Abstract
Acute myocardial infarction (AMI) is a common cause of death in numerous countries. Understanding the molecular mechanisms of the disease and analyzing potential biomarkers of AMI is crucial. However, specific diagnostic biomarkers have thus far not been fully established and candidate regulatory targets for AMI remain to be determined. In the present study, the AMI gene chip dataset GSE48060 comprising blood samples from control subjects with normal cardiac function (n=21) and patients with AMI (n=26) was downloaded from Gene Expression Omnibus. The differentially expressed genes (DEGs) between the AMI and control groups were identified with the online tool GEO2R. The co-expression network of DEGs was analyzed by calculating the Pearson correlation coefficient of all gene pairs, mutual rank screening and cutoff threshold screening. Subsequently, the Gene Ontology (GO) database was used to analyze the genes' functions and pathway enrichment of genes in the most important modules was performed. Kyoto Encyclopedia of Genes and Genomes (KEGG) Disease and BioCyc were used to analyze the hub genes in the module to determine important sub-pathways. In addition, the expression of hub genes was confirmed by reverse transcription-quantitative PCR in AMI and control specimens. In the present study, 52 DEGs, including 26 upregulated and 26 downregulated genes, were identified. As key hub genes, three upregulated genes (AKR1C3, RPS24 and P2RY12) and three downregulated genes (ACSL1, B3GNT5 and MGAM) were identified from the co-expression network. Furthermore, GO enrichment analysis of all AMI co-expression network genes revealed functional enrichment mainly in ‘RAGE receptor binding’ and ‘negative regulation of T cell cytokine production’. In addition, KEGG Disease and BioCyc analysis indicated functional enrichment of the genes RPS24 and P2RY12 in ‘cardiovascular diseases’, of AKR1C3 in ‘cardenolide biosynthesis’, of MGAM in ‘glycogenolysis’, of B3GNT5 in ‘glycosphingolipid biosynthesis’ and of ACSL1 in ‘icosapentaenoate biosynthesis II’. In conclusion, the hub genes AKR1C3, RPS24, P2RY12, ACSL1, B3GNT5 and MGAM are potential markers of AMI, and have potential application value in the diagnosis of AMI.
Keywords: acute myocardial infarction, differentially expressed genes, co-expression network analysis, hub genes, functional enrichment analysis
Introduction
Despite significant progress in vascular remodeling strategies, drug therapy, cardiac rehabilitation algorithms and organ transplantation, acute myocardial infarction (AMI), as a cardiovascular disease, remains a major cause of morbidity and mortality worldwide (1). There are numerous risk factors for AMI, including smoking, obesity, high serum cholesterol, hypertension and diabetes, which may partly predict AMI, but they are not sufficient to provide an acute diagnosis (2,3). Despite countless efforts, the prevention and treatment of this disease remains a major challenge for scientists. At present, this disease has become the major and most common threat to human life (4,5). Therefore, it is urgent to reveal the pathogenic mechanisms of AMI and develop novel treatment strategies.
Early detection of AMI contributes to early treatment interventions and may significantly reduce mortality (6). Numerous studies have investigated potential molecular biomarkers for AMI detection; certain genes and proteins, such as monocyte-platelet aggregation, cardiac fatty acid binding protein and troponin I, have been established as effective markers for the diagnosis of AMI (6-8). In addition, a large number of microRNAs (miRs) are considered to be key markers for AMI, including circulating miR-26a-1(9), miR-17-5p (10) and miR-23a (11). However, interactions between these molecules and altered pathways were rarely reported, and the molecular pathogenesis of AMI has remained largely elusive. Suresh et al (12) established gene expression profiles through microarrays, and compared the differentially expressed genes (DEGs) between AMI samples and normal samples, as well as the dysregulated pathways involving these DEGs. However, they only focused on pathways and recurring events. The regulatory correlation between these genes was not further investigated. Therefore, a study by Gao et al (13) reanalyzed the GSE48060 microarray dataset from the above study and performed protein-protein interaction (PPI) network analysis and transcription factor network analysis after identifying DEGs in AMI samples. Although they predicted several key genes in the progression of AMI, such as C-C chemokine ligand 5, BCL3 and nuclear receptor coactivator 7, subchannel analysis and co-expression network analysis were not performed (13).
Co-expression network analysis is a useful method that has been widely used for gene expression to identify key disease-related modules (14,15). For instance, using co-expression network analysis, Saris et al (16) selected two large co-expression modules related to amyotrophic lateral sclerosis. By analyzing the gene expression pattern, Azuaje et al (17) determined the weighted correlation network analysis (WGCNA) in myocardial infarction to determine the potential role of collagen 5 α2 and its transcription pattern. Malki et al (18) constructed a gene co-expression network to identify the expression of nerve tumor abdominal wall antigen 1 and ubiquitin-specific peptidase 9, X-linked, and indicated that the most significant modules are related to depression and drug treatment response. A co-expression network analysis identified spleen tyrosine kinase as a candidate oncogene for small cell lung cancer (19). Zhao et al (20), through a co-expression network analysis combined with methylation data analysis, determined that dedicator of cytokinesis factor 2 (DOCK2), DOCK8 and IgG Fc fragments, low-affinity IIa, receptor may represent potential therapeutic targets. Therefore, co-expression network analysis may be used for the analysis of AMI chip data.
In the present study, the gene chip dataset GSE48060 from Suresh et al (12) was reanalyzed and co-expression network analysis, enrichment analysis and PPI analysis were performed. In addition, in terms of DEG screening, compared with Zhang et al (21), in addition to a co-expression network and PPI analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) Disease and BioCyc analyses were also performed on DEGs to deeply investigate gene functions. The present study aimed to reveal the molecular basis of the occurrence and development of AMI, and provide novel and more accurate potential biomarkers for the detection and treatment of AMI.
Materials and methods
Data resource and differential expression analysis
The dataset GSE48060, consisting of 47 microarray expression profiles, was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The sample data were obtained from the peripheral blood of 26 patients without recurrence (AMI samples) and 21 controls with normal heart function (control samples). The platform of the dataset was GPL570 (HG-u133pulus2; Affymetrix human genome U133). The DEGs were analyzed by the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) in the AMI and control groups, which were selected according to a threshold of |log2FC| ≥0.5.
AMI specimens
Between January 1, 2020 and May 30, 2020, 31 cases and 31 controls of whole blood samples were collected from the Cardiovascular Department of Internal Medicine of Central Hospital of Karamay (Karamay, China). The blood samples were collected from first-time AMI patients within 48 h post-MI and controls (with a normal echocardiogram). Patients that had history of cardiovascular disease, or had clinical or biochemical evidence of other comorbidities were excluded. Data of the patients and controls are provided in Table SI.
Co-expression analysis
To construct a co-expression network, the Pearson correlation coefficient (PCC) values of all combinations of the 28,492 unique probes from the GSE48060 series substrate were calculated. Gene pairs with P≤0.05 were selected and the PCC threshold was set to 0.88, corresponding to the 99th percentile of the random PCC distribution as described above (22). According to a previous study (23), the mutual rank (MR) value between gene pairs was also calculated as another value of co-expression to further reduce the number of false positives. Only gene pairs with an absolute MR <10 were considered to be important connections for the co-expression network. The calculation was performed by Bioconductor in R (R version 3.6.2, https://cran.r-project.org/bin/windows/base/old/3.6.2/). To extract the acute myocardial infarction subnet dataset, 2 steps were taken from the guide gene to extract the vicinity of the network, as previously described by Mutwil et al (24). Cystoscope software (https://cytoscape.org/) was used to illustrate the network.
Gene Ontology (GO) analysis
GO functional enrichment analysis (25,26) was performed to obtain the genes associated with AMI and to determine their functional terms in the cellular component (CC), biological process (BP) and molecular function (MF) categories. The functional terms for the GO enrichment analysis and expression coherence (EC) analysis were retrieved from the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/). For the GO enrichment analysis, the significant differences of GO enrichment by DEGs and co-expressed network genes were evaluated against a background set consisting of 52 genes and 256 genes, respectively. With multiple test correlations, FDR <0.05 for BP, MF and CC were set as significance thresholds.
KEGG Disease and BioCyc analysis
KEGG Disease and BioCyc analysis of DEGs and co-expression network genes were performed through using the KEGG Orthology-Based on KOBAS 2.0 (27-30), the results of the enrichment were analyzed by Fisher's exact test, using P≤0.05 as the significance threshold.
Reverse transcription-quantitative (RT-q)PCR
Complete RNA was extracted from the whole blood with TRIzol® reagent (Sangon Biotech Co., Ltd.) and all mRNA was subjected to RT and qPCR using a PrimeScript® RT Master Mix Perfect Real Time kit (Takara Bio Inc.) and SYBR Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.), respectively, according to the manufacturers' protocol. qPCR was performed on an Applied Biosystems 7900HT Real-Time System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The program was as follows: 2 min at 95˚C followed by 40 cycles of 20 sec at 95˚C and 30 sec at 60˚C. The genes were amplified using the following specific primers: i) aldo-keto reductase family 1 member C3 (AKR1C3) gene, 5'-GAGACAAACGATGGGTGGACC-3' and 5'-TGGAACTCAAAAACCTGCACG-3'; ii) purinergic receptor P2Y12 (P2RY12) gene, 5'-CACTGCTCTACACTGTCCTGT-3' and 5'-AGTGGTCCTGTTCCCAGTTTG-3'; iii) ribosomal protein S24 (RPS24) gene, 5'-ATGAACGACACCGTAACTATCCG-3' and 5'-CCGAATTTCTGTCTTAGGCACTG-3'; iv) acyl-CoA synthetase long chain family member 1 (ACSL1) gene, 5'-CGACGAGCCCTTGGTGTATTT-3' and 5'-GGTTTCCGAGAGCCTAAACAA-3'; v) UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 5 (B3GNT5) gene, 5'-TTCAAGACTTTTGGATTGGTCGT-3' and 5'-CGGCTGTGTAGTCAGGGTAAG-3'; vi) maltase-glucoamylase (MGAM) gene, 5'-GCTCAGTGTTCTTCTGCTTGT-3' and 5'-CGTTGTCCTAGCATGTGTGGTA-3'; vii) glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, 5'-GGGAAACTGTGGCGTGAT-3' and 5'-GAGTGGGTGTCGCTGTTGA-3'. The results were analyzed using the 2-ΔΔCq method (31).
ELISA
The collected blood samples were centrifuged at 22˚C and 3,000 x g for 10 min. The supernatants were collected into 1.5 ml tubes and preserved at -80˚C for subsequent analysis. The concentrations of AKR1C3, P2RY12, RPS24, ACSL1, B3GNT5 and MGAM were interpolated from a standard curve using linear regression analysis, according to the instructions of the ELISA kits (AKR1C3, cat. no. JM-1169H1; P2RY12, cat. no. JM-1165H1; RPS24, cat. no. JM-1162H1; ACSL1, cat. no. JM-1172H1; B3GNT5, cat. no. JM-1159H1; MGAM, cat. no. JM-1188H1; JingMei Biotechnology Co., Ltd.). The absorbance was measured at a wavelength of 450 nm using an ELx800 microplate spectrophotometer (BioTek Instruments, Inc.).
Statistical analysis
ELISA and RT-qPCR experimental data were expressed as the mean ± standard deviation. Statistical significance was evaluated using Student's t-test using GraphPad Prism 6.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Data preprocessing and DEG screening
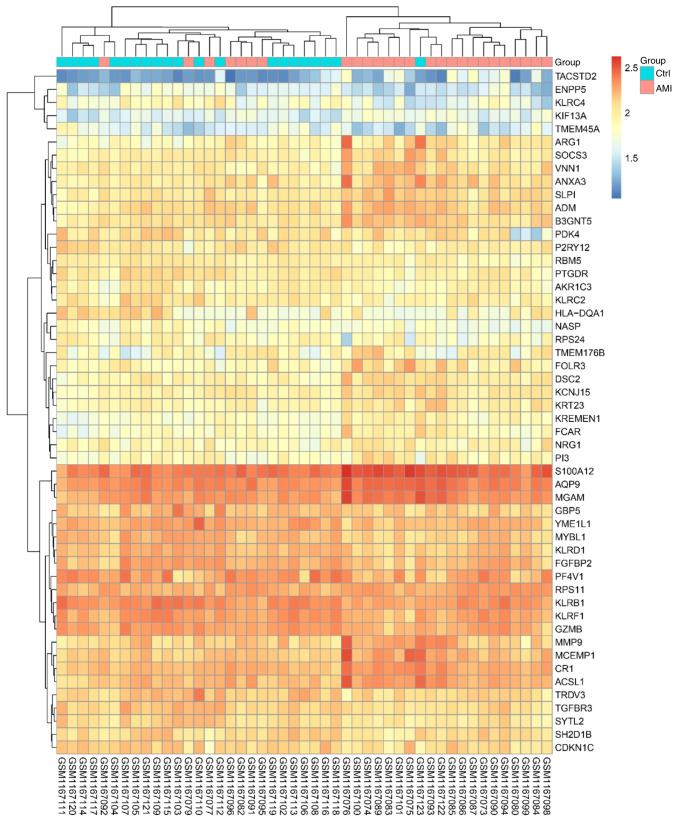
After data preprocessing, an expression matrix of 24,277 genes was obtained from 47 samples (Table SII). Under the threshold of |log2FC| ≥0.5, a total of 52 DEGs were selected for subsequent analysis, including 26 upregulated genes and 26 downregulated genes. The heat map displaying the results of the gene expression cluster analysis indicated that these DEGs were able to clearly distinguish the two samples (Fig. 1 and Table SIII), indicating that these DEGs were suitable for further use in the subsequent analysis.
Figure 1.
Heat map of differentially expressed genes in AMI. Columns of the heat map represent individual samples with patients with AMI on the left-hand side and controls on the right-hand side, while rows represent all differentially expressed probe-sets. The heat map's color gradient represents relative expression, with upregulated genes displayed in red and downregulated genes in blue. AMI, acute myocardial infarction; Ctrl, control.
Functional enrichment of DEGs
GO analysis of 52 DEGs indicated that ‘MHC class I protein complex binding’, ‘cellular response to prostaglandin D stimulus’ and ‘natural killer cell-mediated immunity’ were significantly enriched terms (Fig. S1 and Table SIV). Furthermore, KEGG Disease analysis was performed on the 52 obtained DEGs and the results suggested that the DEGs were mainly involved in cardiovascular diseases (Table SV). In addition, BioCyc analysis revealed that these DEGs are mainly related to cardenolide biosynthesis, glycosphingolipids biosynthesis, fatty acid activation and glycogenolysis.
Co-expression network analysis
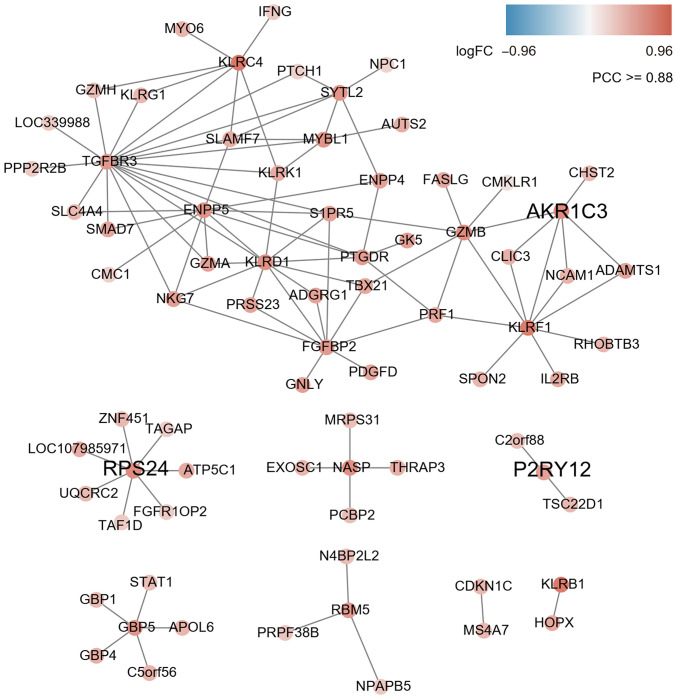
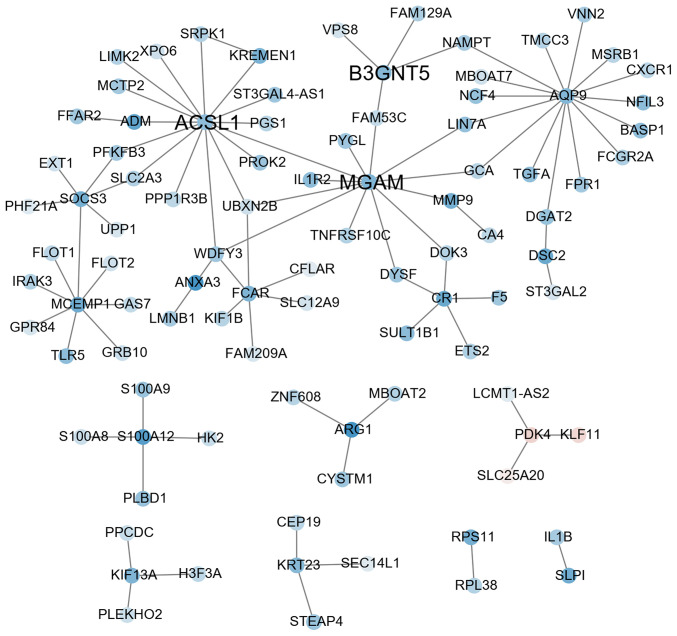
According to the character string database, the PCC of all gene pairs was calculated and those with P<0.05 were selected. Finally, 256 genes co-expressed with DEGs were obtained through calculating the MR (≤10) and PCC ≥0.88 in each pair of genes (as cutoff). The results of the co-expression network analysis revealed that 8 gene modules were upregulated (marked in red; Fig. 2) and 8 gene modules were downregulated (marked in blue; Fig. 3). Among them, the expression levels of AKR1C3, RPS24 and P2RY12 as key hub genes were upregulated (Fig. 2 and Table SIII) and ACSL1, B3GNT5 and MGAM were downregulated (Fig. 3 and Table SIII).
Figure 2.
Co-expression subnetwork constructed using three genes upregulated in AMI as guide genes (AKR1C3, RPS24 and P2RY12). A link between two nodes indicates a direct interaction with P<0.05, PCC ≥0.88 and MR ≤10. The subnetwork vicinity is extracted by taking two steps out from each guide gene. AMI, acute myocardial infarction; PCC, Pearson correlation coefficient; MR, mutual rank; AKR1C3, aldo-keto reductase 1 C3; RPS24, ribosomal protein S24; P2RY12, purinergic receptor P2Y12.
Figure 3.
Co-expression subnetwork constructed using three genes downregulated in AMI as guide genes (ACSL1, B3GNT5 and MGAM). ACSL1, acyl-CoA synthetase long chain family member 1; B3GNT5, UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 5; MGAM, maltase-glucoamylase; FC, fold change; PCC, Pearson correlation coefficient..
Functional enrichment of genes in co-expression networks
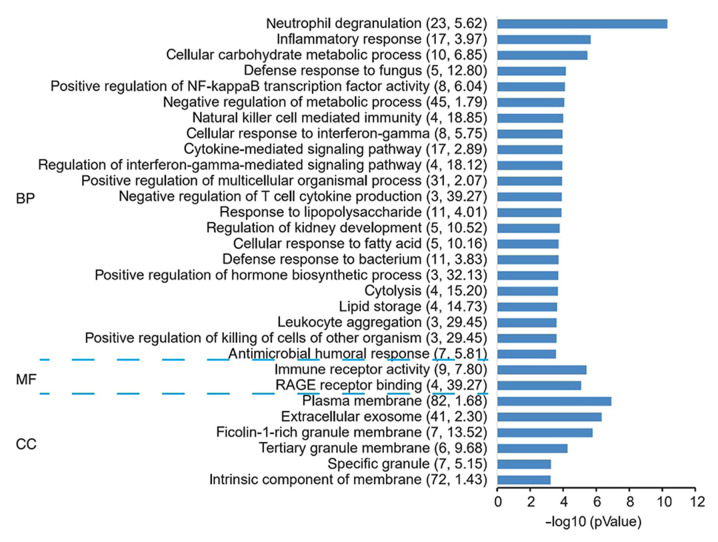
Through analysis of the genes in this module by GO, it was revealed that ‘RAGE receptor binding’, ‘positive regulation of killing of cells of other organism’, ‘leukocyte aggregation’ and ‘negative regulation of T-cell cytokine production’ were significantly enriched terms (Fig. 4 and Table SVI).
Figure 4.
GO term analysis in the categories BP, MF and CC for the co-expression network genes for acute myocardial infarction. The negative logarithm of the P-value (x-axis) indicates the significance of the gene set belonging to predefined categories under the co-expression network gene background. The y-axis represents each GO category. The gene number and EC value of the category in the subnetwork are presented in brackets. GO, Gene Ontology; BP, biological process; MF, molecular function; CC, cellular component; EC, expression coherence.
Furthermore, KEGG Disease and BioCyc analysis was performed and a co-expression network was constructed, containing 256 genes. A total of three co-expression subnets centered on upregulated genes (AKR1C3, RPS24 and P2RY12) and three co-expression subnets centered on downregulated genes (ACSL1, B3GNT5 and MGAM) were obtained. The genes with upregulated expression had significant associations with functions such as ‘cardiovascular diseases’ and ‘cardenolide biosynthesis’ (Table I). The downregulated genes had significant associations with functions such as ‘glycogenolysis’, ‘glycosphingolipids biosynthesis’ and ‘fatty acid activation’ (Table I). Of note, all of them also participated in the enrichment of the 52 DEGs obtained by the KEGG and BioCyc analysis in the present study. This suggested that certain genes that have not been previously reported to be involved in AMI may be mined from reported microarray data and novel interactions between these genes may also be identified from the connections in the network. AMI development is a distinct biological event and co-expression network analysis may have the greatest potential for identifying gene interactions in AMI.
Table I.
KEGG Disease and BioCyc analysis of co-expressed genes with differentially expressed genes.
| Term | Database | Input number | Background number | P-value | Input |
|---|---|---|---|---|---|
| Cardiovascular diseases | KEGG Disease | 5 | 342 | 0.00375 | IFNG|HK2|RPS24| F5|P2RY12 |
| Triacylglycerol biosynthesis | BioCyc | 3 | 26 | 0.000086 | MBOAT2|DGAT2| MBOAT7 |
| Glycogenolysis | BioCyc | 2 | 7 | 0.000305 | MGAM|PYGL |
| CDP-diacylglycerol biosynthesis | BioCyc | 2 | 17 | 0.00142 | MBOAT2|MBOAT7 |
| Super pathway of glycosphingolipids biosynthesis | BioCyc | 2 | 25 | 0.002869 | ST3GAL2| B3GNT5 |
| Fructose 2,6-bisphosphate synthesis | BioCyc | 1 | 5 | 0.017499 | PFKFB3 |
| Allopregnanolone biosynthesis | BioCyc | 1 | 6 | 0.020386 | AKR1C3 |
| Pyrimidine ribonucleosides degradation | BioCyc | 1 | 6 | 0.020386 | UPP1 |
| Coenzyme A biosynthesis | BioCyc | 1 | 6 | 0.020386 | PPCDC |
| Cardenolide biosynthesis | BioCyc | 1 | 6 | 0.020386 | AKR1C3 |
| Globo-series glycosphingolipids biosynthesis | BioCyc | 1 | 8 | 0.026134 | ST3GAL2 |
| Lacto-series glycosphingolipids biosynthesis | BioCyc | 1 | 8 | 0.026134 | B3GNT5 |
| Chondroitin sulfate biosynthesis (late stages) | BioCyc | 1 | 9 | 0.028995 | CHST2 |
| Ganglio-series glycosphingolipids biosynthesis | BioCyc | 1 | 9 | 0.028995 | ST3GAL2 |
| Androgen biosynthesis | BioCyc | 1 | 11 | 0.034693 | AKR1C3 |
| Neolacto-series glycosphingolipids biosynthesis | BioCyc | 1 | 13 | 0.040358 | B3GNT5 |
| Icosapentaenoate biosynthesis II (metazoa) | BioCyc | 1 | 13 | 0.040358 | ACSL1 |
| BMP signaling pathway | BioCyc | 1 | 13 | 0.040358 | SMAD7 |
| Gamma-linolenate biosynthesis | BioCyc | 1 | 14 | 0.043178 | ACSL1 |
| Fatty acid activation | BioCyc | 1 | 15 | 0.04599 | ACSL1 |
| Arachidonate biosynthesis III (metazoa) | BioCyc | 1 | 16 | 0.048793 | ACSL1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Expression of hub genes in AMI specimens
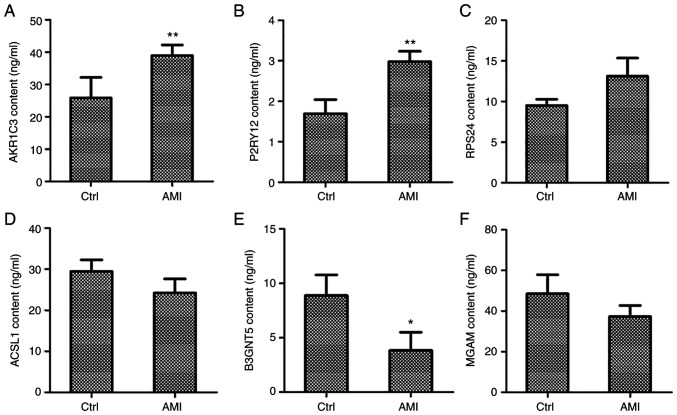
To further investigate the mRNA and protein levels of hub genes in patients with AMI, the protein levels of AKR1C3, RPS24, P2RY12, ACSL1, B3GNT5 and MGAM were assessed in 31 AMI patients (20 males and 11 females, age from 49 to 82, medians: 70) and 31 normal individuals (17 males and 14 females, age from 46 to 83, medians: 68) of whole blood samples using ELISA. As depicted in Fig. 5, AKR1C3 and P2RY12 content were significantly increased in patients with AMI relative to that in the normal adjacent group (P<0.01; Fig. 5A and B), while the increase in RPS24 content was not significant (Fig. 5C). The contents of ACSL1 and MGAM were slightly decreased in patients with AMI (Fig. 5D and F), but only the B3GNT5 content was significantly decreased (P<0.05; Fig. 5E). Further verification at the mRNA levels by RT-qPCR indicated that the relative expression of AKR1C3, RPS24 and P2RY12 was significantly increased in patients with AMI relative to that of the normal adjacent group (P<0.01; Fig. S2A-C). By contrast, the relative expression of ACSL1, B3GNT5 and MGAM was significantly decreased in patients with AMI (P<0.01; Fig. S2D-F). These findings were consistent with the data of the microarray expression profiles.
Figure 5.
Co-expression network genes were analyzed in whole blood collected from patients with AMI and controls with a normal echocardiogram via ELISA. Protein levels of three upregulated genes, namely (A) AKR1C3, (B) P2RY12 and (C) RPS24, and three downregulated genes, namely (D) ACSL1, (E) B3GNT5 and (F) MGAM, were measured by ELISA. *P<0.05, **P<0.01 vs. Ctrl. AMI, acute myocardial infarction; Ctrl, control. AKR1C3, aldo-keto reductase family 1 member C3; P2RY12, purinergic receptor P2Y12; RPS24, ribosomal protein S24; ACSL1, acyl-CoA synthetase long chain family member 1; B3GNT5, UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 5; MGAM, maltase-glucoamylase.
Discussion
AMI is a serious cardiovascular disease that poses a serious threat to human life; it may cause congestive heart failure and malignant arrhythmias, leading to high morbidity and mortality (32). Although thrombolysis and percutaneous coronary intervention may improve the prognosis of patients with AMI, numerous patients eventually develop heart failure or arrhythmia due to unknown etiology (33). Looking for potential diagnostic biomarkers of AMI and possible regulatory targets may help reduce the mortality of AMI. The systematic biological analysis of gene expression profiles provides a useful method for elucidating the possible mechanisms of myocardial infarction from the perspective of gene regulation (34). Using gene expression profiles, sufficient information concerning changes in gene expression associated with disease may be obtained. Kiliszek et al (35) performed microarray methods to demonstrate that, during ST-elevation myocardial infarction, numerous genes exhibit altered expression, including those involved in platelet function, lipid/glucose metabolism and atherosclerotic plaque stability. Based on the systematic level of gene expression profiling, gene co-expression network analysis may be used as an alternative method for analyzing profiling data to gain a deeper understanding of the molecular regulatory mechanisms of heart disease caused by myocardial infarction (7,36). The present study used GO, pathway enrichment, KEGG Disease and BioCyc analysis methods to explore the molecular mechanisms of myocardial infarction-induced heart disease.
Gene co-expression network analysis has also been used to study changes in transcriptome expression patterns in complex diseases (37,38). Compared with the standardized analysis of DEGs (the purpose of which is to detect individual genes associated with disease), co-expression network analysis aims to identify higher associations between gene products. In addition, its algorithm may markedly simplify the multiple testing problems that are unavoidable in the gene-centric standard microarray expression profiling data analysis method (39). Therefore, it is a powerful system analysis method, focusing on the relevant functions of the network module.
Using the above-mentioned bioinformatics methods, GO analysis, pathway function network and gene module changes of patients with AMI were analyzed to explore the potential diagnostic biomarkers and possible regulatory targets of AMI. Based on the gene expression profile (12), a gene co-expression network was constructed and co-expression network analysis was used to detect peripheral blood gene modules when AMI occurred. In the present study, AKR1C3, RPS24 and P2RY12 were increased and ACSL1, B3GNT5 and MGAM were decreased in patients with AMI, which was also confirmed by RT-qPCR and ELISA, highlighting their potential role as biomarkers of cardiac illness. It was observed that the upregulated genes AKR1C3, RPS24 and P2RY12 were mainly involved in cardenolide biosynthesis and cardiovascular diseases. Accumulating evidence suggests that AKR1C3 has an important role in hormone-dependent and hormone-independent cancers (40). RPS24c facilitates tumor angiogenesis via the RPS24c/MVIH/PGK1 pathway in colorectal cancer (41). P2RY12 is a gene encoding P2Y12 receptor present on platelets, which has an essential role in potentiating platelet responses initiated by other activators such as thromboxane and thrombin (42). Mutations in the P2RY12 gene have been demonstrated to be associated with bleeding disorders (43). To date, research has focused on the effects of P2RY12 variants in response to clopidogrel and thrombotic diseases.
In addition, the three genes whose expression was downregulated, ACSL1, B3GNT5 and MGAM, were associated with glycogenolysis, glycosphingolipids biosynthesis and fatty acid activation. ACSL1 exists in cells of the liver, heart and fat, as is considered to affect activation fatty acid synthesis of triglycerides through the peroxisome proliferator-activated receptor γ pathway (44). B3GNT5 has been suggested as the key glycosyltransferase in the biosynthesis of the (neo-)lacto series of glycosphingolipid in regulating malignancy of glioblastoma multiforme (45). This result implies that the etiology of AMI is associated with genes related to intravascular disease, immune response and brain-derived factor regulation system, suggesting that ACSL1, B3GNT5 and MGAM may be also used as biomarkers for gene expression in early AMI. Previous studies have researched DEGs in AMI; however, differential expression may not mean that any of these genes are suitable biomarkers.
Although the present analysis is powerful, the present study has certain limitations. First, the analytic data originate from a microarray chip and not from RNA sequencing. Of note, microarray chips are not able to discover new genes. Furthermore, the sample size of the dataset was relatively small. Thus, certain potential targets of AMI may have been missed in the present analysis. In addition, although hub genes were identified and a co-expression network analysis was performed, the hierarchical processes between them have remained to be fully elucidated. A potent biomarker of AMI should have high sensitivity and specificity to reduce false positives or false negatives. However, the present study did not evaluate the sensitivity and specificity of these genes. In order to identify potential biomarkers of AMI, further studies with larger sample sizes are still warranted.
In conclusion, the present findings provide important information for the prediction of molecular events related to AMI, as well as potential biomarkers for detection and prevention. The hub genes AKR1C3, RPS24, P2RY12, ACSL1, B3GNT5 and MGAM may be considered biomarkers to assess the severity of heart disease and may be promising therapeutic targets to develop novel treatments.
Supplementary Material
Acknowledgements
The authors are grateful to Dr Yong Ye from Shanghai East Hospital Tongji University for his professional statistical advice.
Funding Statement
Funding: This study was supported by research grants from the Chinese National Natural Science Foundation (grant nos. 81900245, 81770395 and 81800224).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZD and XL designed the study. ZH, RL, HH, XD, GL, YW, YJ and SX analyzed the data. ZD, GL, YW and YJ wrote the manuscript. ZD, XL and SX revised the manuscript. XL and ZH confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Patients with AMI and controls provided written informed consent prior to use their blood samples in this study. This study was approved by the Ethics Committee of the Central Hospital of Karamay (Karamay, China; ethics approval code: YL-2020-2).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet. 2017;389:197–210. doi: 10.1016/S0140-6736(16)30677-8. [DOI] [PubMed] [Google Scholar]
- 2.Castro-Dominguez Y, Dharmarajan K, McNamara RL. Predicting death after acute myocardial infarction. Trends Cardiovasc Med. 2018;28:102–109. doi: 10.1016/j.tcm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Shibata T, Kawakami S, Noguchi T, Tanaka T, Asaumi Y, Kanaya T, Nagai T, Nakao K, Fujino M, Nagatsuka K, et al. Prevalence, clinical features, and prognosis of acute myocardial infarction attributable to coronary artery embolism. Circulation. 2015;132:241–250. doi: 10.1161/CIRCULATIONAHA.114.015134. [DOI] [PubMed] [Google Scholar]
- 4.Henry P, Lamhaut L, Delmas C, Belle L. Can we still die from acute myocardial infarction in 2020? Reflex mobile cardiac assistance unit or local team for ECMO implantation? Arch Cardiovasc Dis. 2019;112:733–737. doi: 10.1016/j.acvd.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Abdelaziz HK, Patel B, Chalil S, Choudhury T. COVID-19 pandemic and acute myocardial infarction: Management protocol from a british cardiac centre. Crit Pathw Cardiol. 2020;19:55–57. doi: 10.1097/HPC.0000000000000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson S, Rosengren A, Young K, Jennings E. Mortality and morbidity trends after the first year in survivors of acute myocardial infarction: A systematic review. BMC Cardiovasc Disord. 2017;17(53) doi: 10.1186/s12872-017-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Ma C, Gu J, Yu M. Potential biomarkers of acute myocardial infarction based on weighted gene co-expression network analysis. Biomed Eng Online. 2019;18(9) doi: 10.1186/s12938-019-0625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bavia L, Lidani KCF, Andrade FA, Sobrinho MIAH, Nisihara RM, de Messias-Reason IJ. Complement activation in acute myocardial infarction: An early marker of inflammation and tissue injury? Immunol Lett. 2018;200:18–25. doi: 10.1016/j.imlet.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Xue S, Zhu W, Liu D, Su Z, Zhang L, Chang Q, Li P. Circulating miR-26a-1, miR-146a and miR-199a-1 are potential candidate biomarkers for acute myocardial infarction. Mol Med. 2019;25(18) doi: 10.1186/s10020-019-0086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue S, Liu D, Zhu W, Su Z, Zhang L, Zhou C, Li P. Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p are novel biomarkers for diagnosis of acute myocardial infarction. Front Physiol. 2019;10(123) doi: 10.3389/fphys.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukauskas T, Mickus R, Cereskevicius D, Macas A. Value of serum miR-23a, miR-30d, and miR-146a biomarkers in ST-Elevation myocardial infarction. Med Sci Monit. 2019;25:3925–3932. doi: 10.12659/MSM.913743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suresh R, Li X, Chiriac A, Goel K, Terzic A, Perez-Terzic C, Nelson TJ. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J Mol Cell Cardiol. 2014;74:13–21. doi: 10.1016/j.yjmcc.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Qi GX, Guo L, Sun YX. Bioinformatics analyses of differentially expressed genes associated with acute myocardial infarction. Cardiovasc Ther. 2016;34:67–75. doi: 10.1111/1755-5922.12171. [DOI] [PubMed] [Google Scholar]
- 14.Xiang S, Huang Z, Wang T, Han Z, Yu CY, Ni D, Huang K, Zhang J. Condition-specific Gene Co-Expression network mining identifies key pathways and regulators in the brain tissue of Alzheimer's disease patients. BMC Med Genomics. 2018;11 (Suppl 6)(S115) doi: 10.1186/s12920-018-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma C, Lv Q, Teng S, Yu Y, Niu K, Yi C. Identifying key genes in rheumatoid arthritis by weighted gene Co-Expression network analysis. Int J Rheum Dis. 2017;20:971–979. doi: 10.1111/1756-185X.13063. [DOI] [PubMed] [Google Scholar]
- 16.Saris CG, Horvath S, van Vught PW, van Es MA, Blauw HM, Fuller TF, Langfelder P, DeYoung J, Wokke JH, Veldink JH, et al. Weighted gene co-expression network analysis of the peripheral blood from amyotrophic lateral sclerosis patients. BMC Genomics. 2009;10(405) doi: 10.1186/1471-2164-10-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azuaje F, Zhang L, Jeanty C, Puhl SL, Rodius S, Wagner DR. Analysis of a gene co-expression network establishes robust association between Col5a2 and ischemic heart disease. BMC Med Genomics. 2013;6(13) doi: 10.1186/1755-8794-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malki K, Tosto MG, Jumabhoy I, Lourdusamy A, Sluyter F, Craig I, Uher R, McGuffin P, Schalkwyk LC. Integrative mouse and human mRNA studies using WGCNA nominates novel candidate genes involved in the pathogenesis of major depressive disorder. Pharmacogenomics. 2013;14:1979–1990. doi: 10.2217/pgs.13.154. [DOI] [PubMed] [Google Scholar]
- 19.Udyavar AR, Hoeksema MD, Clark JE, Zou Y, Tang Z, Li Z, Li M, Chen H, Statnikov A, Shyr Y, et al. Co-expression network analysis identifies spleen tyrosine kinase (SYK) as a candidate oncogenic driver in a subset of small-cell lung cancer. BMC Syst Biol. 2013;7 (Suppl 5)(S1) doi: 10.1186/1752-0509-7-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Cai W, Su S, Zhi D, Lu J, Liu S. Screening genes crucial for pediatric pilocytic astrocytoma using weighted gene coexpression network analysis combined with methylation data analysis. Cancer Gene Ther. 2014;21:448–455. doi: 10.1038/cgt.2014.49. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Liu W, Liu X, Qi J, Deng C. Biomarkers identification for acute myocardial infarction detection via weighted gene Co-expression network analysis. Medicine (Baltimore) 2017;96(e8375) doi: 10.1097/MD.0000000000008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aya K, Suzuki G, Suwabe K, Hobo T, Takahashi H, Shiono K, Yano K, Tsutsumi N, Nakazono M, Nagamura Y, et al. Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS One. 2011;6(e26162) doi: 10.1371/journal.pone.0026162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–260. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutwil M, Ruprecht C, Giorgi FM, Bringmann M, Usadel B, Persson S. Transcriptional wiring of cell wall-related genes in arabidopsis. Mol Plant. 2009;2:1015–1024. doi: 10.1093/mp/ssp055. [DOI] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. Gene Ontology Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Shah M, Patel B, Tripathi B, Agarwal M, Patnaik S, Ram P, Patil S, Shin J, Jorde UP. Hospital mortality and thirty day readmission among patients with non-acute myocardial infarction related cardiogenic shock. Int J Cardiol. 2018;270:60–67. doi: 10.1016/j.ijcard.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Song Y, Xu JJ, Tang XF, Wang HH, Jiang P, Jiang L, Liu R, Zhao XY, Gao LJ, et al. Relationship between thrombolysis in myocardial infarction risk index and the severity of coronary artery lesions and long-term outcome in acute myocardial infarction patients undergoing percutaneous coronary intervention. Zhonghua Xin Xue Guan Bing Za Zhi. 2018;46:874–881. doi: 10.3760/cma.j.issn.0253-3758.2018.11.011. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 34.Khodayari S, Khodayari H, Amiri AZ, Eslami M, Farhud D, Hescheler J, Nayernia K. Inflammatory microenvironment of acute myocardial infarction prevents regeneration of heart with stem cells therapy. Cell Physiol Biochem. 2019;53:887–909. doi: 10.33594/000000180. [DOI] [PubMed] [Google Scholar]
- 35.Kiliszek M, Szpakowicz A, Franaszczyk M, Pepinski W, Waszkiewicz E, Skawronska M, Ploski R, Niemcunowicz-Janica A, Budnik M, Poludniewska D, et al. The 9p21 Polymorphism is linked with atrial fibrillation during acute phase of ST-segment elevation myocardial infarction. Heart Vessels. 2016;31:1590–1594. doi: 10.1007/s00380-015-0774-x. [DOI] [PubMed] [Google Scholar]
- 36.Niu X, Zhang J, Zhang L, Hou Y, Pu S, Chu A, Bai M, Zhang Z. Weighted gene co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front Genet. 2019;10(1214) doi: 10.3389/fgene.2019.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choobdar S, Ahsen ME, Crawford J, Tomasoni M, Fang T, Lamparter D, Lin J, Hescott B, Hu X, Mercer J, et al. Assessment of network module identification across complex diseases. Nat Methods. 2019;16:843–852. doi: 10.1038/s41592-019-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Q, Ren J, Hou J, Wang G, Ju L, Xiao Y, Gong Y. Co-expression network analysis identified candidate biomarkers in association with progression and prognosis of breast cancer. J Cancer Res Clin Oncol. 2019;145:2383–2396. doi: 10.1007/s00432-019-02974-4. [DOI] [PubMed] [Google Scholar]
- 39.Tang J, Kong D, Cui Q, Wang K, Zhang D, Gong Y, Wu G. Prognostic genes of breast cancer identified by gene Co-expression network analysis. Front Oncol. 2018;8(374) doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penning TM. AKR1C3 (type 5 17β-hydroxysteroid dehydrogenase/prostaglandin F synthase): Roles in malignancy and endocrine disorders. Mol Cell Endocrinol. 2019;489:82–91. doi: 10.1016/j.mce.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue W, Youjun W, Kun X, Yingjie Z, Gang L, Shiyan X, Fuquan W. RPS24c isoform facilitates tumor angiogenesis via promoting the stability of MVIH in colorectal cancer. Curr Mol Med. 2020;20:388–395. doi: 10.2174/1566524019666191203123943. [DOI] [PubMed] [Google Scholar]
- 42.Shankar H, Garcia A, Prabhakar J, Kim S, Kunapuli SP. P2Y12 receptor-mediated potentiation of thrombin-induced thromboxane A2 generation in platelets occurs through regulation of Erk1/2 activation. J Thromb Haemost. 2006;4:638–647. doi: 10.1111/j.1538-7836.2006.01789.x. [DOI] [PubMed] [Google Scholar]
- 43.Hassani Idrissi H, Hmimech W, El Khorb N, Akoudad H, Habbal R, Nadifi S. Does i-T744C P2Y12 polymorphism modulate clopidogrel response among moroccan acute coronary syndromes patients? Genet Res Int. 2017;2017(9532471) doi: 10.1155/2017/9532471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tingting L, Xiangdong L, Heyu M, Lili C, Fanbo M. ACSL1 affects Triglyceride Levels through the PPARγ pathway. Int J Med Sci. 2020;17:720–727. doi: 10.7150/ijms.42248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong HY, Park SY, Kim HJ, Moon S, Lee S, Lee SH, Kim SH. B3GNT5 is a novel marker correlated with stem-like phenotype and poor clinical outcome in human gliomas. CNS Neurosci Ther. 2020;26:1147–1154. doi: 10.1111/cns.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.