Abstract
Fluorescence-based amplified fragment length polymorphism (fbAFLP) is a novel assay based on the fluorescent analysis of an amplified subset of restriction fragments. The fbAFLP assay involves the selective PCR amplification of restriction fragments from a total digest of genomic DNA. The ligation of adapters with primer-specific sites coupled with primers containing selective nucleotides allowed the full potential of PCR to be realized while maintaining the advantages of restriction endonuclease analysis. Fluorescence-based fragment analysis with polyacrylamide gel electrophoresis provides the accurate band sizing required for homology assessment. The large number of phylogenetically informative characters obtained by fbAFLP is well suited for cluster analysis and database development. The method demonstrated excellent reproducibility and ease of performance and interpretation. We typed 30 epidemiologically well-characterized isolates of vancomycin-resistant enterococci from an outbreak in a university hospital by fbAFLP. Clustering of fbAFLP data matched epidemiological, microbiological, and pulsed-field gel electrophoresis data. This study demonstrates the unprecedented utility of fbAFLP for epidemiological investigation. Future developments in standardization and automation will set fbAFLP as the “gold standard” for molecular typing in epidemiology.
Epidemiological investigation involves the collection and analysis of both epidemiologic and microbiologic data. Several techniques have been used in medical microbiology for acquiring information on the spread of pathogenic bacteria within the hospital environment and outside in the community. Among these, molecular typing has gained popularity and is frequently used to support and/or initiate epidemiological investigation. Accurate study of epidemic bacterial isolates is aided with the determination of genetic relatedness among isolates such that both differences and similarities are found.
A universal and highly discriminatory typing system designed to track the dissemination of genomes over time and space is not best served by the confines of any one particular sequence. Consequently, “genome-scanning” techniques such as restriction endonuclease analysis (REA) have become preferred among epidemiologists. Biotechnology provides numerous restriction enzymes that allow such analyses without a priori knowledge of the genome being investigated. Macrorestriction analysis of genomic DNA followed by pulsed-field gel electrophoresis (PFGE) is built on this philosophy and has become the “gold standard” for molecular typing. However, PFGE is limited in its resolving power (5), and this contributes to difficulties with gel-to-gel and interlaboratory reproducibility (25).
Consequently, several novel methods for DNA fingerprinting of medically important bacteria have received considerable attention for their suitability in epidemiological studies. Amplification techniques have been attempted for monitoring overall genome organization with procedures such as random amplification of polymorphic DNA (RAPD). However, PCR-directed genome scanning with such low-stringency PCR approaches have several drawbacks, including reproducibility (20, 21). Despite their speed and convenience, these approaches do not offer a suitable alternative to REA-based techniques such as restriction fragment length polymorphism (RFLP) analysis or PFGE.
We present here an evaluation of fluorescence-based amplified fragment length polymorphism (fbAFLP) analysis for its potential usefulness in epidemiological investigation. The original AFLP method was developed by Vos et al. in 1995 (27), and its application to bacterial DNA fingerprinting has evolved significantly in the past few years (1–4, 6, 10, 16, 22, 24). The fbAFLP technique introduced here includes the addition of fluorescence-based PCR fragment detection with polyacrylamide gel electrophoresis (PAGE). This fluorescence-based analysis of AFLP fragments allows fbAFLP exceptionally accurate homology assessment, which is absolutely critical for database production and tree building. The utility of the fbAFLP method was evaluated with a select panel of well-characterized isolates from an outbreak of vancomycin-resistant enterococci (VRE).
MATERIALS AND METHODS
Epidemiology.
VRE first appeared at the University of Texas Medical Branch (UTMB) Hospital from a patient in the Surgery Intensive Care Unit (SICU) on 11 February 1994. Application of control measures, including weekly surveillance cultures of patients and environment, barrier isolation of colonized patients, and environmental decontamination guided by culture results, cleared VRE from the intensive care units in the UTMB Hospital in October 1997. No VRE were recovered from clinical or surveillance cultures in the Burn Intensive Care Unit (BICU) in the UTMB Hospital from February 1994 to June 1996. In June 1996, VRE first appeared in the BICU, resulting in an outbreak that was cleared from that unit in July 1997. Once cleared, this unit remained free of VRE.
Seventy-four isolates of VRE were recovered from clinical cultures and cultures of rectal swabs from patients in the BICU, human immunodeficiency virus (HIV), and SICU wards from 1994 to 1997 in the UTMB Hospital. To evaluate fbAFLP for molecular typing in hospital epidemiology, 9 of 39 VRE isolates were selected from the SICU isolated between 1994 and 1997, and 13 of 22 isolates were selected from the BICU during the outbreak in 1996. Six additional epidemiologically unrelated isolates were included from the HIV ward, a surgery floor, and Children's Hospital. Isolates from an EKG lead and the biopsy channel of a bronchoscope from the BICU were also included.
Microbiology.
Swabs taken from patients' rectums and the EKG lead were broken off into tubes of Trypticase soy broth (TSB) containing vancomycin (6 μg/ml) and ciprofloxacin (8 μg/ml). Sterile saline flushed through the biopsy channel of the bronchoscope was inoculated into TSB. Cloudy broths were subcultured onto Enterococcosel agar (Becton Dickinson, Cockeysville, Md.) containing vancomycin (6 μg/ml) and ciprofloxacin (8 μg/ml). Microorganisms which hydrolyzed esculin were subcultured onto blood agar for confirmation and further work-up. A PYR disc was used for presumptive identification of enterococci. The API20 STREP (bioMérieux, Marcy-l'Étoile, France) identification system was used for specific identification. A Synergy Quad plate (Remel, Lenexa, Kans.) was used to confirm resistance to vancomycin and to detect high-level aminoglycoside resistance with gentamicin at 500 μg/ml and streptomycin at 2,000 μg/ml. The Etest (AB Biodisk, Piscataway, N.J.) was used to determine the MICs of vancomycin, teicoplanin, and ciprofloxacin. Clinical isolates of VRE were obtained from the Clinical Microbiology Laboratory where the VRE were identified by standard methods. Briefly, the isolates were first tested with a PYR disk. If the PYR test was positive, the isolate was also tested with ampicillin and vancomycin by disk diffusion. The isolate was then identified to species using the Vitek GPI card (bioMérieux, Vitek, Inc., Hazelwood, Mo.). Isolates were stored on glass beads coated with TSB with 0.1% agar at −70°C. Beads were removed from the freezer vials and inoculated onto blood agar plates in preparation for analysis.
PFGE.
PFGE was performed using Bio-Rad's GenePath Group 1 kits. A single colony from a 24-h isolate was grown overnight in TSB under aerobic conditions at 37°C. Cells were collected and resuspended in cell suspension buffer. The suspension was mixed with lysozyme-lysostaphin and embedding agarose, and plugs were made. Plugs were placed in lysis buffer with additional lysozyme-lysostaphin and incubated for 3 h at 37°C without agitation. The plugs were washed, placed in proteinase K solution, and incubated overnight at 50°C without agitation. Plugs were washed four times with wash buffer for 30 to 60 min at room temperature on a rocker.
DNA was digested overnight with 25 U of SmaI at 25°C. DNA was separated on an agarose gel using the GenePath instrument (based on the pulsed field method of contoured clamped homogenous electric field). The run time was 20 h. S. aureus ATCC 8325, which was used to test the sample plug preparation procedure and the restriction digestion procedure, was included on each gel. Gels were stained with ethidium bromide.
The PFGE patterns were visually inspected. Isolates were considered to be different strains if their PFGE patterns differed by more than four bands (7; R. V. Goering, unpublished data). New strain types with isolates displaying ≥5 band differences were assigned a unique letter (i.e., A-, B-, etc.). Each isolate, which displayed one to four band differences from the type strain, were assigned a unique number within each group (i.e., A1, A2, etc.). In this manner, all unique PFGE profiles were assigned to unique PFGE types.
fbAFLP: genomic DNA isolation.
A total of 5 ml of brain heart infusion broth were inoculated with a single colony of a VRE isolate and incubated at 37°C overnight. Then, 1 ml of this culture was pelleted and digested with 2.5 mg of lysozyme (Sigma-Aldrich [Sigma], Oakville, Ontario, Canada) and 100 μg of lysostaphin (Sigma) per ml in 50 mM EDTA. Following incubation at 37°C for 60 min, total genomic DNA was extracted from bacterial cells using the Promega Wizard Genomic DNA Isolation kit (Fisher Scientific, Ltd. [Fisher], Nepean, Ontario, Canada), according to the manufacturer's protocol. The yield of DNA was estimated by ethidium bromide fluorescence following electrophoresis through 1.0% agarose gels.
fbAFLP: restriction and ligation.
Approximately 20 to 50 ng of genomic DNA was digested with 2 U of HindIII (New England Biolabs, Inc. [NEB], Mississauga, Ontario, Canada) and 0.4 U of MboI (Gibco-BRL [Gibco], Burlington, Ontario, Canada), along with 1 mM dithiothreitol (Gibco) for 5 h at 37°C in a total volume of 20 μl. The enzymes were then inactivated by incubating the reaction at 65°C for 20 min. Next, 5 μl of a master mix containing 2.5 pmol of HindIII adapter (Genosys Biotechnologies [Genosys], Woodlands, Tex.), 25 pmol of MboI adapter (Genosys), 0.5 U of T4 DNA ligase (Gibco), 25 pmol of ATP (Amersham Pharmacia Biotech, Inc. [Pharmacia], Baie d'Urfé, Quebec, Canada), and 1.25 μg of bovine serum albumin (NEB) was added to each restriction reaction before incubation at 22°C for 6 h. The sequences of the adapters are listed in Table 1, and their utility in AFLP have been previously described in the scientific literature (12, 27). All DNA modification incubations were performed on a PE-9600 thermal cycler (PE Applied Biosystems [PE], Mississauga, Ontario, Canada). Ligation reactions were diluted 1/5 in Optima Ultrapure water (Fisher) and then stored at −20°C for the selective amplification reaction.
TABLE 1.
Selective primer and adapter sequences for the HindIII/MboI fbAFLP
| Identifier | Sequencea |
|---|---|
| HindIII recognition site | 5′-A↓AGCTT-3′ |
| MboI recognition site | 5′-↓GATC-3′ |
| HindIII adapter | 5′-AGC TGG TAC GCA GTC TAC |
| CC ATG CGT CAG ATG CTC-5′ | |
| MboI adapter | 5′-GAT CCT CAG GAC TCA T |
| GA GTC CTG AGT AGC AG-5′ | |
| HindIII-0-FAMb | 5′-GAC TGC GTA CCA GCT T-3′ |
| MboI-AC | 5′-GAT GAG TCC TGA GGA TCA C-3′ |
| HindIII-0-HEXb | 5′-GAC TGC GTA CCA GCT T-3′ |
| MboI-CTG | 5′-GAT GAG TCC TGA GGA TCC TG-3′ |
Underlined nucleotides correspond to 5′ overhangs utilized in the ligation.
Fluorescently labeled.
fbAFLP: selective amplification.
Parallel reactions containing identical template DNAs were set up with two different selective primer sets which are differentiated with a unique fluorescent label (Table 1). The primer sets were HindIII-0-FAM with MboI-AC and HindIII-0-HEX with MboI-CTG (Genosys). PCRs were performed in 20-μl volumes containing 2.0 μl of 10× PCR Buffer II (PE), 1.5 mM MgCl2 (PE), 200 μM concentrations of each deoxynucleoside triphosphate (Pharmacia), 250 nM concentrations of either HindIII-0-FAM or HindIII-0-HEX primer, 1.5 μM concentrations of either MboI-AC or MboI-CTG primer, 0.4 U of AmpliTaq DNA polymerase (PE), 88 ng of Taqstart antibody (Clontech Laboratories, Inc., Palo Alto, Calif.), and 5 μl of the diluted ligation. The thermal cycling regime was performed on a PE-9600 cycler as follows: denaturation at 94°C for 2 min (1 cycle), followed by 12 cycles of denaturation for 20 s at 94°C, annealing for 30 s starting at 65°C with 1°C decrement/cycle, and extension for 2 min at 72°C, followed with an additional 20 cycles of denaturation for 20 s at 94°C, annealing for 30 s at 53°C, and extension for 2 min at 72°C, with a final extension at 60°C for 30 min (1 cycle), and then the reaction was held at 4°C. Selective amplification mixtures were stored until fragment analysis at −20°C.
fbAFLP: fragment analysis.
Amplified PCR products were electrophoresed through 4% polyacrylamide denaturing gels with a well-to-read distance of 36 cm for 2.75 h. Detection and size estimation of fluorescently labeled DNA fragments was facilitated by using the ABI Prism 377 DNA Sequencer (PE), along with the 377 Collection 1.1 (PE) and Genescan Analysis 3.1 software (PE). Parallel reactions, each containing a population of PCR products with a different fluorescent label, were pooled and run simultaneously with Genescan-500 TAMRA size standard (PE) in the same lane. This allowed accurate sizing up to 500 bp, as well as the ability to survey all DNA fragments generating the fbAFLP fingerprint originating from two different primer sets at the same time. After analysis of the raw data in ABI Genescan software, comparisons of electropherograms allowed identification of polymorphic bands. Each fbAFLP profile was compared in turn to a standard fbAFLP fingerprint. The fbAFLP fragments were transcribed into a data matrix, thereby providing binary documentation of the homology assessment found with electropherogram comparisons. Rows in the matrix represent individual isolates, while columns represent fragment sizes. A “1” is used to indicate the presence of a band, while a “0” is used to indicate its absence. Two matrices are generated and are specific for each primer set. Once all scoring was complete the two matrices were combined. This data manipulation was facilitated with MacClade version 3.05 (Sinauer Associates, Sunderland, Mass.).
fbAFLP: distance-based phylogenetic analysis.
Distance matrices were calculated from the data matrix by using genetic distance calculated as follows: GDxy = (Nx + Ny)/(Nx + Ny + Nxy), where GDxy is the genetic distance between isolate x and isolate y, Nx is the number of bands present in the fbAFLP fingerprint from isolate x, Ny is the number of bands present in the fbAFLP fingerprint from isolate y, and Nxy is the total number of bands present in the fbAFLP fingerprints in both isolate x and y. Tree reconstruction was done using the neighbor-joining method. Confidence in clustering was assessed by using bootstrap analysis with 100 replications. The construction of the tree based on the calculated distance matrix and bootstrap analysis was accomplished with TREECON for Windows v1.2 (26).
fbAFLP: scoring and classification.
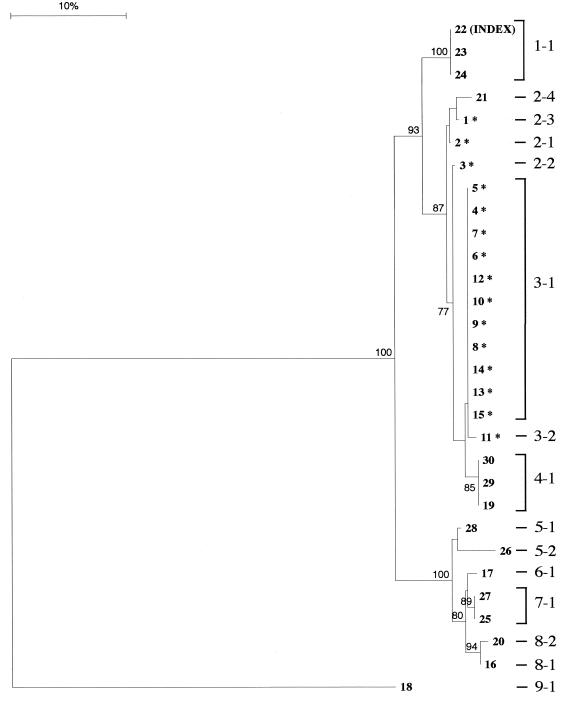
We have developed a scoring protocol that ensures accurate homology assessment and tree building (unpublished data). The protocol involves a reiterative process of fragment and phylogenetic analysis. Subsequently, each unique fbAFLP fingerprint was assigned a unique identification number (i.e., the fbAFLP ID). fbAFLP IDs consist of two parts, cluster and type, separated by a hyphen. The numbers that precede the hyphen correspond to the cluster IDs and were determined based on the clusters found within the neighbor-joining tree with bootstrap values ≥75%. (Fig. 1). Additional factors, such as branch length and the number of isolates within such clusters, were also taken into consideration for more meaningful assignment of cluster ID. The number following the hyphen is a unique identifier particular to each cluster and identifies the assigned type ID. Isolates were assigned a unique fbAFLP type ID if they displayed any polymorphism to other isolates within the cluster. The fbAFLP IDs are intended to be informative but nevertheless are only used as a guide since the phylogram (Fig. 1) displays more-definitive relationships.
FIG. 1.
Phylogram from distance-based phylogenetic analysis of fbAFLP data. The bootstrap values from 100 replicates are shown at each respective node. Bootstrap values of >75% are displayed and represent the amount of support contained in the fbAFLP data matrix for the clustering arrangements depicted in the tree. Branch lengths graphically depict the number of band differences between isolates. The scale correlates branch length to percent genetic difference. The fbAFLP ID assignment is annotated to the right of the phylogram.
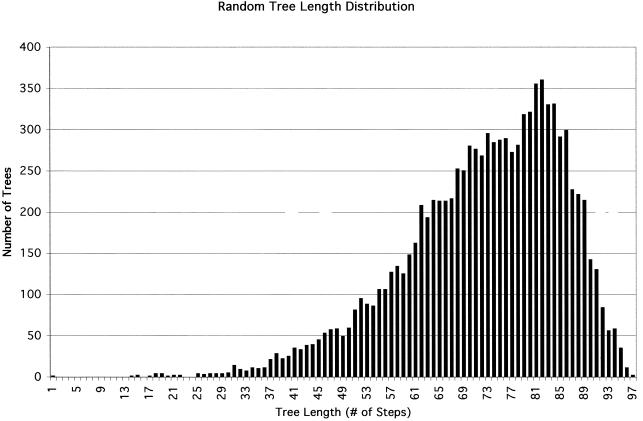
Random tree length distribution.
A graph was generated containing the tree length distribution of 10,000 randomly sampled trees from a set of all possible trees using the data matrix. This statistical test was used to measure the amount of phylogenetic signal in the data matrix and to assess its applicability to cluster analysis. The g1 statistic measures the degree of skewness and was generated by the RANDOM TREES command of the PAUP version 3.1.1 computer program (Illinois Natural History Survey, Champaign, Ill.).
RESULTS
Epidemiology.
Isolate 22 is from the index case of VRE isolated on 11 February 1994. VRE isolates from the SICU are represented with codes 22 to 30 (Table 2). Application of control measures, including weekly surveillance cultures of patients and environment, barrier isolation of colonized patients, and environmental decontamination guided by culture results, cleared VRE from the intensive care units in the UTMB hospitals in October 1997.
TABLE 2.
VRE isolates
| Code | Epidemiological data
|
Phenotype | Susceptibility datac
|
Molecular typing data
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml)
|
Gent | Str | ||||||||||
| Specimen date (mo/day/yr) | Patient locationa | Specimen | Isolateb | Van | Teic | Cip | PFGE type | fbAFLP ID | ||||
| 22 | 2/11/94 | B/SICU | Blood | C | VanA | 256 | 32 | 32 | S | R | A1 | 1-1 |
| 23 | 6/13/94 | A/SICU | Stool | S | VanA | 256 | 32 | 128 | S | R | A1 | 1-1 |
| 24 | 12/1/94 | A/SICU | Rectum | S | VanA | 256 | 32 | 64 | S | R | A1 | 1-1 |
| 2 | 11/18/96 | BICU | EKG lead | S | VanA | >256 | >256 | >32 | R | R | B5 | 2-1 |
| 3 | 11/22/96 | BICU | Rectum | S | VanA | >256 | >256 | >32 | R | R | B5 | 2-2 |
| 1 | 10/7/96 | BICU | Rectum | S | VanA | >256 | >256 | >32 | R | R | B5 | 2-3 |
| 21 | 6/16/97 | PEDI | Peritoneal fluid | C | VanA | >256 | >256 | >32 | R | R | B3 | 2-4 |
| 4 | 11/21/96 | BICU | Neck wound | C | VanA | >256 | >256 | >32 | R | R | B5 | 3-1 |
| 5 | 6/6/96 | BICU | BALd | C | VanA | >256 | 64 | >32 | R | R | B1 | 3-1 |
| 6 | 6/12/96 | BICU | Knee wound | C | VanA | >256 | >256 | >32 | R | R | B1 | 3-1 |
| 7 | 6/11/96 | BICU | Rectum | S | VanA | 256 | 64 | >32 | R | R | B1 | 3-1 |
| 8 | 6/11/96 | BICU | Rectum | S | VanA | >256 | 256 | >32 | R | R | B1 | 3-1 |
| 9 | 6/11/96 | BICU | Bronchoscope | S | VanA | >256 | >256 | >32 | R | R | B1 | 3-1 |
| 10 | 6/11/96 | BICU | Rectum | S | VanA | >256 | 256 | >32 | R | R | B2 | 3-1 |
| 12 | 7/16/96 | BICU | Rectum | S | VanA | >256 | 32 | >32 | R | R | B5 | 3-1 |
| 13 | 6/11/96 | BICU | Rectum | S | VanA | >256 | 64 | >32 | R | R | B5 | 3-1 |
| 14 | 6/26/96 | BICU | Rectum | S | VanA | >256 | 64 | >32 | R | R | B5 | 3-1 |
| 15 | 12/9/96 | BICU | Rectum | S | VanA | >256 | 128 | >32 | R | R | B3 | 3-1 |
| 11 | 6/17/96 | BICU | Wound | C | VanA | >256 | 64 | >32 | R | R | B4 | 3-2 |
| 19 | 4/13/97 | 10B | Wound | C | VanA | >256 | 256 | >32 | R | R | C1 | 4-1 |
| 29 | 2/13/97 | B/SICU | Rectum | S | VanA | >256 | 64 | >32 | R | R | C2 | 4-1 |
| 30 | 4/23/97 | B/SICU | Rectum | S | VanA | >256 | 128 | >32 | R | R | C1 | 4-1 |
| 28 | 5/20/96 | A/SICU | Urine | C | VanA | >32 | >256 | >32 | S | R | D1 | 5-1 |
| 26 | 8/14/95 | B/SICU | Rectum | S | VanB | 16 | 0.25 | >128 | S | R | D2 | 5-2 |
| 17 | 4/10/97 | CECU | Rectum | S | VanB | >256 | 1 | >32 | S | R | G | 6-1 |
| 25 | 4/4/95 | B/SICU | Blood | C | VanB | 256 | 0.25 | >128 | S | R | D1 | 7-1 |
| 27 | 2/6/96 | A/SICU | Rectum | S | VanB | 16 | 0.25 | 128 | S | R | E | 7-1 |
| 16 | 3/31/97 | PICU | Wound | C | VanA | >256 | >256 | >32 | S | R | F | 8-1 |
| 20 | 4/18/97 | 10B | Wound | S | VanA | >256 | 128 | >32 | S | R | D3 | 8-2 |
| 18 | 4/14/97 | SURG | Wound | C | VanB | >256 | 1 | >32 | R | R | H | 9-1 |
BICU, burn intensive care unit; SURG, surgery floor; 10B, HIV ward; PEDI, pediatric floor (Children's Hospital); A/SICU, A section of SICU; B/SICU, B section of SICU; CECU, chronic extended care unit (Children's Hospital); PICU, pediatric intensive care unit (Children's Hospital).
S, surveillance isolate; C, clinical isolate.
Van, vancomycin; Teic, teicoplanin; Cip, ciprofloxacin; Gent, gentamicin; Str, streptomycin; R, resistant; S, sensitive.
BAL, bronchoalveolar lavage fluid.
In June 1996, VRE first appeared in the BICU, resulting in an outbreak that was cleared from that unit in July 1997. Once cleared, this unit remained free of VRE. These outbreak isolates are represented with codes 1 to 15.
The additional VRE isolates were found in other wards of the UTMB Hospitals in 1997 and include codes 16 to 21. These isolates were somewhat sporadic, with no strong epidemiological links with the outbreak strain of the BICU. VRE isolates, with codes 16 and 17, were isolated in the first half of 1997 from two patients in the Children's Hospital. One isolate was recovered from a pediatric patient transferred from another hospital. Pediatric patients are in a separate hospital connected to the other UTMB Hospitals where all the other isolates were recovered from patients, an EKG lead, and a bronchoscope. There is no epidemiological evidence to support the exchange of any isolates between Children's Hospital and any of the other UTMB hospitals.
Microbiology.
The isolates were Enterococcus faecium of either the VanA or the VanB phenotype. All isolates recovered from the BICU were of the VanA phenotype. Isolates before June 1996 had vancomycin MICs ranging from 16 to 256 μg/ml. All but one isolate in the BICU had vancomycin MICs of >256 μg/ml. However, teicoplanin MICs were variable and ranged from 32 to >256 μg/ml. VRE isolates at the onset of the outbreak in the SICU in 1994 were susceptible to gentamicin. Isolates with high-level gentamicin resistance first appeared in the institution at the onset of the outbreak in the BICU in June 1996. Sensitivity to gentamicin was only seen outside the BICU. The susceptibility data are summarized in Table 2.
PFGE.
The molecular typing by PFGE found 15 unique profiles. These 15 PFGE profiles were categorized into 8 groups represented by A to H in Table 2. These groups contain PFGE profiles that differ by no more than four bands. Groups B, C, and D were further subdivided with four or fewer band differences.
The index case, isolate 22, was called A1 and shared an identical PFGE profile with the other VanA isolates in the SICU found in 1994. Group A profiles were never found again. Isolates of VRE recovered from patients in the SICU in 1995 and 1996 were typed into PFGE groups D and E. Group D, after May 1996, did not appear until 1997, when a similar but not identical PFGE profile appeared in the HIV ward. The appearance of VRE in the BICU of the UTMB hospitals produced PFGE profiles grouped into type B. PFGE types B1 to B5 were found exclusively in the 15 isolates from the BICU. Group B later appeared in a patient in Children's Hospital in June 1997. This isolate had the same PFGE pattern (B3) as that recovered from a patient in the BICU 6 months earlier. The remainder of the VRE isolates outside the BICU found in 1997 had PFGE profiles of C, D, F, G, or H.
fbAFLP.
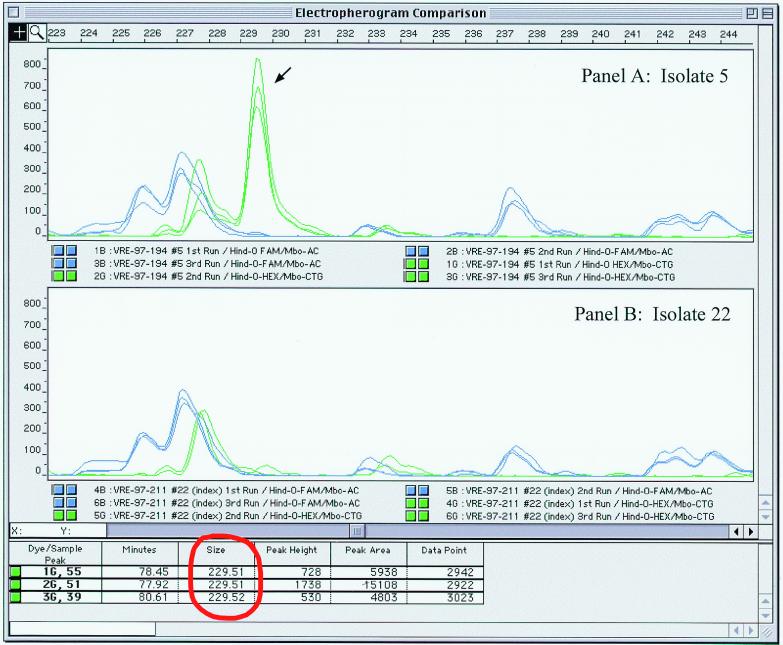
All VRE isolates were taken from pure culture to fbAFLP fingerprint on three separate occasions to assess reproducibility of the fbAFLP method. The resulting fbAFLP fingerprints from each separate run produced identical profiles. There was small variation in peak heights but this did not result in the gain or loss of any information. The reproducibility of fbAFLP banding patterns is graphically depicted with each superimposed electropherogram in Fig. 2, and the precision of band sizing for each run is displayed in the figure for one of the peaks (identified with arrow).
FIG. 2.
Screen capture of electropherogram analysis using Genescan software. Panel A displays the 233- to 243-bp portion of the profiles from isolate 5 (fbAFLP ID 3-1). Panel B contains the same size range for isolate 22 (fbAFLP ID 1-1). Each panel contains all three fbAFLP fingerprints from the same isolates superimposed. The FAM-labeled primer set is indicated with the blue electropherogram. The HEX-labeled primer set is indicated in green. The fbAFLP band sizing data for the peak indicated with the arrow is circled in red.
fbAFLP fingerprinting patterns of the 30 VRE isolates produced a total of 221 bands. Of these, 126 were from the FAM primer set using two selective nucleotides, and 95 were from the HEX primer set with three selective nucleotides. Examination of the fbAFLP patterns by electropherogram comparison resulted in the identification of 169 polymorphic bands (98 from the FAM set and 71 from the HEX set), with the remaining 52 of the 221 bands invariant for this collection of isolates. The 169 polymorphic bands subtype the 30 VRE isolates with 15 unique fbAFLP profiles.
There were 27 informative polymorphic bands (shared among two or more isolates) which contributed to clustering patterns using distance-based phylogenetic analysis shown in Fig. 1. Cluster analysis grouped the 30 isolates into 9 clusters, which correlate well with the eight PFGE groups. Groups A, C, E, F, G, and H correlated with fbAFLP cluster types 1, 4, 6, 7, 8, and 9, respectively. PFGE groups B and D were further subdivided with fbAFLP into cluster types 2 and 3 and types 5, 7, and 8, respectively. The split of these groups was substantiated with high bootstrap values. Discordant results between the two molecular typing methods include isolates grouped in fbAFLP cluster type 7-1 but separated by PFGE into groups D and E. However, both were isolated from patients in the SICU and both were of the VanB phenotype. Similarly, fbAFLP cluster type 8 was shared by single isolates from PFGE groups D and F. These isolates were recovered from patients on different units within a 3-week time period, and both had the VanA phenotype.
Figure 3 shows the results of the statistical test from the fbAFLP data matrix demonstrating the presence of strong phylogenetic signal. The g1 statistic (−0.79) indicates that the data set contains good phylogenetic signal and suggests that cluster analysis is appropriate with the fbAFLP data. Additionally, the shape of the distribution indicates that the signal-to-noise ratio is low and confirms that the homology assessment produced during fragment analysis and matrix construction was accurate.
FIG. 3.
Graph of random tree length distribution analysis on fbAFLP data.
DISCUSSION
PFGE is currently considered the gold standard for molecular typing of isolates recovered from patients and the environment in the course of investigation and control of nosocomial outbreaks (19). For this reason, the validity of the fbAFLP method for molecular typing has been based on its concordance with PFGE, epidemiological, and susceptibility data. The utility of fbAFLP for epidemiological investigation was critically evaluated with assessment of reproducibility, discriminatory power and typeability, suitability for database development and cluster analysis, and critical evaluation against PFGE.
fbAFLP validation.
The validation of the fbAFLP method for epidemiological investigation is based on a selection of 30 VRE isolates with comprehensive epidemiological, microbiological, and molecular typing data. These isolates are representative samples from a period which encompass the introduction to elimination of VRE within the intensive care units in the UTMB hospitals.
Epidemiological investigation of a VRE outbreak within the BICU of UTMB hospitals identified 15 isolates from surveillance and clinical samples. PFGE and antibiogram data identified the outbreak to be due to highly related strains of VRE (PFGE type B). Analysis of fbAFLP data correctly identified the outbreak isolates and grouped them together using cluster analysis. The outbreak strains of the BICU are indicated with an asterisk next to the isolate number in Fig. 1. These strains were classified as fbAFLP cluster types 2 and 3. Only one isolate within fbAFLP cluster 2, type 2-4 (isolate 21) was from outside the BICU. This isolate was similarly identified by PFGE to be genetically related to the outbreak strains (PFGE type B3). The genotyping data for isolate 21 is further supported by susceptibility data concordant with that of other strains in type B.
Three additional isolates that share antibiogram profiles with the outbreak strains are isolates 19, 29, and 30. Both molecular typing methods grouped these isolates together as PFGE type C and fbAFLP ID 4-1. The phylogenetic analysis of fbAFLP data clustered these isolates with fbAFLP cluster types 2 and 3. The clustering of these isolates (fbAFLP types 2, 3, and 4) onto one branch within the phylogram separate all but one of the high-level gentamicin-resistant isolates from the sensitive isolates. This cluster is supported with a bootstrap value of 87%. The only other gentamicin-resistant isolate (isolate 18, fbAFLP type 9-1) shows 135 band differences from other fbAFLP profiles. PFGE also found marked differences, and it identified the only isolate classified as H. The fit of fbAFLP data presented in the phylogram to both PFGE and antibiogram data is virtually perfect. Clustering not only identified the outbreak strains but also showed their relationship to other isolates within the hospital. Such analysis will provide a new level of investigation as these data become available during hospital outbreaks.
fbAFLP reproducibility.
Although the reproducibility of the AFLP technique is reported to be very high (12, 27), we independently validated fbAFLP's reproducibility. Three independent runs from each isolate produced 90 profiles without aberration. Slight variations in peak height had no effect on the numbers or sizes of the PCR products. This result concurs with success we have experienced with gel-to-gel reproducibility. We routinely include positive controls with each fbAFLP batch and have produced hundreds of identical fbAFLP profiles over the years (data not shown). Our optimism for good interlaboratory reproducibility with fbAFLP is based on the accurate homology assessment possible with fbAFLP. We have demonstrated that the accuracy in band sizing, number, and characteristics of fbAFLP bands, coupled with databasing through matrix construction, supersede problems leading to inconsistent results between gels. The logic follows that the gel-to-gel reproducibility of fbAFLP should lead to lab-to-lab reproducibility. It appears from our experience and other published works (1, 2, 4, 6, 22, 24) that the gel-to-gel reproducibility is unquestionable. However, future studies are needed for interlaboratory comparison. Standardization of enzyme-primer combinations will make fbAFLP as effective for interlaboratory comparison as DNA sequence data is today.
fbAFLP discriminatory power and typeability.
Both PFGE and fbAFLP subtyped the 30 VRE isolates into 15 distinct profiles. However, we feel that this may be somewhat biased toward PFGE since the number of isolates included in this study is limited and partially selected based on PFGE profiles. Previous works have demonstrated that the discriminatory power of AFLP is considerably higher than that of PFGE (4) and is far superior to DNA-DNA hybridization or cellular fatty acid analysis (12). To date, the published body of work on AFLP indicates that it has excellent typeability (1–4, 6, 10–16, 22, 24). To that extent, AFLP has been successfully applied to numerous plants, animals, and bacteria. AFLP with its numerous restriction enzyme and selective primer combinations provides unprecedented typeability which is not limited by the complexity nor the origin of DNAs.
fbAFLP suitability for database development and cluster analysis.
An often overlooked but essential criterion for microbial typing is the ability to compare fingerprints over space and time. The fbAFLP method presented here with its fluorescence-based fragment analysis and PAGE provides accurate band sizing, which is absolutely necessary for proper homology assessment. Accurate homology assessment is essential for reliable database development and cluster analysis.
In order to proceed with an appropriate cluster analysis of a particular data set, phylogenetic signal needs to be distinguished from random noise (8). Several investigators mistakenly cite confidence in their molecular data and generated trees without first establishing the phylogenetic signal (18). This becomes very important when homology assessment is questionable such as when one cannot be certain two bands are truly the same or different. This problem is exacerbated with highly divergent strains because through sheer chance two highly variable profiles may share band sizes which are not necessarily homologous bands. Falsely identified homologous bands would mistakenly depict genetic relatedness. fbAFLP is less susceptible to misalignment or miscalls in homology since it produces a large number of bands with a large portion being absolutely conserved, along with very precise band sizing. The high phylogenetic signal measured by the random tree length distribution proves that fbAFLP produced accurate homology assessment and consequently reliable database development and cluster analysis. The use of differently labeled primers for each pair aid in this respect as well. Certainly, several electrophoresis systems will provide accurate sizing capabilities for the conversion of raw data into binary form (15). Nevertheless, several features of fbAFLP are not lost on laboratories without this equipment (1–3, 6, 10–14, 16, 22, 24).
Most of the currently available molecular typing methods, such as PFGE, are comparative only (23). Comparative methods will only allow delineation of isolates closely related from those markedly different in genomic background. However, it is often not sufficient to simply differentiate isolates as same or different. The monitoring of clonal spread and prevalence in populations over extended periods of time requires library typing systems such as fbAFLP with its utility in database production and cluster analysis. This is important for epidemiological studies, where there is a considerable need to understand the genetic relatedness among isolates in outbreak investigations.
It has been argued that such resolution is not required for outbreak investigation (23). However, this study demonstrates that the benefits of good homology assessment and cluster analysis are not wasted. Furthermore, cutoffs based on the number of band differences between isolates do not provide a sound approach to true phylogenetic investigation. It is time epidemiological investigation with molecular typing took a chapter from the phylogeneticist's rulebook. It would be biologically inaccurate to establish the phylogeny of a species on a preset number of base substitutions in 16S ribosomal DNA sequences. For years, phylogenies have been based on cluster analysis of well-aligned sequences with confidence measurements using bootstrap values. This rationale works equally well in epidemiology, as demonstrated here.
Finally, the addition of fluorescence-based analysis to AFLP makes fbAFLP analysis very amenable to automation. Electropherogram comparison, data matrix construction, and phylogenetic analysis are all very suited to automated computer analysis since they have mathematical and objective rules, which can be converted into algorithms. The power of developing databases with fbAFLP will be extended as software engineering works toward the incorporation of these algorithms into a unified software package.
PFGE versus fbAFLP.
Restriction enzyme analysis of whole genomic DNA is a very appropriate and biologically sound approach for molecular typing. Molecular typing by both PFGE and fbAFLP are built on this fundamental technique of whole genomic fingerprinting using the discriminatory power of restriction enzymes. Neither method utilizes all of the information from the restriction digest. PFGE cannot reliably resolve all fragments, and fbAFLP amplifies a subset of the restriction fragments with its selective nucleotide principle. Nuances to each method emerge with the actual restriction enzymes used and approach to detection of the restriction fragments.
PFGE utilizes a limited number of extremely rare cutters such as SmaI and requires microgram quantities of unsheared DNA. fbAFLP, on the other hand, requires as little as 20 ng of DNA, and its unconstrained combination of endonucleases (rare and frequent cutter pairs) is much more forgiving with sheared DNA since the resulting restriction fragments are generally <500 bp and are not detected directly. This flexibility of combinations of restriction enzymes permits extended utility of fbAFLP for DNA fingerprinting and addresses organism-specific issues such as GC content. The quantity and quality of DNA needed for fbAFLP permit typing of autolysogenic and difficult-to-culture microorganisms (2). Furthermore, each stage of fbAFLP is amenable to automation, and results could potentially be obtained quicker with fbAFLP than with PFGE in the near future.
Significant differences between the methodologies are apparent with gel electrophoresis. PFGE is typically done with agarose using a contour-clamped homogeneous electric field, which allows the physical separation of very large DNA molecules. Both the sieving properties of agarose and the imprecision of band sizing with external size standards limit the resolving power of PFGE (5). However, the fbAFLP method with PAGE, internal size standards, calibration curves, and equal well-to-read distances for all fragments provided by ABI 377 DNA Sequencer and Genescan software precisely size all fragments within ±1 bp of actual length (17).
The methods also significantly differ with the number of fragments generated within each DNA fingerprint. PFGE profiles typically contain 8 to 20 sizable bands per isolate, whereas the fbAFLP characters are only limited by the number of selective nucleotides and primer sets used. The restriction-primer system chosen here produced fbAFLP fingerprints with over 180 sizable bands per VRE isolate.
The advantage fbAFLP exhibits for total fragment numbers and precise sizing takes fbAFLP beyond molecular typing into library typing. The phylogenetically informative characters produced by fbAFLP facilitate computer storage, database development, and cluster analysis. There is no need with fbAFLP databases to rerun any of the previous profiles on the same gel, which is often necessary for other methods such as PFGE. Unquestionably, as the numbers of different DNA fingerprints increase beyond the loading capacity of runs, within-gel comparisons are simply not feasible. This stresses the importance of accurate band sizing and data matrix construction.
Conclusion.
This study shows similar power of discrimination between the present gold-standard PFGE and a newer method, fbAFLP, in the analysis of a nosocomial VRE outbreak. The fbAFLP typing method is a robust technique for displaying large numbers of DNA polymorphisms that are very appropriate for epidemiological surveillance. AFLP's potential typeability of all microorganisms becomes exceptionally attractive, as more labs seek this sort of flexibility. The addition of fluorescence-based fragment analysis with PAGE extends the utility of fbAFLP to reliable database development and cluster analyses. With more laboratories fingerprinting organisms, it is important to have the capability of comparing organisms from the past with those analyzed at other laboratories. Although the fbAFLP technique may appear to be complex, we found it to be fast and reproducible. Standardization and automation of both chemistry and analysis will make fbAFLP databases a powerful tool for monitoring the nosocomial and community spread of bacterial pathogens.
ACKNOWLEDGMENTS
We thank Kathy Keller and Olga Hill for their dedication on the fbAFLP bench.
REFERENCES
- 1.Aarts H J M, van Lith L A J T, Keijer J. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett Appl Microbiol. 1998;26:131–135. doi: 10.1046/j.1472-765x.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- 2.Boumedine K S, Rodolakis A. AFLP allows the identification of genomic markers of ruminant Chlamydia psittaci strains useful for typing and epidemiological studies. Res Microbiol. 1998;149:735–744. doi: 10.1016/s0923-2508(99)80020-5. [DOI] [PubMed] [Google Scholar]
- 3.Clerc A, Manceau C, Nesme X. Comparison of randomly amplified polymorphic DNA with amplified fragment length polymorphism to access genetic diversity and genetic relatedness within genospecies III of Pseudomonas syringae. Appl Environ Microbiol. 1998;64:1180–1187. doi: 10.1128/aem.64.4.1180-1187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerner-Smidt P, Graves L M, Hunter S, Swaninathan B. Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages. J Clin Microbiol. 1998;36:1318–1323. doi: 10.1128/jcm.36.5.1318-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson J R, Slater E, Xerry J, Tompkins D S, Owen R J. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J Clin Microbiol. 1998;36:2580–2585. doi: 10.1128/jcm.36.9.2580-2585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goering R V. The molecular epidemiology of nosocomial infection: an overview of principles, application, and interpretation. In: Specter S, Bendinelli M, Friedman H, editors. Rapid detection of infectious agents. New York, N.Y: Plenum Publishing Co.; 1998. pp. 140–142. [Google Scholar]
- 8.Hillis D M, Huelsenbeck J P. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- 9.Huelsenbeck J P. Tree length distribution skewness: an indicator of phylogenetic information. Syst Zool. 1991;40:257–270. [Google Scholar]
- 10.Huys G, Kämpfer P, Altwegg M, Kersters I, Lamb A, Coopman R, Lüthy-Hottenstein J, Vancanneyt M, Janssen P, Kersters K. Aeromonas popoffii sp. nov., a mesophilic bacterium isolated from drinking water production plants and reservoirs. Int J Syst Bacteriol. 1997;47:1165–1171. doi: 10.1099/00207713-47-4-1165. [DOI] [PubMed] [Google Scholar]
- 11.Janssen P, Maquelin K, Coopman R, Tjernberg I, Bouvet P, Kersters K, Dijkshoorn L. Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol. 1997;47:1179–1187. doi: 10.1099/00207713-47-4-1179. [DOI] [PubMed] [Google Scholar]
- 12.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 13.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koeleman J G M, Parlevliet G A, Dijkshoorn L, Savelkoul P H M, Vandenbroucke-Grauls C M J E. Nosocomial outbreak of multi-resistant Acinetobacter baumannii on a surgical ward: epidemiological and risk factors for acquisition. J Hosp Infect. 1997;37:113–123. doi: 10.1016/s0195-6701(97)90181-x. [DOI] [PubMed] [Google Scholar]
- 15.Koeleman J G M, Stoof J, Biesmans D J, Savelkoul P H M, Vandenbroucke-Grauls C M J E. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998;36:2522–2529. doi: 10.1128/jcm.36.9.2522-2529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J-J, Kuo J, Ma J. A PCR-based DNA fingerprinting technique: AFLP for molecular typing of bacteria. Nucleic Acids Res. 1996;24:3649–3650. doi: 10.1093/nar/24.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayrand P E, Corcoran K P, Ziegle J S, Robertson J M, Hoff L B, Kronick M N. The use of fluorescence detection and internal lane standards to size PCR products automatically. Appl Theor Electrophor. 1992;3:1–11. [PubMed] [Google Scholar]
- 18.Mishler B D. Cladistic analysis of molecular and morphological data. Am J Phys Anthropol. 1994;94:143–156. doi: 10.1002/ajpa.1330940111. [DOI] [PubMed] [Google Scholar]
- 19.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penner G A, Bush A, Wise R, Domier L, Kasha K, Laroche A, Scoles G, Molnar S J, Fedak G. Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. PCR Methods Appl. 1993;2:341–345. doi: 10.1101/gr.2.4.341. [DOI] [PubMed] [Google Scholar]
- 21.Power E G M. RAPD typing in microbiology; a technical review. J Hosp Infect. 1996;34:247–265. doi: 10.1016/s0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- 22.Sloos J H, Janssen P, van Boven C P A, Dijkshoorn L. AFLP typing of Staphylococcus epidermidis in multiple sequential cultures. Res Microbiol. 1998;149:221–228. doi: 10.1016/s0923-2508(98)80082-x. [DOI] [PubMed] [Google Scholar]
- 23.Struelensm M J, De Gheldre Y, Deplano A. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect Control Hosp Epidemiol. 1998;19:565–569. doi: 10.1086/647874. [DOI] [PubMed] [Google Scholar]
- 24.Valsangiacomo C, Baggi F, Gaia V, Balmelli T, Peduzzi R, Piffaretti J-C. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J Clin Microbiol. 1995;33:1716–1719. doi: 10.1128/jcm.33.7.1716-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Belkum A, van Leeuwen W, Kaufmann M E, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O'Brein F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J Clin Microbiol. 1998;36:1653–1659. doi: 10.1128/jcm.36.6.1653-1659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 27.Vos P, Hogers R, Bleeker M, Renijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuliper M, Zabaeu M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]