ABSTRACT
To investigate the presence and location of erm(T) in clinical Streptococcus suis isolates and explore the transmission ability and fitness cost of erm(T)-carrying mobile genetic elements among S. suis isolates, MICs were determined by broth microdilution. The presence of erm(T) in S. suis was detected by PCR. The genetic environment of erm(T) in S. suis was explored by whole-genome sequencing (WGS) analysis. Intraspecies and interspecies transmission were examined by electrotransformation. The fitness cost associated with the carriage of an erm(T)-harboring plasmid or an integrative and conjugative element (ICE) was examined by competition experiments. Of 237 nonduplicate strains, erm(T) was detected in 2 S. suis strains (SC262-ST954 and SC117-ST1314), with its location on a 5,125-bp plasmid in S. suis SC262 and on a 64,013-bp ICESsuSC117 in S. suis SC117, respectively. Both the erm(T)-carrying plasmid pSC262 and the ICESsuSC117 were transmissible by transformation. Plasmid pSC262 can replicate and express macrolide-lincosamide resistance in heterologous hosts, including S. aureus and S. pneumoniae. Both the erm(T)-carrying plasmid and the ICE posed a fitness cost to the host S. suis isolate. To the best of our knowledge, this is the first report of the macrolide-lincosamide-streptogramin B resistance gene erm(T) in S. suis. Its location on a plasmid or an ICE will aid in its transmission. The low detection rate of erm(T) gene among the S. suis population might be due to the fitness cost of the erm(T)-carrying plasmid and ICE.
IMPORTANCE Macrolide and lincosamide resistance due to the presence of erm(T) have posed a challenge for the treatment of Gram-positive pathogens. Although the low detection rate of erm(T) gene among the S. suis population due to the fitness cost of the erm(T)-carrying plasmid and ICE, the presence of erm(T) in S. suis and its potential transmission to other Gram-positive pathogens will be of important significance.
KEYWORDS: Streptococcus suis, erm(T), resistance, macrolide, lincosamide, fitness cost
INTRODUCTION
Streptococcus suis is an important Gram-positive pathogen in the swine industry and also an emerging zoonotic pathogen in humans. Antimicrobial treatment is one of the most important measures to control the S. suis infections (1, 2). Penicillins, macrolides, lincosamides, fluoroquinolones, and tetracyclines are often used as first-line treatments. However, antimicrobial resistance has been emerging during the past years (3), which poses a great challenge not only to the swine industry but also to the public health.
Macrolide resistance is commonly mediated by erm genes and complemented by mef and msr genes (4, 5). The erm(T) gene was first identified in Lactobacillus reuteri; since then, it has been described in various Gram-positive organisms, including the genera Lactobacillus, Streptococcus, Staphylococcus, Enterococcus, and Erysipelothrix, and also in Gram-negative organisms, such as Glaesserella (Haemophilus) parasuis (6–11). In most cases, the erm(T) gene was located on broad host-range plasmids of variable sizes. In Staphylococcus aureus, the plasmid-borne erm(T) gene was flanked by two copies of IS431 elements (12), or together with other antimicrobial resistance genes, such as tet(L) and/or dfrK, flanked by two copies of ISSau10 (13). In addition, the erm(T) gene was also identified in the chromosomal DNA of Streptococcus gallolyticus subsp. pasteurianus, where it was flanked by two copies of IS1216V-like elements (14).
To date, the information about erm(T) in S. suis is still limited. Therefore, this study was initiated to analyze the presence and location of erm(T) in clinical S. suis isolates. In addition, the transferability and fitness cost of erm(T) among S. suis isolates were explored.
RESULTS AND DISCUSSION
Plasmid- and ICE-borne erm(T) genes were identified in S. suis.
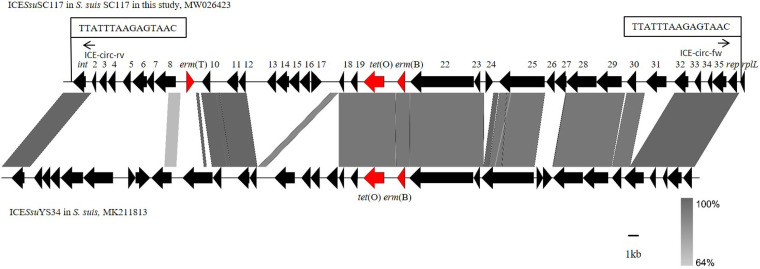
In our study, 2 erm(T)-positive S. suis isolates, SC262-ST954 and SC117-ST1314, out of 237 nonduplicate isolates were identified. In S. suis SC262, erm(T) was located on a small plasmid of 5,125 bp, which is similar to previously described erm(T)-carrying plasmids, such as pER29 from Erysipelothrix rhusiopathiae (KM576795), pCCH208 from S. agalactiae (KJ778678), p5580 from S. dysgalactiae (HE862394), pUR2940, pKKS25, and pUR3912 from S. aureus (HF583292, FN390947, and HF677199), pFS39 from Glaesserella parasuis (KC405064), and p121BS from Lactobacillus sp. (AF310974) (Fig. 1). The previous study identified a complete translational attenuator immediately upstream of the erm(T) gene on the plasmid pRW35 which consisted of two pairs of inverted-repeat sequences of 12 bp each and a reading frame for a regulatory peptide of 19 amino acids (aa) (15). Comparison of the erm(T) regulatory region of pSC262 with that of plasmid pRW35 (EU192194) revealed that the erm(T) regulatory region of pSC262 had many point mutations compared with that of pRW35, and only ribosomal binding sites RBS2 and IR2 could match perfectly (Fig. S1). The rep gene of plasmid pSC262 was compared with those deposited in NCBI GenBank, and a 58.85% identity with that of plasmid pPTDrAP from S. aureus was found, which may point toward the across-genus dissemination potential of this plasmid. As shown in Fig. 2, erm(T) is located on an ICE in S. suis SC117, designated ICESsuSC117, which belongs to the ICESa2603 family of ICEs and has a size of 64,013 kb. ICESa2603 is a 54-kb ICE originally found in S. agalactiae 2603V/R (16). An ICE that carries an integrase gene closely related to intICESa2603, defined as having >60% gene or protein homology, and has significant sequence alignment (60% nucleic acid identity of core genes) and syntonic core structure was classified as a member of the ICESa2603 family (17). The nucleic acid homology between intICESsuSC117 and intICESa2603 is 94.55%. ICESsuSC117 had 95.90% identity and 55.00% coverage rate with the ICESa2603. In addition, it has a core structure similar to that of ICESa2603. Therefore, ICESsuSC117 was classified into the ICESa2603 family. The DNA sequence of ICESsuSC117 was compared with those deposited in the GenBank, and the BLASTn result indicated that it had 95.42% identity and 60.00% coverage rate with the ICESsuYS34 in S. suis (MK211813). The 65.361 kb ICESsuYS34 in the S. suis strain carried the resistance genes erm(B) and tet(O) but not erm(T). ICESsuSC117 was inserted at rplL locus, which is one of the common insertion hot spots of mobile genetic elements (MGEs) in S. suis, forming perfect 15 bp target site duplications at both termini (5′-TTATTTAAGAGTAAC-3′). The 15 bp sequence (TTATTTAAGAGTAAC) at the 3′ end of rplL, the insertion hot spot of ICEs, is highly conserved in streptococci (18). To verify the formation of circular ICESsuSC117 structures, specific primers (ICE-circ-fw/ICE-circ-rv) were designed, and then a 2,537 bp amplicon was detected, which confirmed the ability of ICESsuSC117 to excise from the S. suis chromosomal DNA and to form a circular intermediate. Similarly, the erm(T) upstream regulatory region of ICESsuSC117 was compared with that of plasmid pRW35. The results indicated that the erm(T) upstream regulatory region of ICESsuSC117 had 5 bp point mutations and 1 bp insertion compared to pRW35 in the regulatory peptide open reading frame (ORF). This 1 bp insertion resulted in a frameshift mutation, which extended the reading frame for the regulatory peptide from 19 aa to 28 aa (Fig. S2). The results of the test for inducible clindamycin resistance showed that S. suis strains SC262 and SC117 were resistant to both erythromycin and clindamycin, which revealed that the expression of erm(T) in pSC262 and ICESsuSC117 was constitutive.
FIG 1.
The structural comparison of the erm(T) gene in plasmid DNA from S. suis in this study with that from E. rhusiopathiae, S. agalactiae, S. dysgalactiae, S. aureus, G. parasuis, and Lactobacillus sp. Resistance genes are shown in red and other genes are shown in black.
FIG 2.
The structural comparison of erm(T)-carrying ICESsuSC117 in S. suis in this study with ICESsuYS34 in S. suis (MK211813) described previously. The locations of the PCR primers for the detection of circularizable forms of the ICE are indicated by arrows. The perfect 15 bp target site duplications at both termini are shown in boxes. Regions with more than 90% nucleotide sequence identity are shaded gray.
The erm(T) gene can be transmissible.
Transformation experiments indicated that both the erm(T)-carrying plasmid pSC262 and ICESsuSC117 are transmissible. The transformants P1/7+pSC262 and P1/7+ICESsuSC117 displayed the elevated MICs to the respective antimicrobial agents compared with those of the recipient strain (Table 1). WGS analysis indicated that the erm(T)-carrying ICESsuSC117 was entirely integrated into the rplL gene in the recipient strain, with 15 bp target duplications (5′-TTATTTAAGAGTAAC-3′) immediately up- and downstream of ICESsuSC117 (Fig. 3). The recipient S. suis P1/7 (ST1) and the donor S. suis SC117 (ST1314) were distinguished by multilocus sequence type (MLST).
TABLE 1.
MICs of the erm(T)-carrying strains, the recipient strain S. suis P1/7, S. aureus, and their transformants
| Strain | MIC (mg/L)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| FFC | ERY | LIN | CLI | GEN | CHL | TET | SPE | |
| SC262 | 64 | 512 | 128 | 64 | <1 | 32 | 32 | 2 |
| SC117 | 16 | 512 | 128 | 64 | 2 | 32 | 64 | >512 |
| P1/7 | <1 | <1 | <1 | <1 | 2 | <1 | <1 | 16 |
| P1/7+pSC262 | <1 | 512 | 128 | 64 | 2 | <1 | <1 | 16 |
| P1/7+ICESsuSC117 | <1 | 512 | 128 | 64 | 2 | <1 | 32 | 16 |
| RN4220 | 4 | <1 | <1 | <1 | <1 | 4 | <1 | 16 |
| RN4220+pSC262 | 4 | 512 | 128 | 64 | <1 | 4 | <1 | 16 |
| D39 | <1 | <1 | <1 | <1 | 2 | <1 | <1 | 2 |
| D39+pSC262 | <1 | 512 | 128 | 64 | 2 | <1 | <1 | 2 |
FFC, florfenicol; ERY, erythromycin; LIN, lincomycin; CLI, clindamycin; GEN, gentamicin; CHL, chloramphenicol; TET, tetracycline; SPE, spectinomycin.
FIG 3.
The structural comparison between the recipient S. suis P1/7 and the transformant S. suis P1/7+ICESsuSC117. The 15 bp target sites are shown in boxes. Regions with more than 70% nucleotide sequence identity are shaded gray. Resistance genes are shown in red, rplL genes are shown in blue, and other genes are shown in black.
Furthermore, pSC262 was successfully transferred into the recipient strain S. aureus RN4220 (RN4220+pSC262) and Streptococcus pneumoniae D39 (D39+pSC262) by electrotransformation (19), confirmed by antimicrobial susceptibility testing (AST) (Table 1) and PCR. Compared with the recipient strains, the transformants RN4220+pSC262 and D39+pSC262 displayed elevated MICs of erythromycin, clindamycin, and lincomycin (Table 1), which indicated that an erm(T)-carrying plasmid can replicate and express macrolide-lincosamide resistance in heterologous hosts, including S. aureus and S. pneumoniae.
Fitness cost analyses.
The growth kinetics of P1/7, P1/7+pSC262, and P1/7+ICESsuSC117 in the antibiotic-free Todd-Hewitt broth (THB) were determined (Fig. 4A). The results showed no significant difference for the strains in the absence of selective pressure. However, competition experiments offered a more discriminative and precise measurement of fitness. From the second day on, an obvious decrease in the proportion of pSC262-carrying and ICESsuSC117-carrying strains was observed. At the 7th generation, the pSC262-carrying strain could not be detected (Fig. 4B), and the ICESsuSC117-carrying strain disappeared at the third generation (Fig. 4C). These findings suggests that the erm(T)-carrying strain had a fitness cost compared to S. suis P1/7, which will allow a susceptible strain to outcompete the resistant strain in the absence of a macrolide.
FIG 4.
Fitness cost of pSC262 and ICESsuSC117 in S. suis in this study. (A) Comparison of the growth kinetics of strains S. suis P1/7, S. suis P1/7+pSC262, and S. suis P1/7+ICESsuSC117. (B) Growth competition between S. suis P1/7 and S. suis P1/7+pSC262. (C) Growth competition between S. suis P1/7 and S. suis P1/7+ICESsuSC117. The initial ratio of each strain in competition assay was 1:1.
The acquisition of resistance is generally thought to be accompanied by a fitness cost to the bacterium (20). Spread and maintenance of a resistance gene are directly linked to the fitness cost associated with the gene expression. The constitutive expression of erm(T) in both plasmid pSC262 and ICESsuSC117 observed in this study will increase the fitness cost of erm(T)-carrying mobile genetic elements in these S. suis isolates, which may explain the low detection rate of the erm(T) gene in the S. suis population.
MATERIALS AND METHODS
Bacterial strains and AST.
A total of 237 nonduplicate S. suis strains were isolated and identified from individual diseased pigs in three provinces (Henan, Shanxi, and Guangdong) in China during 2010 to 2016. S. suis P1/7 and S. pneumoniae D39 served as the recipient strain in the transfer experiments (19). All strains were cultivated in THB at 37°C; the medium was supplemented with erythromycin (10 mg/L) for the selection of macrolide-resistant isolates. AST was performed by broth microdilution according to the recommendations given in the EUCAST breakpoint tables for interpretation of MICs and zone diameters, version 11.0 (21). The following antimicrobial agents were tested: florfenicol, erythromycin, lincomycin, clindamycin, gentamicin, chloramphenicol, tetracycline, and spectinomycin. Streptococcus pneumoniae ATCC 49619 served as the quality control strain. Simultaneously, the test for inducible clindamycin resistance in two erm(T)-positive S. suis isolates SC262 and SC117 was performed as described in CLSI document M100 to check whether the expression of erm(T) was inducible or constitutive (22).
PCR analysis.
The erm(T) gene was detected in the S. suis strains by PCR using the primers erm(T)-fw, 5′-ATTGGTTCAGGGAAAGGTC-3′, and erm(T)-rv, 5′-TGGATGAAAGTATTCTCTAGGG-3′, and an annealing temperature of 53.5°C. The presence of circular intermediates in S. suis SC117 was detected by PCR using the primers ICE-circ-fw, 5′-TTGAACAGCCTAAAAGTGCCA-3′, and ICE-circ-rv, 5′-GTAAAGACCAAACAAAGACTCCAG-3′, and an annealing temperature of 59.0°C (Table S1).
WGS analysis.
Whole-genome DNA of SC117 and SC262 was sequenced using the PacBio RS and Illumina MiSeq platforms (Shanghai Personal Biotechnology Co., Ltd., China). The PacBio sequence reads were assembled with HGAP4 and CANU (version 1.6) and then corrected by the Illumina MiSeq reads with pilon (version 1.22). The prediction of ORFs and their annotations were performed using Glimmer 3.0.
Intraspecies transformation.
The transformation experiments were performed as described in a previous study (23). The peptide (GNWGTWVEE) was used as a pheromone for the transformation. The detailed protocols for the transformation were as follows. The recipient strain P1/7 was grown to exponential phase at 37°C under 5% CO2. Then, the logarithmic P1/7 strains were diluted 1:50 into Todd-Hewitt broth supplemented with yeast extract (THY) medium and grown to an optical density at 600 nm (OD600) between 0.035 and 0.058 at 37°C without shaking. The donor DNA (chromosomal DNA, 1 μg or plasmid, 1.2 μg) and synthetic peptide (250 μM) were added to the 100 μL bacterial samples. After 2 h of incubation at 37°C under 5% CO2, the samples were diluted, plated in THA plates with 5% sheep blood and 10 mg/L erythromycin, and incubated at 37°C overnight. Colonies were further confirmed by AST, 16s RNA sequencing, and MLST following harmonized protocols (http://pubmlst.org/) (Table S1).
Interspecies transformation.
To investigate the replication ability of the erm(T)-carrying plasmid pSC262 in heterologous hosts, transformation assays were performed. Plasmid DNA was extracted by using the Qiagen plasmid extraction midi kit (Qiagen, Hilden, Germany) according to the following procedure. After the S. suis SC262 was suspended in buffer P1, lysozyme was added at a final concentration of 20 g/mL, and the mixture was incubated for 2 h at 37°C before buffer P2 was added. Transfer of the purified plasmid DNA was attempted with S. aureus RN4220 and S. pneumoniae D39 by electrotransformation. The transformants were selected on brain heart infusion (BHI) agar supplemented with 10 μg/mL erythromycin.
Fitness cost experiments.
The growth kinetics of S. suis P1/7 and the two transformants P1/7+pSC262 and P1/7+ICESsuSC117 were determined. Cultures were grown for 24 h at 160 rpm and 37°C, and the absorbance at 600 nm was measured every hour.
The fitness cost of the plasmid pSC262 was determined by three independent competition experiments between P1/7 and P1/7+pSC262, and the fitness cost of the ICESsuSC117 was determined between P1/7 and P1/7+ICESsuSC117, as described previously (24). Strains were grown in THB for 16 h at 37°C. Then, 1 × 108 CFU of P1/7 was mixed with 1 × 108 CFU of P1/7+pSC262 or P1/7+ICESsuSC117, respectively. The mixtures were grown at 37°C and 160 rpm and diluted at 1:100 to fresh THB every 12 h. Before every dilution, samples were taken and plated onto antibiotic-free and erythromycin-containing THA plates simultaneously. The number of colonies growing on erythromycin plates was the number of drug-resistant bacteria in the mixed culture system. The number of colonies on the antibiotic-free plate minus the number of colonies on the erythromycin plate is the number of susceptible bacteria in the mixed culture system.
Data availability.
The sequences of the plasmid pSC262 and the ICESsuSC117, determined in this study, have been deposited in GenBank under accession numbers CP06178 and MW026423, respectively.
ACKNOWLEDGMENTS
This work was supported by grants from the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (No.18IRTSTHN020) and the German Federal Ministry of Education and Research (BMBF) under project number 01KI1727D as part of the Research Network Zoonotic Infectious Diseases.
Footnotes
Supplemental material is available online only.
Contributor Information
Dexi Li, Email: lidexi2006@126.com.
Xiang-Dang Du, Email: xddu@henau.edu.cn.
Aude A. Ferran, INTHERES
REFERENCES
- 1.Segura M, Zheng H, de Greeff A, Gao GF, Grenier D, Jiang Y, Lu C, Maskell D, Oishi K, Okura M, Osawa R, Schultsz C, Schwerk C, Sekizaki T, Smith H, Srimanote P, Takamatsu D, Tang J, Tenenbaum T, Tharavichitkul P, Hoa NT, Valentin-Weigand P, Wells JM, Wertheim H, Zhu B, Gottschalk M, Xu J. 2014. Latest developments on Streptococcus suis: an emerging zoonotic pathogen: part 1. Future Microbiol 9:441–444. doi: 10.2217/fmb.14.14. [DOI] [PubMed] [Google Scholar]
- 2.Huong VT, Ha N, Huy NT, Horby P, Nghia HD, Thiem VD, Zhu X, Hoa NT, Hien TT, Zamora J, Schultsz C, Wertheim HF, Hirayama K. 2014. Epidemiology, clinical manifestations, and outcomes of Streptococcus suis infection in humans. Emerg Infect Dis 20:1105–1114. doi: 10.3201/eid2007.131594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yongkiettrakul S, Maneerat K, Arechanajan B, Malila Y, Srimanote P, Gottschalk M, Visessanguan W. 2019. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet Res 15:5. doi: 10.1186/s12917-018-1732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuermberger E, Bishai WR. 2004. The clinical significance of macrolide-resistant Streptococcus pneumoniae: it’s all relative. Clin Infect Dis 38:99–103. doi: 10.1086/380126. [DOI] [PubMed] [Google Scholar]
- 5.Cai Y, Kong F, Gilbert GL. 2007. Three new macrolide efflux (mef) gene variants in Streptococcus agalactiae. J Clin Microbiol 45:2754–2755. doi: 10.1128/JCM.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tannock GW, Luchansky JB, Miller L, Connell H, Thode-Andersen S, Mercer AA, Klaenhammer TR. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100–63. Plasmid 31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead TR, Cotta MA. 2001. Sequence analyses of a broad host-range plasmid containing ermT from a tylosin-resistant Lactobacillus sp. isolated from swine feces. Curr Microbiol 43:17–20. doi: 10.1007/s002840010253. [DOI] [PubMed] [Google Scholar]
- 8.Vandendriessche S, Kadlec K, Schwarz S, Denis O. 2011. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide–lincosamide –streptogramin B resistance gene erm(T) in Belgian hospitals. J Antimicrob Chemother 66:2455–2459. doi: 10.1093/jac/dkr348. [DOI] [PubMed] [Google Scholar]
- 9.DiPersio LP, DiPersio JR, Frey KC, Beach JA. 2008. Prevalence of the erm(T) gene in clinical isolates of erythromycin-resistant group D Streptococcus and Enterococcus. Antimicrob Agents Chemother 52:1567–1569. doi: 10.1128/AAC.01325-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu CW, Zhang AY, Yang CM, Pan Y, Guan ZB, Lei CW, Peng LY, Li QZ, Wang HN. 2015. First report of macrolide resistance gene erm(T) harbored by a novel small plasmid from Erysipelothrix rhusiopathiae. Antimicrob Agents Chemother 59:2462–2465. doi: 10.1128/AAC.00228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SS, Sun J, Liao XP, Liu BT, Li LL, Li L, Fang LX, Huang T, Liu YH. 2013. Co-location of the erm(T) gene and blaROB-1 gene on a small plasmid in Haemophilus parasuis of pig origin. J Antimicrob Chemother 68:1930–1932. doi: 10.1093/jac/dkt112. [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Sanz E, Kadlec K, Feßler AT, Zarazaga M, Torres C, Schwarz S. 2013. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother 57:3275–3282. doi: 10.1128/AAC.00171-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadlec K, Schwarz S. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:915–918. doi: 10.1128/AAC.01091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai JC, Hsueh PR, Chen HJ, Tseng SP, Chen PY, Teng LJ. 2005. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob Agents Chemother 49:4347–4350. doi: 10.1128/AAC.49.10.4347-4350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, Bartkus JM, Lynfield R, Van Beneden C, Beall BW, Active Bacterial Core Surveillance Team. 2008. Plasmid-borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother 52:1140–1143. doi: 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci USA 99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Liang Y, Guo D, Shang K, Ge L, Kashif J, Wang L. 2016. Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs in swine and bovine isolates in China. Front Microbiol 7:55. doi: 10.3389/fmicb.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. 2010. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl Environ Microbiol 76:7957–7965. doi: 10.1128/AEM.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang JR, Veening JW. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13:931. doi: 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 21.EUCAST. 2021. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. https://www.eucast.org/clinical_breakpoints/.
- 22.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI, Wayne, PA. [Google Scholar]
- 23.Zaccaria E, van Baarlen P, de Greeff A, Morrison DA, Smith H, Wells JM. 2014. Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS One 9:e99394. doi: 10.1371/journal.pone.0099394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Dong W, Ma J, Zhang Y, Pan Z, Yao H. 2019. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol 14:207–222. doi: 10.2217/fmb-2018-0279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum01657-21_Supplemental_File_1.docx, DOCX file, 0.02 MB (19KB, docx)
Supplemental material. Download Spectrum01657-21_Supplemental_File_2.docx, DOCX file, 0.25 MB (255.3KB, docx)
Supplemental material. Download Spectrum01657-21_Supplemental_File_3.docx, DOCX file, 0.3 MB (285.8KB, docx)
Data Availability Statement
The sequences of the plasmid pSC262 and the ICESsuSC117, determined in this study, have been deposited in GenBank under accession numbers CP06178 and MW026423, respectively.