Abstract
To gain insight on the significance of human T-cell lymphotropic virus type 1 (HTLV-1) indeterminate serological reactivities, we studied villagers of South Cameroon, focusing on a frequent and specific HTLV-1 Gag indeterminate profile (HGIP) pattern (gag p19, p26, p28, and p30 without p24 or Env gp21 and gp46). Among the 102 sera studied, 29 from all age groups had a stable HGIP pattern over a period of 4 years. There was no epidemiological evidence for sexual or vertical transmission of HGIP. Seventy-five percent of HGIP sera reacted positively on MT2 HTLV-1-infected cells by immunofluorescence assay. However, we could not isolate any HTLV-1 virus or detect the presence of p19 Gag protein in cultures of peripheral blood mononuclear cells obtained from individuals with strong HGIP reactivity. PCR experiments conducted with primers for HTLV-1 and HTLV-2 (HTLV-1/2 primers) encompassing different regions of the virus did not yield HTLV-1/2 proviral sequences from individuals with HGIP. Using 11 peptides corresponding to HTLV-1 or HTLV-2 immunodominant B epitopes in an enzyme-linked immunosorbent assay, one epitope corresponding to the Gag p19 carboxyl terminus was identified in 75% of HGIP sera, while it was recognized by only 41% of confirmed HTLV-1-positive sera. A positive correlation between HTLV-1 optical density values and titers of antibody to Plasmodium falciparum was also demonstrated. Finally, passage of sera through a P. falciparum-infected erythrocyte-coupled column was shown to specifically abrogate HGIP reactivity but not the HTLV-1 pattern, suggesting the existence of cross-reactivity between HTLV-1 Gag proteins and malaria-derived antigens. These data suggest that in Central Africa, this frequent and specific Western blot is not caused by HTLV-1 infection but could instead be associated with P. falciparum infection.
Human T-cell lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia (48) and of tropical spastic paraparesis/HTLV-l associated myelopathy (20). Currently, 15 to 20 million individuals are estimated to be infected by HTLV-1. Most cases are described in highly endemic areas such as southern Japan, intertropical Africa, and the Caribbean and surrounding regions. By contrast, low HTLV-1 seroprevalence rates are usually observed in nontropical areas (2, 12). Early seroepidemiological reports highlighted the high prevalence of HTLV-1 infection in Africa (6, 7, 14–17, 36, 54, 58) and Melanesia (3, 52, 60). However, most of these reports were based only on first-generation enzyme-linked immunosorbent assay (ELISA) tests which were shown to be sensitive but not specific for the detection of HTLV-1 antibodies (11, 18). Since then, stringent Western blot (WB) criteria have been proposed by the World Health Organization and the Centers for Disease Control and Prevention for HTLV-1/2 seropositivity (1). Subsequent analyses of many sera collected from tropical regions led to a high percentage of indeterminate WB exhibiting different HTLV patterns (27, 57).
These indeterminate sera frequently show reactivity to isolated gag-encoded proteins (8, 21). As a consequence, it appears that a large number of early studies performed in tropical areas overestimated the true HTLV-1 seroprevalence (56). Thus, it was suggested that persons from South America, Melanesia, and Africa whose serum exhibits different isolated Gag reactivities did not have genuine HTLV-1 or HTLV-2 infections (19, 21, 22, 43). By contrast, in Europe and in the United States, such indeterminate reactivities were found among blood donors or more recently in a series of patients suffering from multiple sclerosis, but at a much lower frequency (13, 25, 26, 32, 55). Strikingly, a genuine HTLV-1 virus was recently isolated and sequenced from one of these patients whose serum showed this indeterminate HTLV seroreactivity (59).
Nonetheless, for the vast majority of the indeterminate samples originating from tropical areas, it is hypothesized that this indeterminate reactivity was either the result of sequence homologies between Gag epitopes of HTLV-1 and other proteins or caused by an HTLV-1-related virus or rare cases of HTLV-1 transient infection (21). However, the data supporting most of these predictions are still lacking. Recently, using computer analyses, several peptides of the HTLV-1 matrix protein (Gag p19) were shown to have homology with some human proteins and or infectious agents (4, 5, 21–23, 31, 37, 40, 44–47, 50, 53). As an example, antibodies to the blood stage antigens of Plasmodium falciparum were suggested to cross-react with an HTLV p19 epitope, leading to the presence of HTLV indeterminate reactivities seen with specimens from the Philippines, Papua New Guinea, Indonesia, and Brazil, all regions where malaria is endemic (22, 31, 50, 51). Such results, as well as the high frequency of HTLV seroindeterminate reactivity seen in Central Africa, led us to undertake a serological and virologic study of Central African individuals whose sera exhibited such HTLV-1 Gag reactivities on WB. Among all the miscellaneous indeterminate WB profiles, we focused on a peculiar pattern that we previously defined as the HTLV-1 Gag indeterminate profile (HGIP) (40). This profile is the most frequent profile seen in Central Africa. HGIP exhibits intense WB reactivities and has a pattern closely related to a complete HTLV-1 seroreactivity (p19, p26, p28, p32, p36, and p53, but not p24 or any env-encoded glycoproteins, gp21 and gp46 peptide K55 or MTA-1) (21, 40). To unravel the origin of such reactivities, a survey was undertaken between 1990 and 1994 in a community in South Cameroon, Central Africa, where malaria is hyperendemic and the HGIP profile is common. The purposes of this survey were (i) to search for epidemiological evidence of a transmissible agent by studying the familial presence of the HGIP profile; (ii) to isolate a (retro)virus or to detect the presence of an HTLV-1 gag-related sequence in the peripheral blood mononuclear cells (PBMCs) of subjects with HGIP; (iii) to define HTLV-1/2 linear epitopes which could be recognized by these sera and to determine whether antibodies present in HTLV-1-positive sera also recognized these peptides; and (iv) to explore the possible immunological cross-reactivities between HTLV-1 antigens and the blood stage antigens of P. falciparum.
MATERIALS AND METHODS
Study population.
Blood specimens were collected from 102 individuals living in different villages of South Cameroon, a tropical rain forest region of Central Africa where malaria is hyperendemic. For each subject, an aliquot of serum was obtained from 10 to 20 ml of venipuncture and kept frozen (−20°C) until HTLV-1 and HTLV-2 serological screening.
Of these 102 subjects, 76 belonged to seven families and 26 were unrelated. For each family, genealogical trees were drawn. In 1990 to 1992, 82 of the 102 individuals included in the present study were serologically tested for HTLV-1/2 using nonstringent WB criteria (36). Of those tested, 41 were originally considered HTLV-1 infected (36).
Informed consent was obtained from all the subjects, and human experimentation guidelines were followed in the conduct of this study. Furthermore, each of the individuals tested underwent a medical examination and was referred to the local medical facilities if necessary.
Serological tests.
Two different tests were used, according to the manufacturer's instructions, to screen for the presence of HTLV-1 and HTLV-2 antibodies in the sera: an ELISA (Platelia HTLV-1 new; Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France), which contains disrupted virion, and an indirect immunofluorescence assay (IFA) using MT2 and C19 for HTLV-1- and HTLV-2-producing cells, respectively (dilution of the sera 1:10). Two investigators independently read each slide. IFA was also used to titer HTLV-1 antibodies. A second ELISA test containing only synthetic peptides was also used (HTLV-1/2 ELISA; Genelabs Diagnostic, Singapore, Singapore). For confirmation, a WB assay (HTLV2-3 Diagnostic Biotechnology, Singapore, Singapore) was performed on all sera. This kit contains disrupted HTLV-1 virion, a recombinant envelope protein (rgp21), MTA-1, an HTLV-1-specific peptide corresponding to residues 169 to 209 of the gp46 glycoprotein, and K55, an HTLV-2-specific peptide corresponding to residues 162 to 205 of gp46 (27, 28). Stringent WB criteria were used, and a serum was considered HTLV-1 positive only if it exhibited antibodies against rgp21, MTA-1, p19, and p24. A serum was considered negative if no bands were present and indeterminate when partial reactivities were encountered. HGIP reactivity was defined by reactivities against p19, p26, p28, and p53 but without any reactivity against p24 and Env peptide (40). For each commercial kit, i.e., ELISA as well as WB, commercially available positive and negative controls in the kit were used and the run was discarded if optical density values exceeded specified ranges for the controls. For the IFA experiments, each well was seeded with 75% CEM cells (not infected) and 25% MT2 or C19 cells (HTLV-1 or HTLV-2 infected). HTLV-1-positive as well as HTLV-1- or HTLV-2-negative sera were used as controls for each experiment.
ELISA with synthetic peptides.
Using published HTLV-1 and HTLV-2 B-cell epitope sequences (24, 27), several peptides were selected for synthesis. Their designation, origin, and sequence are shown in Table 1. Solid-phase peptide synthesis and peptide ELISA were performed as previously reported (24). Serum samples were first tested on plates coated with a mixture of eight different peptides (HTLV-1 H, T, V, A, and Gag-1 and HTLV-2 H, O, and T at a dilution of 1:50). They were then tested against each of the 11 individual peptides at the same dilution. The cutoff level for positivity was determined as the mean absorbency obtained with 18 HTLV-seronegative controls obtained from the same Cameroonian region plus three standard deviations.
TABLE 1.
HTLV-1/2 peptides used for ELISA
| Peptide | Gene (amino acids) | Amino acid sequence |
|---|---|---|
| gag1p19 | p19 (88–101) | IQTQAQIPSRPAPP |
| gag 1A | p19 (102–117) | PPSSPTHDPPDSDPQI |
| HTLV-1 pol3 | pol (487–502) | KQILSQRSFPLPPPHK |
| HTLV-1 H | env gp46 (176–199) | INTEPSQLPPTAPPLLPHSNLDHI |
| HTLV-1 T | env gp46 (190–212) | LLPHSNLDHILEPSIPWKSKLLT |
| HTLV-1 V | env gp46 (240–262) | VLYSPNVSVPSSSSTPLLYPSLA |
| HTLV-2 O | env gp46 (85–106) | IKKPNRQGLGYYSPSYNDPCSL |
| HTLV-2 H | env gp46 (172–195) | ITSEPTQPPPTSPPLVHDSDLEHV |
| HTLV-2 T | env gp46 (185–208) | PLVHDSDLEHVLTPSTSWTTKILK |
| HTLV-1 tax 23 | p40 tax (321–350) | HEPQISPGGLEPPSEKHFRE |
| HTLV-1 rex 1 | p27 rex (1–20) | MPKTRRRPRRSQRKRPPTPW |
Antibodies to blood stage P. falciparum-derived antigens.
Titers were determined by a standard IFA (34). Briefly, slides were coated with P. falciparum (Palo Alto FUP/CB strain)-infected erythrocytes (3.5% parasitemia, 0.5% hematocrit) and air dried. They were incubated with serial serum dilutions (1:50 to 1:12,800) for 30 min at 37°C, and incubated with fluorescein isothiocyanate-labeled secondary anti-human immunoglobulin G (IgG) antibody (Dako, Roskilde, Denmark).
Absorption of antibodies onto a P. falciparum immunoadsorbant column.
To determine whether antibodies against P. falciparum-derived antigens cause HGIP reactivity, antisera were absorbed onto an immobilized P falciparum extract. Briefly, enriched P. falciparum schizonts (FUP/CB strain) were resuspended in 5 volumes of 0.1 M NaHCO3 (pH 8.3) and kept for 15 min on ice. After a 30-min centrifugation at 12,000 × g, the extract was dialyzed for 3 h against the coupling buffer. Forty-five milligrams of protein (3 mg/ml) was coupled to 1.5 g of a cyanogen bromide-activated Sepharose 4B (Pharmacia, Piscataway, N. J.) under conditions recommended by the supplier. The coupling efficiency was 100% as determined by protein assay of the flowthrough fraction. The remaining active groups were blocked as recommended by the manufacturer. The column was then stored at 4°C in 0.1 M Tris-HCl (pH 8)–0.5 M NaCl buffer with 0.05% sodium azide. As a negative control, a second column was made using the same conditions with uninfected erythrocytes. Sera were diluted 1:50 in 500 μl of phosphate-buffered saline (PBS) and adsorbed onto 100 μl of either the P. falciparum column or the uninfected erythrocyte column for 30 min at room temperature on a rocking platform. After centrifugation of the column, an aliquot of the supernatant was stored at 4°C. The column was washed three times with PBS, and 500 μl of 0.1 M glycine (pH 2.5) was added for 5 min at room temperature. Finally, 25 μl of 2 M Tris was added, and the antibodies were dialyzed overnight in PBS at 4°C. An HTLV-1 WB assay (HTLV2-3 Diagnostic Biotechnology) was used to test the different fractions following the manufacturer's instructions except that the sera, including positive controls, were diluted 1:250 instead of 1:50.
Virus isolation.
PBMCs were separated in Cameroon and sent frozen on dry ice to France. In nine cases (five HTLV-1 and four HGIP), the PBMCs were immediately put in culture and maintained in a 37°C humidified 5% CO2 air atmosphere, with biweekly changes of RPMI 1640 medium (Whittaker Bioproducts, Brussels, Belgium) supplemented with 20% heat-inactivated fetal calf serum, 20 U of interleukin-2 (IL-2; Boehringer, Mannheim, Germany) per ml, 1% l-Gln, and 1% penicillin-streptomycin (Flow Labs, Glasgow, Scotland). During the first 3 days, the cells were stimulated with phytohemagglutinin (PHA; Difco) at 2 μg/106 cells. For coculture experiments, fresh cord blood cells were stimulated with PHA and then added to patient PBMCs (ratio, 1:1) after 4 days of culture. An IFA was performed on different cells obtained from either HTLV-1 or HGIP individuals after 7 weeks of culture or coculture in order to detect viral antigen expression. Either mouse monoclonal antibodies directed against HTLV-1 p19, or p24 (Cambridge Biotech), polyclonal sera from HTLV-1-infected individuals, or sera obtained directly from the HGIP individuals were used. Production of the p19 core antigen in the culture supernatant was measured every week by an antigen capture ELISA test that detects HTLV-1/2 as well as simian T lymphotropic virus type 1 (STLV-1) p19 (Retro-tek; HTLV p19 Antigen ELISA Cellular Products). According to the manufacturer, the sensitivity of the kit for the major HTLV-1 core antigen Gag p19 is 25 pg/ml.
PCR.
High-molecular-weight DNA was extracted in a P3 facility in Cameroon, where HTLV-1 DNA has never been amplified nor cloned. Briefly, following lysis in Tris-EDTA (TE) (pH 7.5)–sodium dodecyl sulfate (10%)–proteinase K–NaCl, the DNA was extracted with phenol, phenol-chloroform, and phenol-chloroform-isoamyl alcohol. It was then precipitated with 3 M sodium acetate and 100% ethanol, washed, and resuspended in TE. PCR was carried out as previously described (19, 38). Each reaction contained 1.5 μg of DNA, 0.2 mM deoxynucleoside triphosphate mix (Boehringer), 10 μl of 10 reaction buffer (Perkin Elmer Cetus), 0.1 μM each oligonucleotide primer (Pharmacia, Piscataway, N. J.), and 2.5 U of Taq DNA polymerase (Perkin Elmer Cetus) in a total volume of 100 μl. The sequences of HTLV-1/2-specific primers and appropriate probes were as follows. For the gag region PCR (HTLV-1-specific primers) we used gag949not, 5′TTTGAGCGGCCGCACCCGGTCCCTCCAGTTACGAT3′ (sense), and gag1244eco, 5′ACTAGAATTCTCATTTGCCATGGGCGATGGTT3′ (antisense). The probe was gag1056 (5′ACTTAGAATTCCCGGGGTATCCTTTTGGGA3′). gag region seminested PCR (HTLV-1-specific primers): gag949not (see sequence above) and gag1244eco (see sequence above) as outer primers followed by gag949not (5′TTTGAGCGGCCGCACCCGGTCCCTCCAGTTACGAT3′) as sense primer and gag1056 (ACTTAGAATTCCCGGGGTATCCTTTTGGGA) as antisense inner primer.
For the pol region (primers amplifying both HTLV-1 and HTLV-2) we used Pol3-4 (CACATCTGGCAAGGCGACATTAC) (sense) and SK111 (5′GTGGTGGATTTGCCATCGGGTTTT3′) (antisense). The probe used was SK110 (5′CCCTACAATCCCACCAGCTCAG).
For the tax region (HTLV-1-specific primers), the primers Rmtax1/Rmtax2 and the probe Probe tax were used as previously described (38). Another series of PCRs were conducted using KKPX1 and KKPX2 as primers and KKPXs (HTLV-1 specific) and SK45 (HTLV-1/HTLV-2) as probes (39).
For the β—globin gene, PCO4 (5′CAACTTCATCCACGTTCACC3′) (sense) and GH2-0 (5′GAAGAGCCAAGGACAGGTAC3′) (antisense) were used.
For all the PCR experiments, the amplification mixtures were made in a room physically separated from the laboratory, and positive displacement pipettes were used. For each PCR run, at least one positive control (i.e., DNA extracted from a known HTLV-1-positive individual) and one negative DNA (i.e., DNA extracted from an HTLV-seronegative blood donor) were used. Moreover, a tube was kept free of DNA to check for possible carryover. Following denaturation at 94°C for 5 min, the reaction mixtures containing DNA were cycled 45 times at 94°C for 1 min, 54°C for (β-globin), 55°C (tax), or 58°C (gag, pol, and LTR) for 1 min, and 72°C for 2 min. An extension of 2 s per cycle was included as well as an extension of 10 min on the last cycle. For the seminested PCR, the first fragment was amplified, and 2 μl of the initial PCR mixture was used for the second PCR run. Amplified DNA was size fractionated by 1.5% agarose gel electrophoresis and transferred overnight on a nylon membrane, then hybridized with a [γ−32P] dATP-end-labeled internal corresponding probe. Nylon membranes were exposed at −80°C on a film (Hyperfilm MP; Amersham) for 24 h and for 7 days.
Statistical analyses.
The association between the titer of anti-P. falciparum antibodies and HTLV enzyme immunoassay (EIA) optical density values (Platelia HTLV-1 new) was assessed using linear regression (PROC REG; Statistical Analysis System, Cary, N.C.). Since the anti-P. falciparum titer was measured using serial twofold dilutions, the log (base 2) anti-P. falciparum titer was entered as the independent variable. The natural logarithm of the optical density of the HTLV EIA was the dependent variable.
RESULTS
Antibodies to HTLV antigens.
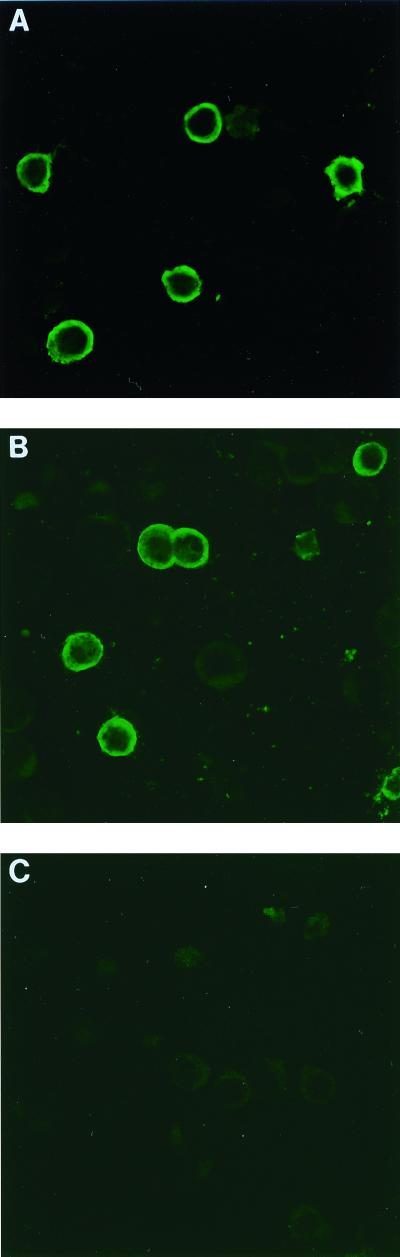
Serum specimens (n = 102) were tested by ELISA to determine the presence of antibodies to HTLV-1 or HTLV-2 antigens. Using the Platelia test, 50 of 102 sera (49%) scored positive. However, when further tested with the new-generation Genelabs ELISA 3.0 kit, which contains only synthetic Env gp21 and gp46 peptides and proteins, only 16 of 102 (15.70%) sera scored positive. All specimens were further tested with an HTLV-1 and an HTLV-2 IFA (dilution 1:10). This showed that 43 of 102 (42%) and 27 of 102 (26%) sera were reactive on MT2 and C19 cells, respectively. WB analysis, performed on all samples, demonstrated the presence of 13 truly seroreactive HTLV-1-infected individuals, no HTLV-2 positive, 20 HTLV negative, and 69 HTLV subjects with an HTLV-indeterminate WB profile. Among the 69 sera with indeterminate profile, 29 (42%) reacted with p19, p26, p28, and p53 without any reactivity against p24 Gag or Env peptides. This profile was recently defined as an HGIP (40). A typical example is shown in Fig. 1. While 22 of the 29 HGIP sera (75.8%) were considered positive with the IFA test on MT2 cells (Fig. 2), in some cases with high titers (up to 1:5,120), only 5 of 29 (17.2%) samples were positive on C19 cells at the same 1:10 dilution. These results allowed us to estimate 100% sensitivity for the Platelia ELISA, the Genelabs ELISA, and the IFA test for the detection of HTLV-1 antibodies. By contrast, the specificity was 55, 96.6, and 66%, respectively, using stringent WB criteria.
FIG. 1.
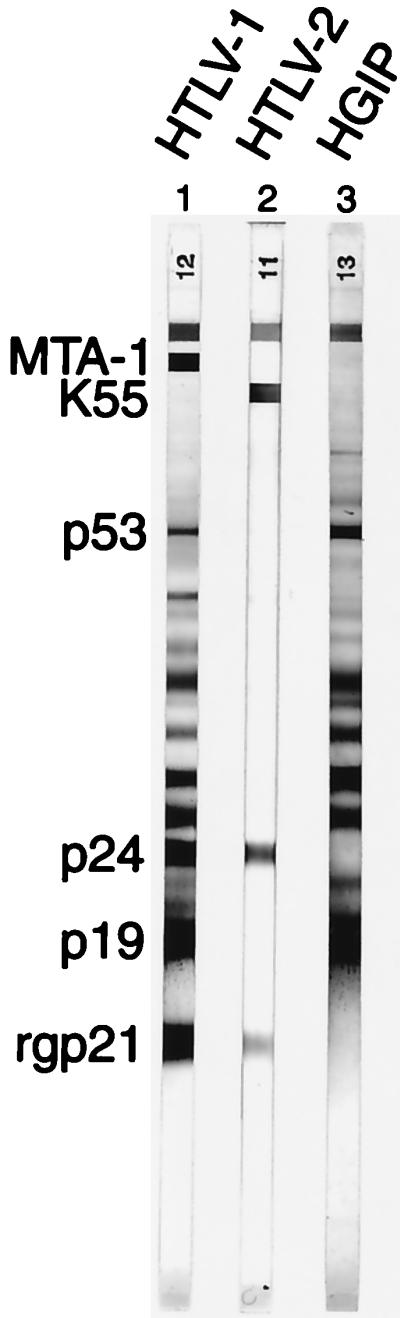
WB (HTLV2-3; Diagnostic Biotechnology) which contains disrupted HTLV-1 virions, a recombinant gp21 (rg21) protein, as well as MTA-1 (amino acids 169 to 209) and K55 (amino acids 162 to 205) which are gp46 HTLV Env-specific peptide of HTLV-1 and HTLV-2, respectively, were used. Representative WB obtained with sera from individuals infected with HTLV-1 (lane 1) or HTLV-2 (lane 2) or exhibiting an HGIP WB pattern (lane 3).
FIG. 2.
Indirect IFA with (A) an HTLV-1 serum, (B) an HGIP serum, and (C) a control serum. MT-2 (HTLV-1 producing) and CEM (negative control) cells were split and acetone fixed at a ratio of 1:4. The serum is used at a 1:40 dilution. Results are representative of at least five independent experiments.
Analysis of the WB profile of 82 sera obtained 4 years after the initial screening did not reveal any major modification of the profiles: there were no seroconversions of an HGIP profile to a complete HTLV-1-seroreactive profile. However, one patient lost the HGIP and became HTLV seronegative by WB, and one previously negative patient seroconverted to HGIP. Epidemiological analysis of the HGIP pattern revealed no evidence supporting transmission of a potential causative agent related to HTLVs. First, there was no increase in HGIP prevalence with age, as is commonly seen for HTLV-1 and HTLV-2 and other vertically and sexually transmitted viruses in endemic populations. HGIP and HTLV-1 prevalence were as 32 and 0%, respectively, in those aged 0 to 20 years, 27 and 11.5% in those aged 21 to 50 years, and 27.7 and 39% in those aged 50 years and older. Thirteen of 42 (30.9%; mean age, 31 years) males had HGIP, compared to 16 of 60 (26.6%; mean age, 30.5 years) in females. Second, although HGIP appeared to randomly affect both members of a few mother-child or husband-wife pairs, there were too few cases for a formal familial analysis. There were also several children with HGIP for whom neither parent had HGIP as well as women with HGIP for whom neither the husband nor the mother had HGIP.
ELISA with different HTLV-1- or HTLV-2-encoded synthetic peptides.
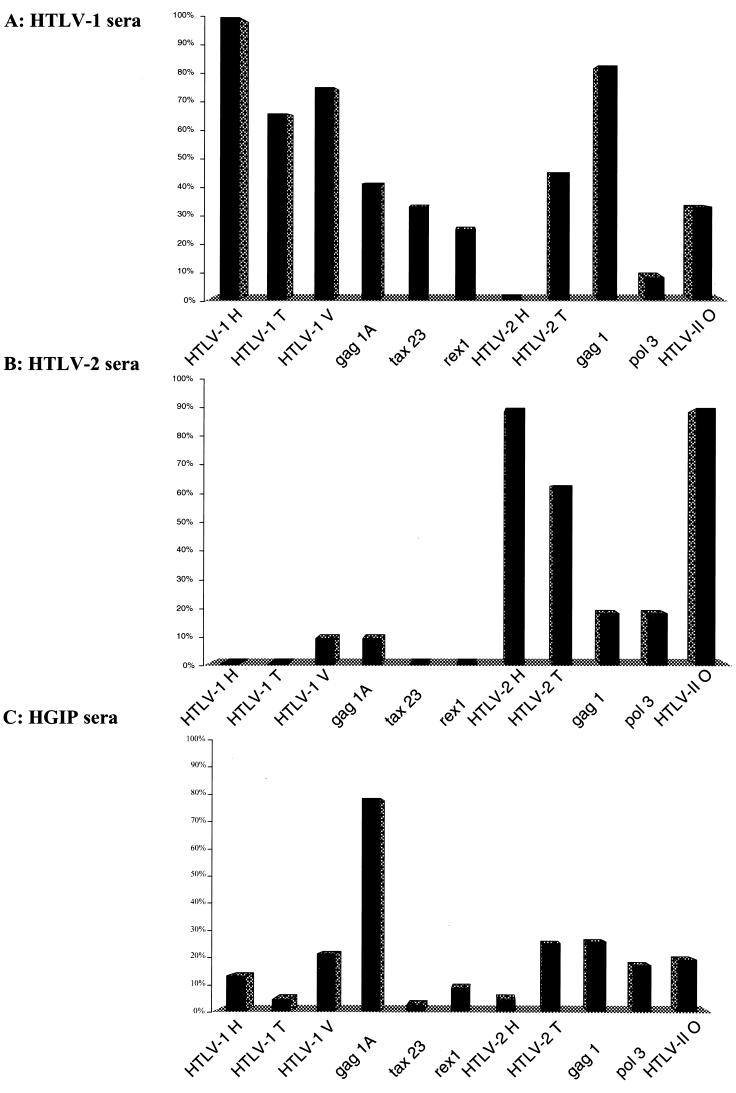
Twelve HTLV-1, 29 HTLV-indeterminate, including 26 HGIP, and 18 HTLV-1/2-negative sera from Cameroon were tested. Furthermore, 11 HTLV-2-positive sera from Amerindian and Gabonese villagers were also used as controls. A preliminary experiment was conducted to test these sera on plates which contained five different HTLV-1 peptides (Henvgp46 Tenvgp46, Venvgp46, Aenvgp21, and gag1p19) and three HTLV-2 (Henvgp46, Oenvgp46, and Tenvgp46). All HTLV-1 sera and all but two HTLV-2 sera of African origin (both with low antibody titers as determined by IFA on C19 cells) were detected as positive. These peptides and others (see Table 1 for a list) were further tested separately, with and without bovine serum albumin (BSA) coupling. The results are summarized in Fig. 3. Sixty-six to 100% of HTLV-1 sera recognized the various HTLV-1 Env peptides. By comparison, HTLV-2 and HGIP sera reacted poorly against these HTLV-1 peptides (0 to 21%). HTLV-2 Env peptides were well recognized by HTLV-2 sera (63 to 90% depending on the peptides). As previously described, Tax, Rex, and Pol peptides were not as efficiently recognized by HTLV-1 or HTLV-2 sera (29). Finally, HGIP sera did not efficiently recognize the same peptides as those recognized by the antibodies present in HTLV-1 and HTLV-2 sera.
FIG. 3.
Immune responsiveness to 11 immunodominant epitopes from the Gag, Pol, Env, Tax, and Rex proteins of HTLV-1 or HTLV-2 in patients with (A) HTLV-1 (n = 12), (B) HTLV-2 (n = 11), and (C) HGIP (n = 26) WB profiles. As controls, 18 HTLV-1/2-negative sera from the same area of Cameroon were used. Results are expressed as percent of sera above the cut off value determined as the mean absorbancy obtained with 18 HTLV—seronegative controls obtained from the same Cameroonian region plus three standard deviations. These results are representative of two independent experiments.
The results obtained with the Gag peptides differed depending on the group of sera tested. While gag-1A (C-terminal part of p19) was recognized by more than 78% of HGIP sera, it reacted with only 41% of HTLV-1-positive sera (Fig. 3A and C). The opposite result was obtained with the gag1p19 peptide (20 versus 80%) (Fig. 3A and C).
Viral isolation.
PBMCs from five HTLV-1-seropositive individuals were cultured for at least 8 weeks in the presence of IL-2. Four long-term cultures expressing HTLV-1 antigens, as detected by IFA and by the presence of p19gag antigen in the culture supernatant (data not shown), were further obtained. The cell surface phenotype determined by flow cytometry analysis was demonstrated to be of T-cell lineage, with expression of CD2, CD5, CD25, and HLA-DR, without B-cell markers and with expression of either CD4 or CD8 (data not shown). Despite culture and coculture attempts, no HTLV-1-related virus was isolated, and no long-term cell lines were established from cells obtained from any of the four HGIP individuals whose sera also presented a positive IFA titer on MT2 cells (1:160 to 1:2,560). An IFA test conducted after 7 weeks of culture of such HGIP peripheral blood lymphocytes using either autologous HGIP serum or an HTLV-1 serum chosen for its high antibody titer, did not detect any HTLV-1 antigen expression (data not shown). Finally, no HTLV-1 p19-related protein was detected in eight successive culture supernatants from each of the four HGIP cultures tested after 5 weeks of culture (data not shown).
Detection of HTLV DNA sequences in PBMCs.
DNA was available from 88 individuals (11 HTLV-1, 23 HGIP, 37 indeterminate with other WB profiles, and 17 seronegative). A control PCR using a β-globin primer pair demonstrated that cellular DNA was amplifiable for all samples. PCR experiments were conducted for each of these samples to search for any presence of HTLV-1-related sequences. Three different specific primer sets encompassing parts of the gag, pol, and tax genes of the HTLV-1 and HTLV-2 genomes were used (Table 2). None of the 17 seronegative or 37 HTLV-indeterminate specimens reacted with any of the three primer-probe combinations. By contrast, all but two (C22-1 and D5-1) HTLV-1 samples gave a positive signal after hybridization with the specific probes. PCR analysis of DNAs extracted from PBMCs obtained from 23 individuals with HGIP failed to amplify any product with either primer-probe combination. The same negative results were obtained using a seminested PCR protocol encompassing the gag region on five HGIP DNA samples. These samples were chosen from individuals whose sera exhibited the highest antibody titers, assuming that these persons were at highest risk of carrying an HTLV-related agent. In contrast, we obtained positive signals using the sensitive technique for all the HTLV-1 DNAs tested, including the two samples that did not give a signal using simple PCR.
TABLE 2.
Detection of HTLV-1 gene sequences in PBMCs by PCR
| Donor status | No. of samples giving indicated result/no. tested
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
gag
|
pol
|
taxa
|
semi-nested gag
|
taxb
|
β-Globin
|
Total tested | |||||||
| − | + | − | + | − | + | − | + | − | + | − | + | ||
| HTLV negative | 17 /17 | 0 /17 | 17 /17 | 0 /17 | 17 /17 | 0 /17 | 5 /5 | 0 /5 | 6 /6 | 0 /6 | 0 /17 | 17 /17 | 17 |
| HTLV indeterminate | 37 /37 | 0 /37 | 37 /37 | 0 /37 | 37 /37 | 0 /37 | ND | ND | 6 /6 | 0 /6 | 0 /37 | 37 /37 | 37 |
| HGIP | 23 /23 | 0 /23 | 23 /23 | 0 /23 | 23 /23 | 0 /23 | 5 /5 | 0 /5 | 5 /5 | 0 /5 | 0 /23 | 23 /23 | 23 |
| HTLV-1 positive | 2 /11 | 9 /11 | 2 /11 | 9 /11 | 2 /11 | 9 /11 | 0 /5 | 5 /5 | 0 /5 | 5 /5 | 0 /11 | 11 /11 | 11 |
| Total | 79 | 9 | 79 | 9 | 79 | 9 | 10 | 5 | 17 | 5 | 0 | 88 | 88 |
Rmtax1/Rmtax2.
KKPX1/KKPX2.
Finally, we extracted again the DNA of 22 samples (five HGIP, six HTLV-1/2 indeterminate, six HTLV-1/2 seronegative, and five HTLV-1). Using primers corresponding to highly conserved regions of the tax gene which allow the detection of all known primate T lymphotropic virus types, we performed additional independent PCR experiments followed by hybridization with either HTLV-1 or HTLV-1/HTLV-2-specific probes. All five HTLV-1 samples were scored as positive, but none of the HTLV-1/2-seronegative, HTLV-1/2-indeterminate, or HGIP DNAs gave a positive signal.
Correlation between antibodies to HTLV-1 and malarial titers.
All but one of the 102 sera tested had anti-P. falciparum antibodies, with an average IFA titer of 1:2,560. The strength of HTLV EIA (Platelia HTLV new kit) reactivity, as represented by the natural logarithm of the optical density value, was significantly correlated with the log2 anti-P. falciparum antibody titer by linear regression (intercept = −2.1821, beta = 0.1684, R2 = 0.06, P = 0.01). Therefore, a positive correlation between positive HTLV-1 ELISA optical density results and titers of antibody to P. falciparum was demonstrated.
Inhibition of HGIP profile after incubation with a P. falciparum-infected erythrocyte lysate.
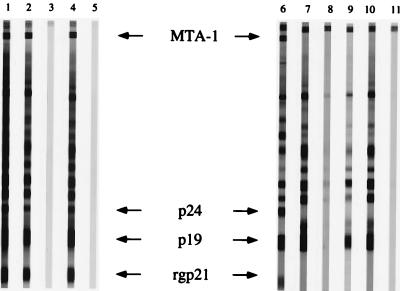
Based on a previous report (31), competitive inhibition experiments were designed to determine the interactions between the blood stage of malarial antigens with antibodies present in HTLV-1-positive or HGIP specimens from some of the Cameroonian subjects. Incubation of three different HTLV-1-positive sera with infected or uninfected erythrocyte lysate prior to HTLV-1 WB always yielded to similar results. A representative example is shown in Fig. 4 (lanes 1 to 5). The antibody binding to HTLV-1-specific antigens (lane 1) was not adsorbed onto the P. falciparum (lane 2) or control (lane 4) erythrocyte columns. No reactivity was recovered upon elution of bound antibodies to the column (lanes 3 and 5). By contrast, the reactivity of all four HGIP specimens that were tested was completely inhibited after incubation on the P. falciparum-infected erythrocyte-coupled column. A representative example is shown in lanes 7 and 8. The antibodies eluted from the P. falciparum column had a typical HGIP profile on the HTLV-1 WB (lane 9). The specificity of the reaction was assessed by using a column prepared with uninfected erythrocytes onto which no reacting antibodies were absorbed (lanes 10 and 11).
FIG. 4.
Competitive inhibition of HTLV-1 or HGIP antibodies with a Sepharose column loaded with P. falciparum-infected or noninfected erythrocytes. Lanes 1 and 6, HTLV-1 serum from Cameroon; lane 2, same serum after incubation with a Sepharose column loaded with P. falciparum-infected erythrocytes; lane 3, reactivity of the eluted antibodies; lane 4, same serum after incubation with a Sepharose column loaded with noninfected erythrocytes; lane 5, reactivity of the eluted antibodies; lane 7, HGIP serum from Cameroon; lane 8, same serum after incubation with a Sepharose column loaded with P. falciparum-infected erythrocytes; lane 9, reactivity of the eluted antibodies; lane l0, same serum after incubation with a Sepharose column loaded with noninfected erythrocytes; lane 11, reactivity of the eluted antibodies. This result is representative of three independent experiments.
Possible cross-reactivity between Exp-1 protein of P. falciparum and anti-HTLV-1 antibodies.
To test for possible antigenic cross-reactivity between HTLV-1 p19 and the P. falciparum Exp-1-derived protein (49), an anti-Exp-1 monoclonal antibody and a polyclonal anti-Exp-1 serum were tested in an HTLV-1 WB analysis. Despite several attempts at different dilutions, we were not able to detect any HGIP reactivity. However, and as reported previously (49), we detected a GD21 band with the polyclonal anti-Exp-1 serum (data not shown). In a control experiment, the same monoclonal sera reacted strongly with P. falciparum-infected erythrocytes in an IFA test (data not shown).
DISCUSSION
The HTLV WB seroindeterminate frequency varies according to HTLV-1/2 endemicity, i.e., to the geographical area studied. Among blood donors in areas of low endemicity (Europe and the United States), the seroindeterminate WB patterns consist of faint isolated Gag reactivity (2, 12, 32). They occur at a frequency similar to true HTLV-1 seropositivity (ranging from 0 to 0.022% among blood donors) (26). In such populations, the HGIP appears to be very rare (26). Although some uncertainty remains, WB-indeterminate blood donors are generally counseled that they are not infected with HTLV (9, 13, 25, 35, 55). By contrast, in tropical areas such as Central Africa, Melanesia, and some regions of southeast Asia and South America, the prevalence rate of the indeterminate WB reactivities is high, representing in some cases more than 50% of all WB profiles (8, 30). Of the indeterminate WB patterns, HGIP makes up a large proportion. In the present study, HGIP represented the most common WB pattern, with 42% of the seroindeterminate, namely, 28% of the total population of the villagers tested. Therefore, in several previous reports, misclassification (due to nonstringent WB criteria) of such HGIP as true HTLV-1 seropositive led not only to an overestimation of the global HTLV-1 seroprevalence rate, but also to some bizarre epidemiological findings (36). As an example, the findings for some children initially considered HTLV-1 seropositive but born of HTLV-1-seronegative mothers led to speculation about modes of transmission other than breast-feeding (36). In light of the present findings, one can assume that these infants were not HTLV-1 infected but had most probably presented an HGIP reactivity.
The current study yielded several new insights on the significance of such HGIP in Central Africa, and several conclusions can be drawn.
(i) The epidemiological analysis of the demographic characteristics and familial occurrence of the HGIP pattern failed to reveal patterns consistent with sexual or vertical transmission of a putative infectious agent, in contrast to previously published studies of WB- and/or PCR-confirmed HTLV-1 (42). Instead of increasing steadily with age, HGIP prevalence was roughly constant. HGIP was equally prevalent among males and females, instead of the previously reported higher HTLV-1 prevalence among women in most endemic areas (41). These data are consistent with a previous epidemiological study of HGIP in Cameroon (40), but are unique in showing a lack of familial aggregation of HGIP. The results are also consistent with other studies which showed no evidence for HTLV-1 infection in WB-indeterminate U.S. blood donors (9, 10, 25, 32) but are unique in showing no evolution of HGIP WB patterns over a long follow-up time and in the African setting of the study.
(ii) Previous studies demonstrated that Tax primers are highly sensitive to detect HTLV-1, HTLV-2, STLV-1, STLV-2, and PTLV-L (39, 55). The lack of detection of any HTLV-1/2 proviral sequences by PCR (even when performing a seminested PCR) as well as the absence of p19 in the supernatant of short-term cultures of PBMCs obtained from HGIP individuals and the inability to establish long-term cell lines suggest that there was no HTLV-1 provirus and no transforming agent at a detectable level in the PBMCs of such individuals. These results strongly suggest that 22 of 38 sera considered HTLV-1 positive in earlier seroepidemiological studies using nonstringent WB criteria (36) were in fact HGIP specimens.
By contrast, HTLV-1 proviral DNA could easily be detected and long-term cultures of T cells frequently established from PBMCs collected from the majority of the HTLV-1-seropositive individuals living in the same area. This reinforces the interpretation that these HGIP do not derive from infection by an HTLV-1-like virus (at least in the PBMCs), but rather from serological cross-reactivities. As mentioned above, there is only one report of the isolation of an HTLV-1 virus from an African-American female suffering from multiple sclerosis with an HGIP seroreactivity (59).
(iii) Our peptide-based ELISA results clearly indicate that the antibodies present in HGIP sera and in HTLV-1 sera do not recognize the same Gag epitopes. This result again strongly suggests that these seroreactivities do not reflect a true HTLV-1 infection. Interestingly our results obtained with the gag1p19 and the gag-1A peptides show some differences from those of Lal et al. (33). These authors reported 90% seroreactivity with gag-1A peptide versus 5% with gag1p19 when using HTLV-1 sera. However, it is worth noting that due to high background technical problems, we did not use the same ELISA procedure. Our slight modification in the ELISA protocol (elimination of BSA) could be an explanation for the observed differences. In addition, the sera used by Lal et al. (33) were collected in the United States and Japan, many of them from symptomatic carriers with possible high specific anti-HTLV-1 titers, whereas our sera were collected in Central Africa, where HTLV-1-infected asymptomatic individuals also have very high non HTLV-1-specific Ig titers.
(iv) Our adsorption experiments strongly suggest that, at least in central Africa, HGIP reactivities could be due to anti-P. falciparum antibodies. The fact that all tested cases of HGIP WB reactivities were abolished after absorption onto a P. falciparum immunoabsorbant and recovered after acid elution is a strong argument in favor of the hypothesis that HGIP WB reactivity is to be attributed to anti-P. falciparum antibodies. Furthermore, the correlation between the log (base 2) anti-P. falciparum titer and logarithm EIA absorbency indicates that the former may be responsible for false-positive tests using the latter assay on a population basis. However, the rather low R2 value indicates poor prediction of any one EIA absorbance value on the basis of that individual's anti-P. falciparum titer. Hence, a higher prevalence of false-positive HTLV-1 EIA tests may be expected in populations with higher anti-P. falciparum titers, but confirmation of individual high EIA values in these areas will remain necessary.
While we were able to test the previously suggested hypothesis of the Exp-1 protein as the source of HGIP (49, 50), we did not observe an HGIP reactivity on an HTLV-1 WB using anti-Exp-1 mouse antibodies. Thus, we are unable to confirm this hypothesis. However, it is unlikely that the large number of the different antigens detected by HGIP sera derive from cross-reactivity with a single P. falciparum protein. P. falciparum expresses a large number of proteins during its development in humans. WB analysis of P. falciparum blood stage extracts using sera from malaria-endemic areas usually generates different complex multiple band patterns. In fact, the large number of serological specificities characteristic of malaria-immune sera may provide the basis of reactivity on multiple HTLV-1-derived antigens.
ACKNOWLEDGMENTS
This work was financially supported by Agence Nationale de Recherches sur le SIDA (ANRS) and the French Ministry of Cooperation. R. Mahieux was a CANAM Fellow.
We thank Emmanuelle Perret for her technical assistance during the microscopy experiments, Joao Aguiar for the mouse anti-Exp-1 antibodies, Vincent Foumane and Emmanuel Tina Abada for their technical assistance during the collecting of the samples, and Wilfrid Mahieux for his help during the editing of the manuscript.
REFERENCES
- 1.Anonymous. Acquired immunodeficiency syndrome (AIDS). Proposed WHO criteria for interpreting results from western blot assays for HIV-1, HIV-2, and HTLV-I/HTLV-II. Wkly Epidemiol Rec. 1990;65:281–283. [PubMed] [Google Scholar]
- 2.Anonymous. Seroepidemiology of the human T-cell leukaemia/lymphoma viruses in Europe. The HTLV European Research Network. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:68–77. doi: 10.1097/00042560-199609000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Asher D M, Goudsmit J, Pomeroy K L, Garruto R M, Bakker M, Ono S G, Elliot N, Harris K, Askins H, Eldadah Z, et al. Antibodies to HTLV-I in populations of the southwestern Pacific. J Med Virol. 1988;26:339–351. doi: 10.1002/jmv.1890260402. [DOI] [PubMed] [Google Scholar]
- 4.Banki K, Maceda J, Hurley E, Ablonczy E, Mattson D H, Szegedy L, Hung C, Perl A. Human T-cell lymphotropic virus (HTLV)-related endogenous sequence, HRES-1, encodes a 28-kDa protein: a possible autoantigen for HTLV-I gag-reactive autoantibodies. Proc Natl Acad Sci USA. 1992;89:1939–1943. doi: 10.1073/pnas.89.5.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biggar R J, Gigase P L, Melbye M, Kestens L, Sarin P S, Bodner A J, Demedts P, Stevens W J, Paluku L, Delacollette C, et al. ELISA HTLV retrovirus antibody reactivity associated with malaria and immune complexes in healthy Africans. Lancet. 1985;2:520–523. doi: 10.1016/s0140-6736(85)90461-1. [DOI] [PubMed] [Google Scholar]
- 6.Biggar R J, Neequaye J E, Neequaye A R, Ankra-Badu G A, Levine P H, Manns A, Taylor M, Drummond J, Waters D. The prevalence of antibodies to the human T lymphotropic virus (HTLV) in Ghana, West Africa. AIDS Res Hum Retroviruses. 1993;9:505–511. doi: 10.1089/aid.1993.9.505. [DOI] [PubMed] [Google Scholar]
- 7.Biggar R J, Saxinger C, Gardiner C, Collins W E, Levine P H, Clark J W, Nkrumah F K, Blattner W A. Type-I HTLV antibody in urban and rural Ghana, West Africa. Int J Cancer. 1984;34:215–219. doi: 10.1002/ijc.2910340212. [DOI] [PubMed] [Google Scholar]
- 8.Bonis J, Preux P M, Nzisabira L, Letenneur L, Muhirwa G, Buzingo T, Kamuragiye A, Preux C, Ngoga E, Dumas M, et al. HTLV-I in Burundi (east Africa): lack of reactivity to the HTLV-I immunodominant envelope epitope. J Acquir Immune Defic Syndr. 1994;7:1099–1100. [PubMed] [Google Scholar]
- 9.Busch M P, Laycock M, Kleinman S H, Wages J W, Jr, Calabro M, Kaplan J E, Khabbaz R F, Hollingsworth C G. Accuracy of supplementary serologic testing for human T-lymphotropic virus types I and II in US blood donors. Retrovirus Epidemiology Donor Study. Blood. 1994;83:1143–1148. [PubMed] [Google Scholar]
- 10.Busch M P, Switzer W M, Murphy E L, Thomson R, Heneine W. Absence of evidence of infection with divergent primate T-lymphotropic viruses in United States blood donors who have seroindeterminate HTLV test results. Transfusion. 2000;40:443–449. doi: 10.1046/j.1537-2995.2000.40040443.x. [DOI] [PubMed] [Google Scholar]
- 11.Cossen C, Hagens S, Fukuchi R, Forghani B, Gallo D, Ascher M. Comparison of six commercial human T-cell lymphotropic virus type I (HTLV-I) enzyme immunoassay kits for detection of antibody to HTLV-I and -II. J Clin Microbiol. 1992;30:724–725. doi: 10.1128/jcm.30.3.724-725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courouce A M, Pillonel J, Lemaire J M, Maniez M, Brunet J B. Seroepidemiology of HTLV-I/II in universal screening of blood donations in France. AIDS. 1993;7:841–847. doi: 10.1097/00002030-199306000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Cowan E P, Nemo G J, Williams A E, Alexander R K, Vallejo A, Hewlett I K, Lal R B, Dezzutti C S, Gallahan D, George K, Pancake B A, Zucker-Franklin D, McCurdy P R, Tabor E. Absence of human T-lymphotropic virus type I tax sequences in a population of normal blood donors in the Baltimore, MD/Washington, DC, area: results from a multicenter study. Transfusion. 1999;39:904–909. doi: 10.1046/j.1537-2995.1999.39080904.x. [DOI] [PubMed] [Google Scholar]
- 14.Delaporte E, Dupont A, Peeters M, Josse R, Merlin M, Schrijvers D, Hamono B, Bedjabaga L, Cheringou H, Boyer F, et al. Epidemiology of HTLV-I in Gabon (Western Equatorial Africa) Int J Cancer. 1988;42:687–689. doi: 10.1002/ijc.2910420509. [DOI] [PubMed] [Google Scholar]
- 15.Delaporte E, Peeters M, Durand J P, Dupont A, Schrijvers D, Bedjabaga L, Honore C, Ossari S, Trebucq A, Josse R, et al. Seroepidemiological survey of HTLV-I infection among randomized populations of western central African countries. J Acquir Immune Defic Syndr. 1989;2:410–413. [PubMed] [Google Scholar]
- 16.de The G, Gessain A, Gazzolo L, Robert-Guroff M, Najberg G, Calender A, Peti M, Brubaker G, Bensliman A, Fabry F, et al. Comparative seroepidemiology of HTLV-I and HTLV-III in the French West Indies and some African countries. Cancer Res. 1985;45:4633s–4636s. [PubMed] [Google Scholar]
- 17.Dumas M, Houinato D, Verdier M, Zohoun T, Josse R, Bonis J, Zohoun I, Massougbodji A, Denis F. Seroepidemiology of human T-cell lymphotropic virus type I/II in Benin (West Africa) AIDS Res Hum Retroviruses. 1991;7:447–451. doi: 10.1089/aid.1991.7.447. [DOI] [PubMed] [Google Scholar]
- 18.Gallo D, Yeh E T, Moore E S, Hanson C V. Comparison of four enzyme immunoassays for detection of human T-cell lymphotropic virus type 2 antibodies. J Clin Microbiol. 1996;34:213–215. doi: 10.1128/jcm.34.1.213-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garin B, Gosselin S, de The G, Gessain A. HTLV-I/II infection in a high viral endemic area of Zaire, Central Africa: comparative evaluation of serology, PCR, and significance of indeterminate western blot pattern. J Med Virol. 1994;44:104–109. doi: 10.1002/jmv.1890440119. [DOI] [PubMed] [Google Scholar]
- 20.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;2:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 21.Gessain A, Mahieux R, de The G. HTLV-I “indeterminate” Western blot patterns observed in sera from tropical regions: the situation revisited. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:316–319. [PubMed] [Google Scholar]
- 22.Hayes C G, Burans J P, Oberst R B. Antibodies to human T lymphotropic virus type I in a population from the Philippines: evidence for cross-reactivity with Plasmodium falciparum. J Infect Dis. 1991;163:257–262. doi: 10.1093/infdis/163.2.257. [DOI] [PubMed] [Google Scholar]
- 23.Haynes B F, Robert-Guroff M, Metzgar R S, Franchini G, Kalyanaraman V S, Palker T J, Gallo R C. Monoclonal antibody against human T cell leukemia virus p19 defines a human thymic epithelial antigen acquired during ontogeny. J Exp Med. 1983;157:907–920. doi: 10.1084/jem.157.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horal P, Hall W W, Svennerholm B, Lycke J, Jeansson S, Rymo L, Kaplan M H, Vahlne A. Identification of type-specific linear epitopes in the glycoproteins gp46 and gp21 of human T-cell leukemia viruses type I and type II using synthetic peptides. Proc Natl Acad Sci USA. 1991;88:5754–5758. doi: 10.1073/pnas.88.13.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khabbaz R F, Heneine W, Grindon A, Hartley T M, Shulman G, Kaplan J. Indeterminate HTLV serologic results in U.S. blood donors: are they due to HTLV-I or HTLV-II? J Acquir Immune Defic Syndr. 1992;5:400–404. [PubMed] [Google Scholar]
- 26.Kwok S, Lipka J J, McKinney N, Kellogg D E, Poiesz B, Foung S K, Sninsky J J. Low incidence of HTLV infections in random blood donors with indeterminate western blot patterns. Transfusion. 1990;30:491–494. doi: 10.1046/j.1537-2995.1990.30690333477.x. [DOI] [PubMed] [Google Scholar]
- 27.Lal R B. Delineation of immunodominant epitopes of human T-lymphotropic virus types I and II and their usefulness in developing serologic assays for detection of antibodies to HTLV-I and HTLV-II. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:S170–S178. doi: 10.1097/00042560-199600001-00026. [DOI] [PubMed] [Google Scholar]
- 28.Lal R B, Brodine S, Kazura J, Mbidde-Katonga E, Yanagihara R, Roberts C. Sensitivity and specificity of a recombinant transmembrane glycoprotein (rgp21)-spiked western immunoblot for serological confirmation of human T-cell lymphotropic virus type I and type II infections. J Clin Microbiol. 1992;30:296–299. doi: 10.1128/jcm.30.2.296-299.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal R B, Giam C Z, Coligan J E, Rudolph D L. Differential immune responsiveness to the immunodominant epitopes of regulatory proteins (tax and rex) in human T cell lymphotropic virus type I-associated myelopathy. J Infect Dis. 1994;169:496–503. doi: 10.1093/infdis/169.3.496. [DOI] [PubMed] [Google Scholar]
- 30.Lal R B, Lipka J J, Foung S K, Hadlock K G, Reyes G R, Carney W P. Human T lymphotropic virus type I/II in Lake Lindu Valley, Central Sulawesi, Indonesia. J Acquir Immune Defic Syndr. 1993;6:1067–1068. [PubMed] [Google Scholar]
- 31.Lal R B, Rudolph D, Alpers M P, Sulzer A J, Shi Y P, Lal A A. Immunologic cross-reactivity between structural proteins of human T- cell lymphotropic virus type I and the blood stage of Plasmodium falciparum. Clin Diagn Lab Immunol. 1994;1:5–10. doi: 10.1128/cdli.1.1.5-10.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lal R B, Rudolph D L, Coligan J E, Brodine S K, Roberts C R. Failure to detect evidence of human T-lymphotropic virus (HTLV) type I and type II in blood donors with isolated gag antibodies to HTLV-I/II. Blood. 1992;80:544–550. [PubMed] [Google Scholar]
- 33.Lal R B, Rudolph D L, Griffis K P, Kitamura K, Honda M, Coligan J E, Folks T M. Characterization of immunodominant epitopes of gag and pol gene-encoded proteins of human T-cell lymphotropic virus type I. J Virol. 1991;65:1870–1876. doi: 10.1128/jvi.65.4.1870-1876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Scanf C, Fandeur T, Morales-Betoulle M E, Mercereau-Puijalon O. Plasmodium falciparum: altered expressions of erythrocyte membrane-associated antigens during antigenic variation. Exp Parasitol. 1997;85:135–148. doi: 10.1006/expr.1996.4121. [DOI] [PubMed] [Google Scholar]
- 35.Lipka J J, Young K K, Kwok S Y, Reyes G R, Sninsky J J, Foung S K. Significance of human T-lymphotropic virus type I indeterminant serological findings among healthy individuals. Vox Sang. 1991;61:171–176. doi: 10.1111/j.1423-0410.1991.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 36.Louis J P, Gardon J, Trebucq A, Hengy C, Louis F J, Migliani R, Rey J L, Delaporte E. Epidemiological features of retroviral infection by HTLV-1 in central Africa. Bull Soc Pathol Exot. 1993;86:163–168. [PubMed] [Google Scholar]
- 37.Mager D L, Freeman J D. Human endogenous retroviruslike genome with type C pol sequences and gag sequences related to human T-cell lymphotropic viruses. J Virol. 1987;61:4060–4066. doi: 10.1128/jvi.61.12.4060-4066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahieux R, de The G, Gessain A. The tax mutation at nucleotide 7959 of human T-cell leukemia virus type 1(HTLV-1) is not associated with tropical spastic paraparesis/HTLV-1-associated myelopathy but is linked to the cosmopolitan molecular genotype. J Virol. 1995;69:5925–5927. doi: 10.1128/jvi.69.9.5925-5927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahieux R, Pecon-Slattery J, Gessain A. Molecular characterization and phylogenetic analyses of a new, highly divergent simian T-cell lymphotropic virus type 1 (STLV-1marc1) in Macaca arctoides. J Virol. 1997;71:6253–6258. doi: 10.1128/jvi.71.8.6253-6258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mauclere P, Le Hesran J Y, Mahieux R, Salla R, Mfoupouendoun J, Abada E T, Millan J, de The G, Gessain A. Demographic, ethnic, and geographic differences between human T cell lymphotropic virus (HTLV) type I-seropositive carriers and persons with HTLV-I Gag-indeterminate Western blots in Central Africa. J Infect Dis. 1997;176:505–509. doi: 10.1086/514071. [DOI] [PubMed] [Google Scholar]
- 41.Murphy E L, Figueroa J P, Gibbs W N, Brathwaite A, Holding-Cobham M, Waters D, Cranston B, Hanchard B, Blattner W A. Sexual transmission of human T-lymphotropic virus type I (HTLV-I) Ann Intern Med. 1989;111:555–560. doi: 10.7326/0003-4819-111-7-555. [DOI] [PubMed] [Google Scholar]
- 42.Murphy E L, Figueroa J P, Gibbs W N, Holding-Cobham M, Cranston B, Malley K, Bodner A J, Alexander S S, Blattner W A. Human T-lymphotropic virus type I (HTLV-I) seroprevalence in Jamaica. I. Demographic determinants. Am J Epidemiol. 1991;133:1114–1124. doi: 10.1093/oxfordjournals.aje.a115824. [DOI] [PubMed] [Google Scholar]
- 43.Nerurkar V R, Miller M A, Leon-Monzon M E, Ajdukiewicz A B, Jenkins C L, Sanders R C, Godec M S, Garruto R M, Yanagihara R. Failure to isolate human T cell lymphotropic virus type I and to detect variant-specific genomic sequences by polymerase chain reaction in Melanesians with indeterminate western immunoblot. J Gen Virol. 1992;73:1805–1810. doi: 10.1099/0022-1317-73-7-1805. [DOI] [PubMed] [Google Scholar]
- 44.Palker T J, Scearce R M, Ho W, Copeland T D, Oroszlan S, Popovic M, Haynes B F. Monoclonal antibodies reactive with human T cell lymphotropic virusI (HTLVI) p19 internal core protein: cross-reactivity with normal tissues and differential reactivity with HTLV types I and II. J Immunol. 1985;135:247–254. [PubMed] [Google Scholar]
- 45.Palker T J, Singer K H, Vahlne A. Characterization of an antigen shared by human thymic epithelium and human T cell leukemia virus p19 Gag protein. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;11:10–19. doi: 10.1097/00042560-199601010-00002. [DOI] [PubMed] [Google Scholar]
- 46.Perl A, Rosenblatt J D, Chen I S, DiVincenzo J P, Bever R, Poiesz B J, Abraham G N. Detection and cloning of new HTLV-related endogenous sequences in man. Nucleic Acids Res. 1989;17:6841–6854. doi: 10.1093/nar/17.17.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picchio G R, Bare P, Savignano R, Perez-Bianco R, Yamashita M, Hayami M. HTLV-I/II indeterminate serology and natural killer cell expansion. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:428–431. doi: 10.1097/00042560-199608010-00016. [DOI] [PubMed] [Google Scholar]
- 48.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porter K R, Aguiar J, Richards A, Sandjaya B, Ignatias H, Hadiputranto H, Ridley R G, Takacs B, Wignall F S, Hoffman S L, Hayes C G. Immune response against the exp-1 protein of Plasmodium falciparum results in antibodies that cross-react with human T-cell lymphotropic virus type 1 proteins. Clin Diagn Lab Immunol. 1998;5:721–724. doi: 10.1128/cdli.5.5.721-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porter K R, Anthony R L, Solihin A, Hayes C G. Mapping of a human T-lymphotropic virus type I gag protein epitope that cross-reacts with anti-Plasmodium falciparum antibodies. J Med Virol. 1995;45:469–474. doi: 10.1002/jmv.1890450419. [DOI] [PubMed] [Google Scholar]
- 51.Porter K R, Liang L, Long G W, Bangs M J, Anthony R, Andersen E M, Hayes C G. Evidence for anti-Plasmodium falciparum antibodies that cross-react with human T-lymphotropic virus type I proteins in a population in Irian Jaya, Indonesia. Clin Diagn Lab Immunol. 1994;1:11–15. doi: 10.1128/cdli.1.1.11-15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders R C, Levin A, Anian G, Webber I, Lee H, Swanson P, Diwan A, Desowitz R, Blattner W A, Alpers M P. HTLV-I antibody studies in villagers in East Sepik Province, Papua New Guinea. Arch Virol. 1990;114:27–35. doi: 10.1007/BF01311009. [DOI] [PubMed] [Google Scholar]
- 53.Sato A, Isaka Y, Morita F, Ishii A, Goto Y, Imai J, Igarashi H, Yoshie O, Hinuma Y. Human sera from varicella-zoster virus (VZV) infections cross-react with human T cell leukaemia virus type 1 (HTLV-1): common epitopes in VZV gene 22 protein and HTLV-1 p19 gag protein. J Gen Virol. 1992;73:2969–2973. doi: 10.1099/0022-1317-73-11-2969. [DOI] [PubMed] [Google Scholar]
- 54.Saxinger W, Blattner W A, Levine P H, Clark J, Biggar R, Hoh M, Moghissi J, Jacobs P, Wilson L, Jacobson R, et al. Human T-cell leukemia virus (HTLV-I) antibodies in Africa. Science. 1984;225:1473–1476. doi: 10.1126/science.6089348. [DOI] [PubMed] [Google Scholar]
- 55.Soldan S S, Graf M D, Waziri A, Flerlage A N, Robinson S M, Kawanishi T, Leist T P, Lehky T J, Levin M C, Jacobson S. HTLV-I/II seroindeterminate Western blot reactivity in a cohort of patients with neurological disease. J Infect Dis. 1999;180:685–694. doi: 10.1086/314923. [DOI] [PubMed] [Google Scholar]
- 56.Srivastava B I, Gonzales C, Loftus R, Fitzpatrick J E, Saxinger C W. Examination of HTLV-I ELISA-positive leukemia/lymphoma patients by western blotting gave mostly negative or indeterminate reaction. AIDS Res Hum Retroviruses. 1990;6:617–627. doi: 10.1089/aid.1990.6.617. [DOI] [PubMed] [Google Scholar]
- 57.Tuppin P, Makuwa M, Guerma T, Bazabana M M, Loukaka J C, Jeannel D, M'Pele P, de The G. Low HTLV-I/II seroprevalence in pregnant women in Congo and a geographic cluster of an HTLV-like indeterminate Western blot pattern. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;11:105–107. doi: 10.1097/00042560-199601010-00014. [DOI] [PubMed] [Google Scholar]
- 58.Verdier M, Denis F, Sangare A, Barin F, Gershy-Damet G, Rey J L, Soro B, Leonard G, Mounier M, Hugon J. Prevalence of antibody to human T cell leukemia virus type 1 (HTLV-1) in populations of Ivory Coast, West Africa. J Infect Dis. 1989;160:363–370. doi: 10.1093/infdis/160.3.363. [DOI] [PubMed] [Google Scholar]
- 59.Waziri A, Soldan S S, Graf M D, Nagle J, Jacobson S. Characterization and sequencing of prototypic human T-lymphotropic virus type 1 (HTLV-1) from an HTLV-1/2 seroindeterminate patient. J Virol. 2000;74:2178–2185. doi: 10.1128/jvi.74.5.2178-2185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagihara R, Jenkins C L, Alexander S S, Mora C A, Garruto R M. Human T lymphotropic virus type I infection in Papua New Guinea: high prevalence among the Hagahai confirmed by western analysis. J Infect Dis. 1990;162:649–654. doi: 10.1093/infdis/162.3.649. [DOI] [PubMed] [Google Scholar]