ABSTRACT
Long noncoding RNAs (lncRNAs) function as microregulatory factors that influence gene expression after a variety of pathogenic infections, and they have been extensively studied in the past few years. Although less attention has been paid to lncRNAs in lower vertebrates than in mammals, current studies reveals that lncRNAs play a vital role in fish stimulated by pathogens. Here, we discovered a new lncRNA, termed MIR2187HG, which can function as a precursor of a small RNA, miR-2187-3p, with regulatory functions in the miiuy croaker (Miichthys miiuy). Upon Siniperca chuatsi rhabdovirus (SCRV) virus infection, the expression levels of MIR2187HG were remarkably enhanced. Elevated MIR2187HG expression can act as a pivotally negative regulator that participates in the innate immune response of teleost fish to inhibit the intracellular TANK-binding kinase 1 (TBK1)-mediated antiviral signaling pathways, which can effectively avoid excessive immunity. In addition, we found that SCRV could also utilize MIR2187HG to enhance its own numbers. Our results not only provide evidence regarding the involvement of the lncRNAs in response to viruses in fish but also broaden our understanding of the function of lncRNAs as precursor microRNAs (miRNAs) in teleost fish for the first time.
IMPORTANCE SCRV infection upregulates MIR2187HG levels, which in turn suppresses SCRV-triggered type I interferon production, thus promoting viral replication in miiuy croaker. Notably, MIR2187HG regulates the release of miR-2187-3p, and TBK1 is a target of miR-2187-3p. MIR2187HG could acquire from miR-2187-3p the function of inhibiting TBK1 expression and subsequently modulate TBK1-mediated NF-κB and IRF3 signaling. The collective results suggest that the novel regulation mechanism of TBK1-mediated antiviral response during RNA viral infection was regulated by MIR2187HG. Therefore, a new regulation mechanism for lncRNAs to regulate antiviral immune responses in fish is proposed.
KEYWORDS: long noncoding RNA, TBK1, fish, immune regulation, microRNA
INTRODUCTION
Viruses parasitize the host cell through a large number of replications after infecting the organism. Then, the host resists the invasion of the virus for survival through receptor recognition and immune response. Each pathogen has specific components that are recognized by pattern recognition receptors (PRRs). Among the PRRs that recognize pathogen-associated molecular patterns (PAMPs), the Toll-like receptor (TLR) family and retinoic acid-inducible gene I-like receptor (RLR) family have received considerable attention due to their particular identification of pathogens. In the TLR family of teleost fish, TLR1, TLR2, TLR5, and TLR9 are putative sensors for bacterial ligands in innate immunity, and TLR3 and TLR22 act as putative sensors for viral ligands, which is slightly different from TLRs in humans (1). TLR22 is a nonmammalian TLR that mainly exists in teleost fish, and it responds to extracellular double-stranded (dsRNA) viruses (2). The RLR family mainly includes retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5, and laboratory of genetics and physiology 2, and they all have a special DExD/H-box RNA helicase domain in their structure, which is involved in the binding of viral RNA (3). During the virus-triggered innate immune response, these virus-specific receptors can act as viral RNA sensors and initiate a series of signaling cascades. Upon virus invasion, receptors are urgently mobilized. Then, TANK-binding kinase 1 (TBK1), a downstream nodal protein, is activated and induces the activation of interferon regulatory factor 3 (IRF3) and NF-κB, ultimately leading to an interferon (IFN) antiviral immune response and inhibiting viral replication (4–6). However, excessive activation of innate immune response signals mediated by TBK1 will disrupt host immune homeostasis and may even cause chronic autoimmune diseases. Therefore, extensive research is needed on how to regulate the TBK1-mediated antiviral immune response appropriately.
TBK1 is a vital immune-related kinase that participates in the production of type I IFN to prevent the invasion of pathogenic microorganisms. Thus, TBK1 activation should be tightly regulated to trigger the proper type I IFN production and maintain immune homeostasis. TBK1 can be regulated in multiple ways, including acetylation (7) and ubiquitination (8). In mammals, several regulatory factors regulate the activity of TBK1 in innate immune responses. For example, Mindbomb 2 modulates TBK1 ubiquitination and activation upon RNA virus infection (9). RNF128 functions as a regulator of TBK1 ubiquitination and downstream type I IFN expression of innate signaling induced by different stimuli (10). TBK1 has been reported in several species of teleost fish. For example, zebrafish FGFR3 negatively regulates the production of IFN in the innate immune response by inhibiting the activity of TBK1 during Spring viraemia of carp virus (SVCV) infection (11). Additionally, fish TBK1 has been revealed to be regulated by viruses, which could inhibit the host type I IFN responses and thus facilitate viral replication (12). Given that TBK1 is the core kinase that mediates IFN production in fish, the regulatory mechanism of the TBK1 antiviral pathway in fish should be elucidated.
MicroRNAs (miRNAs), a set of noncoding single-stranded RNAs with a length of about 20 to 24 nucleotides, function as important regulatory factors involved in gene expression at the posttranscriptional level. miRNAs mainly regulate gene expression through bases complementary to the seed sequence of the 3′ untranslated region (UTR) of the targeted gene (13). Approximately half of miRNAs are embedded in the introns of protein-coding genes and are transcribed with the transcription of the encoded protein; these are classified as intronic miRNAs. These miRNAs are cleaved into 70- to 80-bp precursor miRNAs (pre-miRNAs) in the nucleus by the endonuclease Drosha, which are further processed in the cytoplasm to form mature miRNAs (14, 15). However, some miRNAs are embedded in long noncoding RNAs (lncRNAs) and transcribed. These lncRNAs can indirectly regulate mRNA expression by acting as a primary miRNA precursor with regulatory functions (16–18). For example, H19 provides miRNA-containing hairpins for miR-675-3p and miR-675-5p, and it may obtain different functions from these two miRNAs (19). Many studies on the involvement of miRNAs derived from lncRNAs in various biological processes have been conducted in humans. However, the regulation of innate antiviral responses in teleost fish is still not understood, so further research is needed.
lncRNAs are a class of functional noncoding transcripts with a length greater than 200 nucleotides which have attracted considerable attention due to their involvement in gene expression. lncRNAs are capable of regulating mRNAs through different biological mechanisms. Some lncRNAs act as competing endogenous RNAs (ceRNAs), regulating mRNA expression by competing with mRNA for miRNA (20, 21), while others act as host genes that harbor miRNAs, which are responsible for producing mature miRNAs and inhibiting the expression of target mRNA (22, 23). It has been widely reported that in humans, lncRNAs act as host genes involved in regulating miRNA targets. For instance, MIR503HG indirectly regulates Bcl-2 by promoting the cotranscription of miRNA-503 to participate in high-glucose-induced proximal tubular cell apoptosis, providing a new target for diabetic nephropathy treatment (24). The lncRNA H19 and its product miR-675 could repress prostate cancer metastasis by targeting TGFBI (25). However, whether this regulatory mechanism exists in teleost fish remains unknown.
As a lower vertebrate, fish are an excellent biological model to study the origin of innate immunity. Innate immune response is the most fundamental defense mechanism for fish to identify PAMPs and is of great significance in resistance to the invasion and transfer of pathogens (26). Siniperca chuatsi rhabdovirus (SCRV), as one of the most important pathogenic viruses, has caused serious economic losses to the aquaculture industry and even caused food safety problems (27). Therefore, the antiviral response mechanism of teleost fish infected by viruses needs to be investigated. Here, we identified a new immune-related lncRNA, MIR2187HG, that is abnormally highly expressed in the miiuy croaker upon SCRV infection. miR-2187-3p has been found to target TBK1 and inhibit TBK1-mediated antiviral responses in the miiuy croaker. We found that MIR2187HG can act as the host gene of miR-2187-3p to suppress the production of SCRV-triggered type I IFN and inflammatory cytokines by targeting TBK1, thereby facilitating SCRV replication. Importantly, MIR2187HG could modulate the antiviral innate immune response through the NF-κB and IRF3 signaling pathways. To the best of our knowledge, this study is the first to demonstrate that lncRNA functions as pre-miRNA involved in regulatory networks in teleost fish (28, 29), which provides a new strategy to avoid excessive immunization. Our data not only provide new insights into the understanding of the importance of lncRNAs in the antiviral response but also reveal the lncRNA-miRNA network in teleost fish.
RESULTS
Identification of MIR2187HG.
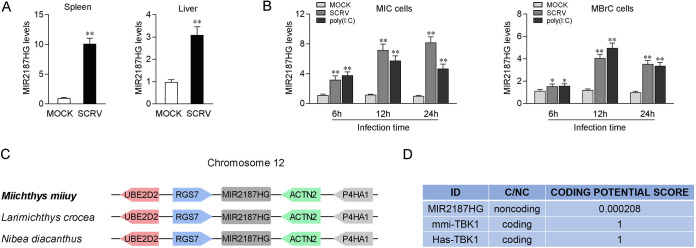
lncRNAs can play a regulatory role as miRNA host genes and affect the host’s immune signal pathway through an miRNA targeting mechanism (30). To investigate the function of lncRNAs in teleost fish antiviral immune responses, transcriptome sequencing (RNA-Seq) analysis was performed by comparing the differential expression of lncRNAs between the non-SCRV-treated group and the SCRV-treated group (GenBank accession number PRJNA685924), and it showed that there were 897 differentially expressed lncRNAs (21). We focused on MIR2187HG, which was significantly upregulated upon SCRV treatment. To further verify the RNA-Seq results regarding the significant abundance of MIR2187HG, quantitative real-time PCR (qPCR) analysis of MIR2187HG expression was performed in miiuy croaker SCRV-treated spleen and liver samples. The results showed that SCRV remarkably increases the expression of MIR2187HG, especially in spleen tissue (Fig. 1A). Then, we determined whether the virus-triggered upward trend of MIR2187HG existed in vitro. Miichthys miiuy intestinal cells (MICs) and M. miiuy brain cells (MBrCs) were treated with SCRV or poly(I·C) for 6, 12, and 24 h. Then, we measured the levels of MIR2187HG by qPCR. As expected, MIR2187HG was remarkably increased upon SCRV infection, and MIR2187HG in MICs was most highly upregulated, at 7- to 8-fold, compared with that in untreated cells (Fig. 1B). These results suggested that MIR2187HG is an immune-related lncRNA.
FIG 1.
Characterization of MIR2187HG. (A) Miiuy croakers were untreated or treated with SCRV. After 24 h infection, the expression levels of MIR2187HG in spleen (left) and liver (right) samples were measured by qPCR. (B) MICs (left) and MBrCs (right) were treated with SCRV or poly(I·C) at the indicated times, and then the expression levels of MIR2187HG were measured by qPCR. (C) Schematic of the MIR2187HG locus. MIR2187HG is located on miiuy croaker chromosome 12. (D) MIR2187HG was predicted to be a noncoding RNA. The RNA sequences of MIR2187HG were put into the Coding Potential Calculator (CPC) program, which predicted noncoding RNAs. mmi-TBK1, Miichthys miiuy TBK1 gene; Has-TBK1, Homo sapiens TBK1 gene. Data are means and standard errors (SE) from three independent triplicate experiments. **, P < 0.01; *, P < 0.05.
To characterize the complete sequence of MIR2187HG, single-molecule full-length transcript sequencing was conducted, and the results demonstrated that MIR2187HG had a length of 9,376 bp and was located on Miichthys miiuy, Larimichthys crocea, and Nibea diacanthus chromosome 12 between the genes RGS7 and ACTN2 (Fig. 1C) with a poly(A) tail (GenBank accession number MW771288). Furthermore, consistent with MIR2187HG being a noncoding RNA (ncRNA), no putative protein was conserved in all species, and the CPC (coding potential calculator) computational algorithm predicted that MIR2187HG has a very low coding potential (Fig. 1D).
MIR2187HG is associated with host antiviral immunity.
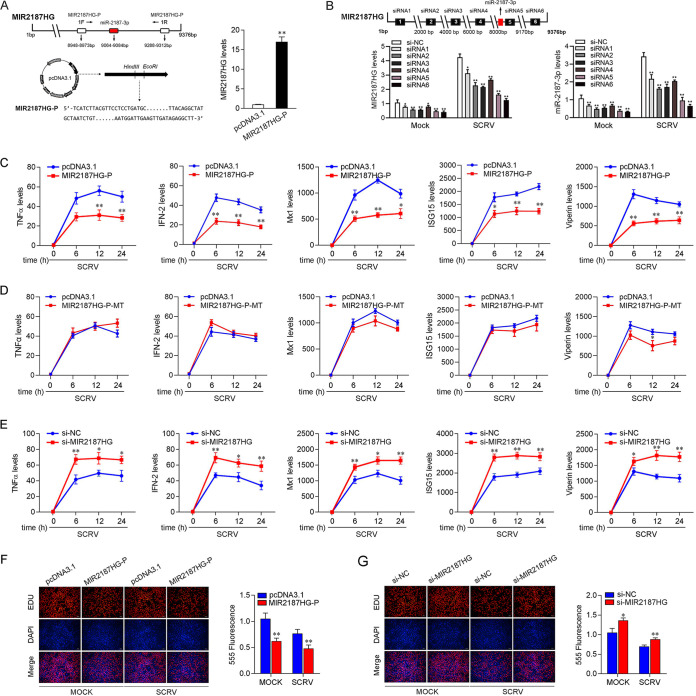
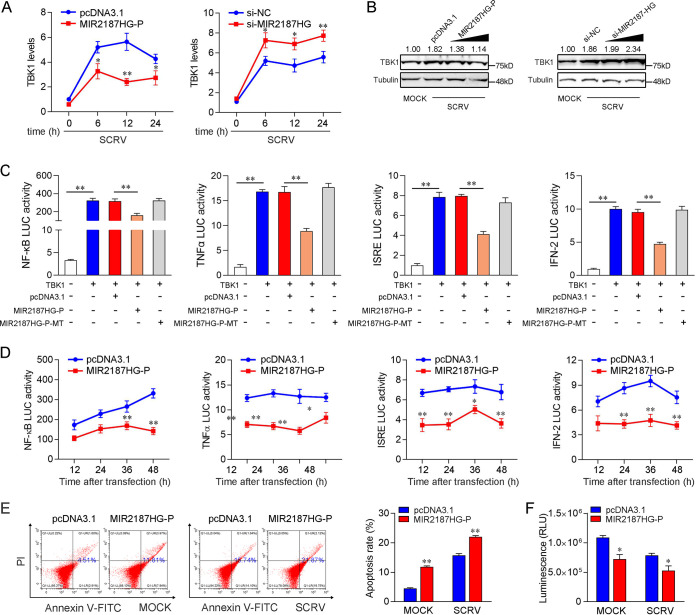
To investigate the biological functions of MIR2187HG, the expression plasmid of MIR2187HG was generated and named MIR2187HG-P. Given that the sequence of MIR2187HG is more than 9,000 bp, only 341 bp of this sequence was selected during the construction of the plasmid (containing the generation site of miR-2187-3p). As shown in Fig. 2A, the MIR2187HG-P overexpression plasmid significantly increased MIR2187HG-P expression levels. To determine whether MIR2187HG acts as an lncRNA to exert its function, we designed six small interfering RNAs (siRNA1, siRNA2, siRNA3, siRNA4, siRNA5, and siRNA6) of MIR2187HG and found that siRNA6 had the best knockdown effect (∼40% reduction) compared with the negative control of small interfering RNA of MIR2187HG (si-NC). siRNA6 was designed on the MIR2187HG sequence except for the pre-miR-2187 sequence. We also tested the knockdown efficiency of these siRNAs on MIR2187HG after SCRV infection. The results showed that siRNA6 had the best inhibitory effect on MIR2187HG. At the same time, we also detected the effect of these siRNAs on the expression of miR-2187-3p. The results showed that these siRNAs inhibited miR-2187-3p to various degrees. In addition, siRNA6 of MIR2187HG was used in all subsequent experiments and designated si-MIR2187HG (Fig. 2B). Then, we explored the role of MIR2187HG in the immune response through overexpression of MIR2187HG-P and knockdown of MIR2187HG in MICs upon SCRV infection. Antiviral genes and inflammatory cytokines are important indicators for detecting antiviral responses, so the effect of MIR2187HG on antiviral genes and inflammatory cytokines was assayed. As shown in Fig. 2C and D, the overexpression of MIR2187HG-P mediated the reduction of some genes, including tumor necrosis factor alpha (TNF-α), IFN-2, Mx1, ISG15 and viperin in MICs upon different SCRV infection times (Fig. 2C), and the overexpression of MIR2187HG-P-MT did not mediate the reduction of these genes (Fig. 2D). In contrast, the knockdown of MIR2187HG increased the expression levels of these genes in MICs receiving SCRV treatment (Fig. 2E). Moreover, ethynyldeoxyuridine (EdU) assays were used to detect the effect of MIR2187HG on cell proliferation rates. As shown in Fig. 2F and G, the overexpression of MIR2187HG-P plasmid induced cell cycle arrest and decreased the cell proliferation rate, whereas transfection with si-MIR2187HG increased cell proliferation. Of note, si-MIR2187HG can promote cell proliferation. On the whole, these data suggest that MIR2187HG acts as a negative regulator to modulate the antiviral response and cell proliferation in MICs.
FIG 2.
MIR2187HG decreases host antiviral immunity. (A) Schematic diagram of MIR2187HG expression plasmid construction (left) and the effect of the MIR2187HG-P expression plasmid on endogenous MIR2187HG expression (right). MICs were transfected with pcDNA3.1 vector or MIR2187HG-P expression plasmid for 48 h; then, MIR2187HG expression was determined by qPCR. (B) Location schematic diagram of si-MIR2187HG and screening of si-MIR2187HG efficiency. The six siRNAs of MIR2187HG were transfected into MICs for 48 h, and then MIR2187HG and miR-2187-3p expression was measured by qPCR after SCRV infection. (C and D) Effect of the expression plasmid of MIR2187HG-P or MIR2187HG-P-MT on TNF-α, IFN-2, Mx1, ISG15, and viperin expression during SCRV infection. MICs were transfected with pcDNA3.1 vector or MIR2187HG-P (C) or MIR2187HG-P-MT (D) expression plasmid for 24 h and then treated with SCRV for 6 h, 12 h, or 24 h; subsequently, expression of the indicated genes was determined by qPCR. (E) MICs were transfected with si-NC or si-MIR2187HG for 24 h. The cells were treated with SCRV for different times. Then, the expression of TNF-α, IFN-2, Mx1, ISG15, and viperin was analyzed by qPCR. (F and G) Cell proliferation was assessed by EdU assays in MICs transfected with pcDNA3.1 vector or MIR2187HG-P expression plasmid (F) and si-MIR2187HG or si-NC (G) to determine the effect of MIR2187 after SCRV infection. MICs were transfected with si-NC, si-MIR2187HG, pcDNA3.1 vector, or IRL for 24 h and then treated with SCRV for 24 h. A cell proliferation assay was performed. Bar, 20 μm. Values are means and SE from triplicate experiments. **, P < 0.01, and *, P < 0.05 compared to results with the control.
MIR2187HG regulates the release of miR-2187-3p.
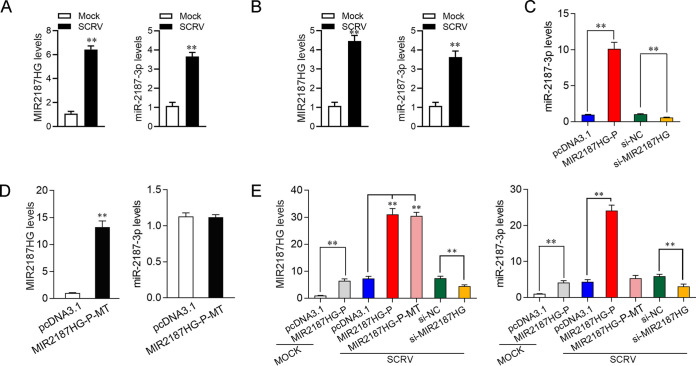
Numerous studies have shown that lncRNA can play a regulatory role as an miRNA host gene and that the expression of lncRNAs is positively or negatively related to miRNA (30). However, this molecular mechanism has not been reported in fish. MIR2187HG, one of the lncRNAs with significantly upregulated expression during SCRV treatment in teleost fish, may be involved in the immune response. A previous study reported that miR-2187-3p was enhanced upon SCRV infection and participated in the antiviral innate immune response (31). Through sequence alignment, we found that miR-2187-3p is an exact match to MIR2187HG, so we further studied the relationship between MIR2187HG and miR-2187-3p. First, we examined the expression of MIR2187HG and miR-2187-3p in MIC and MBrCs undergoing SCRV infection. As shown in Fig. 3A and B, we found that MIR2187HG and miR-2187-3p could be enhanced significantly upon SCRV infection whether in MICs (Fig. 3A) or in MBrCs (Fig. 3B); however, the expression level of miR-2187-3p was lower than that of MIR2187HG.
FIG 3.
MIR2187HG regulates the expression of miR-2187-3p. (A and B) MIR2187HG and miR-2187-3p could be elevated simultaneously upon RNA virus infection. The expression levels of MIR2187HG and miR-2187-3p in MICs (A) and MBrCs (B) stimulated with SCRV for 24 h were determined. (C) The expression profiles of miR-2187-3p by transfection of MIR2187HG or si-MIR2187HG in MICs were detected. MICs were transfected with pcDNA3.1, MIR2187HG, si-NC or si-MIR2187HG for 48 h, and then the expression of miR-2187-3p was detected by qPCR. (D) MIR2187HG-P-MT abrogates production of miR-2187-3p. MICs were transfected with pcDNA3.1 or mutated MIR2187HG plasmid for 48 h, and then the expression of MIR2187HG and miR-2187-3p was detected by qPCR. (E) The effect of MIR2187HG, MIR2187HG-P-MT, and si-MIR2187HG on MIR2187HG and miR-2187-3p upon SCRV infection was tested. MICs were transfected with pcDNA3.1, MIR2187HG-P, MIR2187HG-P-MT, si-NC, or si-MIR2187HG for 24 h and treated with SCRV for 24h, and then the expression levels of MIR2187HG and miR-2187-3p were analyzed by qPCR. GAPDH was used as a negative control for MIR2187HG, and 5.8S was used as a negative control for miR-2187-3p. Values are means and SE from triplicate experiments. **, P < 0.01 compared to results with the control.
Figure 2A and B show that MIR2187HG and si-MIR2187HG enhanced or weakened MIR2187HG levels, respectively. Here, to further verify the relationship between MIR2187HG and miR-2187-3p, we detected the effect of gain and loss of MIR2187HG on miR-2187-3p expression. Figure 3C shows that increasing the levels of MIR2187HG by transfecting MICs with the MIR2187HG expression vector specifically induced the expression levels of miR-2187-3p, suggesting that the processing of miR-2187-3p from MIR2187HG was increased by overexpressing MIR2187HG. At the same time, we found that the expression of miR-2187-3p was significantly reduced when MIR2187HG was silenced, which coincided exactly with the result that MIR2187HG-P overexpression led to increased miR-2187-3p levels. This result suggested that MIR2187HG may be the host gene of miR-2187-3p. To further determine whether MIR2187HG is the host gene of miR-2187-3p, we constructed mutated MIR2187HG-P, which contained a deficient miR-2187-3p sequence. As shown in Fig. 3D, MIR2187HG-P mutant (MIR2187HG-P-MT) transfection did not change the level of MIR2187HG but decreased the expression of miR-2187-3p. This result confirmed that MIR2187HG is the host gene of miR-2187-3p and positively regulates miR-2187-3p expression. Moreover, we detected the expression of MIR2187HG and miR-2187-3p after transfection of pcDNA3.1, MIR2187HG-P, MIR2187HG-P-MT, si-NC, or si-MIR2187HG in MICs for 24 h and treatment with SCRV stimulus for 24 h. As shown in Fig. 3D, miR-2187-3p and MIR2187HG were highly expressed after transfection with MIR2187HG-P in MICs undergoing SCRV treatment. Importantly, upon SCRV infection, the miR-2187-3p level did not change when the MIR2187HG-P-MT was transfected into MICs. These results showed that regardless of viral stimulation, miR-2187-3p is derived from MIR2187HG and miR-2187-3p expression is regulated by MIR2187HG.
TBK1 is a direct target of miR-2187-3p.
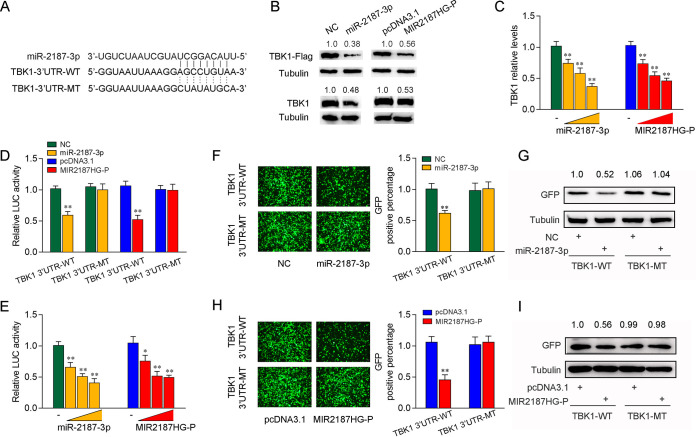
miRNAs affect various biological processes at the posttranscriptional level by targeting the 3′ UTR of mRNA. For this reason, we predicted the targets of miR-2187-3p (receptor-interacting protein 1 [RIP1], TNF receptor associated factor 6 [TRAF6], mitogen-activated protein kinase kinase 4 [MAP2K4], Toll-like receptor 13 [TLR13], signal transducer and activator of transcription 2 [STAT2], serum/glucocorticoid regulated kinase 1 [SGK1], mitogen-activated protein kinase kinase kinase 5 [MP3K5], mixed-lineage kinase domain-like protein [MLKL], interleukin-8 [IL-8], TANK-binding kinase 1 [TBK1], MBL-associated serine protease 1 [MASP1], transforming growth factor β3 [TGF-β3], CREB-regulated transcription coactivator 2 [CRTC2], suppressor of cytokine signaling 5 [SOCS5], and protein kinase N3 [PKN3]) and found that the potential target gene TBK1 is reported to be involved in the antiviral immunity (32). Therefore, we analyzed the sequence of the TBK1 3′ UTR to study whether miR-2187 can directly interact with it and found that there is a binding site for miR-2187. Then, we constructed the TBK1 3′ UTR luciferase plasmid and the mutant plasmid with the mutated miR-2187-3p-binding site (Fig. 4A). To detect the effect of miR-2187-3p on TBK1 expression, we constructed the expression plasmid of TBK1 and transfected it with miR-2187-3p, NC MIR2187HG plasmid, or pcDNA3.1 vector into epithelioma papulosum cyprini (EPC) cells. As shown in Fig. 4B, overexpressed miR-2187-3p led to a decrease in TBK1 protein level, and MIR2187HG plays the same role as miR-2187-3p in inhibiting the expression of TBK1. Meanwhile, we detected the role of miR-2187-3p and MIR2187HG in endogenous TBK1 expression. The results showed that TBK1 expression was decreased significantly after the transfection of miR-2187-3p or MIR2187HG-P. We also overexpressed miR-2187-3p or MIR2187HG-P at a range of concentrations in MICs and found that both miR-2187-3p and MIR2187HG-P reduced the expression level of TBK1 at the mRNA level in a dose-dependent manner (Fig. 4C). These results preliminarily demonstrated the inhibitory effect of miR-2187-3p on TBK1 expression at the mRNA level.
FIG 4.
miR-2187-3p targets the 3′ UTR of miiuy croaker TBK1. (A) Schematic diagram of the predicted target sites of miR-2187-3p in the 3′ UTR of TBK1. miR-2187-3p-binding sites in TBK1 wild-type form (TBK1 3′-UTR-WT) and the mutated form (TBK1 3′-UTR-MT) are shown. (B) miR-2187-3p regulates TBK1 expression. EPC cells were cotransfected with the IRAK4 expression plasmid with a Flag tag, along with miR-2187-3p or NC (top). After 48 h, TBK1 expression was determined by Western blotting. MICs were transfected with miR-2187-3p or NC (bottom). After 48 h, TBK1 expression was determined by Western blotting. (C) miR-2187-3p suppresses the mRNA expression of TBK1. MICs were cotransfected with miR-2187-3p, NC, MIR2187HG-P, or pcDNA3.1 At 48 h posttransfection, the expression of TBK1 was determined by qPCR. (D) miR-2187-3p targets the 3′ UTR of TBK1. EPC cells were transfected with NC, miR-2187-3p, pcDNA3.1, or MIR2187HG plasmid along with TBK1 3′-UTR-WT or TBK1 3′-UTR-MT for 24 h, and then the luciferase activity was determined. The luciferase activity was measured and normalized to Renilla luciferase activity. (E) The concentration gradient assay was performed with miR-2187-3p mimics and MIR2187HG-P plasmid. The miR-2187-3p (0, 25, 50, and 100 nM), NC (100,75, 50, and 0 nM), MIR2187HG-P (0, 50, 100, and 200 ng) or pcDNA3.1 (200, 150, 100, and 0 ng) were cotransfected with TBK1 3′-UTR-WT into EPC cells. After 48 h posttransfection, the luciferase activity was determined. Luciferase activity was normalized to Renilla luciferase activity. (F and G) miR-2187-3p could downregulate GFP expression. EPC cells were cotransfected with the wild type of mVenus-TBK1 or the mutated type, together with NC or miR-2187-3p. At 48 h posttransfection, the fluorescence intensity (F) and the GFP expression (G) were evaluated by a Varioskan Lux multifunctional microplate reader (Thermo Fisher Scientific) and Western blotting, respectively. Bar, 20 μm; original magnification, ×10. (H and I) Analysis of MIR2187HG negatively regulating GFP expression performed by fluorescence intensity (H) and protein detection (I). Data are averages from three independent experiments. **, P < 0.01, and *, P < 0.05 compared to results with the control.
To further clarify that this negative mechanism is realized through the complementation of miR-2187-3p with seed sequences in the 3′ UTR, a dual-luciferase reporter assay was performed. The luciferase reporter assays showed that miR-2187-3p overexpression led to a marked decrease in of luciferase activity of the wild-type TBK1 3′ UTR in EPC cells. However, no change in luciferase activity was observed in the mutant TBK1 3′ UTR, which contains mutations in the putative binding sites (Fig. 4D). Similar data were observed after MIR2187HG cotransfection. Then, concentration gradient assays of miR-2187-3p and MIR2187HG-P were performed to detect TBK1 3′ UTR luciferase activity in EPC cells. The results showed that TBK1 luciferase activity was steadily declined regardless of miR-2187-3p or MIR2187HG-P overexpression (Fig. 4E). In addition, we inserted the wild or a mutated form of the TBK1 3′ UTR into mVenus-C1 vector and examined whether cotransfection with miR-2187-3p could suppress the levels of green fluorescent protein (GFP). As shown in Fig. 4F and G, the results revealed that miR-2187-3p could significantly inhibit the levels of GFP, indicating an interaction between miR-2187-3p and TBK1. Similar results were observed after MIR2187HG-P cotransfection (Fig. 4H and I). Taken together, our results strongly indicate that TBK1 is the direct target of miR-2187-3p and MIR2187HG.
MIR2187HG exerts functions acquired from miR-2187-3p.
Considering that miR-2187-3p could regulate TBK1-mediated antiviral pathways, we investigated whether MIR2187HG, the host gene of miR-2187-3p, can modulate the NF-κB and IRF3 signaling pathways through the functionality obtained from miR-2187-3p. As shown in Fig. 5A, the results of qPCR analysis showed that the increased expression of TBK1 induced by SCRV was attenuated to a certain extent by MIR2187HG, and the inhibitory effect was most obvious when SCRV was stimulated for 12 h. In contrast, MIR2187HG knockdown caused an increase in the expression level of TBK1 after SCRV infection (Fig. 5A). Protein detection showed that the SCRV-induced increase of TBK1 protein level was decreased by MIR2187HG, while MIR2187HG silencing promoted the expression of TBK1 (Fig. 5B). This result indicated that MIR2187HG could regulate TBK1 expression at the posttranscriptional level. To determine whether MIR2187HG could regulate TBK1-mediated signaling, dual-luciferase reporter assays were performed, and the results are similar to the identification of miR-2187-3p. As can be seen from Fig. 5C, MIR2187HG-P inhibited the luciferase reporter activity induced by NF-κB, TNF-α, interferon-stimulated response element (ISRE), and IFN-2 reporter genes, which were activated by TBK1, while MIR2187HG-P-MT could not inhibit the activity of these reporter genes. Then, the time gradient assays were conducted, and dual-luciferase reporter assays were measured at the indicated time points. As expected, MIR2187HG inhibited TBK1-mediated NF-κB and IRF3 signaling pathways (Fig. 5D). Then, cell apoptosis and cell activity assays were performed. The results showed that MIR2187HG increased the proportion of apoptotic cells and suppressed the activity of MICs (Fig. 5E). Collectively, these results demonstrated that MIR2187HG can function as an miR-2187 precursor regulating TBK1 at multiple levels and modulating the cell growth and activity (Fig. 5F). These functions exerted by MIR2187HG are similar to those of mature miR-2187-3p, which suggested that MIR2187HG obtained functions from miR-2187-3p.
FIG 5.
MIR2187HG is able to negatively regulate NF-κB and IRF3 signaling pathways. MIR2187HG inhibits the mRNA level of endogenous TBK1 upon SCRV infection. qPCR assays were performed to determine the expression levels of TBK1 in MICs transfected with MIR2187HG-P, pcDNA3.1, si-MIR2187HG, or si-NC at different SCRV infection times. (B) MIR2187HG suppresses the protein expression of endogenous TBK1 upon SCRV infection. MICs were cotransfected with MIR2187HG-P or pcDNA3.1 at different concentrations and si-MIR2187HG or si-NC at different concentrations for 24 h. Then the cells were treated with SCRV for 24 h, and the expression of TBK1 was determined by Western blotting. (C) MIR2187HG inhibits TBK1-activated luciferase activity. EPC cells were cotransfected with pcDNA3.1, MIR2187HG-P, or MIR2187HG-P-MT, a TBK1 expression plasmid, and a pRL-TK vector, along with an NF-κB, TNF-α, ISRE, or IFN-2 reporter gene, to investigate the regulatory effect of MIR2187HG on NF-κB and IRF3 signals. (D) Relative luciferase activity of the indicated reporters in EPC cells after cotransfection with pcDNA3.1 or MIR2187HG-P was determined. EPC cells were cotransfected with pcDNA3.1 or MIR2187HG-P plasmid, TBK1 expression plasmid, pRL-TK vector, together with NF-κB, TNF-α, ISRE, or IFN-2 luciferase reporter genes, and then the luciferase activity was measured over time, as indicated. (E) The effect of MIR2187HG on cell apoptosis was analyzed by flow-cytometric cell apoptosis assays. The SCRV treatment was performed after transfection with MIR2187HG-P or pcDNA3.1 at 24 h. (F) Effect of MIR2187HG on cell viability. MICs were transfected with MIR2187HG-P or pcDNA3.1 for 24 h and then SCRV infected for 24 h. A cell viability assay was performed. Error bars indicate the SE of data from three independent experiments. **, P < 0.01, and *, P < 0.05, compared to results with the control.
miR-2187-3p inhibits TBK1-mediated antiviral signaling pathways.
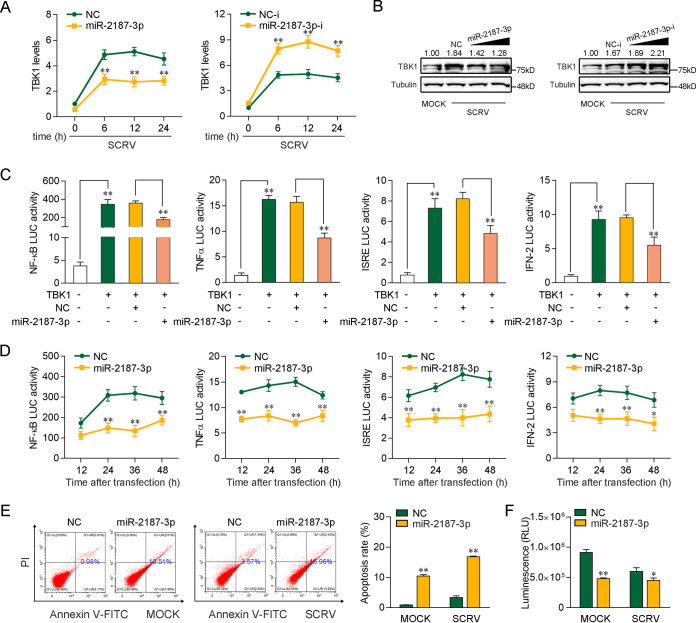
Given that miR-2187-3p targets and regulates TBK1, we then tested whether miR-2187-3p is able to regulate TBK1 expression under SCRV infection. As shown in Fig. 6A, upon SCRV infection, miR-2187-3p overexpression led to a decrease in the expression levels of TBK1 in MICs, whereas miR-2187-3p inhibition enhanced TBK1 expression. Then, we tested whether miR-2187-3p regulates TBK1 expression at the protein level under SCRV treatment. As can be seen from Fig. 6B, the results of Western blot analysis showed that the TBK1 protein level was significantly enhanced after SCRV treatment. Enhanced TBK1 was decreased by miR-2187-3p. In contrast, miR-2187-3p-i increased TBK1 expression indirectly by counteracting miR-2187-3p. Next, we tested whether miR-2187-3p regulates the TBK1-mediated signaling pathway. As shown in Fig. 6C, the results indicated that TBK1 could activate NF-κB, ISRE, TNF-α, and IFN-2 luciferase reporter genes. However, transfection of miR-2187-3p mimics markedly suppressed the activation of NF-κB, TNF-α, ISRE, and IFN-2 luciferase reporter genes induced by TBK1 overexpression compared with negative-control mimics (Fig. 6C). Then, time gradient assays were conducted, and dual-luciferase reporter assays were measured at the indicated time points. As expected, miR-2187-3p inhibited TBK1-mediated NF-κB and IRF3 signaling pathways (Fig. 6D). Cell apoptosis analysis showed that miR-2187-3p overexpression increased the rate of apoptotic cells with or without SCRV infection (Fig. 6E). Furthermore, we tested the effects of miR-2187-3p overexpression on MIC viability. The results showed that miR-2187-3p overexpression decreased cell viability, especially in SCRV-treated cells (Fig. 6F). Taken together, the data suggest that miR-2187-3p acts as a negative modulator in regulating TBK1-mediated signaling pathway, as well as playing a role in cell viability.
FIG 6.
miR-2187-3p can negatively regulate TBK1-mediated antiviral signaling. (A) miR-2187-3p inhibits the mRNA level of endogenous TBK1 upon SCRV infection. qPCR assays were performed to determine the expression levels of TBK1 in MICs transfected with miR-2187-3p, NC, miR-2187-3p-i, or NC-i and infected with SCRV for different times. (B) miR-2187-3p suppresses the protein expression of endogenous TBK1 upon SCRV infection. MICs were cotransfected with miR-2187-3p or NC at different concentrations and miR-2187-3p-i or NC-i at different concentrations for 24 h. Then, the cells were treated with SCRV for 24 h, and the expression of TBK1 was determined by Western blotting. (C) miR-2187-3p inhibits TBK1-activated luciferase activity. EPC cells were cotransfected with NC or miR-2187-3p, TBK1 expression plasmid, and pRL-TK vector, along with the NF-κB, TNF-α, ISRE, or IFN-2 reporter gene, to investigate the regulatory effect of miR-2187-3p on NF-κB and IRF3 signals. (D) Relative luciferase activity of indicated reporters in EPC cells after cotransfection with NC or miR-2187-3p was determined. EPC cells were cotransfected with NC or miR-2187-3p, the TBK1 expression plasmid, and the pRL-TK vector, together with the NF-κB, TNF-α, ISRE, or IFN-2 luciferase reporter gene, and then the luciferase activity was measured over time, as indicated. (E) The effect of miR-2187-3p on cell apoptosis was analyzed by flow-cytometric cell apoptosis assays. The SCRV treatment was performed after transfection with NC or miR-2187-3p at 24 h. (F) Effect of miR-2187-3p on cell viability. MICs were transfected with NC or miR-2187-3p for 24 h and then infected with SCRV for 24 h. A cell viability assay was performed. Error bars indicate the SE for data from three independent experiments. **, P < 0.01, and *, P < 0.05, compared to results with the control.
MIR2187HG enhances SCRV replication.
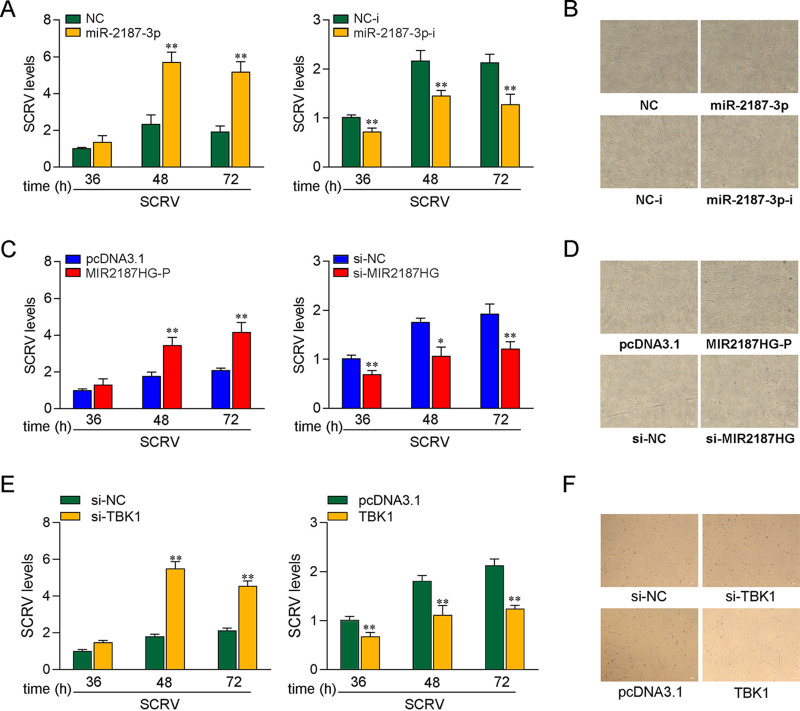
In order to explore the biological significance of MIR2187HG upregulation in SCRV-induced host cells, we tested the effect of MIR2187HG on SCRV replication in MICs. Before examining the effect of MIR2187HG on virus proliferation, we first tested whether mature miR-2187-3p could regulate virus replication. By measuring intracellular SCRV levels in the infected MICs, we found that miR-2187-3p overexpression increased SCRV replication, while miR-2187-3p inhibition decreased SCRV replication (Fig. 7A). Consistent with the results of the qPCR analysis, as shown in the Fig. 7B, MICs treated with miR-2187-3p showed a significant cytopathic effect (CPE) compared with the control group, but the pathological effect was alleviated with the addition of miR-2187-3p-i. At the same time, the effect of MIR2187HG on SCRV replication was determined. The results indicated that MIR2187HG can also promote SCRV replication (Fig. 7C) and increase the proportion of CPE (Fig. 7D). Moreover, the effect of TBK1 on SCRV replication was determined. The results indicated that TBK1 can inhibit SCRV replication (Fig. 7E) and increase the proportion of CPE (Fig. 7F). Collectively, these data indicate that host lncRNA MIR2187HG can promote SCRV replication.
FIG 7.
lncRNA MIR2187HG promotes SCRV replication. (A) miR-2187-3p enhanced SCRV replication. MICs were transfected with NC, miR-2187-3p, NC-i, or miR-2187-3p-i for 24 h and then treated with SCRV (MOI = 5). After different SCRV treatment times, the intracellular SCRV level was determined by qPCR. (B) miR-2187-3p enhanced the cytopathic effect. MICs were transfected with NC, miR-2187-3p, NC-i, or miR-2187-3p-i for 24 h and infected with SCRV for 24 h. Then, cells were observed for morphological changes. (C) MIR2187HG promoted SCRV replication. MICs were transfected with pcDNA3.1, MIR2187HG-P, si-NC, or si-MIR2187HG for 24 h and then treated with SCRV (MOI = 5). After different SCRV treatment times, the intracellular SCRV level was determined by qPCR. (D) MIR2187HG enhanced cytopathic effect. MICs were transfected with pcDNA3.1, MIR2187HG-P, si-NC, or si-MIR2187HG for 24 h and infected with SCRV for 24 h. Then, cells were observed for morphological changes. (E) TBK1 inhibited SCRV replication. MICs were transfected with pcDNA3.1, TBK1, si-NC, or si-TBK1 for 24 h and then treated with SCRV (MOI = 5). After different SCRV treatment times, the intracellular SCRV level was determined by qPCR. (F) TBK1 enhanced the cytopathic effect. MICs were transfected with pcDNA3.1, TBK1, si-NC, or si-TBK1 for 24 h and infected with SCRV for 24 h. Then, cells were observed for morphological changes. All data are means and SE from at least three independent triplicate experiments. **, P < 0.01, and *, P < 0.05, versus the controls.
MIR2187HG regulating TBK1 is widely found in other teleost fish.
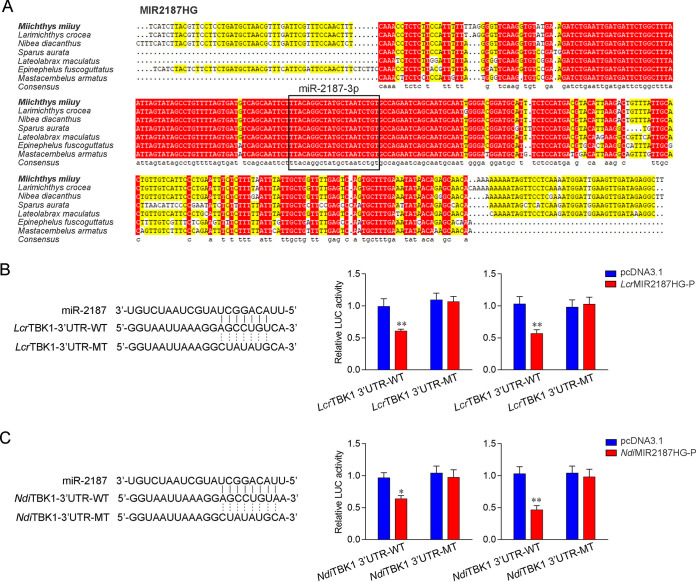
To address the generality of our findings, we first examined the sequence alignment of MIR2187HG from different fish species. Interestingly, as shown in Fig. 8A, mature miR-2187-3p displayed a high degree of conservation in multiple fish. Furthermore, the miR-2187-3p binding site in the TBK1 3′ UTR also displayed a high degree of conservation in other fish (Fig. 8B and C). To obtain direct evidence that MIR2187HG could regulate the TBK1 3′ UTR across species, luciferase reporter genes were generated by cloning the TBK1 3′ UTR of L. crocea and N. diacanthus into pmir-GLO vector (devoid of a miR-2187-3p binding site) as a negative control. At the same time, we constructed the MIR2187HG expression plasmid of L. crocea and N. diacanthus. Strikingly, the luciferase activities decreased when LcrMIR2187HG-P or NdiMIR2187HG-P was cotransfected with the wild-type L. crocea TBK1 3′ UTR or N. diacanthus TBK1 3′ UTR into EPC cells, respectively. However, transfection with their mutant types showed no effect on the luciferase activity (Fig. 8B and C, middle). The same experiment was carried out in HEK293 cells, and we obtained similar results (Fig. 8B and C, right), which shows that MIR2187HG regulates TBK1, which is widespread in fish. In summary, these results indicate that miR-2187-3p is highly conserved among different vertebrate groups and that its precursor, MIR2187HG, can also exert an inhibitory effect to regulate the expression of TBK1 genes in other species.
FIG 8.
lncRNA MIR2187HG regulating the TBK1 gene is widely found in other fish. (A) Sequence alignment of MIR2187HG from various species where it is present; miR-2187-3p sequences are shown in the black box. (B and C) Schematic diagram of the predicted target sites of miR-2187-3p in the 3′ UTRs of L. crocea TBK1 (B) and N. diacanthus TBK1 (C) (left). (B) EPC cells (middle) and HEK293 cells (right) were transfected with pcDNA3.1 or LcrMIR2187HG-P along with the wild-type L. crocea TBK1 3′ UTR (LcrTBK1 3′-UTR-WT) or the mutant (LcrTBK1 3′-UTR-MT) for 24 h, and then the luciferase activity was determined. (C) EPC cells (middle) and HEK293 cells (right) were transfected with pcDNA3.1 or NdiMIR2187HG-P, along with the wild-type N. diacanthus TBK1 3′ UTR (NdiTBK1 3′-UTR-WT) or the mutant type (NdiTBK1 3′-UTR-MT) for 24 h, and then the luciferase activity was determined. Data are averages from three independent experiments. Error bars indicate SE for data from three independent experiments. **, P < 0.01; *, P < 0. 05.
DISCUSSION
Viral diseases have become one of the biggest threats restricting the development of the aquatic industry. To resist the invasion and proliferation of pathogens, hosts have developed a variety of defense mechanisms that recognize viruses. Similar to mammals, teleost fish respond to virus invasion by activating a series of signaling cascades, inducing type I IFN production and leading to innate antiviral responses (33). In the present study, lncRNAs exert the functionality of miRNAs to participate in antiviral immunity of fish. We found that fish MIR2187HG is the developmental reservoir of miR-2187-3p and positively regulates the expression of miR-2187-3p. Our data show that miR-2187-3p targets the TBK1 gene, suppresses TBK1-mediated signaling, and acts as a negative regulator of the innate antiviral responses. Additionally, MIR2187HG functions as pre-miR-2187, the overexpression of which can suppress the level of TBK1 and inhibit TBK1-triggered antiviral innate immunity, thereby promoting SCRV replication.
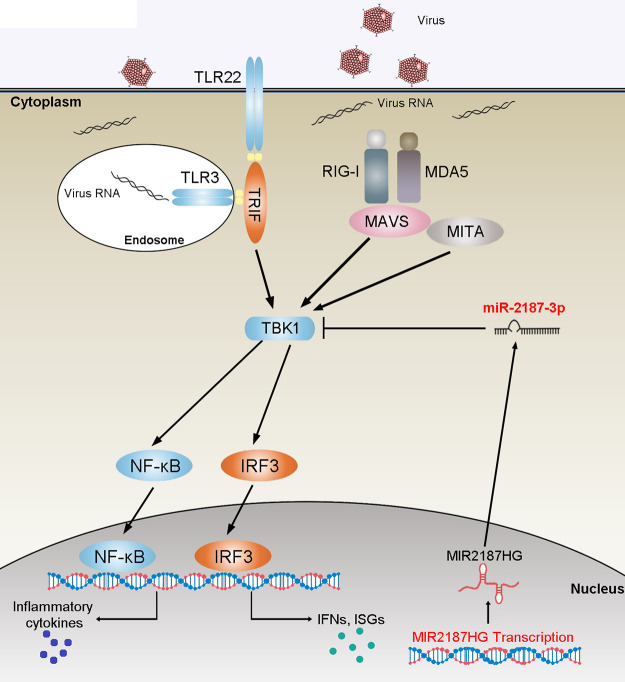
TBK1 has been identified as a critical mediator of innate antiviral responses that induces the activation of IRF3 and accelerates the production of type I IFN in the TLR- and RLR-triggered antiviral pathway. Therefore, the regulation mechanism of TBK1-mediated signal transduction has been extensively studied. A recent study showed that miR-155-5p could act as a negative regulator involved in TBK1/IRF3 signaling by targeting TBK1 during rotavirus infection (34). Other studies showed that E3 ubiquitin ligase inhibits the phosphorylation level of IRF3 and the production of IFN I by regulating the homeostasis of the TBK1/IKKi complex, thereby negatively regulating the antiviral immune response (35). Meanwhile, viruses have evolved mechanisms to regulate TBK1-mediated signal transduction. For example, porcine epidemic diarrhea virus (PEDV) nsp15 can disrupt the IFN response by inducing the RNA degradation of TBK1 and IRF3 (36). Zika virus NS5 protein antagonizes type I IFN production by preventing TBK1 activation (37). In fish, studies have confirmed that TBK1 serves as a crucial mediator to participate in the antiviral response (11, 38, 39). The present study demonstrated that TBK1 is activated by two antiviral signaling pathways, TLR3/22-TRIF and RIG-I-MAVS, mediating signal transmission and participating in the RNA virus-induced immune regulation network in miiuy croaker. Further research showed that MIR2187HG with a pre-miRNA regulatory role functions as a vital regulator of TBK1 and negatively regulates the TBK1-mediated signaling pathways (Fig. 9).
FIG 9.
Mechanism of the regulatory network and function of TBK1. (A) Fish TBK1 could induce innate antiviral responses by recruiting NF-κB and IRF3 to trigger inflammatory cytokines and antiviral factor activation upon SCRV infection. miR-2187-3p targets TBK1 and represses TBK1-mediated antiviral responses. MIR2187HG could regulate the release of miR-2187-3p and function as a precursor of miR-2187-3p to indirectly inhibit TBK1 expression, thereby avoiding excess immune responses to maintain the stable, appropriate immune response. The lncRNA-miRNA-mRNA regulatory networks may be widespread in vertebrate species.
ncRNAs, an abundant type of RNA transcript encoded by eukaryotes, play an essential role in regulating gene expression at the posttranscriptional level. As one of the main members of ncRNAs, miRNAs have also received much attention due to their regulatory functions. Since the discovery of let-7 and lin-4 miRNAs as regulators involved in the development of Caenorhabditis elegans, researchers have gradually studied the functions of miRNAs in various biological processes in many model organisms. Recently, many studies conducted on miRNAs in lower vertebrates, such as fish (40), are also increasingly intensive. Existing studies have shown that miiuy croaker miR-3570 can target MyD88 and MAVS, thereby regulating anti-inflammatory and antiviral responses, respectively (41, 42). miR-210 can downregulate the expression of MITA and modulate the SCRV-mediated antiviral innate response of miiuy croaker (43). Also, miR-144 and miR-217 modulate NF-κB-mediated immune signaling by targeting NOD1 (44). In the present study, we found that miR-2187-3p derived from MIR2187HG could regulate the expression of TBK1 by binding to the TBK1 3′ UTR, thereby regulating the immune response mediated by TBK1. Collectively, these results indicate that MIR2187HG-derived miR-2187-3p inhibits the expression of TBK1 and the signaling pathway mediated by TBK1.
miRNAs are highly conserved in sequence among different vertebrate groups, and their functions are also conserved to a certain extent. Compared with that of miRNA, the evolutionary conservatism of lncRNAs is relatively poor. Approximately 14,599 lncRNAs identified in the large yellow croaker have been subjected to BLAST analysis, and only 18 conserved lncRNAs were found compared with zebrafish lncRNAs (45). However, the conservatism of lncRNAs is relative, and for species with close relatives, lncRNAs are still somewhat conserved. In this study, we examined the sequence of MIR2187HG and found that MIR2187HG is relatively conserved across several fish species (Fig. 8A). As comparative analysis of genes across species can be a powerful tool for studying their functions, the relative conservation of the MIR2187HG sequence indicated the conservation of its function, suggesting an essential and irreplaceable role of lncRNAs in biological processes of teleost fish.
A growing number of studies on the lncRNA-miRNA interaction network have been conducted, but most previous studies focused on lncRNAs acting as ceRNAs to promote innate immunity, which is in contrast to the inhibitory role played by miRNA in the immune response. The lncRNAs IRL and NARL modulate innate immunity through the downregulation of miRNA-dependent targets in fish (46, 47). Moreover, a new regulatory mechanism of lncRNAs involved has been discovered. lncRNAs can be negatively involved in immune regulation mechanism as miRNA precursors to avoid excessive immunity and maintain immune balance. For example, the lncRNA H19 acts not only as a sponge of the let-7 family but also as a precursor RNA through splicing to produce two mature miRNAs, miR-675-3p and miR-675-5p, and they can suppress growth of the placenta before birth and the Igf1r gene when H19 functions as pre-miR-675. (19, 48, 49). In patients with glioma, MIR155HG is a favorable factor for malignant progression, mesenchymal transition, and poor prognosis and shows cancer-promoting activity by deriving miR-155-5p/-3p (50). Although lncRNA as an miRNA precursor exerts an regulatory role in mammals, it is still very rare in teleost fish. In the present study, MIR2187HG was upregulated upon SCRV infection. Subsequently, we found that overexpression of MIR2187HG in miiuy croaker cells could significantly increase the level of miR-2187-3p, while miR-2187-3p could further inhibit the expression of antiviral genes and inflammatory cytokines by targeting TBK1. These results indicated that miiuy croaker MIR2187HG can act as a negative regulator in the fish antiviral immune response and is an effective molecule to control immunity intensity.
In this study, we found a new lncRNA named MIR2187HG in miiuy croaker and found that it can negatively regulate the antiviral immune response. Subsequently, we found that miR-2187-3p can negatively regulate TBK1-triggered NF-κB and IRF3 signaling pathways. Further studies found that MIR2187HG can produce miR-2187-3p by acting as a precursor of miR-2187-3p and then indirectly inhibit the TBK1-mediated antiviral immune response; this process can maintain immune homeostasis and immune balance. This study is the first to determine the molecular regulation function of lncRNAs as pre-miRNA in teleost fish and provides theoretical support for lncRNA being responsible for regulating protein-encoding genes in teleost fish. Finally, our data provide new insights into the lncRNA negative-feedback regulation of the antiviral innate response in fish and enriches the knowledge of host-virus interactions.
MATERIALS AND METHODS
Animals and immune challenge.
Miiuy croakers were obtained from Zhoushan Fisheries Research Institute. Fish were acclimated at 25°C for at least 6 weeks in a recirculating water tank system filled with air-pumped seawater before experiments. The challenge was performed as described previously (30, 51). Briefly, each miiuy croaker (about 50 g) was injected with 0.2 mL phosphate-buffered saline (PBS; control) or SCRV (multiplicity of infection [MOI] = 5) intraperitoneally. Afterward, fish were sacrificed at different time points, and the spleen and liver tissues were collected for RNA extraction. All animal experimental procedures were performed in accordance with the National Institutes of Health′s Guide for the Care and Use of Laboratory Animals (52), and the experimental protocols were approved by the Research Ethics Committee of Shanghai Ocean University (no. SHOU-DW-2018-047).
Cell culture and transfection.
EPC cells were maintained in medium 199 (HyClone) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin at 28°C in 5% CO2 (53). Miichthys miiuy intestinal cells (MICs) and M. miiuy brain cells (MBrCs) were cultured in L-15 medium (HyClone) supplemented with 15% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 26°C. The MIC lines and MBrC lines are stored in our laboratory, and their preparation process was as described elsewhere (54). For stimulation experiments, MICs and MBrCs were challenged with poly(I·C) (10 μg/mL) or SCRV (MOI = 5) and harvested at different times for RNA extraction.
Transient transfection of cells with miRNA mimics, miRNA inhibitors, or siRNA was performed in 24-well plates using Lipofectamine RNAiMAX (Invitrogen), and cells were transfected with DNA plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer′s instructions. For functional analyses, the expression plasmid or empty plasmid (500 ng) and miRNA mimics (100 nM), miRNA inhibitor (100 nM) or siRNA (100 nM) were transfected into cells in culture medium and then harvested for further detection. For luciferase experiments, miRNA mimics (50 nM) or miRNA inhibitor (50 nM) and pmir-GLO (50 ng per well) containing the wild or mutated plasmid of the TBK1 3′ UTR were transfected into cells.
Plasmid construction.
To construct the TBK1 3′UTR reporter vector, the 3′ UTR of the miiuy croaker TBK1 gene, as well as L. crocea and N. diacanthus TBK1 3′ UTRs, were amplified using PCR and cloned into the pmir-GLO luciferase reporter vector (Promega, USA). To construct the MIR2187HG expression plasmid, MIR2187HG from the above-mentioned species was cloned into the pcDNA3.1 vector, and the resulting construct was named MIR2187HG-P. The mutated MIR2187HG with deficiency mutations in the mature miR-2187-3p region and the mutated reporter plasmids with point mutations in the miR-2187-3p binding site were synthesized using a Mut Express II fast mutagenesis kit, V2, with specific primers (Table 1). Meanwhile, the M. miiuy TBK1 3′ UTR luciferase gene was inserted into mVenus-C1 (Invitrogen, USA), which included the sequence of enhanced green fluorescent protein (GFP). In addition, the TBK1 expression plasmid was constructed as previously described (32). The correct construction of the plasmids was verified by Sanger sequencing and extraction of plasmids from DH5α Chemically Competent Cell (Vazyme) with an Endo Free plasmid DNA miniprep kit (Tiangen Biotech).
TABLE 1.
PCR primers
| Primer | Sequence (5′–3′) |
|---|---|
| MIR2187HG-RT-F | TCATCTTACGTTCCTCCTG |
| MIR2187HG-RT-R | AAGCCTCTATCAACTTCAATC |
| TBK1-qRT-F | AGAGCACCACCAACTACCT |
| TBK1-qRT-R | ACTTTGACGGCATACAGG |
| Mx1-qRT-F | GCTGCTTGTTTACTCCCA |
| Mx1-qRT-R | ACCTGCATCATCTCCCTC |
| ISG15-qRT-F | TGAACGGACAGAAGACGC |
| ISG15-qRT-R | TGAGGAATACCTGCATGG |
| miR-2187-3p-qRT-F | GCAGCGAACCATTATTTGC |
| miR-2187-3p-qRT-R | TCCAGTTTTTTTTTTTTTTTAAAGCAG |
| β-actin-qRT-F | GTGATGAAGCCCAGAGCA |
| β-actin-qRT-R | CGACCAGAGGCATACAGG |
| 5.8s-qRT-F | AACTCTTAGCGGTGGATCA |
| 5.8s-qRT-R | GTTTTTTTTTTTTTTTGCCGAGTG |
| Mx1-qRT-F | GCTGCTTGTTTACTCCCA |
| Mx1-qRT-R | ACCTGCATCATCTCCCTC |
| TNFα-qRT-F | CGCCACCACGCTCTTCTG |
| TNFα-qRT-R | GCCATTGGCCAGGAGGGC |
| IFN-2-qRT-F | GCTCTGCCTTCCCTGCTA |
| IFN-2-qRT-R | CAGTTGACTCCGCCCTCT |
| TBK1-3′UTR-WT-NheI-F | CTAGCTAGCAAGGAGGAGATGGAGGGAGT |
| TBK1-3′UTR-WT-XbaI-R | TGCTCTAGACCGGCAACAATGAAGTGAGT |
| TBK1-3′UTR-MT-F | AAAGGCTATATGCATGGTAAAATTATATCTGTCTGAACATAAATG |
| TBK1-3′UTR-MT-R | ACCATGCATATAGCCTTTAATTACCTTCACATGAGTTTATTAA |
| TBK1-KpnI-F | GACGACAAGAAGCTTGGTACCATGCAGAGCACCACCAACTACC |
| TBK1-XbaI-R | TATAGAATAGGGCCCTCTAGACGGCAACAATGAAGTGAGTAACA |
| GFP-TBK1-3′UTR-F | CGGGGTACCGCTAGAAGGAGGAGATGGAGGGAGT- |
| GFP-TBK1-3′UTR-R | CGCGGATCCCCGGCAACAATGAAGTGAGT |
| GFP-TBK1-3′UTR-MT-F | TATCGGCATCCACTCCATACGGTCTGCTTGGTCC |
| GFP-TBK1-3′UTR-MT-R | ATGGAGTGGATGCCGATACAAAAAGCTTCATCTCACAGTCG |
| NdiTBK1-3′UTR-WT-NheI F | CTAGCTAGCAGGATGGTGCCAAAGTA |
| NdiTBK1-3′UTR-WT-XhoI R | CCGCTCGAGAAGCCGTTAAAGTTCAG |
| NdiTBK1-3′UTR-MT-F | AAAGGGCTGATGTATGGTAAAATGGTATTTGTCTGAACAT |
| NdiTBK1-3′UTR-MT-R | ACCATACATCAGCCCTTTAATTACCTTCACATGAGTTCAT |
| LcrTBK1-3′UTR-WT-NheI F | TGTTTAAACGAGCTCGCTAGCCAGTTTAATTAGATTATTTAACTTTGACGTT |
| LcrTBK1-3′UTR-WT-XhoI R | CAGGTCGACTCTAGACTCGAGTAACTCAGTTATTGATTATTGATCGGC |
| LcrTBK1-3′UTR-MT-F | AAGGCTATATGCATGGGGAAATGACATTTGTCTG |
| LcrTBK1-3′UTR-MT-R | CCCATGCATATAGCCTTTAATTACCTTCACGAGTTTATTCA |
| MIR2187HG-HindIII-F | ACTATAGGGAGACCCAAGCTTTCATCTTACGTTCCTCCTGATGC |
| MIR2187HG-EcoRI-R | TGATGGATATCTGCAGAATTCAAGCCTCTATCAACTTCAATCCATT |
| MIR2187HG-MT-F | GTCAGCAATTCTGCCAGAATCAGCAATGCAATT |
| MIR2187HG-MT-R | TCTGGCAGAATTGCTGACATCACTAAAACAGGC |
| LcrMIR2187HG-HindIII-F | ACTATAGGGAGACCCAAGCTTTCATCTTACGTTCCTCCTGATGC |
| LcrMIR2187HG-EcoRI-R | TGATGGATATCTGCAGAATTCCCTCTATCAACTTCAATCCATTTTTG |
| NdiMIR2187HG-HindIII-F | ACTATAGGGAGACCCAAGCTTTCATCTTACGTTCCTCCTGATGC |
| NdiMIR2187HG-EcoRI-R | TGATGGATATCTGCAGAATTCAAGCCTCTATCAACTTCAATCCATT |
miR-2187-3p target gene identification.
The potential downstream targets of miR-2187-3p targets were predicted using Targetscan, miRanda, and MicroInspector algorithms (55–57). Predictions were ranked based on the predicted efficacy of targeting as calculated using the context and scores of the sites.
Oligonucleotides.
miR-2187-3p mimics (miR-2187-3p), miR-2187-3p inhibitors (miR-2187-3p-i), the MIR2187HG small interfering RNA (si-MIR2187HG), and the negative control (Nc or si-NC) were synthesized and purified from GenePharma (Shanghai, China). Sequences are as follows: for miR-2187-3p mimics, 5′-UUACAGGCUAUGCUAAUCUGU-3′ (sense); for miR-2187-3p inhibitors, 5′-ACAGAUUAGCAUAGCCUGUAA-3′ (chemically modified by 2′-Ome); for si-MIR2187HG-1, 5′-GAUGUGUAAUAAUGUGCAATT-3′; for si-MIR2187HG-2, 5′-GCAAGAUUAUGGAUUUCUATT-3′; for si-MIR2187HG-3, 5′-GGCUAAUGAUGUUAGAGUUTT-3′; for si-MIR2187HG-4, 5′-GGAAUAUUCUGGAGUUUAATT-3′; for si-MIR2187HG-5, 5′-UGCUAAUCUGUGCCAGAAUTT-3′; for si-MIR2187HG-6, 5′-GAUAGAGGCUUUCUGUUAATT-3′; for the negative control (NC or si-NC), 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense); for the negative-control inhibitor, 5′-CAGUACUUUUGUGUAGUACAA -3′.
Dual-luciferase reporter assays.
For miRNA target identification, the oligonucleotide of miR-2187-3p mimics or negative control (NC), MIR2187HG plasmid, or pcDNA3.1 vector was transfected with the TBK1 3′ UTR wild-type (WT) or TBK1 3′ UTR MT plasmid into EPC cells. At 48 h posttransfection, the activity of luciferase was detected using a dual-luciferase reporter assay system (Promega). To determine the functional regulation of TBK1, MICs were cotransfected TBK1 expression plasmid, together with NF-κB, ISRE, IFN-2, and TNF-α luciferase reporter gene plasmids, pRL-TK Renilla luciferase plasmid, and either miR-2187-3p mimics or negative controls. At 48 h posttransfection, the cells were lysed for reporter activity using the dual-luciferase reporter assay system (Promega). All the luciferase activity values were compared with the Renilla luciferase. Transfection of each construct was performed in triplicate in each assay. Ratios of Renilla luciferase readings to firefly luciferase readings were taken for each experiment, and triplicates were averaged.
RNA extraction and real-time PCR.
Total RNA was isolated with TRIzol reagent (Invitrogen), and the cDNA was synthesized using the FastQuant RT kit (Tiangen), which includes DNase treatment of RNA to eliminate genomic contamination. The expression patterns of each gene were determined by using SYBR premix ExTaq (TaKaRa). The small RNA was extracted by using a miRcute miRNA isolation kit (Tiangen), and a miRcute miRNA FirstStrand cDNA synthesis kit (Tiangen) was used for reverse transcription of miRNAs. The expression of miR-2187-3p was analyzed by using the miRcute miRNA qPCR detection kit (Tiangen). Real-time PCR was performed in an Applied Biosystems QuantStudio 3 (Thermo Fisher Scientific). β-Actin and 5.8S rRNA were employed as endogenous controls for mRNA/lncRNA and miRNA, respectively. Primer sequences are displayed in Table 1.
Western blotting.
Cellular lysates were generated by using 1× SDS-PAGE loading buffer. Proteins were extracted from cells, measured with the bicinchoninic acid (BCA) protein assay kit (Vazyme), then subjected to SDS-PAGE (10%), and transferred to polyvinylidene difluoride (PVDF; Millipore) membranes by semidry blotting (Bio-Rad Trans Blot Turbo system). The membranes were blocked with 5% BSA. Protein was blotted with different antibodies. The antibody against TBK1 was diluted at 1:500 (Boster), anti-Flag and anti-tubulin monoclonal antibodies were diluted at 1:2,000 (Sigma), and the anti-GFP monoclonal antibody was diluted at 1:1,000 (Sigma). The results were representative of three independent experiments. The immunoreactive proteins were detected by using WesternBright enhanced chemiluminescence (ECL; Advansta). Digital imaging was performed with a cold charge-coupled device (CCD) camera.
Cell viability and proliferation.
Cell viability was measured 48 h after transfection in SCRV-treated MICs with CellTiter-Glo luminescent cell viability assays (Promega) according to the manufacturer’s instructions. At the same time, cell proliferation was measured with a BeyoClick EdU cell proliferation kit following the manufacturer’s instructions (Beyotime). All the experiments were performed in triplicate.
Flow-cytometric analysis of apoptosis.
The apoptotic assay was conducted using an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Beyotime) and analyzed by flow cytometry (Beckman). The ratio of early to late apoptotic cells was determined to calculate the apoptotic rate (58).
Statistical analysis.
All experiments were replicated at least three times. The relative gene expression data were acquired using the 2−ΔΔCT method, and comparisons between groups were analyzed by one-way analysis of variance (ANOVA) followed by Duncan′s multiple-comparison tests (59). Data are means and standard errors (SE).
Data availability.
The RNA-Seq data for the non-SCRV-treated group and the SCRV-treated group have been deposited in the BioProject database under accession number PRJNA685924. The lncRNA MIR2187HG of the miiuy croaker has been deposited in GenBank under accession number MW771288.1.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31822057) and the National Key Research and Development Project (2018YFD0900503).
We declare no conflict of interest.
Contributor Information
Tianjun Xu, Email: tianjunxu@163.com.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Pietretti D, Wiegertjes GF. 2014. Ligand specificities of Toll-like receptors in fish: indications from infection studies. Dev Comp Immunol 43:205–222. 10.1016/j.dci.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, Matsumoto M, Seya T. 2008. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J Immunol 181:3474–3485. 10.4049/jimmunol.181.5.3474. [DOI] [PubMed] [Google Scholar]
- 3.Zhao YY, Sun XF, Nie XL, Sun LW, Tang TS, Chen DH, Sun QM. 2012. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog 8:e1003086. 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 5.Ji-Seung Y, Hiroki K, Akashi F. 2014. Sensing viral invasion by RIG-I like receptors. Curr Opin Microbiol 20:131–138. 10.1016/j.mib.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-M, Maniatis T. 2003. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 7.Tang JL, Yang Q, Xu CH, Zhao H, Liu YL, Liu CY, Zhou Y, Gai DW, Pei RJ, Wang Y, Hu X, Zhong B, Wang YY, Chen XW, Chen JZ. 2021. Histone deacetylase 3 promotes innate antiviral immunity through deacetylation of TBK1. Protein Cell 12:261–278. 10.1007/s13238-020-00751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Liu H, Zhang K, Meng Q, Hu L, Zhang Y, Xiang Z, Li J, Yang Y, Chen Y, Cui S, Tang H, Pei H, Bu Z, Weng C. 2020. Ubiquitin-conjugating enzyme 2S enhances viral replication by inhibiting type I IFN production through recruiting USP15 to deubiquitinate TBK1. Cell Rep 32:108044. 10.1016/j.celrep.2020.108044. [DOI] [PubMed] [Google Scholar]
- 9.Ye JS, Kim N, Lee KJ, Nam YR, Lee U, Joo CH. 2014. Lysine 63-linked TANK-binding kinase 1 ubiquitination by mindbomb E3 ubiquitin protein ligase 2 is mediated by the mitochondrial antiviral signaling protein. J Virol 88:12765–12776. 10.1128/JVI.02037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G, Liu B, Li Z, Wu H, Wang P, Zhao K, Jiang G, Zhang L, Gao C. 2016. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol 17:1342–1351. 10.1038/ni.3588. [DOI] [PubMed] [Google Scholar]
- 11.Liu SB, Lu LF, Lu XB, Li S, Zhang YA. 2019. Zebrafish FGFR3 is a negative regulator of RLR pathway to decrease IFN expression. Fish Shellfish Immunol 92:224–229. 10.1016/j.fsi.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Li C, Liu J, Zhu F, Wei S, Huang Y, Huang X, Qin Q. 2020. Palmitic acid promotes virus replication in fish cell by modulating autophagy flux and TBK1-IRF3/7 pathway. Front Immunol 11:1764. 10.3389/fimmu.2020.01764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Truscott M, Islam AB, Lightfoot J, López-Bigas N, Frolov MV. 2014. An intronic microRNA links Rb/E2F and EGFR signaling. PLoS Genet 10:e1004493. 10.1371/journal.pgen.1004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang L, Han C, Yang G, Li H, Li T, Yang S, Liang N, Zhong R, Jia L, Zhu D, Zhang Y. 2020. miR-378 and its host gene Ppargc1β exhibit independent expression in mouse skeletal muscle. Acta Biochim Biophys Sin (Shanghai) 52:883–890. 10.1093/abbs/gmaa061. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. 2014. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5:2318–2329. 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. 2014. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9:e86295. 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, Xu Y, Gorospe M, Wang JY. 2016. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 36:1332–1341. 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Cullen BR. 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13:313–316. 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu Q, Xu T, Zheng W, Chang R, Zhang L. 2020. Long noncoding RNA AANCR modulates innate antiviral responses by blocking miR-210-dependent MITA downregulation in teleost fish, Miichthys miiuy. Sci China Life Sci 64:1131–1148. 10.1007/s11427-020-1789-5. [DOI] [PubMed] [Google Scholar]
- 21.Chu Q, Xu T, Zheng W, Chang R, Zhang L. 2020. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PLoS Pathog 16:e1008670. 10.1371/journal.ppat.1008670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910. 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhir A, Dhir S, Proudfoot NJ, Jopling CL. 2015. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22:319–327. 10.1038/nsmb.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao X, Fan QL. 2020. LncRNA MIR503HG promotes high-glucose-induced proximal tubular cell apoptosis by targeting miR-503-5p/Bcl-2 pathway. Diabetes Metab Syndr Obes 13:4507–4517. 10.2147/DMSO.S277869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. 2014. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 281:3766–3775. 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 26.Magnadóttir B. 2006. Innate immunity of fish (overview). Fish Shellfish Immunol 20:137–151. 10.1016/j.fsi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang QY, Gui JF. 2015. Virus genomes and virus-host interactions in aquaculture animals. Sci China Life Sci 58:156–169. 10.1007/s11427-015-4802-y. [DOI] [PubMed] [Google Scholar]
- 28.Haque S, Kaushik K, Leonard VE, Kapoor S, Sivadas A, Joshi A, Scaria V, Sivasubbu S. 2014. Short stories on zebrafish long noncoding RNAs. Zebrafish 11:499–508. 10.1089/zeb.2014.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Zhang X, Li J, Huang S, Xiang S, Hu X, Liu C. 2018. Comprehensive analysis of coding-lncRNA gene co-expression network uncovers conserved functional lncRNAs in zebrafish. BMC Genomics 19:73–85. 10.1186/s12864-018-4458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen LNT, Nguyen LN, Zhao J, Schank M, Dang X, Cao D, Khanal S, Chand Thakuri BK, Lu Z, Zhang J, Li Z, Morrison ZD, Wu XY, El Gazzar M, Ning S, Wang L, Moorman JP, Yao ZQ. 2021. Long non-coding RNA GAS5 regulates T cell functions via miR21-mediated signaling in people living with HIV. Front Immunol 12:601298. 10.3389/fimmu.2021.601298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao W, Chang R, Sun Y, Xu T. 2021. MicroRNA-2187 modulates the NF-κB and IRF3 pathway in teleost fish by targeting TRAF6. Front Immunol 12:647202. 10.3389/fimmu.2021.647202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang R, Chu Q, Zheng W, Zhang L, Xu T. 2020. The Sp1-responsive microRNA-15b negatively regulates rhabdovirus-triggered innate immune responses in lower vertebrates by targeting TBK1. Front Immunol 11:625828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou J, Secombes CJ. 2011. Teleost fish interferons and their role in immunity. Dev Comp Immunol 35:1376–1387. 10.1016/j.dci.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Ran Z, Jiang Q, Hu N, Yu B, Zhu L, Shen L, Zhang S, Chen L, Chen H, Jiang J, Chen D. 2019. Vitamin D alleviates rotavirus infection through a microRNA-155-5p mediated regulation of the TBK1/IRF3 signaling pathway in vivo and in vitro. Int J Mol Sci 20:3562. 10.3390/ijms20143562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Li R, Tan Z, Shi J, Fu Y, Song Y, Zhu M, Zhang L, Huang J. 2020. E3 ubiquitin ligase ASB8 negatively regulates interferon via regulating TBK1/IKKi homeostasis. Mol Immunol 121:195–203. 10.1016/j.molimm.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Zhang H, Shi Z, Chen J, Li M, Shi H, Shi D, Guo L, Feng L. 2020. Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses 12:599. 10.3390/v12060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S, Yang S, He J, Guest JD, Ma Z, Yang L, Pierce BG, Tang Q, Zhang YJ. 2019. Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation. Virol 527:180–187. 10.1016/j.virol.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Lu LF, Li ZC, Zhang C, Zhou XY, Zhou Y, Zhang YA. 2019. Zebrafish MVP recruits and degrades TBK1 to suppress IFN production. J Immunol 202:559–566. 10.4049/jimmunol.1801325. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Yan CZ, Liu J, Yan J, Feng H. 2019. SIKE of black carp is a substrate of TBK1 and suppresses TBK1-mediated antiviral signaling. Dev Comp Immunol 90:157–164. 10.1016/j.dci.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Chu Q, Xu T. 2020. MicroRNA regulation of Toll-like receptor, RIG-I-like receptor and Nod-like receptor pathways in teleost fish. Rev Aquacult 12:2177–2193. 10.1111/raq.12428. [DOI] [Google Scholar]
- 41.Chu Q, Sun Y, Cui J, Xu T. 2017. MicroRNA-3570 modulates the NF-κB pathway in teleost fish by targeting MyD88. J Immunol 198:3274–3282. 10.4049/jimmunol.1602064. [DOI] [PubMed] [Google Scholar]
- 42.Xu TJ, Chu Q, Cui JX, Bi DK. 2018. Inducible microRNA-3570 feedback inhibits the RIG-I dependent innate immune response to rhabdovirus in teleost fish by targeting MAVS/IPS-1. J Virol 92:e01594-17. 10.1128/JVI.01594-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu TJ, Chu Q, Cui JX. 2018. Rhabdovirus-inducible microRNA-210 modulates antiviral innate immune response via targeting STING/MITA in fish. J Immunol 201:982–994. 10.4049/jimmunol.1800377. [DOI] [PubMed] [Google Scholar]
- 44.Chu Q, Bi D, Xu T. 2020. MicroRNA negatively regulates NF-κB-mediated immune responses by targeting NOD1 in the teleost fish, Miichthys miiuy. Sci China Life Sci 64:803–815. 10.1007/s11427-020-1777-y. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Zhou T, Chen B, Bai H, Bai Y, Zhao J, Pu F, Wu Y, Chen L, Shi Y, Ke Q, Zheng W, Chen J, Xu P. 2020. Identification and expression analysis of long non-coding RNA in large yellow croaker (Larimichthys crocea) in response to Cryptocaryon irritans infection. Front Genet 11:590475. 10.3389/fgene.2020.590475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng W, Chu Q, Xu T. 2021. Long noncoding RNA IRL regulates NF-κB-mediated immune responses through suppression of miR-27c-3p-dependent IRAK4 downregulation in teleost fish. J Biol Chem 296:100304. 10.1016/j.jbc.2021.100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng W, Chu Q, Xu T. 2021. The long noncoding RNA NARL regulates immune responses via microRNA-mediated NOD1 downregulation in teleost fish. J Biol Chem 296:100414. 10.1016/j.jbc.2021.100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, Chassat T, Bourette RP, Bourhis X, Adriaenssens E. 2015. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 6:29209–29223. 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. 2012. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14:659–665. 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Wang Y, Yu T, Nie E, Hu Q, Wu W, Zhi T, Jiang K, Wang X, Lu X, Li H, Liu N, Zhang J, You Y. 2017. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro Oncol 19:1195–1205. 10.1093/neuonc/nox017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Chu Q, Chang R, Xu T. 2020. Inducible microRNA-217 inhibits NF-κB- and IRF3-driven immune responses in lower vertebrates through targeting TAK1. J Immunol 205:1620–1632. 10.4049/jimmunol.2000341. [DOI] [PubMed] [Google Scholar]
- 52.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 53.Liu L, Hu Y, Shen YF, Wang GX, Zhu B. 2017. Evaluation on antiviral activity of coumarin derivatives against spring viraemia of carp virus in epithelioma papulosum cyprini cells. Antiviral Res 144:173–185. 10.1016/j.antiviral.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Su H, Chang R, Zheng W, Sun Y, Xu T. 2021. microRNA-210 and microRNA-3570 negatively regulate NF-κB-mediated inflammatory responses by targeting RIPK2 in teleost fish. Front Immunol 12:617753. 10.3389/fimmu.2021.617753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis BP, Shih I-h, Jones-Rhoades MW, Bartel DP, Burge CB. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798. 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 56.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2:e363. 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rusinov V, Baev V, Minkov IN, Tabler M. 2005. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 33:W696–W700. 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jing ZT, Liu W, Wu SX, He Y, Lin YT, Chen WN, Lin XJ, Lin X. 2018. Hepatitis B virus surface antigen enhances the sensitivity of hepatocytes to Fas-mediated apoptosis via suppression of AKT phosphorylation. J Immunol 201:2303–2314. 10.4049/jimmunol.1800732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-Seq data for the non-SCRV-treated group and the SCRV-treated group have been deposited in the BioProject database under accession number PRJNA685924. The lncRNA MIR2187HG of the miiuy croaker has been deposited in GenBank under accession number MW771288.1.