Abstract
The performance and characteristics of Roche COBAS AMPLICOR HIV-1 MONITOR version 1.5 (CA MONITOR 1.5) UltraSensitive (usCA MONITOR 1.5) and Standard (stCA MONITOR 1.5) procedures, Organon Teknika NucliSens HIV-1 RNA QT with Extractor (NucliSens), and Bayer Quantiplex HIV RNA version 3.0 (bDNA 3.0) were compared in a multicenter trial. Samples used in this study included 460 plasma specimens from human immunodeficiency virus (HIV) type 1 (HIV-1)-infected persons, 100 plasma specimens from HIV antibody (anti-HIV)-negative persons, and culture supernatants of HIV-1 subtype A to E isolates diluted in anti-HIV-negative plasma. Overall, bDNA 3.0 showed the least variation in RNA measures upon repeat testing. For the Roche assays, usCA MONITOR 1.5 displayed less variation in RNA measures than stCA MONITOR 1.5. NucliSens, at an input volume of 2 ml, showed the best sensitivity. Deming regression analysis indicated that the results of all three assays were significantly correlated (P < 0.0001). However, the mean difference in values between CA MONITOR 1.5 and bDNA 3.0 (0.274 log10 RNA copies/ml; 95% confidence interval, 0.192 to 0.356) was significantly different from 0, indicating that CA MONITOR 1.5 values were regularly higher than bDNA 3.0 values. Upon testing of 100 anti-HIV-negative plasma specimens, usCA MONITOR 1.5 and NucliSens displayed 100% specificity, while bDNA 3.0 showed 98% specificity. NucliSens quantified 2 of 10 non-subtype B viral isolates at 1 log10 lower than both CA MONITOR 1.5 and bDNA 3.0. For NucliSens, testing of specimens with greater than 1,000 RNA copies/ml at input volumes of 0.1, 0.2, and 2.0 ml did not affect the quality of results. Additional factors differing between assays included specimen throughput and volume requirements, limit of detection, ease of execution, instrument work space, and costs of disposal. These characteristics, along with assay performance, should be considered when one is selecting a viral load assay.
The human immunodeficiency virus (HIV) type 1 (HIV-1) RNA level in plasma is a useful biological marker for assessing disease prognosis and outcome of antiretroviral therapy (8, 24, 31). The commercialization of HIV viral load assays has allowed for widespread and routine monitoring of viral RNA levels in infected persons. Currently, monitoring of HIV viral load along with CD4+-T-lymphocyte counts is considered standard medical practice for patient management (6). Several methodologies are available for the quantification of HIV-1 RNA in plasma, namely, reverse transcription (RT)-PCR and nucleic acid sequence-based amplification (NASBA), both of which are nucleic acid amplification techniques, and the branched-DNA (bDNA) technique, which is based on signal amplification (34).
Potent combination antiretroviral therapies often reduce plasma HIV RNA levels below the limit of detection of commercial assays (17, 18). In 1997 and 1998 and in 1998 and 1999, 42 and 46%, respectively, of specimens analyzed in the HIV viral load testing program of the Canadian province of Quebec had RNA levels below 500 copies/ml, as determined by Quantiplex HIV RNA version 2.0 (bDNA 2.0) (Chiron Corporation, Emeryville, Calif.) (27). The use of a new-generation viral load assay would provide more informative data for patient specimens with RNA levels below 500 copies/ml.
In the present study, we conducted a multicenter comparative evaluation of COBAS AMPLICOR HIV-1 MONITOR version 1.5 (CA MONITOR 1.5) UltraSensitive (usCA MONITOR 1.5) and Standard (stCA MONITOR 1.5) procedures (Roche Diagnostic Systems, Branchburg, N.J.), NucliSens HIV-1 RNA QT with Extractor (NucliSens) (Organon Teknika, Durham, N.C.), and Quantiplex HIV RNA version 3.0 (bDNA 3.0) (Bayer Diagnostics, East Walpole, Mass.), which involve RT-PCR, NASBA, and bDNA technologies, respectively (7, 35, 37). We evaluated the inter- and intrarun, interlot, and intersite reproducibilities of, the specificities of, and the quantification of HIV-1 non-subtype B strains by these methods. The quantitative relationships between the methods were compared. Finally, the effect of different input volumes on the reproducibility of results obtained by NucliSens was investigated.
MATERIALS AND METHODS
Patient population and consent.
Blood specimens were collected from 460 anti-HIV-1-positive individuals monitored at several specialized clinics and hospitals in the Canadian province of Quebec. Patients were adults, and an estimated 15% were treatment naive. Specimens were requested as part of usual follow-up and treatment monitoring from October to December 1998. For testing specificity, blood was obtained from 100 HIV antibody (anti-HIV)-negative volunteers. Informed consent was obtained from all participants.
Test sites.
The evaluation was carried out at four sites: three Canadian university hospitals (Hôpital Saint-Luc, Montreal, Quebec; Royal Victoria Hospital, Montreal, Quebec; and Centre Hospitalier de l'Université Laval, Sainte-Foy, Quebec) and the Laboratoire de Santé Publique du Québec, Sainte-Anne-de-Bellevue, Quebec, Canada (LSPQ). Testing for reproducibility and for comparison of the quantitative relationships between the methods was carried out at the three hospital sites. Each assay was evaluated at two of the three hospital sites. Specificity, non-B subtype HIV-1 strain, and NucliSens sample volume testing were performed at the LSPQ. Laboratory personnel carrying out the assays were trained by each manufacturer prior to the evaluation.
Specimen collection and processing.
EDTA-treated plasma samples were centrifuged within 6 h of collection at the three hospital test sites. Specimens were frozen at −70°C and forwarded on dry ice anonymously to the LSPQ, where they were coded for the study, thawed once, and allocated into volumes and quantities appropriate for each assay. Specimens were frozen again at −70°C. They were sent on dry ice to the testing sites and stored at −70°C until testing. Tests were performed on separate aliquots.
Non-subtype B isolates.
The non-subtype B isolates originated from the LSPQ (three subtype A, one subtype D, and one unknown) and the Henry M. Jackson Foundation Research Laboratory, Washington, D.C. (one isolate each of subtypes A, C, D, E, and F). The latter were obtained via the Ontario Ministry of Health, Toronto, Ontario, Canada. The subtypes of four of the LSPQ isolates were determined by sequencing analysis at the Laboratory Center for Disease Control, Ottawa, Ontario, Canada. The subtype of the other isolate is unknown. This strain was isolated from an infant born to an HIV-infected mother of African origin. Two consecutive blood specimens drawn shortly after birth from this infant were found PCR negative for proviral DNA by the AMPLICOR HIV-1 test (Roche) but were culture positive.
Isolates were propagated in cell cultures by infection of a pool of peripheral blood mononuclear cells from three seronegative donors using standard methods (5). Culture supernatants were clarified, allocated into appropriate volumes, and frozen at −70°C. For initial standardization, the supernatants were used to spike anti-HIV-negative plasma, and the samples were tested by bDNA 2.0. For each isolate, three dilutions in anti-HIV-negative plasma were prepared. The spiked plasma was allocated into appropriate volumes and frozen at −70°C until testing.
Quantification of HIV-1 RNA.
Quantification of HIV-1 RNA was performed following each manufacturer's instructions.
(i) RT-PCR.
stCA MONITOR 1.5 has stated lower and upper detection limits of 400 and 750,000 RNA copies/ml, respectively, and uses a plasma volume of 0.2 ml. usCA MONITOR 1.5 has stated lower and upper detection limits of 50 and 75,000 RNA copies/ml, respectively, and uses a plasma volume of 0.5 ml. usCA MONITOR 1.5 differs from stCA MONITOR 1.5 by an initial high-speed centrifugation step used to concentrate the virus particles and the volume in which the ethanol-precipitated viral RNA is reconstituted prior to RT-PCR. RNA is reconstituted in 0.4 and 0.1 ml of specimen diluent for stCA MONITOR 1.5 and usCA MONITOR 1.5, respectively. Other than specimen preparation, the remaining steps of both procedures are identical. Amplification and detection were performed with an automated CA analyzer (11). Amplification on the CA analyzer is performed using rosettes or A-rings consisting of 12 A-tubes. Two A-rings, or 24 samples, can be processed at a time. In this study, one negative and two positive controls were processed per a maximum of 24 samples. CA MONITOR 1.5 is an updated and automated version of the U.S. Food and Drug Administration-approved AMPLICOR HIV-1 MONITOR version 1.0. CA MONITOR 1.5 is licensed for use in Canada.
(ii) NASBA.
NucliSens has a reported dynamic range of 80 to 8,000,000 RNA copies/input. In this study, testing was performed using an input of 2 ml of plasma for a sensitivity of 40 RNA copies/ml unless otherwise stated. Samples with reportable values below 40 RNA copies/ml, which can be obtained under optimal assay conditions, were included in the analyses. Viral RNA was isolated by use of the NucliSens Extractor (38). The Extractor is an automated system for the extraction of nucleic acids from serum or plasma for use with the silica-based isolation method (3). The instrument is composed of 10 separate extraction stations for maximum processing of 10 samples per run. Electrochemiluminescence detection of amplified products was performed using the NucliSens Reader. NucliSens is licensed for use with the manual nucleic acid extraction procedure in Canada.
(iii) bDNA.
bDNA 3.0 has a reported quantification range of 50 to 500,000 RNA copies/ml and uses 1 ml of plasma. A maximum of 84 samples can be processed per plate. bDNA 2.0 has a reported dynamic range of 500 to 800,000 RNA copies/ml and requires 2 ml of plasma, for a maximum of 42 samples per plate. Both assays were performed using semiautomated Quantiplex bDNA system 340. Two plates may be processed at a time with system 340 (29). bDNA 3.0 and bDNA 2.0 are licensed for use in Canada.
Reproducibility.
Specimens from 90 patients were evaluated for each of the inter- and intrarun, intersite, and interlot reproducibility studies. The RNA levels for these 90 specimens were distributed as follows: 50, 25, and 15 had values below 500, from 500 to 10,000, and above 10,000 copies/ml, respectively, as determined by bDNA 2.0. Different specimens were tested by each method in the inter- and intrarun, intersite, and interlot studies. For RT-PCR, specimens were initially tested by usCA MONITOR 1.5, and all specimens yielding values above 400 RNA copies/ml were retested by stCA MONITOR 1.5. The inter- and intrarun reproducibility was evaluated with specimens tested once in each of three runs at two sites (45 specimens/site). Since the specimens were tested only once in each run, the value obtained will contain both interrun and intrarun sources of variability. The interlot reproducibility was evaluated with specimens tested once with each of two lots at two sites (45 specimens/site/lot), and the intersite reproducibility was evaluated with specimens tested once at each of two sites (90 specimens/site). Specimens were included in the calculation when all replicates were within the lower and upper quantitative limits of each assay.
Quantitative relationships.
To compare the quantitative relationships between the methods, 89 specimens were tested once with all three assays at two sites (44 or 45 specimens/site). For these specimens, 55, 19, and 15 had RNA levels below 500, from 500 to 10,000, and above 10,000 copies/ml, respectively, as determined by bDNA 2.0. For RT-PCR, specimens were tested by usCA MONITOR 1.5, and only those yielding values above 75,000 RNA copies/ml (n = 8) were retested by stCA MONITOR 1.5.
Specificity.
The specificity was evaluated by testing specimens from 100 anti-HIV-negative persons in four runs (25 specimens/run) using two lots (two runs/lot). For RT-PCR, specificity was evaluated using usCA MONITOR 1.5.
Variable input volumes and NucliSens.
The effect of input volume was evaluated with 11 specimens tested at inputs of 0.1, 0.2, and 2.0 ml. These specimens had RNA levels above 1,000 copies/ml.
Relative costs of disposal.
Costs of disposal were evaluated per clinical sample or reportable result. Costs for usCA MONITOR 1.5 were determined with two A-rings, or 24 A-tubes, for 21 reportable results (21 samples and 3 controls). Costs for NucliSens were evaluated with 20 samples, or two runs of 10 samples, for 20 reportable results. Costs for bDNA 3.0 were determined with one run of one plate (84 samples, 9 standards, and 3 controls) for 84 reportable results. The costs of disposable items were those incurred at the LSPQ.
Statistical analysis.
The variability between replicate tests for each method was described using both the standard error of measurement (SEM) for the log10-transformed values and the coefficient of variation for values for RNA copies per milliliter (14). The SEMs obtained for the four tests (usCA MONITOR 1.5, stCA MONITOR 1.5, NucliSens, and bDNA 3.0) were compared by use of the Bartlett test. When a significant result at a 5% level was obtained, two-by-two comparisons were performed by applying Bonferroni's correction. The normality of the distribution was verified by the Shapiro-Wilk test. Orthogonal (Deming) regression on the log10-transformed values was used to compare the relationships between methods. Pearson correlation coefficients and differences in values between methods were determined for the log10-transformed data.
RESULTS
Reproducibility studies. (i) Inter- and intrarun.
The inter- and intrarun reproducibility was evaluated with specimens that were tested once in three different runs. Each assay was evaluated at two sites. bDNA 3.0 displayed the highest reproducibility (P < 0.001), with a SEM of 0.09 log10 RNA copies/ml (Table 1). stCA MONITOR 1.5 and NucliSens yielded higher SEM values, 0.19 and 0.18 log10 RNA copies/ml, respectively. An intermediate SEM of 0.13 log10 RNA copies/ml was observed for usCA MONITOR 1.5. For the Roche assays, the level of reproducibility of usCA MONITOR 1.5 was higher than that of stCA MONITOR 1.5 (P < 0.001). For both NucliSens and bDNA 3.0, a difference in reproducibility was observed between sites (P < 0.001), indicating that the latter two assays may be more difficult to master than CA MONITOR 1.5.
TABLE 1.
Inter- and intrarun, interlot, and intersite reproducibilities of viral load assays
| Reproducibility | Parametera | Result at the indicated siteb for the following assay:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| usCA MONITOR 1.5
|
stCA MONITOR 1.5
|
NucliSens
|
bDNA 3.0
|
||||||||||
| Site A | Site B | Combined | Site A | Site B | Combined | Site A | Site C | Combined | Site B | Site C | Combined | ||
| Inter- and intrarun | SEM | 0.14 | 0.12 | 0.13 | 0.21 | 0.17 | 0.19 | 0.22 | 0.14 | 0.18 | 0.11 | 0.06 | 0.09 |
| % CV | 28.8 | 25.0 | 26.9 | 0.22 | 37.0 | 38.2 | 40.9 | 22.2 | 31.2 | 22.5 | 13.1 | 18.2 | |
| n | 23 | 24 | 47 | 0.23 | 19 | 41 | 28 | 30 | 58 | 28 | 24 | 52 | |
| Interlot | SEM | 0.12 | 0.13 | 0.13 | 0.14 | 0.12 | 0.13 | 0.19 | 0.12 | 0.16 | 0.13 | 0.09 | 0.12 |
| % CV | 23.0 | 24.1 | 23.6 | 21.2 | 21.4 | 21.3 | 30.1 | 22.5 | 26.1 | 25.0 | 17.6 | 21.9 | |
| n | 24 | 25 | 49 | 24 | 20 | 44 | 27 | 26 | 53 | 33 | 24 | 57 | |
| Intersite | SEM | 0.14 | 0.20 | 0.22 | 0.12 | ||||||||
| % CV | 23.8 | 30.5 | 29.6 | 21.3 | |||||||||
| n | 51 | 44 | 54 | 55 | |||||||||
SEM was calculated from log10 RNA copies per milliliter. Percent CV is the mean value calculated from RNA copies per milliliter. n is number of samples included in the analysis.
Sites A, B, and C are the three hospital test sites.
(ii) Interlot.
The interlot reproducibility was evaluated with specimens that were tested once with two different lots, each lot being evaluated at two sites. The reproducibility between lots was similar for all four tests evaluated, with SEM values ranging from 0.12 to 0.16 log10 RNA copies/ml (Table 1). No significant difference was observed between sites (P > 0.05/4, or 0.0125, with Bonferroni's correction). stCA MONITOR 1.5 performed better in the interlot study than in the inter- and intrarun study, while bDNA 3.0 performed better in the inter- and intrarun study than in the interlot study.
(iii) Intersite.
The intersite reproducibility was evaluated with specimens that were tested once at two sites. It must be noted that in the inter- and intrarun study, a difference in the level of reproducibility was observed between sites for NucliSens and bDNA 3.0. Therefore, in the intersite study, the SEM values for these two assays might be biased due to site performance and, as a consequence, their comparison with RT-PCR could be misleading. Nevertheless, the SEM of 0.12 log10 RNA copies/ml observed for bDNA 3.0 was lower (P < 0.001) than the SEM values observed for NucliSens and stCA MONITOR 1.5 but not usCA MONITOR 1.5 (P > 0.05). For the Roche assays, the SEM of 0.14 log10 RNA copies/ml observed for usCA MONITOR 1.5 was not significantly lower than the SEM of 0.20 log10 RNA copies/ml observed for stCA MONITOR 1.5 (P > 0.05/6, or 0.0083, with Bonferroni's correction).
Relationship between methods.
All three assays were found to be significantly correlated (for r values, P < 0.0001; Table 2). The relationship of values between methods was studied using Deming regression analysis. In none of the comparisons were the intercepts significantly different from 0 (Table 2). In a comparison of CA MONITOR 1.5 with NucliSens, the slope of 1.069 (95% confidence interval [CI], 0.952 to 1.186) was not significantly different from 1, and in a comparison of NucliSens with bDNA 3.0, the slope of 1.135 (95% CI, 1.001 to 1.268) was near the limit of significance. However, in a comparison of CA MONITOR 1.5 with bDNA 3.0, the slope of 1.153 (95% CI, 1.069 to 1.237) was significantly different from 1, indicating that the values obtained by CA MONITOR 1.5 were regularly higher than those obtained by bDNA 3.0. A mean difference of 0.274 log10 RNA copies/ml (95% CI, 0.194 to 0.354) was observed between the values for CA MONITOR 1.5 and bDNA 3.0. This value was significantly different from 0, confirming that CA MONITOR 1.5 values were commonly higher than those of bDNA 3.0. The equation fitting the regression line indicated that the difference in values between assays was small at low RNA levels and that the difference increased as the viral load increased (Table 2). The measures differed by <0.5 log10 RNA copies/ml (level of clinical significance) in 75% of the results. The mean difference between NucliSens and bDNA 3.0 values was also significantly different from 0, but to a lesser extent than the difference observed between CA MONITOR 1.5 and bDNA 3.0 (Table 2). Similarly, the divergence of values between assays increased as the viral load increased. Finally, the values for CA MONITOR 1.5 and NucliSens differed by <0.5 log10 RNA copies/ml for 86.3% of the measures, consistent with the observation that the values obtained by these two assays displayed the highest agreement.
TABLE 2.
Relationship of log10-transformed RNA copies per milliliter between assays
| Assays compared | No. of samples | Correlation (95% Cl) | Deming regression equation | Intercept (95% Cl) | Slope (95% Cl) | Mean difference, log10 ± 1.96 SD (95% Cl) | % Showing a <0.5- log10 difference |
|---|---|---|---|---|---|---|---|
| CA MONITOR 1.5 vs NucliSens | 51 | 0.942a (0.836–1.048) | y = 1.069x − 0.129 | −0.129 (−0.552–0.293) | 1.069 (0.952–1.186) | 0.109 ± 0.746 (0.005–0.213) | 86.3 |
| CA MONITOR 1.5 vs bDNA 3.0 | 52 | 0.969a (0.893–1.044) | y = 1.153x − 0.246 | −0.246 (−0.582–0.089) | 1.153 (1.069–1.237) | 0.274 ± 0.579 (0.194–0.354) | 75.0 |
| NucliSens vs bDNA 3.0 | 50 | 0.936a (0.829–1.043) | y = 1.135x − 0.301 | −0.301 (−0.807–0.207) | 1.135 (1.001–1.268) | 0.161 ± 0.759 (0.054–0.268) | 72.0 |
| usCA MONITOR 1.5 vs stCA MONITOR 1.5 | 85 | 0.918a (0.874–0.962) | y = 0.929x + 0.230 | 0.230 (−0.095–0.554) | 0.929 (0.842–1.017) | −0.029 ± 0.432 (−0.249–0.191) | 97.6 |
P, <0.0001.
To study the relationship between usCA MONITOR 1.5 and stCA MONITOR 1.5, the individual values obtained in the inter- and intrarun study were compared. The two procedures were significantly correlated (r = 0.918), and Deming regression analysis indicated that the intercept and slope were not significantly different from 0 and 1, respectively (Table 2). Furthermore, the mean difference in values was not significantly different from 0, confirming that the two procedures yielded equivalent measures. Only two (2.4%) of the values differed by more than 0.5 log10 RNA copies/ml.
The comparison of the relationship between methods included 55 specimens which displayed <500 RNA copies/ml when tested by bDNA 2.0. Nineteen were quantified by all three methods, while 19 were not quantified by any. Of the remaining 17 specimens, 13, 7, and 3 were quantified by NucliSens, usCA MONITOR 1.5, and bDNA 3.0, respectively. The 13 specimens quantified by NucliSens included 1 with 19 RNA copies/ml and 2 others with between 40 and 50 RNA copies/ml (Table 3). It is noteworthy that for the 17 specimens not quantifiable by all three assays, some of the measures varied by threefold or more. For example, four specimens (G to J; Table 3) not quantified by two of the assays yielded RNA measures significantly higher threefold or more) in the third assay. Overall, NucliSens, usCA MONITOR 1.5, and bDNA 3.0 quantified 58.2, 47.3, and 40% of the 55 specimens, respectively.
TABLE 3.
HIV-1 RNA copies per milliliter for 17 specimens below the lower limit of detection in one or two of the viral load assays
| Specimen | No. of RNA copies/ml detected by:
|
||
|---|---|---|---|
| usCA MONITOR 1.5 | NucliSens | bDNA 3.0 | |
| A | <50 | 19 | <50 |
| B | <50 | 40 | <50 |
| C | <50 | 44 | <50 |
| D | <50 | 56 | <50 |
| E | <50 | 60 | <50 |
| F | <50 | 94 | <50 |
| G | <50 | 150 | <50 |
| H | <50 | 290 | <50 |
| I | <50 | 470 | <50 |
| J | 294 | <40 | <50 |
| K | 70 | <40 | <50 |
| L | <50 | 210 | 57 |
| M | 140 | <40 | 149 |
| N | 509 | <40 | 80 |
| O | 57 | 160 | <50 |
| P | 66 | 85 | <50 |
| Q | 132 | 160 | <50 |
Specificity.
The specificity of each assay was assessed by testing 100 anti-HIV-negative volunteers. usCA MONITOR 1.5 and NucliSens showed 100% specificity, while bDNA 3.0 displayed 98% specificity. The two samples quantified by bDNA 3.0 were from different runs and lots and had values of 189 and 1,033 RNA copies/ml. Both specimens were retested, and only the specimen initially showing 189 RNA copies/ml remained quantifiable, at 55 RNA copies/ml.
Quantification of HIV non-subtype B isolates.
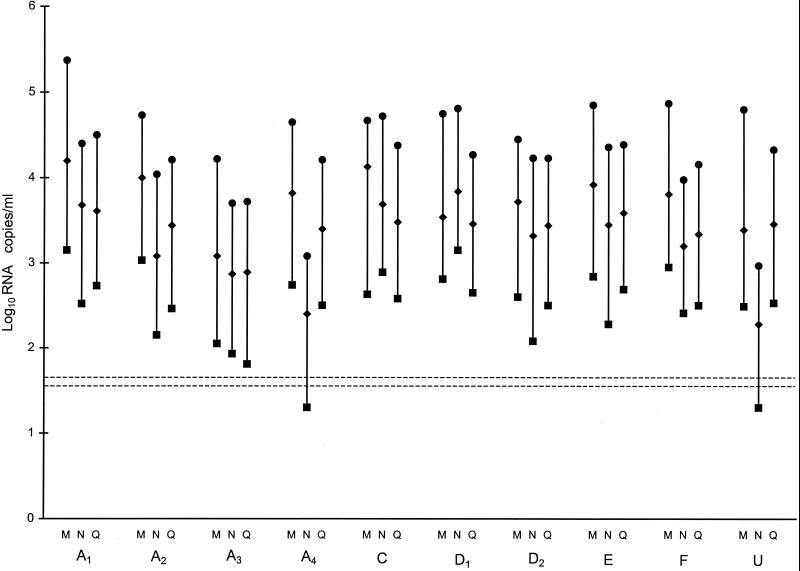
All three methods quantified the 10 viral isolates (Fig. 1). Two of the isolates (A4 and U; Fig. 1) were quantified at 1 log10 lower by NucliSens than by the other two methods. For these two isolates, the lowest dilution tested was below the quantifiable limit of NucliSens. The mean difference between CA MONITOR 1.5 and bDNA 3.0 for the 30 results was 0.360 log10 RNA copies/ml (95% CI, 0.163 to 0.556). This value was similar to the mean difference observed for the clinical specimens of which, in North America, nearly all are expected to be of subtype B (Table 2). The mean differences were 0.553 log10 RNA copies/ml (95% CI, 0.357 to 0.750) between CA MONITOR 1.5 and NucliSens and −0.175 log10 RNA copies/ml (95% CI, −0.372 to 0.022) between NucliSens and bDNA 3.0 for 28 of the 30 quantifiable results. Both mean differences were significantly different from those observed for the clinical specimens (Table 2), accountable mainly by the two specimens quantified at 1 log10 lower by NucliSens.
FIG. 1.
RNA concentrations of 10 HIV-1 non-subtype B isolates determined at three dilutions: 100 (●), 101 (♦), and 102 (▪). RNA levels of each isolate were assayed by all three methods, represented by the one-letter codes M (CA MONITOR 1.5), N (NucliSens), and Q (bDNA 3.0). The panel included four subtype A isolates (A1 to A4), two subtype D isolates (D1 and D2), single isolates of subtypes C, E, and F (C, E, and F), and an isolate of unknown subtype (U). Horizontal broken lines indicate the lower limits of detection of 50 copies/ml for usCA MONITOR 1.5 and bDNA 3.0 and of 40 copies/ml for NucliSens.
NucliSens and variable input volumes.
A SEM of 0.15 log10 RNA copies/ml and a coefficient of variation of 29.1% were observed for 11 specimens each tested at input volumes of 0.1, 0.2, and 2.0 ml. Mean RNA levels for the three measures ranged from 2,267 to 516,667 copies/ml (median, 23,333 copies/ml). The SEM and percent CV were similar to those observed in the inter- and intrarun reproducibility study (Table 1). In the latter study, specimens were tested three times using a fixed volume of 2 ml. The data indicate that volume input does not affect the reproducibility of results.
Relative costs of disposal and throughput.
Costs of viral load tests are extensive and can influence the selection of an assay. Hidden costs, such as those of disposable items, can have an impact on the overall cost. Costs of disposal for bDNA 3.0 were three to four times lower than those for both usCA MONITOR and NucliSens per reportable result (Table 4). Another parameter affecting cost is labor. The throughput of bDNA 3.0 for testing 84 specimens/plate was twice those of both usCA MONITOR 1.5 and NucliSens (Table 4).
TABLE 4.
Characteristics of viral load assays
| Assay | Sample vol (ml) | Quantitative limita
|
% of RNA values above upper limit of detectionb | No. of samples/run | Maximun no. of reportable resultsc | Ease of execution | Costs of disposable items/reportable result (timesd) | |
|---|---|---|---|---|---|---|---|---|
| Upper | Lower | |||||||
| usCA MONITOR 1.5 | 0.5 | 75,000 | 50 | 16.9 | 12 | 30 | Moderate | 3.8 |
| NucliSens | 0.01–2.0 | 8,000,000 | 80 | 0.3 | 10 | 30 | Less moderatee | 4.5 |
| bDNA 3.0 | 1.0 | 500,000 | 50 | 0.9 | 96 | 84 | Moderate | 1 |
Copies per milliliter for usCA MONITOR 1.5 and bDNA 3.0; copies per input for NucliSens.
According to RNA levels obtained for 19,347 specimens tested between 1 April 1999 and 31 March 2000 in the Canadian province of Quebec.
Per day for usCA MONITOR 1.5 and NucliSens and per 1.5 days for bDNA 3.0.
Relative to bDNA 3.0.
The detection procedure is the labor-intensive phase, as it necessitates numerous pipetting steps.
DISCUSSION
Treatment of HIV-infected patients with combination therapy is efficient in reducing HIV replication, opportunistic infections, and mortality (6). It is generally believed that suppression of HIV-1 RNA levels in plasma to low levels (below 20 to 50 copies/ml) is important for prevention of viral rebound and development of resistance (15, 20, 32). Earlier viral load assays were able to quantify RNA levels down to 400 or 500 copies/ml (30, 39). However, recent viral load assays with limits of detection of down to 40 or 50 copies/ml have become available.
In the present study, three different commercially available assays for quantifying HIV-1 RNA in plasma down to 40 or 50 copies/ml were evaluated to assess their reproducibility, specificity, relationship of values, non-subtype B quantification, and general characteristics. We found that, overall, bDNA 3.0 displayed a higher level of reproducibility than CA MONITOR 1.5 and NucliSens. A difference in performance between sites was observed in the inter- and intrarun reproducibility study with NucliSens and bDNA 3.0 but not with CA MONITOR 1.5. If only the data from the site offering the best performance were used to compare SEM values between assays, NucliSens would show a level of reproducibility equivalent to that of usCA MONITOR 1.5, while bDNA 3.0 would show a higher level of reproducibility than the other assays. It is noteworthy that NucliSens and bDNA 3.0 are less automated than CA MONITOR 1.5. Nevertheless, all three viral load assays contain numerous steps requiring the operator's utmost attention. The difficulty in establishing proficiency likely reflects the technical complexity of the viral load assays.
The three HIV viral load assays were found to be highly correlated. However, differences in agreements between assays were observed. A difference of 0.45 log10 RNA copies/ml has been previously reported for Standard AMPLICOR MONITOR 1.0 and bDNA 2.0 (30). In contrast, mean differences of 0.072 and 0.036 log10 RNA copies/ml have been recently reported for Sensitized AMPLICOR MONITOR 1.0 and bDNA 3.0 and for UltraSensitive AMPLICOR (usCA) MONITOR 1.5 and bDNA 3.0, respectively (13, 21). In our study, a mean difference of 0.274 log10 RNA copies/ml was observed between CA MONITOR 1.5 and bDNA 3.0. Our data indicate an improvement in agreement between CA MONITOR 1.5 and bDNA 3.0 over previous versions, but not as close an agreement as those observed by Highbarger et al. (21) and Elbeik et al. (13). This result suggests that the CA analyzer may generate RNA values slightly higher than those generated by the manual procedure. The highest agreement was observed between CA MONITOR 1.5 and NucliSens, with a mean difference of 0.109 log10 RNA copies/ml. This finding is in agreement with that of a previous comparison of Standard AMPLICOR MONITOR 1.0 and NucliSens (12). An intermediate value of 0.161 log10 RNA copies/ml was observed for NucliSens and bDNA 3.0. Ginocchio et al. (16) have reported a difference of 0.323 log10 between NucliSens and a modified bDNA assay having a lower limit of detection of 500 copies/ml. Our results indicate an improvement in agreement between NucliSens and bDNA 3.0 compared to the previous bDNA version. The increase in agreement between bDNA 3.0 and both CA MONITOR 1.5 and NucliSens is due to the two- to fourfold-higher quantification values of bDNA 3.0 over bDNA 2.0 (13, 23). The percentage of specimens showing a difference of <0.5 log10 RNA copies/ml between assays ranged from 72 to 86%. However, 97.6% of specimens displayed a difference of <0.5 log10 RNA copies/ml when tested with the two CA MONITOR 1.5 procedures. Considering the above data and that the difference in RNA measures between assays varies from patient to patient, it is still recommended that CA MONITOR 1.5, NucliSens, and bDNA 3.0 not be used as alternatives to patient monitoring.
The RNA values measured by usCA MONITOR 1.5 and stCA MONITOR 1.5 in their common linear range (400 to 75,000 copies/ml) were equivalent. For our population, it is estimated that 44% of specimens would show RNA levels below 400 copies/ml and that 17% would display RNA levels above 75,000 copies/ml if tested with CA MONITOR 1.5 (27). A minority of specimens are from treatment naive patients (15% in this study), and information on treatment is not always routinely available. Therefore, for our population, it would be more beneficial to test specimens with usCA MONITOR 1.5. Specimens yielding values above 75,000 copies/ml could then be retested with stCA MONITOR 1.5. Furthermore, usCA MONITOR 1.5 displayed less variation in RNA measures than stCA MONITOR 1.5.
The relationship between methods was studied using 55 specimens displaying <500 copies/ml in bDNA 2.0. For these specimens, NucliSens at an input volume of 2 ml showed the highest sensitivity. However, some specimens not quantified by NucliSens were quantified by usCA MONITOR 1.5 and bDNA 3.0. In addition, there was considerable disagreement between values for some specimens quantifiable by one or two of the assays, as they often varied by more than three-fold (Table 3). This result indicates that variability between assays increases as values approach the lower detection limit.
In this study, we evaluated the specificity of the three methods by testing 100 anti-HIV-negative persons. usCA MONITOR 1.5, NucliSens, and bDNA 3.0 displayed specificities of 100, 100, and 98%, respectively. Although 100% specificity was obtained in our study with usCA MONITOR 1.5 and NucliSens, false-positive reactions have been reported by others using RT-PCR, NASBA, and bDNA methods (10, 19, 33). Viral load tests have been developed for quantification of HIV RNA levels in anti-HIV-positive patients and have not been validated for diagnostic purposes. Thus, regardless of the method used, viral load test results used for establishing a diagnosis of HIV infection are still not recommended and must be confirmed with appropriate serologic tests.
The genomes of HIV-1 display sequence variability and have been divided into groups M, N, and O (22). Group M is most prevalent and is further subdivided into at least nine subtypes: A to D, F to H, J, and K. Subtype B is found primarily in North America and Europe, while non-subtype B strains are predominant in Africa and Asia (4). Nonetheless, cocirculation of distinct HIV-1 subtypes has been reported in numerous geographical locations, including North America (26, 36). The accurate quantification of the different HIV subtypes is warranted, since an underestimation of the HIV viral load can lead to suboptimal patient management. Although all methods could quantify the 10 viral isolates tested in this study, NucliSens quantified to 1 log10 lower a subtype A isolate and an isolate believed to be of a subtype other than subtype B. Our results are in agreement with those of others who have found subtype A RNA levels to be underestimated by NucliSens (9, 28). AMPLICOR MONITOR 1.0 has been reported to underestimate HIV RNA levels for some non-subtype B isolates (2, 25, 28). In this study, CA MONITOR 1.5 efficiently quantified all non-subtype B viral isolates, supporting the recent finding of the improvement of version 1.5 over version 1.0 for quantification of HIV non-B subtypes (2, 25, 28).
We found that the reproducibility of results for NucliSens was not affected by differences in input volume. A similar finding has been recently reported for NASBA HIV QT, which is an earlier version of NucliSens (40). The ability to use a larger input volume, such as 2 ml, has the advantage of increased sensitivity, while the use of a smaller input volume is an advantage for testing neonates. At a sample input volume of 0.1 ml, the lower detection limit would be 800 copies/ml; HIV RNA levels after the first week of life are often found to exceed this value (1).
The three assays evaluated can be used to assess HIV-1 viral load in the follow-up of infected patients. The choice of an assay can be influenced by characteristics such as specimen throughput and volume requirements, ease of execution, instrument work space, and costs (Table 4). The throughput of bDNA 3.0 was twice that of usCA MONITOR 1.5 and NucliSens. However, the effect of throughout on labor costs and instrument work space will vary depending on the number of specimens processed. For example, for throughputs of less than 50 specimens per week, the required technical time and instrumentation for both usCA MONITOR 1.5 and NucliSens are equivalent to that of bDNA 3.0. On the other hand, for throughputs of 250 specimens per week, both usCA MONITOR 1.5 and NucliSens require more technical time and instrumentation than bDNA 3.0. We found the NucliSens Extractor to be a great improvement over the manual extraction method, which is quite labor-intensive. However, the throughput was not found to be substantially increased over that of manual extraction. In addition, a substantial amount of guanidium thiocyanate waste accumulated by use of the 9-ml nucleic acid isolation procedure, which necessitated special disposal requirements. Both NucliSens and stCA MONITOR 1.5 are better adapted for testing neonates, as they require less specimen volume than bDNA 3.0. Costs of kits, labor, and disposable items may differ for each assay, and all three need to be considered for assessing overall costs. Costs of disposal can have an impact, as they were found to be three to four times higher per reportable result for usCA MONITOR 1.5 and NucliSens than for bDNA 3.0. Individual laboratory requirements and assay characteristics and performance should be considered in selecting a viral load assay.
ACKNOWLEDGMENTS
We thank Camillia Beaudin, Ginette Breton, Jasmine Chamberland, Susan Fenn, Micheline Lortie, Diane McAleer, Lise Piché, and Lise Roberge for technical assistance and Marie-Claude Guertin from the CHUM Research Center for statistical analysis. We also thank Carol Major and Christiane Claessens for providing non-subtype B HIV-1 isolates. We are grateful to the physicians, patients, and laboratory personnel for providing the clinical specimens used in this study.
This study was supported by Roche Diagnostic Systems, Organon Teknika, and Bayer Diagnostics.
REFERENCES
- 1.Abrams E J, Weedon J, Steketee R W, Lambert G, Bamji M, Brown T, Kalish M L, Schoenbaum E E, Thomas P A, Thea D M the New York City Perinatal HIV Transmission Collaborative Study Group. Association of human immunodeficiency virus (HIV) load early in life with disease progression among HIV-infected infants. J Infect Dis. 1998;178:101–108. doi: 10.1086/515596. [DOI] [PubMed] [Google Scholar]
- 2.Alaeus A, Lilja E, Herman S, Spadoro J, Wang J, Albert J. Assay of plasma samples representing different HIV-1 genetic subtypes: an evaluation of new versions of the Amplicor HIV-1 monitor assay. AIDS Res Hum Retrovir. 1999;15:889–894. doi: 10.1089/088922299310593. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodine S K, Mascola J R, Weiss P J, Ito S I, Porter K R, Artenstein A W, Garland F C, McCutchan F E, Burke D S. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet. 1995;346:1198–1199. doi: 10.1016/s0140-6736(95)92901-0. [DOI] [PubMed] [Google Scholar]
- 5.Burgard M, Mayaux M-J, Blanche S, Ferroni A, Guihard-Moscato M-L, Allemon M-C, Ciraru-Vigneron N, Firtion G, Floch C, Guillot F, Lachassine E, Vial M, Griscelli C, Rouzioux C the HIV Infection in Newborns French Collaborative Study Group. The use of viral culture and p24 antigen testing to diagnose human immunodeficiency virus infection in neonates. N Engl J Med. 1992;327:1192–1197. doi: 10.1056/NEJM199210223271702. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter C C J, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society—USA Panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 7.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L-P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombs R W, Welles S L, Hooper C, Reichelderfer P S, D'Aquila R T, Japour A J, Johnson V A, Kuritzkes D R, Richman D D, Kwok S, Todd J, Jackson J B, DeGruttola V, Crumpacker C S, Kahn J for the AIDS Clinical Trials Group (ACTG) 116B/117 Study Team and the ACTG Virology Committee Resistance and HIV-1 RNA Working Groups. Association of plasma human immunodeficiency virus type 1 RNA level with risk of clinical progression in patients with advanced infection. J Infect Dis. 1996;174:704–712. doi: 10.1093/infdis/174.4.704. [DOI] [PubMed] [Google Scholar]
- 9.de Baar M P, van der Schoot A M, Goudsmit J, Jacobs F, Ehren R, van der Horn K H M, Oudshoorn P, de Wolf F, de Ronde A. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J Clin Microbiol. 1999;37:1813–1818. doi: 10.1128/jcm.37.6.1813-1818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Mendoza C, Holguín A, Soriano V. False positives for HIV using commercial viral load quantification assays. AIDS. 1998;12:2076–2077. doi: 10.1097/00002030-199815000-00022. [DOI] [PubMed] [Google Scholar]
- 11.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 12.Dyer J R, Pilcher C D, Shepard R, Schock J, Eron J J, Fiscus S A. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:447–449. doi: 10.1128/jcm.37.2.447-449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbeik T, Charlebois E, Nassos P, Kahn J, Hecht F M, Yajko D, Ng V, Hadley K. Quantitative and cost comparison of ultrasensitive human immunodeficiency virus type 1 RNA viral load assays: Bayer bDNA Quantiplex versions 3.0 and 2.0 and Roche PCR Amplicor Monitor version 1.5. J Clin Microbiol. 2000;38:1113–1120. doi: 10.1128/jcm.38.3.1113-1120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleiss J L. The design and analysis of clinical experiments. New York, N.Y: John Wiley & Sons, Inc.; 1986. p. 11. [Google Scholar]
- 15.Gallant J E. Strategies for long-term success in the treatment of HIV infection. JAMA. 2000;283:1329–1334. doi: 10.1001/jama.283.10.1329. [DOI] [PubMed] [Google Scholar]
- 16.Ginocchio C C, Tetali S, Washburn D, Zhang F, Kaplan M H. Comparison of levels of human immunodeficiency virus type 1 RNA in plasma as measured by the NucliSens nucleic acid sequence-based amplification and Quantiplex branched-DNA assays. J Clin Microbiol. 1999;37:1210–1212. doi: 10.1128/jcm.37.4.1210-1212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 18.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A for the AIDS Clinical Trials Group 320 Study Team. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 19.Havlichek D H, Jr, Hage-Korban E. False-positive HIV diagnosis by HIV-1 plasma viral load testing. Ann Intern Med. 1999;131:794. doi: 10.7326/0003-4819-131-10-199911160-00027. [DOI] [PubMed] [Google Scholar]
- 20.Havlir D V, Hellmann N S, Petropoulos C J, Whitcomb J M, Collier A C, Hirsch M S, Tebas P, Sommadossi J P, Richman D D. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA. 2000;283:229–234. doi: 10.1001/jama.283.2.229. [DOI] [PubMed] [Google Scholar]
- 21.Highbarger H C, Alvord W G, Jiang M K, Shah A S, Metcalf J A, Lane H C, Dewar R L. Comparison of the Quantiplex version 3.0 assay and a sensitized Amplicor Monitor assay for measurement of human immunodeficiency virus type 1 RNA levels in plasma samples. J Clin Microbiol. 1999;37:3612–3614. doi: 10.1128/jcm.37.11.3612-3614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodroski J. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 23.Manegold C, Krempe C, Jablonowski H, Kajala L, Dietrich M, Adams O. Comparative evaluation of two branched-DNA human immunodeficiency virus type 1 RNA quantification assays with lower detection limits of 50 and 500 copies per milliliter. J Clin Microbiol. 2000;38:914–917. doi: 10.1128/jcm.38.2.914-917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 25.Michael N L, Herman S A, Kwok S, Dreyer K, Wang J, Christopherson C, Spadoro J P, Young K K Y, Polonis V, McCutchan F E, Carr J, Mascola J R, Jagodzinsk L L, Robb M L. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved Amplicor HIV-1 Monitor test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montpetit M. HIV-1 subtype A in Canada. AIDS Res Hum Retrovir. 1995;11:1421–1422. doi: 10.1089/aid.1995.11.1421. [DOI] [PubMed] [Google Scholar]
- 27.Murphy D G, Gonin P, Fauvel M. Reproducibility and performance of the second-generation branched-DNA assay in routine quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:812–814. doi: 10.1128/jcm.37.3.812-814.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nkengasong J N, Kalou M, Maurice C, Bile C, Borget M-Y, Koblavi S, Boateng E, Sassan-Morokro M, Anatole-Ehounou E, Ghys P, Greenberg A E, Wiktor S Z. Comparison of NucliSens and Amplicor Monitor assays for quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma of persons with HIV-1 subtype A infection in Abidjan, Côte d'Ivoire. J Clin Microbiol. 1998;36:2495–2498. doi: 10.1128/jcm.36.9.2495-2498.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolte F S. Branched DNA signal amplification for direct quantitation of nucleic acid sequences in clinical specimens. Adv Clin Chem. 1998;33:201–235. doi: 10.1016/s0065-2423(08)60209-7. [DOI] [PubMed] [Google Scholar]
- 30.Nolte F S, Boysza J, Thurmond C, Clark W S, Lennox J L. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:716–720. doi: 10.1128/jcm.36.3.716-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien W A, Hartigan P M, Daar E S, Simberkoff M S, Hamilton J D for the VA Cooperative Study Group on AIDS. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann Intern Med. 1997;126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 32.Raboud J M, Montaner J S, Conway B, Rae S, Reiss P, Vella S, Cooper D, Lange J, Harris M, Wainberg M A, Robinson P, Myers M, Hall D. Suppression of plasma viral load below 20 copies/ml is required to achieve a long-term response to therapy. AIDS. 1998;12:1619–1624. doi: 10.1097/00002030-199813000-00008. [DOI] [PubMed] [Google Scholar]
- 33.Rich J D, Merriman N A, Mylonakis E, Greenough T C, Flanigan T P, Mady B J, Carpenter C C J. Misdiagnosis of HIV infection by HIV-1 plasma viral load testing:a case series. Ann Intern Med. 1999;130:37–39. doi: 10.7326/0003-4819-130-1-199901050-00007. [DOI] [PubMed] [Google Scholar]
- 34.Saag M S, Holodniy M, Kuritzkes D R, O'Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 35.Segondy M, Ly T-D, Lapeyre M, Montes B. Evaluation of the NucliSens HIV-1 QT assay for quantitation of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1998;36:3372–3374. doi: 10.1128/jcm.36.11.3372-3374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan P S, Do A N, Ellenberger D, Pau C-P, Paul S, Robbins K, Kalish M, Storck C, Schable C A, Wise H, Tetteh C, Jones J L, McFarland J, Yang C, Lai R B, Ward J W. Human immunodeficiency virus (HIV) subtype surveillance of African-born persons at risk for group O and group N HIV infections in the United States. J Infect Dis. 2000;181:463–469. doi: 10.1086/315254. [DOI] [PubMed] [Google Scholar]
- 37.Sun R, Ku J, Jayakar H, Kuo J-C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Buul C, Cuypers H, Lelie P, Chudy M, Nübling M, Melsert R, Nabbe A, Oudshoorn P. The NucliSens™ Extractor for automated nucleic acid isolation. Infusionsther Transfusionsmed. 1998;25:147–151. [Google Scholar]
- 39.Vandamme A-M, Schmit J-C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, De Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV Monitor test. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 40.Witt D J, Kemper M, Stead A, Ginocchio C C, Caliendo A M. Relationship of incremental specimen volumes and enhanced detection of human immunodeficiency virus type 1 RNA with nucleic acid amplification technology. J Clin Microbiol. 2000;38:85–89. doi: 10.1128/jcm.38.1.85-89.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]