ABSTRACT
A comprehensive analysis and characterization of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection model that mimics non-severe and severe coronavirus disease 2019 (COVID-19) in humans is warranted for understating the virus and developing preventive and therapeutic agents. Here, we characterized the K18-hACE2 mouse model expressing human (h)ACE2 in mice, controlled by the human keratin 18 (K18) promoter, in the epithelia, including airway epithelial cells where SARS-CoV-2 infections typically start. We found that intranasal inoculation with higher viral doses (2 × 103 and 2 × 104 PFU) of SARS-CoV-2 caused lethality of all mice and severe damage of various organs, including lung, liver, and kidney, while lower doses (2 × 101 and 2 × 102 PFU) led to less severe tissue damage and some mice recovered from the infection. In this hACE2 mouse model, SARS-CoV-2 infection damaged multiple tissues, with a dose-dependent effect in most tissues. Similar damage was observed in postmortem samples from COVID-19 patients. Finally, the mice that recovered from infection with a low dose of virus survived rechallenge with a high dose of virus. Compared to other existing models, the K18-hACE2 model seems to be the most sensitive COVID-19 model reported to date. Our work expands the information available about this model to include analysis of multiple infectious doses and various tissues with comparison to human postmortem samples from COVID-19 patients. In conclusion, the K18-hACE2 mouse model recapitulates both severe and non-severe COVID-19 in humans being dose-dependent and can provide insight into disease progression and the efficacy of therapeutics for preventing or treating COVID-19.
IMPORTANCE The pandemic of coronavirus disease 2019 (COVID-19) has reached nearly 240 million cases, caused nearly 5 million deaths worldwide as of October 2021, and has raised an urgent need for the development of novel drugs and therapeutics to prevent the spread and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). To achieve this goal, an animal model that recapitulates the features of human COVID-19 disease progress and pathogenesis is greatly needed. In this study, we have comprehensively characterized a mouse model of SARS-CoV-2 infection using K18-hACE2 transgenic mice. We infected the mice with low and high doses of SARS-CoV-2 to study the pathogenesis and survival in response to different infection patterns. Moreover, we compared the pathogenesis of the K18-hACE2 transgenic mice with that of the COVID-19 patients to show that this model could be a useful tool for the development of antiviral drugs and therapeutics.
KEYWORDS: COVID-19, K18-hACE2, SARS-CoV-2, infectious disease, lung infection, mouse model
INTRODUCTION
The global pandemic of coronavirus disease 2019 (COVID-19) caused by the extremely contagious RNA severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to nearly 240 million cases and nearly 5 million deaths worldwide as of October 2021 (https://www.worldometers.info/coronavirus/). The median time to development of symptoms is 5.1 days after exposure to SARS-CoV-2 (1). The median time from onset to clinical recovery for mild cases is approximately 2 weeks, with 3–6 weeks for patients with severe or advanced disease (2). The main symptoms, such as fever or chills, cough, shortness of breath, difficulty breathing, and sore throat normally ease after recovery; however, 3-17% of patients, especially the elderly and individuals with cancer and metabolic and immune disorders, develop rapid viral replication and severe lung damage, resulting in severe disease with greatly increased risk of death (3–6). Many groups are trying to understand this rapid disease progression, and studies suggest that it may involve an impaired immune response (7–9). A mouse model that appropriately mirrors the progression of the human disease should help the development of vaccines or therapeutics for COVID-19 and could be used as a conceptual basis for rapid response to future viral pandemics.
During infection with SARS-CoV-2, the coronavirus spike (S) glycoprotein promotes SARS-CoV-2 entry into host cells via the host receptor angiotensin converting enzyme 2 (ACE2) (10). K18-hACE2 transgenic mice, which were originally developed to study the infection of SARS-CoV, express human ACE2, the receptor used by SARS-CoV-2 to gain entry into cells (11). The human keratin 18 (K18) promoter is responsible for directing expression to airway epithelial cells, where respiratory infections usually begin. Recent research reported that this SARS-CoV-2 K18-hACE2 mouse infection model develops severe interstitial pneumonia with high viral copy numbers into the lungs and immune cell infiltration into the alveoli (12–16). A comprehensive analysis and characterization of this model will be useful for studying the mechanisms of pathogenesis by SARS-CoV-2 and developing preventive, therapeutic, and vaccinal agents.
In the current study, we evaluated K18-hACE2 transgenic mice in response to multiple infectious doses, using multiple tissues, to gain a better understanding of the response to SARS-CoV-2 infection. We also evaluated the response of K18-hACE2 mice to high-dose re-infection after recovery from a low-dose infection.
RESULTS
The K18-hACE2 model is a lethal infection model for SARS-CoV-2 infection.
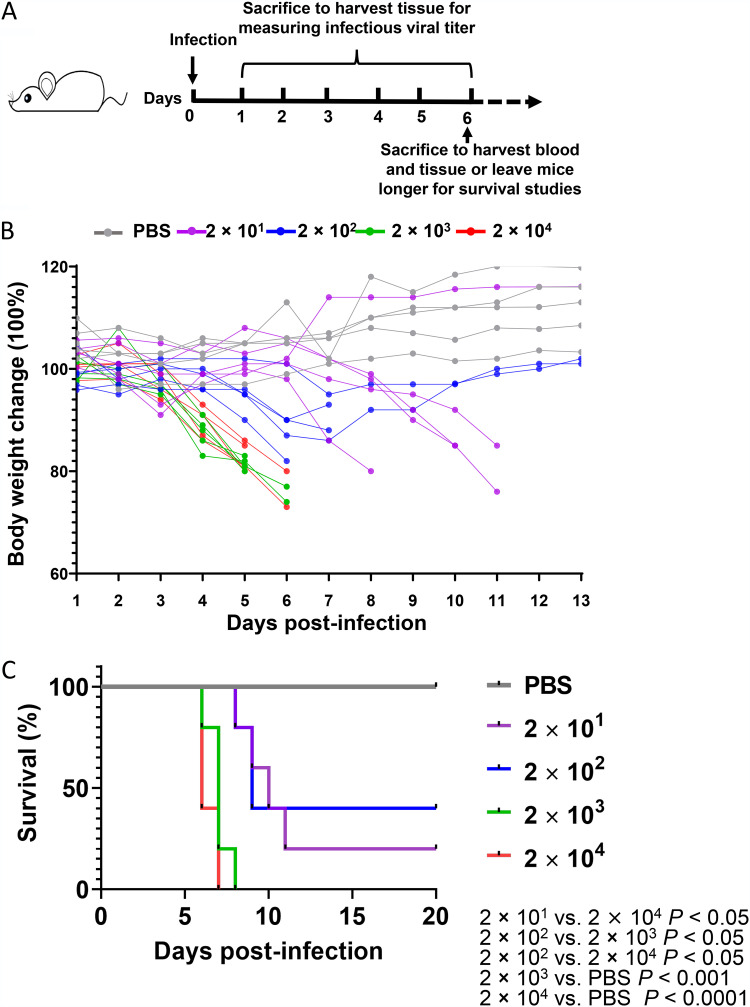
To understand the effects of SARS-CoV-2 viral dosage on pathogenesis and survival, we established an infection model using the K18-hACE2 transgenic mice, in which hACE2 expression is driven by the epithelial cell-specific promoter K18 in C57BL/6 mice (11). The mice were infected with SARS-CoV-2 WA-1/US (BEI Resources) at 2 × 101 PFU (lowest dose), 2 × 102 PFU (low dose), 2 × 103 PFU (high dose), 2 × 104 PFU (highest dose), or PBS vehicle control via intranasal inoculation. Mice were monitored for body weight change and survival or euthanized at various days postinfection (p.i.) to measure viral copy numbers and infectious viral loads as well as to determine pathology (Fig. 1A). We found that mouse body weight decreased in mice infected with 2 × 103 PFU and 2 × 104 PFU by day 4 p.i., whereas mice infected with 2 × 101 PFU and 2 × 102 PFU showed a less dramatic body weight decrease; a body weight decrease of 20% occurred at days 5–6 in the higher-dose groups, but took until day 10–11 for 80% of the mice in the low-dose groups (Fig. 1B). Up to 30% of the mice from the lower-dose groups started to gain weight by day 4 (for 2 × 101 PFU, lowest) and day 8 (for 2 × 102 PFU, low) (Fig. 1B), indicating a recovery from the viral infection in low-dose groups. Consistent with body weight data, 90% of the mice infected with 2 × 103 PFU (high) or 2 × 104 PFU (highest) died approximately at day 7, whereas 50% of the mice infected with 2 × 101 PFU or 2 × 102 PFU died approximately at day 10 (Fig. 1C). One or two mice out of five infected with 2 × 101 PFU or 2 × 102 PFU, respectively, and none infected with 2 × 103 PFU or 2 × 104 PFU, recovered and survived until the end of follow-up on day 20 p.i. (Fig. 1C).
FIG 1.
K18-hACE2 mouse infection model with high and low doses of SARS-CoV-2. (A) Experimental scheme of the K18-hACE2 mouse infection model. Mice were intranasally infected with 2 × 101, 2 × 102, 2 × 103, or 2 × 104 PFU virus per mouse. Blood samples were collected at 6 days postinfection. Tissue samples were collected from day 1 to day 6 postinfection. Mouse body weights (B) and survival (C) were monitored daily for 13 days. Each dot represents one mouse at the indicated time point.
Expression distribution of viral genes in the tissues of K18-hACE2 mice infected with high or low doses of SARS-CoV-2.
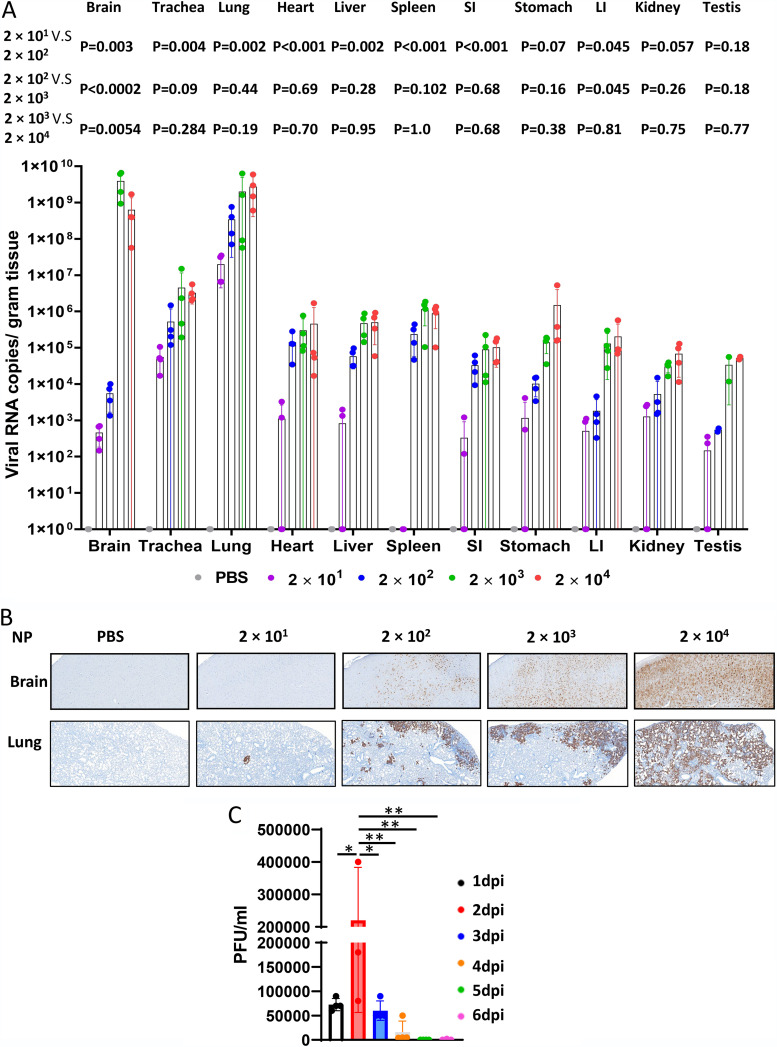
To further investigate the viral infection pattern, we used reverse transcriptase (RT)-PCR to measure viral spike protein RNA levels in the brain, trachea, lung, liver, spleen, small intestine, stomach, large intestine, kidney, and testis. The mean viral copy numbers in tissues, where the dose-dependent effect is expected, were similar for the high and highest two doses in all tissues. Interestingly, mice infected with 2 × 101 PFU did not present virus in the spleen (Fig. 2A). We observed three types of dose-response relationships. In the stomach, there was a stepwise dose-response relationship. In the heart, liver, spleen, and small intestine, we observed a plateau, with the 3 higher dose groups having similar viral copy numbers. In the brain, large intestine, and testis, mice infected with 2 × 101 PFU and 2 × 102 PFU had similar viral copy numbers, and mice infected with 2 × 103 PFU and 2 × 104 PFU had similar viral copy numbers, but much higher than those of the two lower doses. This indicates that there are tissue-specific factors modulating the effect of dose on viral copy number. Viral RNA levels were high in the lungs of animals in all dose groups (Fig. 2A), consistent with the notion that the lung is an important site of infection, especially at lower viral doses. Viral RNA levels were also very high in the brain of mice infected with 2 × 103 PFU or 2 × 104 PFU. We used immunohistochemistry (IHC) to measure the expression of SARS-CoV-2 nucleocapsid protein (NP) in formalin-fixed, paraffin-embedded (FFPE) tissues, which confirmed the infection pattern in the lung and brain of the K18-hACE2 mice infected with a series of increasing viral doses, demonstrating that viral spread in tissues was also dose-dependent (Fig. 2B). We also measured the infectious virus titer of the lung harvested from mice infected with the highest 2 × 104 PFU dose at different time points p.i., by a plaque assay on homogenized tissues. The infectious virus titer peaked at day 2 p.i. and then decreased in a time-dependent manner, reaching almost complete virus clearance at day 5 p.i. (Fig. 2C). Our data are consistent with a previous report using a higher dose (16).
FIG 2.
Viral quantification in mice after SARS-CoV-2 infection. (A) Viral RNA levels are shown for the brain, trachea, lung, heart, liver, spleen, small intestine (SI), stomach, large intestine (LI), kidney, and testis. (B) Viral nucleocapsid protein (NP) was detected in the brain and lung of the mice infected with high and low doses of SARS-CoV-2 (scale bar, 40 μm). (C) Infectious virus loads in the lung at different time points. Mice were infected with 2 × 104 PFU at day 0. Lungs were collected and homogenized at indicated time points. The infectious virus loads were measured by a plaque assay. Data are presented as mean ± standard deviations. Statistical analyses were performed by one-way ANOVA with P values corrected for multiple comparisons by the Bonferroni test (n = 3 or 4 mice). *, P < 0.05; **, P < 0.01.
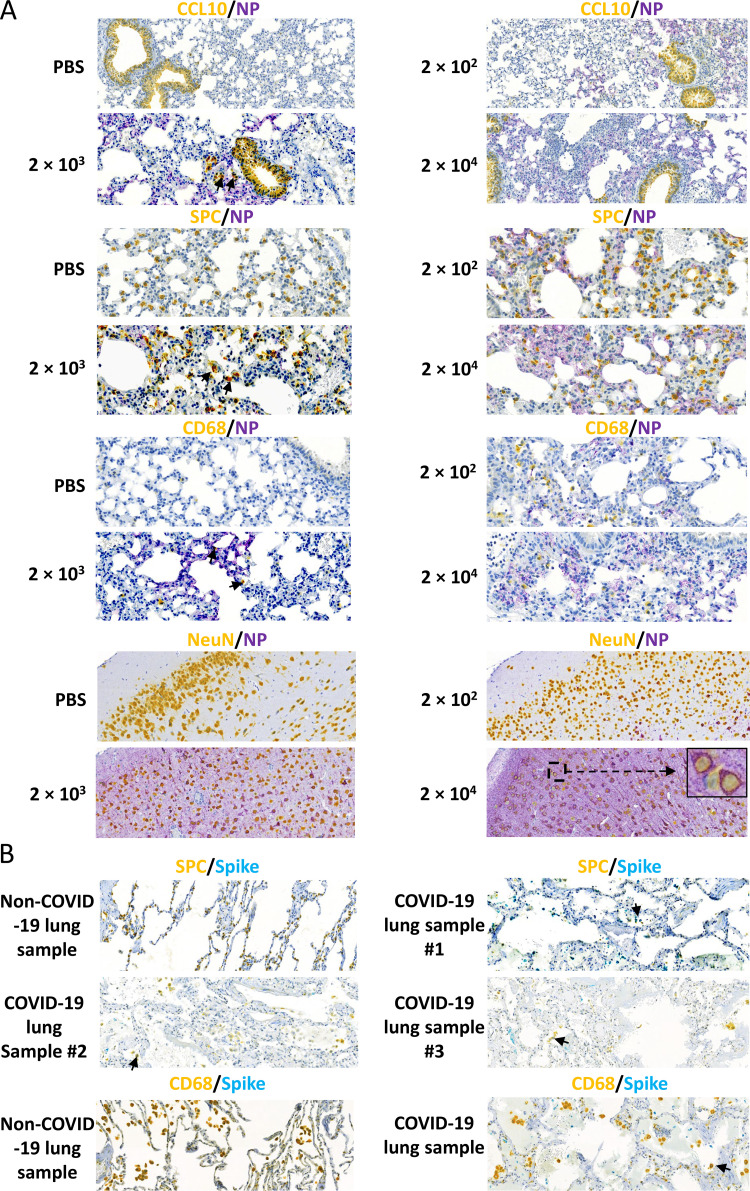
To evaluate lung cell types that are susceptible to SARS-CoV-2 infection, we performed double IHC staining on samples from mice infected with different doses of SARS-CoV-2. We double stained lung sections for viral NP and CCL10, which is the marker of club (clara) cells, or surfactant protein C (SPC), the marker of alveolar epithelial type 2 cells, respectively. We found that some club cells and many alveolar type 2 cells co-localized with NP, suggesting they are susceptible to SARS-CoV-2 infection (Fig. 3A). Macrophages are a major immune cell population in the lung, where they act as a first-line defense against invading pathogens. We therefore used CD68 as a macrophage marker to examine if SARS-CoV-2 could infect tissue-resident macrophages in the lung. We noticed that that CD68 positive cells migrated dramatically to the lung in response to SARS-CoV-2 infection, although only a few macrophages stained positive for NP, suggesting that it could be possible for SARS-CoV-2 to suppress immune response via infection of lung macrophages (Fig. 3A). Consistent with the finding in the K18-hACE2 mouse model, in postmortem lung samples from COVID-19 patients, a few viral spike proteins (teal) were colocalized with alveolar epithelial type 2 cells (yellow) and CD68-positive cells (yellow) (Fig. 3B). These results are in line with another SARS-CoV-2 infection mouse model that uses CRISPR/Cas9 knock-in of hACE2 into Exon 2 of the mAce2 gene, as well as COVID-19 patient samples, which show SARS-CoV-2 in macrophages of pulmonary alveolus (14, 17). Given the high levels of SARS-CoV-2 in the brain, we examined whether neurons were infected with SARS-CoV-2. We found strong overlap between staining for the neuronal marker Neu and staining for NP (Fig. 3A).
FIG 3.
Viral distribution in mice after SARS-CoV-2 infection. (A) Representative images show double staining of NP (purple) with lung club (Clara) cells and alveolar type 2 cells using the markers CCL10 (yellow) and SPC (yellow), respectively, macrophages using the CD68 marker (yellow) (20× magnification for each), and neurons using the NeuN marker (yellow) in mice infected with high-dose SARS-CoV-2 (10× magnification, with 100× magnification for the boxed area in the 2 × 104 PFU group). Black arrows indicate double staining cells. (B) Representative images show double staining of the spike protein (teal) with alveolar type 2 cells using SPC (yellow) as a marker and macrophages using CD68 (yellow) as a marker in COVID-19 patient postmortem samples (20× magnification). Black arrows indicate double staining cells.
Furthermore, human lung alveolar epithelial type II cells were injured significantly by SARS-CoV-2 virus infection (Fig. 3B). This was identified by the reduction of the number and stain intensity of SPC positive cells (yellow) in the infected lung of the postmortem samples compared to the non-infected lung (Fig. 3B). In COVID-19 lung sample #1, which has the highest virus amount (teal), the SPC positive cells are almost completely lost, whereas in the other lungs, SPC positive cells are low and have fallen off from the epithelium into the alveolar lumen (Fig. 3B). Our results indicated that the K18-hACE2 mice could be a suitable model for mimicking the infections of the COVID-19 patients.
Histopathology of tissues of K18-hACE2 mice infected with SARS-CoV-2.
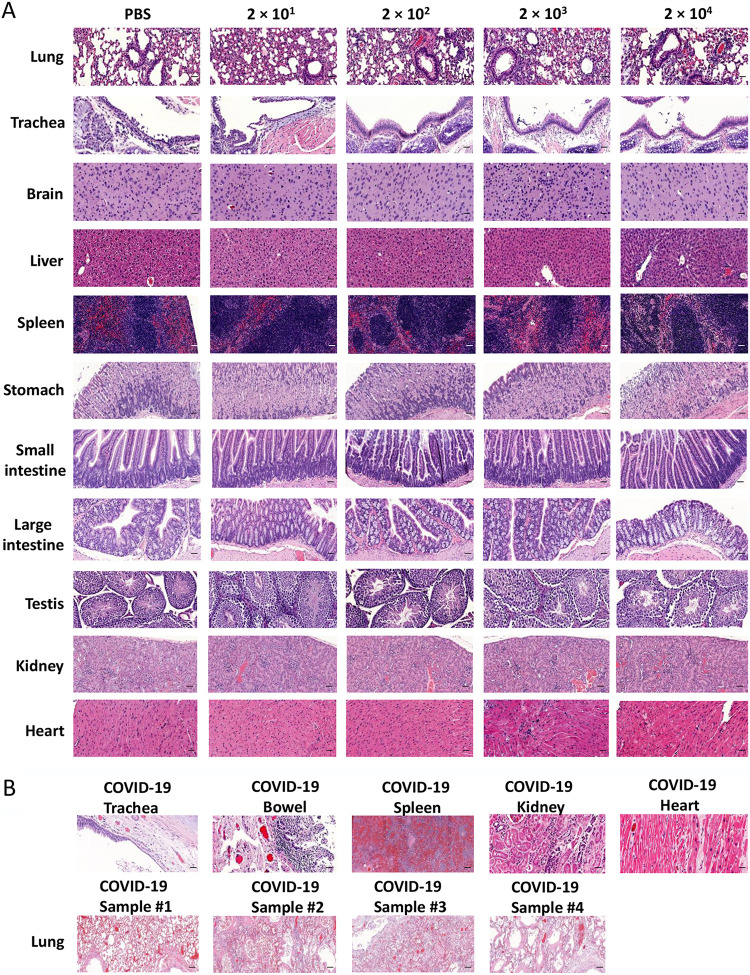
To assess disease severity, H&E staining was performed for histopathological evaluation in various organs from K18-hACE2 mice infected with a series of increasing doses of SARS-CoV-2. As expected, lung tissues showed the most severe damage. Mice in the low-dose groups showed an average of 30–60% more alveolar congestion and consolidation compared to uninfected mice (Fig. 4A). Some lung tissues showed alveolar hemorrhage as well as lymphocytic pneumonitis with alveolar thickening and peripheral parenchymal collapse. Lung damage in the high-dose groups was even more extensive, consisting of ∼20% alveolar collapse with ruptured septa, as well as thickened alveolar and intra-alveolar septa. Parenchymal consolidation was evident in 50% of the tissue, along with interstitial inflammation and pneumonitis (Fig. 4A). Similarly, trachea tissues showed mild epithelial damage in the lower-dose groups and more severe epithelial damage in the higher-dose groups. Consistent with the brain showing high viral gene expression for the higher-dose groups (Fig. 2A), brain tissues showed mild congestion in some mice from the low-dose groups, with more extensive congestion in the high-dose groups. In liver tissues, the low-dose groups showed several foci of spotty and patchy necrosis (focal perivenular), Kupffer cell hyperplasia, and focal portal inflammation; the high-dose groups also showed Kupffer cell hyperplasia along with congestion, reactive change, apoptotic hepatocytes, and focal lobular inflammation (Fig. 4A). In the spleen, the low-dose groups showed red pulp congestion while the high-dose groups also showed lymphoid hyperplasia and mild extramedullary hematopoiesis. The stomach and small and large intestines in the low-dose groups appeared normal but in the high-dose groups, the stomach showed reactive changes, diffuse epithelial sloughing, and chronic inflammation along the myenteric plexus, while the small and large intestine showed myenteric plexus inflammation. The testis in the low-dose groups also appeared normal but showed focal tubular damage and congestion in the high-dose groups. In the kidney, both the low-dose and high-dose groups showed cortical congestion, tubular damage, and focal tubular collapse. Meanwhile, both the low-dose and high-dose groups showed ischemic change and disarray in the heart (Fig. 4A). These findings indicate that SARS-CoV-2 infection damages multiple tissues, with a dose-dependent effect in most tissues.
FIG 4.
Pathological changes in multiple tissues of K18-hACE2 mice infected with the indicated dose of SARS-CoV-2 or postmortem tissue from COVID-19 patients. (A) Tissue damage in the brain, trachea, lung, heart, liver, spleen, small intestine, stomach, large intestine, kidney, and testis of K18-ACE2 mice after SARS-CoV-2 infection (scale bar, 40 μm). (B) Tissue damage in trachea, bowel, spleen, kidney, heart, and lung in COVID-19 patient postmortem samples (scale bar, 40 μm).
To compare the pathological findings in the mouse model to COVID-19 patients, we also examined postmortem samples of COVID-19 patients. We identified congestion and inflammation in multiple organs, as well as epithelial damage and necrosis, which was consistent with our findings in the SARS-CoV-2-infected K18-hACE2 mouse model (Fig. 4B).
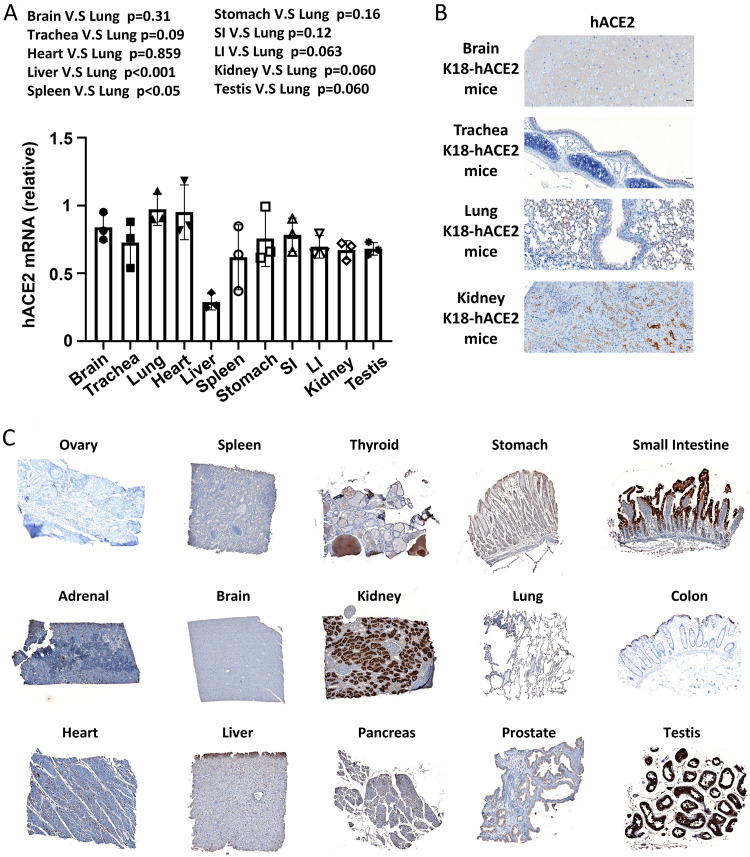
Expression of hACE2 in K18-hACE2 mice and human tissues.
To better understand the infection pattern and virus distribution in this mouse model, we measured hACE2 RNA expression levels in various tissues of K18-hACE2 mice by RT-PCR. We found that, compared to the expression of hACE2 in the lung, there was also high expression in the brain, trachea, heart, stomach, small intestine, large intestine, kidney, and testis; whereas there was lower expression in the liver and spleen (Fig. 5A). We also confirmed the hACE2 expression in the brain, trachea, lung, and kidney of the k18-hACE2 mice at the protein level by IHC (Fig. 5B). The expression of hACE2 in the brain, trachea, and lung tissues in mice is consistent with the finding that they had the highest viral copy numbers (Fig. 5B vs. Fig. 2A), further confirming the K18-hACE2 mouse is a good model for studying in vivo SARS-CoV-2 infection. We also examined hACE2 expression in a human tissue array (Fig. 5C). We found that hACE2 is not only highly expressed in the lung but also in the stomach, small and large intestine, kidney, and testis. However, the expression of ACE2 in the human brain was not as high as in the brain of K18-hACE2 mice; this could be due to the insertion of the K18 promoter, which confers higher expression of hACE2 in the mouse model (Fig. 5C). This finding is consistent with another transgene driven by a human K18 regulatory element in mice (18).
FIG 5.
Tissue distribution of hACE2 in K18-hACE2 mice and human samples. (A) Detection of hACE2 RNA in multiple tissues of K18-hACE2 mice by RT-qPCR. (B) hACE2 expression in the brain, trachea, lung, and kidney of K18-hACE2 mice by IHC. (C) Human tissue array IHC staining for the hACE2 protein.
Protective role of sera from previously infected mice and resistance of low-dose-infected surviving mice to high-dose rechallenge in the K18-hACE2 model.
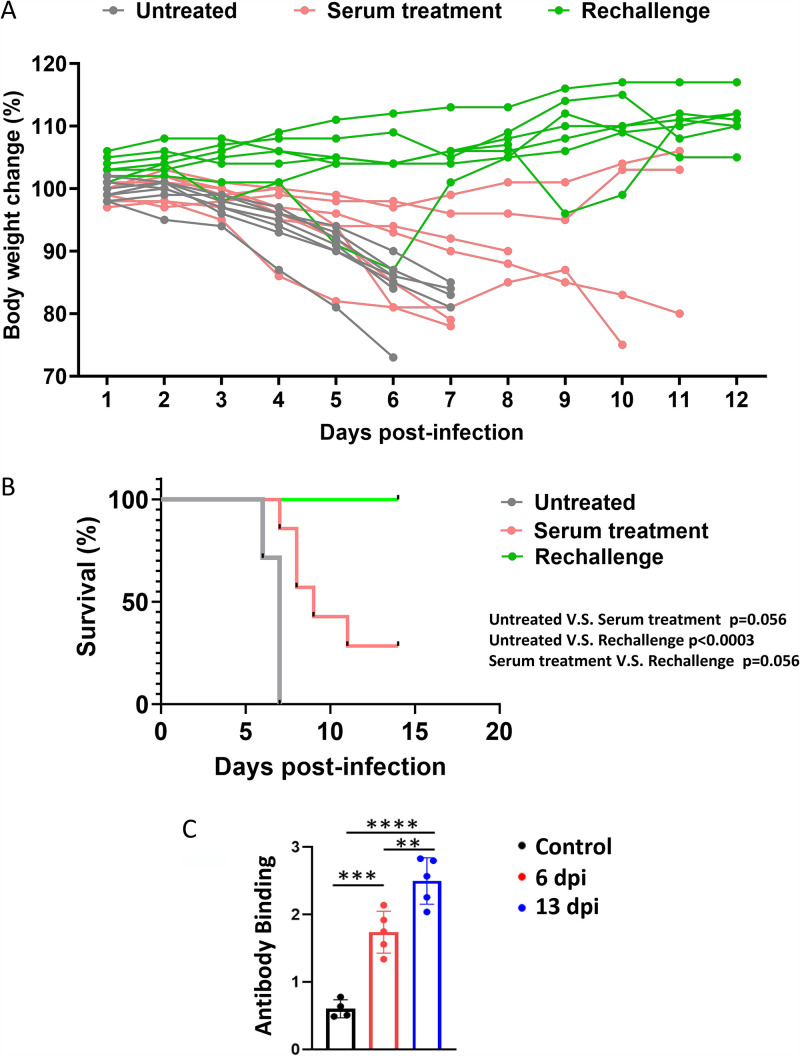
To evaluate if the sera from infected mice could protect the mice from viral infection, sera were collected from mice infected with 2 × 104 PFU SARS-CoV-2 at day 6 p.i. Collected sera were injected intravenously (i.v.) into virus-naive mice. Two days later, mice receiving a prior dose of sera were intranasally infected with 2 × 104 PFU SARS-CoV-2, followed by an immediate i.v. injection of second dose of sera. Body weights of treated versus control mice were monitored daily. Mice treated with sera from infected mice showed a delayed and slower body weight drop compared to the untreated group (Fig. 6A), suggesting sera from infected mice had a protective effect on newly infected mice. Sera from infected mice did not significantly protect but show a trend against death after virus rechallenge, which might be due to the little amount of sera that we could collect to have a large enough sample size of transfused mice for the sera protection experiments (Fig. 6B). Another reason for the moderate protective effect could be that the neutralizing antibody titer at day 6 may be low. To investigate this possibility, mice were infected with a lower dose, 1 × 103 PFU, so that some mice could survive long enough to collect sera. We collected sera at days 6 and 13 p.i. and measured relative anti-spike antibody levels by ELISA. Our data indeed showed that anti-spike antibody levels at day 6 p.i. were significantly lower than those at day 13 p.i. (Fig. 6C). These data indicate that the small amount of neutralizing antibody in sera collected at day 6 p.i. may not be enough for protecting mice receiving passive transfer of the sera.
FIG 6.
Protective role of sera from previously infected mice and resistance of low dose-infected surviving mice to high-dose rechallenge in the K18-hACE2 model. Body weights (A) and survival (B) of mice treated with sera from previously infected mice and prior low dose-infected mice being challenged with a high dose of SARS-CoV-2. Mice with serum protection were infected with 2 × 104 PFU per mouse SASR-CoV-2 24 h after being infused with sera from mice infected with 2 × 104 PFU virus per mouse for 6 days. For the rechallenge, mice were infected with the low dose of 2 × 101 PFU or 2 × 102 PFU per mouse SASR-CoV-2 for 2 weeks and then were rechallenged with the high dose of 2 × 104 PFU per mouse SASR-CoV-2. Survival of mice was analyzed by the Kaplan–Meier method and the log-rank test. (C) Mice were infected with 1 × 103 PFU at day 0. Sera in the peripheral blood were collected via tail vein bleeding 6 days postinfection (dpi) or right before mice were sacrificed at 13 dpi. Relative anti-spike antibody levels were measured by ELISA. Data are presented as mean ± standard deviations. Statistical analyses were performed by one-way ANOVA with P values corrected for multiple comparisons by the Bonferroni test (n = 4 or 5 mice). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In addition, we rechallenged the mice that survived 2 × 101 or 2 × 102 PFU infection with 2 × 104 PFU virus when they recovered to their initial body weight. We found that all mice survived the high-dose virus re-challenge without showing a substantial body weight drop, suggesting that recovery from low-dose infection conferred antiviral activity that protected against rechallenge with high dose virus (Fig. 6B). These results suggest that patients who survive from SARS-CoV-2 infection should be more resistant to the next infection with the same SARS-CoV-2 strain than people without prior infection.
DISCUSSION
The recent outbreak of SARS-CoV-2 that has claimed the lives of nearly 5 million people to date has led to an urgent need for a mouse infection model that accurately mirrors inoculation, development, and progression of the human disease termed COVID-19. The K18-hACE2 model was developed to study coronavirus infection (11). In the current study, we performed a comprehensive analysis of viral distribution and tissue pathology based on a range of inoculation doses and compared the results to observations in postmortem samples from COVID-19 patients. We found a dose-response relationship between infectious dose and loss of body weight, viral titer, tissue pathology, and mortality. Notably, there was not a uniform relationship between infectious dose and viral copy numbers, with some tissues accumulating virus at lower while others at higher copy number. We evaluated hACE2 levels in K18-hACE2 mice and found similarity to ACE2 levels in humans. We also showed that challenge with a low dose of SARS-CoV-2 protected against the lethality of rechallenge with a higher dose. Overall, our results indicate that low-dose infection in the K18-hACE2 model mimics non-severe COVID-19, while infection with higher doses mimics severe COVID-19.
Due to the unprecedented impact of the COVID-19 pandemic, many models of human COVID-19 are under investigation. In addition to the K18-hACE2 model, other mouse models have been described. Hassan et al. used replication-defective adenoviruses encoding human ACE2-tranduced mice to infect BALB/c mice, followed by SARS-CoV-2 infection, to model COVID-19 (15). Following infection by 105 focus-forming units (FFU) of SARS-CoV-2, the body weight of mice was maintained but did not drop, therefore it seems that the model is not a lethal model. Using CRISPR/Cas9 technology to knock-in hACE2, Sun et al. established a model (14). In response to 4 × 105 PFU of SARS-CoV-2, the animals experienced robust viral replication in multiple tissues and interstitial pneumonia, but no obvious clinical symptoms or mortality; only 10% of aged mice lost their weight at day 3 p.i. and recovered. With the low incidence rate and very mild symptoms, this model is not ideal to recapitulate human COVID-19. This might be due to the nature of the knock-in with only one copy of hACE2 systemically existing in mice. Qin and colleagues developed a hACE2 transgenic mouse model with a mouse ACE2 promoter for SARS and used the model to study SARS-CoV-2 infection (13, 19). Infected by 1 × 105 TCID50 of SARS-CoV-2, the mouse body weights dropped at day 1 and continued until day 5, and then almost completely recovered on day 14 without lethality, suggesting that this model at least does not mimic severe COVID-19 patients. This might be due to lower promoter activity or gene expression of murine (m)ACE2 compared to the K18 promoter (19). Ostrowski et al. developed transgenic mice using the promoter of the human FOXJ1, a transcription factor required for differentiation of ciliated epithelial cells in the airway, to drive hACE2 expression (20). Jiang and colleagues used this model to study SARS-CoV-2 (12). Infected with 3 × 104 PFU SARS-CoV-2, among 10 mice, 2 had no infection, 4 had no drop in body weight, and 4 had body weight drops with 3 deaths and 1 recovery (12). In contrast, for the K18-hACE2 model that we characterized here, with the dose of 2 × 103 PFU, which is lower than the doses used in all above mouse models, all mice succumbed to infection with body weight drops and mortality of 100% by day 8. With lower dose infection such as 2 × 101 and 2 × 102 PFU, 30% of the mice recovered. Collectively, these suggest that the K18-hACE2 model is the most sensitive model for COVID-19, with ability to recapitulate aspects of human disease from both non-severe and severe COVID-19 patients. Our results are supported by those from other groups (16, 21–24). This might be due to unique aspects of the K18-hACE2 model: 1) hACE2 is used, where in contrast other models including golden hamster, ferret, cat, Chinese tree shrew, and even mouse models with an endogenous ACE2 promoter recapitulate human expression levels (25); 2) multiple copies of hACE2 are placed in the murine genome (11), whereas one copy of hACE2 may not be ideal, evidenced by the knock-in study by Sun et al. (14); and 3) the human K18 promoter is likely stronger than the promoters in other models, such as the human FOXJ1 in the study by Jiang et al. (12). Apart from the animal models mentioned above, some mouse-adapted models of SARS-CoV-2 were also used for studying pathogenesis of SARS-CoV-2. In these models, recombinant mouse-adapted strains of SARS-CoV-2 bind the mouse ACE2 for viral entry; however, it is possible that accumulated mutations may not occur in humans (26–29).
The pathological damage in the lung and brain in the K18-hACE2 mouse infection model is similar to the clinical symptoms of COVID-19 patients, suggesting that this mouse model can recapitulate SARS-CoV-2 infection in humans. However, the K18-hACE2 mouse model was originally generated by inserting hACE2 in the mouse genome under the human K18 promoter, which may not result in the exact distribution of ACE2 expression as in humans. In fact, we observed high expression levels of hACE2 in mouse brains, correlating with a higher virus titer in the brain compared to most other organs or tissues; however, in the postmortem samples of humans, the hACE2 expression is very low. Nonetheless, brain infections with SARS-CoV-2 and deaths of patients from brain-specific COVID-19 have been frequently reported (30–32). Interestingly, our study showed that SARS-CoV-2 can infect neurons in an animal model, which is consistent with previous studies using human neuron organoids (33–35). It will be interesting to know whether this neuronal infection causes taste and smell loss, commonly found in patients with COVID-19, as neuronal circuits respond to gustatory and olfactory cues (36).
In summary, we characterized the K18-hACE2 model for COVID-19 in comparison to human postmortem samples. The model shows dose-dependent sensitivity to SARS-CoV-2 infection. Infection with low doses recapitulates the disease observed in non-severe COVID-19 patients, while infection with higher doses recapitulates the disease observed in patients with severe COVID-19. The K18-hACE2 humanized COVID-19 mouse model is excellent to study COVID-19 and develop preventive and therapeutic drugs, as well as vaccines, for coronavirus diseases.
MATERIALS AND METHODS
Ethics statement.
Mouse-model studies were performed in an animal biosafety level 3 (ABSL3) facility. The animal protocol of these studies was reviewed and approved by the institutional animal care and use committee (IACUC) of Northern Arizona University (protocol #20-005) and the committee of Texas Biomedical Research Institute (protocol #1718MU). Human postmortem COVID-19 patient samples were previously described (37) and were collected under the approval of the institutional review board.
Viruses, cells, and mice.
The SARS-CoV-2 strain used in the mouse model was SARS-CoV-2/human/USA/WA-CDC-WA1/2020, which was obtained from the Biodefense and Emerging Infections Research Resources (BEI Resources). The viruses were amplified using Vero-E6 cells (ATCC). Vero-E6 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Cells were inoculated with virus at a multiplicity of infection (MOI) of 0.001 and cultured for 96 h. Then the supernatant was collected and titrated using a plaque assay.
The K18-hACE2 transgenic mice, in which the human keratin 18 (KRT18) promoter was used to direct human ACE2 expression, were purchased from The Jackson Laboratory.
Mouse infection.
Female and male 6- to 8-week-old K18-hACE2 transgenic mice under the C57BL/6J background were anesthetized and intranasally (i.n.) infected with SARS-CoV-2 virus at a dose of 2 × 101, 2 × 102, 2 × 103, or 2 × 104 PFU/mouse. The uninfected control mice were inoculated with phosphate-buffered saline (PBS). All the mice were observed and weighed daily. Peripheral blood was collected on day 6 from high dose groups and used for serum protection experiments. The mice were euthanized at day 6 postinfection, and the tissues were collected for further analysis.
Viral rechallenge model.
The mice that survived i.n. infection with 2 × 101 PFU and 2 × 102 PFU of virus were rechallenged with 2 × 104 PFU virus i.n. 2 weeks after the initial infection when they recovered to the initial body weight. The body weight was continuously monitored.
Serum protection model.
Sera of the mice infected by 2 × 104 PFU virus were collected at day 6 postinfection. Half of the sera were intraperitoneally injected into the non-infected mice 2 days before the mice were i.n. infected with 2 × 104 PFU virus. The other half of the sera were intraperitoneally administered right before the infection.
Tissue preparation to detect viral copy number.
Mouse tissues were collected, weighed, and immediately homogenized using an electric homogenizer (Thomas Scientific). After centrifugation at 1200g for 10 min, the supernatant was isolated and used to detect viral RNA copy number by real-time PCR (see below).
Viral RNA copy number detection.
Viral RNA was isolated from homogenized tissues using the PureLink RNA minikit (Invitrogen). A one-step RT-PCR kit (Bio-Rad) was used to detect the viral RNA using Applied Biosystems QuantStudio 12K Flex real-time PCR system with the following cycling protocol: reverse transcription at 50°C for 10 min; hot start at 95°C for 10 min; and 40 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 30 s. The primer sequences were CoV2-S_19F (5′ -GCTGAACATGTCAACAACTC- 3′) and CoV2-S_143R (5′ -GCAATGATGGATTGACTAGC- 3′), which were designed to target a 125 bp region of the SARS-CoV-2 spike protein (38). The standard samples were serial 10-fold dilutions of a known copy number of the HKU1 virus. The results were normalized and expressed as genome equivalent copies per gram of tissue.
Infectious viral load measurement.
Lungs from infected mice were collected and homogenized. Confluent monolayers of Vero E6 cells were infected with serial dilutions of supernatants obtained from the lung homogenates. The immuno-plaque assay with the SARS-CoV-2 NP antibody was performed to measure the viral load.
Measurement of anti-spike antibody.
His-tagged full-length SARS-CoV-2 S protein (Cat. # 40589-V08B1, Sino Biological) was used as a coating protein. The plate was incubated with mouse serum samples for 2 h. The HRP-conjugated goat anti-mouse IgG antibody (Cat# 05-4220, Invitrogen) was used as the detecting antibody. Absorbance was measured at 450 nm by a Multiskan FC Microplate Photometer (Fisher Scientific).
Human ACE2 RNA quantification.
Total RNA from the indicated tissues of the K18-hACE2 mice was isolated using the PureLink RNA minikit (Invitrogen), and cDNA was synthesized using SuperScript Reverse Transcriptase (Thermo Fisher). Human ACE2 expression was examined using the following primers: F: CGAAGCCGAAGACCTGTTCTA; R: GGGCAAGTGTGGACTGTTCC, under the following PCR conditions: 98°C for 30 s, followed by 40 cycles of 98°C for 15 s, 62°C for 30 s, and 72°C for 60 s.
Immunohistochemistry and HandE staining.
Tissues were harvested from the infected K18-hACE2 mice and immediately fixed in 10% neutral buffered formalin. Dehydration, clearing, and paraffinization were performed on a Tissue -Tek VIP Vacuum Infiltration Processor (SAKURA). The samples were embedded in paraffin using a Tissue-Tek TEC Tissue Embedding Station (SAKURA). Samples were then sectioned at 5 μm and put on positively charged glass slides. The slides were deparaffinized, rehydrated, and stained with Modified Mayer’s Hematoxylin and Eosin Y Stain (America MasterTech Scientific) on an H&E Auto Stainer (Prisma Plus Auto Stainer, SAKURA) according to standard laboratory procedures.
Single or double IHC stains were performed on Ventana Discovery Ultra (Ventana Medical Systems, Roche Diagnostics, Indianapolis, USA) IHC Auto Stainer. Briefly, slides were loaded on the machine, and deparaffinization, rehydration, endogenous peroxidase activity inhibition, and antigen retrieval were first performed. For single IHC staining, primary antibodies were incubated with DISCOVERY anti-Rabbit HQ, followed by DISCOVERY anti-HQ-HRP incubation. For double IHC staining, two antigens were sequentially detected and heat inactivation was used to prevent antibody cross-reactivity between the same species. Following each primary antibody incubation, DISCOVERY anti-Rabbit HQ or NP or DISCOVERY anti-Mouse HQ or NP and DISCOVERY anti-HQ-HRP or anti-NP-AP were incubated. The stainings were visualized with DISCOVERY ChromoMap DAB Kit, DISCOVERY Yellow Kit, DISCOVERY Teal Kit, or DISCOVERY Purple Kit, and accordingly counterstained with hematoxylin (Ventana) and coverslipped. The following primary antibodies used were: SPIKE (40150-T62, Sino Biological, at 1:2000), NP (NB100-56576, NOVUS, 1/100), hACE2 (AMAB91262, Sigma, at 1:1000), CCL10 (SC-365992#, Santa Cruz at 1/5000), Pro-SPC (AB37386, Millipore at 1/500), CD68 (ab125212, abcam, at 1/100), and NeuN (24307, cell signaling, at 1/100).
Statistical analysis.
Comparison of 2 groups was performed using Student’s two-tailed t test (for unpaired samples) or a paired t test (for paired samples). Multiple groups were compared using one-way or two-way ANOVA, and P values were adjusted for multiple comparisons by Holm’s procedure or the Bonferroni test. A P value of less than 0.05 was considered statistically significant. Kaplan-Meier analysis was performed on the survival curves.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (NS106170, AI129582, CA247550, CA223400, CA210087, CA163205, and P30CA033572), the Leukemia and Lymphoma Society (1364-19), and The California Institute for Regenerative Medicine (DISC2COVID19-11947). We thank BEI Resources, NIAID at NIH for providing SARS-CoV-2/human/USA/WA-CDC-WA1/2020. Part of the research reported in this publication includes work performed in the Pathology Core at City of Hope supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572.
J. Yu, M.A. Caligiuri, P. Keim, B. Barker, H. Mead, J.B. Torrelles, L. Martinez-Sobrido, and W. Dong conceived and designed the project. P. Keim and B. Barker supervised experiments conducted in the laboratories. W. Dong, H. Mead, L. Tian, J.-G. Park, J.I. Garcia, S. Jaramillo, D. Kollath, V. Coyne, N. Stone, A. Jones, J. Zhang, and A. Li performed experiments and/or data analyses. M. Milanes-Yearsley reviewed the pathology changes. W. Dong, J. Yu, T. Barr, H. Mead, L. Tian, L.-S. Wang, and M.A. Caligiuri wrote, reviewed and/or revised the paper. J. Yu and M.A. Caligiuri acquired funding. All authors discussed the results and commented on the manuscript.
We declare that we have no conflicts of interest.
Contributor Information
Michael A. Caligiuri, Email: mcaligiuri@coh.org.
Jianhua Yu, Email: jiayu@coh.org.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. 2020. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 172:577–582. 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2021. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19).
- 3.Gibson PG, Qin L, Puah SH. 2020. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust 213:54–56.e1. 10.5694/mja2.50674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steenblock C, Schwarz PEH, Ludwig B, Linkermann A, Zimmet P, Kulebyakin K, Tkachuk VA, Markov AG, Lehnert H, de Angelis MH, Rietzsch H, Rodionov RN, Khunti K, Hopkins D, Birkenfeld AL, Boehm B, Holt RIG, Skyler JS, DeVries JH, Renard E, Eckel RH, Alberti K, Geloneze B, Chan JC, Mbanya JC, Onyegbutulem HC, Ramachandran A, Basit A, Hassanein M, Bewick G, Spinas GA, Beuschlein F, Landgraf R, Rubino F, Mingrone G, Bornstein SR. 2021. COVID-19 and metabolic disease: mechanisms and clinical management. Lancet Diabetes Endocrinol 9:786–798. 10.1016/S2213-8587(21)00244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Sawalha AH, Lu Q. 2021. COVID-19 and autoimmune diseases. Curr Opin Rheumatol 33:155–162. 10.1097/BOR.0000000000000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saad MA, Alfishawy M, Nassar M, Mohamed M, Esene IN, Elbendary A. 2021. COVID-19 and autoimmune diseases: a systematic review of reported cases. Curr Rheumatol Rev 17:193–204. 10.2174/1573397116666201029155856. [DOI] [PubMed] [Google Scholar]
- 7.Brodin P. 2021. Immune determinants of COVID-19 disease presentation and severity. Nat Med 27:28–33. 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 8.Blot M, Bour JB, Quenot JP, Bourredjem A, Nguyen M, Guy J, Monier S, Georges M, Large A, Dargent A, Guilhem A, Mouries-Martin S, Barben J, Bouhemad B, Charles PE, Chavanet P, Binquet C, Piroth L, the LYMPHONIE study group. 2020. The dysregulated innate immune response in severe COVID-19 pneumonia that could drive poorer outcome. J Transl Med 18:457. 10.1186/s12967-020-02646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, Mencarini J, Caporale R, Peruzzi B, Antonelli A, Trotta M, Zammarchi L, Ciani L, Gori L, Lazzeri C, Matucci A, Vultaggio A, Rossi O, Almerigogna F, Parronchi P, Fontanari P, Lavorini F, Peris A, Rossolini GM, Bartoloni A, Romagnani S, Liotta F, Annunziato F, Cosmi L. 2020. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest 130:4694–4703. 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCray PB, Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang HJ, Baric RS, Zhou P, Yang XL, Shi ZL. 2020. Pathogenesis of SARS-CoV-2 in Transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58.e8. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 14.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124–133.e4. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Jr, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS. 2020. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182:744–753.e4. 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allue-Guardia A, Olmo-Fontanez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, 2nd, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Jr, Dick EJ, Jr, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, Abbott DA, Donnelly HK, Donayre A, Goldberg IA, Klug ZM, Borkowski N, Lu Z, Kihshen H, Politanska Y, Sichizya L, Kang M, Shilatifard A, Qi C, Lomasney JW, Argento AC, Kruser JM, Malsin ES, Pickens CO, Smith SB, Walter JM, Pawlowski AE, Schneider D, Nannapaneni P, Abdala-Valencia H, Bharat A, Gottardi CJ, Budinger GRS, Misharin AV, Singer BD, Wunderink RG, The NU SCRIPT Study Investigators. 2021. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 590:635–641. 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow YH, O'Brodovich H, Plumb J, Wen Y, Sohn KJ, Lu Z, Zhang F, Lukacs GL, Tanswell AK, Hui CC, Buchwald M, Hu J. 1997. Development of an epithelium-specific expression cassette with human DNA regulatory elements for transgene expression in lung airways. Proc Natl Acad Sci USA 94:14695–14700. 10.1073/pnas.94.26.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XH, Deng W, Tong Z, Liu YX, Zhang LF, Zhu H, Gao H, Huang L, Liu YL, Ma CM, Xu YF, Ding MX, Deng HK, Qin C. 2007. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med 57:450–459. [PubMed] [Google Scholar]
- 20.Ostrowski LE, Hutchins JR, Zakel K, O'Neal WK. 2003. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol Ther 8:637–645. 10.1016/S1525-0016(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 21.Arce VM, Costoya JA. 2021. SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell Mol Immunol 18:513–514. 10.1038/s41423-020-00616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreau GB, Burgess SL, Sturek JM, Donlan AN, Petri WA, Mann BJ. 2020. Evaluation of K18-hACE2 mice as a model of SARS-CoV-2 infection. Am J Trop Med Hyg 103:1215–1219. 10.4269/ajtmh.20-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yinda CK, Port JR, Bushmaker T, Owusu IO, Avanzato VA, Fischer RJ, Schulz JE, Holbrook MG, Hebner MJ, Rosenke R, Thomas T, Marzi A, Best SM, de Wit E, Shaia C, van Doremalen N, Munster VJ. 2020. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. bioRxiv 10.1101/2020.08.11.246314. [DOI] [PMC free article] [PubMed]
- 25.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera R, Poon LLM, Nicholls JM, Peiris M, Yen HL. 2020. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583:834–838. 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun S, Gu H, Cao L, Chen Q, Ye Q, Yang G, Li RT, Fan H, Deng YQ, Song X, Qi Y, Li M, Lan J, Feng R, Guo Y, Zhu N, Qin S, Wang L, Zhang YF, Zhou C, Zhao L, Chen Y, Shen M, Cui Y, Yang X, Wang X, Tan W, Wang H, Wang X, Qin CF. 2021. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Commun 12:5654. 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leist SR, Dinnon KH, 3rd, Schafer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183:1070–1085.e12. 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinnon KH, 3rd, Leist SR, Schafer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang K, Zhang Y, Hui X, Zhao Y, Gong W, Wang T, Zhang S, Yang Y, Deng F, Zhang Q, Chen X, Yang Y, Sun X, Chen H, Tao YJ, Zou Z, Jin M. 2021. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67:103381. 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sepehrinezhad A, Shahbazi A, Negah SS. 2020. COVID-19 virus may have neuroinvasive potential and cause neurological complications: a perspective review. J Neurovirol 26:324–329. 10.1007/s13365-020-00851-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhammad S, Petridis A, Cornelius JF, Hanggi D. 2020. Letter to editor: severe brain haemorrhage and concomitant COVID-19 infection: a neurovascular complication of COVID-19. Brain Behav Immun 87:150–151. 10.1016/j.bbi.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao XY, Jin WL. 2020. The COVID-19 pandemic: consideration for brain infection. Neuroscience 437:130–131. 10.1016/j.neuroscience.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramani A, Pranty AI, Gopalakrishnan J. 2021. Neurotropic effects of SARS-CoV-2 modeled by the human brain organoids. Stem Cell Rep 16:373–384. 10.1016/j.stemcr.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellegrini L, Albecka A, Mallery DL, Kellner MJ, Paul D, Carter AP, James LC, Lancaster MA. 2020. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell 27:951–961.e5. 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang BZ, Chu H, Han S, Shuai H, Deng J, Hu YF, Gong HR, Lee AC, Zou Z, Yau T, Wu W, Hung IF, Chan JF, Yuen KY, Huang JD. 2020. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res 30:928–931. 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper KW, Brann DH, Farruggia MC, Bhutani S, Pellegrino R, Tsukahara T, Weinreb C, Joseph PV, Larson ED, Parma V, Albers MW, Barlow LA, Datta SR, Di Pizio A. 2020. COVID-19 and the chemical senses: supporting players take center stage. Neuron 107:219–233. 10.1016/j.neuron.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong W, Wang J, Tian L, Zhang J, Mead H, Jaramillo SA, Li A, Zumwalt RE, Whelan SPJ, Settles EW, Keim PS, Barker BM, Caligiuri MA, Yu J. 2021. FXa cleaves the SARS-CoV-2 spike protein and blocks cell entry to protect against infection with inferior effects in B.1.1.7 variant. bioRxiv 10.1101/2021.06.07.447437. [DOI]
- 38.Stone NE, Jaramillo SA, Jones AN, Vazquez AJ, Martz M, Versluis LM, Raniere MO, Nunnally HE, Zarn KE, Nottingham R, Ng KR, Sahl JW, Wagner DM, Knudsen S, Settles EW, Keim P, French CT. 2021. Stenoparib, an inhibitor of cellular poly(ADP-ribose) polymerase, blocks replication of the SARS-CoV-2 and HCoV-NL63 human coronaviruses in vitro. mBio 12:e03495-20. [DOI] [PMC free article] [PubMed] [Google Scholar]