Abstract
Purpose
Colorectal cancer (CRC) is one of the common cancers with a high mortality rate worldwide. In Iran, there has been a trend of increased incidence of colorectal cancer in the last three decades that necessitates the early diagnosis. Genetic factors have an influential role in its etiology along with the conventional risk factors such as age, diet, and lifestyle. Results from GWAS have shown significant associations between SMAD7 gene variants and risk of CRC. This study aimed to assess the association of certain polymorphisms as well as haplotypes of this gene and risk of colorectal cancer.
Methods and materials
This study was designed as a case–control association study. After obtaining ethical approval and informed consent, blood samples from 209 patients with colorectal cancer were collected and DNA was extracted. Four variants: rs4939827, rs34007497, rs8085824 and rs8088297 were genotyped using ARMS-PCR method.
Results
SMAD7 rs4939827 in the recessive and co-dominant models was associated with colorectal cancer risk [TT/CT + CC: OR = 2.90, 95%CI (1.38–6.09), p = 0.005; CC + TT/CT: OR = 1.66, 95%CI (1.00–2.75), p = 0.01]. Haplotype analysis indicated that some SNP combinations including two for-SNPs haplotypes of T-T-C-C and T-C-C-A were significantly associated with CRC risk.
Conclusion
Based on the identified association of SMAD7 gene variations and haplotypes with colorectal cancer risk in our population, genetic variations in this gene region may have a role in CRC development. This data may shed light on the genetic predisposition of CRC which involves different pathways including TGF-β.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-021-01150-3.
Keywords: Colorectal cancer, GWAS, SMAD7, Polymorphisms, Association study
Introduction
Cancer is a heterogenic disease and is one of the most significant health problems in developed, industrialized, and developing countries [1]. Colorectal cancer is the most common malignancy among gastrointestinal malignancies, and it accounts for 13% of all digestive tract malignancies [2]. Based on epidemiologic reports, 60% of CRCs occur in developed countries. However, this figure is continuously decreasing. It is in contrast to developing countries, particularly, Asian countries, where the incidence has been increasing during the two recent decades [3]. This disease has the third and fourth rank among cancers in men and women in incidence and mortality respectively [4]. Over the past three decades, in Iran, colorectal cancer incidence has been increased and reported as the third and fourth most common cancer type in males and females, respectively [5].
Several risk loci in Genome Wide Association Studies (GWAS) studies have indicated that they are associated with CRC risk. Some of these are involved in TGF-β signaling pathway. In this pathway, one of the most common polymorphisms associated with CRC risk is located on SMAD7 gene [6].
It has been reported that the transforming growth factor beta signaling pathway controls some processes such as tumor initiation, cell proliferation, invasion and metastasis. This pathway has been considered as both a tumor suppressor pathway and a promoter of tumor invasion and progression [6]. SMADs are known as receptors and intracellular signal transducers mediating TGF-β signaling pathway [7]. SMAD7 is a kind of inhibitory SMADs, and it is able to binned TGF-β receptor type I, so it acts as a negative regulator of the TGF-β signaling pathway. Therefore the TGF-β signaling is remarkably decreased by the act of SMAD7, which lead to increasing cancer risk. [8, 9].
One of the main problems in CRC is the late diagnosis, therefore, there is an increasing need in molecular studies which lead to the recognition of biological markers for early diagnosis [10]. Due to multifactorial nature of colorectal cancer, identifying genetic and environmental factors involved in its pathogenesis is would help to enhance our understanding for a better management of the disease [11]. The molecular variations related to a particular disorder may result in a new view for a preventive strategy. The development of genome characterization could result in advances in personalized medicine. Single Nucleotide Polymorphisms (SNP) have been shown as appropriate markers for forecasting susceptibility to various diseases such as colorectal cancer and can be considered as a road to personalized medicine [12].
Several studies have reported that influence of risk alleles on CRC risk, owing to allele distributions or specific Linkage Disequilibrium (LD) structure and specific genetic and environmental backgrounds, in various population is different. Therefore, identification of these variations could be a valuable tool for risk prediction in our population [13, 14]. In the present study, we conducted a case–control study to investigate the potential association of SMAD7 gene polymorphisms with the risk of colorectal cancer in an Iranian population. Moreover, we assessed the association of these markers with the clinical features and tumor characteristics.
Methods and materials
Study populations
The study was performed in accordance with Declaration of Helsinki and relevant guidelines by the institutional ethics committee. The study was approved by the ethics committee of Mashhad University of Medical Sciences under the ethical approval number of (IR.MUMS.SM.REC.1396.234). The inclusion criteria determined which are as follows: 1- Any patients diagnosed with sporadic CRC who had Age ≥ 40 years. With consideration of the following exclusion criteria. 1-Familial CRC /strong family history of colorectal cancer were excluded. 2- Known genetic susceptibility syndromes (e.g., Lynch Syndrome, FAP) were excluded. 3- Inability to provide informed consent. In this case–control study, a total of 409 Iranian individuals in a period of time between 2016 and 2018 were enrolled. The case group included 209 patients with sporadic CRC diagnosis confirmed with positive colonoscopy and pathology result, and the control group consisted of 200 healthy controls who had normal colonoscopy results. We collected a 5-ml peripheral venous blood sample from each subject (all healthy controls and patients) after getting written informed consent.
SNP selection DNA isolation and genotyping
In our study, selection of polymorphisms was performed based on several criteria; including validation of the association in numerous GWAS studies which denotes a strong association with colorectal cancer risk in different populations. We also considered selecting SNPs that are located in the same region to be able to perform haplotype analysis to examine the overall effect of these polymorphisms. We also considered selecting markers with an acceptable MAF and heterozygosity to achieve the highest possible study power. Therefore, based on these criteria, we selected four polymorphisms to have a more comprehensive picture of a possible association of SMAD7 gene with CRC. Three of them were previously reported to be associated with colorectal cancer and are located in the intronic region (rs4939827, rs34007497, rs8085824) and rs8088297 is located in 3′UTR. Minor allele frequencies of all SNPs were > 5%, and average heterozygosity was more than 0.3. Information related to the SNPs was obtained from the National Center of Biotechnology Information (NCBI) SNP database (www.ncbi.nlm.nih.gov/ SNP, dbSNP BUILD 156).
Collection of peripheral blood was done from each participant in EDTA coated vials and kept at − 80 °C. Genomic DNA extracted by the standard salting-out method. The quality of extracted DNA confirmed using 1% agarose gel electrophoresis containing Green Viewer. The concentration and purity of DNA were also evaluated utilizing Epoch™ Microplate Spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA). ARMS (The Amplification-Refractory Mutation System) method was applied as the main genotyping technique. Primers were designed using Primr1 and WASP primer online software (www.primer1.soton.ac.uk/primer1.html, www.bioinfo.biotec.or.th/WASP/). The specifications of primers and set up protocols have been presented in Table 1. PCR reactions were performed in 10 µl total volume, comprising 1 µl of DNA (200 ng), 5 µl of Taq DNA Polymerase 2 × Master Mix (Ampliqon), 0.3 µl (10 μM) of each primer for rs34007497 and rs8085824. 1.3 µl (10 μM) forward prime, 1.6 µl (10 μM) reverse primer and 1 µl (10 μM) of each control primer for rs4939827. And also 1 µl (10 μM) of each primer for rs8088297.
Table 1.
Information for SMAD7 SNPs primers
| SNP | Primer sequence | Primer type | Tm | PCR product size | |
|---|---|---|---|---|---|
| rs4939827 | CTCATCCAAAAGAGGACAC | Forward inner (C Allele) | 54 | 140 bp | 270 bp |
| GTTTTCTGATTAACTCGCAGT | Reverse outer | ||||
| ATTCACAAGGACCCTTGCT | Forward outer | 58 | 167 bp | ||
| AGGGAGCTCTGGGGTCATA | Reverse inner (T Allele) | ||||
| rs34007497 | AGACTCCTGCCTGCTCCTCTC | Forward inner (C Allele) | 64 | 203 bp | 553 bp |
| TCCCCAGAAGGTGTGATGTCCA | Reverse outer | ||||
| AGTGAGTGGAGTATGGACCTGAGATCG | Forward outer | 64 | 393 bp | ||
| GGGGAGCTGGAGGGGACAGCCAC | Reverse inner (G Allele) | ||||
| rs8085824 | GTCCCCCTTCTCCCCTGCC | Forward inner (C Allele) | 62 | 191bp | |
| CGTCCCCCTTCTCCCCTGCT | Forward inner (T Allele) | ||||
| CCTTACCAGAATCACCCCG | Reverse outer | ||||
| rs8088297 | TAATAGCAGAGCTTAACACACAAGA | Forward inner (A Allele) | 62 | 136 bp | 309 bp |
| TCACAGTATTGCTCACCCAGT | Reverse outer | ||||
| TGAGCTAAGAACAGTGTCGAAGTA | Forward outer | 70 | 217 bp | ||
| TCAGCACTGCCTGCTCCTAG | Reverse inner (C Allele) |
Statistical analysis
Deviations from Hardy–Weinberg equilibrium was assessed by using the Chi-square test. Also The chi-square test was done for evaluation of allele and genotype frequencies between cases and control groups. Odds Ratios (OR) and corresponding 95% confidence intervals (95% CI) were calculated to estimate the conferred risk by the risky alleles. Results were also adjusted for potential confounders such as age, gender and BMI in the logistic regression analysis. To compare age and BMI between two groups (patients and healthy people), the Student’s t-test was performed. Associations between mentioned SNPs and CRC risk was evaluated for three different genetic models (dominant, recessive and allelic model) analysis. In addition, haplotype analysis was performed using PHASE v.2.1.1 software. The LD has been calculated by 2LD software version 1.00. All the statistical analyses were conducted using IBM SPSS Statistics for Windows, Version 16.0 (IBM, USA). p < 0.05 was considered to indicate statistical significance.
Results
Baseline characteristics
A total of 209 colorectal cancer cases and 200 controls were included in this study. The frequency distributions of baseline characteristics and histological parameters are presented in Table 2. In the healthy control group, the number of males and females was equal. No significant differences was indicated in the distribution of sex (p = 0.36) and age (p = 0.19) between cases and controls. Additionally, we investigated the correlations between BMI and the results showed a significant difference between cases and controls (p < 0.001). Regarding tumor status, the proportion of which patients were graded as “well-differentiated” was 11.0%, while moderate and poorly differentiated were 42.5% and 31.1% respectively.
Table 2.
Baseline characteristics of patients (cases) and healthy (control) groups and tumor features
| Characteristic | Cases | Controls | p-value | OR (95%CI) |
|---|---|---|---|---|
| Gender | ||||
| Female | 95(45.5%) | 100(50%) | Ref | |
| Male | 114(54.5%) | 100(50%) | 0.358 | 0.83 (0.56–1.23) |
| Age of diagnosisa (year) | 56.84 ± 13.6 | 57.35 ± 8.4 | 0.192 | 0.99 (0.97–1.01) |
| Smoking status | ||||
| Positive | 19(9.0%) | 29(14.5%) | Ref | |
| Negative | 156(74.6%) | 171(85.5%) | 0.179 | 0.65 (0.35–1.21) |
| Family history | ||||
| Positive | 35(16.7%) | 40(20.0%) | Ref | |
| Negative | 131(62.6%) | 160(80.0%) | 0.321 | 0.78 (0.48–1.27) |
| BMI (Kg/m2)b | 25.5 ± 4.8 | 27.8 ± 4.9 | < 0.001 | 0.90 (0.86–0.95) |
| Tumor location | ||||
| Rectum | 75 (35.8) | |||
| Colon | 59 (28.2) | |||
| Sigmoid | 43 (20.5) | |||
| Grade | ||||
| Poor | 65 (31.1) | |||
| Moderate | 89 (42.5) | |||
| Well | 23 (11.0) |
All statistically significant P-values are shown in bold
aMean ± SD
bBody Mass Index
Association between polymorphisms and the colorectal cancer risk
Table 3 included summarized data of the genotypic and allelic frequencies of four variants in the both case and control groups. The control group genotype distribution followed the Hardy–Weinberg equilibrium. We found a significant association between the rs4939827 SNP and increased CRC risk [p = 0.02, OR = 2.49, 95% CI (1.13–4.49)]. This SNP was associated with CRC risk in the recessive and co-dominant genetic models [TT/CT + CC: OR = 2.90, 95% CI (1.38–6.09), p = 0.005; CC + TT/CT: OR = 1.66, 95% CI (1.00–2.75), p = 0.01].
Table 3.
Distribution of SMAD7 genotypes and allele frequency among colorectal cancer patients and controls
| Genetic model (HWE) | Genotype | Cases (n = 209) | Controls (n = 200) | p-value | OR (95%CI) | p-valueadj.* | OR (95%CI) adj.* |
|---|---|---|---|---|---|---|---|
| rs4939827 (0.1) | CC | 89(42.6%) | 78(40.0%) | Ref | |||
| CT | 83(39.7%) | 101(51.8%) | 0.12 | 0.72(0.47–1.09) | 0.294 | 0.75(0.43–1.29) | |
| TT | 37(17.7%) | 16(8.2%) | 0.036 | 2.02(1.04–3.92) | 0.024 | 2.49(1.13–4.49) | |
| Dominant model | TT + CT | 120 (57.4%) | 117 (60.0%) | Ref | |||
| CC | 89(42.6%) | 78(40.0%) | 0.598 | 1.11(0.75–1.65) | 0.920 | 1.03(0.62–1.70) | |
| Recessive model | CC + CT | 172 (82.3%) | 179 (91.8%) | Ref | |||
| TT | 37(17.7%) | 16(8.2%) | 0.006 | 2.40(1.29–4.48) | 0.005 | 2.90(1.38–6.09) | |
| Multiplicative model | C | 157(37.6%) | 133(34.1%) | Ref | |||
| T | 261(62.4%) | 257(65.9%) | 0.350 | 0.87 (0.65–1.16) | 0.189 | 0.78 (0.54–1.13) | |
| Co-dominant model | CT | 83(39.7%) | 101(51.8%) | Ref | |||
| CC + TT | 126 (60.3%) | 94 (48.2%) | 0.015 | 1.63 (1.10–2.42) | 0.049 | 1.66 (1.00–2.75) | |
| rs34007497 (0.98) | CC | 135 (64.6%) | 126 (63.0%) | Ref | |||
| CG | 52 (24.9%) | 66 (33.0%) | 0.168 | 0.73(0.47–1.14) | 0.183 | 0.68 (0.39–1.20) | |
| GG | 22 (10.5%) | 8 (4.0%) | 0.029 | 2.56 (1.10–5.97) | 0.327 | 1.71 (0.558–5.04) | |
| Recessive model | CC + CG | 187 (89.5%) | 192 (96.0%) | Ref | |||
| GG | 22 (10.5%) | 8 (4.0%) | 0.015 | 2.82(1.22–4.50) | 0.227 | 1.93 (0.66–5.59) | |
| Dominant model | GG + CG | 74 (35.4%) | 74 (37.0%) | Ref | |||
| CC | 135 (64.6%) | 126 (63.0%) | 0.737 | 1.07(0.71–1.60) | 0.379 | 0.79 (0.47–1.33) | |
| Multiplicative model | C | 96 (22.9%) | 82 (20.5%) | Ref | |||
| G | 322 (77.1%) | 318 (79.5%) | 0.393 | 1.16(0.83–1.63) | 0.788 | 0.94 (0.61–1.46) | |
| Co-dominant model | CC + GG | 157 (75.1%) | 134 (67.0%) | Ref | |||
| CG | 52 (24.9%) | 66 (33.0%) | 0.071 | 0.67 (0.44–1.03) | 0.135 | 0.65 (0.37–1.14) | |
| rs8085824 (0.53) | TT | 58 (28.9%) | 77 (38.5%) | Ref | |||
| CT | 97 (48.3%) | 100 (50.0%) | 0.013 | 0.48 (0.27–0.86) | 0.092 | 0.53 (0.26–1.11) | |
| CC | 46 (22.8%) | 23 (11.5%) | 0.002 | 2.06 (1.45–4.86) | 0.086 | 1.95 (0.91–4.20) | |
| Recessive model | CT + TT | 155 (77.1%) | 177 (88.5%) | Ref | |||
| CC | 46 (22.9%) | 23 (11.5%) | 0.042 | 1.54 (1.02–2.34) | 0.477 | 1.21 (0.71–2.07) | |
| Dominant model | CC + CT | 143 (71.1%) | 123 (61.5%) | Ref | |||
| TT | 58 (28.9%) | 77 (38.5%) | 0.003 | 0.44 (0.25–0.75) | 0.067 | 0.52 (0.26–1.05) | |
| Multiplicative model | T | 189 (56.4%) | 213 (45.6%) | Ref | |||
| C | 146 (43.6%) | 254 (54.4%) | 0.003 | 0.65 (0.49–0.86) | 0.150 | 0.76 (0.53–1.10) | |
| Co-dominant model | CC + TT | 104 (51.7%) | 100 (50.0%) | Ref | |||
| CT | 97 (48.3%) | 100 (50.0%) | 0.727 | 0.93 (0.63–1.38) | 0.537 | 0.85 (0.51–1.42) | |
| rs8088297 (0.67) | CC | 3 (1.4%) | 3 (1.5%) | Ref | |||
| AC | 15 (7.2%) | 33 (16.5%) | 0.367 | 0.35 (0.08–2.52) | 0.306 | 0.35 (0.05–2.62) | |
| AA | 191 (91.4%) | 164 (82.0%) | 0.853 | 1.16 (0.23–5.85) | 0.645 | 1.55 (0.23–10.33) | |
| Recessive model | CC + AC | 18 (8.6%) | 36 (18.0%) | Ref | |||
| AA | 191 (91.4%) | 164 (82.0%) | 0.006 | 2.33 (1.27–4.26) | 0.203 | 1.62 (0.77–3.40) | |
| Dominant model | AA + AC | 206 (98.6%) | 197 (98.5%) | Ref | |||
| CC | 3 (1.4%) | 3 (1.5%) | 0.957 | 0.96 (0.19–4.79) | 0.587 | 0.59 (0.09–3.92) | |
| Multiplicative model | C | 21 (5.02%) | 39 (9.7%) | Ref | |||
| A | 397 (94.98%) | 361 (90.3%) | 0.011 | 2.04 (1.18–3.54) | 0.319 | 1.41 (0.72–2.75) | |
| Co-dominant model | CC + AA | 194 (92.8%) | 167 (83.5%) | Ref | |||
| AC | 15 (7.2%) | 33 (16.5%) | 0.004 | 0.39 (0.20–0.74) | 0.129 | 0.54 (0.24–1.20) |
All statistically significant P-values are shown in bold
*Adjusted for age, sex and BMI
The results showed a significant association between the other three polymorphisms and colorectal cancer.rs34007497 was associated with the risk of disease in recessive genetic model (GG/CC + CG, p = 0.01). rs8085824 indicated the association with colorectal cancer in dominant (CC + CT/TT, p = 0.003), recessive (CC/CT + TT, p = 0.04) and allelic (T/C, p = 0.003) genetic models. Furthermore, rs8088297 was associated with colorectal cancer risk in recessive (CC + AC/AA: OR = 2.33, p = 0.006), co-dominant (AC/CC + AA, p = 0.004) and allelic (A/C, p = 0.01) genetic models. Although after adjustment for age, gender and BMI as a confounder factor, there was no association between these three polymorphisms and colorectal cancer risk. Electrophoresis results of polymorphisms genotyping are shown in Fig. 1.
Fig. 1.
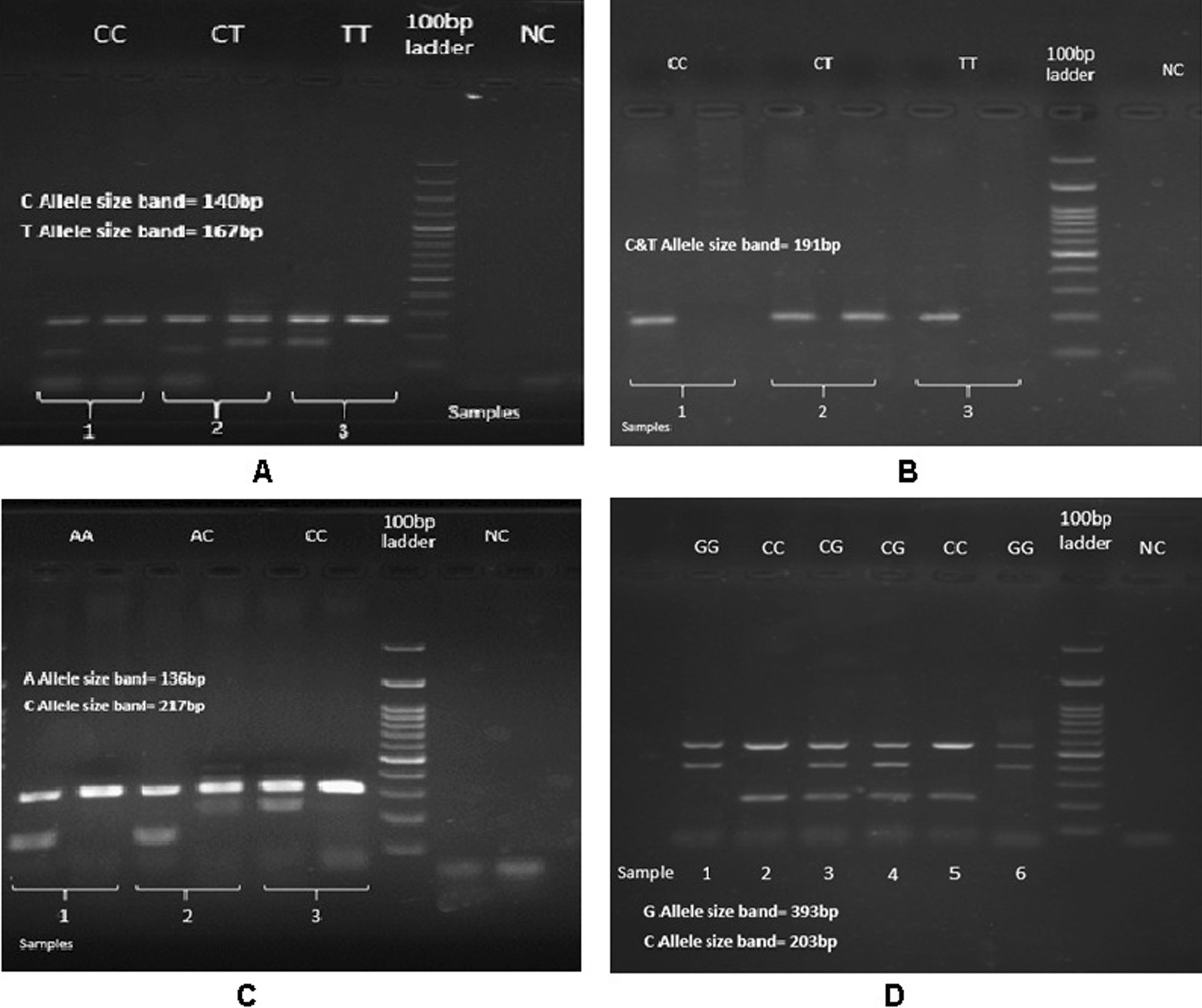
Agarose gel electrophoresis (2%) showing various genotypes of four polymorphisms as follows: A, Represent rs4939827, sample one; CC genotype (140 bp), sample 2; CT genotype (140, 167 bp) and Sample 3; TT genotype (167 bp). B, Represent rs8085824, sample one; CC genotype, sample 2; CT genotype and Sample 3; TT genotype(C & T Allel size band 191 bp). C, Represent rs8088297, sample one; AA genotype (136 bp), sample 2; AC genotype (136, 217 bp) and Sample 3; CC genotype (217 bp). D, Represent rs34007497, sample 1, 6; GG genotype (393 bp), sample 3, 4; CG genotype (203, 393 bp) and Sample 2, 5; CC genotype (203 bp)
Haplotype analysis showed two haplotypes, T-C haplotype of rs8085824- rs34007497, as well as T-C and C-T haplotypes of rs8085824- rs4939827 were associated with CRC risk. In addition, C-C haplotype of rs8085824- rs4939827 conferred a decreased risk of CRC. On the other hand, in haplotypes consisting of rs8085824 - rs34007497- rs4939827 the distribution of three haplotypes C-C-T, C-C-C and T-C-C were different between patients and healthy groups. Also, A-T-C and A-C-T haplotypes of rs8088297 - rs8085824- rs4939827 revealed an elevated risk of CRC. The results have been shown in details in Table 4. In four-SNP haplotype, three haplotypes including T-T-C-C, T-C-C-A and T-C-C-C were associated with risk of CRC. No significant association with the risk of CRC show in other haplotypes which are shown in (Additional file 1 - haplotypes distribution; Table S1).
Table 4.
Distribution of associated haplotypes with risk of colorectal cancer
| Polymorphisms | Haplotype | Case | Control | p-value adj* | OR (95%CI) adj.* |
|---|---|---|---|---|---|
| rs8085824-rs34007497 | Others | 289 (69.1%) | 310 (77.5%) | Ref | |
| T-C | 129 (30.9%) | 90 (22.5%) | 0.029 | 1.55 (1.05–2.95) | |
| rs8085824-rs4939827 | Others | 332 (79.4%) | 362 (90.5%) | Ref | |
| T-C | 86 (20.6%) | 38 (9.5%) | 0.005 | 2.05 (1.24–3.42) | |
| Others | 368 (88.0%) | 373 (93.2%) | Ref | ||
| C-T | 50 (12.0%) | 27 (6.8%) | 0.001 | 2.67 (1.51–4.72) | |
| Others | 243 (58.1%) | 173 (43.3%) | Ref | ||
| C-C | 175 (41.9%) | 227 (56.7%) | 0.002 | 0.56 (0.40–0.80) | |
| rs8088297-rs8085824-rs34007497 | Others | 292 (69.9%) | 312 (78.0%) | Ref | |
| A-T-C | 126 (30.1%) | 88 (22.0%) | 0.047 | 1.49 (1.00–2.22) | |
| rs8088297-rs8085824-rs4939827 | Others | 329 (78.7%) | 364 (91.0%) | Ref | |
| A-T-C | 89 (21.3%) | 36 (9.0%) | 0.001 | 2.35 (1.42–3.89) | |
| Others | 369 (88.3%) | 373 (93.2%) | Ref | ||
| A-C-T | 49 (11.7%) | 27 (6.8%) | 0.002 | 2.52 (1.42–4.46) | |
| Others | 262 (62.7%) | 198 (49.5%) | Ref | ||
| A-C-C | 156 (37.3%) | 202 (50.0%) | 0.002 | 0.57 (0.40–0.82) | |
| rs8085824-rs34007497-rs4939827 | Others | 357 (85.4%) | 371 (92.7%) | Ref | |
| T-C-C | 61 (14.6%) | 29 (7.3%) | 0.002 | 2.38 (1.36–4.15) | |
| Others | 374 (89.5%) | 374 (93.5%) | Ref | ||
| C-C-T | 44 (10.5%) | 26 (6.5%) | 0.004 | 2.40 (1.33–4.35) | |
| Others | 269 (64.4%) | 198 (49.5%) | Ref | ||
| C-C-C | 149 (35.6%) | 202 (50.5%) | 0.001 | 0.54 (0.38–0.77) | |
| rs4939827-rs34007497-rs8085824-rs8088297 | Others | 358 (85.6%) | 373 (93.2%) | Ref | |
| T-T-C-C | 60 (14.4%) | 27 (6.8%) | 0.002 | 2.42 (1.37–4.27) | |
| Others | 374 (89.5%) | 375 (93.8%) | Ref | ||
| T-C-C-A | 44 (10.5%) | 25 (6.2%) | 0.001 | 2.63 (1.45–4.75) | |
| Others | 275 (65.8%) | 221 (55.2%) | Ref | ||
| T-C-C-C | 143 (34.2%) | 179 (44.8%) | 0.009 | 0.61 (0.43–0.88) |
All statistically significant P-values are shown in bold
*Adjusted for age, sex and BMI
Discussion
The role of TGF-β signaling has proven to be important in homeostasis, cell differentiation and tumor suppression [15, 16]. Additionally, it is a pleiotropic cytokine which has a dual function in cancer development: it acts as a tumor suppressor and a tumor promoter in the early stages and late stages [17]. The production of TGF-β increased in various tumor types, such as CRC [18]. Regulation of TGF-β by SMAD7 is critical to preserve gastrointestinal homeostasis. SMAD7 as an intracellular antagonist of TGF-β signaling binned to the receptor compound and act as an obstacle for initiation of downstream signaling proceedings [19, 20]. SMAD7 interacts with activated TGF-β type I receptor, thereby blocking the phosphorylation and activation of SMAD2 thus resulting in lack of formation of SMAD2/SMAD4 complexes and ultimately inhibiting binding to transcription factors and expression of target genes by preventing of entering the complex (SMAD2/SMAD4) into the nucleus [6]. The influence of SMAD7 expression has been proven in CRC progression. High-level expression of SMAD7 mRNA in CRC cell lines was reported to boost cell growth and prevent apoptosis via a mechanism dependent on the suppression of TGF-β signaling pathway [21]. Several investigations have examined numerous genetic variations within SMAD7 gene (on 18q21). Broderick et al. performed a genome-wide association study which revealed that the SMAD7 intron 3 was highly polymorphic, harboring one of the associated SNP- rs4939827 [22]. However, most of the association studies have been performed on European Caucasian populations. Because of differences in the incidence of CRC and the allele distribution of SNPs across populations, this study investigated four candidate variants to examine the association of these SNPs and CRC risk in an Iranian population. We also evaluated the association of these polymorphisms with clinicopathological characteristics such as age, sex, tumor location, tumor grade and family history.
The three polymorphisms rs4939827 (C > T), rs34007497 (C > G) and rs8085824 (C > T) are located in the intronic region of the SMAD7 gene. Our results suggest a significant association in the genotype distribution of rs4939827 between cases and controls. Previous studies have also mainly shown the association between this SNP and the risk of CRC [22–24]. Our study demonstrated that rs4939827 TT genotype increases the risk of CRC. The results of our study were in line with a meta-analysis by Huang Y. et al. including 37 studies on rs4939827 (48,751 cases and 61,529 controls) [25]. They found an association between this SNP and colorectal cancer risk with a 15% increase in the risk of disease. Although in our study, we did not find any significant association between the above mentioned polymorphisms and CRC susceptibility, even when stratifying the samples by age, gender, tumor site or family history. Mates and colleagues demonstrated that carriers of the T allele had an almost fivefold greater risk of developing rectal cancer compared with colon cancer [26]. Regarding the function of selected polymorphisms, none of four SNP in this study has a specific function.
At first, we found a statistically significant association between the rs34007497, rs8085824 and rs8088297 with the risk of CRC in our study.
However, after adjustment for some confounder factors (age, sex and BMI) in different genetic models the associations of these three polymorphisms did not remain significant. Therefore, we can conclude that these three markers were not associated independently with colorectal cancer risk.
rs8088297 (A > C) is located in the 3′UTR region of the SMAD7 gene. Since The 3′ and 5′-UTRs of mRNAs have a regulating role, so it is crucial. mRNA folding is affected by sequence alterations in the UTR regions. SNPs in 3′-UTRs can change target recognition of microRNAs by disrupting sequence complementarity [27]. Super-enhancers define a cell’s identity as large enhancer clusters which in several studies approximately whole of the human SMAD7 gene categorized as a super-enhancer in colon crypts, sigmoid colon and fetal intestine [28].
Haplotype-based association analysis has numerous advantages over one-SNP approach. First, the loss of information, imputable to bi-allelic rather than multi-allelic loci, can be overcome by assessing haplotypes rather than single-locus tests. Second, methods based on haplotypes are regarded as more powerful than those based on single markers. Third, haplotype-based methods can potentially detect associations of cis-interactions SNPs that are among nearby SNPs. In light of these aforementioned benefits, we performed a haplotype analysis [29]. In our study we found a significant association of several haplotype combinations [(rs8085824-rs34007497- rs4939827); (rs8088297-rs8085824-rs4939827); (rs8088297-rs8085824-rs34007497); (rs8085824- rs34007497); (rs8085824-rs4939827); (rs4939827-rs34007497-rs8085824-rs8088297)] with CRC risk. It can reflect the effect of multiple SNPs on the risk of the disease which remained significant even after adjustment for confounders (age, sex, BMI). Therefore, the haplotype analysis further illustrated the pivotal role of these variations in combination with each other where they can confer some degrees of risk.
As mentioned before, we selected a variety of polymorphisms on SMAD7 genes in different positions to have an overview of the whole gene. One of the strengths of this study is the evaluation of several markers across the gene and haplotype analysis which can increase the study power. To the best of our knowledge, none of the previous studies looked at haplotypes of this gene in terms of association with CRC. Moreover, replication of association of the rs4939827 With CRC in our population can strengthen the evidence of the effect of this genetic risk factor on CRC which can be used in clinical risk assessment, especially in direct-to-consumer genetic testing.
The development of genome characterization could result in advances in personalized medicine. Variations have been shown as appropriate markers for forecasting susceptibility to various diseases such as colorectal cancer and can be considered as a road to personalized medicine.
Conclusion
To the best of our knowledge, this is the first case–control study which has assessed the impact of these four SNPs on clinicopathological features and CRC risk in an Iranian population. Our results suggest that the rs4939827 is associated with CRC susceptibility in the Iranian population. Future large population-based studies are needed to confirm the accuracy of findings and to explore gene–environment interactions as well. Ultimately, future studies looking at the functional activity of these variations and their influence on tumorigenesis would help us to realize the principal mechanisms involved in CRC development.
Supplementary Information
Additional file 1. Table S1. Distribution of SMAD7 Haplotypes among Colorectal Cancer Patients and Controls.
Acknowledgements
The authors gratefully thank the study participants for their contributions and commitment to this project.
Authors’ contributions
M.A., L.H.: Performed laboratory tests, data analysis and drafting the initial manuscript. F.A.J, A.K.S., M.A.K., A.F.P., S.A.A.D., A.A., A.B., and H.M.M.: helped with sample recruitment, data collection and data analysis. L.G.: supervised the project and managed clinical investigation. A.P.: Designed and conceived the study, supervised the project, edited and finalized the study. All authors read and approved the final manuscript.
Funding
This work was based on the Master of Science thesis of Ms. Maryam Alidoust and was financially supported by Mashhad University of Medical Sciences (Grant No: 951659).
Availability of data and material
The datasets created during the current study are not publicly accessible due to the possibility of compromising the privacy of individuals. According to the written approval forms accepted by the Ethics Committee of the Mashhad University of Medical Sciences (MUMS), the data will only be available to researchers within project. The data would be available upon request from the corresponding authors (according to the MUMS rules and regulations).
Declarations
Ethics approval and consent to participate
The study was performed in accordance with Declaration of Helsinki and relevant guidelines by the institutional ethics committee. The study was approved by the ethics committee of Mashhad University of Medical Sciences under the ethical approval number of (IR.MUMS.SM.REC.1396.234). In addition, informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors report that there are no conflicts of interest in this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Leila Hamzehzadeh, Asma Khorshid Shamshiri, Fahimeh Afzaljavan and Ladan Goshayeshi have contributed equally to this work.
Contributor Information
Ladan Goshayeshi, Email: GoshayeshiL@mums.ac.ir.
Alireza Pasdar, Email: a.pasdar@abdn.ac.uk, Email: pasdara@mums.ac.ir.
References
- 1.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. 2018;68(1):7–30. [DOI] [PubMed]
- 3.Dolatkhah R, Somi MH, Bonyadi MJ, Asvadi Kermani I, Farassati F, Dastgiri S. Colorectal cancer in Iran: molecular epidemiology and screening strategies. J Cancer Epidemiol. 2015;2015:643020. [DOI] [PMC free article] [PubMed]
- 4.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Dolatkhah R, Somi MH, Kermani IA, Ghojazadeh M, Jafarabadi MA, Farassati F, et al. Increased colorectal cancer incidence in Iran: a systematic review and meta-analysis. BMC Public Health. 2015;15:997. doi: 10.1186/s12889-015-2342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-beta signalling. Biochem J. 2011;434(1):1–10. doi: 10.1042/BJ20101827. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Yang XX, Hu NY, Sun JZ, Li FX, Li M. A risk-associated single nucleotide polymorphism of SMAD7 is common to colorectal, gastric, and lung cancers in a Han Chinese population. Mol Biol Rep. 2011;38(8):5093–5097. doi: 10.1007/s11033-010-0656-3. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Li Y, Gao LB, Wang YY, Zhou B, Lv ML, et al. Vitamin D receptor gene polymorphisms and the risk of colorectal cancer in a Chinese population. Dig Dis Sci. 2009;54(3):634–639. doi: 10.1007/s10620-008-0375-y. [DOI] [PubMed] [Google Scholar]
- 11.Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203. doi: 10.1016/j.clcc.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Mias GI, Snyder M. Personal genomes, quantitative dynamic omics and personalized medicine. Quant Biol (Beijing, China) 2013;1(1):71–90. doi: 10.1007/s40484-013-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64(4):203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- 14.Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, et al. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet. 2005;13(5):677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Zhang LJ, Zhang HR, Tian GF, Tian J, Mao XL, et al. Tumor-derived transforming growth factor-beta is critical for tumor progression and evasion from immune surveillance. Asian Pac J Cancer Prev. 2014;15(13):5181–5186. doi: 10.7314/APJCP.2014.15.13.5181. [DOI] [PubMed] [Google Scholar]
- 16.Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Papageorgis P, Stylianopoulos T. Role of TGFβ in regulation of the tumor microenvironment and drug delivery (review) Int J Oncol. 2015;46(3):933–943. doi: 10.3892/ijo.2015.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;16 Spec No 1:R14–20. [DOI] [PMC free article] [PubMed]
- 19.Hong S, Lee H-J, Kim SJ, Hahm K-B. Connection between inflammation and carcinogenesis in gastrointestinal tract: focus on TGF-beta signaling. World J Gastroenterol. 2010;16(17):2080–2093. doi: 10.3748/wjg.v16.i17.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40(5):631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grau AM, Datta PK, Zi J, Halder SK, Beauchamp RD. Role of Smad proteins in the regulation of NF-kappaB by TGF-beta in colon cancer cells. Cell Signal. 2006;18(7):1041–1050. doi: 10.1016/j.cellsig.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39(11):1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 23.Ho JW, Choi Sc, Lee Yf, Hui TC, Cherny SS, Garcia-Barceló MM, et al. Replication study of SNP associations for colorectal cancer in Hong Kong Chinese. British journal of cancer. 2011;104(2):369–75. [DOI] [PMC free article] [PubMed]
- 24.Song Q, Zhu B, Hu W, Cheng L, Gong H, Xu B, et al. A common SMAD7 variant is associated with risk of colorectal cancer: evidence from a case-control study and a meta-analysis. PLoS ONE. 2012;7(3):e33318-e. [DOI] [PMC free article] [PubMed]
- 25.Huang Y, Wu W, Nie M, Li C, Wang L. SMAD7 polymorphisms and colorectal cancer risk: a meta-analysis of case-control studies. Oncotarget. 2016;7(46):75561–75570. doi: 10.18632/oncotarget.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mates IN, Jinga V, Csiki IE, Mates D, Dinu D, Constantin A, et al. Single nucleotide polymorphisms in colorectal cancer: associations with tumor site and TNM stage. J Gastrointestin Liver Dis. 2012;21(1):45–52. [PubMed] [Google Scholar]
- 27.Zheng J, Jiang L, Zhang L, Yang L, Deng J, You Y, et al. Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis. 2012;33(12):2409–2416. doi: 10.1093/carcin/bgs307. [DOI] [PubMed] [Google Scholar]
- 28.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N, Zhang K, Zhao H. Haplotype-association analysis. Adv Genet. 2008;60:335–405. doi: 10.1016/S0065-2660(07)00414-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Distribution of SMAD7 Haplotypes among Colorectal Cancer Patients and Controls.
Data Availability Statement
The datasets created during the current study are not publicly accessible due to the possibility of compromising the privacy of individuals. According to the written approval forms accepted by the Ethics Committee of the Mashhad University of Medical Sciences (MUMS), the data will only be available to researchers within project. The data would be available upon request from the corresponding authors (according to the MUMS rules and regulations).


