ABSTRACT
The development of mouse models for coronavirus disease 2019 (COVID-19) has enabled testing of vaccines and therapeutics and defining aspects of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogenesis. SARS-CoV-2 disease is severe in K18 transgenic mice (K18-hACE2 Tg) expressing human angiotensin-converting enzyme 2 (hACE2), the SARS-CoV-2 receptor, under an ectopic cytokeratin promoter, with high levels of infection measured in the lung and brain. Here, we evaluated SARS-CoV-2 infection in hACE2 knock-in (KI) mice that express hACE2 under an endogenous promoter in place of murine ACE2 (mACE2). Intranasal inoculation of hACE2 KI mice with SARS-CoV-2 WA1/2020 resulted in substantial viral replication within the upper and lower respiratory tracts with limited spread to extrapulmonary organs. However, SARS-CoV-2-infected hACE2 KI mice did not lose weight and developed limited pathology. Moreover, no significant differences in viral burden were observed in hACE2 KI mice infected with B.1.1.7 or B.1.351 variants compared to the WA1/2020 strain. Because the entry mechanisms of SARS-CoV-2 in mice remain uncertain, we evaluated the impact of the naturally occurring, mouse-adapting N501Y mutation by comparing infection of hACE2 KI, K18-hACE2 Tg, ACE2-deficient, and wild-type C57BL/6 mice. The N501Y mutation minimally affected SARS-CoV-2 infection in hACE2 KI mice but was required for viral replication in wild-type C57BL/6 mice in a mACE2-dependent manner and augmented pathogenesis in the K18-hACE2 Tg mice. Thus, the N501Y mutation likely enhances interactions with mACE2 or hACE2 in vivo. Overall, our study highlights the hACE2 KI mice as a model of mild SARS-CoV-2 infection and disease and clarifies the requirement of the N501Y mutation in mice.
IMPORTANCE Mouse models of SARS-CoV-2 pathogenesis have facilitated the rapid evaluation of countermeasures. While the first generation of models developed pneumonia and severe disease after SARS-CoV-2 infection, they relied on ectopic expression of supraphysiological levels of human ACE2 (hACE2). This has raised issues with their relevance to humans, as the hACE2 receptor shows a more restricted expression pattern in the respiratory tract. Here, we evaluated SARS-CoV-2 infection and disease with viruses containing or lacking a key mouse-adapting mutation in the spike gene in hACE2 KI mice, which express hACE2 under an endogenous promoter in place of murine ACE2. While infection of hACE2 KI mice with multiple strains of SARS-CoV-2 including variants of concern resulted in viral replication within the upper and lower respiratory tracts, the animals did not sustain severe lung injury. Thus, hACE2 KI mice serve as a model of mild infection with both ancestral and emerging SARS-CoV-2 variant strains.
KEYWORDS: SARS-CoV-2, lung infection, mouse model, pathogenesis
INTRODUCTION
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, multiple models of infection and pathogenesis have been developed in mice. Because human angiotensin-converting enzyme 2 (hACE2), but not murine ACE2 (mACE2), can engage and bind the ancestral SARS-CoV-2 spike protein, several strategies were established for expressing hACE2 in mice. One of these models, the K18-hACE2 transgenic mouse (K18-hACE2 Tg), in which hACE2 is expressed under an ectopic cytokeratin K18 promoter, is commonly used and provides a stringent model of severe infection allowing rapid evaluation of vaccines and monoclonal antibody-based therapies in vivo (1–4). However, expression of the hACE2 transgene in these mice is nonphysiological with both a high number of hACE2 transgene insertions as well as ectopic expression on cells that do not normally express ACE2. This is best exemplified by SARS-CoV-2 infection in the brains of K18-hACE2 Tg mice, which does not reflect the central nervous system (CNS) involvement seen in humans or other animals such as hamsters or nonhuman primates (5–8). Because expression of hACE2 in K18-hACE2 Tg mice is independent of the transcriptional regulation that normally governs ACE2 levels, some aspects of infection or transmission likely are not modeled in a physiologically relevant manner. Therefore, development of additional mouse models is necessary to understand aspects of viral pathogenesis and tropism.
In this study, we evaluated SARS-CoV-2 infection of mice that express hACE2 under an endogenous promoter in place of mACE2 (hACE2 knock-in [KI] mice). Human ACE2 was expressed in the nasal turbinate, lung, duodenum, and kidney. After intranasal inoculation of hACE2 KI mice, high levels of viral RNA were detected in nasal turbinates and lungs with limited spread to extrapulmonary organs, including the brain and gastrointestinal tract. Despite productive infection of the respiratory tract, weight loss was not observed, and lung inflammation was limited. Furthermore, we established the impact of the N501Y mutation, a substitution naturally present in both mouse-adapted SARS-CoV-2 strains and variants of concern, in multiple murine models.
RESULTS
Human ACE2 expression in hACE2 KI and K18-hACE2 transgenic mice.
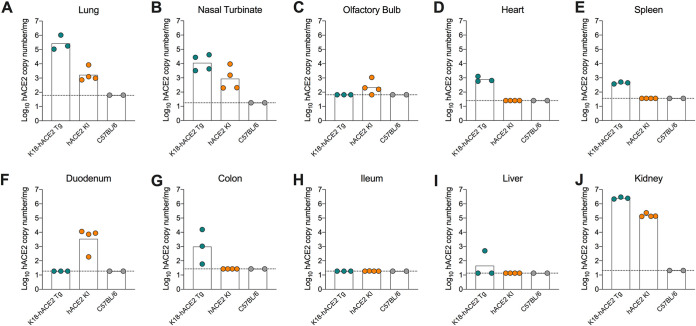
As previously described, hACE2 KI mice were generated (9) with hACE2 cDNA inserted in-frame and immediately downstream of the endogenous initiation codon of mouse Ace2 in exon 2. In addition to replacing mACE2, hACE2 expression is driven by the endogenous Ace2 regulatory elements. Using gene- and species-specific reverse transcription-quantitative PCR (RT-qPCR) primers, we detected hACE2 mRNA expression in the lung, nasal turbinate, kidney, duodenum, and olfactory bulb, but not in the colon, ileum, heart, spleen, or liver of hACE2 KI mice (Fig. 1A to J). This gene expression pattern overlaps but is not entirely consistent with transcriptomic and protein staining data in humans, which show ACE2 expression predominantly in the respiratory tract, kidney, and small intestine (10, 11). Single cell RNA sequencing analysis estimates that Ace2-expressing cells comprise only 1% to 6% of the total population in the nasal mucosa and respiratory tract of humans and nonhuman primates (12–14). We attempted to stain for hACE2 in the lungs using RNA in situ hybridization or immunohistochemistry but were unable to detect hACE2 mRNA or protein in hACE2 KI mice possibly due to the relatively low level of expression or the limit of detection of the assays. Indeed, in the lung and nasal turbinate, hACE2 mRNA expression was 10- to 100-fold higher in K18-hACE2 Tg mice than in hACE2 KI mice.
FIG 1.
hACE2 expression in hACE2 KI and K18-hACE2 transgenic (Tg) mice. (A to J) Expression levels of hACE2 mRNA in the lung (A), nasal turbinate (B), olfactory bulb (C), heart (D), spleen (E), duodenum (F), colon (G), ileum (H), liver (I), and kidney (J) of naive 6-week-old female hACE2 KI, K18-hACE2, or wild-type (WT) C57BL/6J mice (two experiments, n = 2 to 4).
Infection of hACE2 KI mice with SARS-CoV-2 variants of concern.
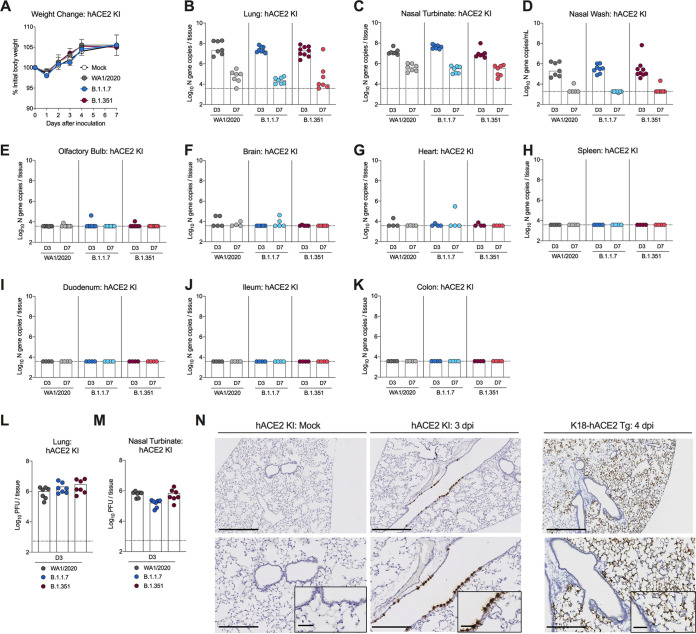
Six-week-old homozygous hACE2 KI C57BL/6 mice were inoculated via an intranasal route with 105 focus-forming units (FFU) of SARS-CoV-2 WA1/2020 or B.1.1.7 or B.1.351 variants of concern (VOC). No significant weight loss was observed following infection with any of the strains (Fig. 2A), but high levels of viral RNA were detected in the lung, nasal wash, and nasal turbinate at both 3 and 7 days postinfection (dpi), with very low levels present in the olfactory bulb and brain of some animals (Fig. 2B to F). Viral RNA was not detected in the heart, spleen, brain, or gastrointestinal tract tissues (Fig. 2G to K). High levels of infectious virus were also recovered from the lungs and nasal turbinates of hACE2 KI mice at 3 dpi by plaque assay (Fig. 2L to M). The tissues supporting SARS-CoV-2 infection in this model overlap with the pattern of hACE2 mRNA expression (Fig. 1) except for the duodenum, which expresses hACE2 but did not support viral infection. No significant differences in viral burden were observed in hACE2 KI mice infected with B.1.1.7, B.1.351, or WA1/2020 strains.
FIG 2.
SARS-CoV-2 infection in hACE2 KI mice. (A to E) Six-week-old female and male hACE2 KI mice were inoculated via intranasal route with PBS or 105 FFU of WA1/2020, B.1.1.7, or B.1.351 SARS-CoV-2 strains. (A) Weight change following SARS-CoV-2 or mock infection (mean ± standard error of the mean [SEM]; n = 6 mice per group, two experiments). (B to K) Viral RNA levels in the lung (B), nasal turbinate (C), nasal wash (D), olfactory bulb (E), brain (F), heart (G), spleen (H), duodenum (I), ileum (J), or colon (K) were analyzed at 3 and 7 dpi by RT-qPCR (n = 6 to 8, two experiments, one-way analysis of variance [ANOVA] with Dunnett’s test comparing titers at days 3 and 7 between respective SARS-CoV-2 variants; all not significant). The dotted lines indicate the limit of detection of the assay. (L and M) Infectious virus levels in the lung (L) and nasal turbinate (M) were analyzed at 3 dpi by plaque assay on Vero-hACE2-TMPRSS2 cells (n = 7, two experiments). (N) SARS-CoV-2 RNA in situ hybridization of lung sections in hACE2 KI mice following mock infection or after intranasal inoculation with 105 FFU of WA1/2020 D614G/N501Y at 3 dpi or in K18-hACE2 mice following intranasal inoculation with 2.5 × 104 PFU of SARS-CoV-2 WA1/2020. Images show low-power magnification (top; bars, 500 μm), medium-power magnification (bottom; bars, 250 μm), and high-power magnification (insets in the bottom panels; bars, 100 μm). Images are representative of n = 4 per group.
To define the tropism of SARS-CoV-2 in hACE2 KI mice, we stained lung sections for viral RNA using in situ hybridization (Fig. 2N). At 3 dpi, expression of SARS-CoV-2 RNA was localized to epithelial cells of the larger airways with little viral RNA detected in the lung parenchyma. In comparison, infection of K18-hACE2 Tg mice showed abundant viral RNA in lung pneumocytes with less infection of the airway-lining cells. While both types of hACE2-expressing mice support SARS-CoV-2 infection, the site of replication in the lung differs, which likely is due to promoter-driven variation in hACE2 expression on different cell types.
Inflammation in the lung following SARS-CoV-2 infection.
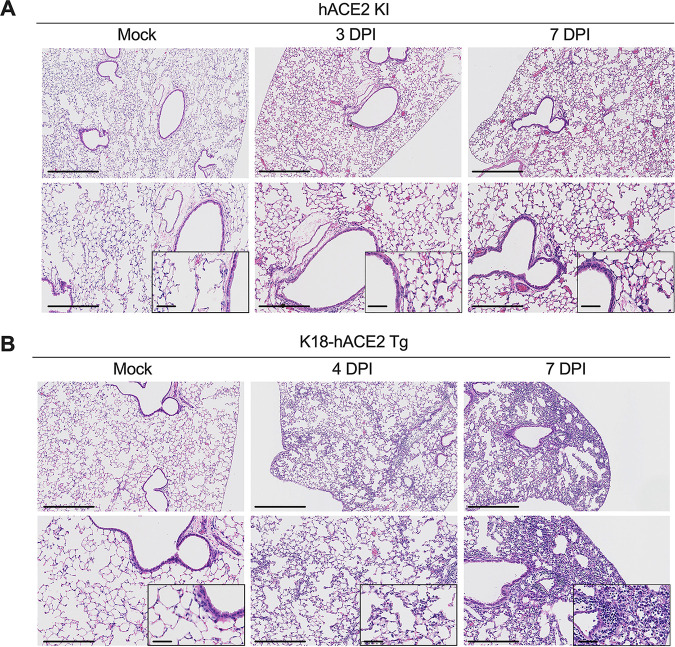
To assess histopathological and inflammatory changes in the lung following SARS-CoV-2 infection in the hACE2 KI model, we analyzed hematoxylin-and-eosin-stained sections from mock-infected animals or mice infected with WA1/2020 at 3 and 7 dpi (Fig. 3A). Following SARS-CoV-2 infection, a mild increase in immune cells and erythrocytes in the alveolar spaces was associated with vascular congestion. However, the airway architecture was largely preserved, consistent with a study reporting minimal pathology in the lungs of heterozygous hACE2 KI mice following infection with SARS-CoV-2 (9). In contrast, infection of K18-hACE2 Tg showed severe and progressive inflammation with abundant immune cells in the alveolar and interstitial locations of the lung parenchyma that was accompanied by edema, fibrin deposition, and lung consolidation (Fig. 3B).
FIG 3.
Histopathological analysis of SARS-CoV-2 infection in hACE2 KI and K18-hACE2 mice. (A) Hematoxylin-and-eosin staining of lung sections from hACE2 KI mice following mock infection or after intranasal inoculation with 105 FFU of WA1/2020 at 3 and 7 dpi. (B) Hematoxylin-and-eosin staining of lung sections from K18-hACE2 mice following mock infection or after intranasal inoculation with 2.5 × 104 PFU of SARS-CoV-2 WA1/2020 at 4 and 7 dpi. Images show low-power (top; bars, 500 μm), medium-power (bottom; bars, 250 μm), and high-power (insets in the bottom panels; bars, 100 μm) magnification. Images are representative of n = 4 per group.
An excessive proinflammatory host response to SARS-CoV-2 infection contributes to pulmonary disease, and distinct cytokine profiles correlate with coronavirus disease 2019 (COVID-19) severity (15–17). Consistent with the mild lung pathology in the hACE2 KI model, relatively few of the proinflammatory cytokines and chemokines (e.g., eotaxin, interleukin 2 [IL-2], IL-15, CXC chemokine ligand 9 [CXCL9], CXCL10, CC chemokine ligand 3 [CCL3], CCL4, tumor necrosis factor alpha [TNF-α], and vascular endothelial growth factor [VEGF]) tested showed statistically significant increases at either 3 or 7 days after SARS-CoV-2 infection (Table 1). Thus, despite sustained SARS-CoV-2 replication in the lungs of hACE2 KI mice, a more limited inflammatory response accompanies infection compared to that seen previously in K18-hACE2 mice (1). Because SARS-CoV-2 infection in hACE2 KI mice is largely restricted to the upper airways (Fig. 2F), differences in inflammation and disease severity between the K18-hACE2 and hACE2 KI mice may be due to model-dependent expression of hACE2 and infection tropism.
TABLE 1.
Cytokine and chemokine analysis following SARS-CoV-2 infection in the lungs of hACE2 KI mice
| Cytokine or chemokine |
Cytokine or chemokine protein levela |
|||||||
|---|---|---|---|---|---|---|---|---|
| Mock |
3 dpi |
7 dpi |
||||||
| Mean (pg/mL) |
SD | Mean (pg/mL) |
SD | Significance compared to mock |
Mean (pg/mL) |
SD | Significance compared to mock |
|
| Eotaxin | 59.9 | 1.6 | 224.3 | 41.1 | *** | 240.4 | 10.9 | *** |
| G-CSF | 1.0 | 0.2 | 1.7 | 1.0 | ns | 0.9 | 0.1 | ns |
| GM-CSF | 5.3 | 0.1 | 9.1 | 5.5 | ns | 4.1 | 0.9 | ns |
| IFN-γ | 2.0 | 0.1 | 3.0 | 0.9 | ns | 2.4 | 0.8 | ns |
| IL-1α | 63.0 | 1.8 | 83.8 | 23.9 | ns | 80.3 | 56.5 | ns |
| IL-1β | 1.1 | 0.1 | 1.8 | 0.5 | ns | 0.8 | 0.1 | ns |
| IL-2 | 14.8 | 1.1 | 3.7 | 1.6 | ** | 6.6 | 3.9 | * |
| IL-3 | 0.3 | 0.0 | 1.6 | 1.4 | ns | 0.3 | 0.1 | ns |
| IL-4 | 0.1 | 0.0 | 1.9 | 2.2 | ns | 0.2 | 0.0 | ns |
| IL-5 | 0.5 | 0.0 | 4.1 | 4.3 | ns | 0.3 | 0.1 | ns |
| IL-6 | 0.4 | 0.0 | 3.0 | 2.2 | ns | 0.8 | 0.1 | ns |
| IL-7 | 1.2 | 0.0 | 2.0 | 0.6 | ns | 1.7 | 0.2 | ns |
| IL-9 | 42.1 | 1.1 | 46.6 | 19.6 | ns | 43.2 | 5.8 | ns |
| IL-10 | 5.0 | 0.4 | 3.5 | 2.0 | ns | 2.9 | 1.2 | ns |
| IL-12p40 | 9.7 | 0.1 | 5.9 | 4.5 | ns | 7.7 | 4.1 | ns |
| IL-12p70 | ND | ND | 1.2 | 0.4 | NDt | 1.2 | ND | NDt |
| IL-13 | ND | ND | 0.8 | 0.6 | NDt | 0.6 | 0.1 | NDt |
| IL-15 | 4.7 | 0.1 | 16.2 | 6.9 | * | 8.6 | 2.6 | ns |
| IL-17 | 0.1 | 0.0 | 0.2 | 0.2 | ns | 0.1 | 0.0 | ns |
| CXCL10 | 15.5 | 0.9 | 176.5 | 53.5 | ** | 68.7 | 8.5 | ns |
| CXCL1 | 122.3 | 13.7 | 85.4 | 57.3 | ns | 94.2 | 66.6 | ns |
| LIF | 0.7 | 0.0 | 2.0 | 0.9 | ns | 1.1 | 0.2 | ns |
| LIX | 217.2 | 5.3 | 212.6 | 135.0 | ns | 239.4 | 91.4 | ns |
| CCL2 | 26.3 | 1.3 | 114.4 | 73.2 | ns | 48.3 | 5.8 | ns |
| M-CSF | 4.8 | 0.7 | 7.7 | 1.2 | ns | 6.2 | 1.0 | ns |
| CXCL9 | 2.9 | 0.7 | 456.2 | 685.6 | * | 164.2 | 34.3 | * |
| CCL3 | 594.9 | 24.6 | 79.4 | 30.4 | * | 349.1 | 240.8 | ns |
| CCL4 | 0.6 | 0.0 | 53.7 | 11.1 | *** | 27.2 | 7.5 | * |
| CXCL2 | 133.6 | 14.1 | 221.9 | 61.3 | ns | 147.3 | 9.7 | ns |
| CCL5 | 37.6 | 8.6 | 58.8 | 8.6 | ns | 86.0 | 16.8 | ns |
| TNF-α | 11.9 | 0.8 | 1.4 | 0.6 | *** | 2.6 | 2.5 | *** |
| VEGF | 79.3 | 4.3 | 192.1 | 29.8 | ns | 266.1 | 70.7 | ** |
Cytokine and chemokine protein levels in the lungs. n = 2 mock, n = 4 for all other groups. Statistical significance was measured by one-way ANOVA with Dunnett’s test comparing protein levels at day 3 or 7 after SARS-CoV-2 WA1/2020 infection to mock infection and is indicated as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05, ns, not significant. SD, standard deviation; ND, not detected; NDt, not determined.
Impact of the N501Y mutation on SARS-CoV-2 infection in mice.
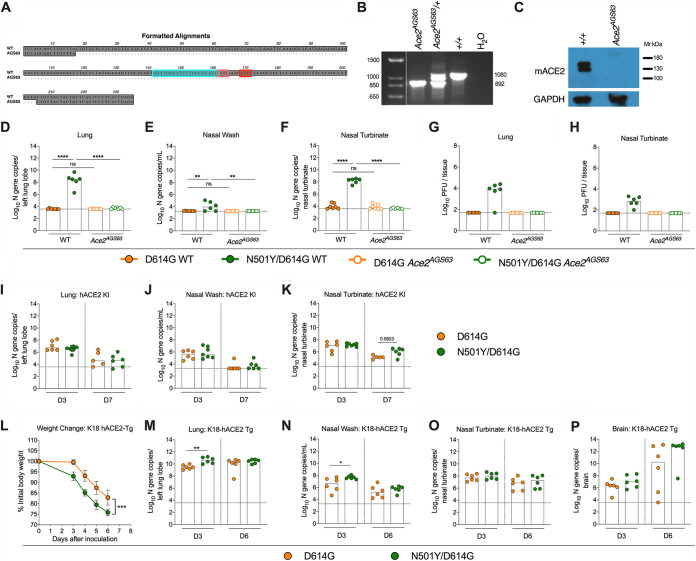
In addition to mouse models engineered to express hACE2, multiple groups have adapted SARS-CoV-2 strains that allow productive infection in mice by serial passage of the virus in vivo. Several key mutations that enable infection in laboratory strains of mice (e.g., BALB/c, 129S2, or C57BL/6) have been identified in mouse-adapted strains (18–23) including an N501Y mutation in the spike protein that is also present naturally in B.1.1.7, B.1.351, and P.1 SARS-CoV-2 VOC. Although it is presumed that the N501Y mutation directly mediates binding to mACE2 to facilitate productive infection in the mouse, this has not been definitively shown in vivo. Given that mouse-adapted strains have other mutations aside from the N501Y mutation, this substitution may not be fully responsible for adaptation or may facilitate viral replication through an alternative receptor. To dissect the requirement of the N501Y mutation for mACE2, we inoculated newly generated loss-of-expression Ace2AGS63 mice (Fig. 4A and B) or wild-type C57BL/6J controls with isogenic WA1/2020 SARS-CoV-2 strains containing the D614G mutation (WA1/2020 D614G) or both the D614G and N501Y mutation (WA1/2020 N501Y/D614G). Loss of the ACE2 protein was confirmed by Western blotting of the kidney homogenates from Ace2AGS63 mice (Fig. 4C). Inoculation of wild-type C57BL/6J mice with WA1/2020 N501Y/D614G but not WA1/2020 D614G resulted in robust infection as judged by viral RNA levels in the lungs, nasal washes, and nasal turbinates (Fig. 4D to F). In contrast, Ace2AGS63 mice inoculated via an intranasal route with WA1/2020 D614G or WA1/2020 N501Y/D614G did not support viral replication. Consistent with these results, infectious virus was recovered from the lungs and nasal turbinates of wild-type but not Ace2AGS63 C57BL/6J mice inoculated with WA1/2020 N501Y/D614G (Fig. 4G and H).
FIG 4.
Impact of the N501Y mutation on SARS-CoV-2 infection in K18-hACE2 and Ace2AGS63 mice. (A) Sequence alignment of wild-type (WT) C57BL/6 (top) and Ace2AGS63 (bottom) alleles, highlighting the guide sequence (cyan), protospacer adjacent motif (pink), and the start codon (red). Ace2AGS63 mice contain a 188-bp deletion surrounding the start codon in exon 2 of the Ace2 gene. (B) Amplification of genomic DNA from WT (+/+), heterozygote, and Ace2AGS63 homozygous mice. (C) Western blot analysis of ACE2 protein expression in the kidneys of WT and Ace2AGS63 mice. (D to F) Seven-week-old male and female ACE2-deficient (Ace2AGS63) mice or wild-type C57BL/6J controls were inoculated intranasally with 105 FFU of WA1/2020 D614G or WA1/2020 D614G/N501Y strains. Tissues were collected at 3 dpi. Viral RNA levels in the lung (D), nasal wash (E), and nasal turbinate (F) were measured (n = 6 or 7, two experiments, one-way ANOVA with multiple comparisons between the indicated groups: *, P < 0.05, **, P < 0.01). (G and H) Infectious virus levels in the lung (G) and nasal turbinate (H) were measured at 3 dpi by plaque assay on Vero-E6 cells (n = 6 or 7, two experiments). (I to K) Six-week-old male and female hACE2 KI mice were inoculated intranasally with 105 FFU of WA1/2020 D614G or WA1/2020 D614G/N501Y strain. Tissues were collected at 3 and 7 dpi. Viral RNA levels in the lung (I), nasal wash (J), and nasal turbinate (K) were measured (n = 7 or 8, two experiments, Mann-Whitney test between the indicated groups). (L to P) Seven-week-old female K18-hACE2 mice were inoculated via the intranasal route with 105 FFU of WA1/2020 D614G or WA1/2020 D614G/N501Y strain. Tissues were collected at 3 and 6 dpi. (L) Weight change following SARS-CoV-2 infection (mean ± SEM; n = 6 mice per group, two experiments; unpaired t test of area under the curve: ****, P < 0.0001). Viral RNA levels in the lung (M), nasal wash (N), nasal turbinate (O), and brain (P) were measured (n = 6, two experiments, Mann-Whitney test between the indicated groups: *, P < 0.05, **, P < 0.01). Dotted line indicates the limit of detection of the assay. ns, not significant.
As the N501Y substitution increases the affinity of spike protein binding to ACE2 and confers a replication advantage in hamsters and primary human airway epithelial cells (24–26), we determined its impact in hACE2 KI mice by inoculating 6-week-old female animals via the intranasal route with either WA1/2020 D614G or WA1/2020 N501Y/D614G. Viral titers in the lungs and nasal washes generally were similar at 3 and 7 dpi between mice inoculated with the two viruses (Fig. 4I to K). Nonetheless, we did observe a trend toward an increase in viral RNA levels at 7 dpi in the nasal turbinates of mice inoculated with WA1/2020 N501Y/D614G compared to WA1/2020 D614G. Future in vivo competition or transmission studies may be needed to distinguish the impact of the N501Y mutation in hACE2 KI mice.
In addition to expression of hACE2 as a transgene under the cytokeratin promoter, K18-hACE2 mice also express mACE2 from its endogenous promoter. Accordingly, we hypothesized that infection of K18-hACE2 mice with viruses containing the N501Y adaptive mutation might enhance pathogenesis through utilization of both mACE2 and hACE2 receptor. K18-hACE2 mice infected with WA1/2020 N501Y/D614G lost weight more rapidly and sustained higher viral titers in the lung and nasal washes at 3 dpi, but not in the brain or nasal turbinates, compared to infection with the WA1/2020 D614G virus (Fig. 4L to P). The increased infectivity and pathogenicity of WA1/2020 N501Y/D614G in K18-hACE2 mice could be due to enhanced binding to hACE2 independent of adaptation to mACE2. However, differences in infectivity between WA1/2020 N501Y/D614G and WA1/2020 D614G in hACE2 KI mice, which lack mACE2 expression entirely, were minimal, which suggests that enhanced interaction with hACE2 is not the principal explanation. Regardless, our experiments establish an in vivo requirement for mACE2 in the adaptation of SARS-CoV-2 strains containing the N501Y mutation to mice.
DISCUSSION
The rapid generation of small animal models has been instrumental in testing antiviral countermeasures as well as furthering our understanding of SARS-CoV-2 pathogenesis. However, at present, no animal model fully recapitulates all aspects of COVID-19, and the continued development of models is necessary to address the diverse spectrum of biology and pathophysiology encompassed by SARS-CoV-2 infection. In this study, we evaluated the homozygous hACE2 KI mouse as a model of SARS-CoV-2 infection in comparison to the K18-hACE2 Tg model. Both models supported viral infection of the upper and lower respiratory tracts with roughly equivalent viral titers in the nasal turbinates. However, K18-hACE2 Tg mice had substantially higher viral titers (∼1,000-fold) in the lung compared to the hACE2 KI model with greater infection of the alveolar pneumocytes that was not apparent in the hACE2 KI model where infection appeared restricted to epithelial cells of the larger airways. Whereas infection of K18-hACE2 Tg mice is accompanied by profound lung pathology, significant weight loss, viral spread to extrapulmonary organs, and mortality (1, 27–29), infection of hACE2 KI mice showed more limited lung inflammation without overt clinical disease or substantial viral spread beyond the respiratory tract. Thus, although both models rely upon hACE2 expression to sustain productive SARS-CoV-2 infection, our study highlights the differences in outcome and infection tropism, which likely reflects the site and amount of hACE2 expression. Consistent with this idea, two additional models of SARS-CoV-2 infection and pathogenesis in mice have been generated by using adenovirus (Adv) (30, 31) or adeno-associated virus (AAV) (32) to transduce cells in the respiratory tract with hACE2. Mice transduced with Adv or AAV encoding hACE2 support SARS-CoVo2 replication in the lung and show evidence of moderate to severe pulmonary pathology but do not develop mortality seen in K18-hACE2 Tg mice, possibly because of the more restricted hACE2 receptor expression in the respiratory tract.
Our finding that homozygous hACE2 KI mice supported SARS-CoV-2 infection in the upper and lower respiratory tracts with minimal disease or lung pathology is generally consistent with prior reports with heterozygous hACE2 KI mice (9) or in a different hACE2 KI mouse strain (31). However, we detected little, if any, viral RNA in the brains of hACE2 KI mice, which contrasts with a previous report following intranasal inoculation (31); the differences in results could be explained by the differential presence of furin cleavage site mutations in viral stocks (33), subtle variation between the two hACE2 KI mouse strains, or animal facility differences that impact the microbiome and gastrointestinal tract effects on the blood-brain barrier (34). In the lungs of hACE2 KI mice, viral infection was restricted to cells of the larger airways with no apparent infection in the alveoli of the lung. Although we were unable to identify hACE2-expressing cells in the lungs using immunohistochemistry or in situ hybridization, knock-in mice that coexpressed tdTomato and hACE2 reported hACE2 expression on club cells of the bronchiolar epithelium but not in the more abundant type I alveolar pneumocytes (31).
Based on human transcriptomic and proteomic studies, ACE2 is expressed at low levels in the respiratory tract (10, 11, 35). Indeed, single cell RNA sequencing studies estimated that only 1% to 6% of cells express ACE2 in the respiratory tract, principally in airway epithelial cells, type II pneumocytes, and nasal goblet cells (13, 36, 37). A large meta-analysis demonstrated an increase in the expression of ACE2 and the entry factor protease TMPRSS2 with age, in men, and with smoking (38), which correlated with findings in mice showing increased ACE2 expression with age and exposure to cigarette smoke and vaping (39, 40). Because infection is confined largely to the airway and not lung parenchyma, the hACE2 KI mouse could provide a model of self-limited infection, mild COVID-19 disease, and possibly transmission. Moreover, because these mice are on a C57BL/6 background, they can be crossed to other transgenic mice or combined with treatments to explore how specific immune functions or risk factors (e.g., diabetes, age, or obesity) modulate SARS-CoV-2 pathogenesis in the setting of physiological hACE2 expression.
We also determined the impact of the N501Y mutation in multiple murine models. The adaptation of SARS-CoV-2 through serial passage in vivo has established key mutations that enable infection in mice. All SARS-CoV-2 strains that are adapted to mice contain mutations in the spike protein (20, 22, 41), and many encode an N501Y mutation (18, 21, 23) that is also present naturally in B.1.1.7, B.1.351, and P.1 SARS-CoV-2 VOC. While biochemical data and experiments in cell lines demonstrated increased binding affinity and infectivity of the N501Y spike for mouse ACE2 (23, 25, 42), it had not yet been demonstrated in vivo that the N501Y mutation alone is sufficient to promote infection in mice since all mouse-adapted strains contain additional mutations. Introduction of the N501Y mutation into the WA1/2020 D614G SARS-CoV-2 backbone was sufficient to enable viral replication in wild-type C57BL/6 mice in a mACE2-dependent manner. Notwithstanding this point, it is likely that additional mutations present in mouse-adapted SARS-CoV-2 strains enhance pathogenesis beyond that observed with the WA1/2020 N501Y virus alone.
The enhanced SARS-CoV-2 pathogenesis in K18-hACE2 Tg mice that is seen with viruses containing N501Y mutations could be due to either the ability of the virus to engage both hACE2 and mACE2 or increased binding to hACE2 (24, 43). As the N501Y mutation had a minimal impact in the hACE2 KI model, which does not express mACE2, following infection with WA1/2020 N501Y/D614G virus or the N501Y-containing VOC B.1.1.7 and B.1.351, it appears that an enhanced interaction with hACE2 is not the principal explanation. Although multiple studies have highlighted the increased affinity of the N501Y spike for hACE2, conflicting reports exist as to whether this mutation translates to higher infectivity in vivo (24, 26). Future virus competition studies in vivo in hACE2 KI mice may be needed to fully determine the biological impact of the N501Y mutation on pathogenesis.
MATERIALS AND METHODS
Cells.
Vero-TMPRSS2 cells (44) were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.3), 1 mM sodium pyruvate, 1× nonessential amino acids, and 100 U/ml of penicillin-streptomycin. Vero-TMPRSS2 cells were supplemented with 5 μg/mL of blasticidin.
Viruses.
The WA1/2020 recombinant strain with substitutions (D614G or N501Y/D614G) were obtained from an infectious cDNA clone of the 2019n-CoV/USA_WA1/2020 strain and described previously (45). The WA1/2020, B.1.1.7, and B.1.351 SARS-CoV-2 isolates were obtained from nasopharyngeal isolates and have been reported previously (3, 30, 46). All viruses were passaged once in Vero-TMPRSS2 cells and subjected to next-generation sequencing as described previously (46) to confirm the introduction and stability of substitutions.
Biosafety.
This study was approved by the office of Environmental Health and Safety at Washington University School of Medicine and the Icahn School of Medicine at Mount Sinai prior to the initiation of experiments. All SARS-CoV-2 experiments were performed in approved biosafety level 3 (BSL-3) facilities by personnel equipped with powered air-purifying respirators.
Mice.
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (47). The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Heterozygous K18-hACE2 C57BL/6J mice [strain 2B6.Cg-Tg(K18-ACE2)2Prlmn/J], hACE2 KI mice [B6.129S2(Cg)-Ace2tm1(ACE2)Dwnt/J], and wild-type (WT) C57BL/6 mice were obtained from The Jackson Laboratory. Ace2AGS63 loss-of-function mice were generated by injecting a ribonucleoprotein (RNP) complex of 20 ng/µL Alt-R CRISPR-Cas9 single guide RNA (sgRNA), guide sequence GGATGGGATCTTGGCGCACG, PAM: GGG (IDT) and 20 ng/µL Cas9 protein (IDT, Alt-R S.p.-Cas9 nuclease-V3, catalog number 1081058) into the male pronuclei of one-cell stage C57BL/6J mouse embryos. After injection, the embryos were returned to the oviducts of pseudopregnant Swiss-Webster females that had been mated the day before with vasectomized Swiss-Webster males. Resulting pups were characterized by deep sequencing and PCR. Mice were genotyped by PCR using the primers GAGGGAGAGGATGGATAGCTT-5′F and TGCTGCCTTCAGTAAACCCC-3′R and the PCR conditions 30 s at 95°C, 30 s at 55°C, and 4 min at 72°C. To confirm loss of the ACE2 protein, kidneys collected from adult WT C57BL/6 and Ace2AGS63 mice were homogenized (MP FastPrep-24, 6 m/s, 30 s) in radioimmunoprecipitation assay (RIPA) buffer (Boston BioProducts) supplemented with Complete Protease Inhibitor Cocktail (Roche). Following protein quantification by Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific), 80 μg of total protein was loaded into 10% Mini-Protean TGX Precast Protein Gels (Bio-Rad), electrophoretically separated, transferred to 0.2-μm polyvinylidene difluoride (PVDF) membranes (Millipore Sigma) using wet electroblotting, and blocked in 5% nonfat powdered milk (Boston BioProducts) in Tris-buffered saline (TBS) containing 0.1% Tween 20 (Fisher Bioreagents). Membranes were blotted against mACE2 (R&D Systems, MAB3437, clone 460502) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling Technology, catalog number 88845) and visualized using a horseradish peroxidase (HRP)-conjugated anti-rat IgG (Scientific Laboratory Supplies, code NA935). KwikQuant Western Blot Detection kit (Kindle Biosciences, catalog number R1004) was used for detection.
Animals were housed in groups and fed standard chow diets. Six- to eight-week-old male and female mice were either administered 2.5 × 104 PFU of SARS-CoV-2 (K18-hACE2) or 105 FFU of SARS-CoV-2 (hACE2 KI, Ace2−/−, and WT C57BL/6) via intranasal administration while under anesthesia induced by ketamine and xylazine.
Focus-forming assay.
Serial dilutions of viral stocks were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (wt/vol) methylcellulose in minimum essential medium (MEM) supplemented with 2% FBS. Plates were harvested 30 h later by removing overlays and fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57, and SARS2-71 (48) anti-S antibodies and HRP-conjugated goat anti-mouse IgG (Sigma, catalog number 12-349) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Measurement of viral burden and hACE2 expression.
Tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1 ml of DMEM medium supplemented with 2% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using the MagMax mirVana Total RNA isolation kit (Thermo Scientific) on a Kingfisher Flex extraction robot (Thermo Scientific). RNA was reverse transcribed and amplified using the TaqMan RNA-to-CT 1-Step kit (ThermoFisher). Reverse transcription was carried out at 48°C for 15 min followed by 2 min at 95°C. Amplification was accomplished over 50 cycles as follows: 95°C for 15 s and 60°C for 1 min. The number of copies of SARS-CoV-2 N-gene RNA in samples was determined using a previously published assay (49). Briefly, a TaqMan assay was designed to target a highly conserved region of the N gene (forward primer, ATGCTGCAATCGTGCTACAA; reverse primer, GACTGCCGCCTCTGCTC; probe, 5,6-carboxyfluorescein [56-FAM]/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/). This region was included in an RNA standard to allow for copy number determination to 10 copies per reaction. The reaction mixture contained final concentrations of primers and probe of 500 and 100 nM, respectively.
Vero-ACE2-TMPRSS2 (50) or Vero-E6 cells were seeded at a density of 2.5 × 105 cells per well in flat-bottom 12-well tissue culture plates. The following day, medium was removed and replaced with 200 μL of 10-fold serial dilutions of clarified tissue homogenates diluted in DMEM supplemented with 2% FBS. One hour later, 1 mL of methylcellulose overlay was added. Plates were incubated for 72 h and then fixed with 4% paraformaldehyde (final concentration) in phosphate-buffered saline for 20 min. Plates were stained with 0.05% (wt/vol) crystal violet in 20% methanol and washed twice with distilled, deionized water before scoring.
For hACE2 expression, RNA was treated with DNase (Thermo Scientific) following the manufacturer’s protocol. RNA levels were quantified as described above with the primer/probe set for hACE2 (IDT assay: Hs.PT.58.27645939), compared to an RNA standard curve, and normalized per milligram of tissue.
Cytokine and chemokine protein measurements.
Lung homogenates were incubated with Triton X-100 (1% final concentration) for 1 h at room temperature to inactivate SARS-CoV-2. Homogenates then were analyzed for cytokines and chemokines by Eve Technologies Corporation (Calgary, AB, Canada) using their Mouse Cytokine Array/Chemokine Array 44-Plex (MD44) platform.
Histology and RNA in situ hybridization.
Animals were euthanized before harvest and fixation of tissues. The left lung was tied off at the left main bronchus and collected for viral RNA analysis. The right lung was then inflated with ∼1.2 mL of 10% neutral buffered formalin using a 3-mL syringe and catheter inserted into the trachea. Tissues were embedded in paraffin, and sections were stained with hematoxylin and eosin. RNA in situ hybridization was performed using the RNAscope 2.5 HD Assay (Brown kit) according to the manufacturer’s instructions (Advanced Cell Diagnostics). Briefly, sections were deparaffinized and treated with H2O2 and Protease Plus before probe hybridization. Probes specifically targeting hACE2 (catalog number 848151) or SARS-CoV-2 spike sequence (catalog number 848561) were hybridized followed by proprietary signal amplification and detection with 3,3′-diaminobenzidine. Tissues were counterstained with Gill’s hematoxylin. An uninfected mouse was used as a negative control and stained in parallel. Images were captured using a Nanozoomer (Hamamatsu) instrument at the Alafi Neuroimaging Core at Washington University.
Statistical analysis.
Statistical significance was assigned when P values were <0.05 using Prism version 8 (GraphPad). Specific tests and multiple comparison corrections are indicated in the figure legends.
Data availability.
All data supporting the findings of this study are found within the paper and are available from the corresponding author upon request.
ACKNOWLEDGMENTS
This study was supported by the NIH (R01 AI157155, R35-HL145242, and R01 AI130591) and the Defense Advanced Research Project Agency (HR001117S0019 and HR0011-507 19-2-0020). This study was also partly funded by CRIPT (Center for Research on Influenza Pathogenesis and Transmission), a NIAID-funded Center of Excellence for Influenza Research and Response (CEIRR, contract 75N93021C00014), by a Fast Grant of the Mercatus Center, and by the generous support of the JPB Foundation, the Open Philanthropy Project (research grant 2020-215611 [5384]), and anonymous donors to A.G.-S. E.S.W. is supported by F30 AI152327. J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. S.Y. received funding from Swiss National Foundation (SNF) Postdoc Mobility fellowship (P400PB_199292). M.J.H. is supported by the Department of Defense (PR190726), the Cystic Fibrosis Foundation, Bebermeyer Fund, Hardy Trust, and Schaefer Fund.
We gratefully acknowledge the support of Cathleen (Cat) Lutz and Jackson Laboratories for supplying the hACE2 KI mice and Pei Yong Shi for providing the recombinant D614G and N501Y/D614G WA1/2020 viruses. We also thank Randy Albrecht and Richard Cadagan for support with the BSL-3 facility and procedures and for technical assistance, respectively, at Icahn School of Medicine at Mount Sinai (ISMMS).
E.S.W., R.E.C., J.B.C., M.S., S.Y., and F.A. performed mouse experiments. E.S.W. and S.Y. analyzed viral burden. M.J.H. performed histological analysis of lung sections. M.B.U. generated the Ace2AGS63 mice. E.S.W. and M.S.D. wrote the initial draft, with the other authors providing editorial comments.
M.S.D. is a consultant for Inbios, Vir Biotechnology, and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. M.J.H. is the founder and equity-holder in NuPeak Therapeutics and is a member of the Data Safety Monitoring Board for AstraZeneca. The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, and Merck, all of which is unrelated to this study. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, and Pfizer. M.B.U. contributed to this article as an employee of Mount Sinai, and the views expressed do not necessarily represent the views of Regeneron Pharmaceuticals Inc.
Contributor Information
Michael S. Diamond, Email: mdiamond@wustl.edu.
Stacey Schultz-Cherry, St. Jude Children’s Research Hospital.
REFERENCES
- 1.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An D, Li K, Rowe DK, Diaz MCH, Griffin EF, Beavis AC, Johnson SK, Padykula I, Jones CA, Briggs K, Li G, Lin Y, Huang J, Mousa J, Brindley M, Sakamoto K, Meyerholz DK, McCray PB, Jr, Tompkins SM, He B. 2021. Protection of K18-hACE2 mice and ferrets against SARS-CoV-2 challenge by a single-dose mucosal immunization with a parainfluenza virus 5-based COVID-19 vaccine. Sci Adv 7:eabi5246. doi: 10.1126/sciadv.abi5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen RE, Winkler ES, Case JB, Aziati ID, Bricker TL, Joshi A, Darling TL, Ying B, Errico JM, Shrihari S, VanBlargan LA, Xie X, Gilchuk P, Zost SJ, Droit L, Liu Z, Stumpf S, Wang D, Handley SA, Stine WB, Jr, Shi PY, Davis-Gardner ME, Suthar MS, Knight MG, Andino R, Chiu CY, Ellebedy AH, Fremont DH, Whelan SPJ, Crowe JE, Jr, Purcell L, Corti D, Boon ACM, Diamond MS. 2021. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassan AO, Shrihari S, Gorman MJ, Ying B, Yuan D, Raju S, Chen RE, Dmitriev IP, Kashentseva E, Adams LJ, Shi P-Y, Fremont DH, Curiel DT, Alter G, Diamond MS. 2021. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. bioRxiv 10.1101/2021.05.08.443267. [DOI] [PMC free article] [PubMed]
- 5.Maiese A, Manetti AC, Bosetti C, Del Duca F, La Russa R, Frati P, Di Paolo M, Turillazzi E, Fineschi V. 2021. SARS-CoV-2 and the brain: a review of the current knowledge on neuropathology in COVID-19. Brain Pathol doi: 10.1111/bpa.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai-Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori SI, Takeda M, Mitsuya H, Krammer F, Suzuki T, Kawaoka Y. 2020. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E. 2020. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, Fentener van Vlissingen M, Fouchier R, de Swart R, Koopmans M, Haagmans BL. 2020. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, Halwe NJ, Ulrich L, Trüeb BS, Fan X, Hoffmann B, Wang L, Thomann L, Lin X, Stalder H, Pozzi B, de Brot S, Jiang N, Cui D, Hossain J, Wilson MM, Keller MW, Stark TJ, Barnes JR, Dijkman R, Jores J, Benarafa C, Wentworth DE, Thiel V, Beer M. 2021. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- 10.Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, Chandiramohan A, Tiessen N, Cao Q, Dvorkin-Gheva A, Revill S, Miller MS, Carlsten C, Organ L, Joseph C, John A, Hanson P, Austin RC, McManus BM, Jenkins G, Mossman K, Ask K, Doxey AC, Hirota JA. 2020. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J 56:2001123. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. 2020. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16:e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. 2020. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH, II, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, et al. 2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181:1016–1035.e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brann DH, Tsukahara T, Weinreb C, Lipovsek M, Van den Berge K, Gong B, Chance R, Macaulay IC, Chou HJ, Fletcher RB, Das D, Street K, de Bezieux HR, Choi YG, Risso D, Dudoit S, Purdom E, Mill J, Hachem RA, Matsunami H, Logan DW, Goldstein BJ, Grubb MS, Ngai J, Datta SR. 2020. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci Adv 6:eabc5801. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, Manne S, Chen Z, Huang YJ, Reilly JP, Weisman AR, Ittner CAG, Kuthuru O, Dougherty J, Nzingha K, Han N, Kim J, Pattekar A, Goodwin EC, Anderson EM, Weirick ME, Gouma S, Arevalo CP, Bolton MJ, Chen F, Lacey SF, Ramage H, Cherry S, Hensley SE, Apostolidis SA, Huang AC, Vella LA, UPenn COVID Processing Unit, Betts MR, Meyer NJ, Wherry EJ. 2020. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IMPACT Team, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel TA, Merkling SH, Treluyer JM, Veyer D, Mouthon L, Blanc C, Tharaux PL, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathnasinghe R, Jangra S, Cupic A, Martínez-Romero C, Mulder LCF, Kehrer T, Yildiz S, Choi A, Mena I, De Vrieze J, Aslam S, Stadlbauer D, Meekins DA, McDowell CD, Balaraman V, Richt JA, De Geest BG, Miorin L, Krammer F, Simon V, García-Sastre A, Schotsaert M. 2021. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv doi: 10.1101/2021.01.19.21249592. [DOI] [Google Scholar]
- 19.Huang K, Zhang Y, Hui X, Zhao Y, Gong W, Wang T, Zhang S, Yang Y, Deng F, Zhang Q, Chen X, Yang Y, Sun X, Chen H, Tao YJ, Zou Z, Jin M. 2021. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leist SR, Dinnon KH, III, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183:1070–1085.e1012. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muruato A, Vu MN, Johnson BA, Davis-Gardner ME, Vanderheiden A, Lokugmage K, Schindewolf C, Crocquet-Valdes PA, Langsjoen RM, Plante JA, Plante KS, Weaver SC, Debbink K, Routh AL, Walker D, Suthar MS, Xie X, Shi PY, Menachery VD. 2021. Mouse adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. bioRxiv doi: 10.1101/2021.05.03.442357. [DOI] [PMC free article] [PubMed]
- 22.Dinnon KH, III, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li XF, Li J, Zhang NN, Yang X, Chen S, Guo Y, Zhao G, Wang X, Luo DY, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin CF, Zhou Y. 2020. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, Ku Z, An Z, Scharton D, Schindewolf C, Menachery VD, Shi PY, Weaver SC. 2021. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv 10.1101/2021.03.08.434499. [DOI] [PMC free article] [PubMed]
- 25.Niu Z, Zhang Z, Gao X, Du P, Lu J, Yan B, Wang C, Zheng Y, Huang H, Sun Q. 2021. N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Signal Transduct Target Ther 6:284. doi: 10.1038/s41392-021-00704-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, Zhang Y, Li T, Liu S, Zhang M, Zhao C, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Liu J, Liang H, Shi Y, Shen Y, Xie L, Zhang L, Qu X, Xu W, Huang W, Wang Y. 2021. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 184:2362–2371.e2369. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, II, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Jr, Dick EJ, Jr, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden JW, Cline CR, Zeng X, Garrison AR, Carey BD, Mucker EM, White LE, Shamblin JD, Brocato RL, Liu J, Babka AM, Rauch HB, Smith JM, Hollidge BS, Fitzpatrick C, Badger CV, Hooper JW. 2020. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight 5:e142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Jr, Perlman S. 2021. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Jr, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS. 2020. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124−133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, Ring A, Wilen CB, Iwasaki A. 2020. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, Zhang L, Bopp N, Schindewolf C, Vu M, Vanderheiden A, Winkler ES, Swetnam D, Plante JA, Aguilar P, Plante KS, Popov V, Lee B, Weaver SC, Suthar MS, Routh AL, Ren P, Ku Z, An Z, Debbink K, Diamond MS, Shi PY, Freiberg AN, Menachery VD. 2021. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang W, Zhu H, Feng Y, Guo R, Wan D. 2020. The impact of gut microbiota disorders on the blood-brain barrier. Infect Drug Resist 13:3351–3363. doi: 10.2147/IDR.S254403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, Bolling MC, Dijkstra G, Voors AA, Osterhaus AD, van der Voort PH, Mulder DJ, van Goor H. 2020. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J Pathol 251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, HCA Lung Biological Network. 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. 2020. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 38.Muus C, Luecken MD, Eraslan G, Sikkema L, Waghray A, Heimberg G, Kobayashi Y, Vaishnav ED, Subramanian A, Smillie C, Jagadeesh KA, Duong ET, Fiskin E, Triglia ET, Ansari M, Cai P, Lin B, Buchanan J, Chen S, Shu J, Haber AL, Chung H, Montoro DT, Adams T, Aliee H, Allon SJ, Andrusivova Z, Angelidis I, Ashenberg O, Bassler K, Bécavin C, Benhar I, Bergenstråhle J, Bergenstråhle L, Bolt L, Braun E, Bui LT, Callori S, Chaffin M, Chichelnitskiy E, Chiou J, Conlon TM, Cuoco MS, Cuomo ASE, Deprez M, Duclos G, Fine D, Fischer DS, Ghazanfar S, Gillich A, Giotti B, et al. 2021. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med 27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naidu V, Zeki AA, Sharma P. 2021. Sex differences in the induction of angiotensin converting enzyme 2 (ACE-2) in mouse lungs after e-cigarette vapor exposure and its relevance to COVID-19. J Invest Med 69:954–961. doi: 10.1136/jim-2020-001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masso-Silva JA, Moshensky A, Shin J, Olay J, Nilaad S, Advani I, Bojanowski CM, Crotty S, Li WT, Ongkeko WM, Singla S, Crotty Alexander LE. 2021. Chronic E-cigarette aerosol inhalation alters the immune state of the lungs and increases ACE2 expression, raising concern for altered response and susceptibility to SARS-CoV-2. Front Physiol 12:649604. doi: 10.3389/fphys.2021.649604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montagutelli X, Prot M, Jouvion G, Levillayer L, Conquet L, Reyes-Gomez E, Donati F, Albert M, van der Werf S, Jaubert J, Simon-Lorière E. 2021. A mouse-adapted SARS-CoV-2 strain replicating in standard laboratory mice. bioRxiv 10.1101/2021.07.10.451880. [DOI]
- 42.Sun S, Gu H, Cao L, Chen Q, Ye Q, Yang G, Li R-T, Fan H, Deng Y-Q, Song X, Qi Y, Li M, Lan J, Feng R, Guo Y, Zhu N, Qin S, Wang L, Zhang Y-F, Zhou C, Zhao L, Chen Y, Shen M, Cui Y, Yang X, Wang X, Tan W, Wang H, Wang X, Qin C-F. 2021. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. bioRxiv 10.1101/2020.11.10.377333. [DOI] [PMC free article] [PubMed]
- 43.Tian F, Tong B, Sun L, Shi S, Zheng B, Wang Z, Dong X, Zheng P. 2021. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife 10:e69091. doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. 2020. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O’Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE, Corti D, Virgin HW, Ellebedy AH, Shi P-Y, Diamond MS. 2021. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 48.Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ. 2021. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29:477–488.e474. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case JB, Bailey AL, Kim AS, Chen RE, Diamond MS. 2020. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanBlargan LA, Adams LJ, Liu Z, Chen RE, Gilchuk P, Raju S, Smith BK, Zhao H, Case JB, Winkler ES, Whitener BM, Droit L, Aziati ID, Bricker TL, Joshi A, Shi PY, Creanga A, Pegu A, Handley SA, Wang D, Boon ACM, Crowe JE, Jr, Whelan SPJ, Fremont DH, Diamond MS. 2021. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity 54:2399–2416.e6. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are found within the paper and are available from the corresponding author upon request.