for microarray analysis. The mice were continued to be followed for an additional 34 days and the experiment was terminated, and livers isolated for microarray analysis (phase III). Fig. 1B shows the changes in the inguinal white fat adipose tissue (i-WF) isolated at 3 different time points (70, 77, and 94 days). The mice maintained on the CHFD for the full 94 days had significant i-WF (6.5 gm) which remained constant from day 70 to day 94. Mice on the THFD diet had the least i-WF (<1.5 gm) which increased rapidly when the diets were switched to CHFD, such that by day 94 the i-WF was around 5 gm (233% change). The CHFD mice went from 6.2 gm to 4.5 gm (day 85) to 3 gm at day 94 when switched to the THFD diet. White fat from mice on the CD diet averaged about 2.2 gm from day 70 to day 94.
The liver showed similar changes as did the weights and i-WF when diets were switched (Fig. 1C). Unlike the i-WF the changes were more modest (1.2 gm to 1.6 gm ~33% difference), but an increase was seen when changing from the THFD to CHFD and a similar decrease was observed for the CHFD to THFD.
3.2. Liver and adipose tissue genes are regulated by tempol
At days 70, 85, and 119 animals were euthanized, and RNA isolated from livers for microarray analysis. Fig. 2 shows the results from the combined time points with the CHFD group on the left (CHFD-D70 (I), THFD-CHFD-D85 (II), and THFD-CHFD-D119 (III)) and the THFD group on the right (THFD-D70 (I), CHFD-THFD-D85(II), and CHFD-THFD-D119 (III)). There were some 598 annotated genes selected which were at least 2.5-fold up or down and used for hierarchical clustering (Fig. 2). It was clear that large clusters of genes were downregulated in the CHFD group. It is important to note that there were 2 key differences when comparing CHFD versus THFD at day 70: 1) the presence of Tempol in the THFD group and 2) the large difference in the adipose tissue present in CHFD vs THFD (6.5 vs 1.5 grams, respectively). Several, smaller clusters of genes of interest were identified. Cluster A includes genes involved in fatty acid synthesis (Fasn, Elovl6, and Me1) while the other clusters (B-D) included genes known to control appetite and fatty acid uptake and synthesis/oxidation (Lipocalin-2, Vanin1, Hadha, Hadhb, and Hsdl2) [31–34]. The most interesting changes occurred when the diets were switched (CHFD-THFD-D85 (II) and THFD-CHFD-D85 (II)) since at this point the weights of the animal (and the i-WF) were very close in both groups (35 grams, Fig. 1A, B). Table S1 shows the genes which underwent the greatest changes when comparing the CHFD-THFD-D85 (II) group to the THFD-CHFD-D85 (II) group. It was found that the genes involved in FA synthesis (Fasn, Me1, and Evovl6) were rapidly increased in the group which was switched from THFD to CHFD while the opposite happened in the CHFD to THFD. Conversely, a gene involved in appetite suppression (Lipocalin-2) was up-regulated in the THFD group as well as other genes involved in FA β-oxidation (Vanin1 and Hadha). Overall, it appears that the rapid loss of weight and adipose tissue after switching to a Tempol diet occurs because of an increase in FA β-oxidation, while in the group switched to the CHFD the liver gene profile showed an activation of FA synthesis.
Fig. 2. Liver genes are modulated by Tempol.
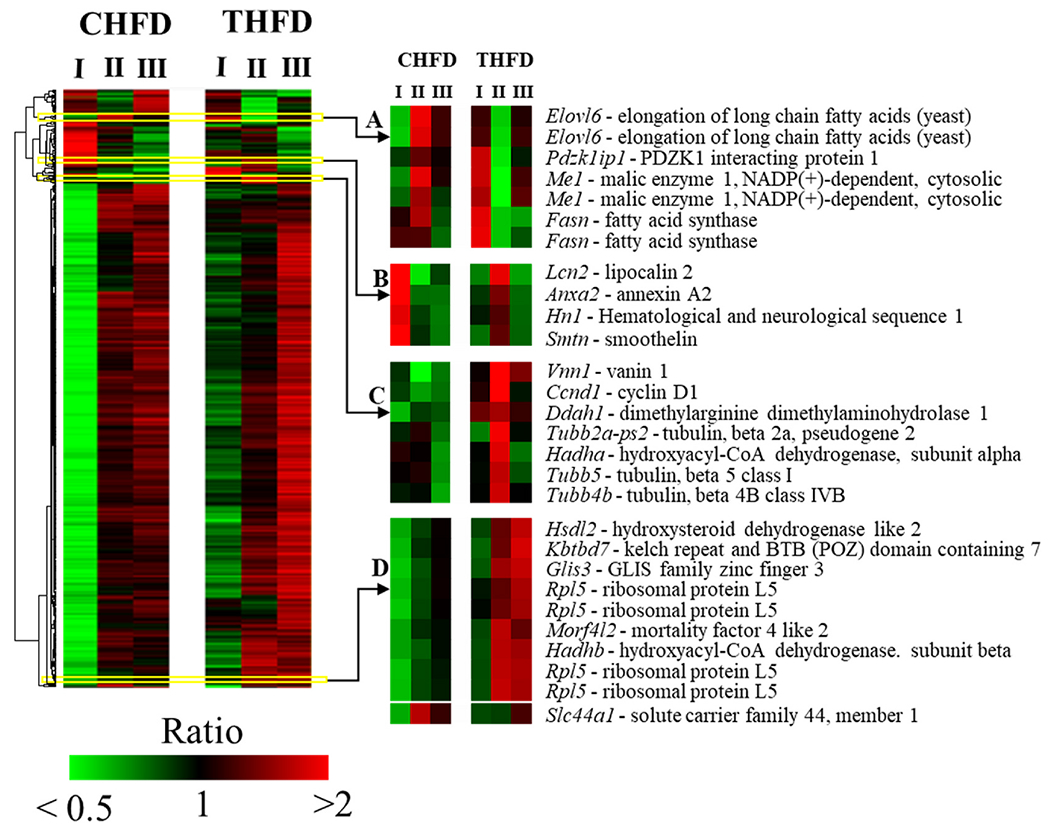
Heat map of 598 features altered by 2.5-fold at one-way ANOVA p < 0.01. Mean centered logarithmic expression ratios are shown in the heat map. Features were clustered by average linkage algorithm using 1-correlation as distance metric. CHFD – control high fat diet. THFD – Tempol + high fat diet. I, II, and III are the time points from Fig. 1A. I – day 70 when the CHFD and THFD fed animal weights were 45g and 25g respectively; II – day 85 when animals reached equal weights by switching CHFD and THFD; III – day 120, continuation of the respective diets until animal weights on CHFD increased to 45g.
More targeted gene array studies were carried out on adipose tissue measuring multiple genes involved in FA oxidation (Fig. 3). It was found that at day 70 in the THFD group versus the CHFD group, a group of adipose tissue genes involved in FA β-oxidation were up-regulated, mostly around 3 to 5 fold, but two genes were sharply increased by 10 to 15 fold (Acsm3 and Acsm5). In addition, when these same genes were found on the microarray chip used in the liver gene analyses, that data were also included in Fig. 3. There was a good correlation between the up-regulation of FA β-oxidation genes in the adipose tissue with the same genes found in the liver.
Fig. 3. Liver and adipose tissue fatty acid metabolism genes are regulated by Tempol.
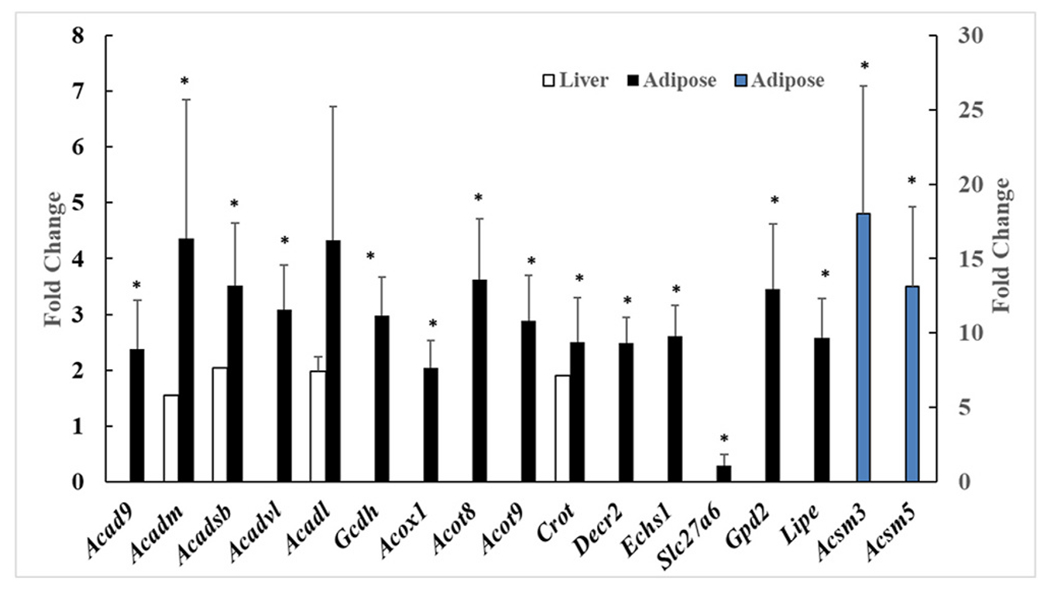
Fold change between Tempol HFD and CHFD mice for both adipose and liver genes involved in fatty acid metabolism. The genes Acsm3 and Acsm5 (blue bars) are measured on the secondary axis to the right. * = p<0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Obesity related proteins are modulated by tempol
ALDH1A1 and truncated FIAF proteins, both involved with fat metabolism [35–42], were detected in adipose tissue after 30 days on the HFD by western blotting and quantitated (Fig. 4A and B). Fig. 4A shows that with THFD ALDH1A1 was significantly reduced by 80% compared to the day 30 CHFD animals (p-value < 0.01). The truncated FIAF protein levels in adipose tissue were also measured, and unlike the ALDH1A1, was increased in the THFD group by 710% compared to the day 30 CHFD group (p-value < 0.01) (Fig. 4B).
Fig. 4. Adipose tissue proteins are influenced by Tempol.
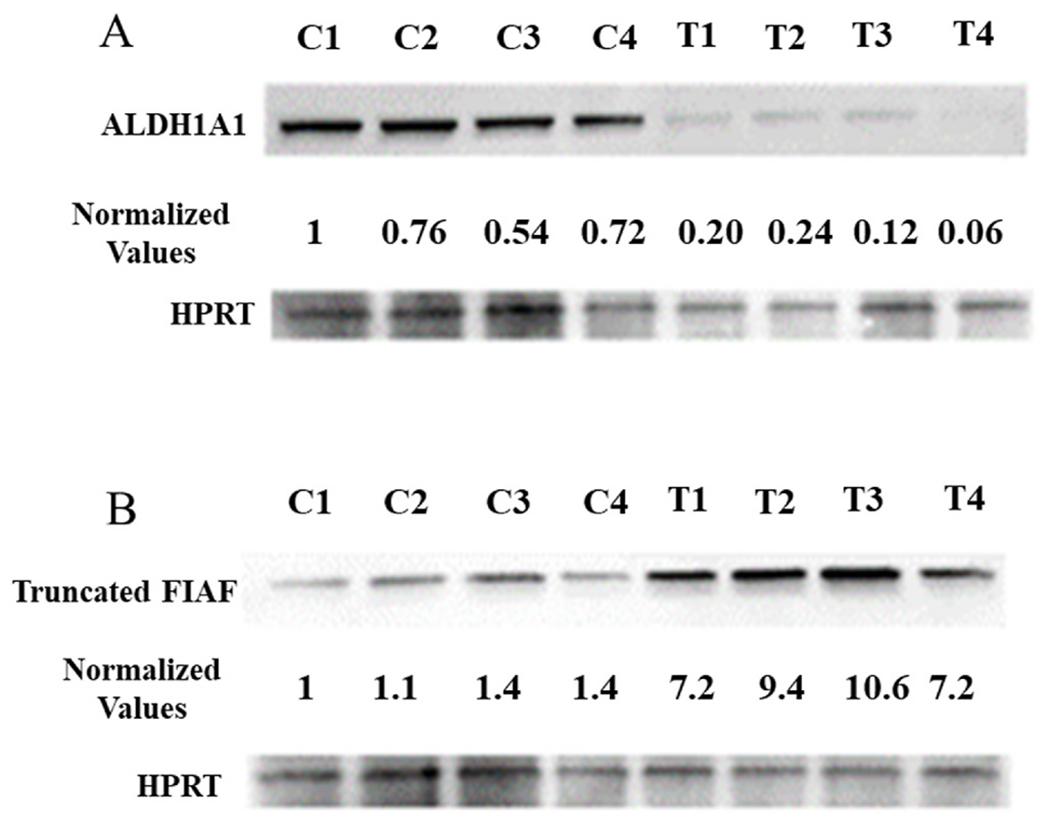
Tempol modulates ALDH1A1 (A) and truncated FIAF (B) protein expressions in the adipose tissue of mice fed HFD. Adipose tissue was collected after 30 days on either a CHFD (C) or a THFD (T) diet. Normalized values were derived by correcting the optical density of each band by the corresponding loading control.
3.4. Bile acids in the liver and serum are altered by tempol
It was clear that Tempol was exerting a significant impact on processes that increased FA oxidation and the storage and utilization of FA by the mice in both the liver and adipose tissue. One group of compounds which are known to strongly affect both these pathways are the bile acid/salts – the primary bile acids produced by the liver (cholic acid (CA) and chenodeoxycholic acid (CDCA)) and also the secondary bile acids which are produced by the gut microbiome (deoxycholic acid (DCA) and lithocholic acid (LCA) [19,21,22]. Fig. 5A and B shows the measurement of different bile acid/salts in liver and serum from mice on either CHFD or THFD after 70 days.
Fig. 5. Tempol regulates bile acids in liver and serum.
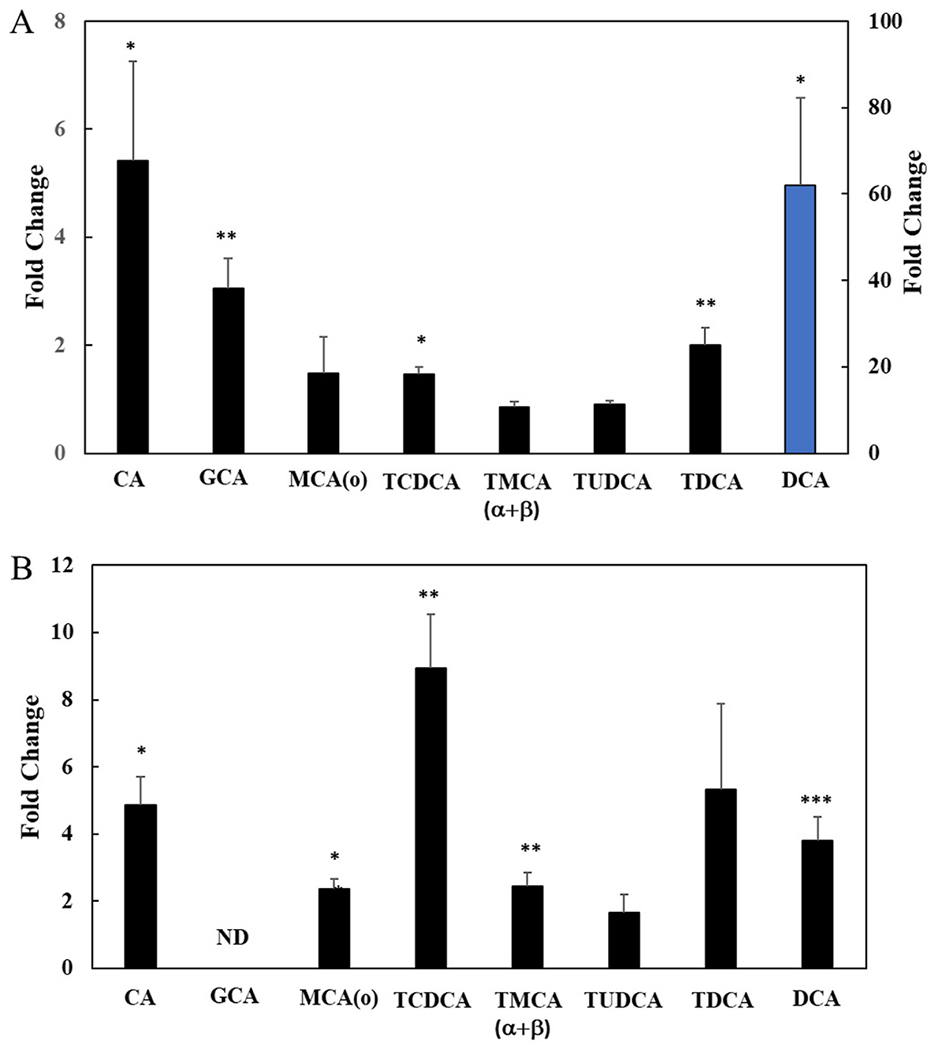
Fold change in bile acids between Tempol fed C3H mice compared to control mice in liver and serum samples (n=3). A) Liver and B) Serum. * = p<0.05, ** = p<0.01 and ***p < 0.001. DCA in panel A (blue bar) is measured with secondary axis to the right. GCA was not detected in serum. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
There was a significant increase in DCA, CA and GCA (glycine conjugated CA) in the liver of the THFD animals (60, 5 and 3-fold, respectively, p<0.05) (Fig. 5A). Other bile compounds were not significantly increased including tauromuricholic acid (α+β) (TMCA (α+β)) which, in an absolute sense, was the highest measured bile acid (BA) present in liver (72 μg/kg). The serum also showed similar elevations of the BA’s measured, although the levels in 4 of 7 BAs were significantly lower than found in the liver (Fig. 5B). Overall, the data suggested that Tempol was having a significant effect on BAs in the liver and DCA was the most impacted by Tempol (Fig. 5A).
3.5. Tempol alters the gut microbiome
Since a secondary bile acid, DCA, a gut microbiome produced compound, was significantly influenced by Tempol we decided to investigate the effect of Tempol on the gut microbiome after 3 weeks on CHFD, 3 weeks on THFD, or 3 weeks or 6 weeks on a conventional plant-based diet (CD) (see schema in Fig. S2). The animals were all maintained on the CD diet for 3 weeks (3wkCD) and then switched to CHFD, THFD, or maintained for 3 more weeks on the CD diet. In addition to the different diets, 4 more groups were also treated with a cocktail of three different antibiotics for 3 weeks using a CD diet and then switched to the CHFD, THFD or maintained on the CD diet so that 3 of the diet groups had been exposed to 6 weeks of antibiotics (CD, CHFD, and THFD). At the end of the time on each diet, the cecum content from each animal was isolated for ribosomal 16S rRNA pyrosequencing and the microbial DNA extracted and quantitated. Fig. 6A shows the total 16S rRNA pool for the 4 diet conditions.
Fig. 6. Gut microbiome population is influenced by Tempol and antibiotics.
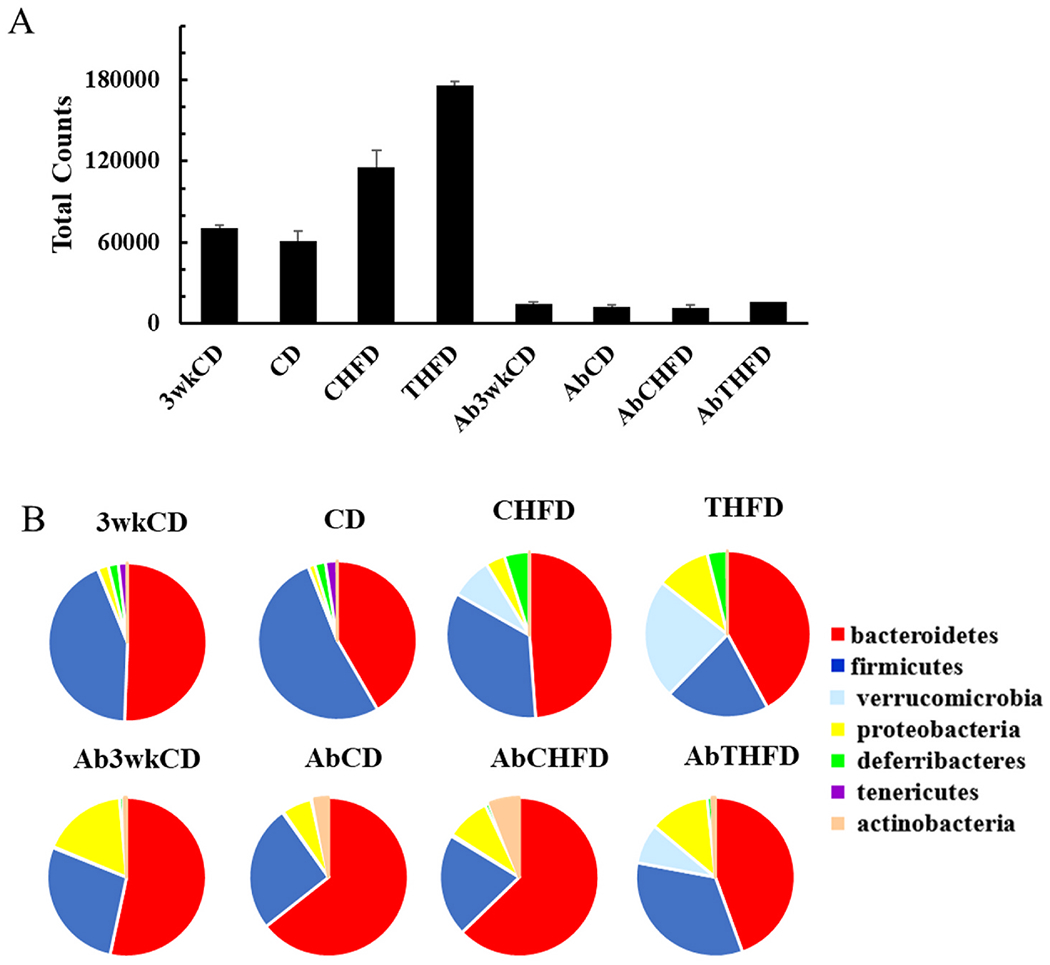
A) The total 16S RNAseq counts determined at the phylum level for control chow at 3 weeks (3wkCD), control chow at 6 weeks (CD), high fat diet (CHFD), high fat diet + Tempol (THFD). The Ab groups were treated with antibiotics for either 3 or 6 weeks with each diet. Error bars are the SEM (N=15). B) Phylum level distribution of the top 7 categories.
As expected, the CD 3 weeks and 6 weeks were very similar while the CHFD diet led to an expansion of the bacterial pool (1.9-fold versus CD). The addition of Tempol (THFD) further increased the bacterial pool significantly (2.75-fold compared to CD). Antibiotic treatment for 3 weeks prior to any diet change and for an additional of 3 weeks post diet change reduced the total 16S rRNA pool by over 80% in the 3 weeks and 6 weeks CD group and by over 84% in the CHFD and THFD group (Fig 6A and Table S2). During the treatment with the antibiotic cocktail the change in weights of the mice in the different diet groups was determined over the last 3 weeks period (Fig. S3). Fig. S3 clearly shows that although there were substantial decreases in the total bacterial loads for all diet groups there was no change in weight increases for animals on the CHFD diet and furthermore, Tempol was still capable of preventing this weight increase. Bacterial composition was initially examined by the effect of diet on the OTU abundances of the 7 phylum level categories (Fig. 6B).
The CD 3 weeks and 6 weeks group had very similar phylum groupings with only small changes in the Firmicutes and Bacteroidetes ratios. The CHFD had significant effects on many of the phylum categories with Firmicutes being much reduced while Verrucomicrobia, Proteobacteria, and Deferribacteres groups increased substantially compared to the CD diet (130, 2.9, and 2.2-fold, respectively). Changes in the Proteobacteria and Deferribactres groups were also increased using THFD (7.8 and 1.8-fold, respectively) but with the Verrucomicrobia showing much greater increased compared to the CD diet (374-fold). Using the antibiotic cocktail significantly changed the phylum distributions with the CD and CHFD having similar distributions, while the THFD diet mice, still had elevated Verrucomicrobia and Proteobacteria when compared to the AbCD group (36 and 2-fold, respectively) (Fig. 6B). The increase in Verrucomicrobia seen in the CHFD group (130-fold) was completely lost with the addition of the antibiotics to both the CHFD and CD groups.
Over 170 species from the OTU’s were identified using the 3 diets (CHFD, THFD, and CD) and various Shannon diversity indexes were calculated and shown in Figs. S4A–C. The CD diet was used as the baseline gut microbiome set and had the greatest Shannon evenness or richness index. The CHFD diet substantially decreased all 3 indexes with regards to the CD diet. Tempol acted to prevent the large decreases in these indexes caused by the CHFD (although not completely). With the addition of antibiotic treatment all 3 indexes were similar across the diets used (Figs. S4D–F).
Fig. S5 shows the principal component (PCA) map of 100 species identified for the 4 different diet groups. It was possible to readily separate the 3 diet groups into distinct clusters while the 3wkCD and CD group clustered very close together (Fig. S5A). However, when antibiotics were included in the diet it was seen that 3 of the groups were quite similar (3wkCD, CD and CHFD), while the Tempol group still segregated from the other 3 groups (Fig. S5B).
Out of 170 species identified by the 16S rRNA sequencing study 50 species were selected which were altered by 5-fold (p<0.001) in any of the high fat diet groups studied (antibiotics included) and used to hierarchically cluster these species to find the influence that Tempol had on specific gut bacteria (Fig. S6). First, the influence of the HFD is apparent in both the CHFD and THFD groups. Second, there was a distinct group of species which was negatively impacted by CHFD (box II) but protected by the inclusion of Tempol (THFD). These species included Lactobacillus intestinalis, Lactobacillus reuteri, Clostridium symbiosum, and Clostridium hathewayi. A small group of bacteria which was elevated by the CHFD diet was further increased with Tempol (Akkermansia muciniphila, Kopriimonas spp., and Bilophila wadsworthia) (box I). Finally, the antibiotic groups demonstrate that most species were strongly decreased by the cocktail of antibiotics (Table S2); however, here again, Tempol did moderate the reduction of several species after 3 weeks of antibiotic treatment.
The overall observation is that the HFD dominated the gut microbiome response and that Tempol amplified increases in certain phyla (Verrucomicrobia and Proteobacteria), while protecting or at least mitigating the loss of other genera such as Clostidrium and Lactobacillus. Fig. 7A examines the relationship between THFD and CHFD diet using the CD group as the gut microbial reference set.
Fig. 7. Relationship between gut microbiome population in CD, CHFD and THFD mice.
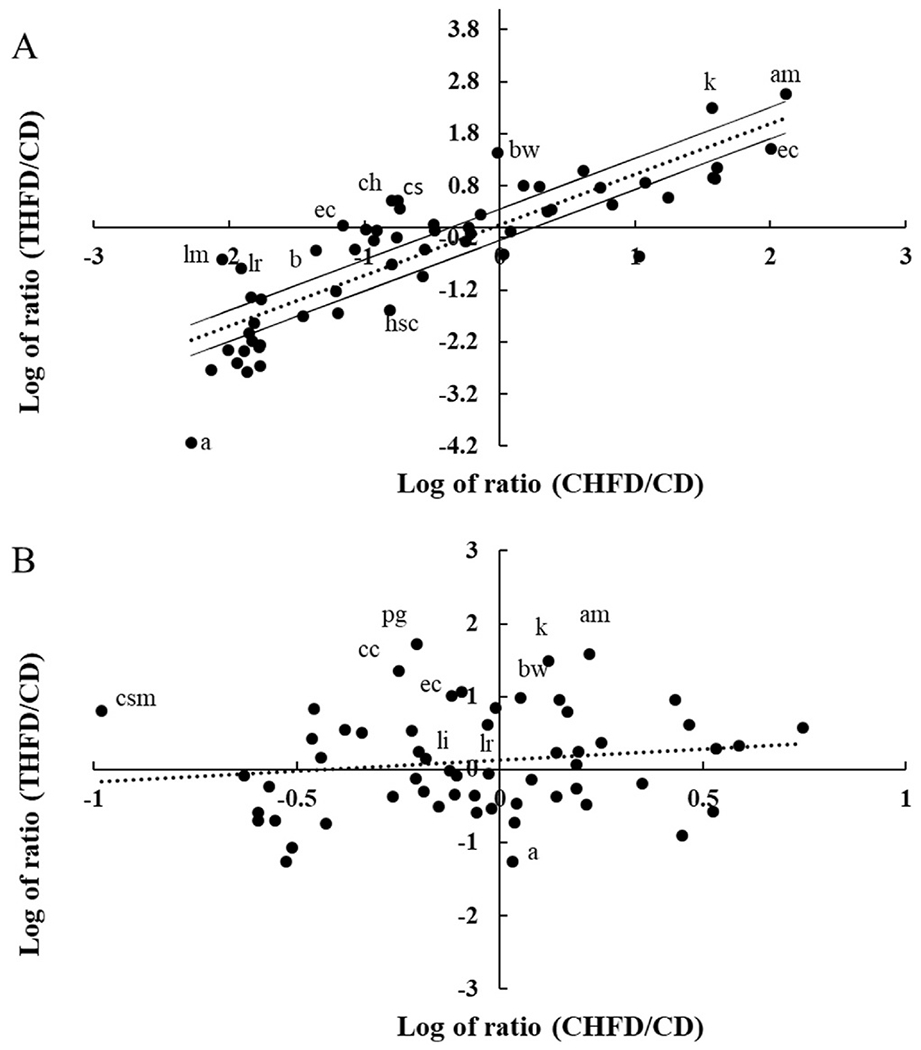
A) Scatter plot of the log ratio of THFD/CD (total counts for species from the Tempol + high fat diet compared to the 6 weeks control chow) vs the log ratio for CHFD/CD (total counts for species on the high fat diet compared to 6 weeks control chow). B) Same conditions as (A) but with anti-biotics added. Dotted line is the linear regression line and solid lines are 2-fold up/down from the calculated regression line. Abbreviations are a: Anaeroplasma spp., ac: Anaerostipes caccae, am: Akkermansia muciniphila, b: Barnesiella spp., bw: Bilophila wadsworthia, cc: Clostridium cocleatum, ch: Clostridium hathewayi, cs: Clostridium symbiosum, csm: Candidatus Stoquefichus massiliensis, ec: Escherichia coli, hs: Hydrogenoanaerobacterium saccharovorans, k: Kopriimonas spp., li: Lactobacillus intestinalis, and lm: Lactobacillus murinus, pg: Parabacteroides goldsteinii.
Since a HFD was used in both groups there was a high degree of correlation between the 2 groups; however, the high fat component could only account for some 72% of the variability (R2 = 72.4%). Clearly, Tempol increased some species above that modulated by the CHFD diet (Akkermansia muciniphila ratio was 374 vs 130 with CHFD), while in a few cases Tempol alone had a substantial effect while high fat alone did not (Bilophila wadsworthia ratio was increased 27-fold with THFD vs 1.0 for the CHFD diet). Overall, out of 78 species with greater than 10 counts, Tempol increased 40 species by greater than 2-fold, caused minimal changes in 20 species, and decreased 18 species by 2-fold or more. When species data from the antibiotic cocktail was plotted, the effect of Tempol was still evident but now there was almost no correlation (R2 = 2.2%) with the high fat component (Fig. 7B). Hence, even while the bacterial numbers were substantially decreased, Tempol still influenced the overall species composition and the weight gain compared with the high fat diet (Fig. S3).
Fig. 8 shows a hierarchal cluster map in all 29 animals for the CHFD and THFD diet groups of 27 of the most altered species caused by Tempol consumption. Genus such as Akkermansia, Kopriimonas, and Bilophila were increased by Tempol compared to the HFD alone (21.9% vs 8.1%, 2.1% vs 0.41%, and 1.8% vs 0.07%, respectively) while Tempol protected Lactobacillius from the decrease observed with CHFD diet (2% vs 0.02%, respectively).
Fig. 8. Most abundant gut microbiome species in CHFD and THFD mice.
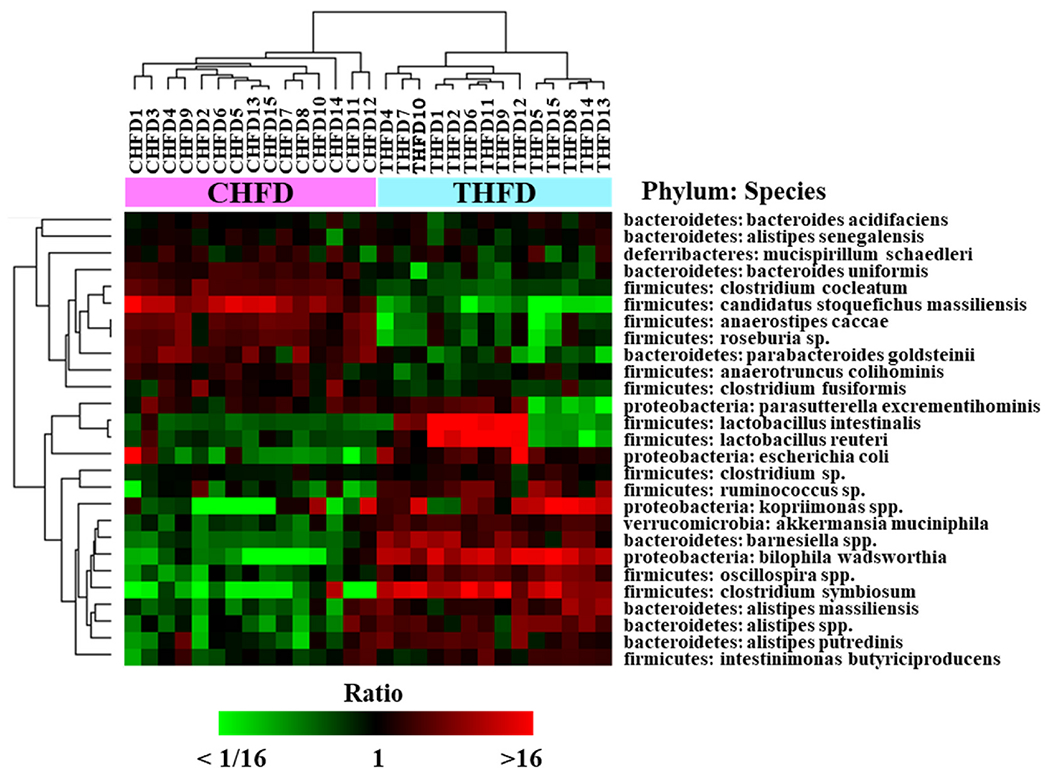
Hierarchical clustering of 27 species highly abundant in CHFD and THFD (maximum abundance of all samples >1.0%). Clustered by average linkage algorithm using 1-correlation as distance. Heat map shows mean centered log2 (abundance %) values.
Fig. S7B (A, B) shows the results for the abundance of the Lactobacillus (Fig. S7A) and Clostridium cluster IV (Fig. S7B). It was found that the high fat diet substantially reduced these genera while the addition of Tempol provided some protection from these decreases. Figs. S8A–G shows the 4 diet groups (CHFD, AbCHFD, THFD, and AbTHFD) and the effect that Tempol had moderating both the effect of the HFD and antibiotics on individual species.
3.6. Tempol restricts weight gain in ovariectomized mice
To examine what role estrogen might play in influencing the weight effects by Tempol, we used ovariectomized mice (which lacked normal physiological levels of estrogen [43]) and placed them on a CD or Tempol-TCD (10 mg/kg) diet. Figure 9S shows the weights of the 2 groups over a 15 week period. It was observed that in the TCD fed ovariectomized mice the weight gain was lower over the 15 weeks treatment duration as compared to the CD fed ovariectomized mice.
4. Discussion
The beneficial effects of the nitroxide Tempol are well documented. Tempol has been shown to be a potent radical scavenger and has biologically relevant effects on limiting the deleterious mutations caused by carcinogens [44], providing radiation protection [8], reducing oxidative stress [12–14], and imparting favorable therapeutic outcomes in a variety of animal models of diseases [45–53].
This study focused on potential mechanism(s) of obesity prevention by Tempol in female C3H mice fed a HF diet. Previously, we carried out in vitro studies with Tempol in 3T3-L1 cells and showed that Tempol reduced differentiation of the 3T3-L1 cells to adipose cells and inhibited lipid storage and adipogenesis factors [54]. These 3T3-L1 changes were only observed when using concentrations of Tempol >0.5 mM and, once the 3T3-L1 cells had formed lipid vesicles, even these high concentrations were unable to reduce the size of the lipid vesicles [54]. Kim et al. also showed that, in vivo, Tempol had profound effects on both the weight and inflammatory cytokine profile of ApoE −/− female mice (17). Both these studies showed that Tempol had significant effects on adipose tissue but the mechanism may involve direct as well as secondary effects of Tempol on metabolic systems.
It has been established that the gut microflora has profound effects on obesity in mice and humans [18,55]. Previous work has investigated the effect of Tempol administered both for 5 days by gavage [19] and for longer than 30 days when added to the drinking water [16]. Both CD and HFDs have been studied [19]. A major difference between this study and previous reports is the use of female C3H/Hen mice compared to the use of male C57BL/6N or C57BL/6J mice [19–22]. One of the mechanisms outlined regarding the anti-obesity effect of Tempol centered on the effect of Tempol on the gut microflora which in turn altered the synthesis and secretion of certain conjugated bile acids [19]. Li et al. [19] showed that the genus Lactobacillus and Clostridium were greatly reduced by Tempol consumption leading to a reduction in BA hydrolases, which in turn led to an increase in the conjugated BAs TMCA-α and TMCA-β. These BAs were shown to be FXR antagonists, which according to these authors, led to an enhancement of fatty acid oxidation and adipose tissue reduction. While this mechanism may be important in male mice, the results in the current study suggest a different group of bile acids may be responsible for the anti-obesity effect of Tempol seen in female mice.
First, we observed in C3H mice that 10 mg/kg Tempol in the HFD results in significantly lower body weight gain, and a significant reduction of adipose tissue. Second, the transition period after switching diets was marked by rapid changes in genes involved in fatty acid synthesis and oxidation both in the liver and adipose tissue directly (Figs. 2 and 3). Tempol also altered several proteins known to be important in lean phenotypes (ALDH1A1) [56] and FIAF [41]. The role of ALDH1A1in obesity has been widely documented [36–38,57–59]. ALDH1A1 is a key protein involved in retinoic acid synthesis in white fat and 1) suppression leads to decreased obesity and a positive metabolic outcome while, 2) HFD up-regulates it [36,38]. The adipokine FIAF is a member of fibrinogen/angiopoietin-like proteins and is a lipoprotein lipase inhibitor [60,61]. It is abundantly present in adipose tissue. FIAF inhibits the activity of Lpl (Lipoprotein lipase), a regulatory enzyme that enhances the absorption of fatty acids and the build-up of adipocyte triglycerides [41]. Hence, an increase in FIAF would be expected to reduce the size of the adipose cells. Finally, plasma FIAF has been reported to be downregulated by chronic HFD feeding [62]. Our observations that THFD decreased ALDH1A (Fig. 4A) and increased FIAF (Fig. 4B) are consistent with the reduction in adipose tissue and therefore a reduction in the overall obesity. Given the importance of BAs reported in other mouse models treated with Tempol, we examined the profiles of BAs in the livers and serum of the THFD versus the CHFD mice (Fig. 5A and B). Unlike the previous report [19], we did not find significant differences in TMCA (α+β), but found differences in the primary bile acid cholic acid (CA) and the secondary bile acid deoxycholic acid (DCA) (Fig. 5A and B). CA is of interest since when it was incorporated into the HFD food and fed long term to male C57BL/6J mice the weight profiles observed were very similar to our Tempol effect (both before and after the diet switching) (Fig. 1A) [63]. Interestingly, Li et al. also noted an increase in CA in the intestine after Tempol administration [19]. In addition, DCA in particular, suggested a strong involvement of the gut microbiome (since conjugated bile acids produced by the liver must first be deconjugated before the dihydroxylation step to produce DCA [64]). Therefore, a major part of this study involved a mapping of the effect of Tempol on the composition of the gut microbiome in female C3H mice.
CHFD diet induced large and significant changes in the gut microbiome as seen at the phylum level in female C3H mice (Fig. 6B). Similar to a previous report [65] the CHFD diet had large effects on the Proteobacteria and Verrucomicrobia phylum at the expense of the Firmicutes phylum (Fig. 6B). The only species in Verrucomicrobia is Akkermansia muciniphila and this species came to represent 8.3% of the total species detected in the CHFD group (Table S3). Although Akkermansia is associated with lean phenotypes and obesity prevention [66], others have also noted that a HFD can result in its increase [65,67,68]. One possibility was the level of unsaturated fatty acids in our high fat diet (12% of the fats used (Table S4)). The addition of Tempol to the HFD led to a remarkable expansion of the Akkermansia such that it accounted for some 23% of the species abundance (Table S3). The CHFD diet also led to a significant reduction in the Lactobacillus genus and the Clostridium cluster IV group (Figs. S7A and B) similar to a finding by Rodríguez-García et al. [68]. An important observation was that Tempol moderated the toxicity of the HFD on the Lactobacillus species (by some 5-fold) (Fig. S7A). Thus, in contrast to the earlier results by Li et al. [19] Tempol administration to female C3H mice was not toxic to the Lactobacillus genus. A regression of the species altered by the THFD against the species in the CHFD demonstrated the strong influence that the HFD had on both groups as some 72% of the variation could be assigned to the high fat variable, but this left some 28% of the variation in which Tempol certainly had a strong influence (Fig. 7A). Three species were strongly altered by Tempol, Akkermansia muciniphilia, Kopriimonas spp, and Bilophila wadsworthia. In fact, since the CHFD had no effect on Bilophila wadsworthia, the increase must have been due to Tempol alone. Others have shown HFDs increase Bilophila, which may account for an increase in inflammatory conditions noted [69]. Tempol, as reported, is a highly effective cyclic antioxidant, thus it may act to reduce some of the undesirable side effects of an expansion of the Bilophila genus. We also found a very interesting association between Akkermansia muciniphila and the secondary bile acid DCA. It has been reported that DCA increases the growth of Akkermansia muciniphila and helps in its survival in the gut [70]. In our experiments, DCA in Tempol treated animals was found to be substantially increased. This increase in DCA may be an important cause of the expansion of Akkermansia when Tempol is present. In addition, DCA is a high affinity ligand for FXR and can activate it for both increases in fatty acid oxidation and metabolism as well to act as an immunoregulator capable of reducing various inflammatory cytokines, thus acting to inhibit IBD (inflammatory bowel disease) [71]. Thus, the combination of a direct antioxidant and the up-regulation of proteins known to control inflammation would be a powerful overall inflammation suppressor.
The cocktail of antibiotics significantly altered the gut microflora, yet Tempol still reduced the weights while the CHFD mice continued to become obese (Fig. S3). In contrast, amongst the three diet groups, the treatment with the antibiotics equalized all three Shannon diversity indexes (Figs. S4D–F). Even under these conditions where the HFD effect on the gut microbiome was greatly reduced, there were still species changed by Tempol (Akkermansia, Kopriimonas, and Bilophila). It is possible that Tempol reduced the efficacy of one of the antibiotics used in this study; however, it should be stressed that a majority of species studied which received the Tempol diet still had a positive antibiotic effect (defined as reducing the total counts of bacteria measured in the diet versus the counts for the diet + antibiotics) (Table S2).
Since female mice were used in this study it seemed appropriate to determine whether estrogen played a role in the significant weight alterations seen in Tempol fed female mice. Estrogen has been reported to interact and activate the trifunctional hydroxyacyl CoA dehydrogenase b subunit (Hadhb) which is involved in fatty acid oxidation by mitochondria [72,73]. Hadhb was identified as one of the genes up-regulated when obese C3H mice were switched to the Tempol diet (Table S1). From Fig. S9 it appears that the normal level of estrogen is not necessary for the Tempol weight effect as the weight gain in ovariectomized mice was reduced by Tempol. Thus, our results in female C3H mice agree with Li et al. [19] in that Tempol acts to prevent adipose tissue expansion primarily at the gut microflora level and may not directly affect metabolic processes in the liver or adipose tissue. Also, similar to what Gonzalez et al. reported, Tempol increased fatty acid β oxidation in the liver [21].
Studies from our laboratory have shown that feeding female C3H mice low dose Tempol in a carbohydrate-based chow over the entire lifespan was not toxic to these mice nor changed chow consumption rate [16]. Indeed, Tempol did act to reduce the incidence of lymphomas after non-lethal whole body radiation exposure [23]. Whether Tempol in HFD is also not toxic over a long time period was not studied in this report. However, it is possible that in addition to its effect on the gut microbiome its strong antioxidant properties may prove beneficial in limiting any deleterious effects caused by alterations in the microbial composition. Additional studies should be carried out to determine if the Tempol effect on bile acids may explain some of the anti-carcinogenesis effects observed in female C3H mice after whole body XRT.
5. Conclusions
Based on our observations, obesity related changes with Tempol were dependent on significant alterations to the gut microbiome. Tempol also altered the bile acid profile increasing specific bile acids (CA and DCA) known to have significant impact on genes involved in fatty acid oxidation and in proteins associated with fatty acid accumulation and metabolism. Ultimately, these changes resulted in the reduction of adipose tissue and obesity. Although Tempol had a significant effect in female mice, the role of estrogen did not appear to be involved with the Tempol mediated effects. Further studies should be done to identify which specific bacterial specie(s) are responsible for the Tempol effect on adipose tissue and whether this effect is gender and strain dependent.
Supplementary Material
Financial support
This research was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute and NIAID Medical Countermeasures against Radiological and Nuclear Threats Program, National Institutes of Health, Bethesda, Maryland 20892.
Abbreviations
- AbCD
Control diet with antibiotics
- AbCHFD
Control high fat diet with antibiotics
- AbTHFD
Tempol high fat diet with antibiotics
- ACSM3
Acyl-CoA Synthetase Medium Chain Family Member 3
- ACSM5
Acyl-CoA Synthetase Medium Chain Family Member 5
- ALDH1A1
Aldehyde dehydrogenase1A1
- BA
Bile acid
- CA
Cholic acid
- CD
Control diet
- CDCA
Chenodeoxycholic acid
- CHFD
Control high fat diet
- DCA
Deoxycholic acid
- Elovl6
Elongation of long chain fatty acids
- FA
Fatty acid
- FASN
Fatty acid synthase
- FIAF/ANGPTL4
Fasting induced adipose factor/angiopoietin-like protein 4
- FXR
Farnesoid X receptor
- HADHA
Hydroxyacyl-CoA dehydrogenase subunit alpha
- HADHB
Hydroxyacyl-CoA dehydrogenase subunit beta
- HFD
High fat diet
- i-WF
Inguinal white fat
- LCA
Lithocholic acid
- LCN2
Lipocalin 2
- Me1
Malic enzyme 1
- THFD
Tempol high fat diet
- TMCA
Tauromuricholic acid
Footnotes
Declaration of competing interest
The authors declare no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2021.12.006.
References
- [1].Goossens GH, The metabolic phenotype in obesity: fat mass, body fat distribution, and adipose tissue function, Obes Facts 10 (3) (2017) 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Piché ME, Tchernof A, Després JP, Obesity phenotypes, diabetes, and cardiovascular diseases, Circ. Res 126 (11) (2020) 1477–1500. [DOI] [PubMed] [Google Scholar]
- [3].Kawai T, Autieri MV, Scalia R, Adipose tissue inflammation and metabolic dysfunction in obesity, Am. J. Physiol. Cell Physiol 320 (3) (2021) C375–c391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jackson VM, Breen DM, Fortin JP, Liou A, Kuzmiski JB, Loomis AK, et al. , Latest approaches for the treatment of obesity, Expet Opin. Drug Discov 10 (8) (2015) 825–839. [DOI] [PubMed] [Google Scholar]
- [5].Sharma G, Prossnitz ER, G-Protein-Coupled estrogen receptor (GPER) and sex-specific metabolic homeostasis, Adv. Exp. Med. Biol 1043 (2017) 427–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sonoda J, Chen MZ, Baruch A, FGF21-receptor agonists: an emerging therapeutic class for obesity-related diseases, Horm. Mol. Biol. Clin. Invest 30 (2) (2017). [DOI] [PubMed] [Google Scholar]
- [7].Mitchell JB, Samuni A, Krishna MC, DeGraff WG, Ahn MS, Samuni U, et al. , Biologically active metal-independent superoxide dismutase mimics, Biochemistry 29 (11) (1990) 2802–2807. [DOI] [PubMed] [Google Scholar]
- [8].Mitchell JB, DeGraff W, Kaufman D, Krishna MC, Samuni A, Finkelstein E, et al. , Inhibition of oxygen-dependent radiation-induced damage by the nitroxide superoxide dismutase mimic, tempol, Arch. Biochem. Biophys 289 (1) (1991) 62–70. [DOI] [PubMed] [Google Scholar]
- [9].Hahn SM, Tochner Z, Krishna CM, Glass J, Wilson L, Samuni A, et al. , Tempol, a stable free radical, is a novel murine radiation protector, Cancer Res. 52 (7) (1992) 1750–1753. [PubMed] [Google Scholar]
- [10].Krishna MC, Grahame DA, Samuni A, Mitchell JB, Russo A, Oxoammonium cation intermediate in the nitroxide-catalyzed dismutation of superoxide, Proc. Natl. Acad. Sci. U. S. A 89 (12) (1992) 5537–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Krishna MC, Russo A, Mitchell JB, Goldstein S, Dafhi H, Samuni A, Do nitroxide antioxidants act as scavengers of O2-. or as SOD mimics? J. Biol. Chem 271 (42) (1996) 26026–26031. [DOI] [PubMed] [Google Scholar]
- [12].Soule BP, Hyodo F, Matsumoto K, Simone NL, Cook JA, Krishna MC, et al. , The chemistry and biology of nitroxide compounds, Free Radic. Biol. Med 42 (11) (2007) 1632–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Soule BP, Hyodo F, Matsumoto K, Simone NL, Cook JA, Krishna MC, et al. , Therapeutic and clinical applications of nitroxide compounds, Antioxidants Redox Signal. 9 (10) (2007) 1731–1743. [DOI] [PubMed] [Google Scholar]
- [14].Wilcox CS, Pearlman A, Chemistry and antihypertensive effects of tempol and other nitroxides, Pharmacol. Rev 60 (4) (2008) 418–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Naz S CJ, Krishna MC, Mitchell JB, Biological applications of nitroxide stable free radicals, in: Ouari O GD (Ed.), Nitroxides: Synthesis, Properties, and Applications, Royal Society of Chemistry, London, 2021, pp. 519–550. [Google Scholar]
- [16].Mitchell JB, Xavier S, DeLuca AM, Sowers AL, Cook JA, Krishna MC, et al. , A low molecular weight antioxidant decreases weight and lowers tumor incidence, Free Radic. Biol. Med 34 (1) (2003) 93–102. [DOI] [PubMed] [Google Scholar]
- [17].Kim CH, Mitchell JB, Bursill CA, Sowers AL, Thetford A, Cook JA, et al. , The nitroxide radical TEMPOL prevents obesity, hyperlipidaemia, elevation of inflammatory cytokines, and modulates atherosclerotic plaque composition in apoE−/− mice, Atherosclerosis 240 (1) (2015) 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI, Obesity alters gut microbial ecology, Proc. Natl. Acad. Sci. U. S. A 102 (31) (2005) 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, et al. , Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity, Nat. Commun 4 (2013) 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cai J, Zhang L, Jones RA, Correll JB, Hatzakis E, Smith PB, et al. , Antioxidant drug tempol promotes functional metabolic changes in the gut microbiota, J. Proteome Res 15 (2) (2016) 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gonzalez FJ, Jiang C, Patterson AD, An intestinal microbiota-farnesoid X receptor Axis modulates metabolic disease, Gastroenterology 151 (5) (2016) 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gonzalez FJ, Jiang C, Xie C, Patterson AD, Intestinal farnesoid X receptor signaling modulates metabolic disease, Dig. Dis 35 (3) (2017) 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mitchell JB, Anver MR, Sowers AL, Rosenberg PS, Figueroa M, Thetford A, et al. , The antioxidant tempol reduces carcinogenesis and enhances survival in mice when administered after nonlethal total body radiation, Cancer Res. 72 (18) (2012) 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chuang YY, Chen Y, Gadisetti, Chandramouli VR, Cook JA, Coffin D, et al. , Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells, Cancer Res. 62 (21) (2002) 6246–6254. [PubMed] [Google Scholar]
- [25].Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. Roy. Stat. Soc. B 57 (1) (1995) 289–300. [Google Scholar]
- [26].Eisen MB, Spellman PT, Brown PO, Botstein D, Cluster analysis and display of genome-wide expression patterns, Proc. Natl. Acad. Sci. U. S. A 95 (25) (1998) 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Koopman R, Schaart G, Hesselink MK, Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids, Histochem. Cell Biol. 116 (1) (2001) 63–68. [DOI] [PubMed] [Google Scholar]
- [28].Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. , Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment, Science 342 (6161) (2013) 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fisher RA, Corbet AS, Williams CB, The relation between the number of species and the number of individuals in a random sample of an animal population, J. Anim. Ecol 12 (1) (1943) 42–58. [Google Scholar]
- [30].Rohart F, Gautier B, Singh A, Ka LC, mixOmics: an R package for ‘omics feature selection and multiple data integration, PLoS Comput. Biol 13 (11) (2017), e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mosialou I, Shikhel S, Luo N, Petropoulou PI, Panitsas K, Bisikirska B, et al. , Lipocalin-2 counteracts metabolic dysregulation in obesity and diabetes, J. Exp. Med 217 (10) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bartucci R, Salvati A, Olinga P, Boersma YL, Vanin 1: its physiological function and role in diseases, Int. J. Mol. Sci 20 (16) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rector RS, Payne RM, Ibdah JA, Mitochondrial trifunctional protein defects: clinical implications and therapeutic approaches, Adv. Drug Deliv. Rev 60 (13–14) (2008) 1488–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kowalik D, Haller F, Adamski J, Moeller G, In search for function of two human orphan SDR enzymes: hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O), J. Steroid Biochem. Mol. Biol 117 (4–5) (2009) 117–124. [DOI] [PubMed] [Google Scholar]
- [35].Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, et al. , Retinaldehyde represses adipogenesis and diet-induced obesity, Nat. Med 13 (6) (2007) 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haenisch M, Treuting PM, Brabb T, Goldstein AS, Berkseth K, Amory JK, et al. , Pharmacological inhibition of ALDH1A enzymes suppresses weight gain in a mouse model of diet-induced obesity, Obes. Res. Clin. Pract 12 (1) (2018) 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mcllroy GD, Delibegovic M, Owen C, Stoney PN, Shearer KD, McCaffery PJ, et al. , Fenretinide treatment prevents diet-induced obesity in association with major alterations in retinoid homeostatic gene expression in adipose, liver, and hypothalamus, Diabetes 62 (3) (2013) 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Haenisch M, Nguyen T, Fihn CA, Goldstein AS, Amory JK, Treuting P, et al. , Investigation of an ALDH1A1-specific inhibitor for suppression of weight gain in a diet-induced mouse model of obesity, Int. J. Obes 45 (2021) 1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xu A, Lam MC, Chan KW, Wang Y, Zhang J, Hoo RL, et al. , Angiopoietin-like protein 4 decreases blood glucose and improves glucose tolerance but induces hyperlipidemia and hepatic steatosis in mice, Proc. Natl. Acad. Sci. U. S. A 102 (17) (2005) 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mandard S, Zandbergen F, Tan NS, Escher P, Patsouris D, Koenig W, et al. , The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment, J. Biol. Chem 279 (33) (2004) 34411–34420. [DOI] [PubMed] [Google Scholar]
- [41].Mandard S, Zandbergen F, van Straten E, Wahli W, Kuipers F, Müller M, et al. , The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity, J. Biol. Chem 281 (2) (2006) 934–944. [DOI] [PubMed] [Google Scholar]
- [42].Janssen AWF, Katiraei S, Bartosinska B, Eberhard D, Willems van Dijk K, Kersten S, Loss of angiopoietin-like 4 (ANGPTL4) in mice with diet-induced obesity uncouples visceral obesity from glucose intolerance partly via the gut microbiota, Diabetologia 61 (6) (2018) 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cousins SW, Marin-Castaño ME, Espinosa-Heidmann DG, Alexandridou A, Striker L, Elliot S, Female gender, estrogen loss, and Sub-RPE deposit formation in aged mice, Invest. Ophthalmol. Vis. Sci 44 (3) (2003) 1221–1229. [DOI] [PubMed] [Google Scholar]
- [44].DeGraff WG, Krishna MC, Kaufman D, Mitchell JB, Nitroxide-mediated protection against X-ray-and neocarzinostatin-induced DNA damage, Free Radic. Biol. Med 13 (5) (1992) 479–487. [DOI] [PubMed] [Google Scholar]
- [45].Deng-Bryant Y, Singh IN, Carrico KM, Hall ED, Neuroprotective effects of tempol, a catalytic scavenger of peroxynitrite-derived free radicals, in a mouse traumatic brain injury model, J. Cerebr. Blood Flow Metabol 28 (6) (2008) 1114–1126. [DOI] [PubMed] [Google Scholar]
- [46].Ghosh MC, Tong WH, Zhang D, Ollivierre-Wilson H, Singh A, Krishna MC, et al. , Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice, Proc. Natl. Acad. Sci. U. S. A 105 (33) (2008) 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kato N, Yanaka K, Hyodo K, Homma K, Nagase S, Nose T, Stable nitroxide Tempol ameliorates brain injury by inhibiting lipid peroxidation in a rat model of transient focal cerebral ischemia, Brain Res. 979 (1–2) (2003) 188–193. [DOI] [PubMed] [Google Scholar]
- [48].Lipman T, Tabakman R, Lazarovici P, Neuroprotective effects of the stable nitroxide compound Tempol on 1-methyl-4-phenylpyridinium ion-induced neurotoxicity in the Nerve Growth Factor-differentiated model of pheochromocytoma PC12 cells, Eur. J. Pharmacol 549 (1–3) (2006) 50–57. [DOI] [PubMed] [Google Scholar]
- [49].Rak R, Chao DL, Pluta RM, Mitchell JB, Oldfield EH, Watson JC, Neuroprotection by the stable nitroxide Tempol during reperfusion in a rat model of transient focal ischemia, J. Neurosurg 92 (4) (2000) 646–651. [DOI] [PubMed] [Google Scholar]
- [50].Schnackenberg CG, Welch WJ, Wilcox CS, Normalization of blood pressure and renal vascular resistance in SHR with a membrane-permeable superoxide dismutase mimetic: role of nitric oxide, Hypertension 32 (1) (1998) 59–64. [DOI] [PubMed] [Google Scholar]
- [51].Schnackenberg CG, Wilcox CS, The SOD mimetic tempol restores vasodilation in afferent arterioles of experimental diabetes, Kidney Int. 59 (5) (2001) 1859–1864. [DOI] [PubMed] [Google Scholar]
- [52].Sledziński Z, Woźniak M, Antosiewicz J, Lezoche E, Familiari M, Bertoli E, et al. , Protective effect of 4-hydroxy-TEMPO, a low molecular weight superoxide dismutase mimic, on free radical toxicity in experimental pancreatitis, Int. J. Pancreatol 18 (2) (1995) 153–160. [DOI] [PubMed] [Google Scholar]
- [53].Thiemermann C, Membrane-permeable radical scavengers (tempol) for shock, ischemia-reperfusion injury, and inflammation, Crit. Care Med 31 (1 Suppl) (2003) S76–S84. [DOI] [PubMed] [Google Scholar]
- [54].Samuni Y, Cook JA, Choudhuri R, Degraff W, Sowers AL, Krishna MC, et al. , Inhibition of adipogenesis by Tempol in 3T3-L1 cells, Free Radic. Biol. Med 49 (4) (2010) 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lundgren P, Thaiss CA, The microbiome-adipose tissue axis in systemic metabolism, Am. J. Physiol. Gastrointest. Liver Physiol 318 (4) (2020) G717–g724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kiefer FW, Vernochet C, O’Brien P, Spoerl S, Brown JD, Nallamshetty S, et al. , Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue, Nat. Med 18 (6) (2012) 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Petrosino JM, Disilvestro D, Ziouzenkova O, Aldehyde dehydrogenase 1A1: friend or foe to female metabolism? Nutrients 6 (3) (2014) 950–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Omran Z, Obesity: current treatment and future horizons, Mini Rev. Med. Chem 17 (1) (2017) 51–61. [DOI] [PubMed] [Google Scholar]
- [59].Blaner WS, Vitamin A signaling and homeostasis in obesity, diabetes, and metabolic disorders, Pharmacol. Ther 197 (2019) 153–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yoon JC, Chickering TW, Rosen ED, Dussault B, Qin Y, Soukas A, et al. , Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation, Mol. Cell Biol 20 (14) (2000) 5343–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yoshida K, Shimizugawa T, Ono M, Furukawa H, Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase, J. Lipid Res 43 (11) (2002) 1770–1772. [DOI] [PubMed] [Google Scholar]
- [62].Kersten S, Mandard S, Tan NS, Escher P, Metzger D, Chambon P, et al. , Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene, J. Biol. Chem 275 (37) (2000) 28488–28493. [DOI] [PubMed] [Google Scholar]
- [63].Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. , Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation, Nature 439 (7075) (2006) 484–489. [DOI] [PubMed] [Google Scholar]
- [64].ţarenac TM, Mikov M, Bile acid synthesis: from nature to the chemical modification and synthesis and their applications as drugs and nutrients, Front. Pharmacol 9 (2018) 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F, Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling, Cell Metabol. 22 (4) (2015) 658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. , Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity, Proc. Natl. Acad. Sci. U. S. A 110 (22) (2013) 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM, et al. , Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice, Sci. Rep 5 (2015), 16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rodríguez-García C, Sánchez-Quesada C, Algarra I, Gaforio JJ, The high-fat diet based on extra-virgin olive oil causes dysbiosis linked to colorectal cancer prevention, Nutrients 12 (6) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, et al. , Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice, Nat. Commun 9 (1) (2018) 2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hagi T, Geerlings SY, Nijsse B, Belzer C, The effect of bile acids on the growth and global gene expression profiles in Akkermansia muciniphila, Appl. Microbiol. Biotechnol 104 (24) (2020) 10641–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ding L, Yang L, Wang Z, Huang W, Bile acid nuclear receptor FXR and digestive system diseases, Acta Pharm. Sin. B 5 (2) (2015) 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhou Z, Zhou J, Du Y, Estrogen receptor alpha interacts with mitochondrial protein HADHB and affects beta-oxidation activity, M111, Mol. Cell. Proteomics 11 (7) (2012), 011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ, MicroRNAs in lipid metabolism, Curr. Opin. Lipidol 22 (2) (2011) 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


