ABSTRACT
The emergence of SARS-CoV-2 created a crucial need for serology assays to detect anti-SARS-CoV-2 antibodies, which led to many serology assays entering the market. A trans-government collaboration was created in April 2020 to independently evaluate the performance of commercial SARS-CoV-2 serology assays and help inform U.S. Food and Drug Administration (FDA) regulatory decisions. To assess assay performance, three evaluation panels with similar antibody titer distributions were assembled. Each panel consisted of 110 samples with positive (n = 30) serum samples with a wide range of anti-SARS-CoV-2 antibody titers and negative (n = 80) plasma and/or serum samples that were collected before the start of the COVID-19 pandemic. Each sample was characterized for anti-SARS-CoV-2 antibodies against the spike protein using enzyme-linked immunosorbent assays (ELISA). Samples were selected for the panel when there was agreement on seropositivity by laboratories at National Cancer Institute’s Frederick National Laboratory for Cancer Research (NCI-FNLCR) and Centers for Disease Control and Prevention (CDC). The sensitivity and specificity of each assay were assessed to determine Emergency Use Authorization (EUA) suitability. As of January 8, 2021, results from 91 evaluations were made publicly available (https://open.fda.gov/apis/device/covid19serology/, and https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/serology-test-evaluation.html). Sensitivity ranged from 27% to 100% for IgG (n = 81), from 10% to 100% for IgM (n = 74), and from 73% to 100% for total or pan-immunoglobulins (n = 5). The combined specificity ranged from 58% to 100% (n = 91). Approximately one-third (n = 27) of the assays evaluated are now authorized by FDA for emergency use. This collaboration established a framework for assay performance evaluation that could be used for future outbreaks and could serve as a model for other technologies.
IMPORTANCE The SARS-CoV-2 pandemic created a crucial need for accurate serology assays to evaluate seroprevalence and antiviral immune responses. The initial flood of serology assays entering the market with inadequate performance emphasized the need for independent evaluation of commercial SARS-CoV-2 antibody assays using performance evaluation panels to determine suitability for use under EUA. Through a government-wide collaborative network, 91 commercial SARS-CoV-2 serology assay evaluations were performed. Three evaluation panels with similar overall antibody titer distributions were assembled to evaluate performance. Nearly one-third of the assays evaluated met acceptable performance recommendations, and two assays had EUAs revoked and were removed from the U.S. market based on inadequate performance. Data for all serology assays evaluated are available at the FDA and CDC websites (https://open.fda.gov/apis/device/covid19serology/, and https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/serology-test-evaluation.html).
KEYWORDS: Emergency Use Authorization, SAR-CoV-2, serology assay, evaluation panel
INTRODUCTION
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes coronavirus disease 2019 (COVID-19) triggered a global pandemic responsible for more than 222 million confirmed infections and 4.6 million deaths between December 2019 and September 2021 (1). In addition to the need for diagnostic assays to detect acute SARS-CoV-2 infection, there was also an urgent need for the development of accurate and reliable serology assays to identify individuals with recent or prior SARS-CoV-2 infections, for seroprevalence studies. Serology assays detect the presence of virus-specific antibodies in blood samples and are critical for population-level surveillance of past infection.
On February 4, 2020, the Secretary of the Department of Health and Human Services (HHS) declared that circumstances existed justifying the authorization of emergency use of in vitro diagnostics for the detection and/or diagnosis of COVID-19 (2). To expedite access to serology assays, the U.S. Food and Drug Administration (FDA) published guidance in March 2020 that included policies where the FDA did not intend to object to serology assay developers marketing their assay without an Emergency Use Authorization (EUA), provided the assay was validated, the FDA was notified of the developer’s intent to market, and the assay’s reports included certain limitations (3). Concerns emerged in some cases regarding inadequate performance, and, in some cases, false claims regarding FDA approval (4, 5). FDA issued a safety communication in April 2020 to provide information about the appropriate use of serology assays for COVID-19 (5). Guidance was then updated in May 2020 to revise the policies with the expectation that serology assay developers submit an EUA request that included documentation of their validation data within a given time frame of their notification to help ensure that the assay’s performance claims had been validated and that it demonstrated adequate performance (6).
Meanwhile, it became clear that a unified, independent federal government effort to consistently evaluate serology assays would be in the public interest and would support the pandemic response. Even when assay developers attempted rigorous validation, access to well-characterized antibody positive and negative samples in the early stages of the pandemic was limited, and this restricted their ability to adequately assess assay sensitivity and specificity. Therefore, in April 2020, a collaborative effort among the FDA, National Cancer Institute (NCI), Centers for Disease Control and Prevention (CDC), National Institute of Allergy and Infectious Diseases (NIAID), National Cancer Institute’s Frederick National Laboratory for Cancer Research (NCI-FNLCR), National Institutes of Health (NIH) Clinical Center, and Biomedical Advanced Research and Development Authority (BARDA), was established to address this identified need. The collaborative program independently evaluated the performance of serology assays using well-characterized panels consisting of serum samples collected from patients infected with SARS-CoV-2 and negative plasma and serum samples collected before the pandemic, which included samples from 10 individuals living with HIV infection. All panel samples were characterized for SARS-CoV-2 seropositivity for total or pan-immunoglobulins (IgG/IgM/IgA) against the spike protein. The seropositive samples were then further characterized for anti-spike IgM and IgG at both the CDC and NCI-FNLCR. Receptor binding domain (RBD) IgG seropositivity was also assessed at NCI-FNLCR.
Here, we report the aggregated study data for 91 SARS-CoV-2 serology assay evaluations, including lateral flow assays, ELISAs, and three automated chemiluminescent immunoassays (CIA). Assay performance was assessed for positive percent agreement (PPA) and negative percent agreement (NPA) using confirmed positive and negative blood samples (serum and plasma), to provide estimates of sensitivity and specificity, respectively. This effort aided the U.S. government’s response to the COVID-19 pandemic by providing rigorous and consistent performance evaluations of SARS-CoV-2 serology assays to help inform FDA regulatory decisions and ensure that assays in the marketplace had acceptable performance (4).
RESULTS
Performance evaluation panel development and characterization.
Three different evaluation panels were assembled using serum and plasma from individuals with a confirmed prior SARS-CoV-2 infection (positives) and from individuals whose samples were collected before December 2019 (negatives) before the COVID-19 pandemic. Several evaluation panels were needed because of the limited sample volumes and aliquots available from each donor and the number of serology assays that needed to be evaluated. Positive samples had a wide range of SARS-CoV-2 spike-specific IgG and IgM antibody titers (Table 1; titers 100 to 6400 or higher). Each panel had similar overall antibody titer composition for IgG and IgM and were matched on a sample-by-sample basis whenever possible. The median number of days post-symptom onset in the panels were 23, 26, and 27 days. The range of days post-symptom onset for each panel was also comparable with 17 being the minimum number of days and 46 the maximum. All negative samples tested negative for SARS-CoV-2 specific antibodies at CDC.
TABLE 1.
Anti-SARS-CoV-2 spike antibody titers for IgG and IgM in the positive samples from three performance evaluation panels
| Antibody titer | IgG |
IgM |
||||
|---|---|---|---|---|---|---|
| Panel 1 | Panel 2 | Panel 3 | Panel 1 | Panel 2 | Panel 3 | |
| 100 | 1 | 0 | 0 | 13 | 12 | 11 |
| 400 | 7 | 6 | 7 | 11 | 11 | 12 |
| 1600 | 12 | 12 | 11 | 6 | 6 | 6 |
| ≥6400 | 10 | 12 | 12 | 0 | 1 | 1 |
| Samplesa (n) | 30 | 30 | 30 | 30 | 30 | 30 |
Samples were tested for anti-SARS-CoV-2 IgG and IgM using CDC ELISA against the spike protein.
Serology assay performance evaluations.
In the current study, 91 serology assay evaluations were performed at either the NCI-FNLCR, NIH-Clinical Center, or CDC. The program focused on lateral flow (n = 78) and ELISA (n = 10) evaluations. In addition, a few automated CIAs (n = 3) were evaluated when the platforms were available at CDC or NIH (Table 2). The target antigens evaluated were summarized in Table 2 and were as follows: spike protein (n = 17), nucleocapsid (n = 5), spike and nucleocapsid (n = 8), or unknown target antigen (n = 61). The number of evaluations per panel were as follows: panel 1 (n = 23), panel 2 (n = 54), and panel 3 (n = 14) (Table S1).
TABLE 2.
SARS-CoV-2 serology assay evaluations grouped by assay type and target antigen
| Evaluationsa | Lateral flow | ELISA | CIAb |
|---|---|---|---|
| Antibody Type | |||
| IgG | 75 | 4 | 2 |
| IgM | 72 | 2 | 0 |
| Pan-Ig | 1 | 3 | 1 |
| IgM/IgA | 1 | 0 | 0 |
| IgM/IgG | 2 | 0 | 0 |
| IgA | 0 | 1 | 0 |
| Target antigens | |||
| Spike | 11 | 5 | 1 |
| Nucleocapsid | 3 | 0 | 2 |
| Spike and nucleocapsid | 7 | 1 | 0 |
| Unknown | 57 | 4 | 0 |
91 total evaluations. Four assays (Zhuhai Livzon Diagnostics Inc. Livzon IgM/IgG Diagnostic kit for IgM/IgG Antibody to Coronavirus (SARS-Cov-2) Lateral Flow; BTNX Inc. COVID-19 IgG/IgM Test Cassettes; Nirmidas Biotech, Inc. COVID-19 (SARS-CoV-2) IgM/IgG Antibody Detection kit; and Jiangsu Well Biotech Co., Ltd. COVID-19 IgM/IgG Rapid Test) were evaluated twice on two separate panels with either the same lot number of assays (n = 3) or a different lot number of the assay (n = 1). These assays were evaluated twice because they represented cases where PPA or NPA were below the recommended performance threshold.
Automated chemiluminescent immunoassays (CIA) is a general term used to describe a variety of automated clinical immunoassays, such as Abbott, Architect i1000 SARS-CoV-2 IgG; Roche Diagnostic Corporation, Elecsys Anti-SARS-CoV-2; and Ortho-Clinical Diagnostics, Inc., VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack.
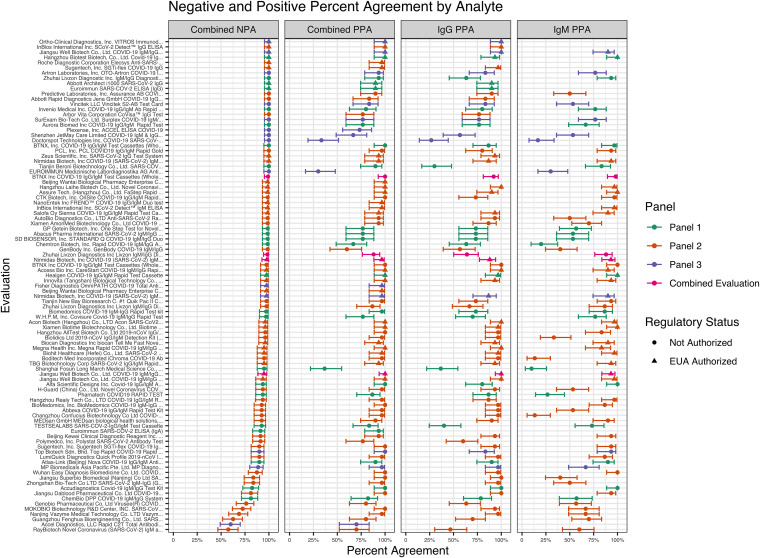
Sensitivity (PPA) and specificity (NPA) estimates for each of the serology assays evaluated for IgG, IgM, or combined analyses are shown in Fig. 1. The combined analysis considered a positive result for any antibody type that was detected positive, and a negative result if all antibodies were detected negative. The assays evaluated showed a wide range of assay sensitivity and specificity for IgG, IgM, and combined detection. Sensitivity ranged from 27% (95% CI: 14%, 44%) to 100% (95% CI: 89%, 100%) for IgG and 10% (95% CI: 4%, 26%) to 100% (95% CI: 89%, 100%) for IgM. Combined sensitivity ranged from 30% (95% CI: 17%, 48%) to 100% (95% CI: 89%, 100%). Combined specificity ranged from 58% (95% CI: 47%, 68%) to 100% (95% CI: 95%, 100%). For the five assays that measured pan-Ig, sensitivity ranged from 73% (95% CI: 56%, 86%) to 100% (95% CI: 89%, 100%), and specificity ranged from 98% (95% CI: 91%, 99%) to 100% (95% CI: 95%, 100%).
FIG 1.
Overall performance characteristics of the 91 evaluations. Combined evaluations were included for four assays that were tested twice. The positive percent agreement (PPA, sensitivity) for each evaluation is presented based on the antibody isotype measured (combined, IgG, and IgM) as well as the combined negative percent agreement (NPA, specificity) for each evaluation. The combined PPA considered a positive result for any antibody type that was detected positive, and a combined NPA considered a negative result if all antibodies tested were negative. The panel (green, panel 1; orange, panel 2; purple, panel 3; pink, combined evaluation) used for the evaluation as well as regulatory status (triangle for EUA Authorized, circle for Not Authorized) is also indicated for each evaluation. Assays were granted EUA, based on results from this independent evaluation as well as other information submitted to FDA for review.
In total, 27 assays that were ultimately issued an EUA demonstrated performance that met the FDA’s recommendations for clinical performance for SARS-CoV-2 serology assays (recommendations for sensitivity [combined PPA ≥90% for IgM/IgG, PPA ≥90% for pan-Ig, PPA for IgM ≥70%, and/or PPA for IgG ≥90%] and specificity [combined NPA ≥93% with a lower bound of the 95% confidence interval greater than 87.8%]) as described in the FDA’s serology EUA template for assay developers (https://www.fda.gov/media/137698/download). Of note, two assays were ultimately issued an EUA that were evaluated twice for a “combined evaluation” as described in Text S1 and shown in Fig. 1. Both assays had a lower bound of the 95% confidence interval for PPA or NPA that was considered acceptable (PPA ≥ 87% with a lower bound of the 95% confidence interval greater than 74.4% for IgG and pan-Ig, and NPA ≥ 93% with a lower bound of the 95% confidence interval greater than 74.4%) with the larger number of unique samples assessed in the combined evaluations (7).
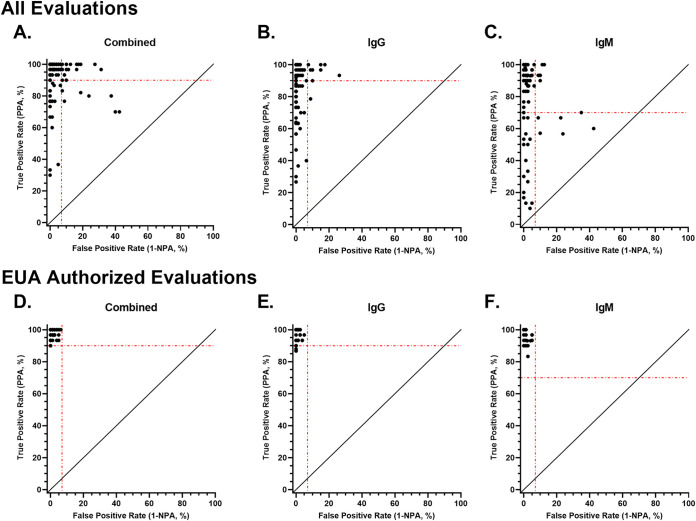
Sensitivity and specificity estimates were assessed together for all assays evaluated (Fig. 2A to C) or only the EUA authorized assays (Fig. 2D to F). The evaluations were examined for combined performance (Fig. 2A), IgG performance (Fig. 2B), and IgM performance (Fig. 2C), if relevant to the antibody isotype detected in each assay. Most of the evaluations were above the recommended sensitivity threshold (90%) or the specificity threshold (7%, 1-NPA, with a lower bound of the 95% confidence interval greater than 87.8%) (Fig. 2A). However, for combined performance, 8 of the evaluations were below both the sensitivity and specificity threshold. A similar trend was noted for the IgG performance and IgM performance assessment, with 1 and 5 evaluations, respectively, falling below the recommended performance thresholds for each antibody isotype. As for the EUA authorized assays, none of the evaluations fell below both the recommended sensitivity and specificity thresholds, whether assessing the combined, IgG, or IgM performance, indicating the EUA authorized assays demonstrated overall strong performance. Of note, six performance evaluations were conducted post-EUA. The EUAs for two of these assays were later revoked due to inadequate performance.
FIG 2.
True positive rate (PPA) compared with false-positive rate (1 - specificity). True positive rate (positive percent agreement [PPA], sensitivity) and false-positive rate (1 - negative percent agreement [NPA], specificity) were determined for all the evaluations or EUA authorized evaluations, respectively, for combined (A and D), IgG (B and E), and IgM (C and F) performance. The dashed lines on each graph represent the performance thresholds for the true positive rate (90% for combined and IgG, and 70% for IgM) and false-positive rate (7% for combined, IgG, and IgM).
Four assays were evaluated twice across two panels for combined analysis with similar results for both panels. For two of the assays (BTNX Inc. and Jiangsu Well Biotech Co, Ltd.), the combined sensitivity was the same for both evaluations (100%); and the combined specificity ranged from 98% to 100% for BTNX Inc. while the second assay ranged from 94% to 100% (Jiangsu Well Biotech Co, Ltd.). For the other two assays (Nirmidas Biotech, Inc. and Zhuhai Livzon Diagnostics Inc.), the combined sensitivity ranged from 93% to 97% for Nirmidas Biotech, Inc, while the other assay ranged from 87% to 93% (Zhuhai Livzon Diagnostics Inc.); the combined specificity for both assays ranged from 98% to 100%.
Sensitivity and specificity performance estimates of the assays were grouped and compared based on the panel used for testing (Fig. S1) and assay type (Fig. S2). The combined sensitivity measures of the assays were lower in panel 1 and panel 3 compared to panel 2 (P = 0.002 and P = 0.033, respectively), and the IgG sensitivity measures of the assays were lower in panel 1 compared to panel 2 (P < 0.001) as shown in Fig. S1A. The IgM sensitivity measures were not significantly different across panels. The combined IgG and IgM specificity measures were consistently higher for panel 3 compared to panel 2 (combined, P = 0.02; IgG, P = 0.007; IgM, P = 0.011) as shown in Fig. S1B. However, there are limited data to formally evaluate overall assay performance across evaluation panels because only four assays were tested with more than one panel. The combined IgG and IgM sensitivity measures of the assays were not significantly different between the different assay types as indicated in Fig. S2A. The combined and IgG specificity estimates were lower for lateral flow assays compared to ELISAs (combined, P = 0.012; IgG, P = 0.036), and a significant decrease in combined specificity estimates were observed for lateral flow assays compared to CIA assays (P = 0.022; shown in Fig. S2B).
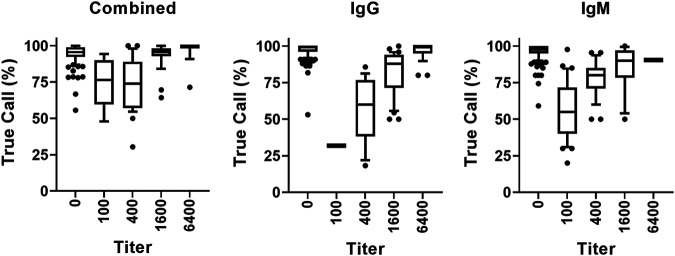
Additionally, to determine if lower assay sensitivity estimates were associated with an inability of the assays to detect lower titer level samples, samples were examined across the titer range for the true call percentage, defined as the percentage of evaluations that accurately identified positive samples from patients that were previously characterized as positive by a prior nucleic acid amplification test and IgM and IgG antibody testing. As shown in Fig. 3, similar results were observed across each analyte (combined, IgG, and IgM), where the true call percentage was the lowest for the samples with lower titers and increased as the titer range increased to the highest titer (6400).
FIG 3.
True call percentage by analyte and titer. For each analyte (combined, IgG, and IgM), the percentage of evaluations that accurately determined a SARS-CoV-2 seropositive sample as being positive regardless of titer (100, 400, 1600, and 6400), and the percentage of evaluations that accurately determined a SARS-CoV-2 seronegative sample (titer = 0) as being negative are illustrated. The bar graphs indicate the median, 25% to 75% range of true call percentages for each titer group value, and the vertical lines represent the 10% to 90% range.
DISCUSSION
Data from the individual serology assay evaluations performed were first made publicly available on May 4, 2020 and have been periodically updated (8). This effort helped ensure that the serology assays that were available in the United States market during the COVID-19 public health emergency demonstrated acceptable performance for detecting anti-SARS-CoV-2 antibodies. Assays with acceptable performance are crucial to determine the percentage of a population that has anti-SARS-CoV-2 antibodies (seroprevalence).
By late April 2020, the market was flooded with serology assays, and mispromotion, misuse, and reports of poor-performing, unauthorized serology assays were increasing (9, 10). Under FDA’s initial March 16, 2020 policy intended to facilitate early access to serology assays for laboratories and health care providers, many SARS-CoV-2 serology assays came to market without regulatory review. Other factors may also have driven unauthorized serology tests, including those with inadequate performance and those with misleading claims, to flood the market. This trans-governmental network was established to independently evaluate the performance of serology assays developed by commercial manufacturers and became more valuable when concerns with poorly performing assays emerged. These independent evaluations also helped support FDA’s regulatory decision-making during the public health emergency, particularly after the policy was updated on May 4, 2020 and more developers began submitting EUA requests for serology assays. More than half of the assays evaluated did not meet the criteria for issuing an EUA. The EUAs for two assays evaluated as part of this program were revoked and were removed from the U.S. market based in part on the inadequate assay performance observed in this study.
FDA considered the totality of scientific information available in its regulatory decision-making, and some assays were not authorized where the recommended performance was met. There are many reasons why assays were not authorized. For example, in some cases, the developer did not conduct additional necessary validation studies (7). Other assays may not have been authorized because assay developers subsequently chose not to pursue authorization for the U.S. market.
Most EUA authorized assays measure antibodies to the viral nucleocapsid and/or spike protein. Now that a progressively larger number of people have been receiving COVID-19 vaccines (11–13), it is important to recognize that the vaccines approved or authorized for use in the United States are designed to generate protective immune responses to the spike protein. Therefore, in the COVID-19 vaccine era, serology results should be interpreted carefully; antibodies to the nucleocapsid protein can indicate recent or past infection, while those against the spike protein may represent an immune response to prior infection and/or to vaccination. However, the currently EUA authorized SARS-CoV-2 serology assays are not authorized to assess the immune response to COVID-19 vaccination, and more research is needed in vaccinated individuals (14). Some of the limitations of this study were access to samples to reach the target sample size, with the desired sample volume, and level of antibody response. In this study, we used three evaluation panels to determine SARS-CoV-2 serology assay performance, and samples from each panel were selected to maintain similar characteristics between panels, including SARS-CoV-2 spike IgM and IgG titer distributions with similar numbers of samples for each titer, days post symptom onset (17 to 46 days), and sample matrix. These panels could also include antibodies that recognize different epitopes and potentially other antigens present on SARS-CoV-2 that were not evaluated. The assay antigens were based on the first SARS-CoV-2 isolate, Wuhan-Hu-1, and the samples in the evaluation panels in this study were from patients with blood collected at the beginning of the pandemic, and they had not been sequenced. New evaluation panels will be needed to evaluate the potential influence of infection with the delta, kappa, and other variants of SARS-CoV-2 on assay performance characteristics. In addition, the sensitivity and specificity estimates were based on serum and plasma samples and may not be indicative of performance with other sample types, such as whole blood. Furthermore, the samples used in this study may not be representative of the antibody profile observed in patient populations. Only three CIA assays were evaluated in this study, which is limited data to formally conclude about overall assay performance for this technology. However, it appears that combined specificity estimates for CIA assays are higher compared to lateral flow assays. In the future, it would be helpful to evaluate further CIA assays to compare performance more thoroughly to lateral flow assays and ELISAs.
In summary, this trans-government collaboration of independent testing helped inform regulatory decision-making and ensured that marketed assays had acceptable performance, which played a critical role in response to the current public health emergency. This rigorous evaluation of SARS-CoV-2 serology assays helped to harmonize assay performance evaluations. This agile trans-governmental partnership demonstrated the power of collaboration to address emergency needs during a pandemic and created a workflow that could help combat new outbreaks in the future. This program showed the value in having an established centralized framework for independent evaluations of assay performance to help ensure high confidence in the evaluation and test performance, provide consistency in the evaluations performed across assays, reduce the burden on assay developers, and facilitate regulatory decision making. Such a program established before outbreaks occur could enable rapid assessments of assay performance during new outbreaks, where the material used to evaluate the assays, such as patient samples or contrived specimens, would depend on what is available at the time the evaluation is taking place. Such a program could also be employed more broadly for other technologies used outside outbreaks.
MATERIALS AND METHODS
Performance evaluation panels.
Three evaluation panels were assembled to assess the performance of commercial assays submitted for independent evaluation. All samples in each panel were blinded to the analysts to avoid bias. Each panel was composed of 30 anti-SARS-CoV-2 antibody-positive serum samples from patients with confirmed SARS-CoV-2 infection by a nucleic acid amplification test (NAAT), as well as 80 antibody-negative plasma and/or serum samples, collected before December 1, 2019. Samples were obtained from multiple sources, collected under approved protocols, and selected to maintain consistency between panels, including sample SARS-CoV-2 spike IgM and IgG titer profile, days post symptom onset (17 to 46 days), and sample matrix (anti-SARS-CoV-2 antibody-positive samples were all serum).
As described previously and in the Text S1, the panel samples were characterized at the CDC and NCI-FNLCR for SARS-CoV-2 IgG and IgM antibody levels against the spike protein, and NCI-FNLCR also tested the samples for IgG antibodies against the RBD (15, 16). All negative samples were assessed at dilutions of 1:100 and 1:400 at CDC using a pan-Ig assay. Positive samples and a subset of negative samples were further evaluated to determine anti-spike IgG and IgM antibody endpoint titers. The CDC pan-Ig, IgG, and IgM ELISAs (17) used the prefusion stabilized ectodomain of the SARS-CoV-2 spike protein expressed in suspension-adapted HEK-293 cells, as described previously (16) and in the Text S1.
Antibody assays evaluated.
Details on the assays evaluated are provided in Table S1, and additional information regarding the evaluation workflow is provided in Text S1. The assay procedure was performed according to the package insert for the respective assay. Due to various constraints, such as instrumentation and other technical requirements, the program focused on evaluating lateral flow assays and ELISAs.
Statistical analyses.
Serology assays submitted for evaluation by this program were evaluated for two performance parameters: (i) sensitivity estimates (positive percent agreement [PPA]), representing the percentage of positive panel samples with a positive assay result; and (ii) specificity estimates (negative percent agreement [NPA]), representing the percentage of negative panel samples with a negative assay result. Additional information on the analysis of assay performance characteristics can be found in the Text S1. Confidence intervals for PPA and NPA were calculated using the Wilson score method (18).
Cross-reactivity with 10 samples from individuals living with HIV was evaluated. If the false-positive rate in samples from HIV-infected samples was high, a 95% confidence interval for the difference in false-positive rates was calculated using Newcombe’s method (19). If cross-reactivity was detected, the 10 HIV+ samples were not included in the calculations of NPA.
Positive predictive value (PPV) and negative predictive value (NPV) were estimated using combined PPA and combined NPA, respectively, and assuming a prevalence of antibody-positive individuals in the population of 5%, based on estimates of prevalence seen in various locations in the United States in spring and early summer 2020. Confidence intervals (95%) for PPV and NPV were estimated using the values from the 95% confidence intervals for combined PPA and combined NPA.
Eighty-seven individual assays were evaluated under this program by January 8th, 2021. Four of the 87 assays, (i) Zhuhai Livzon Diagnostics Inc. Livzon IgM/IgG Diagnostic kit for IgM/IgG Antibody to Coronavirus (SARS-Cov-2) Lateral Flow, (ii) BTNX Inc. COVID-19 IgG/IgM Test Cassettes, (iii) Nirmidas Biotech, Inc. COVID-19 (SARS-CoV-2) IgM/IgG Antibody Detection kit, and (iv) Jiangsu Well Biotech Co., Ltd. The COVID-19 IgM/IgG Rapid Test was evaluated twice for a total of 91 evaluations.
Nonparametric comparisons across panels and assay types were performed for each pair using the Wilcoxon method with GraphPad Prism Version 8.4.3.
Data for each evaluation are available at the FDA and CDC websites (https://open.fda.gov/apis/device/covid19serology/, and https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/serology-test-evaluation.html).
ACKNOWLEDGMENTS
We thank the anonymous blood donors for their generosity during the COVID-19 pandemic. Without their commitment to biomedical research, this study would not have been possible. Critical samples and materials used in the evaluation were provided by the NIH National Institute of Allergy and Infectious Diseases supported BEI Resources Repository (beiresources.org), the Mount Sinai Health System, the Icahn School of Medicine at Mount Sinai, including members of the Departments of Microbiology and Pathology, the Vitalant Research Institute, Centers for Disease Control and Prevention, Boca Biolistics, and Northwestern University. We thank all the analysts at the Serology Laboratory at the Frederick National Laboratory, CDC, and NIH Clinical Center for their time preparing sample aliquots and conducting testing. We thank the FDA Serology Review and Serology Leadership Teams for their time and effort in reviewing EUAs and data obtained from this testing collaboration as well as their commitment to protecting and promoting public health during the COVID-19 pandemic. We thank the OIR-Policy Team at FDA for coordinating the shipment of kits to NCI and tracking testing information. We thank the Office of the Chief Counsel (OCC) at FDA for their support. The use of trade names is for identification purposes only and does not constitute an endorsement by the CDC or the US Department of Health and Human Services. The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract no.75N91019D00024 and HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplemental material is available online only.
Contributor Information
Ligia A. Pinto, Email: pintol@mail.nih.gov.
Tulip Jhaveri, University of Mississippi Medical Center.
REFERENCES
- 1.WHO. 2020. WHO COVID-19 Dashboard. Geneva: World Health Organization. https://covid19.who.int/. [Google Scholar]
- 2.HHS. 2020. Determination of Public Health Emergency, on National Archives. https://www.federalregister.gov/documents/2020/02/07/2020-02496/determination-of-public-health-emergency.
- 3.FDA. 2020. Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised). https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised.
- 4.Shuren J, Stenzel T. 2021. The FDA's Experience with Covid-19 Antibody Tests. N Engl J Med 384:592–594. doi: 10.1056/NEJMp2033687. [DOI] [PubMed] [Google Scholar]
- 5.FDA. 2020. Important Information on the Use of Serological (Antibody) Tests for COVID-19 - Letter to Health Care Providers. https://www.fda.gov/medical-devices/letters-health-care-providers/important-information-use-serological-antibody-tests-covid-19-letter-health-care-providers.
- 6.Shah A, Shuren J. 2020. Insight into FDA’s Revised Policy on Antibody Tests: prioritizing Access and Accuracy. Silver Spring, MD: Food and Drug Administration. [Google Scholar]
- 7.FDA. 2021. In Vitro Diagnostics EUAs. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas.
- 8.FDA. 2020. U.S. Department of Health and Human Services. Food and Drug Administration. Independent Evaluations of COVID-19 Serological Tests. https://open.fda.gov/apis/device/covid19serology.
- 9.Shuren J, Stenzel T. 2020. Covid-19 Molecular Diagnostic Testing - Lessons Learned. N Engl J Med 383:e97. doi: 10.1056/NEJMp2023830. [DOI] [PubMed] [Google Scholar]
- 10.Laurent L. 2020. Theranos Lessons Apply to Covid-19 Antibody Tests., on Bloomberg LP. https://www.bloomberg.com/opinion/articles/2020-05-11/coronavirus-antibody-tests-are-a-wild-west-without-a-sheriff.
- 11.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O'Dell S, Schmidt SD, Swanson PA, 2nd, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, m RNASG . 2020. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med 383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Şahin U, Jansen KU. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 13.Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, Stoop J, Tete S, Van Damme W, Leroux-Roels I, Berghmans PJ, Kimmel M, Van Damme P, de Hoon J, Smith W, Stephenson KE, De Rosa SC, Cohen KW, McElrath MJ, Cormier E, Scheper G, Barouch DH, Hendriks J, Struyf F, Douoguih M, Van Hoof J, Schuitemaker H. 2021. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N Engl J Med 384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FDA. 2021. Antibody Testing Is Not Currently Recommended to Assess Immunity After COVID-19 Vaccination: FDA Safety Communication. https://www.fda.gov/medical-devices/safety-communications/antibody-testing-not-currently-recommended-assess-immunity-after-covid-19-vaccination-fda-safety.
- 15.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. 2020. SARS-CoV-2 Seroconversion in Humans: a Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr Protoc Microbiol 57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman B, Lester S, Mills L, Rasheed MAU, Moye S, Abiona O, Hutchinson GB, Morales-Betoulle M, Krapinunaya I, Gibbons A, Chiang CF, Cannon D, Klena J, Johnson JA, Owen SM, Graham BS, Corbett KS, Thornburg NJ. 2020. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv. doi: 10.1101/2020.04.24.057323. [DOI]
- 18.Garrett P, Lasky F, Meier K. 2008. CLSI. EP12-A2. User Protocol for Evaluation of Qualitative Test Performance; Approved Guideline—Second Edition, vol 28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Altman D, Machin D, Bryant T, Gardner M. 2000. Statistics with Confidence: confidence Intervals and Statistical Guidelines, 2nd Edition. BMJ Books. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum_01564-21_Revised_Supplemental-file.pdf, PDF file, 0.9 MB (730.8KB, pdf)