ABSTRACT
The ability of viruses to evade the host antiviral immune system determines their level of replication fitness, species specificity, and pathogenic potential. Flaviviruses rely on the subversion of innate immune barriers, including the type I and type III interferon (IFN) antiviral systems. Zika virus infection induces the degradation of STAT2, an essential component of the IFN-stimulated gene transcription factor ISGF3. The mechanisms that lead to STAT2 degradation by Zika virus are poorly understood, but it is known to be mediated by the viral NS5 protein that binds to STAT2 and targets it for proteasome-mediated destruction. To better understand how NS5 engages and degrades STAT2, functional analysis of the protein interactions that lead to Zika virus and NS5-dependent STAT2 proteolysis were investigated. Data implicate the STAT2 coiled-coil domain as necessary and sufficient for NS5 interaction and proteasome degradation after Zika virus infection. Molecular dissection reveals that the first two α-helices of the STAT2 coiled-coil domain contain a specific targeting region for IFN antagonism. These functional interactions provide a more complete understanding of the essential protein-protein interactions needed for Zika virus evasion of the host antiviral response and identify new targets for antiviral therapeutic approaches.
IMPORTANCE Zika virus infection can cause mild fever, rash, and muscle pain and in rare cases can lead to brain or nervous system diseases, including Guillain-Barré syndrome. Infections in pregnant women can increase the risk of miscarriage or serious birth defects, including brain anomalies and microcephaly. There are no drugs or vaccines for Zika disease. Zika virus is known to break down the host antiviral immune response, and this research project reveals how the virus suppresses interferon signaling, and may reveal therapeutic vulnerabilities.
KEYWORDS: NS5, STAT2, Zika virus
INTRODUCTION
Interferon (IFN)-family cytokines are recognized as fundamental mediators of innate antiviral responses that restrict virus replication and establish essential tissue-specific and species-specific barriers to virus infection (1). Type I IFN includes the single IFN-β and multiple IFN-α subtypes, which bind to a common receptor, and type III IFN includes IFN-λ1 to -4, which bind to a distinct receptor (2–5). Type I IFN induces the expression of hundreds of antiviral effector genes in nearly all nucleated cell types (6–8), but the type III IFN system is particularly important for protecting mucosal epithelial cells and establishing barrier functions of tissues. Although these two antiviral IFNs use distinct receptors, their intracellular signaling converges on the formation of the IFN-stimulated gene transcription factor ISGF3, a complex of IRF9 along with IFN-activated STAT1 and STAT2 (9). Consequently, these cytokine families activate an overlapping antiviral gene expression program leading to hundreds of IFN-stimulated genes (ISGs) (10–12). The accumulation of ISG products produces an antiviral state that creates a broadly effective barrier that is a hostile environment for virus replication. STAT2 plays an essential role in type I and type III IFN responses and is a master regulator of both tonic and acute ISG transcription and antiviral gene regulation (9, 13, 14). For most viruses, the IFN system provides strong selective pressure leading to the evolution of evasion and antagonism strategies (15–17) that can determine their replication efficiency, host range, virulence, and pathogenicity.
Zika virus (ZV) is a member of the flavivirus family with significant medical consequences for infected patients as well as their unborn offspring. Typically transmitted via Aedes mosquitos, ZV typically causes a mild to moderate illness in adults but can also drive more serious symptoms, including Guillain-Barré syndrome. In the developing fetus, ZV is capable of causing severe microcephaly, brain development defects, ocular anomalies, and other features of neurological impairment (18, 19). There are no current anti-Zika therapies or vaccines available, indicating an urgent need for understanding the mechanisms this virus uses to counteract immunity in its human host and for developing new treatment methods (20).
Like other flaviviruses, ZV has a single-stranded positive-sense RNA genome that is translated as a single polyprotein and processed by host and viral proteases to generate the structural proteins (capsid [C], premembrane [prM], and envelope [E]) and the nonstructural proteins required for host suppression, genome replication, and protein processing (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Several flavivirus NS proteins have been shown to alter host immune recognition or mediate antagonism of host antiviral responses (21). Among these, the NS5 protein has been shown to inhibit IFN signaling by targeting STAT2 (16, 22). Zika virus NS5 protein can target human STAT2 for proteasome-mediated degradation, but the mechanisms leading to STAT2 degradation are poorly understood. The closely related Dengue virus NS5 protein can target STAT2 for destruction by hijacking the cellular UBR4 protein (23), but ZV-mediated STAT2 degradation apparently does not require UBR4 (24). To gain greater insight into the mechanisms by which ZV NS5 targets the IFN response, the protein interactions that lead to STAT2 proteasome targeting were investigated. Results indicate that the STAT2 coiled-coil (CC) domain is necessary and sufficient for mediating ZV NS5 interaction and initiating degradation via the proteasome after ZV infection. Moreover, the first two α-helices of the STAT2 coiled-coil domain constitute a minimal targeting region for ZV IFN antagonism. These functional interactions provide a more nuanced and detailed understanding of the essential protein-protein interactions needed for ZV to evade the host antiviral response, and they represent potential targets for developing anti-ZV therapeutic approaches.
RESULTS
The STAT2 N terminus mediates Zika virus NS5 interaction.
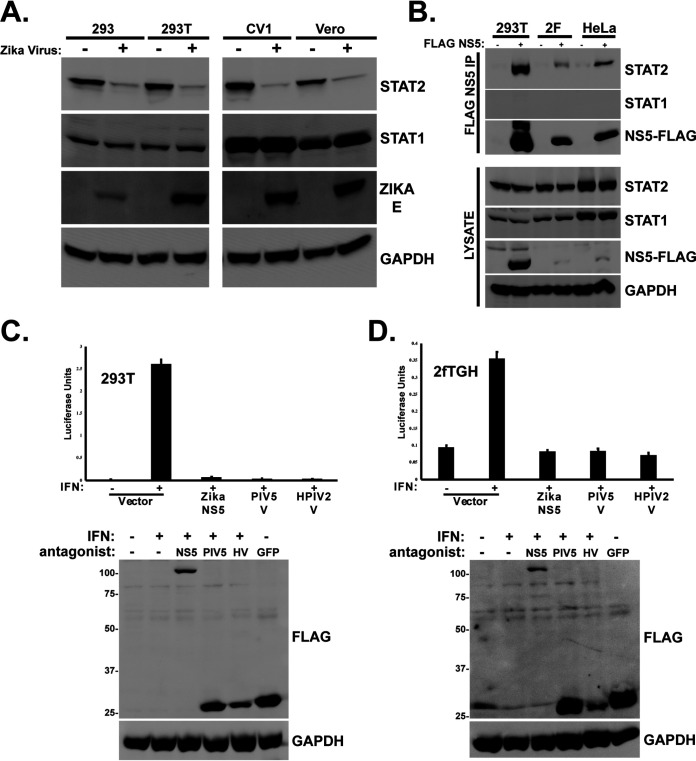
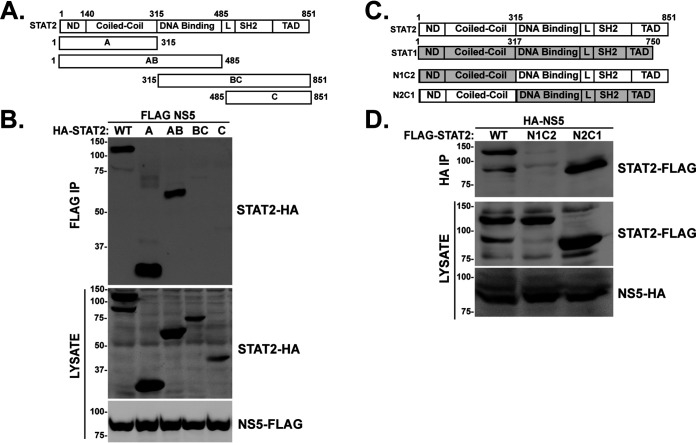
In support of prior investigations of ZV IFN evasion, we observed that infection of cultured primate cells results in loss of STAT2 but has no apparent effect on the closely related STAT1 (Fig. 1A). In agreement with this paradigm, expression of FLAG-tagged ZV NS5 by transient transfection coimmunoprecipitates with endogenous STAT2 (Fig. 1B) and is able to suppress IFN-stimulated transcription (Fig. 1C and D). These results are consistent with previous findings regarding specific STAT2 and IFN targeting by NS5 (16, 22). STAT2 shares the general domain structure of other STAT proteins (25), with an N-terminal domain (ND) adjacent to an extended 4-helix CC domain upstream of the DNA binding domain (DBD), linker domain (LD), SH2 domain, and transcriptional activation domain (TAD) (Fig. 2A and C). To define minimal STAT2 fragments that interact with ZV NS5 and lead to proteasomal degradation, experiments were carried out by expressing ZV NS5 with full-length human STAT2 or a partially overlapping set of STAT2 fragments (Fig. 2A). STAT2 proteins containing the first 315 amino acids, representing the STAT2 ND and CC regions, were able to coprecipitate with NS5, but the DNA-binding domain, linker domain, SH2, and TAD were not (Fig. 2B). In a corresponding experiment, a complementary set of chimeric STAT1/2 proteins with swapped ND and CC domain were assessed for NS5 association (Fig. 2C). Only the chimeric STAT protein containing amino acids 1 to 315 of STAT2 was able to coprecipitate with ZV NS5 (Fig. 2D, N2C1), but the complementary protein with the STAT1 ND and CC was not (Fig. 2D, N1C2). These results suggest that the ND and/or CC of STAT2 serves as the target for NS5 recognition.
FIG 1.
Zika virus STAT2 targeting and degradation requires amino acids 1 to 315. (A) Infection of primate cells with Zika virus results in STAT2 degradation. Cells were infected with 5 PFU/cell ZV MR766 and incubated 48 h prior to lysis and immunoblotting with antiserum for STAT2, STAT1, Zika E protein, or GAPDH. (B) Zika virus NS5 coprecipitates with endogenous STAT2. FLAG-tagged NS5 was expressed by plasmid transfection into the indicated cell lines, and lysates were immunoaffinity purified with FLAG M2 agarose prior to immunoblotting with antiserum for STAT2, STAT1, GAPDH, or FLAG peptide. (C) IFN-stimulated gene expression is blocked by ZV NS5. An ISRE-luciferase reporter gene was transfected into HEK293T cells with empty expression vector or plasmids encoding NS5 or the V proteins from paramyxoviruses PIV5 or HPIV2 and stimulated with 1,000 U/ml IFN-α for 8 h prior to the luciferase assay. Data were normalized to coexpressed Renilla luciferase; data are averages and standard deviations (SD) (n = 3). Immunoblot controls at the bottom were probed with anti-FLAG antiserum to detect viral IFN antagonist expression (ZV NS5, PIV5 V, and HPIV2 V) as indicated. FLAG GFP was included as a negative control and indicator of transfection efficiency. (D) Similar to panel C but using human fibrosarcoma 2fTGH cells. Immunoblot controls below were probed with anti-FLAG antiserum to detect viral IFN antagonist expression (ZV NS5, PIV5 V, and HPIV2 V) as indicated. FLAG GFP was included as a negative control and indicator of transfection efficiency.
FIG 2.
STAT2 N-terminal domains specify recognition by Zika Virus NS5. (A) Diagram of full-length WT STAT2 and STAT2 fragments A, AB, BC, and C with domains and boundaries indicated. (B) STAT2 amino acids 1 to 315 engage ZV NS5. HA-tagged STAT2 proteins were expressed in HEK 293T cells along with FLAG-tagged NS5, and lysates were immunoaffinity purified with FLAG M2 agarose prior to immunoblotting with HA antibody to detect coprecipitating fragments. (C) Diagram of STAT2 (white), STAT1 (gray), and chimeric N1C2 and N2C1, with domain boundaries indicated. (D) STAT2 amino acids 1 to 315 are transferable for NS5 recognition. FLAG-tagged STAT2, N1C2, and N2C1 proteins were expressed along with HA-tagged NS5 and immunoprecipitated with HA-specific antibody prior to immunoblotting with FLAG antiserum to detect coprecipitation.
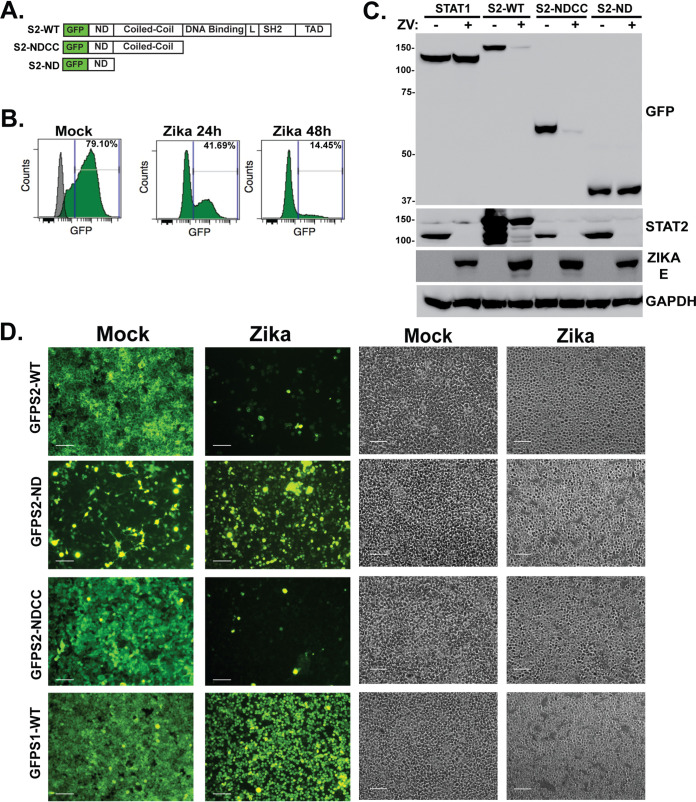
The STAT protein ND and CC domain are known to mediate protein interactions important for signaling, ISGF3 assembly, and cofactor association, and recent structural studies implicated both domains in ZV NS5 interaction (2, 25–30). To evaluate the contribution of the STAT2 domains toward NS5 recognition and degradation in intact cells, stable cell lines were selected expressing full-length STAT2 and its fragments as green fluorescent protein (GFP) fusion proteins (Fig. 3A). The ability of ZV to degrade the full-length, WT STAT2-GFP fusion protein was verified and quantified by flow cytometry. Infection with 5 PFU/cell ZV potently reduced the level of the GFP-STAT2 to 41% by 24 h postinfection (hpi) and to 14% after 48 hpi (Fig. 3B). GFP fusions with the combined STAT2 ND and CC domain (amino acids [aa] 1 to 315) and the isolated STAT2 ND (aa 1 to 140) were similarly expressed and detected by immunoblotting in uninfected cells. Zika virus infection induced degradation of endogenous STAT2 and GFP fusions to full-length WT STAT2 and the NDCC fragment (Fig. 3C). In contrast, neither GFP-STAT1 nor GFP-STAT2-ND fusions were targeted by ZV infection, though the endogenous cellular STAT2 protein remained susceptible to ZV degradation.
FIG 3.
The STAT2 coiled-coil domain is sufficient for Zika virus NS5 recognition and degradation. (A) Diagram of GFP-STAT2 fusion proteins carrying full-length STAT2, NDCC (aa 1 to 315), or ND (aa 1 to 140). (B) Quantification of ZV-mediated GFP-STAT2 degradation. Flow cytometry was used to measure GFP fluorescence in mock-infected or ZV-infected GFP-STAT2-WT-expressing cells. Cells were infected with 5 PFU/cell and analyzed at 24 and 48 hpi. The gray peak overlay indicates the background fluorescence level of control HEK293 cells. (C) Cell lines expressing GFP fusions were infected with ZV and lysates prepared at 48 hpi for immunoblotting with antiserum for GFP, STAT2, ZV E protein, or control GAPDH. (D) Cells expressing the indicated GFP fusion proteins were imaged 48 h after mock infection or ZV infection. GFP fusions with full-length STAT2 and STAT2-NDCC were efficiently degraded, but fusions with STAT2-ND or the negative control, STAT1, were unaffected by ZV infection. Bar = 200 μm. Corresponding phase-contrast images are shown in parallel.
To directly observe the effects of ZV on STAT2 and its fragments, the GFP-STAT2 fusion proteins were visualized with fluorescence microscopy (Fig. 3D). The subcellular distribution of GFP-STAT2 was found to reflect the known cytoplasmic distribution of endogenous STAT2, and fluorescence was dramatically eliminated by ZV infection. Unlike WT STAT2, the GFP-STAT1, GFP-STAT2-NDCC, and GFP-STAT2-ND were not retained in the cytoplasm and were more generally distributed in the cells. Infection with ZV resulted in efficient degradation of GFP-STAT2-NDCC, but GFP-STAT1 protein and GFP-STAT2-ND fluorescence remained intact after ZV infection. These findings indicate that the NDCC is responsible for directing ZV-mediated degradation.
The STAT2 coiled-coil domain is essential and sufficient for ZV recognition and degradation.
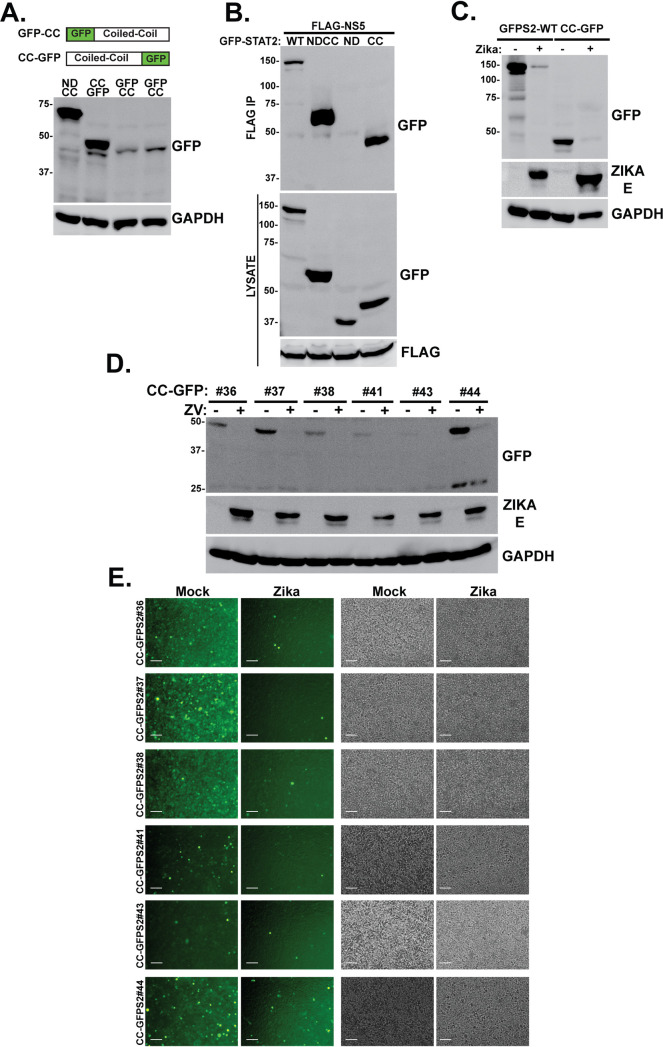
The inability of the ND to mediate GFP fusion protein degradation after ZV infection suggests that it may be dispensable for STAT2 targeting, and the key to STAT2 recognition and degradation may be encoded in the CC domain. To test this hypothesis, GFP fusions to the CC domain were constructed. Visualization of GFP-STAT2-CC degradation by microscopy was confounded by an observed intrinsic instability of both N-terminal and C-terminal GFP fusion proteins, giving rise to low abundance of the desired GFP fusion proteins relative to GFP-NDCC (Fig. 4A). The STAT2-CC-GFP fusion was more stable than GFP-STAT2-CC, and microscopic analysis of several independent clonally selected stable lines supported ZV-dependent degradation, despite their weak overall fluorescent signals (Fig. 4D and E). Despite this complication, immunoblotting was able to be used to evaluate both NS5 protein interaction and fusion protein degradation. Expression of STAT2-CC-GFP fusion protein along with FLAG-tagged NS5 resulted in specific coprecipitation of the STAT2-CC fusion (Fig. 4B). In addition to NS5 interaction, the STAT2-CC fusion also was targeted for degradation after ZV infection (Fig. 4C). These findings indicate that the STAT2 CC domain is an independent domain susceptible to recognition by NS5 and degradation during ZV infection.
FIG 4.
STAT2-CC domain is sufficient for recognition and degradation by ZV NS5. (A) (Top) Diagrams illustrating complementary GFP fusions with the STAT2 CC domain. (Bottom) Immunoblotting with GFP antiserum reveals relative steady-state expression levels of STAT2-CC fusion proteins. (B) STAT2-CC-GFP is recognized by ZV NS5. Coimmunoprecipitation assays were carried out as for Fig. 2B; GFP fusion proteins are indicated. (C) STAT2-CC-GFP is degraded by ZV infection. Cells expressing GFP-STAT2 or STAT2-CC-GFP were mock infected or infected with 5 PFU/cell ZV for 48 h prior to immunoblotting with antisera for GFP, Zika virus E protein, or GAPDH. (D) Immunoblot of lysates from STAT2-CC-GFP clones in HEK293 cells before and after infection with 5 PFU/ml ZV for 48 h. GFP antibody was used to detect STAT2-CC-GFP, and ZV infection was verified with E protein antiserum. (E) Fluorescence (left) and phase-contrast (right) micrographs of STAT2-CC-GFP clones before and after infection with 5 PFU/ml ZV for 48 h. Bar = 200 μm.
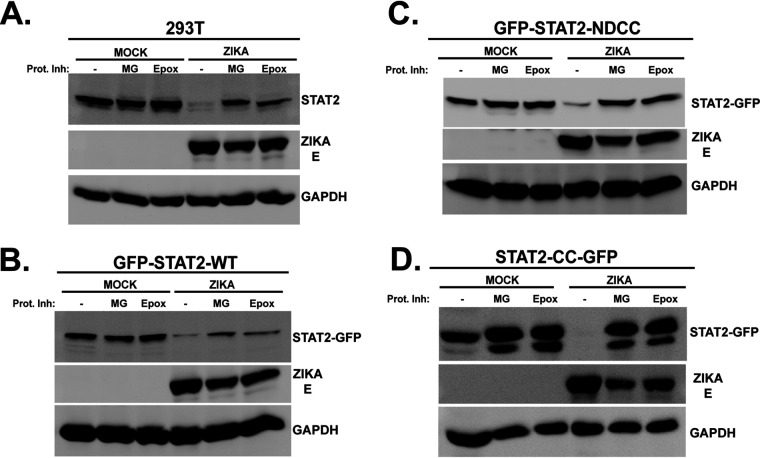
STAT2 coiled-coil domain degradation by Zika virus requires the proteasome.
Zika virus is thought to induce STAT2 degradation via the proteasome (24). To rule out alternate or off-target mechanisms for destabilization of the GFP-STAT2 fusion proteins and to validate the ability of the STAT2 CC domain to act as a discrete degron for degradation by the proteasome, the effects of proteasome inhibitors on ZV-induced degradation was evaluated. ZV infection efficiently eliminated endogenous STAT2 in control cells, and this was inhibited by both MG132 and epoxomicin treatment (Fig. 5A). Similar experiments were conducted with cell lines expressing GFP-STAT2-WT, GFP-STAT2-NDCC, or STAT2-CC-GFP (Fig. 5B to D). All the GFP fusion proteins were efficiently degraded following ZV infection, but they were protected by treatment with proteasome inhibitors. These results demonstrate that the STAT2 CC domain functions as an independent degron that can target a heterologous protein (GFP) for proteasomal degradation during ZV infection.
FIG 5.
STAT2-CC domain degradation requires the proteasome. Control HEK293T cells (A) or cells stably expressing GFP fusions with full-length (WT) STAT2 (B), STAT2-NDCC (C), or STAT2-CC (D) were mock infected or infected with ZV in the presence (+) or absence (−) of the proteasome inhibitor MG132 (MG) or epoxomicin (Epox). Lysates were prepared and probed with antiserum for the STAT2 C terminus, GFP, Zika virus E protein, or GAPDH as indicated.
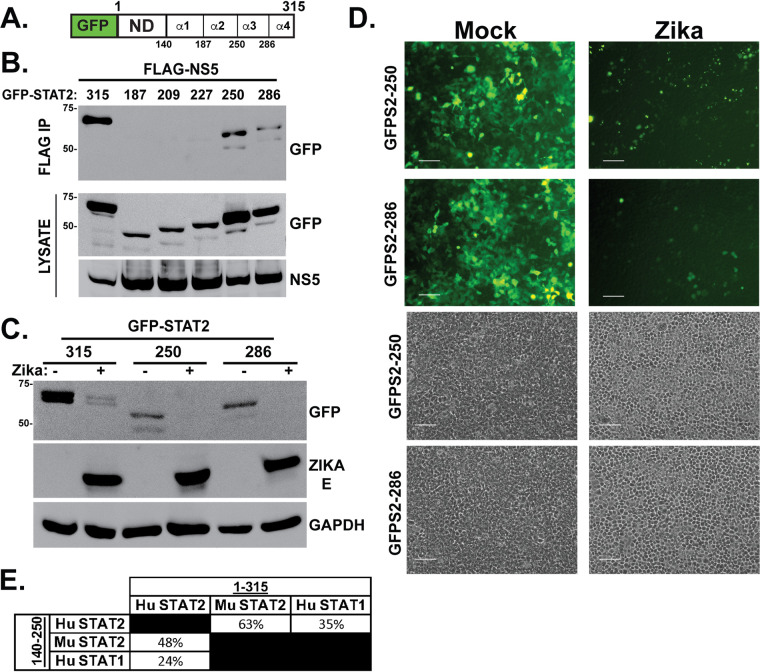
Two helices of the STAT2 coiled-coil domain mediate NS5 interaction and degradation.
The STAT2 CC consists of four α-helices that present a predominantly hydrophilic surface area for interaction with other proteins in signal transduction and gene regulation (31, 32). To further dissect the STAT2 CC interaction and targeting by ZV NS5, cell lines stably expressing proteins with 3-helix (aa 1 to 286), 2-helix (aa 1 to 250), and 1-helix (aa 1 to 187) fragments linked to GFP-ND were compared to GFP-STAT2-NDCC (aa 1 to 315), which has all four helices (Fig. 6A). The fragment containing STAT2 amino acids 1 to 250 (α1,2) retained the ability to coprecipitate with ZV NS5. However, further loss of 23 or more additional amino acids (i.e., the fragments containing aa 1 to 227, 1 to 209, and 1 to 187) prevented NS5 coprecipitation, defining the first two helices of the STAT2 CC as crucial for ZV recognition (Fig. 6B). To determine if this region can also mediate degradation, ZV was used to infect cells expressing GFP-STAT2-286 (α1,2,3) and GFP-STAT2-250 (α1,2). Consistent with their NS5 association, both of these proteins were degraded by ZV infection as determined by immunoblotting (Fig. 6C) as well as fluorescence microscopy (Fig. 6D). These experiments implicate the first two α-helices of the STAT2 CC domain as the essential targeting region for recognition and degradation after ZV infection.
FIG 6.
Zika virus degradation and NS5 interaction require only the first two helices of the STAT2 CC domain. (A) Diagram of GFP-STAT2-NDCC indicating boundaries of 4 α-helices of the STAT2 CC domain. (B) Helices α3 and α4 are dispensable for NS5 interaction. Coimmunoprecipitation assay using the indicated GFP-STAT2-NDCC domain fragments along with FLAG-tagged NS5. (C) Zika virus induces degradation of GFP-STAT2 fragments 1 to 250 and 1 to 286. Immunoblotting was carried out as for Fig. 3C but using the indicated GFP fusion proteins. (D) Zika virus induces degradation of GFP-STAT2 fragments 1 to 250 and 1 to 286; fluorescence microscopy carried out as for Fig. 3D but using cells expressing the indicated GFP fusion proteins. Bar = 200 μm. Corresponding phase-contrast images are shown at the bottom. (E) Comparison of the amino acid sequence identity among human (Hu) and mouse (Mu) STAT2 and human STAT1 in the NDCC (aa 1 to 315) or the first two helices of the CC domain (aa 140 to 250).
DISCUSSION
Virus replication in a host cell requires not only the fundamental processes of unpackaging, genome replication, mRNA transcription, and assembly of new particles but also the passive or active suppression of host antiviral immune responses. For most RNA viruses, the IFN antiviral response system is a primary target of immune suppression, and the IFN-responsive STAT protein, STAT2, is a common target for flaviviruses, including Zika, Dengue, and yellow fever viruses (33).
STAT2 degradation is a conserved feature for these viruses, but the precise mechanisms and cellular partners needed for IFN signaling disruption are not well conserved between virus species. As a result, it is essential to functionally evaluate each viral mechanism systematically to determine its vulnerabilities and idiosyncrasies to inform their exploitation for therapeutic advantage. Zika virus can actively eliminate STAT2 to prevent IFN responses, using the viral nonstructural protein, NS5, to target the typically stable STAT2 protein for efficient proteasome-mediated degradation (24). The experiments described here functionally dissect the interacting regions of ZV NS5 protein and its cellular target, STAT2. Data indicate that the STAT2 N-terminal 315 amino acids are key components for ZV NS5 recognition and degradation, corresponding to the STAT2 N domain (ND) and coiled-coil (CC) domain. Substitution of these 315 amino acids was sufficient to transfer the targeting ability of NS5 to a STAT2-STAT1 chimeric protein. This region is only ∼35% identical between STAT1 and STAT2 (Fig. 6E), accounting for the specificity of NS5 interaction and degradation for STAT2. Expression of the STAT2 ND or CC domain as GFP fusions was used to delineate the minimal region(s) that is sufficient to act as an NS5 target. Experiments revealed that not only did GFP fused to the STAT2 NDCC domains bind to NS5 and rapidly turn over in infected cells but also the CC domain alone (aa 140 to 315) was sufficient to mediate NS5 interaction and Zika-dependent degradation via the cellular proteasome.
Sequential elimination of the four α-helices of the STAT2 CC indicated that helix 1 and helix 2 are essential for NS5 interaction and degradation but helix 3 and helix 4 are dispensable, narrowing the essential region for determining STAT2 targeting specificity to amino acids 140 to 250. STAT2 targeting by ZV is known to be species restricted; the murine STAT2 protein is not degraded in infected cells and does not interact with NS5 (24). Alignment of the human and mouse STAT2 amino acid sequences in the NDCC region (aa 1 to 315) indicates that they are 63% identical, but focusing on amino acids 140 to 250 reveals only 48% sequence identity (Fig. 6E). This lower sequence conservation may account for species-specific STAT2 targeting by ZV NS5. These sequence differences corresponding to differential ZV NS5 targeting support a role for the identified degron as a crucial viral target for highly specific host IFN evasion and therefore represent a novel therapeutic target.
During the course of these investigations, crystallographic and cryo-electron microscopy (cryo-EM) structures of the Zika virus and Dengue virus NS5-STAT2 complex were described that provide further support of our functional analysis (30). The structures illustrate the importance of the CC domain in forming an NS5-STAT2 complex. The two domains of NS5, the RNA-dependent RNA polymerase (RDRP) and the methyltransferase (MT), were both implicated in STAT2 contact. The NS5 RDRP fragment was found to associate with helices 1 and 2, and conserved STAT2 residues F175 and R176 established key contacts with NS5. Atomic models based on the cryo-EM structures show that the STAT2 CC is anchored at an interdomain cleft formed between the NS5 MT and RDRP domains, indicating that both domains of NS5 interact with STAT2. These results strongly support our conclusion that the primary functional ZV target site is the STAT2 CC domain and invoke the importance of amino acids 140 to 250. Interestingly, the same region of the STAT2 CC is required for IRF9 binding to form the IFN-activated ISGF3 transcription complex heterotrimer (32). Functional antagonism between hSTAT2 and ZIKV NS5 in prevention of ISGF3 assembly was observed in competition assays, indicating that multilevel NS5 IFN suppression is determined by the STAT2 CC interaction (30).
The structures also recognized a contact site between the STAT2 ND and the NS5 RDRP fragment, mediated by an invisible linker between the ND and the CC domain. As our results strongly indicate that the STAT2 ND itself does not coprecipitate with NS5 or undergo degradation during ZV infection, the observed ND-RDRP interaction may represent a low-affinity interaction that could participate in other aspects of NS5 activity. Alternatively, the ND engagement of NS5 may reflect a means by which STAT2 counterantagonizes NS5 polymerase action as an antiviral measure.
In summary, the results presented here define a transferable NS5-mediated targeting sequence that allows ZV to overcome IFN antiviral immunity by proteasomal degradation of STAT2. As a key feature of ZV immune escape, this region may be exploited as a target for development of antiviral compounds or other therapeutic strategies.
MATERIALS AND METHODS
Cells, viruses, and inhibitors.
HEK293, Bosc, 293T, CV1, HeLa, 2fTGH, and Vero cells were grown in DMEM (Invitrogen, catalog no. 11965118) supplemented with 10% Cosmic Calf serum (CCS; HyClone, catalog no. 25200114) and 1% penicillin-streptomycin (Invitrogen, catalog no. 15140122). Cells are routinely tested for mycoplasma contamination and regularly restored from early-passage frozen stocks. Zika virus (ATCC, catalog no. VR-84, MR 766 strain) was propagated and titrated in Vero cells. Infection of cells with Zika virus was performed at a multiplicity of infection (MOI) of 5 PFU/cell in serum-free medium. After 1 h, cells were washed, placed in growth medium supplemented with 2% CCS, and harvested 48 h later. The proteasome inhibitors MG132 (20 μM; Selleck Chemicals, catalog no. S2619) and epoxomicin (400 nM; R&D Systems, catalog no. I-110) were incubated with cells for 12 h. Stable cell lines were generated by expressing GFP-STAT2 plasmids in HEK293 cells and selecting with 500 μg/ml G418. Positive clones were ultimately screened by immunoblotting with a GFP antibody, and highly expressing cells were selected by fluorescence-activated cell sorting.
Plasmids and mutagenesis.
To generate hemagglutinin (HA)-tagged NS5, Vero cells were infected with Zika virus MR 766 strain, total RNA was isolated using TRIzol (Invitrogen, catalog no. 15596018), and cDNA was amplified by PCR with gene-specific primers and cloned into a pcDNA3-HA expression vector. pLV_Zika_NS5_Flag was obtained from Vaithi Arumugaswami (Addgene plasmid no. 79639) and used to transfect cells directly for FLAG protein expression.
STAT2 expression vectors, fragments, and STAT1 chimeras were previously described (34–36). To generate GFP fusion proteins, the full-length STAT2 open reading frame (ORF) was amplified by PCR and subcloned into mammalian expression plasmid (pEGFP-C1). Truncations were generated by stop codon insertion mutagenesis with a QuikChange II XL mutagenesis kit (Agilent, catalog no. 200522), and a GFP fusion expressing the STAT2 CC domain alone was constructed by PCR with specific primers to enable cloning into plasmid pEGFP-N1 or pEGFP-C1. All constructs were confirmed by DNA sequencing.
Immunoprecipitations and immunoblotting.
For coimmunoprecipitation experiments, FLAG-tagged and HA-tagged or GFP-tagged plasmids were transfected in HEK293T cells by the calcium phosphate method. At 48 h posttransfection, cells were harvested by first washing with cold phosphate-buffered saline and then lysed with whole-cell extract buffer (WCEB) consisting of 50 mM Tris, 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol (DTT), 2.5 mM sodium vanadate, and protease inhibitors. Cell lysates were then precleared with Sepharose beads. A percentage of the cleared lysates was reserved for analysis, and the remainder was incubated with FLAG M2 affinity beads (Sigma, catalog no. A2220) overnight and washed five times with WCEB. FLAG and HA immunoprecipitates (IPs) were eluted with SDS sample buffer, and FLAG and GFP IPs were eluted with 3× FLAG peptide (ApexBio, catalog no. A6001); IPs were then separated by SDS-PAGE and processed for immunoblotting. For immunoblotting, the separated proteins were transferred to nitrocellulose and probed with commercial primary antibodies recognizing FLAG (Sigma, catalog no. F3165), HA (Sigma, catalog no. H3663), GFP (Santa Cruz Biotechnology, catalog no. sc-9996), STAT2-C (Santa Cruz Biotechnology, catalog no. sc-476), STAT1-C (Santa Cruz Biotechnology, catalog no. sc-345), Zika virus E (Millipore Sigma, catalog no. MABF2046), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, catalog no. sc-47724). Proteins were visualized by enhanced chemiluminescence (PerkinElmer, catalog no. NEL105001EA). Figures show representative images of 3 to 5 biological replicates.
Reporter gene assays.
HEK293 and 2fTGH cells were transfected with a 5× interferon-sensitive response element (ISRE) luciferase reporter gene along with a Renilla luciferase vector and expression vectors for Zika virus NS5, parainfluenza virus type 5 (PIV5) V, and human parainfluenza virus type 2 (HPIV2) V. At 24 h posttransfection, cells were stimulated for 8 h with IFN-α (1,000 U/mL), harvested, and assayed for firefly and Renilla luciferase activities using the dual-luciferase reporter assay system (Promega, catalog no. E1960). Relative luciferase activity was calculated by dividing the firefly luciferase values by those of the Renilla luciferase. Data are plotted as mean values, with error bars representing standard deviations.
Fluorescence microscopy and imaging.
Cell sorting was performed at the Northwestern Flow Cytometry Core Facility on a BD FACSAria SORP system and BD FACSymphony S6 SORP system. Fluorescent images were obtained using a Nikon Eclipse Ti fluorescence microscope equipped with a Photometrics CoolSnap CF2 camera at ×10 magnification. Images were adjusted for brightness and contrast using Adobe Photoshop.
ACKNOWLEDGMENTS
This work was supported by NIH grant R21 AI148949-01 to C.M.H. J.J.L. and G.A. were supported by a Cellular and Molecular Basis of Disease Training Grant (NIH T32 GM008061), and the Northwestern University Flow Cytometry Core Facility is supported by Lurie Cancer Center support grants NCI CA060553, NIH 1S10OD011996-01, and 1S10OD026814-01. The BD FACSAria SORP system and BD FACSymphony S6 SORP system were purchased through the support of NIH 1S10OD011996-01 and 1S10OD026814-01.
Contributor Information
Curt M. Horvath, Email: Horvath@northwestern.edu.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147:258–267. 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Darnell JE, Jr.. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell JE, Jr. 1998. Studies of IFN-induced transcriptional activation uncover the Jak-Stat pathway. J Interferon Cytokine Res 18:549–554. 10.1089/jir.1998.18.549. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE, Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421. 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 5.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80:4501–4509. 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE, Jr.. 1988. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative control. Genes Dev 2:383–393. 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 7.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell JE, Jr.. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci USA 84:6394–6398. 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA 95:15623–15628. 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Au-Yeung N, Horvath CM. 2018. Transcriptional and chromatin regulation in interferon and innate antiviral gene expression. Cytokine Growth Factor Rev 44:11–17. 10.1016/j.cytogfr.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odendall C, Kagan JC. 2015. The unique regulation and functions of type III interferons in antiviral immunity. Curr Opin Virol 12:47–52. 10.1016/j.coviro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou JH, Wang YN, Chang QY, Ma P, Hu Y, Cao X. 2018. Type III interferons in viral infection and antiviral immunity. Cell Physiol Biochem 51:173–185. 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 12.Wack A, Terczyńska-Dyla E, Hartmann R. 2015. Guarding the frontiers: the biology of type III interferons. Nat Immunol 16:802–809. 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus TA, Lau JF, Parisien JP, Horvath CM. 2003. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem 278:13033–13038. 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- 14.Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, Gossenreiter T, Muller M, Novatchkova M, Decker T. 2019. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun 10:2921. 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale M, Jr, Sen GC. 2009. Viral evasion of the interferon system. J Interferon Cytokine Res 29:475–476. 10.1089/jir.2009.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miorin L, Maestre AM, Fernandez-Sesma A, Garcia-Sastre A. 2017. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun 492:587–596. 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22:176–184. 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowen JR, Zimmerman MG, Suthar MS. 2018. Taking the defensive: immune control of Zika virus infection. Virus Res 254:21–26. 10.1016/j.virusres.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130:69–80. 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munjal A, Khandia R, Dhama K, Sachan S, Karthik K, Tiwari R, Malik YS, Kumar D, Singh RK, Iqbal HMN, Joshi SK. 2017. Advances in developing therapies to combat Zika virus: current knowledge and future perspectives. Front Microbiol 8:1469. 10.3389/fmicb.2017.01469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17:1766–1775. 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best SM. 2017. The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J Virol 91:e01970-16. 10.1128/JVI.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aaronson DS, Horvath CM. 2002. A road map for those who don't know JAK-STAT. Science 296:1653–1655. 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 26.Au-Yeung N, Mandhana R, Horvath CM. 2013. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2:e23931. 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majoros A, Platanitis E, Kernbauer-Holzl E, Rosebrock F, Muller M, Decker T. 2017. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol 8:29. 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. 1998. Structure of the amino-terminal protein interaction domain of STAT-4. Science 279:1048–1052. 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE, Jr, Kuriyan J. 2003. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci 12:361–365. 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Thurmond S, Zhou K, Sanchez-Aparicio MT, Fang J, Lu J, Gao L, Ren W, Cui Y, Veit EC, Hong H, Evans MJ, O'Leary SE, Garcia-Sastre A, Zhou ZH, Hai R, Song J. 2020. Structural basis for STAT2 suppression by flavivirus NS5. Nat Struct Mol Biol 27:875–885. 10.1038/s41594-020-0472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827–839. 10.1016/S0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 32.Rengachari S, Groiss S, Devos JM, Caron E, Grandvaux N, Panne D. 2018. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc Natl Acad Sci USA 115:E601–E609. 10.1073/pnas.1718426115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurmond S, Wang B, Song J, Hai R. 2018. Suppression of type I interferon signaling by flavivirus NS5. Viruses 10:712. 10.3390/v10120712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisien JP, Lau JF, Horvath CM. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J Virol 76:6435–6441. 10.1128/jvi.76.13.6435-6441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien JP, Lau JF, Rodriguez JJ, Ulane CM, Horvath CM. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J Virol 76:4190–4198. 10.1128/jvi.76.9.4190-4198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau JF, Nusinzon I, Burakov D, Freedman LP, Horvath CM. 2003. Role of metazoan mediator proteins in interferon-responsive transcription. Mol Cell Biol 23:620–628. 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]