ABSTRACT
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in more than 235 million cases worldwide and 4.8 million deaths (October 2021), with various incidences and mortalities among regions/ethnicities. The coronaviruses SARS-CoV, SARS-CoV-2, and HCoV-NL63 utilize the angiotensin-converting enzyme 2 (ACE2) as the receptor to enter cells. We hypothesized that the genetic variability in ACE2 may contribute to the variable clinical outcomes of COVID-19. To test this hypothesis, we first conducted an in silico investigation of single-nucleotide polymorphisms (SNPs) in the coding region of ACE2. We then applied an integrated approach of genetics, biochemistry, and virology to explore the capacity of select ACE2 variants to bind coronavirus spike proteins and mediate viral entry. We identified the ACE2 D355N variant that restricts the spike protein-ACE2 interaction and consequently limits infection both in vitro and in vivo. In conclusion, ACE2 polymorphisms could modulate susceptibility to SARS-CoV-2, which may lead to variable disease severity.
IMPORTANCE There is considerable variation in disease severity among patients infected with SARS-CoV-2, the virus that causes COVID-19. Human genetic variation can affect disease outcome, and the coronaviruses SARS-CoV, SARS-CoV-2, and HCoV-NL63 utilize human ACE2 as the receptor to enter cells. We found that several missense ACE2 single-nucleotide variants (SNVs) that showed significantly altered binding with the spike proteins of SARS-CoV, SARS-CoV-2, and NL63-HCoV. We identified an ACE2 SNP, D355N, that restricts the spike protein-ACE2 interaction and consequently has the potential to protect individuals against SARS-CoV-2 infection. Our study highlights that ACE2 polymorphisms could impact human susceptibility to SARS-CoV-2, which may contribute to ethnic and geographical differences in SARS-CoV-2 spread and pathogenicity.
KEYWORDS: COVID-19, SARS-CoV, SARS-CoV-2, HCoV-NL63, ACE2, SNP
INTRODUCTION
Coronaviruses (CoVs) are enveloped viruses with positive-sense, single-stranded RNA genomes that belong to the Coronaviridae family and Orthocornavirinae subfamily (1, 2). There are four genera in this subfamily, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (2). In the last 2 decades, three betacoronaviruses have caused outbreaks of severe pneumonia in humans, severe acute respiratory syndrome coronavirus (SARS-CoV) (3), Middle East respiratory syndrome coronavirus (MERS-CoV) (4), and SARS-CoV-2 (5–7), the cause of the ongoing COVID-19 pandemic.
Similar to SARS, the severity of COVID-19 disease is positively correlated with increased age, preexisting comorbidities, and/or other environmental factors (8). Host genetic factors could also contribute to clinical outcomes, as demonstrated for other infectious diseases such as HIV and malaria (9–12). Given the variable incidence and disease severity of COVID-19 in different regions and ethnicities of the world, human genetic variation has attracted increasing attention for its potential role in SARS-CoV-2 transmission and pathogenicity (13, 14). Genome-wide association studies of severe COVID-19 patients have identified several genetic risk factors, including 6 genes found in a region of chromosome 3 (15, 16). In addition, some individuals with life-threatening COVID-19 pneumonia were found to have a deficiency in their type I interferon (IFN)-mediated immune response due to genetic mutations or autoimmune antibodies targeting type I IFN (17, 18).
SARS-CoV-2, as well as SARS-CoV and human coronavirus NL63 (HCoV-NL63), utilizes the human protein ACE2 as a cellular receptor to gain entry into human cells (19–22). The viral spike (S) protein of NL63, SARS-CoV, and SARS-CoV-2 is comprised of an S1 and S2 domain, which are separated by a protease cleavage site. The S1 domain directly and specifically binds the peptidase domain of ACE2 via its receptor-binding domain (RBD), exposing the cleavage site for processing by host proteases. This ultimately leads to S2-mediated virus-host cell membrane fusion in a species-specific manner (23). A number of variations have been observed in the ACE2 gene, some of which have been significantly associated with arterial hypertension (24), diabetes mellitus (25), cerebral stroke, coronary artery disease (26), and ventricular hypertrophy (27, 28). However, it is unknown whether these natural ACE2 variants decrease or increase their affinity for coronavirus S protein and affect the susceptibility of individuals to infection.
In this study, we performed a series of biochemical and functional experiments to assess the impact of ACE2 SNPs on interaction with coronavirus S proteins and SARS-CoV-2 entry in vitro and in vivo. This led to the identification of an SNP, D355N (rs961360700), that potentially protects individuals against SARS-CoV-2 infection. Our study suggests that ACE2 polymorphism may alter human susceptibility to SARS-CoV-2 infection and contribute to ethnic and geographical differences in SARS-CoV-2 spread.
RESULTS
Genetic polymorphisms in the human ACE2 gene could impact interaction with coronavirus spike proteins.
ACE2 is a peptidase that is expressed on the surface of many cell types, including lung epithelial cells, and regulates the renin-angiotensin-aldosterone system (29). ACE2 polymorphisms associated with hypertension (HT) and diabetic heart disease have been previously described (26, 30). Here, we surveyed the Genome Aggregation Consortium Database (gnomAD) (https://gnomad.broadinstitute.org/) and found that human ACE2 is highly polymorphic, with 223 ACE2 single-nucleotide variants (SNVs) that result in missense mutations. We hypothesized that these human ACE2 SNVs could influence susceptibility to SARS-CoV, SARS-CoV-2, and/or HCoV-NL63 and potentially affect disease outcomes. To test this, we chose 12 SNVs that led to amino acid substitutions at or in close proximity to the interface of the SARS-CoV, SARS-CoV-2, or HCoV-NL63 S protein complexed with human ACE2 (Fig. 1A and B) (31–35). Of the 12 ACE2 SNVs, 4 have a mutated residue at the interface with the SARS-CoV-2 or SARS-CoV S protein (T27A [rs781255386], E37K [rs146676783], M82I [rs766996587], and D355N [rs961360700]) (31–33, 35) and 2 at the interface with the NL-63 S protein (E37K [rs146676783] and D355N [rs961360700]) (34).
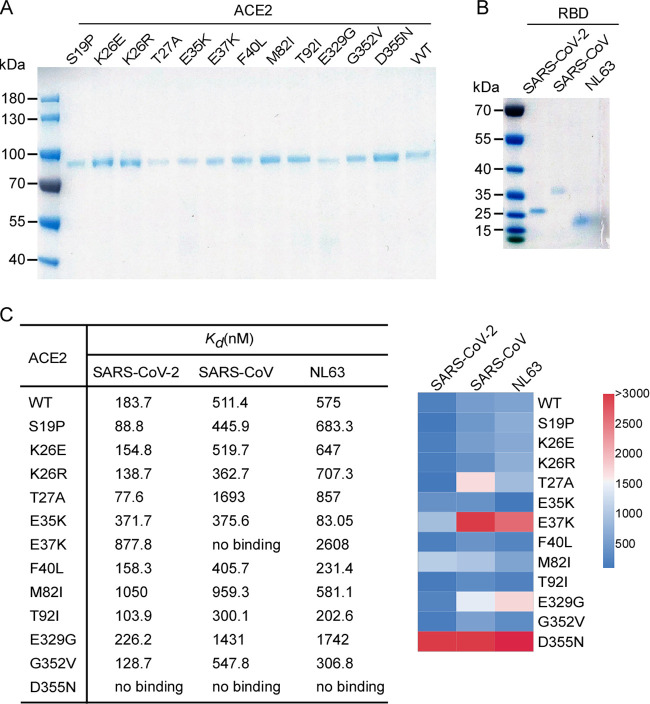
FIG 1.
Schematic representation of the ACE2 molecule and positions of the studied SNP loci. (A) Structures of human ACE2 complexed with the spike proteins of SARS-CoV-2 (PDB code 6M0J), SARS-CoV (PDB code 2AJF), or HCoV-NL63 (PDB code 3KBH). ACE2 and the spike protein of each virus are colored in green and cyan, respectively. The residues of ACE2 at the interface with each spike protein are highlighted. (B) Coding region variants from gnomAD in the genes encoding ACE2 used in this study are indicated. The SNPs and the alteration of the amino acids in this study are shown. (C) Prediction of the interaction of coronavirus spike proteins with ACE2 variants. The ΔΔG for missense mutation was calculated by mCSM-PPI2 with PBD codes 2AJF (SARS-CoV spike complexed with human ACE2), 6M0J (SARS-CoV-2 spike complexed with human ACE2), or 3KBH (NL63-CoV spike complexed with human ACE2) as the model. The individual SNPs are named according to their identification numbers registered at the SNP database (dbSNP). The allele frequency of SNPs are as referenced in gnomAD database.
To elucidate the potential impact of these mutations on viral entry, we predicted the binding affinity of the ACE2 variants and the viral spike proteins. By using the computational platform mCSM-PPI2 (36), we assessed the difference between the binding Gibbs free energy (ΔG) of the wild-type ACE2-spike protein complex with that of each variant ACE2-spike protein complex, thus calculating the ΔΔGwild type–mutant value. According to the mCSM-PPI2 calibration, ΔΔG >1 kcal/mol is a confident indication that a variant has enhanced binding to the viral spike protein relative to the wild-type protein, whereas a ΔΔG of less than −1 kcal/mol indicates a variant has impaired binding. Based on these criteria, most of the ACE2 SNVs exhibited limited alteration in their interaction with the viral spike proteins compared to wild-type ACE2 (Fig. 1C). However, the SNV E37K was predicted to have a significantly decreased interaction with the SARS-CoV-2 spike (ΔΔG, −1.1 kcal/mol) and even more so with the NL63 spike (ΔΔG, −1.6 kcal/mol). G352V and D355N showed dramatically reduced binding with all three spike proteins, and some of these results were consistent with a previous prediction by another calculation method (37). The other variants were considered to have an uncertain impact on binding (Fig. 1C).
Human ACE2 variants bind coronavirus spike proteins with various efficiencies in a cell-based assay.
We next employed a cell-based assay that used flow cytometry to assess the binding of the viral S proteins to human ACE2 variants (38, 39). We cloned the cDNA of each human ACE2 variant (mouse ACE2 was included as a negative control) with a C-terminal FLAG tag into a bicistronic lentiviral vector, pLVX-IRES-zsGreen1. Since this vector contained the fluorescent protein zsGreen1 cloned under an internal ribosomal entry site (IRES) element, we used zsGreen1 expression as a measure of transduction efficiency. We transduced HeLa cells, which lack endogenous ACE2 expression (19), and performed fluorescence-activated cell sorting (FACS) to collect cells with comparable expression levels of the ACE2 variants (data not shown).
Next, we incubated HeLa-ACE2 variant cells with the purified fusion protein consisting of the S1 domain of the coronavirus S proteins being examined (SARS-CoV-2, SARS-CoV, or HCoV-NL63) and an Fc domain of human lgG (S1-Fc) (1 μg/ml). Binding of the fusion proteins to ACE2 was quantified by flow cytometry, and the binding efficiency was calculated as the percentage of S1-Fc-positive cells among ACE2-expressing cells (zsGreen1+) (Fig. 2A). As expected, the S1-Fc proteins of SARS-CoV-2 or NL63 did not bind to HeLa cells expressing mouse ACE2 and showed levels comparable to that of the empty vector control, whereas SARS-CoV S1-Fc exhibited 21% binding efficiency.
FIG 2.

ACE2 variants bind viral spike proteins. (A) HeLa cells transduced with ACE2 variants were incubated with the recombinant S1 domain of the SARS-CoV-2, SARS-CoV, or HCoV-NL63 spike proteins C-terminally fused with Fc (1 μg/ml). Cells were then incubated with goat anti-human IgG (H+L) conjugated to Alexa Fluor 647 followed by flow cytometry analysis. Values are binding efficiencies defined as the percentage of ACE2-expressing cells (zsGreen1+) positive for S1-Fc. Values are means plus standard deviations (SD) (error bars) (n = 3). This experiment was independently repeated three times with similar results. (B) HeLa cells transduced with lentiviruses expressing FLAG-tagged human ACE2 variants were subjected to immunoblotting. Tubulin served as the loading control. This experiment was independently repeated three times with similar results. A representative blot is shown. (C) HeLa cells transduced with lentiviruses (pLVX-IRES-zsGreen1) expressing ACE2 variants as indicated were incubated with rabbit polyclonal antibody (Sino Biological Inc. China; catalog no. 10108-T24) against ACE2. The cells were washed and then stained with 2 μg/ml goat anti-rabbit IgG (H+L) conjugated with Alexa Fluor 568 and DAPI (1 μg/ml). The cell images were captured with a Zeiss LSM 880 confocal microscope. ACE2 on cell surface was shown in the merge images processed by ZEN3.2 software. This experiment was independently repeated three times with similar results, and the representative images are shown. (D) HeLa cells transduced with ACE2 variants were incubated with increasing doses (1 μg/ml, 5 μg/ml, or 10 μg/ml) of the recombinant S1 domain of SARS-CoV-2 (left), SARS-CoV (middle), or HCoV-NL63 (right) spike proteins fused to Fc. The other procedure and binding efficiency were performed as described in panel A. Values are means plus standard deviations (SD) (error bars) (n = 3). This experiment was independently repeated three times with similar results.
For HeLa cells expressing the human ACE2 variants, the S1-Fc proteins exhibited various levels of binding efficiencies. For example, SARS-CoV-2 S1-Fc bound to ACE2 variants at a level similar to that of wild-type ACE2 (range of 82% to 98% versus 93%), with the exception of the D355N variant, which showed dramatically impaired binding (0.5%). Interestingly, SARS-CoV S1-Fc protein bound to some ACE2 variants (K26R, E35K, and T92I) at a significantly higher efficiency (84.5%, 87.3%, and 91.8%, respectively) than the wild-type ACE2 (77.1%). In contrast, other variants such as T27A, E37K, and D355N showed decreased binding affinity, with the latter being the most impaired variant (0.7%).
Of the three coronavirus S proteins tested, the one from HCoV-NL63 bound human ACE2 the least efficiently (71.2%). In addition, it exhibited similar binding efficiencies for most of the ACE2 variants, except for E37K, E329G, and D355N. While E329G bound 3-fold less efficiently than wild-type ACE2 (22.7% versus 71.2%), E37K and D355N were almost completely impaired in binding HCoV-NL63 spike protein (2.6% and 0.5%, respectively).
Collectively, our results showed that ACE2 variants exhibited various binding abilities with the SARS-CoV-2, SARS-CoV, and HCoV-NL63 spike proteins. Some ACE2 variants had virus-specific differences in binding spike protein. D355N was severely impaired in binding all three spike proteins tested. We confirmed that the striking differences in the binding between the ACE2 variants and spike proteins were not due to differing protein expression levels or cell surface localization (Fig. 2B and C). Increasing the concentration of SARS-CoV-2 S1-Fc, but not that of SARS-CoV or HCoV-NL63, did allow for some detection of binding with D355N (5 μg/ml, 7%; 10 μg/ml, 25.7%) (Fig. 2D). Together, these observations suggest that despite using the same cellular receptor, these coronaviruses evolved to utilize distinct amino acids of ACE2 for cell entry.
Binding of ACE2 SNVs with coronavirus spike protein in vitro by SPR analysis.
To further assess the binding of the ACE2 SNVs with the coronavirus spike proteins, we expressed and purified recombinant ACE2 variants, SARS-CoV-2 receptor-binding domain (RBD), SARS-CoV RBD, and HCoV-NL63 RBD (Fig. 3A and B) and directly assayed the protein binding in vitro by surface plasmon resonance (SPR) analysis (Fig. 3C). The dissociation constant (Kd) for wild-type ACE2 binding the SARS-CoV-2 RBD was 183.7 nM, while that of E37K or M82I was about 5- to 6-fold higher (877.8 nM and 1,050 nM, respectively). In line with our cell-based assay, no binding of D355N with SARS-CoV-2 was detected. The Kds of other SNVs with the SARS-CoV-2 RBD ranged from 77 to 371.7 nM (Fig. 3C) and were consistent with the binding efficiencies determined in our cell-based assay (Fig. 2A).
FIG 3.
SPR analysis of ACE2 variants binding viral spike proteins. (A and B) The N-terminal peptidase domain of each human ACE2 (residues Met1-Asp615) variant (A), SARS-CoV-2 RBD (residues Thr333-Pro527), SARS-CoV RBD (residues Arg306-Leu515), or NL63-CoV RBD (residues Gln481-Ile616) (B) were expressed and purified as described in Materials and Methods. The purified proteins were analyzed by SDS-PAGE with Coomassie blue staining. (C) Affinity of ACE2 SNVs for coronavirus spike proteins. Dissociation constant (Kd) values were determined by SPR and are presented as a heatmap according to the indicated color legend. This experiment was independently repeated three times with similar results.
For SARS-CoV RBD, the Kd for wild-type ACE2 was 511.4 nM, while the binding of E37K or D355N was not detected. The Kd for other SNVs ranged from 300 to 1,693 nM, not strikingly different from wild-type ACE2 (Fig. 3C). With respect to binding the HCoV-NL63 RBD, wild-type human ACE2 had a Kd of 575 nM, while E37K and E329G had 5- and 3-fold less affinity (Kds of 2,608 nM and 1,742 nM, respectively). Similar to the other RBD proteins, HCoV-NL63-CoV RBD did not bind the D355N variant (Fig. 3C).
Collectively, our SPR analysis suggests that the ACE2 SNVs possess distinct binding affinities for the three coronavirus RBDs examined. Notably, D355N bound all three coronavirus RBDs with limited affinity in both cell-based and SPR assays.
Molecular dynamic simulations of the interaction between ACE2 variants and SARS-CoV-2 RBD.
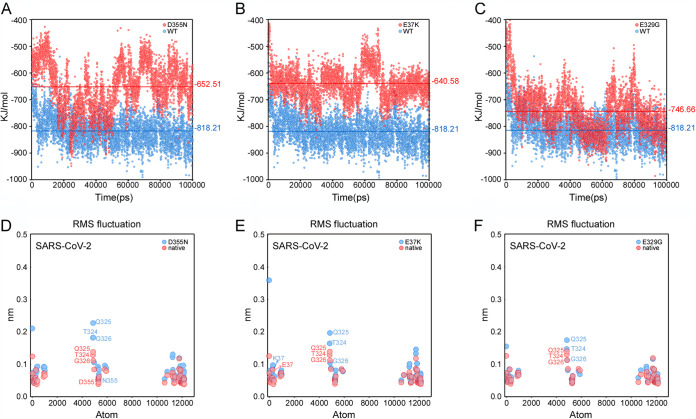
Based on our experimental results, the binding affinities of the D355N, E37K, and E329G variants (meaning ACE2 variants of the ACE2-RBD complexes unless otherwise mentioned) are lower than those of the wild types. To further understand the variations of binding affinities for different mutants from a structural perspective, a series of molecular dynamic (MD) simulations were performed (for details, see Materials and Methods). We first calculated the average nonbonded interaction energies between the mutated residues and RBD (SARS-CoV-2) (data not shown), and the differences between wild-type and mutations were negligible (ranging from 0.1 to 5.5 kJ/mol). Next, we calculated nonbonded interaction energies between ACE2s and RBD, and the differences of average energies between wild type and three variants (D355N, E37K, and E329G) were 166, 178, and 72 kJ/mol, respectively (Fig. 4A to C). These results showed that these substituted residues alone did not affect the binding affinity to RBD, but the overall stability of the systems was significantly affected. To further understand how single substitution affected the stability of the system, Cα root mean square fluctuations (Cα-RMSFs), which is a metric of protein flexibility, were analyzed for the interface residues of D355N, E37K, and E329G variants. Three residues in a small helix, i.e., T324, Q325, and G326, showed great conformational fluctuations in the MD simulations of D355N mutant (Fig. 4D). The small helix was stable in the wild types, while it collapsed to a flexible loop in the D355N variant, suggesting that the small helix may play a crucial role in binding RBDs. Interestingly, this helix was stable in AEC2-only simulations for both the wild type and the D355N variant (similar effects were shown in other mutants as shown in Fig. 4E and F), suggesting that the substitution did not affect the stability of the helix unless RBD was present. Taken together, our MD simulations provided a plausible explanation for the observed various binding affinity for three variants.
FIG 4.
MD simulation results of the wild type and the mutants for different ACE2-RBDs complexes. (A to C) Nonbonded interaction energies between RBD and different mutants as follows for D355N (A), E37K (B), and E329G (C). The blue and red represent interaction energies between ACE2-RBD complexes of wild-type and mutants. Each point represents interaction energy between ACE2 and RBD at a certain time, whereas lines represent the average energies. (D to F) Cα-RMSFs of interface atoms for D355N (D), E37K (E), and E329G (F).
Ability of ACE2 SNVs to mediate entry of virus pseudotyped with coronavirus spike protein.
To assess the functionality of the human ACE2 variants, we evaluated their ability to support the entry of virus pseudotyped with either SARS-CoV-2 or SARS-CoV spike protein. To this end, we produced pseudotyped virus particles containing a firefly luciferase reporter gene and expressing on their surface either vesicular stomatitis virus glycoprotein (VSV-G; positive control) or the spike proteins of SARS-CoV-2 or SARS-CoV. HeLa cells expressing the ACE2 variants were then inoculated with these pseudoparticles, and at 48 h postinoculation, the cells were lysed, and the luciferase activity was monitored as a measure of virus entry. As expected, the VSV-G pseudoparticles readily infected cells independent of which ACE2 variant was expressed (Fig. 5A and B, red columns). Compared to vector-transduced HeLa cells, expression of human ACE2 enhanced the entry of SARS-CoV-2 pseudoparticles by 173-fold (Fig. 5A, blue columns). Most of the ACE2 SNVs mediated SARS-CoV-2 pseudoparticle entry at comparable levels, with the luciferase activity 125- to 269-fold higher than the negative control. However, viral entry was dramatically compromised in cells expressing E329G and D355N, with the luciferase activity increased by only 30- and 10-fold of the negative control, respectively.
FIG 5.

The ability of ACE2 variants to facilitate entry of virus pseudotyped with coronavirus spike proteins. (A and B) HeLa cells transduced with lentiviruses expressing ACE2 orthologs, SNVs, or empty vector were infected with virus pseudotyped with SARS-CoV spike, SARS-CoV-2 spike, or VSV-G and containing a firefly luciferase (Fluc) reporter gene. Intracellular Fluc activity of cell lysates was determined after 48 h of infection. Values are means plus standard deviations (SD) (error bars) (n = 3). This experiment was independently repeated at least three times. **, P < 0.01; ***, P < 0.001. Significance assessed by one-way ANOVA.
Human ACE2 could mediate SARS-CoV pseudoparticle entry, as demonstrated by a 1,000-fold increase in luciferase activity (Fig. 5B, blue columns). Most of the ACE2 SNVs mediated SARS-CoV pseudoparticle entry to a similar extent (700- to 1,200-fold increase over negative control). However, the luciferase activity of cells expressing D355N was severely impaired and only 32-fold higher than the negative control.
Taken together, these findings show that the ACE2 SNVs vary in their ability to support viral entry. In agreement with the data from our other assays, the D355N variant was especially limited in its ability to support SARS and SARS-CoV-2 pseudoparticle entry.
Ability of ACE2 variants to mediate authentic SARS-CoV-2 infection in vitro and in vivo.
As demonstrated above, most of the ACE2 variants were able to bind SARS-CoV-2 spike protein and support efficient entry of pseudovirus particles. However, three of the variants showed suboptimal activity against either all or a subset of the three viruses we tested. Also, some discrepancies were noted between the protein binding and pseudovirus entry assays. For example, E37K was impaired in binding with the SARS-CoV spike protein but mediated efficient entry of SARS-CoV pseudoparticles. In contrast, although E329G showed high binding efficiency for the SARS-CoV-2 spike protein, it supported SARS-CoV-2 pseudoparticle entry significantly less than wild-type ACE2. D355N was highly deficient in interacting with spike of all viruses, both in the binding as well as pseudovirus entry assays (Fig. 2A, Fig. 3C, and Fig. 5).
To further assess E37K, E329G, and D355N variants, we tested their ability to mediate authentic SARS-CoV-2 entry. HeLa cells ectopically expressing individual ACE2 variants were infected with SARS-CoV-2 virus at different doses (multiplicity of infection [MOI], 0.03, 0.1, 0.3, and 1) (Fig. 6A). At 48 h postinfection, the variant-expressing cells were fixed and stained with an antibody directed against the viral nucleocapsid (N) protein, an indicator of virus infection and replication. As expected, HeLa cells expressing mouse ACE2 were not susceptible to SARS-CoV-2 infection, while those expressing wild-type human ACE2 showed high levels of infection. ACE2 SNVs E37K and E329G were comparable to wild-type ACE2 in mediating viral infection, whereas D355N supported significantly lower infection (Fig. 6B).
FIG 6.
The ACE2 variants mediate authentic SARS-CoV-2 virus infection in vitro. (A) HeLa cells transduced with lentiviruses expressing human ACE2 SNVs or mouse ACE2 were infected with various doses of SARS-CoV-2 virus (MOI, 1, 0.3, 0.1, or 0.03). Expression of the viral nucleocapsid (N) protein or ACE2 orthologs was visualized by using the Operetta high content imaging system (PerkinElmer). Viral N protein (red) and ACE2 variant/ortholog (green) are shown. Scale bar, 100 μm. This experiment was independently repeated three times with similar results, and representative images are shown. (B) Images were analyzed and quantified using PerkinElmer Harmony high-content analysis software 4.9. The infection efficiency represents the percentage of SARS-CoV-2 infected cells/ACE2 positive cells (y axis). Values are means plus standard deviations (SD) (error bars) (n = 4). ns, no significance; **, P < 0.01. Significance assessed by one-way ANOVA.
Next, we further evaluated D355N in vivo using a mouse model transduced with replication-defective adenovirus encoding a functional human ACE2 gene which has previously been shown to support productive SARS-CoV-2 infection (38, 40, 41). BALB/c mice were transduced intranasally with recombinant adenovirus expressing human ACE2, the D355N SNV, or a vector control (adenovirus alone) followed by intranasal infection with SARS-CoV-2. After 3 days of SARS-CoV-2 infection, mice were sacrificed and lung tissues collected for viral antigen detection and viral load titration by focus-forming assay (Fig. 7A). To examine the viral antigen spread in the lungs, we performed immunochemical staining with anti-N antibody. Viral N antigen was only well detected in lungs from infected mice transduced with hACE2 and its D355N variant; however, the viral N antigen was less abundant in that of D355N variants (Fig. 7B). Consistent with the N antigen staining, the greatest levels of infectious SARS-CoV-2 virus (about 1 × 106 focus-forming units [FFU]/g of lung tissue) were in lung tissue homogenates from mice transduced with hACE2; viral load in that of D355N transduced mice was moderately reduced (about 1 × 105 FFU/g of lung tissue), whereas virtually none or minimal levels were detected in that of mice transduced with vector (Fig. 7C). Taken together, our results demonstrate that D355N is limited in susceptibility to SARS-CoV-2 infection in vitro and in vivo.
FIG 7.

Ability of ACE2 variants to mediate authentic virus entry in vivo. (A) Schematic representation of the experimental timeline. Wild-type BALB/c mice were transduced with empty recombinant adenovirus (Vector), or recombinant adenovirus expressing wild-type human ACE2 or the D355N variant for 3 days, followed by SARS-CoV-2 challenge. Mice were sacrificed at day 3 postinfection (n = 5 mice per group), and lung tissues were collected for immunostaining with anti-N serum (B) and viral load titration (C). Representative images are shown from n = 5 mice. Scale bar, 200 μm in panel B. Viral load was determined by focus-forming assay. **, P < 0.01. Significance assessed by one-way ANOVA in panel C.
DISCUSSION
Clinical outcomes of SARS-CoV-2 infection range from asymptomatic or mildly symptomatic infections to severe pneumonia, respiratory failure, and even death (23). Epidemiological studies have identified at least three risk factors for severe disease, including being male, of advanced age, and having certain comorbidities (5, 42). Genetic factors have also been linked to the severity of COVID-19, such as polymorphisms in a cluster of genes on chromosome 3 (including the SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1 genes) (15, 16) and genetic deficiency in the type I interferon (IFN) pathway (17). Such findings are of utmost biological and medical importance for understanding the pathogenesis of COVID-19 and development of countermeasures. ACE2 is the receptor for SARS-CoV, SARS-CoV-2, and HCoV-NL63 (19, 21, 22, 43) and is also the major genetic determinant of host range and tissue tropism for these viruses (38, 44–46). ACE2 encodes a metallopeptidase that catalyzes the conversion of angiotensin II to angiotensin, which acts as a vasodilator and exerts important modulatory effects on the cardiovascular system (26). ACE2 SNPs have been found in association with multiple disorders, including essential hypertension (24, 47), dyslipidemia (47), hypertrophic cardiomyopathy (48), and ventricular hypertrophy (27, 28). Therefore, an investigation of the impact of ACE2 polymorphisms on SARS-CoV-2 infection is critical and could pave the way for personalized treatment strategies for COVID-19.
In this study, we evaluated how select ACE2 SNVs compare to the wild-type protein in their binding efficiency with the spike proteins of SARS-CoV, SARS-CoV-2, and NL63-HCoV. We also examined the functionality of these ACE2 variants in mediating infection of virus particles pseudotyped with these different coronavirus spike proteins as well as their ability to support authentic virus infection both in vitro and in vivo. The ACE2 gene is quite polymorphous in human populations, with 223 missense SNVs recorded in the gnomAD database (https://gnomad.broadinstitute.org/). As the structure of viral spike protein complexed with human ACE2 has been solved (31–33), the residues at the interface of the complex are well characterized. We chose 12 SNVs with substitutions at or closest to the complex interface, hypothesizing that these SNVs would have the greatest impact on the spike-ACE2 interaction. We identified several missense ACE2 SNVs that showed significantly altered interaction with the spike proteins of three coronaviruses we tested. The variant D355N (rs961360700) was of particular interest, as it had a significant increase in predicted binding free energy (Fig. 1B), limited binding affinity with spikes in both cell-based and SPR assays (Fig. 2A and Fig. 3C), and reduced susceptibility to authentic SARS-CoV-2 infection in vitro and in vivo (Fig. 6 and 7). These findings provide experimental evidence that a human ACE2 D355N (rs961360700) polymorphism can affect susceptibility to SARS-CoV-2, SARS-CoV, or HCoV-NL63 infection. Based on analysis of the structure of ACE2 complexed with SARS-CoV-2 RBD (31–33), the loop in a beta-sheet (K353-R357) of ACE2 is critical for RBD binding, and the D355N mutation could disrupt the beta-sheet structure, which might explain why D355N mutant is refractory to binding by the SARS-CoV-2 spike protein.
We note that our conclusions are based on experiments performed in cell culture and in an artificial animal model and therefore have certain limitations. It will be important to extend this work to a real-world study which could bridge the gap between genotype, phenotype, and epidemiology. In addition, we only considered the effect of ACE2 SNVs. As the serine protease TMPRSS2 can prime the viral spike for direct fusion with the plasma membrane (43, 49), SNPs in TMPRSS2 might also impact SARS-CoV-2 cell entry and be a useful target of future studies. Then, the ACE2 SNVs receptor activity was evaluated in the absence of TMPRSS2 or other relevant host factors facilitating viral entry; thus, future investigations are necessary to validate the receptor activities of ACE2 SNVs such as D355N in the presence of TMPRSS2 and other relevant host factors. Moreover, SNPs in the 5′ untranslated region (UTR) or other noncoding regions of ACE2 were not included in this study and may also be important in regulating ACE2 transcription or translation, leading to varied ACE2 protein levels in vivo that could affect virus entry and the severity of COVID-19. Due to these limitations, we urge caution not to overinterpret the results of this study.
Of note, SARS-CoV-2 has evolved rapidly in humans, and a variety of genomic changes, including mutations and deletions in the spike protein, have recently been identified (50, 51). Given that genetic variants will continue to arise, it is likely that we will see more genomic changes in the spike protein that might affect the spike interaction with human ACE2. Thus, it will be important to continue testing the interactions of these mutated spike proteins with the ACE2 SNVs. In summary, our study suggests that ACE2 polymorphism could impact human susceptibility to SARS-CoV-2 infection, which contributes to ethnic and geographical differences in SARS-CoV-2 spread and pathogenicity.
MATERIALS AND METHODS
Cell cultures and SARS-CoV-2 virus.
HEK293T cells (American Tissue Culture Collection [ATCC], Manassas, VA, USA; CRL-3216), Vero E6 (Cell Bank of the Chinese Academy of Sciences, Shanghai, China), and HeLa (ATCC CCL-2) were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco, NY, USA) supplemented with 10% (vol/vol) fetal bovine serum (FBS), 10 mM HEPES, 1 mM sodium pyruvate, 1× nonessential amino acids, and 50 IU/ml penicillin-streptomycin in a humidified 5% (vol/vol) CO2 incubator at 37°C. Cells were tested routinely and found to be free of mycoplasma contamination. The SARS-CoV-2 strain nCoV-SH01 (GenBank accession no. MT121215) was isolated from a COVID-19 patient and propagated in Vero E6 cells for use. All experiments involving virus infections were performed in the biosafety level 3 facility of Fudan University following the appropriate regulations.
Plasmids.
The cDNAs encoding ACE2 orthologs (Table S1 in the supplemental material) were synthesized by GenScript and cloned into pLVX-IRES-zsGreen1 vectors (catalog no. 632187; Clontech Laboratories, Inc.) with a C-terminal FLAG tag. ACE2 mutants were generated by QuikChange (Stratagene) site-directed mutagenesis. All of the constructs were verified by Sanger sequencing.
Lentivirus production.
VSV-G-pseudotyped lentiviruses expressing ACE2 orthologs tagged with FLAG at the C terminus were produced by transient cotransfection of the third-generation packaging plasmids pMD2G (Addgene; plasmid no. 12259) and psPAX2 (Addgene; plasmid no. 12260) and the transfer vector with VigoFect DNA transfection reagent (Vigorous) into HEK293T cells. The medium was changed 12 h posttransfection. Supernatants were collected at 24 and 48 h after transfection, pooled, passed through a 0.45-μm filter, and frozen at −80°C.
Protein expression and purification.
The RBDs of SARS-CoV-2, SARS-CoV, NL63-CoV, and the N-terminal peptidase domain of wild-type and human ACE2 SNVs were expressed using the Bac-to-Bac baculovirus system (Invitrogen). Specifically, SARS-CoV-2 RBD (residues Thr333-Pro527), SARS-CoV RBD (residues Arg306-Leu515), or NL63-CoV RBD (residues Gln481-Ile616) with an N-terminal gp67 signal peptide for secretion and a C-terminal 6×His tag for purification was inserted into pFastBac-Dual vector (Invitrogen). The recombinant baculoviruses were generated according to the manufacturer’s instruction to infect Hi5 cells at a density of 2 × 106 cells/ml. After 60 h, the supernatant of cell culture containing the RBD was collected, concentrated, and buffer-exchanged to HBS (10 mM HEPES, pH 7.2, 150 mM NaCl). The recombinant RBD was captured by Ni-nitrilotriacetic acid (Ni-NTA) resin (GE Healthcare) and eluted with 500 mM imidazole in HBS buffer. RBD was then purified by gel-filtration chromatography using the Superdex 200 column (GE Healthcare) preequilibrated with HBS buffer. Fractions containing RBD were collected for further analysis. The N-terminal peptidase domain of each human ACE2 (residues Met1-Asp615) variant was expressed and purified by essentially the same protocol used for RBD.
Surface ACE2 binding assay.
HeLa cells were transduced with lentiviruses expressing the ACE2 SNVs for 48 h. The cells were collected with TrypLE (Thermo; catalog no. 12605010) and washed twice with cold phosphate-buffered saline (PBS). Live cells were incubated with the recombinant proteins, S1 domain of SARS-CoV-2, SARS-CoV, or NL63 spike C-terminally fused with Fc (1 μg/ml) at 4°C for 30 min. After washing, cells were stained with goat anti-human IgG (H+L) conjugated to Alexa Fluor 647 (Thermo; catalog no. A21445; 2 μg/ml) for 30 min at 4°C. Cells were then washed twice and subjected to flow cytometry analysis (Thermo; Attune NxT).
SPR experiments.
SARS-CoV RBD, SARS-CoV-2 RBD, or NL63 RBD was immobilized to a CM5 sensor chip (GE Healthcare) using a Biacore T200 (GE Healthcare) and a running buffer composed of 10 mM HEPES, pH 7.2, 150 mM NaCl, and 0.05% Tween 20. Serial dilutions of ACE2 SNV proteins were flowed through with a concentration ranging from 1,600 to 25 nM. The resulting data were fit to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
MD simulations.
MD simulations were performed by using GROMACS (52) 2018.4 with AMBER99SB force field (53). The X-ray structures of spike receptor-binding domain (RBD) bound with ACE2 complexes of NL63 (PDB ID 3KBH), SARS-CoV (PDB ID 2AJF), and SARS-CoV-2 (PDB ID 6M0J) were used for MD simulations. The initial structures were prepared by Protein Preparation Wizard in Maestro. The simulation system was conducted in an ∼13.7- by 13.7- by 13.7-nm3 cubic box, which consisted of ∼80,000 water molecules and was solvated with a 0.05 M NaCl solution. We performed 5000 steps of energy minimizations with the steepest descent algorithm, followed by 0.1-ns equilibration NVT and NPT ensembles. During equilibrations, the system was coupled to a temperature bath of 300 K using V-rescale thermostat with a time constant 0.1 ps. The pressure coupling was using a coupling constant of 2.0 ps and compressibility of 4.5 × 10−5 bar−1. In the production run, 100-ns MD simulations were performed with the leapfrog algorithm (2-fs time step). All bonds were constrained using LINCS algorithm (54), and long-range electrostatic interactions were using particle mesh Ewald method. The cutoff distance for the Lennard-Jones and electrostatic interaction was set to 10 Å. The temperature and pressure regulation were accomplished via V-rescale thermostat (55) and Berendsen thermostat (56), respectively. The coordinates during simulation were saved every 100 ps. The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) of interface residues alpha carbon coordinates were calculated by GROMACS. The interaction energy was calculated by equation 1 as follows:
where i and j represent the atoms in two groups, and and are electrostatic energy and Lennard-Jones energy between atom i and atom j, respectively. For Lennard-Jones energy calculation, ε is the well depth, σ is the distance at which the intermolecular potential is zero, and represents the distance between atom i and atom j. is the permittivity in vacuum, and q represents the partial charge.
Production of SARS-CoV-2 or SARS-CoV pseudotyped virus.
Pseudoviruses were produced in HEK293T cells by cotransfecting the retroviral vector pTG-MLV-Fluc, pTG-MLV-Gag-pol, and pcDNA3.1 expressing the SARS-CoV spike, SARS-CoV-2 spike, or VSV-G (pMD2.G [Addgene; plasmid no. 12259]) using VigoFect (Vigorous Biotechnology). At 48 h posttransfection, the cell culture medium was collected for centrifugation at 3,500 rpm for 10 min, and then the supernatant was subsequently aliquoted and stored at −80°C for further use. Virus entry was assessed by transduction of pseudoviruses in cells expressing ACE2 variants in 48-well plates. After 48 h, intracellular luciferase activity was determined using the luciferase assay system (Promega; catalog no. E1500) according to the manufacturer’s instructions. Luminescence was recorded on a GloMax Discover system (Promega).
Immunofluorescence staining of viral nucleocapsids.
HeLa cells were transduced with lentiviruses expressing the ACE2 SNVs for 48 h. Cells were then infected with nCoV-SH01 at an MOI of 1 for 1 h, washed three times with PBS, and incubated in 2% FBS culture medium for 48 h for viral antigen staining. Cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100, and incubated with the rabbit polyclonal antibody against SARS-CoV nucleocapsid protein (Rockland; catalog no. 200-401-A50; 1 μg/ml) at 4°C overnight. After three washes, cells were incubated with the secondary goat anti-rabbit antibody conjugated to Alexa Fluor 555 (Thermo; catalog no. A32732; 2 μg/ml) for 2 h at room temperature, followed by staining with 4′,6-diamidino-2-phenylindole (DAPI). Images were collected using an EVOS microscope M5000 imaging system (Thermo; catalog no. AMF5000). Images were processed using the ImageJ program (http://rsb.info.nih.gov/ij/).
Generation and production of recombinant adenovirus expressing ACE2 variants.
cDNA of ACE2 variants with a FLAG tag at the C terminus was cloned into pShuttle. The pShuttle-ACE2 plasmids were then electroporated 440 into BJ5183 AD-1 cells (Agilent), which were pretransformed with pAdEasy-1 to facilitate recombination with the pShuttle-CMV vector. The adenovirus constructs were then transfected into HEK293 cells. The transfected HEK293 cells were maintained until the cells exhibited complete cytopathic effect (CPE) and then harvested and freeze-thawed. The supernatants were serially passaged two more times, with harvest at complete CPE and freeze-thaw. For virus purification, the cell pellets were purified using cesium chloride density gradient ultracentrifugation, and the number of virus particles was determined using a NanoDrop 2000 (Thermo Fisher Scientific). The adenovirus stocks were aliquoted and stored at −80°C.
SARS-CoV-2 infection of adenovirus transduced mice.
Six- to 8-week-old male mice (BALB/c) were transduced intranasally with adenovirus expressing wild-type ACE2, the D355N variant, or empty control (5 × 1010 viral particles per mouse). After 3 days, mice were infected intranasally with SARS-CoV-2 (8 × 104 FFU per mouse) and sacrificed at day 3 postinfection. The lung tissues were harvested for histopathological analysis and virus titration. This animal experiment protocol was approved by the Animal Ethics Committee of the School of Basic Medical Sciences at Fudan University.
Histological analysis.
Lung tissues were harvested and fixed in 4% paraformaldehyde (PFA) for 48 h. Tissues were embedded in paraffin for sectioning. For viral antigen detection, the sections were incubated with house-made mouse anti-SARS-CoV-2 nucleocapsid protein serum (1:5,000) and HRP465-conjugated goat anti-mouse IgG secondary antibody (1:5,000 dilution; Invitrogen). The lung sections from the vector-transduced mouse were used as negative control. The sections were observed under microscope (Olympus, Tokyo, Japan).
Virus load determination by focus-forming assay.
Vero E6 monolayer cells in 96-well plates were inoculated with serially diluted virus for 2 h and then overlaid with methylcellulose for 48 h. Cells were fixed with 4% paraformaldehyde in PBS for 1 h and permeabilized with 0.2% Triton X-100 for 1 h. Cells were stained with homemade mouse anti-SARS-CoV-2 N serum overnight at 4°C and incubated with the secondary goat anti-mouse HRP-conjugated antibody for 2 h at room temperature. The focus-forming unit was developed using TrueBlue substrate (Sera Care; catalog no. 5510-0030).
Statistics analysis.
One-way analysis of variance (ANOVA) with Tukey’s honestly significant difference (HSD) test was used to test for statistical significance of the differences between the different group parameters. P values of less than 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We acknowledge Di Qu, Zhiping Sun, Wendong Han, Gaowei Hu, and other colleagues at the biosafety level 3 laboratory of Fudan University for help with experiment design and technical assistance. We thank Jenna M. Gaska for suggestions and revision of the manuscript.
This work was supported by the National Natural Science Foundation of China (32070153 to Q.D.), Tsinghua University Spring Breeze Fund (2021Z99CFY030 to Q.D.), Beijing Municipal Natural Science Foundation (M21001 to Q.D.), and Start-up Foundation of Tsinghua University (53332101319).
Contributor Information
Xinquan Wang, Email: xinquanwang@tsinghua.edu.cn.
Rong Zhang, Email: rong_zhang@fudan.edu.cn.
Qiang Ding, Email: qding@tsinghua.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Fung TS, Liu DX. 2019. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73:529–557. 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 2.Lai MM. 1990. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol 44:303–333. 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Morens DM, Fauci AS. 2020. Emerging pandemic diseases: how we got to COVID-19. Cell 182:1077–1092. 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YT, Landeras-Bueno S, Hsieh LE, Terada Y, Kim K, Ley K, Shresta S, Saphire EO, Regla-Nava JA. 2020. Spiking pandemic potential: structural and immunological aspects of SARS-CoV-2. Trends Microbiol 28:605–618. 10.1016/j.tim.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med 3:252–253. 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 10.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. 1997. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet 349:1219–1220. 10.1016/S0140-6736(05)62411-7. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 2:1240–1243. 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 12.Kariuki SN, Williams TN. 2020. Human genetics and malaria resistance. Hum Genet 139:801–811. 10.1007/s00439-020-02142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casanova JL, Su HC, Effort CHG, COVID Human Genetic Effort . 2020. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell 181:1194–1199. 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Initiative C-HG. 2020. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet 28:715–718. 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernandez J, Prati D, Baselli G, Asselta R, Grimsrud MM, Milani C, Aziz F, Kassens J, May S, Wendorff M, Wienbrandt L, Uellendahl-Werth F, Zheng T, Yi X, de Pablo R, Chercoles AG, Palom A, Garcia-Fernandez AE, Rodriguez-Frias F, Zanella A, Bandera A, Protti A, Aghemo A, Lleo A, Biondi A, Caballero-Garralda A, Gori A, Tanck A, Carreras Nolla A, Latiano A, Fracanzani AL, Peschuck A, Julia A, Pesenti A, Voza A, Jimenez D, Mateos B, Jimenez BN, Quereda C, Paccapelo C, Gassner C, Angelini C, Cea C, et al. 2020. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med 383:1522–1534. 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeberg H, Paabo S. 2020. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587:610–612. 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 17.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Beziat V, Manry J, Shaw E, Haljasmagi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Le Pen J, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370:eabd4585. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schluter A, Le Voyer T, Khan T, Li J, et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fung TS, Liu DX. 2021. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu Rev Microbiol 75:19–47. 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102:7988–7993. 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B, Guo H, Zhou P, Shi ZL. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Liu C, Guan T, Li Y, Lai Y, Li F, Zhao H, Maimaiti T, Zeyaweiding A. 2019. Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in south Xinjiang. Hypertens Res 42:681–689. 10.1038/s41440-018-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Li Y, Guan T, Lai Y, Shen Y, Zeyaweiding A, Zhao H, Li F, Maimaiti T. 2018. ACE2 polymorphisms associated with cardiovascular risk in Uygurs with type 2 diabetes mellitus. Cardiovasc Diabetol 17:127. 10.1186/s12933-018-0771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Huang W, Su S, Li B, Zhao W, Chen S, Gu D. 2006. Association study of ACE2 (angiotensin I-converting enzyme 2) gene polymorphisms with coronary heart disease and myocardial infarction in a Chinese Han population. Clin Sci (Lond) 111:333–340. 10.1042/CS20060020. [DOI] [PubMed] [Google Scholar]
- 27.Hamming I, Cooper ME, Haagmans BL, Hooper NM, Korstanje R, Osterhaus AD, Timens W, Turner AJ, Navis G, van Goor H. 2007. The emerging role of ACE2 in physiology and disease. J Pathol 212:1–11. 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieb W, Graf J, Gotz A, Konig IR, Mayer B, Fischer M, Stritzke J, Hengstenberg C, Holmer SR, Doring A, Lowel H, Schunkert H, Erdmann J. 2006. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men. Results of the MONICA Augsburg echocardiographic substudy. J Mol Med (Berl) 84:88–96. 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- 29.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. 2000. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res 87:e1–e9. 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 30.Frossard PM, Malloy MJ, Lestringant GG, Kane JP. 2001. Haplotypes of the human renin gene associated with essential hypertension and stroke. J Hum Hypertens 15:49–55. 10.1038/sj.jhh.1001107. [DOI] [PubMed] [Google Scholar]
- 31.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 32.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, Wang Q, Zhou H, Yan J, Qi J. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904.e9. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci USA 106:19970–19974. 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigues CHM, Myung Y, Pires DEV, Ascher DB. 2019. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res 47:W338–W344. 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim H, Baek A, Kim J, Kim MS, Liu J, Nam KY, Yoon J, No KT. 2020. Hot spot profiles of SARS-CoV-2 and human ACE2 receptor protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci Rep 10:16862. 10.1038/s41598-020-73820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Hu G, Wang Y, Ren W, Zhao X, Ji F, Zhu Y, Feng F, Gong M, Ju X, Zhu Y, Cai X, Lan J, Guo J, Xie M, Dong L, Zhu Z, Na J, Wu J, Lan X, Xie Y, Wang X, Yuan Z, Zhang R, Ding Q. 2021. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc Natl Acad Sci USA 118:e2025373118. 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren W, Zhu Y, Wang Y, Shi H, Yu Y, Hu G, Feng F, Zhao X, Lan J, Wu J, Kenney DJ, Douam F, Tong Y, Zhong J, Xie Y, Wang X, Yuan Z, Zhou D, Zhang R, Ding Q. 2021. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog 17:e1009392. 10.1371/journal.ppat.1009392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Jr, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS. 2020. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182:744–753.e4. 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang HJ, Baric RS, Zhou P, Yang XL, Shi ZL. 2020. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58.e8. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhopal SS, Bhopal R. 2020. Sex differential in COVID-19 mortality varies markedly by age. Lancet 396:532–533. 10.1016/S0140-6736(20)31748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F. 2008. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol 82:6984–6991. 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F. 2013. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res 100:246–254. 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F. 2015. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964. 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan Y, Wang T, Li Y, Guan T, Lai Y, Shen Y, Zeyaweiding A, Maimaiti T, Li F, Zhao H, Liu C. 2018. Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Health Dis 17:241. 10.1186/s12944-018-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Merwe L, Cloete R, Revera M, Heradien M, Goosen A, Corfield VA, Brink PA, Moolman-Smook JC. 2008. Genetic variation in angiotensin-converting enzyme 2 gene is associated with extent of left ventricular hypertrophy in hypertrophic cardiomyopathy. Hum Genet 124:57–61. 10.1007/s00439-008-0524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA 117:7001–7003. 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peacock TP, Penrice-Randal R, Hiscox JA, Barclay WS. 2021. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J Gen Virol 102:001584. 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu A, Wang L, Zhou HY, Ji CY, Xia SZ, Cao Y, Meng J, Ding X, Gold S, Jiang T, Cheng G. 2021. One year of SARS-CoV-2 evolution. Cell Host Microbe 29:503–507. 10.1016/j.chom.2021.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. 2005. GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 53.Tian C, Kasavajhala K, Belfon KAA, Raguette L, Huang H, Migues AN, Bickel J, Wang Y, Pincay J, Wu Q, Simmerling C. 2020. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput 16:528–552. 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 54.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. 1997. LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. . [DOI] [Google Scholar]
- 55.Bussi G, Donadio D, Parrinello M. 2007. Canonical sampling through velocity rescaling. J Chem Phys 126:014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 56.Berendsen HJC, Postma JPM, Gunsteren WFv, DiNola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]